Significance
Radial glia (RG) are primary neural stem cells of the cortex, and our current models of human corticogenesis are derived from investigations into the spatiotemporal, transcriptomic, and morphological diversity of human RG. However, it is the progeny of RG, neural intermediate progenitor cells (nIPCs), that directly generate most neurons. As such, nIPCs play an important role in corticogenesis, and yet little is known about human nIPCs. We identified three gaps in our current understanding: when and where human nIPCs appear, what transcriptional subtypes exist, and what their morphological features are. Our results present a more complete view of human nIPCs and serve as a basis for interpreting organoids and other in vitro models of human brain development, evolution, and disease.
Keywords: neuronal, human, cortex, development, progenitor
Abstract
Studies of the spatiotemporal, transcriptomic, and morphological diversity of radial glia (RG) have spurred our current models of human corticogenesis. In the developing cortex, neural intermediate progenitor cells (nIPCs) are a neuron-producing transit-amplifying cell type born in the germinal zones of the cortex from RG. The potential diversity of the nIPC population, that produces a significant portion of excitatory cortical neurons, is understudied, particularly in the developing human brain. Here we explore the spatiotemporal, transcriptomic, and morphological variation that exists within the human nIPC population and provide a resource for future studies. We observe that the spatial distribution of nIPCs in the cortex changes abruptly around gestational week (GW) 19/20, marking a distinct shift in cellular distribution and organization during late neurogenesis. We also identify five transcriptomic subtypes, one of which appears at this spatiotemporal transition. Finally, we observe a diversity of nIPC morphologies that do not correlate with specific transcriptomic subtypes. These results provide an analysis of the spatiotemporal, transcriptional, and morphological diversity of nIPCs in developing brain tissue and provide an atlas of nIPC subtypes in the developing human cortex that can benchmark in vitro models of human development such as cerebral organoids and help inform future studies of how nIPCs contribute to cortical neurogenesis.
The human cerebral cortex comprises a diverse set of neurons that is essential to its functions in perception, judgment, and sensory integration. Furthermore, it is three times larger than that of our closest living relative, the chimpanzee. The abundance and diversity of progenitor cells that emerge during development contribute to this expansion. The cerebral cortex arises from the daughter cells produced by radial glia (RG) that serve as neural stem cells. Ventricular RG (vRG) reside in the ventricular zone (VZ), a progenitor region that lines the ventricles of the developing brain. In mammals, vRG can produce neurons directly, but they more often produce neurons indirectly, via a transit amplifying cell, the neural intermediate progenitor cell (nIPC). nIPCs can divide one or more times before producing neurons (1–4). In rodents, RG produce nIPCs that at early stages of neurogenesis divide in the VZ, but at later stages migrate out of the VZ to divide in a secondary proliferative zone, the inner subventricular zone (iSVZ) (2–5). Clonal analysis has revealed that in the mouse, nIPCs contribute neurons to all cortical layers (5). In addition to vRGs, the developing brains of humans and large brain mammals contain an abundant second type of RG, the outer radial glia (oRG) cells, that also generate nIPCs (1). oRG are most abundant in a third proliferative zone, the outer SVZ (oSVZ), which is considered an expansion of the iSVZ present in large brain mammals (6). Importantly, oSVZ progenitor regions containing oRG cells have been observed in the developing cortex of primates, carnivores, and rodents, including species with gyrencephalic or lissencephalic cortices, indicating a likely adaptation associated with large brain mammals regardless of cortical folding (7–9). In the dorsal cortex of the mouse, the identity of the SVZ-dividing nIPCs is driven by EOMES, a transcription factor enriched in nIPCs, which is sufficient for their generation (10–12). Within the human brain, multiple single cell studies have shown that EOMES expression also marks nIPCs (13, 14, 15). In mice, manipulation of EOMES strongly influences cortical neuron number (10, 11) and loss-of-function mutations in EOMES are typically embryonic lethal (16). Likewise, patients with partial EOMES deletions have developmental disorders, including intellectual disability and autism (17). Despite the relevance of nIPCs to the evolutionary and developmental expansion of human cortex, human nIPCs remain poorly characterized.
The heterogeneity of other progenitors in the developing human and primate cortex, including RG and oligodendrocyte precursor cells, has been tied to temporal, spatial, morphological, and transcriptomic characteristics (1, 18, 19–21). However, similar descriptions of nIPC features are scarce, and previous descriptions of neural progenitor cells in postmortem preterm infant tissue are subject to concerns of tissue preservation (22). Here we analyze the spatiotemporal, transcriptomic, and morphological diversity of EOMES+ nIPCs within developing human cortex.
Results
Intermediate Progenitors Follow Distinct Spatiotemporal Patterns in Early vs. Late Neurogenesis.
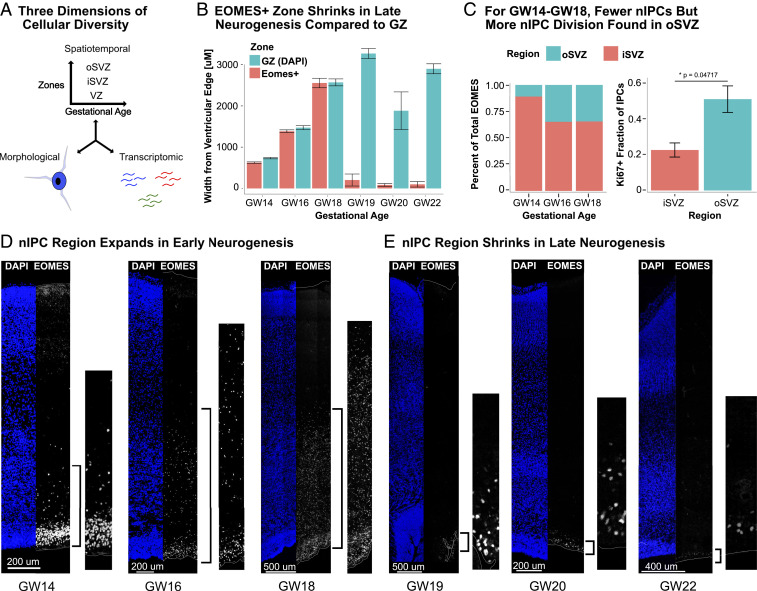
To verify the prevalence and distribution of nIPCs at different stages of human cortical neurogenesis during the second trimester, we performed immunostaining for EOMES, which serves as a marker of nIPCs. From gestational week 10 (GW10) to GW18, the majority of EOMES+ nIPCs form a dense band in the iSVZ, with increasing numbers in the oSVZ (Fig. 1 B–E). Consistent with previous observations that oRGs produce highly proliferative nIPCs (1), we observed that the nIPCs in the oSVZ expressed KI67 more often than those in the iSVZ (Fig. 1C and SI Appendix, Fig. S1). Nevertheless, most nIPCs were found within the iSVZ from GW14 to GW18 (Fig. 1C). Furthermore, staining with SOX2, EOMES, and TBR1 between GW14 and GW18 revealed laminar transcription factor expression in the VZ and iSVZ. SOX2+ cells were present in the VZ, with EOMES+ cells in the adjacent “inner” iSVZ, and TBR1+ cells inset from the EOMES+ cells in the “outer” iSVZ (SI Appendix, Fig. S2). No such pattern was evident in the oSVZ; instead we observed SOX2-, EOMES-, and TBR1-expressing cells mixed throughout the oSVZ. The gradient of transcription factors in the iSVZ, ranging from RG to nIPCs to newborn neurons, suggests that most nIPCs in the iSVZ differentiate into neurons locally and do not migrate into the oSVZ to differentiate. Thus, nIPCs born from the iSVZ may not contribute significantly to the oSVZ nIPC population.
Fig. 1.
(A) The spatiotemporal, transcriptomic, and morphological diversity of nIPCs remains almost entirely unexplored. (B) The GZs expand in thickness during neurogenesis. The EOMES+ area expands until GW19, after which it virtually disappears in the oSVZ and diminishes in the iSVZ, as shown in E. Distance measurements were taken from the ventricle to the outermost EOMES+ cell. Error bars represent technical variations from nine independent measurements of one biological sample. The GW20 measurements represent an average of nine values from both the occipital and prefrontal cortex. Error bars are SD. (C) From GW14 to GW18, most nIPCs are found within the iSVZ, not the oSVZ. However, in the oSVZ, nIPCs costain with KI67 at a higher rate, indicating that these nIPCs are more proliferative (C). The percentage of total EOMES in the oSVZ or iSVZ is the number of EOMES+ nuclei in that region over the total EOMES+ nuclei for a given length along the ventricle. The KI67 fraction represents the percentage of EOMES+ nuclei that also stain for KI67 in a given region for a given length along the ventricle. Error bars are SD. (D) The number of nIPCs expand from GW14 to GW18, during early neurogenesis. (E) Around GW19, the number of nIPCs abruptly declines. A thin band of EOMES+ cells remains in the iSVZ, and few or no nIPCs are found in the oSVZ. This later distribution was evident in three out of four samples from GW19 to GW20 and all three samples after GW20.
Between GW19 and GW21, we observed an abrupt change in both the number and distribution of nIPCs (Fig. 1 B and E). While EOMES+ nuclei largely disappeared from the oSVZ, a thin band of EOMES+ cells remained in the iSVZ (Fig. 1C). This change corresponds to approximately the time period when gliogenesis becomes dominant (23). We found this pattern in two out of three samples at GW19 and GW20, and all three samples at GW21 and GW22. Because this pattern had not been observed before (22), we sought to confirm our observations using a second EOMES antibody that replicated the same pattern (SI Appendix, Fig. S3A).
Because nIPCs are hypothesized to give rise to the majority of neurons during human cortical development (24, 25), we expected that excitatory neuron markers in the germinal zones (GZs) would follow similar spatiotemporal trends and mostly disappear from the oSVZ at the same time as EOMES+ cells. To verify this concordance, we stained for transcription factors NEUROD1, TBR1, and NHLH2. We found that these markers followed the expected spatiotemporal pattern (SI Appendix, Fig. S4), with substantial labeling throughout the germinal zone during peak neurogenesis (GW18) and few cells remaining in the iSVZ afterward. Little to no marker expression was observed in the oSVZ after GW19/GW20. Since five antibodies for EOMES and neuron markers across five samples showed little to no staining in the oSVZ during late neurogenesis, we sought to verify whether the RG scaffold had changed in the oSVZ at this time. Across our samples, markers for RG (HOPX and VIM) as well as intermediate zone fiber tracts (AChE) matched previous descriptions and revealed the expected cytoarchitecture of developing human brain (26–28) (SI Appendix, Fig. S3 B and E). Most neuron marker staining at later ages was in the cortical plate (SI Appendix, Fig. S4), indicating that most newborn excitatory neurons have migrated to the cortical plate at this time. These dynamics indicate that late neurogenesis is marked by a relatively abrupt shift in both the distribution of EOMES-expressing cells and excitatory neurogenesis across the germinal zones.
Analysis of Single-Cell RNA Sequencing Identifies Subtypes of Intermediate Progenitor Cells.
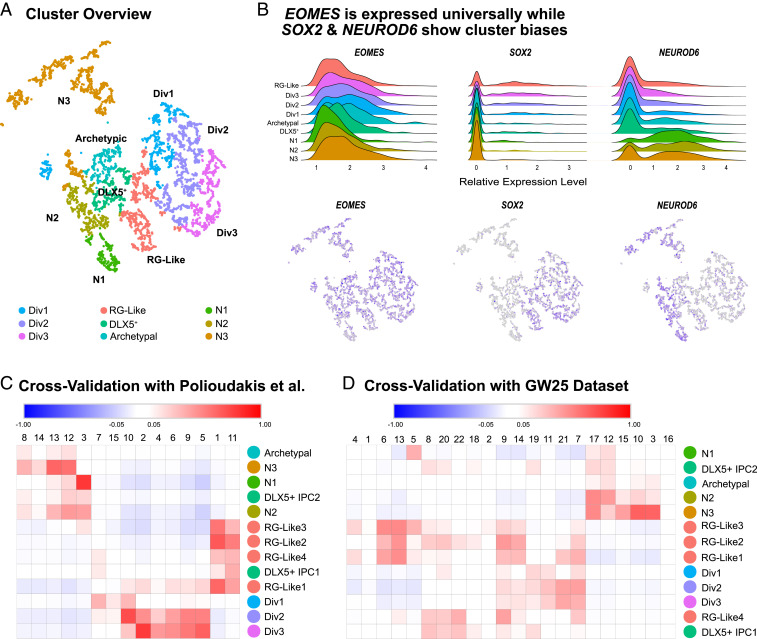
To identify potential transcriptomic subtypes of nIPCs in the developing human cortex, we examined a subclustering of 5,959 Eomes+ cells from a dataset we recently published (29) collected from five independent samples of GW8 to GW22 human cortex (Fig. 2). Noticeably, no EOMES+ cells were detected at GW8. This refined dataset was then normalized, batch corrected, and clustered to identify putative subpopulations of nIPCs. Given the filtering criteria and the comparative homogeneity of nIPCs, we sought to thoroughly verify our clusters and cluster markers by generated statistical thresholds from scrambled data (SI Appendix, Fig. S5). By comparing our clusters to this significance threshold and heuristically combining clusters based on similar marker genes, five clear subtypes or states of nIPCs emerged: three neuronlike nIPC clusters that expressed neurogenic genes; one radial glia-like nIPC (RG-like nIPCs) cluster; and one DLX5+ nIPC cluster (Fig. 2A). The distribution of P values for all these clusters was also significantly different from similarly sized clusters of scrambled cells (Dataset S5 and SI Appendix, Fig. S5). Unbiased clustering can also be influenced by the percentage of reads in each cell related to important cellular processes, like cell cycle phase and transcription, especially when other sources of variation are absent (30, 31). When we removed other cell types, several clusters formed solely around these processes, and we did not include these clusters in our nIPC subtype/state analysis. One cluster, labeled archetypic, had only one marker gene that passed our significance threshold and no evident similarity to another cluster to enable combination. Cell cycle, mitochondrial, and ribosomal genes did not appear to influence identity of this cluster, but the P values of its marker genes were not significantly different from scrambled clusters, leading us to believe that this cluster may be forming because of technical noise. We report it here to inform future single-cell studies.
Fig. 2.
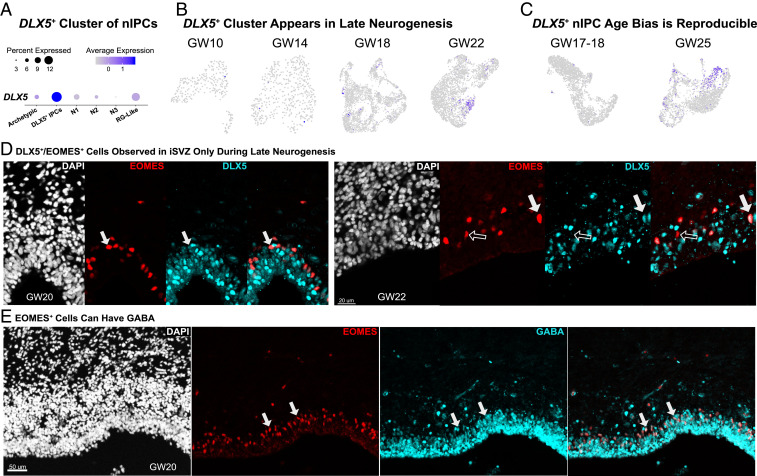
(A) Overview of high-quality clusters in t-distributed stochastic neighbor embedding (tSNE) space. “Div” clusters are the dividing clusters. nIPC clusters (N1–N3) are biased for neuron genes. Archetypal nIPCs express classic nIPC genes, but do not express unique identifying markers. This cluster did not pass the statistical threshold for a biologically meaningful cluster. (B) Within EOMES-expressing nIPCs, SOX2 and NEUROD6 clearly differentiate clusters and guide interpretation. SOX2 is biased toward RG-like and dividing clusters, while NEUROD6 is biased toward neuron-like (N1–N3 nIPC) clusters. (C) Many clusters cross-validated well with specific clusters of EOMES-expressing cells from a previously published GW17–GW18 dataset. N3 and N1 nIPCs validated particularly well. (D) Several clusters showed a near one-to-one relationship with nIPC clusters from a separate validation dataset.
All five nIPC subtype clusters expressed roughly similar amounts of EOMES (Fig. 2B). SOX2 and NEUROD6 were inversely related across the clusters as expected (Fig. 2B). RG-like nIPCs specifically expressed SOX2, while neuron-like nIPC clusters were marked broadly by neuron genes, including NEUROD6 (Fig. 2B). When we analyzed other single-cell datasets (14), we found similar nIPC subtypes as measured by marker gene expression (Fig. 2 C and D). The reproducibility of individual clusters helped validate cluster identities. If clusters represent several unique transcriptomic subtypes, we would expect each cluster to correlate with a different set of clusters in other datasets. Instead, we observed near one-to-one correspondence across two other single-cell datasets from developing human cortex, highlighting the robustness of our nIPC subtype classification.
Validation of Intermediate Progenitor Cell Subtypes throughout Human Neurogenesis.
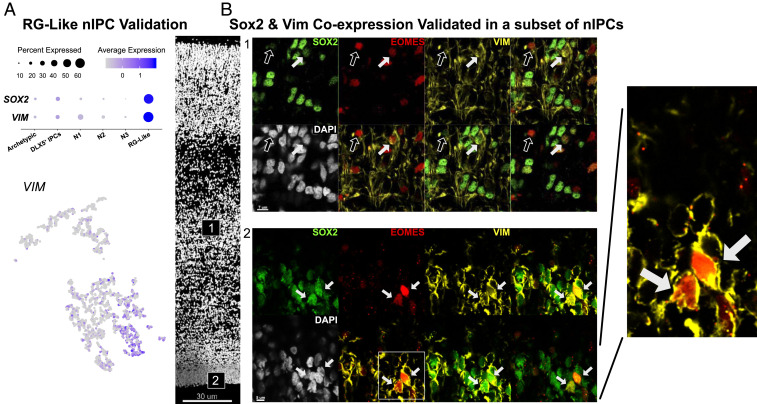
The RG-like nIPCs maintained expression of many RG progenitor genes, including VIM (32) and SOX2 (Fig. 3). Immunostaining for VIM and SOX2 revealed protein coexpression in a subset of EOMES+ nIPCs, validating that these genes identify a transcriptomic subtype of RG-like nIPCs. Because of this persistent RG gene expression, we hypothesized that if the nIPCs were uniquely derived from one specific RG subtype they might also retain subtype-specific gene expression. For example, oRGs are marked by HOPX, while truncated RG are marked by NR4A1. However, while marker genes for RG subtypes were expressed by nIPCs, they did not uniquely mark subclusters of RG-like nIPCs (SI Appendix, Fig. S6). This suggests that nIPCs derived from different RG subtypes may not have significantly different transcriptomes. Additionally, these RG-like nIPCs could be found in both the iSVZ and oSVZ across multiple ages, further suggesting that multiple RG subtypes can produce RG-like nIPCs (Fig. 3).
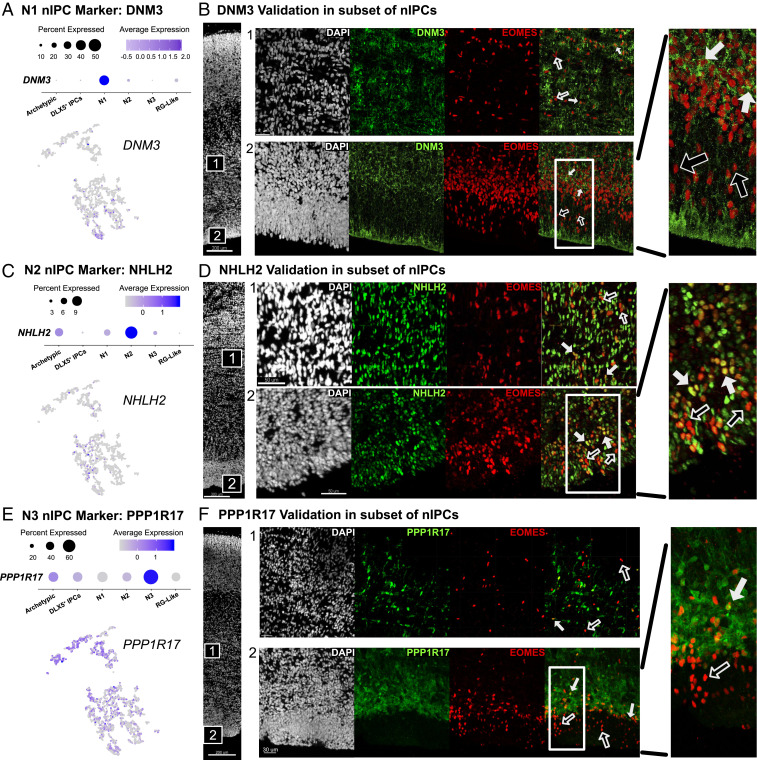
Fig. 3.
(A) N1 nIPCs are marked by DNM3. (B) DNM3 is expressed in a subset of nIPCs within both the iSVZ (2) and oSVZ (1) at GW14. Filled-in arrows represent colocalization and empty arrows represent nuclei with only EOMES. This theme carries through the rest of the figure. (C) N2 nIPCs are marked by strong NHLH2 expression. (D) NHLH2 is expressed in a subset of EOMES+ nuclei in both the iSVZ and the oSVZ. (E) N3 nIPCs are marked by strong PPP1R17 expression. (F) PPP1R17 can also be seen in a specific subset of EOMES+ cells in both the iSVZ and the oSVZ.
In contrast to the uniform cluster of RG-like nIPCs, three discrete types of nIPCs emerged from the dataset, each with a near one-to-one relationship with nIPC clusters from other published datasets (14) (Fig. 2 C and D). These three types, N1, N2, and N3 nIPCs, were uniquely enriched for DNM3, NHLH2, and PPP1R17, respectively. These markers, along with many others, formed gene sets unique to each cluster (Dataset S2). We validated each subtype via immunohistochemistry and found that each marker was expressed in a subset of EOMES+ cells (Fig. 4). PPP1R17 marked the cell bodies of some nIPCs. DNM3, a dynamin motor protein, marked a subset of EOMES+ cells. NHLH2, a neuronal transcription factor, was highly elevated in a subset of nIPC nuclei. Taken together, these analyses suggest that each of these three nIPC clusters could be considered as a discrete biological subtype. All three subtypes were broadly visible across the iSVZ and the oSVZ (Fig. 3). However, within the iSVZ, DNM3, and PPP1R17 nIPCs showed a laminar distribution (Fig. 3 B and F). As previously noted, the iSVZ and OSVZ can be distinguished by a more radial organization of the oSVZ and by the appearance of a fiber layer separating the two zones (6), but also, as shown here, by the denser layer of EOMES- and PPP1R17-positive IPCs in the iSVZ (Fig. 3F). nIPCs closer to the ventricle were low or negative for both markers. In contrast, EOMES+/NHLH2+ cells were distributed across the iSVZ (Fig. 3C). Some of the markers of nIPC subtypes are also expressed by neurons (SI Appendix, Figs. S4 and S7C), consistent with the differentiation of neurons from nIPCs. For example, neurons in the cortical plate also express DNM3 (N1) and NHLH2 (N2). One nIPC subtype marker, PPP1R17 (N3), was uniquely expressed in the germinal zone (SI Appendix, Fig. S7A) where it was also found in cells that did not costain with EOMES (SI Appendix, Fig. S7B). The presence of TBR1 in these PPP1R17+/EOMES− cells suggested that this population may represent neurons derived from N3 nIPCs (SI Appendix, Fig. S7D). These results suggest that some nIPC subtype markers, like PPP1R17, may remain expressed in newborn neurons prior to migration out of the germinal zone.
Fig. 4.
(A) RG-like nIPCs coexpress several classic RG genes including SOX2 and VIM. (B) VIM and SOX2 protein are colocalized with a subset of nIPCs in the iSVZ and the oSVZ. VIM, SOX2, and EOMES triple positive cells are marked by filled-in arrows. EOMES+ nuclei are marked by empty arrows.
While most nIPC subtypes were prevalent during early neurogenesis, DLX5+ nIPCs appeared in late neurogenesis (Fig. 5). When each age within our dataset was clustered independently, a DLX5+ cluster was not evident before GW18, but was clearly distinct at GW22 (Fig. 5B).
Fig. 5.
(A) One cluster of nIPCs expresses DLX5. (B) This DLX5+ cluster appears at later ages in our dataset. The DLX5+ cluster is absent at GW10 and GW14. By GW18, this is a small cluster to the Left, and by GW22, there is a large cluster of DLX5+ cells. This pattern repeats with other datasets. For the GW17–GW18 Polioudakis et al. (16) dataset, a small cluster of DLX5+ cells is visible. (C) Our validation dataset (GW25) confirms a DLX5+ population. (D) DLX5 and EOMES costains a subset of EOMES+ cells at GW20 and GW22. Filled-in arrows mark costaining, and empty arrows mark EOMES staining. (E) EOMES+ cells at GW20 can also be GABA+, suggesting that inhibitory neurons may be generated from these nIPCs at this time. Colabeled cells are marked with filled-in arrows.
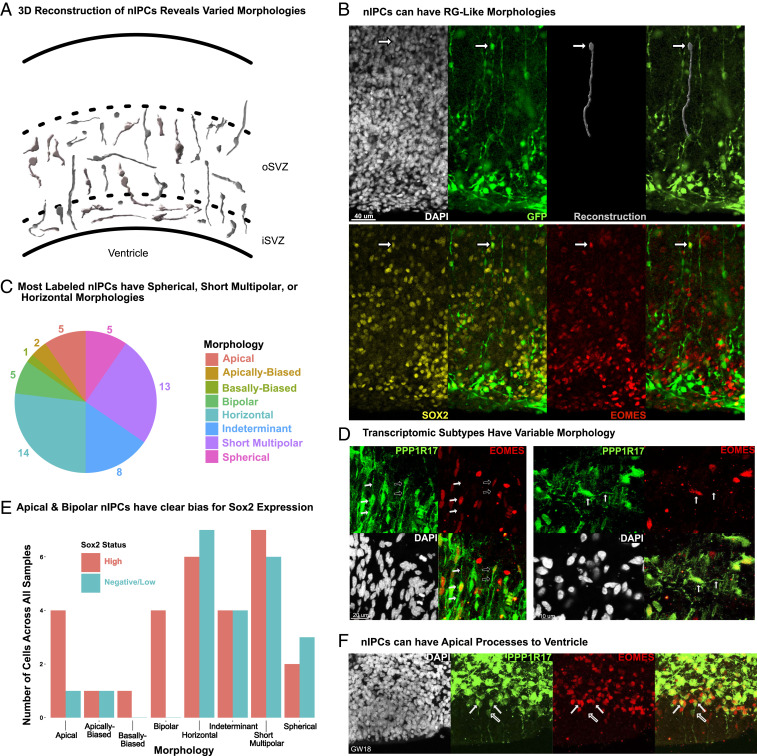
Likewise, we found that DLX5+ cells are present in a very small cluster in a previously published GW17–GW18 dataset (14) (Fig. 5C), while a large DLX5+ nIPC cluster is evident in a separate GW25 dataset (Fig. 5C). When we evaluated DLX5 and EOMES coexpression, our samples from GW20 (n = 3) and later had significant colabeling (Fig. 5D). We did not note such colabeling, or similar ventricular staining, between GW14 and GW18 (n = 3). Noticeably, at GW20, we also observed a proportion of EOMES+ cells labeled by GABA (Fig. 5E), suggesting that these progenitors may give rise to GABAergic neurons (Discussion). These results suggest that unique molecular programs may be active during late neurogenesis that may not be expressed at earlier stages. nIPC morphology does not correlate with transcriptomic subtypes. Many features of the developing nervous system were initially identified based on observations of cellular morphology as typified by the work of Ramón y Cajal (33). RG have been defined by their long radial extensions, while subtypes of ventricular and outer RG are often differentiated by the presence or absence of apical extensions. Transcriptomic differences have recently been related to RG morphology, and these features together help define RG subtypes (13, 26). In contrast, the morphology of nIPCs remains largely undefined. We sought to characterize the morphology of human nIPCs through viral labeling and immunohistochemical approaches (Fig. 6). We began by infecting organotypic human brain slices with adenoviral-CMV-GFP to label dividing progenitors and identified nIPCs by poststaining with EOMES. Reconstruction of EOMES+/GFP+ cells revealed a wide range of morphologies across the iSVZ and oSVZ. Our results clearly indicate the EOMES+ cells can have very long processes, up to 175 μm in length (Dataset S4). Cells with processes less than 30 μm we labeled as either apically biased, basal biased, or short multipolar, depending on the direction and number of extensions. Noticeably, we did not observe nIPCs with only a single long basal process, and only 1 cell out of 53 was basal biased. However, we did observe several nIPCs with long apical or bipolar extensions (Fig. 6B), similar to cells that have been labeled as RG in previous publications (34). Out of 53 total nIPCs, 5 had no processes, and we observed similar numbers of apical or bipolar nIPCs, suggesting that nIPCs are just as likely to have long processes as they are to have none (spherical morphology). In contrast, short multipolar and horizontal nIPCs constituted more than half of nIPCs (Fig. 6C). To control for alterations that could result from organotypic slice culture or viral labeling, we sought to identify nIPC morphologies in thin sections of fixed tissue stained for EOMES and PPP1R17, a marker that labels the cell bodies of N3 nIPCs. Among EOMES+/PPP1R17+ cells we observed horizontal as well as apical and bipolar morphologies, corroborating our previous observations (Fig. 6D). As with our viral labeling approach, we did not find obvious examples of basal PPP1R17+ nIPCs. Our PPP1R17 staining also suggests that morphologies do not correspond to transcriptomic subtypes, since N3 nIPCs could be observed with multiple different morphologies (Fig. 6D). The large morphological diversity across individual nIPCs suggests that caution should be exercised when identifying cell types based on morphology alone.
Fig. 6.
(A) A representation of reconstructed nIPCs based on GFP viral labeling and EOMES costaining. (B) nIPCs can have very long extensions that resemble radial glia. (C) Most labeled nIPCs either had no extensions (spherical), had minimal extensions (less than 30 μm), or had horizontal morphologies with long extensions that ran parallel to the VZ. (D) PPP1R17+/EOMES+ cells have varied morphologies with both apical and basal extensions (filled-in arrows) and with only apical extensions (empty arrows). Horizontal morphologies can also be found. (E) Sox2 expressing nIPCs can have varied morphologies. (F) nIPCs can contact the ventricle. EOMES+ cells with low PPP1R17 expression can have extensions to the ventricular edge. We observed this staining in all samples observed from GW14 to GW18.
To further investigate the possible correspondence between transcriptomic subtypes and morphology, we quantified the SOX2 expression status of our reconstructed nIPCs shown in Fig. 6A. SOX2 marks RG-like nIPCs in humans, and we found that apical and bipolar nIPCs had a clear positive bias for high SOX2 expression. However, Sox2-high cells also had other morphologies and at similar numbers to Sox2-low/negative cells (Fig. 6E). This wide range of observed morphologies suggests that transcriptomic nIPC subtypes may not be reliably identified based on their morphology alone. Lastly, we identified EOMES+ cells with low levels of PPP1R17 in the VZ/iSVZ. These cells were particularly common between GW14 and GW18, and had apical projections extending to the ventricle, a feature characteristic of vRGs (Fig. 6F). These nIPCs could potentially be influenced by factors within the ventricle or the ventricular niche. Our findings suggest that apical surface contact, long apical processes, or long bipolar processes cannot be used as the sole basis for identifying radial glia as these features are also observed across multiple nIPC types.
Discussion
Studies of the progenitor cells underlying human cortical neurogenesis have largely focused on RG cells. Indeed, models of corticogenesis center around spatiotemporal, transcriptomic, and morphological diversity of the RG population, of which there appear to be at least three subtypes in the developing human brain (13, 18, 19, 26, 35). However, neuronal output during neurogenesis also depends significantly on the role of nIPCs. Defining the identity and diversity of nIPCs will be critical to understanding the production of neuronal diversity during normal development as well as alterations to neural populations associated with neurodevelopmental diseases. A comprehensive atlas of human nIPC cell subtypes can help resolve lineage relationships of neuronal and glial cell types and may clarify the intermediate steps by which RG influence neurogenesis and corticogenesis. In our study, we observe RG-like and N1 to N3 nIPC subtypes in both vRG and oRG niches, the iSVZ and oSVZ, respectively, during early neurogenesis. After GW19/GW20, we observe that the oRG niche becomes depleted of nIPCs, with only small numbers remaining in the iSVZ. We find a clear difference in EOMES between the iSVZ and the oSVZ in all samples at all ages, with a clear laminar pattern in the iSVZ, regardless of age, and an abrupt decrease in EOMES across the oSVZ at the end of neurogenesis. Previous studies have also found a shift at this time, but with a different pattern of EOMES staining. Malik et al. reported a diffuse and relatively unstructured pattern across the SVZ, with little to no structural difference between the iSVZ and oSVZ (22). Several experimental details may explain the difference in observation, as Malik et al. relied on tissue collected after a much longer postmortem interval, from patients suffering from serious premorbid conditions, including sepsis, heart failure, respiratory failure, and necrotizing enterocolitis. In contrast, our samples were collected with a postmortem interval of 10 to 20 min. It is difficult to ascertain whether the observed differences result from a greater postmortem interval, a biologically important stress response, or other experimental differences.
We did not observe the diffuse, late neurogenic staining pattern of Malik et al. (22) using two different EOMES antibodies, and multiple neuronal markers that were expressed earlier in the germinal zone. Additionally, we see a concordance between our staining pattern and the single-cell data, with N1, N2, and N3 nIPC clusters enriched for early nIPCs and depleted of late neurogenic nIPCs (after controlling for sample size). Since we observe a dramatic shift in the distribution of nIPCs, late neurogenesis may be a developmentally distinct period from early neurogenesis. We find four nIPC subtypes that dominate early neurogenesis across both the iSVZ and oSVZ and a fifth transcriptomic subtype of nIPC unique to late neurogenesis. These five nIPC subtypes were validated using three independent approaches: internal statistical controls, bioinformatic cross-validation with two other datasets, and immunohistochemistry. The primary transcriptional features appeared to be maturity, with one RG-like IPC, and four more neuron-like nIPC subtypes. The transcriptional features of more neuron-like nIPCs may be evidence that neuron subtype identity starts emerging at the nIPC stage. For example, NHLH2 marks both N2 nIPCs and a specific subset of excitatory neurons in the cortical plate (13). However, the neuronal lineages of N1 and N3 are not as clear, since DNM3 appears to be broadly expressed in the cortical plate, and PPP1R17, an excitatory lineage gene, is expressed only by IPCs and newborn excitatory neurons in the germinal zone (SI Appendix, Fig. S7 A and D) (13). It is tempting to speculate that these two broader subtypes (N1 and N3) may relate to the broader categories of upper layer intracortical and deep layer subcortical projection neurons. It is also possible that the RG-like nIPCs produce the more neuron-like N1, N2, or N3 nIPC subtypes. However, significant follow-up work will be needed to test this hypothesis and elucidate the exact lineage of N1 and N3 nIPCs. Unlike RG, the transcriptomic nIPC subtypes do not appear to be correlated with a particular morphological feature. However, nIPC morphologies do overlap with some morphological patterns typically seen in RG. Our observations help clarify the range of molecular and morphological properties of nIPCs and can be used to identify the functions of nIPC subtypes in future studies.
While the majority of nIPCs are found in the iSVZ, a significant number of nIPCs appear in the oSVZ in early neurogenesis, where we observed higher rates of KI67+ labeling, suggesting high proliferative activity. These results are consistent with previous observations of nIPCs dividing multiple times unlike their mouse counterparts that usually undergo one division (2, 3). These observations suggest that oSVZ nIPCs may be more highly proliferative than their iSVZ counterparts. Surprisingly, this observation was the only clear molecular difference between nIPCs in the iSVZ and the oSVZ, despite evidence that these nIPCs arise from molecularly and morphologically distinct RG subtypes. Cytoarchitecture does not appear to be strongly correlated with gene expression and did not suggest a relationship between a particular nIPC subtype and an RG subtype, though we cannot definitively rule out a possible lineage association. Most transcriptomic nIPC subtypes (RG-like and N1 to N3 nIPCs) were found across both the iSVZ and oSVZ. The only spatial bias appears to be with N1 and N3 nIPCs, both of which showed preferential location within the iSVZ. This pattern changed after GW19, when the spatial distribution of nIPCs dramatically shifts. At this point, a novel DLX5-expressing nIPC appears (Fig. 5 B and C). The absence of nIPCs in the oSVZ and the minimal number of nIPCs in the iSVZ at these later timepoints suggest a change in neurogenesis and coincide temporally with the onset of gliogenesis (23). The near absence of excitatory neuron markers in the proliferative zones at this age supports this notion (SI Appendix, Fig. S2). These observations are also consistent with the model that excitatory neurogenesis is largely dependent on EOMES but suggests that the inverse may not be true. EOMES might be capable of promoting inhibitory neuronal identity as well. The presence of GABA within the iSVZ, and within EOMES+ cells specifically, suggests that GABAergic neurons may arise from nIPCs in the cortex at late stages of neurogenesis. These results are consistent with a previous report of rare DLX+ and GABA+ radial glia in the human dorsal cortex during late neurogenesis (GW20) (36). The exact nature of this late neurogenic period, whether locally produced GABAergic neurons are destined for the cortex or possibly for the olfactory bulb, and the noncanonical role of EOMES and DLX5 will be interesting to explore. In developing rodent cortex, EOMES+ nIPCs are known to contribute to extracortical sites, such as the lateral olfactory bulb tract and ventromedial hypothalamic nucleus, but do not give rise to cortical interneurons (5). A source of cortical GABAergic interneurons in the developing human cortex would represent a species difference between human and mouse.
Here we have conducted an analysis of nIPC morphotypes and attempted to relate them to molecular subtypes. Unlike RG, we find that nIPC morphologies do not relate well to transcriptomic subtypes and may exhibit RG-like shapes, such as long apical and bipolar extensions. However, long range basal processes may be unique to oRG cells, since we did not observe this morphological feature in nIPCs. We frequently observed EOMES+ cells with these morphologies, suggesting caution when classifying progenitor cells by these features (34). The clear bias for high SOX2 expression in morphotypes with long processes indicates that RG-like nIPCs may preserve a RG-like morphology. However, roughly half of our spherical EOMES+ cells were RG-like nIPCs based on SOX2 expression. The diversity of RG-like nIPC morphologies may help explain observations from Betizeau et al., where RG-like cells with long bipolar or apical extensions could be produced by cells without processes that were defined as nIPCs (34). Betizeau et al. also observed cells with bipolar morphology that retracted their processes entirely. Importantly, the above patterns were true only for bipolar and apical morphologies, not basal ones. These observations correlate well with our observations of nIPC morphologies. It is highly likely that RG-like nIPCs with apical and bipolar processes can dynamically change shape. RG-like nIPCs may convert from bipolar or apical to spherical and produce additional RG-like nIPCs, which then extend long, RG-like processes. The potential for such dynamic activities requires future study. Additionally, we observed long horizontal processes in Sox2 low/negative IPCs, suggesting that long processes are not unique to RG-like nIPCs. To validate that nIPC subtypes could take on a wide variety of shapes in vivo, we stained for PPP1R17, which marks the cell bodies of nIPCs and their processes. PPP1R17 is also not highly expressed by RG-like nIPCs (SI Appendix, Fig. S5). We observed PPP1R17+ nIPCs with a wide range of morphologies, corroborating our viral labeling in organotypic cultures.
Additionally, we found that nIPCs can have extensions to the ventricle, a trait traditionally linked to RG. These results indicate that RG morphological properties alone, including apical extensions and long basal processes, cannot be used to differentiate between RG and nIPCs.
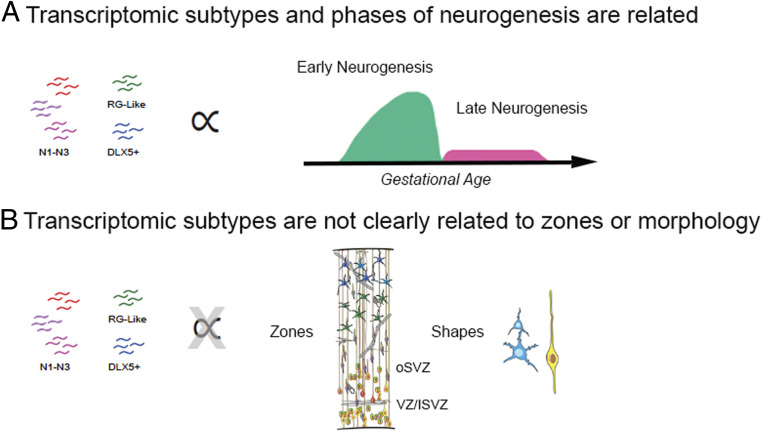
Within the RG population, spatial, molecular, and morphological diversity converge upon clear cellular subtypes. For nIPCs, however, our results indicate that these defined axes of variation do not similarly converge (Fig. 7). While transcriptomic subtypes can be found, they do not relate well with germinal zone niche or morphological features. We also found a dramatic temporal shift of nIPC diversity as neurogenesis shifts to gliogenesis, altering the distribution of nIPCs in the germinal zones and correlating with the appearance of a novel nIPC subtype. These results provide a dynamic roadmap of changing nIPC diversity and characteristics during human brain development and will help inform future studies of the mechanisms by which nIPCs influence neurogenesis.
Fig. 7.
Transcriptomic subtypes of IPCs are distinguished by their stage of differentiation (A) and are not distinguished by location or morphology (B).
Methods
Bioinformatics Analysis.
Our main dataset was published by Bhaduri et al. (29). All initial clustering and marker identification were conducted in Seurat version 2. When noted, some subsequent analysis was conducted in Seurat version 3. Cells were quality controlled by less than 5% mitochondrial input. We filtered for all cells that expressed EOMES above 0.25 after normalization. The comparison dataset was a GW17–GW18 dataset from ref. 14. After filtering, the cells were scaled and batch corrected according to methods previously described (29). We ran the FindAllMarkers command and correlated the specificity. Using this correlation matrix and our clustering of scrambled data, we combined these initial clusters heuristically. The final clusters included one defined entirely by ribosomal genes, and two clusters defined by mitochondrial genes. Three clusters were defined by cell cycle genes and scored as either G2/M or S phase cells by Seurat’s cell cycle scoring function. The remaining clusters were considered high-quality G1 phase clusters and tentatively representative of biological subtypes.
Clusters and Cluster Marker Generated on Scrambled Data.
Both technical noise and biological variance can drive single-cell clustering, which a priori biases differential gene expression calculations. We found that even scrambled data could generate statistically significant differentially expressed cluster markers, and this P value may vary according to cluster size. To evaluate the significance of differentially expressed marker genes, we created a significance threshold of each cluster according to size by generating a distribution of marker P value scrambled data. Cluster markers were evaluated by their corresponding cluster’s P value threshold. In more detail, the original gene expression matrix was scrambled 1,000 times by genes across single cells. This scrambling preserved the distribution of individual gene expression but scrambled any coordinated gene expression that might mark biological nIPC subpopulations. For each iteration, clustering was performed at varying resolution to generate a range of cluster sizes. For each iteration and resolution, we recorded the cluster sizes and identified positively expressed cluster markers using the Seurat command FindAllMarkers. To generate our comparison distribution of P values, we identified all clusters that were within 50 cells of each original cluster size, and for each iteration, we found the resolution for which the most clusters were generated. Using this filtered marker list, we generated a null distribution of false positives using significant (P < 0.05) Benjamini–Hochberg corrected P values. After a negative-log transform, we used the 95th quantile of these P values as our significance threshold. A robust number of significant markers were found in all our final clusters, with exception of archetypic nIPCs (Dataset S2). Given that clustering could be influenced by technical noise, we validated the significance of each cluster by comparing its P value distribution to P values generated from similarly sized clusters of scrambled data, as discussed previously. After conducting an unpaired Mann–Whitney U test without continuity correction, all but one cluster (archetypic) had marker P values that were significantly different from the scrambled dataset (Dataset S3). Code for this work can be found at https://github.com/MPPebworth/HumanIPCs. Viral labeling of slice culture and analysis of organotypic slices of developing human brain tissue were prepared and infected with CMV::GFP adenovirus (Vector Biolabs) as previously described (1, 29). Four to five days after viral infection, the slices were fixed in 4% paraformaldehyde for 1 h at room temperature, and washed two times in phosphate buffered saline. Slices were then stained according to a previously described protocol with antibodies for GFP (1:500, AB13970 Abcam, RRID: AB_300798), Sox2 (1:300, sc-365823 Santa Cruz Biotechnologies, RRID: AB_10842165), and EOMES (1:250 AF6166 R&D Systems, RRID: AB_10569705) (1).
Morphological Analysis.
For morphological analysis, we identified nIPC morphologies based on the number, direction, and length of extensions. When we could track extensions farther than 30 μm toward the basal surface, apical surface, or both surfaces, we designated the nIPC morphology as either basal, apical, or bipolar, respectively. If extensions were less than 30 μm apically, basally, or directionally, morphologies were labeled as either apically biased, basally biased, or as short multipolar. Cellular extensions were measured until they passed by other GFP+ cells and extensions and could not be reliably tracked further. As such, our measurements represent a conservative estimate of nIPC projection length. Some nIPCs had no evident extensions and were labeled as spherical. Finally, for some nIPCs, we could only reconstruct the morphology in one direction because of the presence of other cells, and these cells were labeled as indeterminant. Three-dimensional cell reconstruction was done with the Imaris image analysis software.
Immunohistochemistry.
Developing human brain tissue was prepared for sectioning as previously described (1, 13). Slices were stained with primary antibodies for SOX2 (1:300, sc-365823 Santa Cruz Biotechnologies, RRID: AB_10842165), PPP1R17 (1:150 HPA047819 Atlas Antibodies/Sigma Aldrich, RRID: AB_2680166), EOMES (1:250 AF6166 R&D Systems, RRID: AB_10569705), DNM3 (1:100 GTX23458 GeneTex, RRID: AB_385058), NHLH2 (1:100 HPA055238 Sigma Aldrich, RRID: AB_2682752), DLX5 (1:100 HPA005670 Sigma Aldrich, RRID: AB_1078681), VIM (1:250 AB5733 EMD Millipore, RRID: AB_11212377), SATB2 (1:250 AB92446 Abcam, RRID: AB_10563678), TBR1 (1:100 20932-1-AP Proteintech, RRID: AB_10695502), and NEUROD1 (1:100 AB60704 Abcam, RRID: AB_943491). For TBR1, PerkinElmer’s TSA kit (RRID: AB_2572409) was used to amplify signal, following methods from previously published work (37).
Tissue Collection.
As previously published, developing human brain tissue was obtained and processed according to guidelines approved by the University of California, San Francisco Human Gamete, Embryo, and Stem Cell Research Committee (approval 10-05113) (29). Patient consent was obtained before the collection of all samples used in this study. Samples were collected within 10 to 20 min of elective termination at San Francisco General Hospital and placed in fresh, preoxygenated artificial cerebral spinal fluid on ice for preservation until fixation later that day (<8 h).
Microscopy.
Confocal imaging was conducted using Leica SP5 and Leica SP8s from the microscopy core for the Eli and Edythe Broad Center for Regeneration Medicine and Stem Cell Research. Large tile scans were taken either using similar confocal microscopes or a Keyence BZ-X from the same core facility.
Supplementary Material
Acknowledgments
We acknowledge Arturo Alvarez-Buylla for his helpful feedback and review of the manuscript before submission and Shaohui Wang for her tireless work in sectioning and preparing materials as well as coordinating access to human tissue. Funding for this work has been provided by an NSF Graduate Research Fellowship to M.-P.P. and grants from the National Institute of Neurological Disorders and Stroke and National Institute of Mental Health to A.R.K.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019415118/-/DCSupplemental.
Data Availability
Code data have been deposited in Github Reposity Human IPCs (https://github.com/MPPebworth/HumanIPCs) (38). All study data are included in the article and/or supporting information.
References
- 1.Hansen D. V., Lui J. H., Parker P. R. L., Kriegstein A. R., Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Hevner R. F., From radial glia to pyramidal-projection neuron: Transcription factor cascades in cerebral cortex development. Mol. Neurobiol. 33, 33–50 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Noctor S. C., Martínez-Cerdeño V., Ivic L., Kriegstein A. R., Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Haubensak W., Attardo A., Denk W., Huttner W. B., Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 3196–3201 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasistha N. A., et al., Cortical and clonal contribution of Tbr2 expressing progenitors in the developing mouse brain. Cereb. Cortex 25, 3290–3302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smart I. H. M., Dehay C., Giroud P., Berland M., Kennedy H., Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 12, 37–53 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Moreno F., Vasistha N. A., Trevia N., Bourne J. A., Molnár Z., Compartmentalization of cerebral cortical germinal zones in a lissencephalic primate and gyrencephalic rodent. Cereb. Cortex 22, 482–492 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Fietz S. A., et al., OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Kelava I., et al., Abundant occurrence of basal radial glia in the subventricular zone of embryonic neocortex of a lissencephalic primate, the common marmoset Callithrix jacchus. Cereb. Cortex 22, 469–481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sessa A., Mao C. A., Hadjantonakis A. K., Klein W. H., Broccoli V., Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold S. J., et al., The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 22, 2479–2484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mihalas A. B., et al., Intermediate progenitor cohorts differentially generate cortical layers and require Tbr2 for timely acquisition of neuronal subtype identity. Cell Rep. 16, 92–105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nowakowski T. J., et al., Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polioudakis D., et al., A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron 103, 785–801.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan X., et al., Single-cell transcriptome analysis reveals cell lineage specification in temporal-spatial patterns in human cortical development. Sci Adv. 6, eaaz2978 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lek M.et al.; Exome Aggregation Consortium , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firth H. V., et al., DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betizeau M., et al., Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 80, 442–457 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Pollen A. A., et al., Molecular identity of human outer radial glia during cortical development. Cell 163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rash B. G., et al., Gliogenesis in the outer subventricular zone promotes enlargement and gyrification of the primate cerebrum. Proc. Natl. Acad. Sci. U.S.A. 116, 7089–7094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalebic N., Huttner W. B., Basal progenitor morphology and neocortex evolution. Trends Neurosci. 43, 843–853 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Malik S., et al., Neurogenesis continues in the third trimester of pregnancy and is suppressed by premature birth. J. Neurosci. 33, 411–423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semple B. D., Blomgren K., Gimlin K., Ferriero D. M., Noble-Haeusslein L. J., Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 106-107, 1–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lui J. H., Hansen D. V., Kriegstein A. R., Development and evolution of the human neocortex. Cell 146, 18–36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baala L., et al., Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat. Genet. 39, 454–456 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Nowakowski T. J., Pollen A. A., Sandoval-Espinosa C., Kriegstein A. R., Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91, 1219–1227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Žunić Išasegi I., et al., Interactive histogenesis of axonal strata and proliferative zones in the human fetal cerebral wall. Brain Struct. Funct. 223, 3919–3943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krsnik Ž., Majić V., Vasung L., Huang H., Kostović I., Growth of thalamocortical fibers to the somatosensory cortex in the human fetal brain. Front. Neurosci. 11, 233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhaduri A., et al., Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freytag S., Tian L., Lönnstedt I., Ng M., Bahlo M., Comparison of clustering tools in R for medium-sized 10x Genomics single-cell RNA-sequencing data. F1000 Res. 7, 1297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acosta J. R., et al., Single cell transcriptomics suggest that human adipocyte progenitor cells constitute a homogeneous cell population. Stem Cell Res. Ther. 8, 250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pixley S. K. R., de Vellis J., Transition between immature radial glia and mature astrocytes studied with a monoclonal antibody to vimentin. Brain Res. 317, 201–209 (1984). [DOI] [PubMed] [Google Scholar]
- 33.Ramón y Cajal S., Histologie du système nerveux de l’homme & des vertébrés (Maloine, 1909). [Google Scholar]
- 34.Betizeau M., et al., Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 80, 442–457 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Thomsen E. R., et al., Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods 13, 87–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X., Zecevic N., Dorsal radial glial cells have the potential to generate cortical interneurons in human but not in mouse brain. J. Neurosci. 31, 2413–2420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raju C. S., et al., Secretagogin is expressed by developing neocortical GABAergic neurons in humans but not mice and increases neurite arbor size and complexity. Cereb. Cortex 28, 1946–1958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pebworth M.-P., Human IPCs. GitHub. https://github.com/MPPebworth/HumanIPCs. Deposited 9 July 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code data have been deposited in Github Reposity Human IPCs (https://github.com/MPPebworth/HumanIPCs) (38). All study data are included in the article and/or supporting information.