Abstract
Objectives
Early recognition of coronavirus disease 2019 (COVID-19) severity can guide patient management. However, it is challenging to predict when COVID-19 patients will progress to critical illness. This study aimed to develop an artificial intelligence system to predict future deterioration to critical illness in COVID-19 patients.
Methods
An artificial intelligence (AI) system in a time-to-event analysis framework was developed to integrate chest CT and clinical data for risk prediction of future deterioration to critical illness in patients with COVID-19.
Results
A multi-institutional international cohort of 1,051 patients with RT-PCR confirmed COVID-19 and chest CT was included in this study. Of them, 282 patients developed critical illness, which was defined as requiring ICU admission and/or mechanical ventilation and/or reaching death during their hospital stay. The AI system achieved a C-index of 0.80 for predicting individual COVID-19 patients’ to critical illness. The AI system successfully stratified the patients into high-risk and low-risk groups with distinct progression risks (p < 0.0001).
Conclusions
Using CT imaging and clinical data, the AI system successfully predicted time to critical illness for individual patients and identified patients with high risk. AI has the potential to accurately triage patients and facilitate personalized treatment.
Key Point
• AI system can predict time to critical illness for patients with COVID-19 by using CT imaging and clinical data.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00330-021-08049-8.
Keywords: Coronavirus infections, Helical CT, Disease progression, Deep learning
Introduction
Coronavirus disease 2019 (COVID-19) causes severe respiratory complications, including acute respiratory failure and has put significant strain on healthcare systems worldwide to accommodate the massive influx of critically ill patients. For example, patients with acute-respiratory distress syndrome (ARDS) from COVID-19 often require intubation and intensive care unit (ICU) care, which are resource intensive [1]. Because of the shortage of mechanical ventilators and ICU care, it is crucial to accurately and timely predict COVID-19 patients who will require critical care. More importantly, an early determination of patient prognosis is useful when implementing new treatments such as remdesivir [2], convalescent plasma transfusion [3], ruxolitinib [4], and other emerging therapies, leading to better outcomes [5, 6]. Progression to critical illness in COVID-19 occurs across all patients, especially those with comorbidities such as obesity, cardiovascular disease, chronic lung disease, hypertension, or cancer [7]. However, it remains challenging to predict when a COVID-19 patient will progress to critical illness.
Beyond patient demographics and laboratory parameters, chest CT has been instrumental in distinguishing severe from non-severe cases of COVID-19. For example, chest CT severity score has been developed and shown to correlate disease severity and/or emergent status in COVID-19 patients [8–10]. Patients with severe disease will generally have diffuse multi-lobe involvement, pleural effusion, consolidation, bronchial wall thickening, and poor lung aeration on chest CT [8–12]. Due to the characteristic signs of COVID-19 on chest CT, artificial intelligence (AI) may have utility in ascribing disease severity status and prognosis to patients [13].
Using chest CT and clinical data, this study aimed to develop an artificial intelligence (AI) system to predict future deterioration to critical illness in COVID-19 patients.
Methods
Patient cohorts
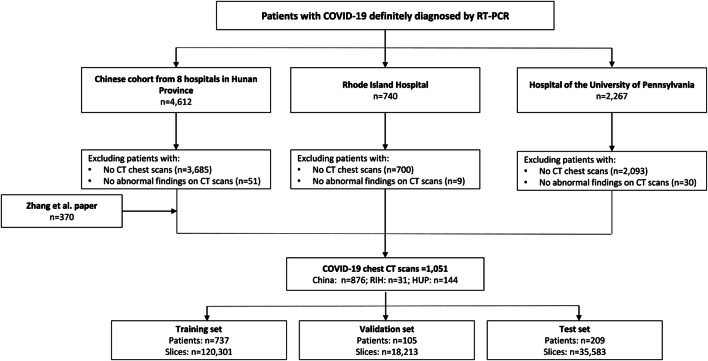
A total of 1,051 patients with confirmed positive COVID-19 by RT-PCR and chest CT imaging suggestive of pneumonia were retrospectively identified from nine hospitals in Hunan Province, China; the Hospital of the University of Pennsylvania in Philadelphia (HUP) in Philadelphia, PA; the Rhode Island Hospital (RIH) in Providence, RI; and open-source data from a previously published paper [13]. Ninety COVID-19 patients without abnormality on chest CT were excluded as they did not develop critical illness and the addition of these patients would have inflated model performance. A diagram illustrating patient inclusion and exclusion criteria is shown in Fig. 1.
Fig. 1.
Patient exclusion inclusion workflow. Abbreviations: CT, computed tomography; RIH, Rhode Island Hospital; HUP, Hospital of the University of Pennsylvania; AI, artificial intelligence; RT-PCR, reverse transcriptase polymerase chain reaction
The identified CT scans were directly downloaded from the hospital Picture Archiving and Communications System (PACS). The open-source data containing chest CT images and clinical metadata of COVID-19 patients confirmed by molecular PCR were directly downloaded from the China National Center for Bioinformation (http://ncov-ai.big.ac.cn/download?lang=en). Four radiologists, each with 5–10 years of experience in thoracic radiology and direct clinical experience with COVID-19 chest CT cases, reviewed and assessed the CT scans. Two radiologists independently reviewed each half of the 1051 cases (F. F. Xie and L. P. Zhu for one half, S. Li and D. Cao for the other half). A third radiologist (Z. Xiong), with 20 years of cardiothoracic imaging experience, helped to resolve differences to reach consensus.
The RT-PCR results were extracted from the patients’ electronic medical records in the hospital information system. The RT-PCR assays were performed using TaqMan One-Step RT-PCR Kits from Shanghai Huirui Biotechnology Co., Ltd. or Shanghai BioGerm Medical Biotechnology Co., Ltd., both of which have had their use approved by the China Food and Drug Administration and the COVID-19 RT-PCR test (Laboratory Corporation of America) for Chinese and US cohorts, respectively. For patients with multiple RT-PCR assays, a positive result on the last performed test was used as a confirmation of diagnosis.
The institutional review boards of all institutions approved this retrospective study, and written informed consent was waived. To avoid any potential breach of confidentiality, the patient data were de-identified and had no linkage to the researchers.
Clinical information
The patient’s age, sex, symptom (presence or absence of fever), white blood cell count, lymphocyte count, comorbidity status (cardiovascular disease, hypertension, chronic obstructive pulmonary disease, diabetes, chronic liver disease, chronic kidney disease, cancer, and human immunodeficiency virus [HIV]), and exposure history with the COVID-19 epicenter and/or another patient with COVID-19 were collected. Utilization of mechanical ventilation and/or intensive medical care and/or progression to death was recorded as well. For all patients, admission and discharge times were also recorded.
The patients were determined to be critical or have severe disease if they reached any of the following endpoints: mechanical ventilation, intensive medical care, and death. If not, they were determined to be non-critical. For critical patients, the timeframe of their progression to critical event was calculated from time of CT to the earliest time of developing one of the above three critical events. A comparison of clinical data among the four patient cohorts is shown in Supplementary Table 1.
AI models
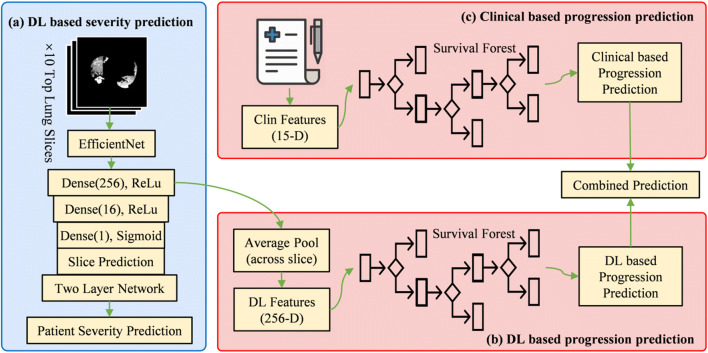
First, the pulmonary tissue and lung parenchymal abnormality from COVID-19 were automatically segmented on CT images by a deep learning (DL) model using deep convolutional neural network. Examples of the automatically segmented pulmonary tissue and lung parenchymal abnormality from COVID-19 are demonstrated in Supplementary Figure 1. Second, DL-based severity prediction models were built using lung and lesion segmentations to determine whether a patient with COVID-19 will develop critical or non-critical illness at the time of CT scan. Third, progression prediction models were built by using DL features extracted from the severity prediction models and clinical data as input to random survival forest to assign risk scores to different subjects. The workflow pipeline is shown in Fig. 2. The detailed descriptions of AI-based lung and lesion segmentation (Supplementary Text 1), AI-based severity prediction models (Supplementary Text 2), and AI-based progression prediction models (Supplementary Text 3) are included in the supporting information.
Fig. 2.
Illustration of our analysis pipeline. The pipeline includes a severity prediction stage and two progression prediction branches. (a) Deep learning (DL)–based severity prediction. The top 10 segmented lung slices by largest area of pathology were used as input to EfficientNet to predict disease severity based on individual slices, and then pooled to predict severity at the patient level. (b) DL-based progression prediction. In this branch, 256-D DL features from the model were aggregated via an average pool layer for each patient. Then, a random survival forest model was optimized based on the DL features to assign risk scores to different subjects. (c) Clinical (Clin) based progression prediction. In this branch, 15 clinical features extracted from demographic recordings were input to another survival forest model to assign risk scores to different subjects. Finally, for each patient, the DL-based prediction and Clin-based prediction were combined to predict progression for each patient
Statistical analysis
Segmentations were evaluated by calculating the Dice similarity coefficient scores and visual examinations. Accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (ROC-AUC) were calculated for the classification models. The 95% confidence intervals on accuracy, sensitivity, and specificity were determined using the adjusted Wald method [14]. C-index for right-censored data was applied to evaluate the performance of progression prediction models [15]. Time-dependent ROC-AUC was calculated from the obtained risk scores and progression information via the Kaplan-Meier method [16] to further evaluate the progression prediction performance.
Data and code availability
In respect of patient confidentiality and consent, the CT images and clinical information datasets analyzed in this study are not available for download. The models generated by these datasets are publicly available at https://www.dropbox.com/sh/g1w13gyoezq36y8/AAC0DvGyuLHdPPXKtvOQ_lTma?dl=0
The DL models for severity prediction were implemented with Keras (version 2.2.5) and Tensorflow (version 1.12.3) and trained with NVIDIA V100 GPUs. The progression prediction models were implemented with Scikit-Learn (version 0.21.3). To allow other researchers to develop their models, the code is publicly available on Github at http://github.com/robinwang08/COVID19.
Results
Patient characteristics
From the 1,051 patients with RT-PCR confirmed COVID-19 and chest CT, 282 patients developed critical illness (aka severe disease), which was defined as requiring ICU admission and/or mechanical ventilation and/or reaching death during their hospital stay. The median time of initial CT acquisition from RT-PCR confirmed COVID-19 diagnosis was 2 days. The distribution of time between CT acquisition and RT-PCR diagnosis is shown in Supplementary Figure 2. The median age of patients who progressed to critical illness was higher than that of non-critical patients (57 vs. 45 years, p < 0.001). The median number of days from admission to critical illness was 0.4 days (range: 0 to 30 days). The clinical characteristics of COVID-19 patients with critical or non-critical illness are shown in Table 1. This cohort was randomly divided into training, validation, and testing sets with a 7:1:2 split ratio to build the severity prediction models and progression prediction models. Patient characteristics across sets can be found in Supplementary Table 2.
Table 1.
Clinical characteristics of critical and non-critical COVID-19 patients
| Critical (n = 282) | Non-Critical (n = 769) | p value | |
|---|---|---|---|
| Age (year) | < 0.001 | ||
| Median ± interquartile range |
57 ± 23, range of 0 to 92 |
46 ± 22, range of 0 to 84 |
|
| < 20 | 18 (6) | 23 (3) | |
| 20–39 | 30 (11) | 246 (32) | |
| 40–59 | 105 (37) | 334 (43) | |
| ≥ 60 | 184 (65) | 166 (22) | |
| Sex | 0.298 | ||
| Male | 154 (55) | 393 (51) | |
| Female | 126 (45) | 372 (48) | |
| Presence of fever | < 0.001 | ||
| Fever | 103 (37) | 325 (42) | |
| No fever | 20 (7) | 156 (20) | |
| White blood cell count | < 0.001 | ||
| Elevated | 45 (16) | 22 (3) | |
| Normal | 79 (28) | 457 (59) | |
| Lymphocyte count | < 0.001 | ||
| Normal | 78 (28) | 193 (25) | |
| Decreased | 45 (16) | 323 (42) | |
| Comorbidities | |||
| Cardiovascular disease | 42 (15) | 37 (5) | < 0.001 |
| Hypertension | 62 (22) | 84 (11) | < 0.001 |
| COPD | 15 (5) | 20 (3) | < 0.001 |
| Diabetes | 36 (13) | 49 (6) | < 0.001 |
| Chronic liver disease | 6 (2) | 18 (2) | 0.453 |
| Chronic kidney disease | 19 (7) | 8 (1) | < 0.001 |
| Malignant tumor | 9 (3) | 8 (1) | < 0.001 |
| HIV | 0 (0) | 0 (0) | 1.000 |
| Outcomes* | |||
| Ventilator | 93 (33) | N/A | |
| Intensive care unit | 112 (40) | N/A | |
| Death | 24 (9) | N/A | |
| Unknown critical** | 152 (54) | N/A | |
| Progression to critical event (days) | |||
| Median | 0.4, range of 0 to 30 | N/A | |
| Day 1 | 165 (59) | N/A | |
| Day 2 | 25 (9) | N/A | |
| Day 3 | 12 (4) | N/A | |
| ≥ Day 4 | 69 (24) | N/A | |
| Progression to discharge (days) | |||
| Median | N/A | 12.0, range of 0 to 46 | |
| 0–4 | N/A | 57 (7) | |
| 5–9 | N/A | 155 (20) | |
| 10–14 | N/A | 230 (30) | |
| ≥ 15 | N/A | 203 (26) | |
| Epidemiologic contact | |||
| Epicenter*** | 14 (5) | 155 (20) | <0.001 |
| COVID-19 patients | 26 (9) | 114 (15) | <0.001 |
*Patients with multiple critical outcomes may be counted in multiple categories
**For patients from public data source [17], the type of critical condition was not specified
***Epidemiologic contact with epicenter includes patients who have visited Wuhan, China, and New York City, New York, USA
COPD chronic obstructive pulmonary disease, HIV human immunodeficiency virus
AI-based segmentation of lung and lesion
Deep learning–based lung and lesion segmentations resulted in mean validation Dice similarity coefficients of 0.97 ± 0.02 and 0.62 ± 0.22, respectively, using manual segmentation as the gold standard. The average volume error for lesion is 27.1% ± 29.0% of the ground truth. Average false positive rate is 48.3% and average false negative rate is 12.6%, indicating that the automatic model tends to overestimate the lesion. Supplementary Figure 3 shows the Bland-Altman plots for the volume differences from the automatic and manual segmentation. Therefore, in order to reduce the impact from automatic segmentation, the generated lung and lesion segmentations were then manually corrected as needed—a total of 115 lesion segmentations (11%) were manually corrected. Examples of manually corrected segmentations are shown in Supplementary Figure 4.
Severity and progression prediction models
The model utilizing the top 10 segmented lung slices based on the largest lesion area for each patient achieved a slice level accuracy of 0.778 (95% CI: 0.760–0.795), sensitivity of 0.710 (95% CI: 0.667–0.751), specificity of 0.796 (95% CI: 0.776–0.815), and a ROC-AUC of 0.832. At the pooled patient level, the model achieved an accuracy of 0.833 (95% CI: 0.776–0.877), sensitivity of 0.622 (95% CI: 0.476–0.749), specificity of 0.890 (95% CI: 0.832–0.930), and a ROC-AUC of 0.856. A summary of the results for the model is shown in Supplementary Table 3 with the ROC-AUC graphs presented in Fig. 3.
Fig. 3.
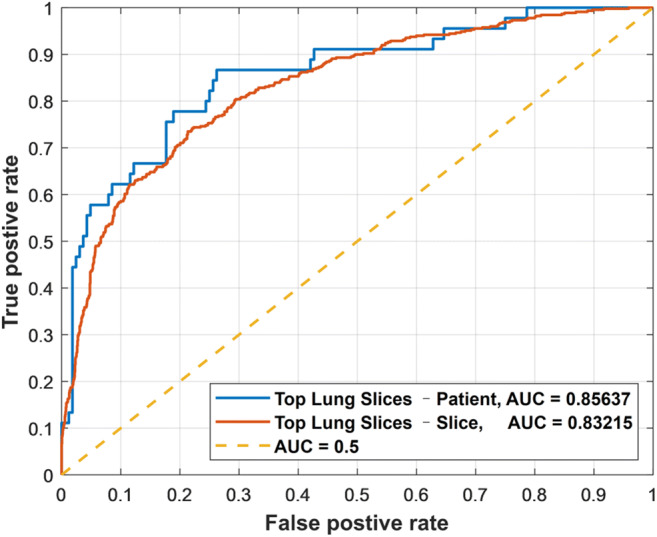
Performance of deep learning severity model in area under receiver operating characteristic curve (ROC-AUC) utilizing top ten segmented lung slices by largest lesion area
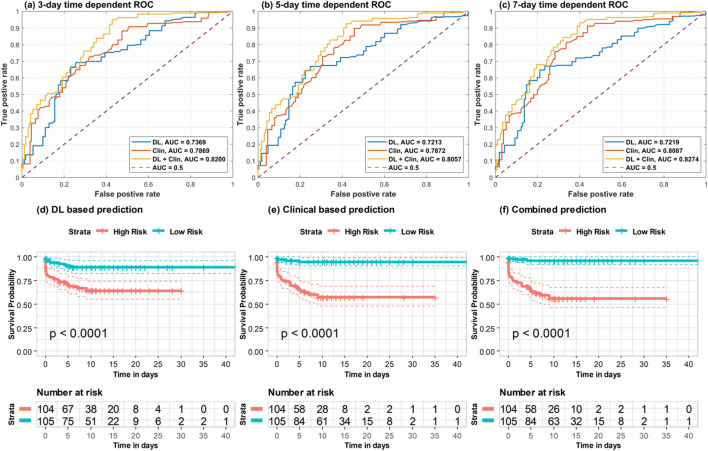
Built to predict the length of time from admission to critical illness, the progression prediction model was optimized on the same data split of severity prediction model and tested on an independent test set. The prediction model achieved a C-index of 0.804, demonstrating success in assigning patients with risk scores consistent with their progression outcomes. As shown in Fig. 4a–c, the progression prediction model achieved time-dependent ROC-AUC of 0.82, 0.81, and 0.83 for prediction of progression risk at cutoff values of 3, 5, and 7 days, respectively. Median of risk scores obtained by the progression prediction model were utilized to stratify patients into high-risk and low-risk subgroups. As indicated by survival curves of the stratified risk groups shown in Fig. 4f, the high-risk and low-risk subgroups had statistically significant difference in the risk of disease progression (p < 0.0001, log-rank test).
Fig. 4.
Time-dependent ROC-AUCs and risk stratified subgroup survival curves based on deep learning (DL) features extracted from top lung slices. a–c Time-dependent ROC curves and AUCs with different cutoff values (3-day, 5-day, and 7-day). d–f The risk-stratified survival curves based on DL-based progression prediction, clinical-based progression prediction, and combined progression prediction. The y-axis is survival probability representing the probability of not progressing to critical event. The “+” in survival curves denotes the censored patient. Risk tables of these stratification results are also listed in the bottom of this figure
Progression prediction models based on DL features and clinical data separately achieved C-index values of 0.719 and 0.774, respectively. As shown in Fig. 4d and e, the median of risk scores obtained by these two progression prediction models also successfully stratified the patients into groups with distinctive critical outcomes (p value < 0.0001, log-rank test). The model based on the CT severity scores as described by Yeun-Chung Chang et al [17] achieved a C-index value of 0.724. The progression prediction and risk stratification performances of the aforementioned methods are summarized in Supplementary Table 4.
Discussion
Early disease detection and treatment has been linked to decreased mortality in COVID-19 patients, especially for those who are severely ill [5, 6]. Approximately 15% of COVID-19 patients develop acute-respiratory distress syndrome and over half of ICU admits develop hypoxia or respiratory exhaustion [18]. Early anticipation of severe disease development is crucial because it allows for timely intervention, which can potentially improve outcomes for critically ill COVID-19 patients [18]. Further, strained resources such as mechanical ventilators or extracorporeal membrane oxygenation (ECMO) machines can be allocated appropriately when patients’ disease trajectories are known. This study developed an AI system based on chest CT and clinical data that predicts COVID-19 disease progression better than clinical data alone. This is relevant and impactful because it demonstrates that AI has the potential to help identify patients at risk for progression to critical illness and affect patient care by integrating data from multiple sources.
AI can help in identifying patients at risk for progression to critical illness within the timeframe for early treatment and improve prognosis. Remdesivir was recently approved for emergency use by the Food and Drug Administration (FDA) and, along with ruxolitinib [4], has shown preliminary promise in reducing recovery time for critically ill patients [19]. Similarly, convalescent plasma transfusion (CPT) has shown efficacy in reducing clinical symptoms and mortality for severely ill COVID-19 patients [3]. Historically, CPT has been used to successfully treat Spanish influenza A (H1N1), severe acute respiratory syndrome (SARS), and Ebola among others [3]. If these results are maintained in future studies, identifying patients with severe disease early on may be necessary to maximize clinical benefit of these treatments.
This chest CT–based AI system has the potential to assist physicians in patient management by enhancing clinical data in the prediction of progression risk. In contrast to previous studies [13, 20, 21], this study was attentive to the exact timespan between the performance of chest CT and the earliest occurrence of critical events (i.e., ICU admission, intubation, or death) using a multi-international cohort of patients from different institutions. Different from existing studies that build prediction models using pattern classification techniques, our AI system was built in a time-to-event (survival) analysis framework that can effectively handle censored data in the risk prediction study. By using the risk prediction AI model based on CT imaging and clinical data, it may be possible to stratify patients into different risk groups for progression to critical illness, assign a critical window for early treatment, and have a more informed timeline for obtaining advanced respiratory support equipment.
The study has several limitations. There was likely patient selection bias secondary to the retrospective and multi-institutional nature of the study. For example, the clinical characteristics differed between the cohorts from USA and China. While the percentage of critical patients was 22% for the cohort form China, it was almost 50% for both cohorts from USA. The data heterogeneity likely reflects the differences in practice pattern. In the Chinese population, the CT scans were used more often for COVID-19 screening, especially during the earlier period of the pandemic. Other explanations include the difference in demographics and disease prevalence. However, our study encompasses data from two hospitals in the USA, nine hospitals in China, and open-source data [13]. The stable performance on an independent held-out test set supported the robustness and generalizability of the model. Additionally, the definition of critical outcome defined herein may encompass more patients than is typical of other COVID severity studies such as respiratory failure, septic shock, or multiple organ dysfunction. The present study focused on patients who went to the ICU, were intubated, or died.
Conclusions
AI based on CT imaging and clinical data has the potential for prediction of risk for future deterioration to critical illness among patients with COVID-19. As this technology is further developed, providers may be able to utilize AI to help designate high-risk patients based on their disease severity and progression risk prediction, enabling them to readily allocate appropriate treatments, equipment, and other necessary resources and to assist with early clinical decision-making.
Supplementary Information
(DOCX 4096 kb)
Abbreviations
- AI
Artificial intelligence
- ARDS
Acute-respiratory distress syndrome
- COVID-19
Coronavirus disease 2019
- CPT
Convalescent plasma transfusion
- DL
Deep learning
- ECMO
Extracorporeal membrane oxygenation
- FDA
The Food and Drug Administration
- HIV
Human immunodeficiency virus
- HUP
The Hospital of the University of Pennsylvania
- ICU
Intensive care unit
- PACS
Picture Archiving and Communications System
- RIH
The Rhode Island Hospital
- ROC-AUC
Area under the receiver operating characteristic curve
- SARS
Severe acute respiratory syndrome
Funding
This study was supported by the Brown COVID-19 Research Seed Award, AWS Diagnostic Development Initiative, RSNA Research Scholar Grant (RSCH2004), and National Institutes of Health/National Cancer Institute R03 grant (R03CA249554) to Dr. Harrison X. Bai, as well as NIH grants (CA223358, DK117297, MH120811, and EB022573) to Dr. Yong Fan. Research reported in this publication was supported by a training grant from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under award number 5T32EB1680 and by the National Cancer Institute (NCI) of the National Institutes of Health under Award Number F30CA239407 to K. Chang. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Dr. Xue Feng currently works in Carina Medical, a for-profit organization that develops clinical products.
Declarations
Guarantor
The scientific guarantor of this publication is Harrison Bai.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• case-control study
• multicenter study
Footnotes
Harrison X. Bai is the first corresponding author and Wei-Hua Liao is the second corresponding author.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Robin Wang and Zhicheng Jiao contributed equally to this work.
Contributor Information
Wei-Hua Liao, Email: owenliao@csu.edu.cn.
Harrison X. Bai, Email: harrison_bai@brown.edu
References
- 1.Weissman DN, Perio MA, Radonovich LJ., Jr COVID-19 and risks posed to personnel during endotracheal intubation. JAMA. 2020;323:2027–2028. doi: 10.1001/jama.2020.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grein J, Ohmagari N, Shin D, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajendran K, Krishnasamy N, Rangarajan J, Rathinam J, Natarajan M, Ramachandran A (2020) Convalescent plasma transfusion for the treatment of COVID-19: systematic review. J Med Virol. In press. 10.1002/jmv.25961 [DOI] [PMC free article] [PubMed]
- 4.Cao Y, Wei J, Zou L et al (2020) Ruxolitinib in treatment of severe coronavirus disease 2019 (COVID-19): a multicenter, single-blind, randomized controlled trial. J Allergy Clin Immunol. In press. 10.1016/j.jaci.2020.05.019 [DOI] [PMC free article] [PubMed]
- 5.Sun Q, Qiu H, Huang M, Yang Y. Lower mortality of COVID-19 by early recognition and intervention: experience from Jiangsu province. Ann Intensive Care. 2020;10:33. doi: 10.1186/s13613-020-00650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyal DK, Mansab F, Iqbal I, Bhatti S. Early intervention likely improves mortality in COVID-19 infection. Clin Med. 2020;20:248–250. doi: 10.7861/clinmed.2020-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan RE, Adab P, Cheng KK. Covid-19: risk factors for severe disease and death. BMJ. 2020;368:m1198. doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 8.Yang R, Li X, Liu H, et al. Chest CT severity score: an imaging tool for assessing severe COVID-19. Radiol Cardiothorac Imaging. 2020;2:2. doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao W, Zhong Z, Xie X, Yu Q, Liu J. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am J Roentgenol. 2020;214:1072–1077. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 10.Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Invest Radiol. 2020;55:327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Ling Y, Bai T, et al. COVID-19 with different severity: a multi-center study of clinical features. Am J Respir Crit Care Med. 2020;201:1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colombi D, Bodini FC, Petrini M et al Well-aerated lung on admitting chest CT to predict adverse outcome in COVID-19 pneumonia. Radiology. In press. 10.1148/radiol.2020201433 [DOI] [PMC free article] [PubMed]
- 13.Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423–1433. doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52(2):119–126. [Google Scholar]
- 15.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61(1):92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 17.Chang Y-C, Yu C-J, Chang S-C, et al. Pulmonary sequelae in convalescent patients after severe acute respiratory syndrome: evaluation with thin-section CT. Radiology. 2005;236(3):1067–1075. doi: 10.1148/radiol.2363040958. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Li R, Wu Z, et al. Therapeutic strategies for critically ill patients with COVID-19. Ann Intensive Care. 2020;10:1–9. doi: 10.1186/s13613-020-00661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Zhao W, Xie X et al (2020) Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. arXiv preprint arXiv:200311988
- 21.Homayounieh F, Ebrahimian S, Babaei R, et al. CT radiomics, radiologists and clinical information in predicting outcome of patients with COVID-19 pneumonia. Radiol Cardiothorac Imaging. 2020;2(4):e200322. doi: 10.1148/ryct.2020200322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 4096 kb)
Data Availability Statement
In respect of patient confidentiality and consent, the CT images and clinical information datasets analyzed in this study are not available for download. The models generated by these datasets are publicly available at https://www.dropbox.com/sh/g1w13gyoezq36y8/AAC0DvGyuLHdPPXKtvOQ_lTma?dl=0
The DL models for severity prediction were implemented with Keras (version 2.2.5) and Tensorflow (version 1.12.3) and trained with NVIDIA V100 GPUs. The progression prediction models were implemented with Scikit-Learn (version 0.21.3). To allow other researchers to develop their models, the code is publicly available on Github at http://github.com/robinwang08/COVID19.