Abstract
It is very difficult to gain a better understanding of the events in human pregnancy that occur during and just after implantation because such pregnancies are not yet clinically detectable. Animal models of human placentation are inadequate. In vitro models that utilize immortalized cell lines and cells derived from trophoblast cancers have multiple limitations. Primary cell and tissue cultures often have limited lifespans and cannot be obtained from the peri-implantation period. We present here two contemporary models of human peri-implantation placental development: extended blastocyst culture and stem-cell derived trophoblast culture. We discuss current research efforts that employ these models and how such models might be used in the future to study the “black box” stage of human pregnancy.
Keywords: placenta, implantation, human, extended blastocyst culture, trophoblast stem cells (TSC), pluripotent stem cells
Human extended embryo culture and stem-cell-derived trophoblast cells offer new insight into peri-implantation stage placental development.
Introduction
The placenta is arguably the most important solid organ in humans. It is essential to the immediate and future health of the pregnant mother and her fetus, as well as the health and well-being of generations to come. Despite this, it is also one of the most poorly understood solid organs. There are several challenges to the study of normal and abnormal placental development and function that have hindered progress. First, while much has been learned about human physiology through the study of animal models, the human placenta is in many respects unique among eutherian mammals. This distinctiveness has placed constraints on the utility of animals such as rodents, which otherwise allow facile genetic manipulation and access to a wide array of molecular reagents and methodologies for the study of many aspects of human placentation. For instance, the human placenta is remarkable in its ability to invade deeply into the maternal decidua, with some placental trophoblast cells reaching at least the inner third of the uterine myometrium and in some disease states being able to extend beyond that region. Defects in this invasion have been closely associated with intrauterine growth restriction and with preeclampsia, a pregnancy disorder seen almost exclusively in humans. Even our closest evolutionary ancestors, the nonhuman primates, show differences in placental morphology and trophoblast interactions with the maternal decidual tissues that, together with cost and access constraints, limit their value as animal models of normal and abnormal human placental form and function.
Added to the limitations of animal models for the study of the human placenta, there are significant restrictions on access to human tissues, particularly placental tissues from early pregnancy. Our most robust understanding of the human placenta comes from specimens obtained at the time of term deliveries. More limited access to placental samples from preterm deliveries is accompanied by a more limited understanding of placental function and dysfunction at this time in pregnancy. Most of our functional knowledge of the placenta from prior to delivery comes from studies of maternal blood and from imaging studies. Still, there remain virtually no reliable serum biomarkers of placental disorders. Additionally, the persistent limitations in placental imaging technologies led to issuance of the 2018 NIH Request for Applications entitled, “Novel Approaches to Safe, Non-invasive, Real Time Assessment of Human Placental Development and Function Across Pregnancy”. Placental tissue samples can be obtained early in the second trimester without interruption of the pregnancy as a byproduct of chorionic villous sampling for prenatal genetic diagnosis, but this procedure is being used less and less as new fetal screening approaches emerge. Placental tissues from the first and early second trimester can be obtained after spontaneous or elective abortions; however, the former specimens may be damaged by the etiology of the loss and the latter no longer may be used in federally funded research.
It is likely that many disorders of placentation, including isolated and recurrent spontaneous pregnancy loss, placenta accreta and percreta, intrauterine growth restriction and preeclampsia, may have their origins very early in pregnancy, possibly even in the preimplantation and peri-implantation periods. Infertility and recurrent implantation failure accompanying assisted reproduction may actually be the earliest manifestations of abnormal preimplantation and peri-implantation placental development. At this point in pregnancy, restrictions on the study of these events in humans are nearly insurmountable, as pregnancy in humans cannot be clinically detected prior to implantation, the time when an embryo begins secreting detectable levels of human chorionic gonadotropin (hCG). For all these reasons, normal and dysfunctional peri-implantation placental development and function in humans must be modeled in vitro. Here we will review two state-of-the-art in vitro approaches: 1) extended human blastocyst culture and 2) human stem cell-derived trophoblast culture and co-culture models, originating from embryonic, placental, or adult cells.
Extended Human Blastocyst Culture
Human implantation is a critical, yet elusive, biological event establishing the first contact of the expanded, hatched blastocyst with the uterine wall. The peri-implantation period is a precarious time in pregnancy, with an estimated 30% of conceptions lost prior to implantation and another 30% of conceptions lost following implantation, but before the missed menstrual period [1]. While a large percentage of these preclinical losses may be due to fetal chromosomal aberrations [2], there are a sizable number of karyotypically normal embryos also failing to implant. For these embryos, pregnancy loss is widely considered the result of a breakdown in communication between the mother and the implanting embryo. This breakdown in crosstalk has been historically difficult to investigate due to logistical and ethical reasons, leading investigators to name this period the “black box of pregnancy” [1].
The first attempt to shed light into the “black box of pregnancy” began with a Carnegie grant in 1913 to the anatomist Franklin P. Mall. Mall had a collection of over 500 human embryos [3] and used this grant to establish the Carnegie Institute of Washington’s embryology department [4]. Much of this research has been memorialized in a collection of historic papers called “Contributions to Embryology” [5]. One of the most meaningful and abiding contributions of the Carnegie Department of Embryology is the collection of samples of fallopian tubes and uteri from women requiring hysterectomies. From these tissues, the fertility researchers John Rock and Arthur Hertig collected 34 embryos ranging from the zygote stage to a 17-day embryo [6]. Similarly, the Cambridge Collection in the UK has a number of specimens collected from hysterectomies performed in early pregnancy [7]. Incredibly, these collections remain as the most informative pieces of evidence of early human embryology, still serving as the foundation of what we know today about human implantation. Mall’s work describes a placenta emerging as a layer of trophectoderm (TE) at the embryonic pole that facilitates invasion into the decidualized endometrium. At the time of implantation, the TE is comprised of two cell types: the progenitor-like cytotrophoblast (CTB) cells and the terminally differentiated, multinucleated primitive syncytium (Figure 1). As it continues to invade, the primitive syncytium becomes more prominent and hollowed, lacunar spaces quickly form that are filled first with uterine secretions and ultimately, maternal blood [6]. Until the villi and intervillous spaces form, these lacunae serve as the syncytial interface between trophoblast and maternal fluids. Beginning at approximately day 13, proliferating CTB begin to form villous structures that push through the primitive syncytium, then fill with fetal mesenchyme to form secondary villi, and finally with fetal blood vessels to form the tertiary villi by 21 days. Mature villi are covered in a single layer of syncytiotrophoblast (STB), where they contact maternal blood in the intervillous space. During this time, the ends of the emerging villi that contact the decidua form a “cytotrophoblastic shell” that gradually thins and becomes more discontinuous as the placenta grows [8]. CTB also begin differentiating toward the extravillous cytotrophoblast (EVT) lineage in the anchoring villi that contact the decidua. These EVT cells are competent to invade into the maternal decidua and break down the extracellular matrix, and eventually the myometrium and spiral arteries [9]. Since the publication of the original works from the Carnegie Institute of Washington and Boyd Collection in the UK, there has been little opportunity to expand our knowledge about this critical phase of pregnancy.
Figure 1.
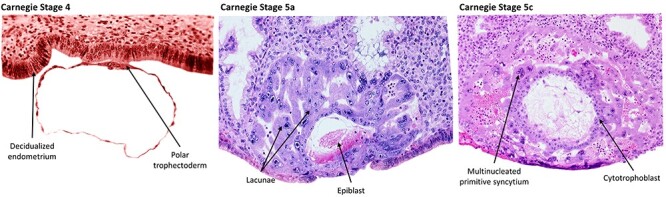
Carnegie images depicting stage 4, 5a, and 5c human embryos. These histological sections from the original Carnegie collection represent approximately days 6–12 postfertilization. At stage 4 the polar trophectoderm attaches and adheres to the decidualized endometrium. By stage 5, the CTB has given rise to a multinucleated primitive syncytium that has begun to invade and form lacunae. Carnegie stage images are from the Virtual Human Embryo Project at Louisiana State University with permission to reprint (http://virtualhumanembryo.lsuhsc.edu).
The acquisition of human embryos for in vitro culture was not possible until the 1980s when human in vitro fertilization became more routine in clinical practice [10]. Soon, extended culture to the blastocyst stage, but not beyond, became customary as a means of selecting embryos more likely to implant, but a variety of restrictions limited experimental studies on such embryos, even those that would otherwise be discarded. There were, however, multiple attempts to culture mouse embryos to a point beyond which implantation would normally have been initiated in vivo. First attempts in mouse involved pre-attachment with the culture of blastocysts for 48 h on transparent bovine lens fiber, to which they attached and began to show some aspects of tissue organization reminiscent of normal development in vivo [11]. Following this initial finding, there were multiple efforts to grow mouse blastocysts for extended periods on a variety of substrata and in an array of media to initiate further development, including placenta formation [12–17]. However, this early work to study placental trophoblast emergence was plagued by poor embryo viability, inadequate culture conditions, and inconsistent descriptions of morphological endpoints. A breakthrough was made in 2014, following the description of a system capable of supporting what appeared to be relatively normal implantation stage mouse embryo development up to embryonic day 8.5 [18]. Within 2 years, a similar culture system proved successful with human blastocysts [19, 20]. This culture system supports human embryo development up to day 13 postfertilization and recapitulates the differentiation of TE in ways that appear consistent with the histological work performed many years earlier on in vivo specimens [3–6].
Specifically, morphological features of the cultured embryos resembled those reported in the Cambridge and Carnegie collections, and the outer cells expressed known molecular markers of trophoblast in sequence. At day 6–7, before blastocyst attachment, TE of the cultured embryos expressed recognized early trophoblast markers (GATA3, CDX2, and KRT7), whereas there was very little expression of the pluripotency marker POU5F1 and hypoblast marker GATA6. Most human blastocysts attached between day 7 and 8 at the polar TE, which is the closest to the inner cell mass (ICM). At day 8, in vitro cultured embryos flattened, and POU5F1 and GATA6 staining was lost in TE. As at day 6, CDX2 remained at very low levels and was restricted to weak nuclear staining of a small subset cells at the periphery. However, KRT7 staining became prominent, suggesting progressive differentiation of trophoblast cells. At day 10, TE cells positive for GATA3 and KRT7 continued to differentiate, with a subset of new cells expressing the STB marker, human chorionic gonadotropin beta (CGB), indicating the formation of the early syncytium. At day 12, CGB staining had increased in intensity and multinucleated cells displaying lacunar structures organized along the periphery of the embryo. A layer of mononucleated cells adjacent to the substratum was also observed [19]. These features closely mimic the Carnegie stage 5b-c histological samples (Figure 1). Collectively, these data suggest that this culture system supports human embryo development past implantation in a reasonably faithful manner and recapitulates many key features of in vivo development including formation of early, primitive placenta.
The transcriptome and DNA methylome landscapes of peri-implantation stage human embryos have recently been described at days 6, 8, 10, 12, and 14 of extended culture. By using known cell type markers, cells were clustered as epiblast, primitive endoderm, trophoblast, or yolk-sac TE [21]. Sublineages of trophoblast could be recognized as distinct populations at approximately day 10. Within this grouping were cells recognized as the undifferentiated CTB, and newly differentiated, human CGB-positive STB that were expressing genes encoding pregnancy-specific glycoproteins, the PSG family [21].
The extent of global DNA methylation was unable to distinguish epiblast, primitive endoderm, and TE at the blastocyst stage. However, beyond the blastocyst stage, the cell types formed three discrete clusters indicative of a divergence of methylation patterns among these cell types during implantation [21]. Also, while methylation levels at the blastocyst stage were low, they increased asynchronously in the different lineages during time associated with implantation in vivo. The median DNA methylation level of CpG islands in the epiblast increased from 26.1% at day 6 to 60% at day 10, whereas the TE methylation levels only increased from 23.5% to 46.3%. Additionally, by day 8, the promoters for the pluripotency regulators POU5F1 and NANOG were specifically methylated in the trophoblast, as might be expected for genes that have been silenced, but not in the epiblast where they continue to be expressed. Conversely, several genes implicated in TE emergence were specifically methylated in the epiblast but not in TE. These results are consistent with the widely held view that DNA methylation plays an important role in cell fate determination by selectively silencing cell-fate genes.
While Zhou et al. [21] provided the transcriptome and methylome landscapes of all the cell lineages during human implantation, work from our laboratory focused exclusively on TE differentiation [22]. By employing single-cell RNA sequencing (RNAseq) and the same extended culture system used by others [19–21, 23], we described the dynamic changes of the transcriptome during TE differentiation from progenitor CTB to two differentiated TE sub-lineages, namely the STB that were CGB-positive and involved in producing placental hormones, and the migratory trophoblast cells (MTB) that were HLA-G-positive (Figure 2) [22]. These MTB, although carrying many EVT markers probably should not be referred to as EVT, as others have done [21, 23], since the villous structures have not appeared by the stage when MTB arise. Instead, these HLA-G+ cells make their first appearance at the periphery of the conceptus around day (D)10 and could be observed subsequently swarming away from the structure. As in earlier studies [19, 20], embryo attachment to the substratum occurred D7–8, an event that was soon followed by the formation of multinucleated syncytium around the conceptus (Figure 2). Syncytium formation reached its zenith, with a more than two-fold increase in hCG production into culture medium from day D10–11. Levels of hormone leveled off between days 11 and 12, even as the embryo continued to grow and develop. For our single cell analyses, the tiny CTB and larger STB components were readily distinguishable, allowing them to be picked individually from the trypsin-loosened D8, 10, and 12 conceptuses, while migratory cells, which were also larger than CTB, were sampled only at D12. The CTB at D8 and 12 had a transcriptome suggesting high proliferative activity, protein synthesis, and energy metabolism, which at D8 is consistent with their inferred role in supplying cells to the STB and MTB lineages and at D12 to the emergence of villous trophoblast. STB gene networks linked to protein transport, steroidogenesis, and migration. These are the cells that must release sufficient amount of hCG to rescue the corpus luteum from destruction, thereby permitting the continued production of progesterone by the ovary and prevention of menses. Without this intervention, the pregnancy would end. The early STB also has a transcriptome suggesting that these cells are invasive, a feature that is probably responsible for the ability of the conceptus to burrow into the endometrium and occupy a niche with access to nutrients necessary for its further growth and development. The MTB had even stronger features of an invasive, migratory phenotype. These cells are likely responsible for deeper and more extensive colonization of the uterine endometrium and may even be responsible for intravascular invasion to form trophoblast plugs in the lumen of spiral arteries before the placental villi form [8, 24] (Figure 3). Clustering analyses have also revealed two additional subsets of cells that were morphologically indistinguishable from CTB yet had transcriptomes with features of STB and MTB, respectively. Cells with a mixed phenotype of CTB, STB, and MTB were not evident, suggesting that CTB cells had already begun to segregate into distinct lineages by D8, yet remained highly proliferative. This lineage bifurcation became more pronounced by D10 when there appeared to be a greater stress on STB formation, while by D12 there was greater weight placed on MTB production. Additionally, at D12 there was an upsurge of undifferentiated, mitotically active CTB, probably, as remarked upon earlier, reflecting the beginnings of villous formation.
Figure 2.
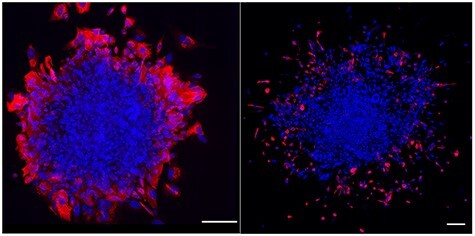
Extended human blastocyst culture. Human embryos grown in extended embryo culture at day 10 demonstrating the syncytiotrophoblast (STB) marker CGB (red, left panel), and day 12 demonstrating the MTB marker HLA-G (red, right panel). Scale bar, 150 μm.
Figure 3.
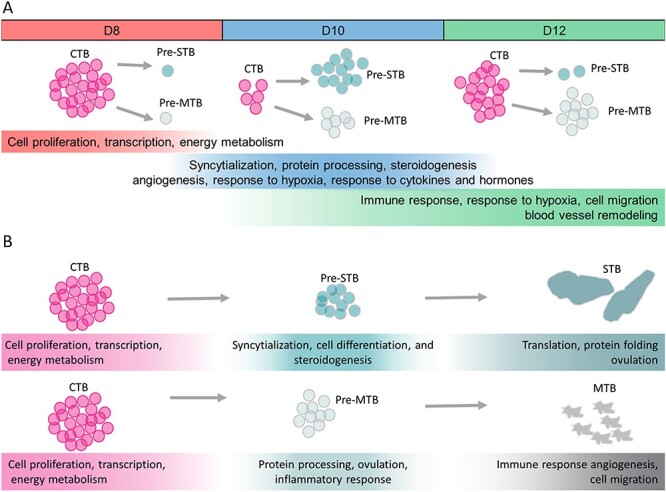
Enriched pathways in cells from the peri-implantation stage placenta. A) Gene ontology (GO) enrichment analysis reveals a transition of pathways enriched in CTB cells collected at different time points during development. B) CTB cells go through a transitory process of differentiation, remaining in the cell cycle while also displaying features of STB and MTB cells before leaving the cell cycle to become terminally differentiated to the STB and MTB lineages. This figure was adapted from ref. 22 with permission.
Another interesting finding revealed by single-cell RNAseq was the presence of several genes related to Type I and Type II interferon (IFN) signaling pathways. Some of these genes were expressed as early as day 8 in CTB, but the number and expression levels of such genes had greatly increased by D12 in association with the emergence of MTB. Despite these changes, there was no evidence for the expression of either Type I IFNs, for example IFN-alpha family members, or of the Type II IFN, interferon-gamma (IFNG), despite the high upregulation, of the interferon-stimulated genes, ISG15 and ISG20. Deeper analysis into the IFN signaling pathway identified a lack of phosphorylated signal transducer and activator of transcription 1 (STAT1), indicating that the IFN-related pathways are probably inactive at this stage, although they may be anticipating an IFN signal coming from the endometrium/decidua. We propose that the upregulated IFN response pathways may act as a safety switch when the embryo is faced with a choice between a receptive or potentially hostile uterine environment. Conceivably, IFN released by cells residing in the endometrium would immediately trigger an IFN response in trophoblast and provide some survival advantage to the embryo and serve as a first line of defense against viral and bacterial infections. Alternatively, high levels of IFN might desensitize IFN pathways and result in implantation failure. High levels of IFNG secreted by natural killer (NK) cells, for example, is associated with miscarriage [25]. In addition, in vitro studies have demonstrated that IFNG has cytotoxic effects on human trophoblast [26].
Single-cell RNAseq has also been used to probe the differentiation of trophoblast in D6 to D10 human peri-implantation stage embryos [23]. The embryo culture medium in these experiments was conditioned by co-culture with human primary endometrial cells, a feature that was presumed to provide enhanced development. The findings of Lv et al. [23] generally corroborated our own, although events appeared slightly quickened. For example, indications of STB emergence were evident at D7 and what they claimed was EVT at D8, although we suspect that the latter were the equivalent of the MTB described in our paper [22].
Finally, Xiang et al. [27] cultured human embryos in an extracellular matrix comprised of Matrigel up to embryonic D14. Single-cell RNAseq was performed on whole embryos rather than just on the primitive placenta, but trophoblast lineages could be clustered separately from others (e.g., epiblast, yolk sac, amnion) through selection with cell-type specific marker genes. As in our own work, at least five main classes of trophoblast were defined, namely CTB (defined further into early and late), early STB, STB, early EVT, and EVT. As in our observations, the early STB and early EVT appear to represent transitory differentiation states where the cells are still mitotically active but have begun to differentiate toward their specified lineages. The CTB, as expected, were concentrated close to the embryonic disk beneath the CGB-positive cells comprising the multinucleated syncytial cells, and, by D12, an outer layer of HLA-G+ cells had emerged, which again likely represent the MTB described by us [22].
Xiang et al. also made the surprising discovery that at least 120 genes encoding polypeptide hormones were expressed by trophoblast, predominantly STB, during the course of embryo development between D8 and D14. Even as early as D8, the emerging STB expressed 31 such genes including CGA, CGB, and placental growth factor (PGF). However, expression declined markedly toward the end of the culture period, possibly reflecting the demise of the STB and/or the loss of viability of the conceptuses as a whole. These observations raise at least two important questions. First, does this inferred early production of polypeptide hormones, in addition to hCG, contribute to the phenomenon of maternal recognition of pregnancy in humans? Second, does the highly motile EVT/MTB, as well as the more sedentary STB, also have role in maintaining the pregnancy in these early critical days when embryonic loss is so high? Finally, Xiang et al. noted that EVT/MTB cells of the later stage embryos expressed genes linked to immune system modulation and angiogenesis, suggesting that, as the embryo continues to develop, these migratory cells are being prepared for final roles in remodeling spiral arteries and mediating interactions with the maternal immune cells that sense a foreigner in the uterine implantation site.
These studies are, of course, only the beginning, but they provide a gratifyingly consistent start to unraveling the early mysteries of the peri-implantation stage in humans, including the timing and features of trophoblast lineage emergence and subsequent differentiation and the formation of the primitive placenta. Additionally, while great strides have been made in developing media that permit human embryos to grow and develop, the present technologies for extended embryo culture are most likely suboptimal and lack the endometrium–conceptus interactions necessary for successful in vivo implantation. As recurrent implantation failure and various placental pathologies plague couples struggling with infertility, it is critical that these new research areas be advanced, allowing us to tease out the factors that promote successful implantation.
Stem-Cell Derived Models of Early Human Placental Development
As described in the preceding section, extended human blastocyst culture has recently provided new opportunities to study peri-implantation placental development in humans. Other in vitro approaches to the study of early placental development have been widely used, but each has significant drawbacks. Several cell lines of placental origin such as HTR8/SVneo, TEV-1, ACH-3P, SGHPL-5, and HIPEC65 have been immortalized from isolated first trimester EVT by genetic manipulation and continue to provide models for human placental research [28–32]. Although the HTR-8/SVneo cell line contains a mixed population of cells [33], it remains the most commonly used to study EVT invasion, proliferation, and regulation [34–36]. Others, including BeWo, JEG-3, and JAr cells, are derived from choriocarcinomas. BeWo cells can be induced to fuse, form STB, and secrete placental hormones such as hCG, human placental lactogen (hPL), progesterone, and estradiol. These cells are frequently used to mimic villous CTB and to study syncytialization, adhesion, and placental endocrine function. JEG-3 cells can spontaneously fuse in vitro as well as release hCG, hPL, and progesterone. They too are used to study syncytialization but have also been frequently used to study proliferation and invasion of CTB. JAr cells have less of a propensity to form STB than BeWo and JEG-3 [37]. Unfortunately, the positions of each of these cell lines in the trophoblast lineage hierarchy remain unclear. Moreover, many of these cell lines have been maintained for decades in culture and likely possess genomes/epigenomes, and thereby phenotypes, that are far removed from the trophoblast lineage from which they were derived.
Primary trophoblast cells can be isolated from the placenta and grown in culture [38, 39]. Primary cell cultures can be obtained with varying success throughout much of pregnancy but are dependent on the availability of placental tissues, which differs for different stages of pregnancy. Samples are most easily obtained after delivery of normal term pregnancies and of pregnancies delivered early for maternal and/or fetal indications. The latter are most often derived from deliveries in the third trimester or very late second trimester of pregnancy. At this point in gestation, however, the study of placental pathologies is often obfuscated by an inability to determine whether abnormalities are a cause or consequence of a given pregnancy complication. Moreover, the invasiveness and number of EVT cells are significantly decreased at term [40], making the study of this trophoblast subpopulation more difficult in the most easily accessed samples. Although the syncytialization process is also attenuated at term, isolated cytotrophoblast cells from both term and preterm human placentas will spontaneously syncytialize in culture over the course of several days so these short-term in vitro approaches using primary CTB are most often used to study trophoblast fusion [41]. Placental villous explants have also been used to study the transport of small molecules by the trophoblast [42]. These in vitro culture systems can allow the study of both invasion and syncytialization of human trophoblast cells and methods have been developed to manipulate these cells genetically [43, 44]. However, for several biological and logistical reasons, access to first trimester and early second trimester placental tissues are often quite limited. Questions of fetal aneuploidy and cause versus effect arise when these placental tissues are obtained after a spontaneous pregnancy loss. For example, it is difficult to determine whether a change detected in the immune microenvironment of decidual tissues isolated after spontaneous pregnancy loss is a cause of the loss versus a local or systemic reaction to the presence of a nonviable pregnancy. Access to tissues from elective terminations of pregnancy is often limited by ethical and legal challenges and these tissues are also typically of unknown ploidy status. Neither source of early human placental tissues will reflect peri-implantation placental development.
Embryonic stem cell (ESC) technologies have been used to develop human trophoblast in vitro. Although the original ESC were obtained from the epiblast layer of the human embryo that develops after differentiation of the TE from the ICM [45], regulatory bodies in the United States have allowed the continued experimental use of selected lines. However, the creation of new human embryonic stem cell (hESC) lines cannot be pursued through use of federal funds in the United States. The derivation of trophoblast from hESC after exposure to the BMP family of growth factors, specifically BMP4, was first described by Xu et al. [46] in 2002. Building upon these results and those of others [47–53], the Roberts laboratory has refined these methods via the inclusion of two additional compounds, the ACTIVIN A signaling inhibitor A83-01, and the FGF2 signaling inhibitor, PD173074 [54, 55]. This combination of BMP4, A83-01, and PD173074 has been termed BAP treatment. Upon exposure to BAP, almost all cells become positive transiently for the transcription factor CDX2 at 24–48 h and have continued expression of the pan-trophoblast marker KRT7 thereafter [54, 55]. These cells can be maintained in culture for approximately 8–9 days before the syncytial regions begin to detach and have allowed both syncytializing and invasive subpopulations to be studied and isolated [55, 56]. Alternatively, BMP4 treatment may be performed in a defined minimal stem cell medium with the WNT inhibitor IWP2, and then directed toward either CGB-positive STB or HLA-G-positive EVT-like cells by manipulating oxygen concentrations [57].
While comparison of RNAseq results from these cells to existing databases has shown BAP cells to be definitive trophoblast, comparison to RNAseq results from CTB and STB isolated from term placenta has revealed differences in the expression levels of many trophoblast-related genes [58, 59]. This led us to hypothesize that BAP cells best modeled the primitive trophoblast arising from the newly implanted human blastocyst. We continue attempts to test this hypothesis using comparisons with primary placental tissues across human gestation and the results of extended blastocyst culture [60].
Interestingly, we have found that BMP4 needs only to be present in the culture medium at most for the first 24 h of BAP culture. Subsequently, exposure to AP supplemented conditions alone drives trophoblast formation [54, 55]. It appears that transient BMP4 exposure may drive the cells to a heightened pluripotent state, since these BMP4-primed cells can be isolated and cultured, and remain pluripotent and genetically stable over many passages [55]. Further, when these BMP-primed cells are exposed to differentiation conditions in the absence of additional BMP, the conversion into trophoblast is more efficient than that seen with parental ESC [55]. In the case of mouse pluripotent stem cells (PSCs), BMP4 augments conversion from a primed to naive status via alterations in SMAD- and lipid-signaling and chromatin modification [61–63]. The mechanisms underlying BMP4 action in human cells have not been characterized; however, dual activities in cell signaling and nuclear architecture are expected to be involved to enable trophoblast differentiation from the primed-type PSCs.
Induced pluripotent stem cells (iPSC) are derived, not from embryos, but rather from somatic cells that have been reprogrammed to an embryonic-like pluripotent state [64] (Figure 4). How similar these cells are to ESC and whether they carry an epigenetic memory of their origins has yet to be resolved [65]. Nevertheless, the epiblast type of human iPSC respond to BMP4 in a manner similar to that of true embryonic epiblast-derived stem cell counterparts and readily form CTB [66] with mixed STB and EVT cells in the culture (Figure 4). Since they are not embryo-derived, their use carries little to no ethical and legal encumbrances. Most-importantly, they can be used for personalized medicine and to study the origins of placental diseases [67].
Figure 4.
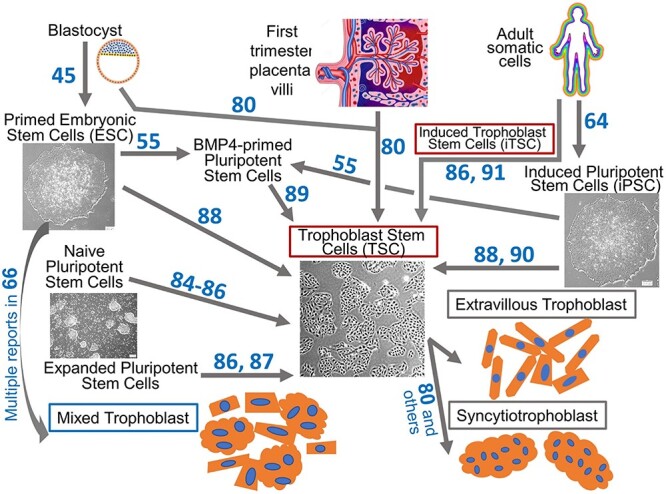
Modeling the human placenta in vitro using stem cells. Various cell type conversions between primary cells, pluripotent stem cells, and trophoblast stem cells. Reference numbers for original studies are shown next to the arrows. Human ESC and iPSC are derived from the blastocyst ICM and various somatic cells, respectively. By exposing undifferentiated ESC and iPSC to BMP4 and signaling inhibitors, the cells differentiate into a mixture of trophoblast subtypes including extravillous trophoblast and multinucleated syncytiotrophoblast [66]. Human trophoblast stem cells (TSC) can be derived from trophoblast cells of both blastocysts and first trimester placental villi. Distinct types of pluripotent stem cells (PSC), primed-, naïve-, expanded-, and BMP4 primed-PSC can be converted into TSC. Induced TSC are generated by exposing reprogrammed cells to TSC culture medium using otherwise standard iPSC generation methods. TSC generate extravillous trophoblast and syncytiotrophoblast with distinct differentiation conditions.
The study of diseases of abnormal placentation such as intrauterine growth restriction and preeclampsia is severely hampered by the biological fact that although the disease may have its origin in the earliest stages of pregnancy, including during peri-implantation placental development, the signs and symptoms of the disease may not arise until many weeks to months later. Use of iPSC-derived trophoblast models allows somatic cells from a patient who has experienced a particular disease to be reprogrammed and then differentiated to trophoblast in vitro by using the aforesaid model that recapitulates the earliest stages of placental development [67]. Sheridan et al. [67] demonstrated reduced trophoblast invasion in iPSC cell lines generated from umbilical cord fibroblasts that originated from 19 cases of early onset preeclampsia (EOPE) pregnancies. Overall differences between the EOPE and control iPSC derived trophoblast were subtle; however, reduced trophoblast invasiveness was observed only under the high (20%) O2 condition and the production of PGF was altered in response to changes in O2 conditions. It is possible that these differences in O2 sensing/protection from oxidative damage and subsequent reductions in trophoblast invasion may be common inciting factors for the abnormal placentation seen in cases of EOPE.
The etiologies of preeclampsia have been hypothesized for centuries. The disorder is most likely not a single disease but rather a common end pathway for multiple disorders that affect placental development. Further, even those groups of women who follow similar pathophysiologic pathways may have differing primary etiologic events as the disorder is almost certainly polygenic [68]. The personalized nature of iPSC should allow for comparisons between EOPE and control groups as well as among EOPE specimens to better understand the breadth of pathophysiologic pathways. This is a first opportunity to detect changes in trophoblast that may initiate the disease process. As outlined in our original description of early comparisons of iPSC derived from EOPE and control pregnancies, this model can be used to assess invasion, oxygen sensitivities, protection from oxidative damage, expression of pro- and anti-angiogenic factors, and the effects of fetal sex on the development of disease. Disease pathophysiology can be assessed at the level of transcription and translation, but also mined for epigenetic and metabolomic differences between EOPE and control trophoblast and, in a personalized approach, among the EOPE pregnancies. We envisage that interactions between these EOPE trophoblast cells and maternal decidual components can be further examined in co-culture experiments by using endometrial epithelial organoids [69, 70], decidualized stroma [71], and isolated decidual immune cells to recapitulate disease-specific maternal–fetal interactions. Use of iPSC should provide new and important insights into the origins of many of the diseases of abnormal placentation, including early and late-onset preeclampsia, intrauterine growth restriction, placenta accreta and percreta, molar pregnancies, gestational diabetes and likely some cases of pregnancy-induced hypertension.
Although it has long been assumed that placental stem cells, also known as trophoblast stem cells (TSC), exist in all placental mammals, particularly during the early stages of placental development, the quest for true human trophoblast stem cells has been unpredictably challenging. TSC populations had previously been isolated from preimplantation blastocysts in a number of animal models (porcine, bovine, rhesus monkey) and both murine and more recently rat TSC have been extensively characterized [72–76]. Several groups have reported on conditions for the isolation and/or derivation of human TSC, although none exhibited all of the self-renewal, pluripotency, and immortality characteristics of a true stem cell population [77–79]. It was not until 2018 that Okae et al. [80] reported culture conditions that allow self-renewal and long-term propagation of human TSC and TSC-derived STB and EVT-like cells (Figure 4). The discovery that EGF, WNT activation, and TGFB inhibition are necessary for the maintenance of human TSC has provided the necessary insights allowing this field to progress.
The conditions employed by Okae et al. [80] for self-renewal of human TSC are similar to those that support self-renewing human trophoblast organoids [81, 82] but substantially different from those that are compatible with mouse TSC culture [83]. Sources for human TSC appear to be limited largely to the blastocyst stage embryo and early 1st trimester placenta of pregnancy [80]. However, multiple attempts are now being made to convert PSCs into TSC, thereby allowing generation of TSC from individuals born from pregnancies with placental diseases and from individuals with diverse genetic backgrounds. Studies with naïve PSC [84–86] and expanded (or extended) PSC [87] indicate that TSC can be generated from these stem cells with the same medium used by Okae et al. [80, 87] (Figure 4). Conversion of primed PSC to TSC failed when these conditions were employed. On the other hand, when primed PSC were treated with BMP4, the TGFβ signaling inhibitor, SB431542 and the signaling sphingolipid, sphingosine 1-phosphate, two types of TSC-like cells, CDX2− and CDX2+, could be isolated. The former resembled the standard TSC of Okae et al. [80], while the latter was inferred to be a more primitive TSC [88]. Our group has generated TSC lines from BMP4-primed PSC cells by directly exposing the cells to the TSC conditions [89]. Others have demonstrated a primed human iPSC line could be converted to TSC after it was allowed to form a cyst-like cell mass on a micromesh substratum [90]. It was concluded that so far unidentified factors released during formation of the cystic mass permitted the lineage switch from pluripotency to TSC. It will be of considerable interest to understand the signaling pathways that govern the relationship between pluripotency status and the trophoblast state and how certain supplements, such as BMP4, contribute to the process. A related study has reported that induced TSC (iTSC) can be generated from human somatic cells by using the standard reprogramming factors for iPSC—POU5F1, SOX2, KLF4, and MYC [86, 91] (Figure 4). Significant in this study was the finding that the iPSC reprogramming process inherently passes through an intermediate stage that has signatures of trophoblast gene expression [91]. Human iTSC can then be established by intervening in this progression by exposing these cells to TSC culture conditions during a specific window of time during reprogramming. These results contrast with mouse iTSC, which can be established from somatic cells by overexpressing the transcription factors, EOMES, GATA3, TFAP2C, and MYC or ETS2 [92, 93]. Although this reprogramming approach has, as yet, not been successful for human somatic cells, it seems likely that it soon will be.
Recent progress is clearly broadening approaches to TSC derivation from a diversity of cell types. Combining a range of TSC sources along with the ability to differentiate such cells to EVT and STB and possibly other, yet poorly characterized, sublineages should allow rapid progress in our understanding of normal and abnormal human trophoblast development and the origins of placental diseases.
Comparison of models and conclusions
Despite years of study, morphologic and functional differences in placentation among animals have combined with drawbacks in many cell and tissue-based in vitro models to keep us profoundly ignorant about the earliest stages of human pregnancy. We are particularly unenlightened about events accompanying implantation and early placentation, when embryonic losses are exceedingly high [1] and when several other diseases of abnormal placentation are thought to arise [94]. Two novel approaches have now made it possible to study some aspects of early human placental development in the laboratory. Extended blastocyst culture opens windows onto events never seen before in humans. This system may be the closest we presently have to mimic in vivo events, but it has several shortcomings. Access to frozen human blastocysts is limited, not so much by the numbers available, but by regulations and restricted funding. Experiments use discarded human blastocysts obtained through in vitro fertilization, which may differ from those fertilized in vivo. For example, some discarded blastocysts are aneuploid embryos that the developmental potential remains unknown. Finally, although it is remarkable that these blastocysts can be cultured in the absence of typical maternal tissues and their resident epithelial, stromal, and immune cells, this same attribute may also be a drawback to the model. To a large extent, these deficiencies might be addressed through the development of co-culture models. For instance, it may be possible to allow extended human blastocyst development on a monolayer of fallopian tubal cells, which would recapitulate the surrounding in vivo environment of very early blastocyst development, or a monolayer of endometrial epithelial or decidualized endometrial cells, or even multilayered endometrial cells or explants, which would mimic the in vivo environment of later human blastocyst development. Both have been utilized in the fairly distant past [95–99] for short-term blastocyst culture to assess clinical application but none have been adopted experimentally. One could even envision staged co-culture of developing human blastocysts, first on fallopian tubal cells, followed by extended culture on endometrial cell-derived substrates, although such an approach may raise new ethical concerns in some. Of course, many of these same events, which occur prior to the clinical recognition of pregnancy, can also be mimicked in vitro by using stem cell models. The ESC/iPSC model is markedly malleable and in our own labs has already been used to study peri-implantation placental development [47, 54–56, 58, 60, 66], Zika virus infection [100], preeclampsia [67], toxicology, and placental immunology [101, 102]. We and others have used it to develop models for human blastocyst implantation. Disadvantages of the model include the inherent difficulties in verifying that trophoblast derived from the BMP4 methodologies truly mimics the primitive leading-edge invasive STB, since primary tissues representing this stage of placental development are only present in a subject prior to or very early after clinical detection of pregnancy and, even if such tissues could be obtained at the earliest clinically detectable timepoint, such access is ethically challenging. Further, the model has fairly short-term viability in two-dimensional culture conditions and experiment to experiment variability, particularly when different iPSC lines are used. The latter impediment can be fairly readily overcome. By applying the ESC/iPSC to the 3D spheroid culture approach, these stem cell-based models can be grown for months as organoids and can mimic maternal–placental interactions with endometrium organoids. Furthermore, and most critically, the use of iPSCs allows study of cells with a variety of genetic backgrounds and use of materials obtained from cases with known placental diseases and matched controls. The latter cannot be studied by using extended blastocyst culture approaches. Only iPSC-based approaches facilitate study of the origins of diseases that do not manifest until late in pregnancy and the use of personalized, precision medicine approaches. For instance, although their pathophysiology involves abnormal placental development and function that arise much earlier in pregnancy, diseases such as preeclampsia and intrauterine growth retardation most often present clinically in the late second or early third trimester. Until recently, this has dramatically limited our understanding of the etiology and early stages of these disorders, as tissues from affected women could only be studied at the end-stages of the pathophysiologic process. Using iPSCs from affected women to study the earliest stages in trophoblast differentiation and growth directly addresses this obstacle. Further, it allows investigators to dissect out the personalized effects of a mother’s specific genetic background and the sex and genetic background of a particular fetus in a specific mother in the development of diseases of abnormal placentation. Human TSC can be derived from ESC, from iPSC, and from either blastocysts or placental tissue obtained from primary placental tissues, although only those obtained during the first trimester [80] while these latter sources can pose ethical challenges, the ability to create TSC lines from tissues obtained after early miscarriage poses no ethical hurdles and is the only way to use these stem cell models to study the underpinnings of early pregnancy loss, which is the most common complication of pregnancy. Use of human TSC derived from either ESC or iPSCs or even primary pregnancy tissues will circumvent some of the interassay variability inherent in differentiation from ESC. This should help to standardize the field and should address inter-experimental variability. Human TSC will likely be used to study a wide array of pregnancy disorders. Particularly enticing are potential applications in metabolic and transcellular transport studies as well as small- and large-scale testing of environmental and pharmacologic toxicities in normal and diseased placentas.
Author contributions
ZJ, RCW, YY, and DJS wrote the manuscript and prepared the figures. ELE, TE, LCS, and RMR critically revised the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgments
The graphical abstract was created with BioRender.com.
Footnotes
† Grant Support: The authors’ research is supported by grants 1R01HD094937 (to RMR, DJS, TE, LCS) and 1R21AI145071 (to DJS, TE) from the National Institutes of Health, and Colorado Center for Reproductive Medicine (CCRM) Internal Research Grant (to RCW, YY).
Contributor Information
J Zhou, Mizzou Institute for Women’s Health Research, Department of Obstetrics, Gynecology and Women’s Health, University of Missouri School of Medicine, Columbia, MO USA; Bond Life Sciences Center, Division of Animal Sciences, University of Missouri, Columbia, MO USA.
R C West, Colorado Center for Reproductive Medicine, Lone Tree, CO USA.
E L Ehlers, Mizzou Institute for Women’s Health Research, Department of Obstetrics, Gynecology and Women’s Health, University of Missouri School of Medicine, Columbia, MO USA.
T Ezashi, Bond Life Sciences Center, Division of Animal Sciences, University of Missouri, Columbia, MO USA.
L C Schulz, Mizzou Institute for Women’s Health Research, Department of Obstetrics, Gynecology and Women’s Health, University of Missouri School of Medicine, Columbia, MO USA.
R M Roberts, Bond Life Sciences Center, Division of Animal Sciences, University of Missouri, Columbia, MO USA.
Y Yuan, Colorado Center for Reproductive Medicine, Lone Tree, CO USA.
D J Schust, Mizzou Institute for Women’s Health Research, Department of Obstetrics, Gynecology and Women’s Health, University of Missouri School of Medicine, Columbia, MO USA.
Conflict of interest
All the authors declare no conflict of interest in this work.
References
- 1. Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: The ‘black box’of early pregnancy loss. Hum Reprod Update 2002; 8:333–343. [DOI] [PubMed] [Google Scholar]
- 2. Philipp T, Philipp K, Reiner A, Beer F, Kalousek D. Embryoscopic and cytogenetic analysis of 233 missed abortions: Factors involved in the pathogenesis of developmental defects of early failed pregnancies. Hum Reprod 2003; 18:1724–1732. [DOI] [PubMed] [Google Scholar]
- 3. Mall FP. Report upon the collection of human embryos at the johns Hopkins university. Anat Rec 2005; 5:343–357. [Google Scholar]
- 4. Buettner KA. Franklin Paine Mall (1862-1917). In: Embryo Project Encyclopedia. Arizona State University. School of Life Sciences. Center for Biology and Society;2007. [Google Scholar]
- 5. Washington CIo . Contributions to embryology. In: Carnegie institution of Washington. Washington, D.C.: The Carnegie Institute of Washington; 1921. [Google Scholar]
- 6. Hertig AT, Rock J, Adams EC. A description of 34 human ova within the first 17 days of development. Am J Anat 1956; 98:435–493. [DOI] [PubMed] [Google Scholar]
- 7. Boyd JD, Hamilton WJ. The human placenta. Heffer: Wiley-Liss, Inc.; 1970. [Google Scholar]
- 8. Burton GJ, Jauniaux E. The cytotrophoblastic shell and complications of pregnancy. Placenta 2017; 60:134–139. [DOI] [PubMed] [Google Scholar]
- 9. Pollheimer J, Vondra S, Baltayeva J, Beristain AG, Knöfler M. Regulation of placental Extravillous trophoblasts by the maternal uterine environment. Front Immunol 2018; 9:2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards RG, Bavister BD, Steptoe PC. Early stages of fertilization in vitro of human oocytes matured in vitro. Nature 1969; 221:632–635. [DOI] [PubMed] [Google Scholar]
- 11. Jenkinson E, Wilson I. In vitro support system for the study of blastocyst differentiation in the mouse. Nature 1970; 228:776–778. [DOI] [PubMed] [Google Scholar]
- 12. Hsu Y-C. Differentiation in vitro of mouse embryos beyond the implantation stage. Nature 1972; 239:200–202. [DOI] [PubMed] [Google Scholar]
- 13. Wiley LM, Pedersen RA. Morphology of mouse egg cylinder development in vitro: A light and electron microscopic study. J Exp Zool 1977; 200:389–402. [DOI] [PubMed] [Google Scholar]
- 14. Hsu Y-C. In vitro development of individually cultured whole mouse embryos from blastocyst to early somite stage. Dev Biol 1979; 68:453–461. [DOI] [PubMed] [Google Scholar]
- 15. Carson DD, Tang J-P, Gay S. Collagens support embryo attachment and outgrowth in vitro: Effects of the Arg-Gly-asp sequence. Dev Biol 1988; 127:368–375. [DOI] [PubMed] [Google Scholar]
- 16. Salomon D, Sherman M. Implantation and invasiveness of mouse blastocysts on uterine monolayers. Exp Cell Res 1975; 90:261–268. [DOI] [PubMed] [Google Scholar]
- 17. Hsu Y-C. Differentiation in vitro of mouse embryos to the stage of early somite. Dev Biol 1973; 33:403–411. [DOI] [PubMed] [Google Scholar]
- 18. Bedzhov I, Leung CY, Bialecka M, Zernicka-Goetz M. In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc 2014; 9:2732. [DOI] [PubMed] [Google Scholar]
- 19. Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH. Self-organization of the in vitro attached human embryo. Nature 2016; 533:251–254. [DOI] [PubMed] [Google Scholar]
- 20. Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, Fogarty NM, Campbell A, Devito LG, Ilic D. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol 2016; 18:700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou F, Wang R, Yuan P, Ren Y, Mao Y, Li R, Lian Y, Li J, Wen L, Yan L. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 2019; 572:660–664. [DOI] [PubMed] [Google Scholar]
- 22. West RC, Ming H, Logsdon DM, Sun J, Rajput SK, Kile RA, Schoolcraft WB, Roberts RM, Krisher RL, Jiang Z. Dynamics of trophoblast differentiation in peri-implantation–stage human embryos. Proc Natl Acad Sci 2019; 116:22635–22644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lv B, An Q, Zeng Q, Zhang X, Lu P, Wang Y, Zhu X, Ji Y, Fan G, Xue Z. Single-cell RNA sequencing reveals regulatory mechanism for trophoblast cell-fate divergence in human peri-implantation conceptuses. PLoS Biol 2019; 17:e3000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moser G, Windsperger K, Pollheimer J, Sousa Lopes SC, Huppertz B. Human trophoblast invasion: New and unexpected routes and functions. Histochem Cell Biol 2018; 150:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Micallef A, Grech N, Farrugia F, Schembri-Wismayer P, Calleja-Agius J. The role of interferons in early pregnancy. Gynecol Endocrinol 2014; 30:1–6. [DOI] [PubMed] [Google Scholar]
- 26. Berkowitz RS, Hill JA, Kurtz CB, Anderson DJ. Effects of products of activated leukocytes (lymphokines and monokines) on the growth of malignant trophoblast cells in vitro. Am J Obstet Gynecol 1988; 158:199–203. [DOI] [PubMed] [Google Scholar]
- 27. Xiang L, Yin Y, Zheng Y, Ma Y, Li Y, Zhao Z, Guo J, Ai Z, Niu Y, Duan K. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 2020; 577:537–542. [DOI] [PubMed] [Google Scholar]
- 28. Feng HC, Choy MY, Deng W, Wong HL, Lau WM, Cheung A, Ngan H, Tsao SW. Establishment and characterization of a human first-trimester extravillous trophoblast cell line (TEV-1). J Soc Gynecol Investig 2005; 12:e21–e32. [DOI] [PubMed] [Google Scholar]
- 29. Graham CH, Hawley TS, Hawley RC, MacDougall JR, Kerbel RS, Khoo N, Lala PK. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res 1993; 206:204–211. [DOI] [PubMed] [Google Scholar]
- 30. Husslein H, Haider S, Meinhardt G, Prast J, Sonderegger S, Knöfler M. Expression, regulation and functional characterization of matrix metalloproteinase-3 of human trophoblast. Placenta 2009; 30:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fournier T, Handschuh K, Tsatsaris V, Evain-Brion D. Involvement of PPARγ in human trophoblast invasion. Placenta 2007; 28:S76–S81. [DOI] [PubMed] [Google Scholar]
- 32. Hiden U, Wadsack C, Prutsch N, Gauster M, Weiss U, Frank H-G, Schmitz U, Fast-Hirsch C, Hengstschläger M, Pötgens A. The first trimester human trophoblast cell line ACH-3P: A novel tool to study autocrine/paracrine regulatory loops of human trophoblast subpopulations–TNF-α stimulates MMP15 expression. BMC Dev Biol 2007; 7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abou-Kheir W, Barrak J, Hadadeh O, Daoud G. HTR-8/SVneo cell line contains a mixed population of cells. Placenta 2017; 50:1–7. [DOI] [PubMed] [Google Scholar]
- 34. Ding G-C, Chen M, Wang Y-X, Rui C, Xu W, Ding H-J, Shi Z-H. MicroRNA-128a-induced apoptosis in HTR-8/SVneo trophoblast cells contributes to pre-eclampsia. Biomed Pharmacother 2016; 81:63–70. [DOI] [PubMed] [Google Scholar]
- 35. Muschol-Steinmetz C, Jasmer B, Kreis N-N, Steinhäuser K, Ritter A, Rolle U, Yuan J, Louwen F. B-cell lymphoma 6 promotes proliferation and survival of trophoblastic cells. Cell Cycle 2016; 15:827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang M, Chen Y, Chen L, Wang K, Pan T, Liu X, Xu W. miR-15b-AGO2 play a critical role in HTR8/SVneo invasion and in a model of angiogenesis defects related to inflammation. Placenta 2016; 41:62–73. [DOI] [PubMed] [Google Scholar]
- 37. Hannan NJ, Paiva P, Dimitriadis E, Salamonsen LA. Models for study of human embryo implantation: Choice of cell lines? Biol Reprod 2010; 82:235–245. [DOI] [PubMed] [Google Scholar]
- 38. Li L, Schust DJ. Isolation, purification and in vitro differentiation of cytotrophoblast cells from human term placenta. Reprod Biol Endocrinol 2015; 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stromberg K, Azizkhan J, Speeg K. Isolation of functional human trophoblast cells and their partial characterization in primary cell culture. In Vitro 1978; 14:631–638. [DOI] [PubMed] [Google Scholar]
- 40. Borbely AU, Sandri S, Fernandes IR, Prado KM, Cardoso EC, Correa-Silva S, Albuquerque R, Knöfler M, Beltrão-Braga P, Campa A. The term basal plate of the human placenta as a source of functional extravillous trophoblast cells. Reprod Biol Endocrinol 2014; 12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss IIIJF. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology 1986; 118:1567–1582. [DOI] [PubMed] [Google Scholar]
- 42. Vaidya SS, Walsh SW, Gerk PM. Application of human placental villous tissue explants to study ABC transporter mediated efflux of 2, 4-dinitrophenyl-S-glutathione. Curr Pharm Biotechnol 2011; 12:814–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haider S, Meinhardt G, Saleh L, Fiala C, Pollheimer J, Knöfler M. Notch1 controls development of the extravillous trophoblast lineage in the human placenta. Proc Natl Acad Sci 2016; 113:E7710–E7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lokossou AG, Toufaily C, Vargas A, Barbeau B. siRNA transfection and EMSA analyses on freshly isolated human villous Cytotrophoblasts. J Vis Exp 2016; 20(115):53995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145–1147. [DOI] [PubMed] [Google Scholar]
- 46. Xu R-H, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 2002; 20:1261–1264. [DOI] [PubMed] [Google Scholar]
- 47. Das P, Ezashi T, Schulz LC, Westfall SD, Livingston KA, Roberts RM. Effects of fgf2 and oxygen in the bmp4-driven differentiation of trophoblast from human embryonic stem cells. Stem Cell Res 2007; 1:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu P, Pan G, Yu J, Thomson JA. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 2011; 8:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schulz L, Ezashi T, Das P, Westfall S, Livingston K, Roberts RM. Human embryonic stem cells as models for trophoblast differentiation. Placenta 2008; 29:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marchand M, Horcajadas JA, Esteban FJ, McElroy SL, Fisher SJ, Giudice LC. Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod 2011; 84:1258–1271. [DOI] [PubMed] [Google Scholar]
- 51. Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Cheng L. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem 2008; 283:24991–25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Erb TM, Schneider C, Mucko SE, Sanfilippo JS, Lowry NC, Desai MN, Mangoubi RS, Leuba SH, Sammak PJ. Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: The role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev 2011; 20:1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 2011; 9:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM, Ezashi T. Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci 2013; 110:E1212–E1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang Y, Adachi K, Sheridan MA, Alexenko AP, Schust DJ, Schulz LC, Ezashi T, Roberts RM. Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure. Proc Natl Acad Sci 2015; 112:E2337–E2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Telugu B, Adachi K, Schlitt J, Ezashi T, Schust D, Roberts R, Schulz L. Comparison of extravillous trophoblast cells derived from human embryonic stem cells and from first trimester human placentas. Placenta 2013; 34:536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horii M, Bui T, Touma O, Cho HY, Parast MM. An improved two-step protocol for trophoblast differentiation of human pluripotent stem cells. Curr Protoc Stem Cell Biol 2019; 50:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yabe S, Alexenko AP, Amita M, Yang Y, Schust DJ, Sadovsky Y, Ezashi T, Roberts RM. Comparison of syncytiotrophoblast generated from human embryonic stem cells and from term placentas. Proc Natl Acad Sci 2016; 113:E2598–E2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y, Zhang Y, Li S, Cui J. Gene expression pattern of trophoblast-specific transcription factors in trophectoderm by analysis of single-cell RNA-seq data of human blastocyst. Funct Integr Genomics 2021; 21:205–214. [DOI] [PubMed] [Google Scholar]
- 60. Karvas RM, McInturf S, Zhou J, Ezashi T, Schust DJ, Roberts RM, Schulz LC. Use of a human embryonic stem cell model to discover GABRP, WFDC2, VTCN1 and ACTC1 as markers of early first trimester human trophoblast. Mol Hum Reprod 2020; 26:425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Onishi K, Tonge PD, Nagy A, Zandstra PW. Local BMP-SMAD1 signaling increases LIF receptor-dependent STAT3 responsiveness and primed-to-naive mouse pluripotent stem cell conversion frequency. Stem cell reports 2014; 3:156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kime C, Sakaki-Yumoto M, Goodrich L, Hayashi Y, Sami S, Derynck R, Asahi M, Panning B, Yamanaka S, Tomoda K. Autotaxin-mediated lipid signaling intersects with LIF and BMP signaling to promote the naive pluripotency transcription factor program. Proc Natl Acad Sci 2016; 113:12478–12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yu S, Zhou C, Cao S, He J, Cai B, Wu K, Qin Y, Huang X, Xiao L, Ye J. BMP4 resets mouse epiblast stem cells to naive pluripotency through ZBTB7A/B-mediated chromatin remodelling. Nat Cell Biol 2020; 22:651–662. [DOI] [PubMed] [Google Scholar]
- 64. Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2007; 2:3081. [DOI] [PubMed] [Google Scholar]
- 65. Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011; 144:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roberts RM, Ezashi T, Sheridan MA, Yang Y. Specification of trophoblast from embryonic stem cells exposed to BMP4. Biol Reprod 2018; 99:212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sheridan MA, Yang Y, Jain A, Lyons AS, Yang P, Brahmasani SR, Dai A, Tian Y, Ellersieck MR, Tuteja G. Early onset preeclampsia in a model for human placental trophoblast. Proc Natl Acad Sci 2019; 116:4336–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Laissue P, Vaiman D. Exploring the molecular aetiology of preeclampsia by massive parallel sequencing of DNA. Curr Hypertens Rep 2020; 22:1–10. [DOI] [PubMed] [Google Scholar]
- 69. Turco MY, Gardner L, Hughes J, Cindrova-Davies T, Gomez MJ, Farrell L, Hollinshead M, Marsh SG, Brosens JJ, Critchley HO. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat Cell Biol 2017; 19:568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fitzgerald HC, Dhakal P, Behura SK, Schust DJ, Spencer TE. Self-renewing endometrial epithelial organoids of the human uterus. Proc Natl Acad Sci 2019; 116:23132–23142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brighton PJ, Maruyama Y, Fishwick K, Vrljicak P, Tewary S, Fujihara R, Muter J, Lucas ES, Yamada T, Woods L. Clearance of senescent decidual cells by uterine natural killer cells in cycling human endometrium. Elife 2017; 6:e31274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tanaka S. Derivation and culture of mouse trophoblast stem cells in vitro. In: Embryonic Stem Cell Protocols. Humana Press, Springer; 2006: 35–44. [DOI] [PubMed] [Google Scholar]
- 73. Kubaczka C, Senner C, Araúzo-Bravo MJ, Sharma N, Kuckenberg P, Becker A, Zimmer A, Brüstle O, Peitz M, Hemberger M. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem cell reports 2014; 2:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hou D, Su M, Li X, Li Z, Yun T, Zhao Y, Zhang M, Zhao L, Li R, Yu H. The efficient derivation of trophoblast cells from porcine in vitro fertilized and parthenogenetic blastocysts and culture with ROCK inhibitor Y-27632. PLoS One 2015; 10:e0142442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vandevoort CA, Thirkill TL, Douglas GC. Blastocyst-derived trophoblast stem cells from the rhesus monkey. Stem Cells Dev 2007; 16:779–788. [DOI] [PubMed] [Google Scholar]
- 76. Huang X, Han X, Uyunbilig B, Zhang M, Duo S, Zuo Y, Zhao Y, Yun T, Tai D, Wang C. Establishment of bovine trophoblast stem-like cells from in vitro-produced blastocyst-stage embryos using two inhibitors. Stem Cells Dev 2014; 23:1501–1514. [DOI] [PubMed] [Google Scholar]
- 77. Bianco K, Leon-Martinez D, Farrell J, Gormley M, McMaster M, Fisher SJ. 620: Human trophoblast progenitor cell (TBPC) lines derived from aneuploid placentas: Studying fundamental aspects of trophoblast biology. Am J Obstet Gynecol 2016; 214:S331. [Google Scholar]
- 78. Chang C-W, Wakeland AK, Parast MM. Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J Endocrinol 2018; 236:R43–R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. James J, Hurley D, Gamage T, Zhang T, Vather R, Pantham P, Murthi P, Chamley L. Isolation and characterisation of a novel trophoblast side-population from first trimester placentae. Reproduction 2015; 150:449–462. [DOI] [PubMed] [Google Scholar]
- 80. Okae H, Toh H, Sato T, Hiura H, Takahashi S, Shirane K, Kabayama Y, Suyama M, Sasaki H, Arima T. Derivation of human trophoblast stem cells. Cell Stem Cell 2018; 22:50–63.e56. [DOI] [PubMed] [Google Scholar]
- 81. Turco MY, Gardner L, Kay RG, Hamilton RS, Prater M, Hollinshead MS, McWhinnie A, Esposito L, Fernando R, Skelton H. Trophoblast organoids as a model for maternal–fetal interactions during human placentation. Nature 2018; 564:263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Haider S, Meinhardt G, Saleh L, Kunihs V, Gamperl M, Kaindl U, Ellinger A, Burkard TR, Fiala C, Pollheimer J. Self-renewing trophoblast organoids recapitulate the developmental program of the early human placenta. Stem cell reports 2018; 11:537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tanaka S, Kunath T, Hadjantonakis A-K, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998; 282:2072–2075. [DOI] [PubMed] [Google Scholar]
- 84. Dong C, Beltcheva M, Gontarz P, Zhang B, Popli P, Fischer LA, Khan SA, Park K-m, Yoon E-J, Xing X. E. Elife 2020; 9:e52504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cinkornpumin JK, Kwon SY, Guo Y, Hossain I, Sirois J, Russett CS, Tseng H-W, Okae H, Arima T, Duchaine TF. Naive human embryonic stem cells can give rise to cells with a trophoblast-like transcriptome and Methylome. Stem cell reports 2020; 15:198–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Castel G, Meistermann D, Bretin B, Firmin J, Blin J, Loubersac S, Bruneau A, Chevolleau S, Kilens S, Chariau C. Induction of human trophoblast stem cells from somatic cells and pluripotent stem cells. Cell Rep 2020; 33:108419. [DOI] [PubMed] [Google Scholar]
- 87. Gao X, Nowak-Imialek M, Chen X, Chen D, Herrmann D, Ruan D, Chen ACH, Eckersley-Maslin MA, Ahmad S, Lee YL. Establishment of porcine and human expanded potential stem cells. Nat Cell Biol 2019; 21:687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mischler A, Karakis V, Mahinthakumar J, Carberry C, San Miguel A, Rager J, Fry R, Rao BM. Two distinct trophectoderm lineage stem cells from human pluripotent stem cells. J Biol Chem 2021; 100386. doi: 10.1016/j.jbc.2021.100386. Epub ahead of print. PMID: 33556374; PMCID: PMC7948510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ezashi T, Zhou J, Alexenko AP, Tian Y, Schulz LC, Schust DJ, Roberts RM. Direct conversion of BMP4-primed pluripotent stem cells into trophoblast stem cells SSR abstract. 2020. https://higherlogicdownload.s3.amazonaws.com/SSR/fbd87d69-d53f-458a-8220-829febdf990b/UploadedImages/Abstract_Book_Complete_Final_2.pdf.
- 90. Li Z, Kurosawa O, Iwata H. Establishment of human trophoblast stem cells from human induced pluripotent stem cell-derived cystic cells under micromesh culture. Stem cell research & therapy 2019; 10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu X, Ouyang JF, Rossello FJ, Tan JP, Davidson KC, Valdes DS, Schröder J, Sun YB, Chen J, Knaupp AS. Reprogramming roadmap reveals route to human induced trophoblast stem cells. Nature 2020; 586:101–107. [DOI] [PubMed] [Google Scholar]
- 92. Kubaczka C, Senner CE, Cierlitza M, Araúzo-Bravo MJ, Kuckenberg P, Peitz M, Hemberger M, Schorle H. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell 2015; 17:557–568. [DOI] [PubMed] [Google Scholar]
- 93. Benchetrit H, Herman S, van Wietmarschen N, Wu T, Makedonski K, Maoz N, Tov NY, Stave D, Lasry R, Zayat V. Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell 2015; 17:543–556. [DOI] [PubMed] [Google Scholar]
- 94. Roberts JM, Gammill HS. Preeclampsia: Recent insights. Hypertension 2005; 46:1243–1249. [DOI] [PubMed] [Google Scholar]
- 95. Teklenburg G, Weimar CH, Fauser BC, Macklon N, Geijsen N, Heijnen CJ, Sousa Lopes SMC, Kuijk EW. Cell lineage specific distribution of H3K27 trimethylation accumulation in an in vitro model for human implantation. PLoS One 2012; 7:e32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yamanaka K, Fujisawa M, Tanaka H, Okada H, Arakawa S, Kamidono S. Significance of human testicular mast cells and their subtypes in male infertility. Hum Reprod 2000; 15:1543–1547. [DOI] [PubMed] [Google Scholar]
- 97. Carver J, Martin K, Spyropoulou I, Barlow D, Sargent I, Mardon H. An in-vitro model for stromal invasion during implantation of the human blastocyst. Hum Reprod 2003; 18:283–290. [DOI] [PubMed] [Google Scholar]
- 98. Menezo Y, Sakkas D, Janny L. Co-culture of the early human embryo: Factors affecting human blastocyst formation in vitro. Microsc Res Tech 1995; 32:50–56. [DOI] [PubMed] [Google Scholar]
- 99. Teklenburg G, Macklon NS. In vitro models for the study of early human embryo-endometrium interactions. Reprod Sci 2009; 16:811–818. [DOI] [PubMed] [Google Scholar]
- 100. Sheridan MA, Yunusov D, Balaraman V, Alexenko AP, Yabe S, Verjovski-Almeida S, Schust DJ, Franz AW, Sadovsky Y, Ezashi T. Vulnerability of primitive human placental trophoblast to Zika virus. Proc Natl Acad Sci 2017; 114:E1587–E1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lee Y-L, Fong S-W, Chen AC, Li T, Yue C, Lee C-L, Ng EH, Yeung WS, Lee K-F. Establishment of a novel human embryonic stem cell-derived trophoblastic spheroid implantation model. Hum Reprod 2015; 30:2614–2626. [DOI] [PubMed] [Google Scholar]
- 102. Roberts RM, Yabe S, Yang Y, Ezashi T. A human stem cell model for creating placental syncytiotrophoblast, the major cellular barrier that limits fetal exposure to xenobiotics. In: Stem Cells in Toxicology and Medicine, vol. 2016, 1st ed. Wiley & Sons; 2016; 179–195. [Google Scholar]


