Significance
Chemical tailoring of canonical bases expands the functionality of DNA in the same manner that posttranscriptional and -translational modifications enhance functional diversity in RNA and proteins. We describe the activities of ten-eleven translocation dioxygenase (TET)-like iron(II)- and 2-oxo-glutarate–dependent 5mC dioxygenases that are encoded by several bacteriophages to enable hypermodification of C5-methyl cytosine bases in their DNA. Phage TETs act on methylation marks deposited within GpC sequences by functionally-associated cytosine 5-methyltransferases. The hydroxymethyl groups installed are further elaborated by tailoring enzymes, thereby decorating the phage DNA with diverse, complex modifications. These modifications are predicted to have protective roles against host defenses during viral infection.
Keywords: bacteriophage, TET, methyltransferase, glycosyltransferase, DNA modification
Abstract
TET/JBP (ten-eleven translocation/base J binding protein) enzymes are iron(II)- and 2-oxo-glutarate–dependent dioxygenases that are found in all kingdoms of life and oxidize 5-methylpyrimidines on the polynucleotide level. Despite their prevalence, few examples have been biochemically characterized. Among those studied are the metazoan TET enzymes that oxidize 5-methylcytosine in DNA to hydroxy, formyl, and carboxy forms and the euglenozoa JBP dioxygenases that oxidize thymine in the first step of base J biosynthesis. Both enzymes have roles in epigenetic regulation. It has been hypothesized that all TET/JBPs have their ancestral origins in bacteriophages, but only eukaryotic orthologs have been described. Here we demonstrate the 5mC-dioxygenase activity of several phage TETs encoded within viral metagenomes. The clustering of these TETs in a phylogenetic tree correlates with the sequence specificity of their genomically cooccurring cytosine C5-methyltransferases, which install the methyl groups upon which TETs operate. The phage TETs favor Gp5mC dinucleotides over the 5mCpG sites targeted by the eukaryotic TETs and are found within gene clusters specifying complex cytosine modifications that may be important for DNA packaging and evasion of host restriction.
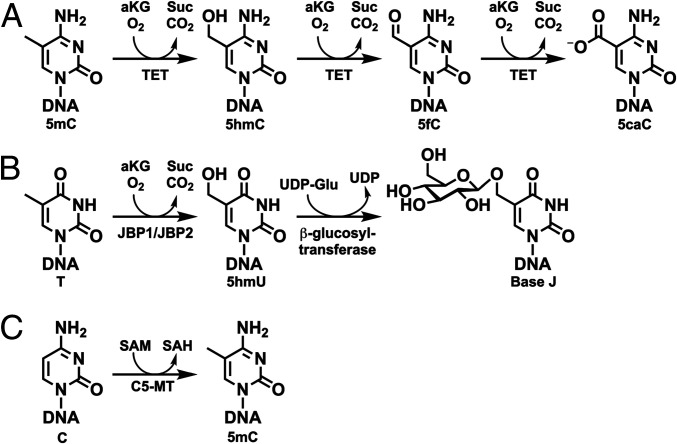
TET/JBPs (ten-eleven translocation/base J binding proteins) are iron(II)- and 2-oxo-glutarate–dependent (Fe/2OG) dioxygenases that hydroxylate the C5-methyl group of pyrimidine bases in DNA (1). In mammals, TET dioxygenase 1, 2, and 3 catalyze the iterative oxidation of the DNA 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxycytosine (5caC) (Fig. 1A) (2–4). These DNA modifications have important roles in epigenetic regulation (5, 6). The closely related Trypanosoma brucei base J-binding protein 1 and 2 (JBP1 and JBP2), with dioxygenase domains highly homologous to those of TET, oxidize thymine (T) on DNA to 5-hydroxymethyluracil (5hmU) in the first step of base J [5-(β-d-glucosyloxymethyl)uracil] biosynthesis (Fig. 1B) (7). Both base J and 5hmU have roles as molecular markers involved in regulation of genetic mechanisms of antigenic variation (8). Genome sequences and phylogenetics confirm the presence of TET/JBP enzymes in all kingdoms of life (1). Intriguingly, though, only eukaryotic orthologs have been characterized to date, despite the fact that these enzymes are thought to have originated in bacteria or phage.
Fig. 1.
(A) Iterative oxidation of 5mC by the mammalian TET dioxygenase. (B) Biosynthetic pathway for base J incorporation into DNA by JBP1/JBP2 and β-glucosyltransferase in T. brucei. (C) Methylation of cytosine by the SAM-dependent C5-MT.
It has been speculated by Aravind and coworkers that phage and bacterial homologs of the TET/JBP enzymes install hydroxy groups upon DNA 5-methylpyrimidine bases as chemically reactive handles for further complex base modifications (9). The resulting hypermodifications are hypothesized to be the result of a veritable arms race: bacterial hosts evolve more advanced restriction systems to identify and eliminate foreign DNA, while phage evolves ever more complex nucleotide biosynthesis to evade them. We became interested in this hypothesis because this role—essentially in the biosynthesis of rare DNA bases—would be a marked departure from the primary role currently assigned to the mammalian TETs, whose three products—5hmC, 5fC, and 5caC—are stable, regulatory epigenetic markers on genomic DNA (gDNA) and undergo no further modification (10, 11). Additionally, the combination of TET and cytosine-C5-methyltransferase (C5-MT) provides an alternative postreplicative pathway to the prereplicative mechanism by which bacteriophages install 5hmC into DNA. T-even coliphages invoke their cytidine hydroxymethyltransferases, hydroxymethylcytidine kinases, and the Escherichia coli nucleotide diphosphate kinase to switch their nucleotide composition (5hmC instead of C) during the evolutionary arms race with their E. coli host (12). Therefore, we investigated metavirome databases for homologs of the characterized eukaryotic TETs. We show here that phage TETs are encoded within gene clusters that specify biosynthetic pathways to hypermodify the pyrimidine bases of DNA. Among the proteins genomically paired with the phage TET is C5-MT, which utilizes S-adenosyl-L-methionine (SAM) to append a methyl group onto the C5-carbon of cytosine, thus supplying TET with its 5mC substrate (Fig. 1C). Hydroxylation of this methyl group by the TET affords the handle for other enzymes encoded in the cluster to further elaborate the 5mC base (SI Appendix, Fig. S1). In certain clusters, the downstream tailoring enzymes include glycosyltransferases (GTs) that append one or two glucose units upon the cytosine-derived base (Fig. 2). Notably, the nucleotide sequence specificity of the cooccurring C5-MTs correlates to their phylogenetic clustering, which is congruent with the clustering of the phage TETs by their amino acid sequences. A GpC dinucleotide preference is prevalent, and a single-step oxidation activity is detected for these viral enzymes.
Fig. 2.
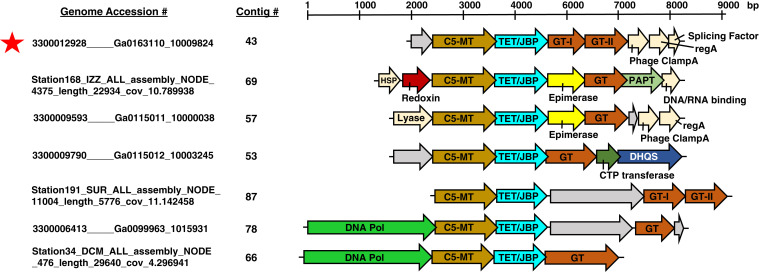
Select viral metagenomic contigs encoding C5-MT, TET/JBP, and GT genes. A genome accession number and a contig number assignment are shown to the left of each contig. A more detailed description of the accession numbering system and database sources is located in SI Appendix, Table S1 and SI Materials and Methods. Color assignments of gene predictions are as follows: C5-MT in brown, TET/JBP in cyan, GT in orange, epimerase in yellow, redoxin in red, DNA polymerase in fluorescent green, CTP transferase in dark green, polyamine aminopropyltransferase (PAPT) in light green, and 3-dehydroquinate synthase-like (DHQS) in dark blue. Genes that are colored beige correspond to phage regulatory proteins. Gray genes are proteins of unknown function. The red asterisk marks contig 43, whose C5-MT, TET/JBP, GT-I, and GT-II genes, were expressed and functionally tested both in vivo and in vitro.
Results
A Bioinformatic Screen for C5-MT/TET–Encoding Contigs of Metavirome Origin.
It has been hypothesized that eukaryotes requisitioned TETs from bacteriophages through the course of evolution and repurposed them to lay epigenetic marks on the genome (9). As an approach to identify possible TET progenitors, we used the BLAST algorithm to search the Global Ocean Virome Project (GOV 2.0) (13) and Joint Genome Institute’s Integrated Microbial Genomes-Virus (IMG/VR) (14) databases for sequences similar to the mammalian and Naegleria gruberi TETs. As TET is highly similar to the T-dioxygenase, JBP, and both target DNA 5-methylpyrimidine bases, we screened our results for assemblies encoding both a C5-MT and a TET. The result was a dataset of 32 metagenomic contigs that harbored high confidence C5-MT and TET coding frames (BLAST E-value cutoff of 1 × 10−4) that are parallel and are separated by no more than five genes (SI Appendix, Fig. S2). Examination of the 32 contigs revealed the C5-MT/TET pair to frequently cluster with one or multiple genes with functional annotations suggesting that they might modify DNA. The most common of these cooccurring genes are GTs (Fig. 2), which have sequence and structural fold similarities to 5hmC-GTs from T-even phage that glucosylate 5hmC in the coliphage DNA and protect it from restriction endonucleases (15, 16). In some cases, the GT gene is accompanied by an open reading frame encoding an apparent nicotinamide-adenine dinucleotide–dependent epimerase from the dehydratase family (e.g., contigs 57 and 69), which is speculated to act on a nucleotide sugar to afford the substrate for the GT (17). Another commonly present gene is the P-loop nucleotide kinase (P-LK) similar to that from ΦW-14, SP10, Vi1, and M6 phages (SI Appendix, Fig. S2) (18). In these phages, the P-LK is believed to synthesize 5-(pyrophosphoryloxymethyl)uracil (5-PPmU) from 5hmU on the pathway to thymine hypermodification (9). An analogous role in phosphorylating the TET-generated 5hmC would be possible for the cases of contigs 49, 76, and 88, with the expectation that the product would be further modified by downstream hypothetical proteins (SI Appendix, Fig. S2). In certain contigs (e.g., 72, 80, and 86), the P-LK gene is accompanied by a DNA base glycosylase-like gene termed “alpha-glutamyl/putrescinyl thymidine pyrophosphorylase” (aG/PT-PPlase), which has been hypothesized to act on 5-PPmU during DNA hypermodification in SP10 and ΦW-14 phages (9). Several contigs contain redoxins specifically related to the alkyl hydroperoxide reductase AhpC family, which act as molecular chaperones to suppress the aggregation of their client proteins under oxidative- and heat-stress conditions (19). Redoxins colocalize with genes for heat-shock proteins (HSPs) in the C5-MT/JBP contigs, possibly implying a role for these enzymes in controlling heat-stress response during early stages of bacteriophage infection and proliferation (20). Several other proteins with roles in phage particle assembly and infection—such as the chromosome-partitioning protein ParB, the viral DNA packaging terminase, lysozyme, splicing factor protein, clamp loader A subunit, and T4 capsid protein—are also observed in the C5-MT/TET contigs (SI Appendix, Fig. S2). These associations might indicate a role for the C5-MT/TET contigs in viral morphogenesis. Finally, the genomic conservation of the C5-MT and TET genes across identified contigs in gene content and in many cases organization and association with higher modification enzymes such as GT and P-LK is possibly linked to horizontal gene exchange of functional modules between various phages, which ultimately contributes to the adaptation of the bacteriophage to challenges in its environment (21–24).
Phylogenetic Analysis of Metavirome C5-MT and TETs.
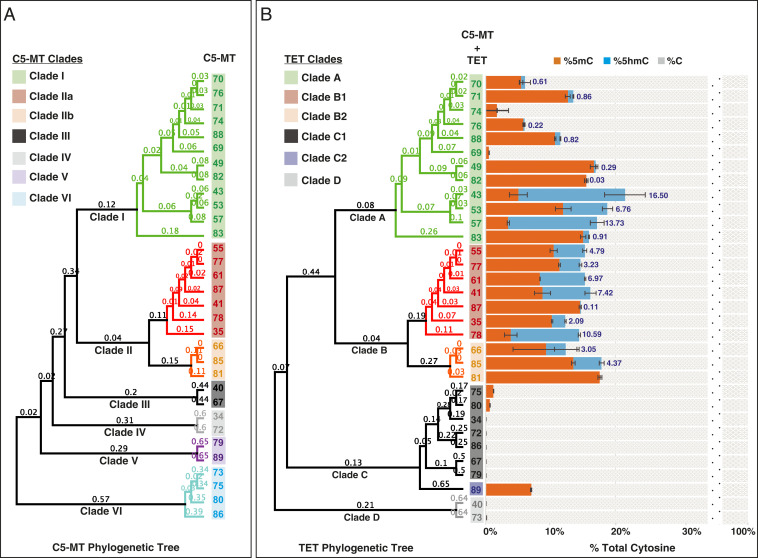
Toward revealing an evolutionary pattern of C5-MT/TET-encoding contigs, we generated and analyzed phylogenetic trees of the C5-MTs and TETs. The C5-MT and TET regions from every gene cluster were extracted, translated, and aligned (SI Appendix, Figs. S3 and S4). A tree was generated for each enzyme, and the orthologs of each protein were sorted into clades (color-coded in the figure) on the basis of tree topology (Fig. 3). Overall, the C5-MT and TET phylogenetic reconstructions share almost identical clade segregations, with a few minor divergences. An especially striking resemblance is seen in clades I and II of the C5-MTs when compared to clades A and B of the TETs. Additionally, common gene neighborhoods are found within these clades. For example, clade I/A members have a cooccurring HSP, redoxin, and either a P-LK or a GT (SI Appendix, Fig. S2). Similarly, clade II/B members have a DNA polymerase gene upstream of the C5-MT in addition to a GT or P-LK gene downstream of TET. No distinct gene-architectural relationship is observed within the other clades.
Fig. 3.
(A) Phylogenetic analysis of C5-MTs by their amino acid sequences. (B) Phylogenetic analysis of TETs by their amino acid sequences (Left) and LC-MS/MS data of percent cytosine species formed in vivo on E. coli gDNA upon coexpression of C5-MT and TET from a specific contig as indicated by the contig number (Right). The data labels reflect the percent 5hmC formed per total cytosines as a result of the activity of the TET tested. Error bars represent the SD, n = 2.
An alignment of the predicted sequences of the hypothetical C5-MTs shows that all conserve the catalytically essential cysteine of this functional class (SI Appendix, Fig. S3). In addition, the sequences within clades I and II share high conservation of both overall sequence and known C5-MT signature motifs, while their domain architectures appear to differ only in the fact that clade I has an additional insert in the region of the target recognition domain (TRD) (25). In contrast, hypothetical C5-MTs of clades III to VI have much less sequence similarity and exhibit significant differences specifically in motifs IX (role in organization of TRD) and X (role in SAM binding) and in the TRD.
The sequence alignment of the phage TETs shows that all members conserve the catalytically essential HXD-H motif that contributes the iron ligands and the Arg that pairs with the C5-carboxylate of 2OG (SI Appendix, Fig. S4) (26). Furthermore, all the phage TETs share a conserved Arg that is presumably equivalent to R1261 from human TET2 (hTET2) and R224 from N. gruberi TET1 (NgTET1), which are shown in the X-ray crystal structures of these two enzymes [Protein Data Bank ID codes: 4NM6 (hTET2) (27) and 4LT5 (NgTET1) (28)] to hydrogen-bond with C1 of 2OG and are strictly conserved among all identified 5mC dioxygenases. Phage TETs also display a conserved Tyr, Val, and Thr, which are presumably analogous to the residues of hTET2 and NgTET1 [(h, Y1902; Ng, F295); (h, V1900; Ng, V293); and (h, T1372; Ng, A212)] that form a hydrophobic pocket in the active site and may be important in substrate selection (5mC vs. 5hmC vs. 5fC) (29, 30).
In Vivo Activities of Predicted Metavirome C5-MTs and TETs.
To test the proposed functions of the hypothetical C5-MT and TET proteins as partners in DNA modification, we coexpressed pairs in T7 Express E. coli cells and used a mass spectrometry-based approach to analyze the bacterial gDNA for cytosine methylation (Fig. 3, orange bars) and hydroxymethylation (Fig. 3, blue bars). Coexpression of any of the C5-MTs from clades I and II with their associated TETs from clades A and B (respectively) led to detectable cytosine methylation of the gDNA and, in most of these cases, some degree of subsequent hydroxylation of the new methyl group. Expression of three C5-MTs from clades V and VI with the associated TETs from clade C led to some cytosine methylation without detectable hydroxylation, while the other members of clades III to VI/C and D displayed no detectable activity. The results establish the 5mC-oxidation activity of the metavirome-derived TETs in clades A and B and provides evidence of DNA 5mC oxidation by a viral enzyme.
Consistent with their inactivity, the C5-MTs from clades III through VI either lack, or have disruptions in, methylation domains VIII, IX, and X, although they do conserve the catalytic cysteine in methylation domain IV (SI Appendix, Figs. S3 and S5). These sequence variations could explain the low (or lack of) methylase activity of the clade III to VI orthologs. The coexpression experiments could have failed to identify 5mC-oxidation activity in TETs associated with the inactive C5-MTs. Therefore, we coexpressed each of the TETs that appeared to be inactive with one of the three most active C5-MTs from clades I and II (C5-MT 43, 41, and 85) and analyzed the gDNA of E. coli for modifications (SI Appendix, Table S2). Very modest activity could be detected for two of these TETs (TET40 and TET89 produce 0.93 and 0.15% 5hmC, respectively) that had otherwise appeared to be inactive when coexpressed with their cognate C5-MT. The rest of clade C1, C2, and D TETs remained inactive toward the 5mC introduced by the efficient heterologous C5-MT. Interestingly, one of the rescued TETs (TET89 from clade C2) was separately found to oxidize ∼10% of total thymine bases in the E. coli gDNA to 5hmU, indicating that it has dual T- and 5mC-oxygenase activities (SI Appendix, Fig. S6). It is conceivable that T might even be the relevant physiological substrate for TET89 considering that more 5hmU product is formed compared to 5hmC (SI Appendix, Fig. S6 and Table S2). This result is interesting, given that no other viral enzyme has previously been shown to hydroxylate T in DNA. The TET sequence alignment reveals no obvious explanation (e.g., lack of conservation of residues known to be involved in substrate recognition or catalysis) for the inactivity of the TET orthologs in clades C and D (SI Appendix, Fig. S4), although it is apparent that the general domain architecture of TETs is better conserved in clades A and B than in clades C and D. Likewise, the structural basis for the dual T and 5mC-oxidation activity of TET89 is also not clear. In general, not much is known about the selectivity of 5-methylpyrimidine dioxygenases for 5mC versus T (31). Therefore, more detailed biochemical and structural investigations will be needed to address these notable observations. We also retested select TETs from clades A and B2 (TET69, 70, 74, and 81) that have shown minimal hydroxylation activity when coexpressed with their cognate C5-MT with the very active C5-MT43, 41, and 85 (SI Appendix, Table S2). Increased hydroxylation was shown for TET69, 70, and 74, suggesting that the original low activity might be related to inefficient methylation by the cognate C5-MT. This is not the case for TET81, which remained inactive upon coexpression of C5-MT 43, 41, and 85 and whose cognate C5-MT is very active. Upon closer examination of contig 81, it seems that the gene immediately upstream of TET81 was misannotated as a gene with undefined function and should have been part of TET81.
Identification of Phage C5-MT and TET Recognition Sequences.
We employed NEBNext enzymatic methyl sequencing (EM-seq) to map regions of the genome methylated by the C5-MTs (32). In this method, gDNA isolated from E. coli cells with the C5-MT gene expressed overnight was used to construct DNA libraries that are subjected to three enzymatic conversion steps, as detailed in SI Appendix, SI Materials and Methods and Fig. S7 (light brown box), to differentiate 5mC from unmethylated cytosines. Additionally, E. coli gDNA from cells expressing both TET and C5-MT was used as input DNA for libraries to identify the subset of methylated sites that were oxidized by phage TETs and, thus, provide information on the recognition sequences of these enzymes (SI Appendix, Fig. S7, blue box). In this case, we applied the recently described protocol for detection of 5hmC modifications (33). The method as applied in this study has been described in SI Appendix, SI Materials and Methods. Control libraries are generated from gDNA obtained from cells transformed with empty vectors. Sequence logo information reflecting C5-MT or TET specificity is generated by applying the Fisher’s exact test to call statistically significant, differentially modified bases in C5-MT–expressed or C5-MT + TET–expressed samples in comparison to no-enzyme control (34).
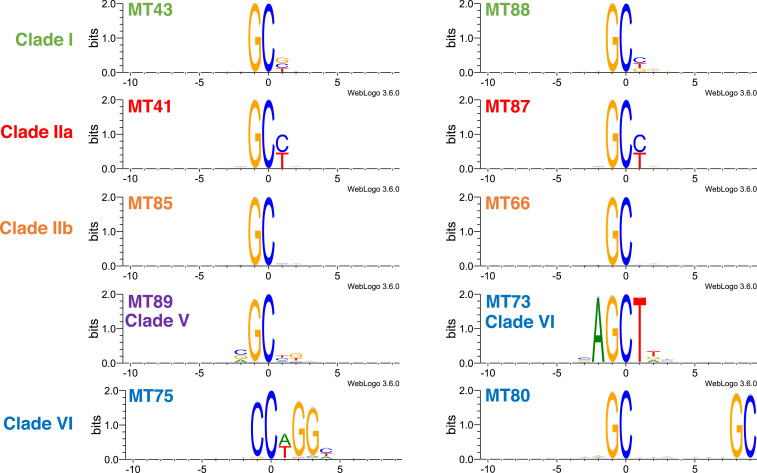
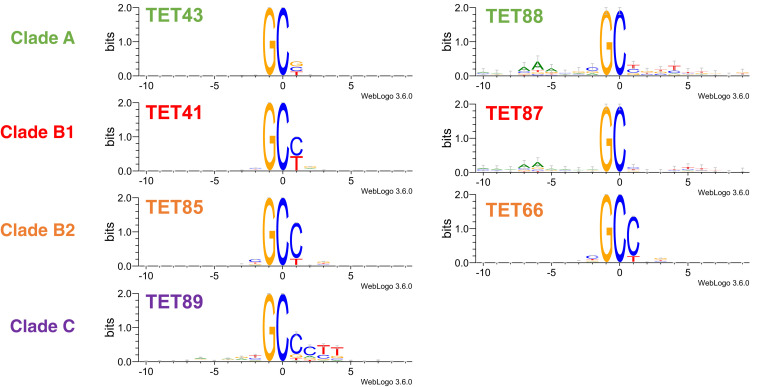
From each of clades I, IIa, and IIb we chose two representative C5-MTs (Fig. 4) with comparably high methylation activity but whose cognate TETs mediate varying levels of subsequent methyl hydroxylation (Fig. 3). For clades III to VI we tested all members that showed methylation activity (Fig. 4); their associated TETs had undetectable 5mC oxidation activity in vivo (Fig. 3). The results reveal 1) that the phage C5-MTs tested generally favor a GpC-containing sequence and 2) that members of clades I, IIa, and IIb are similarly specific for GpC[C/T]. Among these orthologs, clade IIb C5-MT66 and 85 appear to be the most promiscuous in also targeting GpCN (N is any base), and clade IIa C5-MT41 and 87 are the most specific in acting on only GpC[C/T] (SI Appendix, Fig. S8). Clade I C5-MT43 and 88 methylate GpC[C/T/G] but do not target GpCA (SI Appendix, Fig. S8), whereas members of clade VI do not share a common sequence specificity (Fig. 4). C5-MT activity could not be detected in any orthologs from clades III and IV (Fig. 3), and, consequently, their substrate specificity could not be delineated.
Fig. 4.
Sequence logo plots of DNA methylation motifs by C5-MTs. Experimental details pertaining to sequencing and generation of sequence logos are described in SI Appendix, SI Materials and Methods.
When testing TETs for methyl hydroxylation specificity on their DNA substrate, we coexpressed them with their cognate C5-MTs in T7 Express cells and mapped the E. coli gDNA for hydroxymethylated cytosines using EM-seq with the enzymatic oxidation step omitted from the manufacturer’s protocol (SI Appendix, Fig. S7, blue box) (33). From each of clades A, B1, and B2, we selected two members (Fig. 5) that supported different levels of 5hmC production in the initial experiments (Fig. 3) in order to investigate if this variation results from a more stringent sequence specificity or different levels of overall activity. Clade C2 TET89 was the only active 5mC dioxygenase outside of clades A and B (as shown in SI Appendix, Table S2 when coexpressed with clade I and II C5-MTs), so we also examined its methyl hydroxylation specificity when coexpressed with C5-MT43 (Fig. 5). The results show that TETs also favor Gp5mC-containing motifs. This specificity may simply be a consequence of the fact that their activities require prior action of a C5-MT that selectively targets GpC, as demonstrated in the previous section. A deeper bioinformatic analysis of the 5hmC modification levels of all GCN (SI Appendix, Fig. S9), NGC (SI Appendix, Fig. S10), and NGCN sites (SI Appendix, Fig. S11) that are sequenced in the E. coli genome established that the TETs have no strong preference at the −1 position and that selectivity at the +1 position is for either C or T. This selectivity is similar to that shown for C5-MTs (compare SI Appendix, Figs. S9 and S8). TET43 and 88 prefer Gp5mC[C/T/G] sites while TET41 and 87 prefer Gp5mC[C/T] sites. The only difference is that TET85 and 66 divulge a stricter site selectivity (Gp5mC[C/T]) than their cognate C5-MTs, which act on all +1 position bases (compare SI Appendix, Fig. S9 clade B2 to SI Appendix, Fig. S8 clade IIb). The histogram plot (SI Appendix, Fig. S12), which reflects the distribution of GCN sites converted to GhmCN sites at different hydroxymethylation levels, clearly reveals that TET88 and TET87 have the same substrate specificity as TET43 and TET41, respectively (SI Appendix, Fig. S9), suggesting that variation in expression or overall activity rather than differences in sequence selectivity are responsible for the observed variation in modification levels between TET 5mC dioxygenases within the same clade.
Fig. 5.
Sequence logo plots of DNA methyl hydroxylation motifs by TETs when coexpressed with their cognate C5-MT in E. coli. One exception is TET89, which was coexpressed with C5-MT43 of clade I. Experimental details related to sequencing and generation of sequence logos are described in SI Appendix, SI Materials and Methods.
Examining the Activity of Phage TET43 In Vitro.
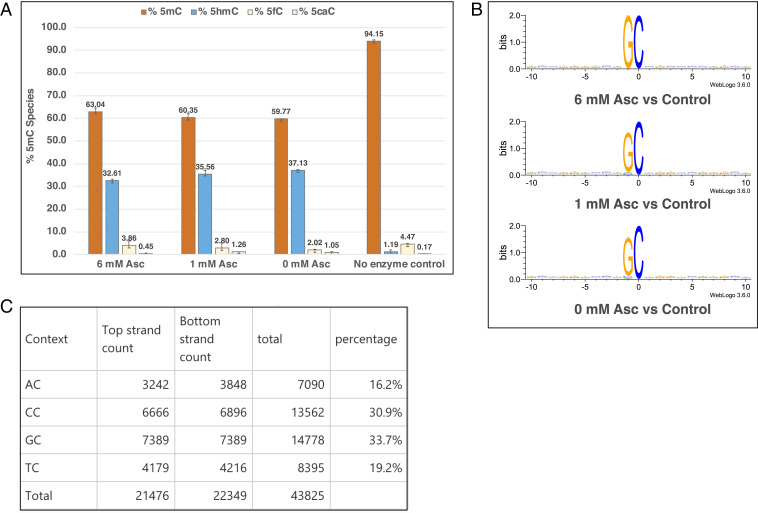
After the identification and in vivo activity screening of non-eukaryotic 5mC dioxygenases, we examined the in vitro activity of the best performing phage TET, TET43. The full-length, C-terminally (histidine)6-tagged phage TET43 was expressed and purified to homogeneity (SI Appendix, Fig. S13) and tested for activity on DNA extracted from Xanthomonas oryzae bacteriophage Xp12, which contains 5mC in place of all Cs in its genome (35). This specific choice of DNA substrate enables the examination of TET43 function on 5mC in all sequence contexts and a comprehensive analysis of the enzyme’s substrate specificity without the bias toward GpC[C/T/G] elements imposed by the cognate C5-MT43. A liquid chromatography–mass spectrometry/mass spectrometry (LC-MS/MS)–based assay was used to detect and quantify 5mC and its oxidized products in the reaction of TET43 with Xp12 DNA. Approximately 35% of all 5mC of Xp12 DNA (6.3 μM 5mC or 144 × 10−3 μM DNA) could be oxidized by TET43 (20 μM) to 5hmC product under conditions of 50 mM MES, pH 6.0, 70 mM NaCl, 5 mM 2OG, and 80 μM Fe(II) with an overnight incubation at 37 °C (Fig. 6A). Unlike mammalian TET2 and NgTET1, TET43 appears to not require ascorbate or any other reducing agent for maximal oxidation activity (Fig. 6A) (31). This observation suggests that the enzyme couples oxidation of 5mC and decarboxylation of 2OG efficiently while effectively maintaining its Fe(II) cofactor in the reduced form.
Fig. 6.
(A) LC-MS/MS data showing 5mC oxidation in vitro on Xp12 gDNA (6 ng/μL or 6.3 μM 5mC) by TET43 (20 μM) in 50 mM MES, pH 6.0, 70 mM NaCl, 5 mM 2OG, and 80 μM Fe(II) and varying concentrations of ascorbate (6, 1, or 0 mM). The reactions were incubated for ∼17 h at 37 °C. Error bars represent the SD, n = 3. (B) Sequence logo plots of Xp12 methyl hydroxylation motifs by TET43. Experimental details are found in SI Appendix, SI Materials and Methods. (C) Calculation of percent NC sites in Xp12 DNA. As Gp5mC constitutes 33.7% of all 5mC in Xp12, the concentration of Gp5mC is calculated to be 2.1 μM.
We mapped the 5mC sites targeted by TET43 on the Xp12 DNA substrate using EM-seq, with the enzymatic oxidation step omitted from the manufacturer’s protocol (see SI Appendix, SI Materials and Methods for details) and found that phage TET43 is specific for Gp5mC (Fig. 6B). We calculated that GC constitutes 34% of total NC sites in Xp12 (Fig. 6C). This prevalence correlates with the percent total oxidized product determined by LC-MS/MS (Fig. 6A). Additionally, the enzyme retains a more relaxed specificity than its corresponding C5-MT43. Perhaps the most intriguing observation in the reaction of phage TET43 on 5mC is that it performs only a single oxidation step under the conditions tested, resulting in the formation of 5hmC but not 5fC or 5caC, in contrast to what has been observed for the eukaryotic TETs from mouse, humans, N. gruberi, and Coprinopsis cinerea (2, 3, 36, 37).
The C5-Cytosine–Hypermodifying Activity of GT43/14-I and II.
To further test whether the gene neighbors of the metavirome TETs in clades A and B are truly a biosynthetic gene cluster that function in hypermodification of cytosine we tested for the GT activity in the Pfam-predicted GT-I and GT-II enzymes encoded within contigs 43 and 14. Contig 43 was chosen because the functions of both the C5-MT and TET it encodes were confirmed both in vivo and in vitro (Figs. 3B and 6A and SI Appendix, Figs. S5 and S14). Contig 14 is from Proteobacteria bacterium TMED261 and has a gene architecture analogous to that of contig 43 (SI Appendix, Fig. S15A) (9). The genome of P. bacterium TMED261 was assembled at the National Center for Biotechnology Information from the publicly available reads from the TARA Oceans Project (38). Its C5-MT and TET proteins have high sequence identity to the corresponding contig 43 proteins (SI Appendix, Fig. S15 B and C) and also mediate methylation followed by methyl hydroxylation of cytosines in vivo (SI Appendix, Fig. S15D). Pairwise alignment of GT14-I and II to their corresponding proteins in contig 43 shows that they are fairly similar (SI Appendix, Fig. S16 A and B). To assay their activities in vitro, we successfully expressed and purified to homogeneity the full-length, C-terminally (His)6-tagged GT43-I, GT14-I, and GT14-II (SI Appendix, Fig. S16C). Unfortunately, we were not able to obtain a soluble form of GT43-II.
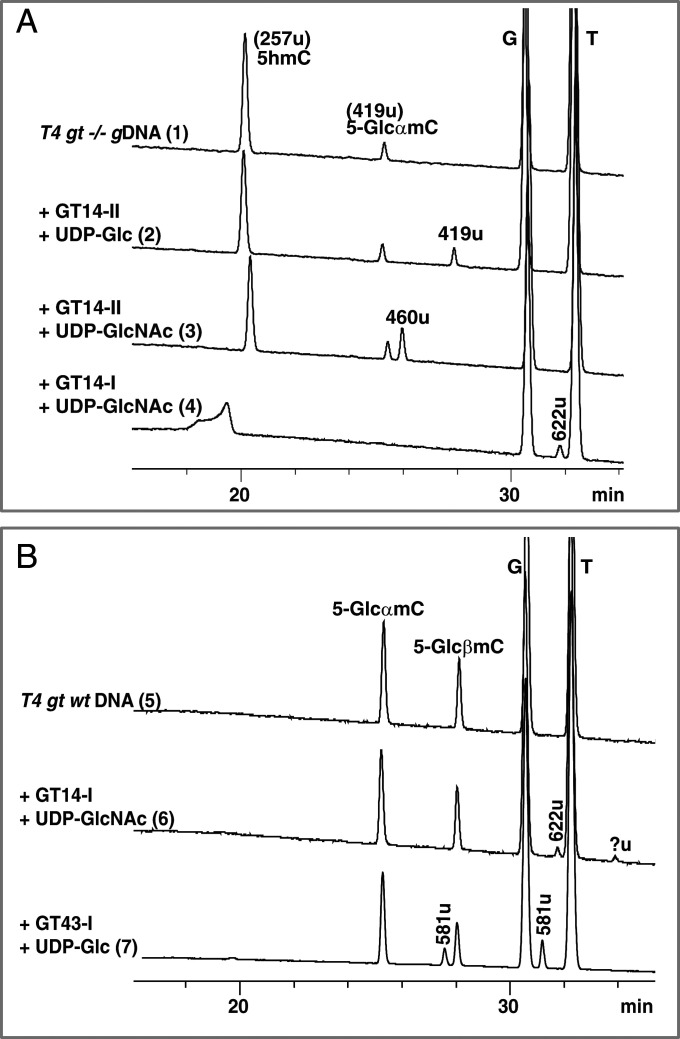
The activities of the soluble GTs were tested on DNA substrates that were extracted from E. coli T4 phage wild type (wt) and T4 phage gt−/−. T4 phage wt has all its cytosines in an α- or β-5-(d-glucosyl)oxymethylcytosine form (70% 5-GlcαmC and 30% 5-GlcβmC) (Fig. 7B, trace 5) (15). T4 phage gt−/− is a double mutant defective in both the α- and β-GT genes; its DNA thus consists mainly of 5hmC (low residual activity of the α-GT results in <4% of the 5hmC content’s being converted to 5-GlcαmC) (Fig. 7A, trace 1) (39, 40). In addition to the 5hmC/5-GlcmC–containing DNA substrate, the assays contained one of a number of uridine diphosphate-sugar donors that could potentially be utilized by the GT as a cosubstrate (Fig. 7 and SI Appendix, Fig. S17 A and B). GT14-II was indeed found to append glucose (Glc) or an N-acetylglucosamine (GlcNAc) upon the 5hmC base (257 u), as confirmed by the appearance of two new species with nominal masses of 419 u (at 29.3 min) and 460 u (at 27.4 min), respectively (Fig. 7A, traces 2 and 3). The Glc group is appended in the β-configuration as it exhibits the same retention time as 5-GlcβmC released from digestion of T4 phage wt gDNA to nucleosides. A decrease in the relative abundance of 5hmC after treatment of T4 phage gt −/− DNA with GT14-II and UDP-sugar validates the conclusion that 5hmC is the sugar acceptor (SI Appendix, Table S3). While the anomeric form of product of GlcNAc transfer has not been determined, we have assumed it as having a β-configuration by analogy with Glc transfer by the same enzyme.
Fig. 7.
LC-MS analysis showing the (A) in vitro activity of purified GTs in the presence of UDP-sugar and T4 phage gt−/− DNA (traces 1 through 4) or (B) T4 phage wt DNA (traces 5 through 7). Nominal masses are labeled for each peak and further described in the text. Experimental details are described in SI Appendix, SI Materials and Methods.
Following the same reasoning above, GT14-I and GT43-I were demonstrated to be C5-cytosine-disaccharide-forming enzymes (Fig. 7B, traces 6 and 7). Treatment of T4 phage wt gDNA with GT14-I and UDP-GlcNAc results in the production of two new peaks: The first is with nominal mass of 622 u at 31.7 min and the second at 33.9 min with a mass signal that is below the sensitivity of detection of the instrument. Examination of the relative abundance of α- and β-anomers of 5-GlcmC (419 u) revealed a decrease in both values for the GT14-I–treated sample (SI Appendix, Table S3, trace 6) when compared to the no-enzyme control (SI Appendix, Table S3, trace 5). Furthermore, subtraction of 419 u (5-GlcmC) from 622 u (new species) yields a mass difference that corresponds exactly to the transfer of a GlcNAc functionality to 5-GlcmC. The evidence thus implies that GT14-I transfers a second sugar to both α- and β-anomeric forms of 5-GlcmC, resulting in the formation of two new anomeric species (Fig. 7B, trace 6). GT43-I behaves similarly to GT14-I in its reactivity with both forms of 5-GlcmC, enabling the transfer of a second sugar to these species. GT14-I favors UDP-Glc as a sugar donor and results in two new products with an identical nominal mass of 581 u (at 28.2 and 31.8 min, respectively) (Fig. 7B and SI Appendix, Table S3, trace 7). Because we were not able to obtain a soluble form of GT43-II, we tested the activity of its crude lysate but obtained no proof that it is a functional enzyme (SI Appendix, Fig. S18). However, judging from the similarities of the GT43-II and GT14-II protein sequences and the gene architectures of contigs 43 and 14, as well as the in vivo evidence detailed below (specifically Fig. 8, trace 13), we anticipate that the latter will behave similarly to GT14-II in its transfer of a sugar moiety from a UDP-sugar donor to 5hmC. It is worth noting that GT43-I, GT14-I, and GT14-II did not show reactivity toward UDP-Gal or UDP-GalNAc cosubstrates (SI Appendix, Fig. S17 A and B), indicating selectivity toward UDP-Glc/GlcNAc sugar forms.
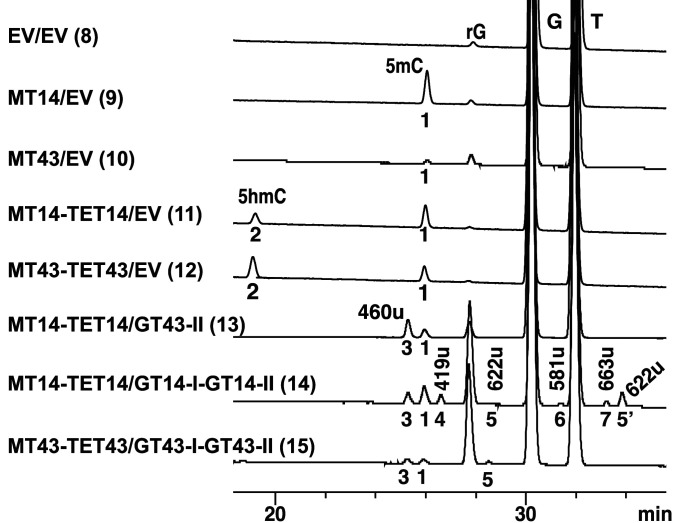
Fig. 8.
LC-MS analysis of in vivo activity of C5-MT, TET, GT-II, and GT-I of contigs 14 and 43. Nominal masses are labeled for each peak and further described in the text. Peak 1 (243 u) corresponds to 5mC. Peak 2 (257 u) corresponds to 5hmC. Peak 3 (460 u) was attributed to 5-GlcNAcßmC, peak 4 to 5-GlcßmC, peaks 5 and 5′ (622 u) to 5-GlcNAcGlcmC, peak 6 (581 u) to 5-GlcGlcmC, and peak 7 (663 u) to 5-GlcNAcGlcNAcmC. Anomeric configurations of disaccharides 581, 622, and 663 u were not determined. EV, empty vector and r, ribo form of the nucleoside.
To investigate the ability of C5-MT, TET, GT-I, and GT-II in contigs 14 and 43 to act collaboratively in vivo, we utilized two compatible duet vectors, pETduet-1 and pACYC-duet-1 (Novagen), to express the four genes in E. coli. We then monitored the formation of a diglycosylated 5mC derivative on the bacterial genome by LC/MS. The results confirm that these enzymes can operate collaboratively with C5-MT to further elaborate its methyl mark on cytosine (Fig. 8, traces 9 and 10, peak 1 [243 u]). TET hydroxylates the methyl group to install a nucleophilic “handle” (Fig. 8, traces 11 and 12, peak 2 [257 u]), GT-II transfers the first sugar (Fig. 8, traces 13, 14 and 15, peaks 3 [460 u] and 4 [419 u]), and GT-I adds the second sugar to the first in the final step (Fig. 8, traces 14 and 15, peaks 5 [622 u], 6 [581 u], and 7 [663 u]). The fact that all nominal masses 419 u (+Glc), 460 u (+GlcNAc), 581 u (+GlcGlc), 622 u (+GlcNAcGlc), and 663 u (+GlcNAcGlcNAc) were found in the assay with GT14-I/GT-14-II confirms that both enzymes can accept UDP-Glc and UDP-GlcNAc as substrates (Fig. 8, trace 14). In contrast, only 460 and 622 u were found in the assay with GT43-I/GT-43-II, suggesting that GT-43-II prefers UDP-GlcNAc (Fig. 8, trace 15). Taken together, the LC/MS data from both the in vitro and in vivo experiments provide strong support to the assignment of the products and functions of C5-MT, TET, GT-II, and GT-I enzymes in contigs 14 and 43.
Discussion
Discovery of a 5mC-Dioxygenase Subfamily from Bacteriophages.
Of the widely spread TET/JBP family, viral and prokaryotic members remain completely unexplored. We have herein unveiled a subfamily of active 5mC dioxygenases from bacteriophage origins and shown their genomic and functional association with genes that are involved in hypermodification of cytosine in DNA at the C5 position. Not surprisingly, these 5mC dioxygenases are closely linked to C5-MTs that are responsible for methylating the cytosine sites on the genome, which are subsequently targeted by TET for hydroxylation. The phylogenetic trees corresponding to 32 Pfam-annotated C5-MTs and TETs show similar hypothetical paths to the present-day enzymes (Fig. 3 and SI Appendix, Fig. S5). This observation, combined with the in vivo evidence establishing the collaborative methylation and methyl hydroxylation functions of C5-MT and TET, respectively, strongly supports a coevolutionary model for these phage enzymes. The C5-MTs mainly recognize a GpC[C/T] consensus sequence, which, according to REBASE, is a specificity not previously shown for any other C5-MT (41). One subclade, IIb, is promiscuous and methylates any GpC dinucleotide (Fig. 4 and SI Appendix, Fig. S8), a specificity which has been previously detected for M.CviPI methyltransferase from Chlorella virus IL-3A (42). In fact, among currently studied prokaryotic C5-MT, the only other dinucleotide-specific methyltransferase besides M.CviPI is M.SssI from Spiroplasma sp. strain MQ1, which recognizes a CpG site (43). According to REBASE, both CviPI and M.SssI are orphan methyltransferases, meaning that they exist alone with no companion restriction endonuclease. In bacteria, such enzymes are mainly involved in gene regulation, whereas in viruses they confer protection from host restriction endonucleases (44). The phage-derived enzymes that we have discovered could have either of these functions. It is thought that eukaryotic cytosine DNA methyltransferases emerged via fusion of new domains to ancestral methyltransferases from bacterial restriction-modification systems and assumed functions in epigenetic regulation (45). In eukaryotes, cytosine methylation is mainly found on CpG. It is speculated that the choice of a palindromic dinucleotide sequence for methylation in eukaryotes is due to the fact that CpG is the simplest sequence that contains the desired base to be methylated in both strands. This facilitates maintenance of information in semiconservative DNA replication, by parental strand instruction of daughter-strand methylation. Also, CpG methylation confers higher stability to double-stranded DNA as a result of improved stacking by the methyl groups in the CpG context (46, 47). Since GpC palindromes harbor the same biophysical properties as CpGs, it is possible that the selectivity of C5-MT to G upstream of C might be the result of similar factors. For clade VI C5-MTs, different specificities are exhibited as compared to enzymes in the other clades and among each other (Fig. 4). Among these specificities, only AGCT has been previously observed—in the Arthobacter luteus (ATCC 21606) M. AluI enzyme (REBASE).
The activities of the 5mC dioxygenases that we have identified are coupled to those of clade I and II C5-MTs and exhibit a similar Gp5mC[C/T] recognition motif (Fig. 5 and SI Appendix, Fig. S9). Because this preference derives from the preference of the C5-MTs, in vitro assays are required to assess the intrinsic preferences for clade A and B TETs. One example, TET43, that was subjected to in vitro biochemical characterization revealed that it selectively oxidizes Gp5mC sites, confirming that it recognizes primarily a dinucleotide motif (Fig. 6B). This specificity is similar, but not identical, to the 5mCpG dinucleotide specificity observed for the eukaryotic TETs. Future experiments aimed toward mining the bacteriophage database for TETs with alternate sequence targets [e.g., (N)5mC(N)] to examine the diversity of these ancestral 5mC dioxygenases would be of considerable interest.
5hmC as the Sole Oxidation Product of TET43.
TET43 deviates from other characterized TETs in its single-step oxidation chemistry on 5mC. In vitro reactions under single-turnover conditions ([TET43] = 20 μM and [Gp5mC] = 2 μM) at 37 °C for ∼17 h achieved complete conversion of Gp5mC sites on Xp12 substrate to 5hmC without detectable formation of higher oxidized species (Fig. 6). By contrast, mammalian TETs 1, 2, and 3, NgTET1, and CcTET have all been shown to perform consecutive oxidations of 5mC to produce 5hmC, 5fC, and 5caC (2, 3, 36, 37). TET from Apis mellifera has been reported to produce only 5hmC, but there has been no further confirmation of this activity (48). More careful biochemical and kinetic studies will be needed to confirm the aforementioned observation concerning phage TET43. If corroborated, it will indicate a new role for TET 5mC dioxygenases in bacteriophages, which must overcome the bacterial host’s defense during infection. Therefore, in situ production of 5hmC and more extensively modified cytosines is likely to be important for evasion of restriction by bacterial endonucleases (49, 50). This would be a departure from what is observed in higher organisms, such as mammals, in which TETs are essential for the production of three stable epigenetic marks (5hmC, 5fC, and 5caC) for the initiation of active demethylation and/or generation of new layers of epigenetic control (31).
C5-Cytosine Hypermodification by GTs in Contigs 43 and 14.
In this study we have shown that GT-II and I from contigs 43 and 14 glycosylate the TET-formed 5hmC in a collaborative fashion, resulting in the formation of mono- and disaccharide-modified C5-methylcytosine (Figs. 7 and 8, and SI Appendix, Fig. S17). GT14-II can use either UDP-Glc or UDP-GlcNAc as a cosubstrate (Fig. 7A, traces 2 and 3 and Fig. 8 trace 14), whereas GT43-II mainly utilizes UDP-GlcNAc (Fig. 8, traces 13 and 15). In appending the second sugar, GT43-I uses UDP-Glc as donor (Fig. 7B, trace 7 and Fig. 8, trace 15). GT14-I employs UDP-GlcNAc in vitro (Fig. 7A, trace 4, Fig. 7B, trace 6, and SI Appendix, Fig. S17A) but is shown to use both UDP-Glc and UDP-GlcNAc in vivo (Fig. 8, trace 14). Both enzymes are able to accept the α- and β-anomers of 5-GlcmC as substrates. The activities of these sugar-modifying enzymes are reminiscent of 5hmC-DNA α- and β-glucosyltransferases from T2, T4, and T6 coliphages, which generate 5-GlcαmC and 5-GlcβmC on the genomes of their phages (15, 16). However, in T-even phages, the hydroxymethyl moieties on cytosine are introduced at the nucleotide level by 2′-deoxycytidylate 5-hydroxymethyltransferase (a thymidylate synthase paralog) as opposed to postreplicatively as in the cases of TET43 and 14 (51, 52). In fact, the action of these two TETs on the DNA polymer to produce a reactive hydroxyl group that is then exploited by GTs mirrors the roles of JBP1 and 2 in trypanosomes, which are involved in the two-step production of base J (Fig. 1B).
The observation of the collaborative functions of C5-MT, TET, GT-II, and GT-I in modifying the DNA polymer as shown for contigs 43 and 14 (Figs. 7 and 8) suggests that bacteriophage developed yet another pathway for modifying its DNA during the evolutionary arms race with their bacterial hosts. These modifications could primarily function to protect the phage DNA from restriction endonuclease attack by their bacterial host (49, 50). It is, however, also possible that modifications introduced by the TET-containing biosynthetic gene clusters play roles in replication and packaging of DNA into the phage head, as evidenced by the association of the TET/JBP genes shown in contigs of Fig. 2 with ParB proteins. Aravind and coworkers have suggested that ParB proteins direct DNA-modification apparatuses to specific chromosomal sites during packaging (9). The colocalization of putative phage lysozyme and clamp A proteins with the biosynthetic genes in some contigs may also suggest that cytosine hypermodifications have function during cell infection. In fact, contig 69 contains a polyaminopropyltransferase gene among the hypothetical hypermodifying enzymes. This enzyme might generate a polyamine moiety to neutralize the charge of the DNA backbone during packaging and/or penetration of the bacterial cell wall similar to what is observed for ΦW-14, which has putrescine appended to thymine and packs DNA with 25% greater density than phage with similar sized head and canonical DNA (53). Finally, it has been proposed previously that sugar-modified cytosines in T-even phage play a regulatory role in controlling phage-specific gene expression, a function that is also conceivable for the TET-encoding gene clusters.
Materials and Methods
Data sources, bioinformatic analysis, in vivo and in vitro functional characterization of C5-MT, TET, and the C5-MT/TET/GT-II/GT-I gene cluster, EM-seq analysis and data processing, protein isolation, DNA substrate preparation, LC/MS, and all materials used in the study are listed in detail in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. Matthew Sullivan and Ahmed Zayed of The Ohio State University for sharing with us a database containing the computational translations of all predicted coding sequences in the GOV 2.0 metavirome dataset. We also thank Drs. V. K. Chaithanya Ponnaluri and Louise Williams for technical help with EM-seq, Dr. Cristian Ruse and Colleen McClung for peptide sequencing services, Drs. James C. Sameulson and Elizabeth A. Raleigh for insightful discussions, and the New England BioLabs sequencing core for Illumina sequencing.
Footnotes
Competing interest statement: S.R.L., Z.S., K.H.O., Y.-J.L., N.D., I.R.C., P.R.W., and L.S. are employees of New England Biolabs, a manufacturer and vendor of molecular biology reagents, including DNA methyltransferases and TET. This affiliation does not affect the authors’ impartiality, adherence to journal standards and policies, or availability of data.
This article is a PNAS Direct Submission. M.R.S. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026742118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Iyer L. M., Tahiliani M., Rao A., Aravind L., Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle 8, 1698–1710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He Y. F., et al., Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito S., et al., Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kriaucionis S., Heintz N., The nuclear DNA base 5-hydroxymethylcytosine is present in brain and enriched in Purkinje neurons. Science 324, 929–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellinger M. W., et al., 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 19, 831–833 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mellén M., Ayata P., Dewell S., Kriaucionis S., Heintz N., MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cliffe L. J., et al., JBP1 and JBP2 are two distinct thymidine hydroxylases involved in J biosynthesis in genomic DNA of African trypanosomes. Nucleic Acids Res. 37, 1452–1462 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borst P., Sabatini R., Base J: Discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 62, 235–251 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Iyer L. M., Zhang D., Burroughs A. M., Aravind L., Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res. 41, 7635–7655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bachman M., et al., 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachman M., et al., 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat. Chem. 6, 1049–1055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker M. J., Lee Y.-J., Weigele P. R., Saleh L., “5-Methylpyrimidines and their modifications in DNA” in Comprehensive Natural Products, Liu H.-W. B., Begley T. P., Eds. (Elsevier, 2020), vol. III, chap. 5.19, pp. 465–488. [Google Scholar]
- 13.Gregory A. C.et al.; Tara Oceans Coordinators , Marine DNA viral macro- and microdiversity from pole to pole. Cell 177, 1109–1123.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Páez-Espino D., et al., IMG/VR: A database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res. 45, D457–D465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehman I. R., Pratt E. A., On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J. Biol. Chem. 235, 3254–3259 (1960). [PubMed] [Google Scholar]
- 16.Wyatt G. R., Cohen S. S., The bases of the nucleic acids of some bacterial and animal viruses: The occurrence of 5-hydroxymethylcytosine. Biochem. J. 55, 774–782 (1953). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoden J. B., et al., Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 36, 6294–6304 (1997). [DOI] [PubMed] [Google Scholar]
- 18.Lee Y. J., et al., Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses. Proc. Natl. Acad. Sci. U.S.A. 115, E3116–E3125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamariah N., Eisenhaber B., Eisenhaber F., Grüber G., Molecular mechanism of the Escherichia coli AhpC in the function of a chaperone under heat-shock conditions. Sci. Rep. 8, 14151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrody E., et al., A bacteriophage-encoded J-domain protein interacts with the DnaK/Hsp70 chaperone and stabilizes the heat-shock factor σ32 of Escherichia coli. PLoS Genet. 8, e1003037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Sousa J. A. M., Pfeifer E., Touchon M., Rocha E. P. C., Causes and consequences of bacteriophage diversification via genetic exchanges across lifestyles and bacterial taxa. Mol. Biol. Evol. 38, 2497–2512 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatfull G. F., Hendrix R. W., Bacteriophages and their genomes. Curr. Opin. Virol. 1, 298–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix R. W., Lawrence J. G., Hatfull G. F., Casjens S., The origins and ongoing evolution of viruses. Trends Microbiol. 8, 504–508 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Lima-Mendez G., Van Helden J., Toussaint A., Leplae R., Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 25, 762–777 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J., Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 17, 2421–2435 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Que L. Jr, One motif–Many different reactions. Nat. Struct. Biol. 7, 182–184 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Hu L., et al., Crystal structure of TET2-DNA complex: Insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto H., et al., Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hashimoto H., et al., Structure of Naegleria Tet-like dioxygenase (NgTet1) in complexes with a reaction intermediate 5-hydroxymethylcytosine DNA. Nucleic Acids Res. 43, 10713–10721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu M. Y., et al., Mutations along a TET2 active site scaffold stall oxidation at 5-hydroxymethylcytosine. Nat. Chem. Biol. 13, 181–187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker M. J., Weigele P. R., Saleh L., Insights into the biochemistry, evolution, and biotechnological applications of the ten-eleven translocation (TET) enzymes. Biochemistry 58, 450–467 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Vaisvila R., et al., EM-seq: Detection of DNA methylation at single base resolution from picograms of DNA. bioRivx [Preprint] (2020). 10.1101/2019.12.20.884692 (Accessed 18 May 2021). [DOI]
- 33.Sun Z., et al., Nondestructive enzymatic deamination enables single-molecule long-read amplicon sequencing for the determination of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Genome Res. 31, 291–300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akalin A., et al., methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo T. T., Huang T. C., Teng M. H., 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae. J. Mol. Biol. 34, 373–375 (1968). [DOI] [PubMed] [Google Scholar]
- 36.Pais J. E., et al., Biochemical characterization of a Naegleria TET-like oxygenase and its application in single molecule sequencing of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 112, 4316–4321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., et al., A TET homologue protein from Coprinopsis cinerea (CcTET) that biochemically converts 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. J. Am. Chem. Soc. 136, 4801–4804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karsenti E.et al.; Tara Oceans Consortium , A holistic approach to marine eco-systems biology. PLoS Biol. 9, e1001177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgopoulos C. P., Isolation and preliminary characterization of T4 mutants with nonglucosylated DNA. Biochem. Biophys. Res. Commun. 28, 179–184 (1967). [DOI] [PubMed] [Google Scholar]
- 40.Revel H. R., Hattman S., Luria S. E., Mutants of bacteriophages T2 and T6 defective in alpha-glucosyl transferase. Biochem. Biophys. Res. Commun. 18, 545–550 (1965). [DOI] [PubMed] [Google Scholar]
- 41.Roberts R. J., Vincze T., Posfai J., Macelis D., REBASE–A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu M., Kladde M. P., Van Etten J. L., Simpson R. T., Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 26, 3961–3966 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renbaum P., et al., Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 18, 1145–1152 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy J., Mahony J., Ainsworth S., Nauta A., van Sinderen D., Bacteriophage orphan DNA methyltransferases: Insights from their bacterial origin, function, and occurrence. Appl. Environ. Microbiol. 79, 7547–7555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyer L. M., Abhiman S., Aravind L., Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 101, 25–104 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Bochtler M., Fernandes H., DNA adenine methylation in eukaryotes: Enzymatic mark or a form of DNA damage? BioEssays 43, e2000243 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Xu G. L., Bochtler M., Reversal of nucleobase methylation by dioxygenases. Nat. Chem. Biol. 16, 1160–1169 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Wojciechowski M., et al., Insights into DNA hydroxymethylation in the honeybee from in-depth analyses of TET dioxygenase. Open Biol. 4, 140110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flodman K., Corrêa I. R. Jr, Dai N., Weigele P., Xu S. Y., In vitro type II restriction of bacteriophage DNA with modified pyrimidines. Front. Microbiol. 11, 604618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loenen W. A., Raleigh E. A., The other face of restriction: Modification-dependent enzymes. Nucleic Acids Res. 42, 56–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schellenberger U., Livi L. L., Santi D. V., Cloning, expression, purification, and characterization of 2′-deoxyuridylate hydroxymethylase from phage SPO1. Protein Expr. Purif. 6, 423–430 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Wilhelm K., Rüger W., Deoxyuridylate-hydroxymethylase of bacteriophage SPO1. Virology 189, 640–646 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Scraba D. G., Bradley R. D., Leyritz-Wills M., Warren R. A., Bacteriophage phi W-14: The contribution of covalently bound putrescine to DNA packing in the phage head. Virology 124, 152–160 (1983). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.