Abstract
A pandemic disease, COVID-19, has caused trouble worldwide by infecting millions of people. The studies that apply artificial intelligence (AI) and machine learning (ML) methods for various purposes against the COVID-19 outbreak have increased because of their significant advantages. Although AI/ML applications provide satisfactory solutions to COVID-19 disease, these solutions can have a wide diversity. This increase in the number of AI/ML studies and diversity in solutions can confuse deciding which AI/ML technique is suitable for which COVID-19 purposes. Because there is no comprehensive review study, this study systematically analyzes and summarizes related studies. A research methodology has been proposed to conduct the systematic literature review for framing the research questions, searching criteria and relevant data extraction. Finally, 264 studies were taken into account after following inclusion and exclusion criteria. This research can be regarded as a key element for epidemic and transmission prediction, diagnosis and detection, and drug/vaccine development. Six research questions are explored with 50 AI/ML approaches in COVID-19, 8 AI/ML methods for patient outcome prediction, 14 AI/ML techniques in disease predictions, along with five AI/ML methods for risk assessment of COVID-19. It also covers AI/ML method in drug development, vaccines for COVID-19, models in COVID-19, datasets and their usage and dataset applications with AI/ML.
Keywords: COVID-19, Pandemic, Artificial intelligence, Machine learning, Systematic review, Research analysis
Introduction
COVID-19, novel coronavirus, was announced in Wuhan, China, in December 2019 as a group of fatal respiratory infections and spread quickly as a pandemic [1]. Coronaviruses are pronounced zoonotic in nature and readily spread amongst people [2]. It is still a burning issue to investigate how it is transferred into animal reserves and others [3]. Because no vaccine and decided medication for COVID-19 found until the beginning of 2021, social distancing was stated as the most effective tactic to control and prevent [4]. In addition to social distancing, quarantine is also a critical part of controlling and avoiding the spread of the virus. According to John Hopkins University, the total confirmed cases is 107.5 million, and global death is over 2.3 million in the world [5]. The most affected ten countries are the United States, Brazil, India, Russia, France, Spain, Italy, Turkey, Germany and Colombia. The COVID-19 pandemic is not only a medical contagious but also an economical contagious [6]. Consequently, it is necessary to build an artificial intelligence-based healthcare system because it can quickly and precisely detect cases and avoid the pandemic.
Artificial intelligence (AI) and machine learning (ML) [7] have been recognized as the most potent and hopeful analytical tools in the healthcare domain [8]. Although many health problems are handled by bioinformaticians and statisticians instead of data scientists, a massive amount of data generated in the healthcare creates a necessity to produce more beneficial tools to distinguish exceptional cases from big data. AI computing performs various cognitive functions like humans in a machine to act or react to input data. On the other hand, classical computing has no autonomous intelligence since it requires a hand-code to react to data [9]. It cannot react when an unpredicted state has occurred. Therefore, AI tools continually adapt their reaction to adjust creating their behaviors. In an AI method, computers are designed to analyze, interpret and solve a problem. In machine learning, one of the principal forms of AI, machines learn reactions to use in the future for the same inputs when they face a particular result.
The applicability of AI/ML for epidemiological research of COVID-19 is explored in the literature. Initially, it identifies the relevant key explanatory variables then uses the dimensionality reduction technique to remove redundant features or information. It utilizes Random forest and gradient boosted machine learning models to measure the relative influence of the explanatory variables. This method also determines the interconnections among key explanatory variables, COVID-19 case and death counts. The study shows that air pollution has a high impact on COVID-19 casualties [10]. COVIDetectioNet [11] is proposed to detect the COVID-19. It uses in-depth features generated from the convolution and fully connected layers of the AlexNet architecture. This method has three steps such as pre-learned in-depth features ensemble, feature selection, and classification. It uses the relief algorithm for feature selection and the support vector machine model for classification. This method uses a tenfold cross-validation method to calculate the accuracy.
Deep learning (DL) models are very effective for time-series datasets. In the literature, the prediction of COVID-19 cases using time series data is discussed with DL techniques. Some models, such as long short-term memory (LSTM), are used to predict the time-series datasets. Integration of a convolutional neural network (CNN) and Long short-term memory (LSTM) detects COVID-19 automatically using X-ray images. CNN is used for deep feature extraction, and detection is performed using LSTM using the extracted features [12]. The sample size is a significant challenge with the existing method. Samples contain multiple disease symptoms is one more challenge of this method. Similarly, the prediction of confirmed cases, deaths and recoveries in 10 major countries affected due to COVID-19 is studied. Autoregressive integrated moving average (ARIMA), Support Vector Machine (SVM), LSTM and bidirectional LSTM can be applied for prediction purposes [13]. The superiority of the models can be measured various performance metrics such as root mean square error, mean absolute error and score. Multiple CNN models like ResNet, Inception net V3, Xception net can be used to detect COVID-19 using chest X-ray scans. The small sample size is the main disadvantage of these methods. Due to overfitting, these methods are unable to produce high accuracy [14].
AI/ML techniques have been widely applied to detect new molecules on the way to ascertain COVID-19. Many data scientists adopt AI tools to discover new medicines for the cure, to use X-rays and computational tomography (CT) scans by image processing, to identify the infectious people [15]. AI tools can also develop tracking software to classify people who breach the quarantine rule. AI-embedded thermal cameras and smartphones are practiced to catch infected patients [16]. In a general manner, AI is utilized to identify, track and predict outbreaks by diagnosing the virus. The drones and robots are used to transport food and medicine to related areas or people [17]. Some researches benefit from AI advantages to develop drugs and prepare vaccines [18, 19].
Chest X-ray images have demonstrated a highly effective screening technique for diagnosing the COVID-19. Various hybrid techniques are adopted to detect the COVID-19. Recently, a hybrid DL called COVID-CheXNet is demonstrated to identify the COVID-19. In the beginning, the contrast X-ray image is enhanced using contrast-limited adaptive histogram equalization, and the noise level is reduced with the help of the Butterworth bandpass filter. It uses two pre-trained models such as ResNet34 and HRNet, to identify the COVID-19. Each model’s score is fused to obtain the final class whether the individual is affected by the COVID-19 or not [20]. Similarly, a transfer learning-based hybrid 2D/3D CNN architecture for COVID-19 detection. It uses a pre-trained VGG16 deep model, a shallow 3D CNN. It is also combined with a depth-wise separable convolution layer (to preserve the valuable features) and a spatial pyramid pooling module (to extract multi-level representations). It uses the dataset with three classes such as COVID-19, pneumonia and normal. It achieves reasonable performance concerning sensitivity, specificity and accuracy [21]. A comprehensive study is performed to understand the automatic detection of COVID-19 based on X-ray images using both machine learning and deep learning models. The method’s novelty is demonstrated using COVID-19 vs. Normal dataset and adopt transfer learning to showcase the accuracy. Experimental results indicate that the ResNet50 model performs better as compared to other pre-trained models [22].
The number of studies on COVID-19 increases day by day because of its popularity and necessity. Researchers need to get a piece of quick information about related studies in this area. In the field of healthcare, AI/ML techniques have been implemented for many applications. For example, because of the availability of MRI, X-ray, and CT images, they have been widely applied for the COVID-19 outbreaks. Although AI/ML applications provide satisfactory solutions to the COVID-19 pandemic, these solutions have a wide diversity in nature. There is no comprehensive study discussing the AI/ML techniques used for the COVID-19 pandemic from different perspectives. Therefore, to fill this scientific gap in the literature, the study’s motivation is to analyze the potential studies using the AI/ML methods [23, 24] for several purposes about the current COVID-19. The study analyzes research on COVID-19 using AI/ML techniques from various perspectives, such as data types, software/tools, applied methods, drug and vaccines. This research’s novelty includes systematically addressing AI/ML techniques as an emerging discipline with tremendous applications in the pandemic. These techniques can be used to understand the nature of this virus and further predict the upcoming issues related to pandemics. This study discusses the significance of AI/ML in resolving the COVID-19 pandemic crisis by examining 264 latest references from seven accessible databases in a systematic way.
Contributions of this study include
This study mainly focuses on different AI/ML techniques that were applied for the COVID-19 outbreak.
This study highlights the reasons for applying AI/ML techniques to the pandemic.
This study explains the data perspective of COVID-19 studies regarding measurement types of study success and data types.
This review research gives direction to researchers about the various repositories available for COVID-19 outbreak so that researchers can easily access.
This study focuses on the current situation of drug and vaccine discovery and how AI/ML methods can help in the drug development.
This study lists various software platforms available to implement AI/ML methods in the COVID-19 outbreak.
This study is structured as follows. The next section gives the research methodology based on seven significant considerations. Research questions, which are critical aspects of the review, are determined. Databases and search strategy are explained together with inclusion and exclusion criteria to select relevant studies. Then data extraction and collections steps are considered. Factors that affect validity to know the strengths and weaknesses of the systematic review are discussed. The subsequent section presents the results and discussions considering defined research questions. Then the limitations of the review are given. Finally, the study is concluded.
Research methodology
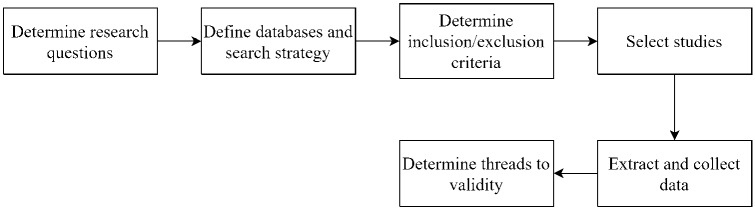
According to Brereton et al. [25], a systematic review of the literature is a method of identifying, evaluating, and interpreting all existing work on a particular research question, subject area or interest. A systematic literature search is conducted with a set of research questions. It aims to answer these questions using a secure, rigorous and auditable methodology [26]. The steps taken in this study are shown in Fig. 1. The process steps in this study are described in the following subsections
Fig. 1.
Systematic literature review flowchart
Research questions
The main objective of this systematic literature review is to describe, analyze and synthesize the studies related to the AI/ML implementations in the COVID-19 outbreak. To obtain a more detailed and comprehensive view of the subject, the overall objective is based on the following six research questions (RQs) with motivations.
RQ 1: What are the most frequently applied AI/ML techniques in COVID-19?
RQ 2: Why AI/ML approaches are applied in COVID-19?
RQ 3: What is the data perspective of studies?
RQ 4: What is the current situation in drugs preparation?
RQ 5: What software platforms are used?
RQ 6: Which data sources can be reached?
Databases and search strategy
Seven online academic search engines were used to find related studies.
ACM Digital Library
ArXiv.org
Elsevier
IEEE Xplore Digital Library
PubMed
Springer
Wiley Online Library
The search string used to facilitate searching in selected libraries have four dimensions with their sub-domains: AI/ML, study objective, COVID-19, and healthcare.
Inclusion/exclusion criteria
After collecting the studies, duplicate articles were removed. If there are more than one studies, only the most complete version was chosen. Later, studies were selected using the following inclusion and exclusion criteria to find answers to identified research questions and identify the most appropriate studies.
Inclusion criteria:
Studies applying at least one AI/ML algorithm
Studies producing solution to at least one of the COVID-19 problem
Studies containing experimental work using COVID-19 datasets
Studies that explicitly address the COVID-19 issue
Studies written in English only
Exclusion criteria:
Studies published before 2019
Extended abstracts and poster work
Studies that mention AI/ML techniques but are not part of the COVID-19 outbreak
Studies that mention COVID-19 techniques but do not use AI/ML techniques
Theoretical studies without application
Study selection
The articles defined by the search terms from the databases were initially considered only metadata (title and summary). All works related to the subject were scanned. However, since the number of studies found was too large, a second selection was made according to the keywords. The keyword is a way of reducing the time needed to develop the classification scheme and to ensure that the plan considers current work [27]. The full text was examined for the suitability of the articles at the end of the second stage. In the third step, reference lists of related articles were scanned to find extra articles. At the end of the final phase, 264 studies were found eligible for the review.
Data extraction
A data extraction form was used to collect relevant data from the selected studies to answer research questions. Selected studies were evaluated three times in different days by different authors.
RQ 1: AI/ML algorithms and techniques used for COVID-19 should be defined.
RQ 2: Objective of AI/ML approaches should be given.
RQ 3: The data type, data size, study reliability should be investigated.
RQ 4: AI architecture for protein structure and drug analysis should be identified.
RQ 5: AI/ML-based software specific to COVID-19 outbreak should be given.
RQ 6: Data sources should be searched with a direct link.
Data collection
The electronic databases include international indexed journals and conferences searched and defined concerning AI/ML approaches against COVID-19. ACM (), arXiv (), Elsevier (), IEEE Xplore (), PubMed (), Springer () and Wiley () databases were scanned. 27 additional studies have been identified by manually searching the reference lists from important studies.
Threads to validity
It is essential to consider the factors that affect validity to know the strengths and weaknesses of a systematic review [28]. The factors are mainly related to study selection, data extraction and researcher bias in this research.
To find out related studies, the seven search engines mentioned above were scanned. However, it may not be possible to have other relevant works on the results. For this threat, reference lists of selected studies were searched manually to find other related studies, and 27 research were added to the list.
Data extraction is one of the most critical tasks in this work. To reduce the likelihood of extracting wrong data, studies were evaluated twice on different days, and the data needed to answer the RQs were collected.
When selecting and extracting data, it is possible to mention researcher bias [29]. It is a useful systematic review method that one researcher selects studies, and another researcher checks them [30]. The studies in this study were evaluated independently by two researchers and tried to prevent the researcher bias.
Results and discussion
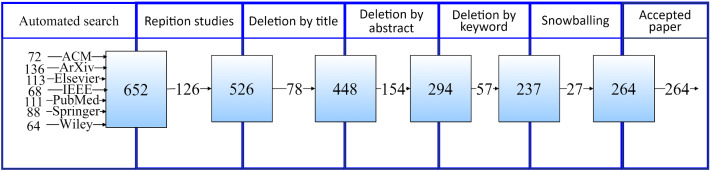
Relevant studies were determined by applying the research strategy and inclusion/exclusion criteria. For the search on the seven electronic databases described above, 652 candidate studies were selected, as shown in Fig. 2. After removing the first three exclusion criteria and the duplicated studies, 526 articles remained. Then a search based on meta-data (title, keywords and abstract) was done. 237 studies were left after unsuitable studies were eliminated according to the title, abstract and keywords. All of the studies were examined in full text. Since no inconvenience was observed, no elimination was done. As a result, 237 studies related to AI/ML implementations against COVID-19 were agreed suitable for examination. After reviewing these studies’ full text, 27 other studies related to the research were added to the sources through reference lists. Thus, 264 articles were selected directly related to the research.
Fig. 2.
Result of the study selection process
In recent years, AI has been widely used in various fields of medicine and healthcare [31–33]. Since the outbreak of COVID-19, researchers were successfully used advanced AI technologies in the COVID-19 battle and were achieved significant progress [34–36]. In this survey, a comprehensive review of the contributions of AI/ML in combating COVID-19 is presented. The main scope of AI/ML in COVID-19 research includes the aspects of epidemic and transmission prediction, diagnosis and detection, drug/vaccine development [37].
RQ 1: What are the most frequently applied AI/ML techniques in COVID-19?
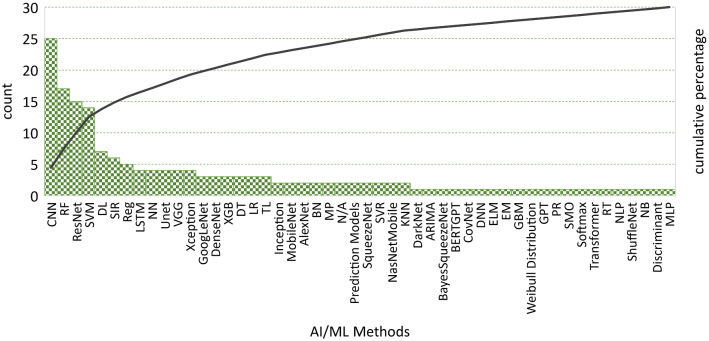
The comparative survey presented in Fig. 3 showed that the convolutional neural network (CNN) model is widely used for medical imaging [38–45]. CNNs are specialized types of neural networks and can be applied to many kinds of data with different dimensions. CNN includes three kinds of layers: convolutional, pooling, and fully connected layers. Convolutional layers constitute the main building blocks of a CNN and summarize the features in an image [46]. CNNs are sensitive to the spatial coherence or local pixel correlations in images. Most of the papers presented in this survey adopted the CNN model because of its high accuracy [47–51]. The results prove that the CNN and deep learning (DL) methods perform best among all the models used in COVID-19 [52, 54–57]. Moreover, CNN was applied together with other methods in many studies such as Unet [58, 59], AlexNet [60] and long short-term memory (LSTM) [61, 62]. ResNet is a pre-trained DL approach that applied more than others [53, 63–67]. However, there some challenges are using CNNs in medical tasks. It is difficult to collect medical images in good quality and sufficient numbers. The availability of labeled data is limited. Collecting and labeling data is a time-consuming process; besides, correctly labeling is critical and depends on specialist experience [68–71]. Random forest (RF) classifier is an ML classifier used by more than 50% of the studies because of its ability to choose the best features for classification [72–78]. SVM is another ML method mostly applied in all scenarios like classification [79–81], prediction [82–84], and diagnosis [85]. Some studies applied more than one pre-trained models and compared their results to find the best method against image recognition [86–89]. Pre-trained networks are composed of two parts. The first part includes a series of convolution and pooling layers, and these layers end with a densely connected classifier. Convolutional feature maps take into consideration of object locations in an input image. On the other hand, densely connected layers at the top of the convolutional base are mostly useless for object detection problems. A pre-trained network is trained on a large dataset, generally on large-scale image classification problems using ResNet, UNet, VGG, Xception, GoogLeNet and XGBoost.
Fig. 3.
AI/ML approaches in COVID-19
Researchers frequently combined AI/ML techniques and advanced statistical methods to increase the effectiveness of the study outcomes [74, 77, 78, 86, 87, 90–94]. Various ML techniques supported many of the COVID-19 studies [72, 95–100]. For example, Mei et al. [76] developed a joint model that uses CNN and ML (SVM and RF) as a classifier. Susceptible–infectious–recovered (SIR) model and its derivatives such as susceptible–infectious–recovered–deceased (SIRD) or susceptible–exposed–infectious–recovered (SEIR) produces acceptable results using case data [101–104]. Some studies proposed intelligent methodologies including some ML techniques to present effective solutions. For example, Mohammed et al. [37] have evaluated and compared by an intelligent methodology of COVID-19 diagnosis models. They have presented a decision matrix that combined a mix of ten evaluation criteria and twelve diagnostic models for COVID-19. The multi-criteria decision-making method is applied to evaluate and benchmark the various diagnostic models for COVID-19. They have selected SVM classifier as the best diagnosis model for COVID-19.
RQ 2: Why AI/ML approaches are applied in COVID-19?
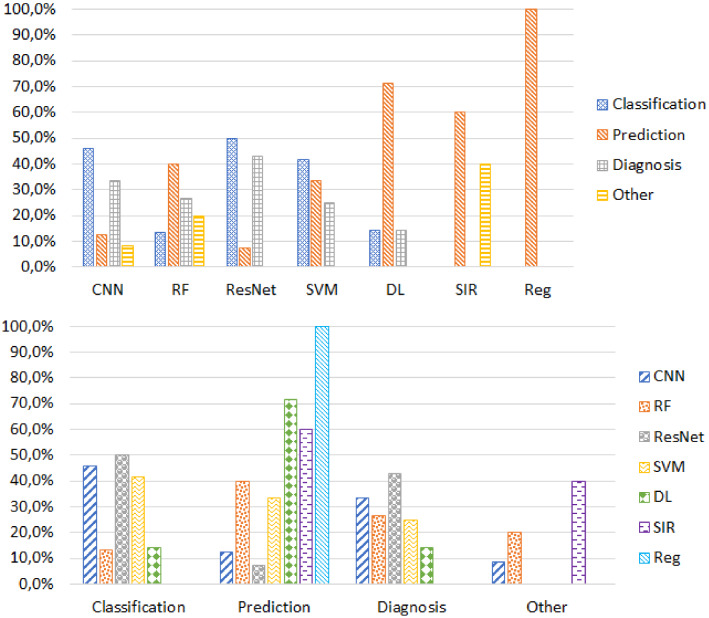
AI/ML techniques were used in the COVID-19 pandemic for (1) classification, (2) prediction, (3) diagnosis and (4) other applications like early warnings and alerts. Classification is the most popular aim for applying AI/ML methods [38, 48, 56, 65, 89, 105]. Review results presented in Fig. 4 indicates that most of the models (almost 50% of studies) used ResNet for classification. Recent advancements in DL led to the potential usage of various CNN architectures. Next to ResNet, some authors attempted the CNN model for classification (45% of studies). Few authors also tried to use traditional ML algorithms like SVM and RF for classification of COVID-19 data.
Fig. 4.
Objectives of AI/ML approaches in COVID-19
Prediction is the second popular objective in AI/ML approaches [106–111]. Regression analysis is a widely accepted model for prediction purposes (100% of studies) [112]. DL models are another popular prediction approach, which was adopted by 70% of studies. One of the most used mathematical models for the COVID-19 pandemic is SIR frameworks. More than 60% of the studies used the SIR framework for prediction [32, 91, 101–104]. Diagnosis is the third popular AI/ML usage purpose [113–117]. RF and SVM techniques were applied for diagnosis of COVID-19 with nearly 25–30% rates, respectively. As DL-based methods, CNN and ResNet, were used to classify, predict, and diagnose purposes. The results produced by this comprehensive review prove that AI methods are a promising mechanism to use for the current scenario of the COVID-19 pandemic.
Other reasons that concluded from the selected studies to apply AI/ML approaches in COVID-19 are given below.
Patient outcome prediction AI tools were developed to predict risk status of contracting the coronavirus. It is critical to know the factors that will put the patients at risk. LSTM is a popular method to predict patient outcome. For example, Obaid et al. [62] proposed a prediction mechanism that uses LSTM to carry this model out on a coronavirus dataset that identified from the records of infections, recovery cases and deaths across the world. Researchers came up with a different proposal to identify the risk factors that will help the clinicians. Some studies proposed models to assess the patients’ severity using the RF and regression model (Reg) [118–120]. Time-series prediction is an important task to predict pandemic diseases. In [121], the authors developed a time series forecasting model using a hybrid machine learning model. Beetle antennae search swarm intelligence algorithm is used for optimization. The proposed model was evaluated using real-time patient data obtained from China by World Health Organization (WHO). The proposed model obtained an score of 0.9763. Table 1 summarizes AI/ML methods for patient outcome prediction.
Table 1.
AI/ML methods for prediction of patient outcome
| Study | Objective | AI/ML approach |
|---|---|---|
| [97] | Identify the monocyte ratio and blood pressure in human body | RF |
| [118] | Predicting hospitalization | RF and Reg |
| [119] | Severity assessment | RF and Reg |
| [120] | Severity assessment | Reg |
| [122] | Identify the high-risk and low-risk patients | Reg |
| [123] | Identify the mortality risk, | XGBoost |
| [124] | Patient risk stratification | CNN |
| [125] | Confirmation of covı cases | LSTM |
XGBoost extreme gradient boosting
AI and ML models are potentially strong to fight with different pandemic (flu, dengue, zika, cholera, ebola, H1N1, influenza, swine fever) with different methods like classification, forecasting, prediction and pattern recognition. AI/ML tools covering these methods to play an essential role in fighting with the deadly disease [126]. Table 2 shows different AI/ML techniques in disease predictions.
Table 2.
AI/ML techniques in disease predictions
| Study | Disease | AI/ML method | Country |
|---|---|---|---|
| [127] | Dengue fever | CTree | Bangladesh |
| [128] | Oyster norovirus | GP | USA |
| [129] | Dengue fever | Reg, NB | India |
| [130] | H1N1 Flu | NN | Japan |
| [131] | Influenza | RF | Iran |
| [132] | Dengue fever | NN | Japan |
| [133] | Swine Fever | RF | China |
| [134] | Asthma exacerbations | NB, SVM | USA |
| [135] | Dementia prediction | SVM | Italy |
| [136] | Diabetes classification | Reg, NN, NB, KNN, RF | Brazil |
| [137–139] | Hepatic fibrosis | NB, RF, KNN, SVM, NN | N/A |
| [140] | Course of depression | Reg | N/A |
CTree classification tree, GP genetic programming, KNN K-nearest neighbors, NB Naive Bayes, NN neural network
Risk assessment of pandemic AI/ML models help to assess the risk of the pandemic. DL-based models were developed to predict the duration of the disease [141, 142], community-level risk assessment [143] and transmission prediction [144]. Early risk assessment of COVID-19 patients helps to reduce mortality. Several ML algorithms were developed in the literature. For example, Heldt et al. [145] proposed a model that extracts the informative clinical features from the data. XGBoost algorithm with 100 trees was trained on the dataset. The proposed model obtained (AUC-ROC) scores from 0.76 to 0.87. Table 3 gives an overview of risk assessment of COVID-19 with AI/ML methods.
Table 3.
AI/ML methods for risk assessment of COVID-19
| Study | Objective | AI/ML technique |
|---|---|---|
| [141] | Predict the duration of the disease | LSTM |
| [142] | Transmission prediction | LSTM, RNN |
| [143] | Community-level risk assessment | GAN |
| [144] | Transmission prediction | TL |
| [146] | Disease monitoring | CNN |
GAN generative adversarial network, RNN recurrent neural networks, TL transfer learning
Workload reduction of health professionals Because the sudden spike of COVID-19-affected patients, healthcare workers have a growing burden. Various AI/ML techniques were proposed for early diagnosis of the disease [147–149]. AI can tackle future challenges and address to reduce the workload of healthcare professionals [150].
Social control With high transmissibility of COVID-19, many countries adopted AI for pandemic management [151] and are successful in reducing the mortality rate. For example, a predictive model for mortality rate in COVID-19 using ML was developed by Booth et al [152]. Model identified the prognostic serum biomarkers in COVID-19 patients. Five serum parameters were used in the data set using a support vector classifier for classification. The proposed model achieved 91% specificity and 91% sensitivity. AI can facilitate the management of contact tracing, quarantine and self-isolation of people, screening for infection [153, 154]. AI-based drones were used to enforce social isolation [155].
Early warnings and alerts AI is a potential tool to fight against COVID-19, and AI-based systems are used in spotting COVID-19 disease outbreaks. Bots based on AI were used to predict the possible outbreak [156, 157]. Before the WHO (World Health Organization) sounded an alarm on the possible outbreak of COVID-19, an AI bot named “BlueDOT” [158] alerted employees’ possible outbreak of a pandemic. A similar bot, called “Health Map”, developed in the USA sounded the alarm for possible outbreak [159].
RQ 3: What is the data perspective of the research?
Table 4 gives the validity measurement types of researches. Most of the studies validated the research results by accuracy [77, 160–163]. Accuracy scores vary from 50 to 100%. However, these results are not the final output of these studies. For example, Elgendi et al. [86] and Hemdan et al. [87] applied various pre-trained AI methods. Whereas Elgendi et al. [86] reached 100% accuracy rates using ResNet-50, DarkNet-53, VGG-19, DenseNet-201, ResNet-18, ResNet-101, and GoogLeNet, Hemdan et al. [87] obtained a 50% accuracy score by InceptionV3. 82% of the research were tested the validity by three measurement types: accuracy, precision and sensitivity [58, 164].
Table 4.
Measurement types of study success
| Measurement | Percentage | Min (%) | Max (%) | Measurement | Percentage | Min | Max (%) |
|---|---|---|---|---|---|---|---|
| Accuracy | 31 | 50 | 100 | Precision | 6 | 79% | 99.29 |
| AUC | 12 | 85 | 99.6 | R squared | 3 | 98% | 99.7 |
| Explained variance | 2 | 99 | 99.7 | RMSE | 1 | 136.547 | |
| Sensitivity | 20 | 0.01% | 99.62 | ||||
| F1-score | 7 | 79 | 98.46 | Specificity | 18 | 70.7% | 99.99 |
AUC area under the curve, RMSE root mean square error
Table 5 represents data types and their statistics. Almost half of the COVID-19 works that benefit from AI/ML techniques analyzed CT images [59, 165–170]. X-ray is the second popular data type with a rate 35% [31, 66, 89, 162, 171–174]. A massive data size scale was used in those studies, ranging from 106 to 16,756 CT images and 50–15,085 X-ray images. Some studies focused on case data such as death and recovery numbers between a specific period [77, 90, 91, 175, 176]. Other data types such as dialogue data [92, 177], genome data [99], symptoms [72], blood data [74, 98] were excluded in Table 5 because they were measured below 5% of the studies.
Table 5.
Data types used in the COVID studies
| Data type | Percentage | Min | Max |
|---|---|---|---|
| CT | 49 | 106 images | 16,756 images |
| X-ray | 35 | 50 images | 15,085 images |
| Case data | 16 | 14 days | 77 days |
RQ 4: What is the current situation in drug preparation?
Due to the rapidly spreading across to the world and the lack of effective treatment options, drug developers have adopted the various strategies to fast track the drug discovery. Whereas some studies applied AI/ML techniques to predict, some of them analyzed the molecular structure of coronavirus because drug discovery is an expansive and lengthy process. Table 6 represents the drug studies against to COVID-19.
Table 6.
AI/ML method in drug development
| Study | Drug type | AI method | AI/ML objective | Potential drugs |
|---|---|---|---|---|
| [178] | SARS-CoV-2 inhibitors | ChemAI | Predict inhibitory effects of molecules | 30,000 top-ranked compounds |
| [179] | Antiviral drugs | MT-DTI | Predict commercially available antiviral drugs | Atazanavir, Remdesivir, and Efavirenz |
| [180] | Antiviral drugs | MT-DTI | Predict binding affinity between drugs and protein target | Remdesivir, Atazanavir, Efavirenz, Ritonavir, Dolutegravir, Kaletra |
| [181] | Anti-COVID-19 drugs | CNN, LSTM, MLP | Generate SMILES strings and molecules | 110 drugs |
| [182] | Targeted proteins of SARS-CoV-2 | DL | Predict binding between drugs and protein | 10 drugs |
| [183] | SARS-CoV-2 drug | NN, NB | Construct drug likelihood prediction model | 3 drugs |
| [184] | 2019-nCoV | DL | Generate new molecular structures for 3CLpro structures | 100 molecules |
The viral main proteinase of coronavirus
AI is a cost-effective and fast tool in drug discovery to fight against COVID-19. Shin et al. [180] proposed a Molecule Transformer Drug Target Interaction (MT-DTI) model that provides low-cost drugs and personalized medicines with multi-layered protein. MT-DTI was also applied to predict commercially available drugs [179]. This is the drug-target interaction model that uses deep learning. The result showed that Atazanavir, Remdesivir, and Efavirenz are suitable to fight against SARS-CoV-2. Hofmarcher et al. [178] proposed a DL model for drug discovery by predicting the inhibitory effects of molecules. Initially, they identified one billion molecules from the ZINC database for screening and ranking, and further molecules were reduced to 30K.
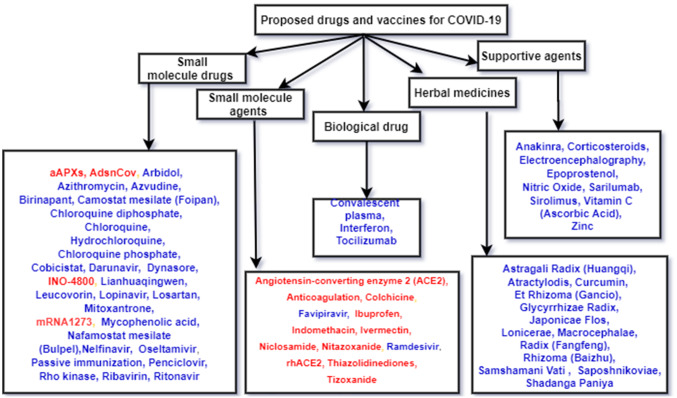
Some studies identified the drug compounds to fight against SARS CoV-2 coronavirus. Kadioglu et al. [183] identified three potential drugs for COVID-19 by adopting in silico methods to identify novel drugs using an AI model based on NB and NN. Hu et al. [182] identified ten drugs as potential inhibitors fight against SARS-CoV-2 by predicting the binding between drugs and protein using DL methods. Figure 5 summaries some candidate drugs or vaccines to treat this disease, which includes small molecule drugs, small molecule agents, herbal medicines and biological products [185–190]. Blue texts show the drug developments, whereas green texts refer vaccine developments.
Fig. 5.
Drugs and vaccines for COVID-19
Both small molecule drugs and small molecule agents are more potential drugs for COVID-19 [191]. Small molecule drugs like Lopinavir/Ritonavir and Ribavirin were used for the antiretroviral activity. On the other hand, Chloroquine phosphate and Arbidol were used to synthesize viral DNA or RNA. Small molecule agents such as Remdisivir, Favipiravir were used as an RdRp inhibitor. Similarly, biological products were used as a monoclonal antibody (Tocilizumab) or passive immunity boosters (Convalescent plasma). Some studies treated the COVID-19 with the help of a combination of drugs such as (hydroxychloroquine, azithromycin), (azithromycin, nitazoxanide), (favipiravir, hydroxychloroquine) and (favipiravir, azithromycin) [192].
Scientists are looking for a vaccine at least 95% effective to stop the pandemic [193]. AI techniques were widely used in the design of vaccines against SARS-CoV-2 [194, 195]. Some studies utilized AI approaches to obtain protein sequences [196] and nucleotide sequences [197]. Epitope prediction using AI/ML techniques were also popular in vaccine development against COVID-19 [196–201].
RQ 5: What software platforms are used?
Practitioners encountered severe challenges in the detection of Ncov-2019 because SAR-CoV-2 viruses spread rapidly. Reverse Transcription Polymerase Chain Reaction (RT-PCR) approach is not applicable due to some obstructions [202]. The shortcomings of RT-PCR can be obviated by analyzing medical images because developing digital technologies help prevent diseases by applying statistics, machine learning, and artificial intelligence models [203]. Table 7 presents several models and software platforms. These models’ capability was provided in a broad range of uses; from disease detection and prediction to social control. Applications involve real-time data analysis for disease detection and diagnosis, treatment monitoring, prediction of cases and mortality, and drugs/vaccines development [204]. Except from the studies in the table, some studies used more than one software such as Python and Excel [205], Python and R [118, 206], MATLAB and Excel [207].
Table 7.
Models in COVID-19 with software platform
| Software | Study | Model | Data source |
|---|---|---|---|
| Python | [207] | SIR, SDM, PA | Worldometers |
| [208] | Regression model | MoHFW, covid19india.org | |
| [209] | Pre-trained CNN | GitHub, Kaggle, Open-I repository | |
| [160] | CT radiomics | GitHub | |
| [205] | Regression model | covid19india.org, WHO | |
| R | [210] | SIRD and SVM | Worldometers |
| [211] | ARIMA, SIR | Johns Hopkins U. | |
| [212] | Regression model | Worldometers | |
| [213] | SIR | Johns Hopkins U. | |
| [214] | Regression model | Worldometer, covid19India.org | |
| [163] | Hybrid model approach | Worldometers, ourworldindata.org | |
| [215] | Regression model | MoHFW, John Hopkins U | |
| [216] | Regression model | WHO, Historical weather | |
| Not Given | [217] | Regression model, MLP | Kaggle |
| [218] | ARIMA, SVM | WHO | |
| [219] | Fractional mathematical model | N/A | |
| [220] | AP, TB | WHO, Worldometers | |
| [221] | Exponential growth model | MoHFW, WHO, covid19india.org | |
| [222] | SIR, Network model | COVID19USA | |
| [223] | Regression model | John Hopkins U |
AP arithmetic projection, ARIMA autoregressive integrated moving average, MoHFW Ministry of Health and Family Welfare, Government of India, MLP Multilayer perceptron, PA propagation analysis, SDM social distancing matrix, TB tree-based model
RQ 6: Which data sources can be reached?
Data are presented as an essential aspect of implementing scientific methods. The research community always follows two approaches: closed source or open source [224]. Closed source is considered for proprietary objects, whereas open source leads to more precious quality, transparency, verifiability, usability [225, 226]. In the COVID-19 pandemic, the open-source approach is considered more effective for mitigating and detecting the virus due to its prior symptoms. It is highlighted that the COVID-19 pandemic needs a collaborative and unified approach along with open-source data, so the scientific community can get transparent and valid research [227, 228]. Different datasets were presented to combat with the COVID-19 pandemic in different ways [224].
Three main types of datasets in COVID-19 were used, textual data, medical data and speech data. Textual data represents dashboard, mobility data, case reports, social media posts and articles. Medical data generally presents diagnosis and screening of COVID-19 patients since medical images consider X-rays, CT scans, ultrasound or MRI (Magnetic Resonance Imaging). Most of the datasets represent CT scans, X-rays, and AI/ML techniques applied to predict resources in the future. Speech datasets help to detect and diagnose by cough sound, breathing rate and stress detection techniques.
Most of the datasets were stored on different repositories, such as Github and Kaggle. Table 8 presents 18 textual datasets, nine medical datasets and seven speech datasets.
Table 8.
Datasets and their details
| Textual data sets | Medical datasets | ||
|---|---|---|---|
| Data sets | Explanation | Data sets | Explanation |
| T1 [229] | Datahub repository | M1 [230] | COVID-19 CT scans of Chinese hospitals with an online repository |
| T2 [231] | Github repository of the data | M2 [232] | Dataset consists of 20 COVID-19 CT scans |
| T3 [233] | Medical community | M3 [234] | Segmentation benchmark |
| T4 [235] | Real-time interactive dashboard | M4 [236] | COVID-19 CT segmentation dataset |
| T5 [237] | Open source datasets | M5 [238] | Images from a repository |
| T6 [239] | crowd-sourced list of open access COVID-19 projects | M6 [240] | 3D CT scans of confirmed cases |
| T7 [241] | Country specific case reports and articles | M7 [242] | COVID-19 positive and suspected patients |
| T8 [243] | Demographic database | M8 [244] | Analyzing radiographical images |
| T9 [245] | Real-time and historical mobility data from Wuhan | M9 [246] | Repository for COVID-19 radiographic images |
| T10 [247] | Real-time data | Speech and audio datasets | |
| T11 [248] | Data sets of Twitter posts | Data sets | Explanation |
| T12 [249] | Data sets of Twitter posts | S1 [250] | Web application for data collection |
| T13 [251] | Web search portal for dataset of scholarly articles | S2 [252] | Open source voice dataset |
| T14 [253] | Google mobility reports | S3 [254] | Collection of the cough data |
| T15 [255] | Data set available on mobility based on user requests to location services | S4 [256] | Collection of the cough data |
| T16 [257] | Web application identifying mobility patterns across the U.S | S5 [258] | Collection of the cough data |
| T17 [259] | Mobility data from Baidu location services | S6 [260] | Data collection for cough data |
| T18 [261] | Google location services | S7 [262] | Repository for the cough data |
Total 18 textual datasets were discussed to show the relevancy of different purposes. These datasets consider COVID-19 case reports, report analysis, mobility data, social media data, scholarly articles, tweets, non-pharmaceutical interventions (NPI). Several studies maintained and shared the epidemiological data of COVID-19 cases in China [225, 263]. COVID-19 case reports include different details like (a) symptoms of the disease, (b) dates of patient admission, date of infection confirmation, travel dates, (c) other information like resources of food [263]. They were presented to analyze the transmission, testing, forecasting and death cases [264–269]. Some studies evaluated and investigated human mobility, travel restriction, social distancing and control measure [270–274]. Social media data and scholarly articles were also collected to present different textual data such as emotions and worries [275–281] and scientific article data from existing studies [282–286]. Tweets also provide collected textual data. Several studies collected twitter datasets to identify the pandemic information from a social aspect and analyze human behavior [278, 279, 287]. NPI is considered as different sets of measures accepted by governments to prevent the COVID-19 pandemic. The NPI effect was analyzed for COVID-19 cases [288]. Mobility datasets are significant to provide the information of infected cases and also helpful to diagnose the response of societies in NPI restrictions. Several open-source datasets provide information with dynamic features.
Medical datasets, which include CT and X-ray images, are essential in diagnosis of COVID-19. Studies based on COVID-19 diagnosis used different datasets for CT-Scan [34, 89, 165, 289–297] and X-ray [20, 78, 87, 298, 299] images by different AI/ML techniques [160, 300, 301]. The study of Sharma and his colleagues [302] distributed the original image dataset into 10% external validation dataset-I and 90% training dataset as Dataset-II. Dataset-I has 35 images, and Dataset-II has 317 images and generated a total of 27 different types of training and validation datasets for chest X-ray images. Out of these datasets, one dataset includes real images, and 26 datasets consist of single augmentation images. All these 27 datasets were used to train and validate the 29 types of chest X-ray classification models. A comprehensive study was performed to understand the performance of automatic detection of COVID-19 based on medical images [22]. This study uses COVID-19 and normal X-ray images and adopts transfer learning to increase the accuracy. To make general framework and avoid overfitting, different training policies are adopted using AdaGrad algorithm. A hybrid deep learning framework COVID-CheXNet has been proposed by Al-Waisy et al. [20] to reduce the load on radiologists and control of the pandemic. This model helps to diagnose the COVID-19 virus in chest X-ray images and is composed of four primary stages: image pre-processing, image classification, features extraction and fusion. Mohammed et al. [22] have proposed an automatic prediction to identify COVID-19 for discriminating automatically between normal and COVID-19 infected people in X-ray images. To accomplish this, they used traditional ML methods such as SVM, NN, DT and kNN techniques. They also applied deep learning models such as ResNet50, MobileNets V2, DarkNet, GoogleNet, and Xception.
Speech or audio datasets help to detect and diagnosis of infection by three different method such as cough sound analysis [303–305], breathing rate analysis [306–309] and stress detection [310–312]. Cough sounds can identify a COVID-19 infected case by applying ML techniques. Breathing rate can be identified by speech, resulting in COVID-19 patient screening. Stress detection also helps to identify the cases that person suffer from mental health issues and symptoms of COVID-19. These methods can be done by remote medical care or smart devices. AI/ML techniques are successfully applied for extracting features and classify new inputs based on model training.
Table 9 gives a tabular and descriptive survey for various open source datasets. This table covers 20 datasets with different data-types such as X-ray, CT Scans, Ultra-sound, case data, tweets, voice data. These datasets were applied different methods with different applications. For example, CNN, SVM and TL were applied for diagnosis [38, 165, 313–315]. Bayesian approach method was applied in community transmission [316–321], while data mining methods [322–327] were used for symptoms identifications. Regression analysis methods [148, 328–331] were used for transmission control analysis.
Table 9.
Dataset applications with AI/ML
| Study | Application | Methods | Database |
|---|---|---|---|
| [332] | COVID-19 diagnosis | DenseNet, TL | Medical |
| [290] | COVID-19 diagnosis | Deep CNN | Medical |
| [87] | COVID-19 diagnosis | Deep learning | Medical |
| [78] | COVID-19 diagnosis | CNN, TL | Medical |
| [31] | COVID-19 diagnosis | CNN | Medical |
| [301] | COVID-19 diagnosis | CNN | Medical |
| [271] | Cases exported from China | Statistical | Medical |
| [266] | Correcting under reported cases | Statistical | Textual |
| [273] | International travel control analysis | Statistical | Textual |
| [274] | COVID-19 transmission control | Regression analysis | Textual |
| [333] | Community transmission | Expectation maximization | Textual |
| [334] | Community transmission | Bayesian approach | Textual |
| [276] | Social dynamics data | Statistical analysis | Textual |
| [335] | Perception and policies | Proposed NLP | Textual |
| [281] | COVID-19 symptom identification | Data mining | Textual |
| [304] | COVID-19 diagnosis | Boosting Trees, SVM | Speech |
| [305] | COVID-19 diagnosis | N/A | Speech |
| [309] | COVID-19 speech analysis | SVM with linear kernel | Speech |
| [279] | Government and Media Tweets | N/A | Textual |
| [277] | Conversation dynamics | N/A | Textual |
Limitations
Some limitations of the current research should be accepted. The research is limited to selected search terms, databases and selection criteria.
This research was conducted in a certain period of time. However, the number of studies on COVID-19 increases day by day because of its popularity and necessity. Because a systematic literature review was presented with this research, it is necessary to limit the research content. To decrease the effect of this situation, the inclusion and exclusion questions were prepared to select the studies published in the research period.
Seven online databases were scanned for the review. However, other databases can be scanned. If the research is to be expanded, the number of databases can be increased.
Apart from selected studies in this research, there are many different studies. It should not be forgotten that some criteria were set for narrowing the research scope. For example, studies that do not mention the algorithm applied in the implementation or do not give details were ignored. Applied AI/ML studies are generally implemented for different purposes without considering COVID-19 problems. Therefore, COVID-19 problems are not explicitly stated in the publications. By evaluating each study individually, it was determined which problem discussed. At this stage, there may be unobserved publications.
Conclusion
This systematic review study investigates 264 studies from seven accessible databases to find answers for six significant research questions. This research aims to explore and organize potential literature so that practitioners, academicians, and researchers can easily access the existing methods, applications, and datasets. The main contribution of this research to identify the AI/ML methods and techniques for disease prediction, measurement and data types, AI/ML method in drug development, available drug and vaccines, and existing models and datasets for the COVID-19 pandemic. CNN, RF, ResNet and SVM approaches are the most used AI/ML approaches against COVID-19. These approaches were applied for various purposes. Classification, prediction and diagnosis are the most popular AI/ML objectives. ResNet applied for classification and diagnosis, whereas regression is used for prediction studies. Apart from these objectives, previous studies benefited from the advantages of AI/ML tools for several additional purposes, such as patient outcome prediction, risk assessment, workload reduction of health professionals, social control and early warnings and alerts. This study concludes that the methods’ success varies widely. Nine major measurement types were considered to evaluate models’ success. Accuracy, sensitivity and specificity were measured 69% of studies. 84% of studies used either CT or X-ray images between 50 and near to 17,000. Case data are the third popular data type with a rate of 16% up to 77 days. Python and R the most preferred software platform to apply AI/ML methods. Some studies used Matlab, Microsoft Excel and more than one software. Data were stored in three main categories, textual, medical, and speech. Because the research has review borders, it has some limitations that were discussed in the study.
This study is most significant for new practitioners and researchers who plan to develop an AI/ML model or drug for COVID-19. They can reuse existing models and drugs rather than design from scratch and save time for doing potential research and future studies. Besides, this research provides a backbone for different aspects such as disease diagnosis and detection, drug and vaccine development, AI/ML models and techniques. The conducted literature provides comprehensive details of AI’s potential and existing contribution to combating the pandemic.
As it is understood from the literature review, many researchers applied CNN models. The main reason can be that they are powerful for the spatial coherence or local pixel correlations in medical images. CNN technique was usually applied for either classification or diagnosis. However, authors should remind aforementioned drawbacks before applying CNN for COVID-19 studies.
For further research, the authors can focus on several points. First of all, researchers can scan other databases such as ERIC, DOAJ and JSTOR. Some additional research questions can be investigated to clarify interesting and meaningful results.
Abbreviation
Table 10 presents the abbreviations used in the study.
Table 10.
Abbreviations used in this study
| Abbr. | Explanation | Abbr. | Explanation |
|---|---|---|---|
| AI | Artificial intelligence | NN | Neural network |
| AP | Arithmetic projection | NPI | Non-pharmaceutical interventions |
| ARIMA | Autoregressive integrated moving average | PA | Propagation analysis |
| AUC | Area under curve | Reg | Regression models |
| CNN | Convolutional neural network | RF | Random forest |
| COVID-19 | Coronavirus disease 2019 | RMSE | Root mean square error |
| CT | Computational tomography | RNN | Recurrent neural networks |
| CTree | Classification tree | RQ | Research questions |
| DL | Deep learning | RT-PCR | Reverse transcription polymerase chain reaction |
| GAN | Generative adversarial network | SDM | Social distancing matrix |
| GP | Genetic programming | SEIR | Susceptible, exposed, infectious, recovered |
| KNN | K-Nearest Neighbor | SIR | Susceptible, infectious, recovered models |
| LSTM | Long short-term memory | SIRD | Susceptible, infectious, recovered, deceased |
| ML | Machine learning | SVM | Support vector machine |
| MLP | Multilayer perceptron | TB | Tree-based |
| MRI | Magnetic resonance imaging | TL | Transfer learning |
| MT-DTI | Molecule transformer drug target interaction | WHO | World Health Organization |
| NB | Naive Bayes | XGBoost | Extreme gradient boosting |
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Onur Dogan, Email: onur.dogan@bakircay.edu.tr.
Sanju Tiwari, Email: tiwarisanju18@ieee.org.
M. A. Jabbar, Email: jabbar.meerja@gmail.com
Shankru Guggari, Email: shankar286@gmail.com.
References
- 1.Wuhan Municipal Health Commission (2019) Report of clustering pneumonia of unknown aetiology in Wuhan City. http://wjw.wuhan.gov.cn/front/web/showDetail/201. Accessed 20 Jan 2020
- 2.Tiwari SM, Gaurav D, Abraham A. COVID-19 outbreak in India: an early stage analysis. Int J Sci Rep. 2020;6(8):332–339. doi: 10.18203/issn.2454-2156.IntJSciRep20203117. [DOI] [Google Scholar]
- 3.Jahanbin K, Rahmanian V, et al. Using twitter and web news mining to predict COVID-19 outbreak. Asian Pac J Trop Med. 2020;13(8):378. doi: 10.4103/1995-7645.279651. [DOI] [Google Scholar]
- 4.Ferguson NM, Laydon D, Nedjati-Gilani G, Imai N, Ainslie K, Baguelin M, Bhatia S, Boonyasiri A, Cucunubá Z, Cuomo-Dannenburg G et al (2020) Impact of non-pharmaceutical interventions (NPIs) to reduce COVID-19 mortality and healthcare demand. Imperial College COVID-19 Response Team
- 5.COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (2020) https://github.com/CSSEGISandData/COVID-19/blob/master/README.md. Accessed 29 July 2020
- 6.Baldwin R (2020) https://bit.ly/3vyXRhH. Accessed 30 May 2020
- 7.Gaurav D, Tiwari SM, Goyal A, Gandhi N, Abraham A. Machine intelligence-based algorithms for spam filtering on document labeling. Soft Comput. 2020;24(13):9625–9638. doi: 10.1007/s00500-019-04473-7. [DOI] [Google Scholar]
- 8.Silver D, Schrittwieser J, Simonyan K, Antonoglou I, Huang A, Guez A, Hubert T, Baker L, Lai M, Bolton A, et al. Mastering the game of go without human knowledge. Nature. 2017;550(7676):354–359. doi: 10.1038/nature24270. [DOI] [PubMed] [Google Scholar]
- 9.Agrebi S, Larbi A (2020) Use of artificial intelligence in infectious diseases. In: Barh D (ed) Artificial intelligence in precision health. Academic Press, pp 415–438
- 10.Chakraborti S, Maiti A, Pramanik S, Sannigrahi S, Pilla F, Banerjee A, Das DN. Evaluating the plausible application of advanced machine learnings in exploring determinant factors of present pandemic: a case for continent specific COVID-19 analysis. Sci Total Environ. 2021;765:142723. doi: 10.1016/j.scitotenv.2020.142723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muammer T. Covidetectionet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl Intell. 2021;51:1213–1226. doi: 10.1007/s10489-020-01888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahid F, Zameer A, Muneeb M. Predictions for COVID-19 with deep learning models of LSTM, GRU and BI-LSTM. Chaos Solitons Fractals. 2020;140:110212. doi: 10.1016/j.chaos.2020.110212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rachna J, Meenu G, Soham T, Jude HD. Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl Intell. 2021;51:1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen TT (2020) Artificial intelligence in the battle against coronavirus (COVID-19): a survey and future research directions, preprint on webpage at arXiv:2008.07343
- 16.Maghdid HS, Ghafoor KZ, Sadiq AS, Curran K, Rabie K (2020) A novel AI-enabled framework to diagnose coronavirus COVID 19 using smartphone embedded sensors: design study, preprint on webpage at arXiv:2003.07434
- 17.Kumar A, Gupta PK, Srivastava A. A review of modern technologies for tackling COVID-19 pandemic. Diabetes Metab Syndr Clin Res Rev. 2020;14(4):569–573. doi: 10.1016/j.dsx.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nguyen D, Gao K, Chen J, Wang R, Wei G (2020) Potentially highly potent drugs for 2019-ncov, preprint on webpage at 10.1101/2020.02.05.936013v1
- 19.Bullock J, Luccioni A, Pham KH, Lam CSN, Luengo-Oroz M. Mapping the landscape of artificial intelligence applications against COVID-19. J Artif Intell Res. 2020;69:807–845. doi: 10.1613/jair.1.12162. [DOI] [Google Scholar]
- 20.Al-Waisy AS, Al-Fahdawi S, Mohammed MA, Abdulkareem KH, Mostafa SA, Maashi MS, Arif M, Garcia-Zapirain B. COVID-chexnet: hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft Comput. 2020;24:1–16. doi: 10.1007/s00500-020-05424-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Khaled Bayoudh AM, Hamdaoui F. Hybrid-COVID: a novel hybrid 2d/3d CNN based on cross-domain adaptation approach for COVID-19 screening from chest X-ray images. Phys Eng Sci Med. 2020;43(4):1415–1431. doi: 10.1007/s13246-020-00957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed MA, Abdulkareem KH, Garcia-Zapirain B, Mostafa SA, Maashi MS, Al-Waisy AS, Subhi MA, Mutlag AA, Le DN. A comprehensive investigation of machine learning feature extraction and classification methods for automated diagnosis of COVID-19 based on X-ray images. Comput Mater Continua. 2020;66(3):3289–3310. doi: 10.32604/cmc.2021.012874. [DOI] [Google Scholar]
- 23.Mishra S, Sagban R, Yakoob A, Gandhi N. Swarm intelligence in anomaly detection systems: an overview. Int J Comput Appl. 2018;43:109–118. [Google Scholar]
- 24.Rahul M, Kohli N, Agarwal R, Mishra S. Facial expression recognition using geometric features and modified hidden Markov model. Int J Grid Util Comput. 2019;10(5):488–496. doi: 10.1504/IJGUC.2019.102018. [DOI] [Google Scholar]
- 25.Brereton P, Kitchenham BA, Budgen D, Turner M, Khalil M. Lessons from applying the systematic literature review process within the software engineering domain. J Syst Softw. 2007;80(4):571–583. doi: 10.1016/j.jss.2006.07.009. [DOI] [Google Scholar]
- 26.Kadi I, Idri A, Fernandez-Aleman J. Knowledge discovery in cardiology: a systematic literature review. Int J Med Inform. 2017;97:12–32. doi: 10.1016/j.ijmedinf.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Petersen K, Feldt R, Mujtaba S, Mattsson M (2008) Systematic mapping studies in software engineering. In: 12th international conference on evaluation and assessment in software engineering (EASE) 12, pp 1–10
- 28.Roberto R, Lima JP, Teichrieb V. Tracking for mobile devices: a systematic mapping study. Comput Graph. 2016;56:20–30. doi: 10.1016/j.cag.2016.02.002. [DOI] [Google Scholar]
- 29.Petersen K, Gencel C (2013) Worldviews, research methods, and their relationship to validity in empirical software engineering research. In: 2013 Joint Conference of the 23rd international workshop on software measurement and the 8th international conference on software process and product measurement. IEEE, pp 81–89
- 30.Kitchenham B, Brereton OP, Budgen D, Turner M, Bailey J, Linkman S. Systematic literature reviews in software engineering: a systematic literature review. Inf Softw Technol. 2009;51(1):7–15. doi: 10.1016/j.infsof.2008.09.009. [DOI] [Google Scholar]
- 31.Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al Emadi N, et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 32.Ardabili SF, Mosavi A, Ghamisi P, Ferdinand F, Varkonyi-Koczy AR, Reuter U, Rabczuk T, Atkinson PM. COVID-19 outbreak prediction with machine learning. Algorithms. 2020;13(10):249. doi: 10.3390/a13100249. [DOI] [Google Scholar]
- 33.Chimmula VKR, Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals. 2020;135:109864. doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M. COVID-19 image data collection: prospective predictions are the future. J Mach Learn Biomed Imaging. 2020;2:1–38. [Google Scholar]
- 35.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kavadi DP, Patan R, Ramachandran M, Gandomi AH. Partial derivative nonlinear global pandemic machine learning prediction of COVID 19. Chaos Solitons Fractals. 2020;139:110056. doi: 10.1016/j.chaos.2020.110056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohammed MA, Abdulkareem KH, Al-Waisy AS, Mostafa SA, Al-Fahdawi S, Dinar AM, Alhakami W, Abdullah B, Al-Mhiqani MN, Alhakami H, et al. Benchmarking methodology for selection of optimal COVID-19 diagnostic model based on entropy and Topsis methods. IEEE Access. 2020;8:99115–99131. doi: 10.1109/ACCESS.2020.2995597. [DOI] [Google Scholar]
- 38.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using detrac deep convolutional neural network. Appl Intell. 2020;51:854–864. doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A. COVID-caps: a capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manapure P, Likhar K, Kosare H. Detecting COVID-19 in X-ray images with keras, tensor flow, and deep learning. Artif Comput Intell. 2020;2(3):1–6. [Google Scholar]
- 41.Asif S, Wenhui Y (2020) Automatic detection of COVID-19 using X-ray images with deep convolutional neural networks and machine learning, preprint on webpage at 10.1101/2020.05.01.20088211v2
- 42.Asnaoui KE, Chawki Y, Idri A (2020) Automated methods for detection and classification pneumonia based on X-ray images using deep learning, preprint on webpage at arXiv:2003.14363
- 43.Ghoshal B, Tucker A (2020) Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection, preprint on webpage at arXiv:2003.10769
- 44.El Asnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2020;39:1–12. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan AI, Shah JL, Bhat MM. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Programs Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge: MIT Press; 2016. [Google Scholar]
- 47.Jin C, Chen W, Cao Y, Xu Z, Zhang X, Deng L, Zheng C, Zhou J, Shi H, Feng J (2020) Development and evaluation of an AI system for COVID-19 diagnosis, preprint on webpage at 10.1101/2020.03.20.20039834v3 [DOI] [PMC free article] [PubMed]
- 48.Karim MR, Döhmen T, Cochez M, Beyan O, Rebholz-Schuhmann D, Decker S (2020) Deepcovidexplainer: explainable COVID-19 diagnosis from chest X-ray images. In: 2020 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 1034–1037
- 49.Shibly KH, Dey SK, Islam MT-U, Rahman MM. COVID faster R-CNN: a novel framework to diagnose novel coronavirus disease (COVID-19) in X-ray images. Inform Med Unlocked. 2020;20:100405. doi: 10.1016/j.imu.2020.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. 2020;140:110122. doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das NN, Kumar N, Kaur M, Kumar V, Singh D (2020) Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays, preprint on webpage at 10.1016/j.irbm.2020.07.001 [DOI] [PMC free article] [PubMed]
- 52.Altan A, Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2d curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals. 2020;140:110071. doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu H, Guo Q, Li M, Wang C, Fang Z, Wang P, Tan J, Wu S, Xiao Y (2020) Host and infectivity prediction of Wuhan 2019 novel coronavirus using deep learning algorithm, preprint on webpage at 10.1101/2020.01.21.914044v4
- 55.Zhu JS, Ge P, Jiang C, Zhang Y, Li X, Zhao Z, Zhang L, Duong TQ. Deep-learning artificial intelligence analysis of clinical variables predicts mortality in COVID-19 patients. J Am Coll Emerg Physicians Open. 2020;1(6):1364–1373. doi: 10.1002/emp2.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hall LO, Paul R, Goldgof DB, Goldgof GM (2020) Finding COVID-19 from chest X-rays using deep learning on a small dataset, preprint on webpage at arXiv:2004.02060
- 57.Jamil M, Hussain I et al (2020) Automatic detection of COVID-19 infection from chest x-ray using deep learning, preprint on webpage at 10.1101/2020.05.10.20097063v1
- 58.Jin S, Wang B, Xu H, Luo C, Wei L, Zhao W, Hou X, Ma W, Xu Z, Zheng Z et al (2020) Ai-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks, preprint on webpage at 10.1101/2020.03.19.20039354v1 [DOI] [PMC free article] [PubMed]
- 59.Voulodimos A, Protopapadakis E, Katsamenis I, Doulamis A, Doulamis N (2020) Deep learning models for COVID-19 infected area segmentation in CT images, preprint on webpage at 10.1101/2020.05.08.20094664v2 [DOI] [PMC free article] [PubMed]
- 60.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK (2020) Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms, preprint on webpage at arXiv:2004.00038
- 61.Alakus TB, Turkoglu I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos Solitons Fractals. 2020;140:110120. doi: 10.1016/j.chaos.2020.110120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Obaid OI, Mohammed MA, Mostafa SA. Long short-term memory approach for coronavirus disease prediction. J Inf Technol Manag. 2020;12:11–21. [Google Scholar]
- 63.Farooq M, Hafeez A (2020) COVID-resnet: a deep learning framework for screening of covid19 from radiographs, preprint on webpage at arXiv:2003.14395
- 64.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al (2020) Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT, preprint on webpage at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233473/
- 65.Pathak Y, Shukla PK, Tiwari A, Stalin S, Singh S (2020) Deep transfer learning based classification model for COVID-19 disease, preprint on webpage at 10.1016/j.irbm.2020.05.003 [DOI] [PMC free article] [PubMed]
- 66.Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) COVID-19 screening on chest X-ray images using deep learning based anomaly detection, preprint on webpage at arXiv:2003.12338
- 67.Castiglioni I, Ippolito D, Interlenghi M, Monti CB, Salvatore C, Schiaffino S, Polidori A, Gandola D, Messa C, Sardanelli F (2020) Artificial intelligence applied on chest X-ray can aid in the diagnosis of COVID-19 infection: a first experience from Lombardy, Italy, preprint on webpage at 10.1101/2020.04.08.20040907v1
- 68.Dogan O, Martinez-Millana A, Rojas E, Sepúlveda M, Munoz-Gama J, Traver V, Fernandez-Llatas C. Individual behavior modeling with sensors using process mining. Electronics. 2019;8(7):766. doi: 10.3390/electronics8070766. [DOI] [Google Scholar]
- 69.Dogan O, Oztaysi B. Genders prediction from indoor customer paths by Levenshtein-based fuzzy KNN. Expert Syst Appl. 2019;136:42–49. doi: 10.1016/j.eswa.2019.06.029. [DOI] [Google Scholar]
- 70.Li X, Pang T, Xiong B, Liu W, Liang P, Wang T (2017) Convolutional neural networks based transfer learning for diabetic retinopathy fundus image classification. In: 2017 10th international congress on image and signal processing, BioMedical engineering and informatics (CISP-BMEI). IEEE, pp 1–11
- 71.Wan S, Liang Y, Zhang Y. Deep convolutional neural networks for diabetic retinopathy detection by image classification. Comput Electr Eng. 2018;72:274–282. doi: 10.1016/j.compeleceng.2018.07.042. [DOI] [Google Scholar]
- 72.Ahamad MM, Aktar S, Rashed-Al-Mahfuz M, Uddin S, Liò P, Xu H, Summers MA, Quinn JM, Moni MA. A machine learning model to identify early stage symptoms of SARS-COV-2 infected patients. Expert Syst Appl. 2020;160:113661. doi: 10.1016/j.eswa.2020.113661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Moraes Batista AF, Miraglia JL, Donato THR, Chiavegatto Filho ADP (2020) COVID-19 diagnosis prediction in emergency care patients: a machine learning approach, preprint on webpage at 10.1101/2020.04.04.20052092v2
- 74.de Freitas Barbosa VA, Gomes JC, de Santana MA, de Almeida Albuquerque JE, de Souza RG, de Souza, RE, dos Santos WP (2020) Heg. IA: an intelligent system to support diagnosis of COVID-19 based on blood tests, preprint on webpage at 10.1007/s42600-020-00112-5
- 75.Iwendi C, Bashir AK, Peshkar A, Sujatha R, Chatterjee JM, Pasupuleti S, Mishra R, Pillai S, Jo O. COVID-19 patient health prediction using boosted random forest algorithm. Front Public Health. 2020;8:357. doi: 10.3389/fpubh.2020.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mei X, Lee H-C, Diao K-Y, Huang M, Lin B, Liu C, Xie Z, Ma Y, Robson PM, Chung M, et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26(8):1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pourghasemi HR, Pouyan S, Heidari B, Farajzadeh Z, Shamsi SRF, Babaei S, Khosravi R, Etemadi M, Ghanbarian G, Farhadi A, et al. Spatial modeling, risk mapping, change detection, and outbreak trend analysis of coronavirus (COVID-19) in Iran (days between February 19 and June 14. Int J Infect Dis. 2020;98(2020):90–108. doi: 10.1016/j.ijid.2020.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Apostolopoulos ID, Mpesiana TA. COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hassanien A, Mahdy LN, Ezzat KA, Elmousalami HH, Ella HA (2020) Automatic X-ray COVID-19 lung image classification system based on multi-level thresholding and support vector machine, preprint on webpage at 10.1101/2020.03.30.20047787v1
- 80.Özkaya U, Öztürk Ş, Barstugan M (2020) Coronavirus (COVID-19) classification using deep features fusion and ranking technique. In: Hassanien A-E, Dey N, Elghamrawy S (eds) Big data analytics and artificial intelligence against COVID-19: innovation vision and approach. Springer, pp 281–295
- 81.Ozturk S, Ozkaya U, Barstugan M (2020) Classification of coronavirus images using shrunken features, preprint on webpage at 10.1101/2020.04.03.20048868v2
- 82.Jiang X, Coffee M, Bari A, Wang J, Jiang X, Huang J, Shi J, Dai J, Cai J, Zhang T, et al. Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity. Comput Mater Continua. 2020;63(1):537–551. doi: 10.32604/cmc.2020.010691. [DOI] [Google Scholar]
- 83.Schwab P, Schütte A, Dietz B, Bauer S (2020) PREDCOVID-19: a systematic study of clinical predictive models for coronavirus disease 2019, preprint on webpage at arXiv:2005.08302
- 84.Song F, Shi N, Liu F, Li S, Li P, Zhang W, Jiang X, Zhang Y, Sun L, Sun L, et al. Combination of four clinical indicators predicts the severe/critical symptom of patients infected COVID-19. J Clin Virol. 2020;128:104431. doi: 10.1016/j.jcv.2020.104431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Elgendi M, Fletcher R, Howard N, Menon C, Ward R (2020) The evaluation of deep neural networks and X-ray as a practical alternative for diagnosis and management of COVID-19, preprint on webpage at 10.1101/2020.05.12.20099481v1
- 87.Hemdan EE-D, Shouman MA, Karar ME (2020) Covidx-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images, preprint on webpage at arXiv:2003.11055
- 88.Loey M, Smarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on gan and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 89.Sethy PK, Behera SK, Ratha PK, Biswas P (2020) Detection of coronavirus disease (COVID-19) based on deep features and support vector machine, preprint on webpage at https://www.preprints.org/manuscript/202003.0300/v2
- 90.Dang Q, Miao R, Yong L (2020) COVID-19 in shang hai: it is worth learning from the successful experience in preventing and controlling the overseas epidemic situation, preprint on webpage at 10.1101/2020.05.13.20100164v1
- 91.Rui M, Qi D, Yong L (2020) A sparse gaussian network model for prediction the growth trend of COVID-19 overseas import case: When can Hong Kong lift the international traffic blockad? Preprint on webpage at 10.1101/2020.05.13.20099978v1
- 92.Yang W, Zeng G, Tan B, Ju Z, Chakravorty S, He X, Chen S, Yang X, Wu Q, Yu Z et al (2020) On the generation of medical dialogues for COVID-19, preprint on webpage at arXiv:2005.05442
- 93.Tuli S, Tuli S, Tuli R, Gill SS. Predicting the growth and trend of COVID-19 pandemic using machine learning and cloud computing. Internet Things. 2020;11:100222. doi: 10.1016/j.iot.2020.100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fong SJ, Li G, Dey N, Crespo RG, Herrera-Viedma E (2020) Finding an accurate early forecasting model from small dataset: a case of 2019-ncov novel coronavirus outbreak, preprint on webpage at arXiv:2003.10776
- 95.Ribeiro MHDM, da Silva RG, Mariani VC, dos Santos Coelho L. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos Solitons Fractals. 2020;135:109853. doi: 10.1016/j.chaos.2020.109853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi F, Xia L, Shan F, Wu D, Wei Y, Yuan H, Jiang H, Gao Y, Sui H, Shen D (2020) Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification, preprint on webpage at arXiv:2003.09860 [DOI] [PubMed]
- 97.Tang Z, Zhao W, Xie X, Zhong Z, Shi F, Liu J, Shen D (2020) Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images, preprint on webpage at arXiv:2003.11988
- 98.Avila E, Kahmann A, Alho C, Dorn M. Hemogram data as a tool for decision-making in COVID-19 management: applications to resource scarcity scenarios. PeerJ. 2020;8:e9482. doi: 10.7717/peerj.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Randhawa GS, Soltysiak MP, El Roz H, de Souza CP, Hill KA, Kari L. Machine learning using intrinsic genomic signatures for rapid classification of novel pathogens: COVID-19 case study. PLoS One. 2020;15(4):e0232391. doi: 10.1371/journal.pone.0232391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khanday AMUD, Rabani ST, Khan QR, Rouf N, Din MMU. Machine learning based approaches for detecting COVID-19 using clinical text data. Int J Inf Technol. 2020;12(3):731–739. doi: 10.1007/s41870-020-00495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kotwal A, Yadav AK, Yadav J, Kotwal J, Khune S. Predictive models of COVID-19 in India: a rapid review. Med J Armed Forces India. 2020;76(4):377–386. doi: 10.1016/j.mjafi.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Postnikov EB. Estimation of COVID-19 dynamics “on a back-of-envelope”: does the simplest sir model provide quantitative parameters and predictions? Chaos Solitons Fractals. 2020;135:109841. doi: 10.1016/j.chaos.2020.109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shao P, Shan Y (2020) Beware of asymptomatic transmission: study on 2019-ncov prevention and control measures based on extended Seir model, preprint on webpage at 10.1101/2020.01.28.923169v1
- 104.Vaid S, McAdie A, Kremer R, Khanduja V, Bhandari M. Risk of a second wave of COVID-19 infections: using artificial intelligence to investigate stringency of physical distancing policies in North America. Int Orthop. 2020;44(8):1581–1589. doi: 10.1007/s00264-020-04653-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. Med Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vaishya R, Javaid M, Khan IH, Haleem A. Artificial intelligence (AI) applications for COVID-19 pandemic. Diabetes Metab Syndr Clin Res Rev. 2020;14:337–339. doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu J, Zhang P, Zhang L, Meng W, Li J, Tong C, Li Y, Cai J, Yang Z, Zhu J et al (2020) Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results, preprint on webpage at 10.1101/2020.04.02.20051136v1
- 108.Yadav M, Perumal M, Srinivas M. Analysis on novel coronavirus (COVID-19) using machine learning methods. Chaos Solitons Fractals. 2020;139:110050. doi: 10.1016/j.chaos.2020.110050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yesilkanat CM. Spatio-temporal estimation of the daily cases of COVID-19 in worldwide using random forest machine learning algorithm. Chaos Solitons Fractals. 2020;140:110210. doi: 10.1016/j.chaos.2020.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perumal V, Narayanan V, Rajasekar SJS. Detection of COVID-19 using CXR and CT images using transfer learning and haralick features. Appl Intell. 2020;51:341–358. doi: 10.1007/s10489-020-01831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Subudhi S, Verma A, Patel AB. Prognostic machine learning models for COVID-19 to facilitate decision making. Int J Clin Pract. 2020;4(12):e13685. doi: 10.1111/ijcp.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chowdhury R, Heng K, Shawon MSR, Goh G, Okonofua D, Ochoa-Rosales C, Gonzalez-Jaramillo V, Bhuiya A, Reidpath D, Prathapan S, et al. Dynamic interventions to control COVID-19 pandemic: a multivariate prediction modelling study comparing 16 worldwide countries. Eur J Epidemiol. 2020;35(5):389–399. doi: 10.1007/s10654-020-00649-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Misra S, Jeon S, Lee S, Managuli R, Jang I-S, Kim C. Multi-channel transfer learning of chest X-ray images for screening of COVID-19. Electronics. 2020;9(9):1388. doi: 10.3390/electronics9091388. [DOI] [Google Scholar]
- 115.Vinod DN, Prabaharan S. Data science and the role of artificial intelligence in achieving the fast diagnosis of COVID-19. Chaos Solitons Fractals. 2020;140:110182. doi: 10.1016/j.chaos.2020.110182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wynants L, Van Calster B, Bonten MM, Collins GS, Debray TP, De Vos M, Haller MC, Heinze G, Moons KG, Riley RD et al (2020) Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ 369 (8242):m1328 [DOI] [PMC free article] [PubMed]
- 117.Yang S, Jiang L, Cao Z, Wang L, Cao J, Feng R, Zhang Z, Xue X, Shi Y, Shan F. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann Transl Med. 2020;8(7):450. doi: 10.21037/atm.2020.03.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qi X, Jiang Z, Yu Q, Shao C, Zhang H, Yue H, Ma B, Wang Y, Liu C, Meng X et al (2020) Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-COV-2 infection: a multicenter study, preprint on webpage at 10.1101/2020.02.29.20029603v1 [DOI] [PMC free article] [PubMed]
- 119.Yue H, Yu Q, Liu C, Huang Y, Jiang Z, Shao C, Zhang H, Ma B, Wang Y, Xie G et al (2020) Machine learning-based CT radiomics method for predicting hospital stay in patients with pneumonia associated with SARS-COV-2 infection: a multicenter study. Ann Transl Med 8(14):859 [DOI] [PMC free article] [PubMed]
- 120.Shi W, Peng X, Liu T, Cheng Z, Lu H, Yang S, Zhang J, Li F, Wang M, Zhang X et al (2020) Deep learning-based quantitative computed tomography model in predicting the severity of COVID-19: a retrospective study in 196 patients. Lancet 9(3):216 [DOI] [PMC free article] [PubMed]
- 121.Zivkovic M, Bacanin N, Venkatachalam K, Nayyar A, Djordjevic A, Strumberger I, Al-Turjman F. COVID-19 cases prediction by using hybrid machine learning and beetle antennae search approach. Sustain Cities Soc. 2021;66:102669. doi: 10.1016/j.scs.2020.102669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feng C, Huang Z, Wang L, Chen X, Zhai Y, Zhu F, Chen H, Wang Y, Su X, Huang S et al (2020) A novel triage tool of artificial intelligence assisted diagnosis aid system for suspected COVID-19 pneumonia in fever clinics, preprint on webpage at 10.1101/2020.03.19.20039099v1
- 123.Yan L, Zhang H-T, Xiao Y, Wang M, Guo Y, Sun C, Tang X, Jing L, Li S, Zhang M et al (2020) Prediction of criticality in patients with severe COVID-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in Wuhan, preprint on webpage at 10.1101/2020.02.27.20028027v2
- 124.Linda W. A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. J Netw Comput Appl. 2020;20:1–12. [Google Scholar]
- 125.Bandyopadhyay SK, Dutta S (2020) Machine learning approach for confirmation of COVID-19 cases: positive, negative, death and release, preprint on webpage at 10.1101/2020.03.25.20043505v1
- 126.Hollister M. AI can help with the COVID-19 crisis-but the right human input is key. World Econ Forum. 2020;30:1–4. [Google Scholar]
- 127.Muurlink OT, Stephenson P, Islam MZ, Taylor-Robinson AW. Long-term predictors of dengue outbreaks in Bangladesh: a data mining approach. Infect Dis Model. 2018;3:322–330. doi: 10.1016/j.idm.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chenar SS, Deng Z. Development of genetic programming-based model for predicting oyster norovirus outbreak risks. Water Res. 2018;128:20–37. doi: 10.1016/j.watres.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 129.Agarwal N, Koti SR, Saran S, Kumar AS. Data mining techniques for predicting dengue outbreak in geospatial domain using weather parameters for new Delhi, India. Curr Sci. 2018;114(11):2281–2291. doi: 10.18520/cs/v114/i11/2281-2291. [DOI] [Google Scholar]
- 130.Koike F, Morimoto N. Supervised forecasting of the range expansion of novel non-indigenous organisms: alien pest organisms and the 2009 h1n1 flu pandemic. Glob Ecol Biogeogr. 2018;27(8):991–1000. doi: 10.1111/geb.12754. [DOI] [Google Scholar]
- 131.Tapak L, Hamidi O, Fathian M, Karami M. Comparative evaluation of time series models for predicting influenza outbreaks: application of influenza-like illness data from sentinel sites of healthcare centers in Iran. BMC Res Notes. 2019;12(1):353. doi: 10.1186/s13104-019-4393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anno S, Hara T, Kai H, Lee M-A, Chang Y, Oyoshi K, Mizukami Y, Tadono T. Spatiotemporal dengue fever hotspots associated with climatic factors in Taiwan including outbreak predictions based on machine-learning. Geospat Health. 2019;14(2):183–194. doi: 10.4081/gh.2019.771. [DOI] [PubMed] [Google Scholar]
- 133.Liang R, Lu Y, Qu X, Su Q, Li C, Xia S, Liu Y, Zhang Q, Cao X, Chen Q, et al. Prediction for global African swine fever outbreaks based on a combination of random forest algorithms and meteorological data. Transbound Emerg Dis. 2020;67(2):935–946. doi: 10.1111/tbed.13424. [DOI] [PubMed] [Google Scholar]
- 134.Finkelstein J, cheol Jeong I. Machine learning approaches to personalize early prediction of asthma exacerbations. Ann N Y Acad Sci. 2017;1387(1):153. doi: 10.1111/nyas.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Battineni G, Chintalapudi N, Amenta F. Machine learning in medicine: performance calculation of dementia prediction by support vector machines (SVM) Inform Med Unlocked. 2019;16:100200. doi: 10.1016/j.imu.2019.100200. [DOI] [Google Scholar]
- 136.Olivera AR, Roesler V, Iochpe C, Schmidt MI, Vigo Á, Barreto SM, Duncan BB. Comparison of machine-learning algorithms to build a predictive model for detecting undiagnosed diabetes-elsa-Brasil: accuracy study. Sao Paulo Med J. 2017;135(3):234–246. doi: 10.1590/1516-3180.2016.0309010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen Y, Luo Y, Huang W, Hu D, Zheng R-Q, Cong S-Z, Meng F-K, Yang H, Lin H-J, Sun Y, et al. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis b. Comput Biol Med. 2017;89:18–23. doi: 10.1016/j.compbiomed.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 138.Shousha HI, Awad AH, Omran DA, Elnegouly MM, Mabrouk M. Data mining machine learning algorithms using il28b genotype and biochemical markers best predicted advanced liver fibrosis in chronic HCV. Jpn J Infect Dis. 2017;71(1):51–57. doi: 10.7883/yoken.JJID.2017.089. [DOI] [PubMed] [Google Scholar]
- 139.Zhou L-Q, Wang J-Y, Yu S-Y, Wu G-G, Wei Q, Deng Y-B, Wu X-L, Cui X-W, Dietrich CF. Artificial intelligence in medical imaging of the liver. World J Gastroenterol. 2019;25(6):672. doi: 10.3748/wjg.v25.i6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dinga R, Marquand AF, Veltman DJ, Beekman AT, Schoevers RA, van Hemert AM, Penninx BW, Schmaal L. Predicting the naturalistic course of depression from a wide range of clinical, psychological, and biological data: a machine learning approach. Transl Psychiatry. 2018;8(1):1–11. doi: 10.1038/s41398-018-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pal R, Sekh AA, Kar S, Prasad DK (2020) Neural network based country wise risk prediction of COVID-19, preprint on webpage at arXiv:2004.00959
- 142.Punn NS, Sonbhadra SK, Agarwal S (2020) COVID-19 epidemic analysis using machine learning and deep learning algorithms, preprint on webpage at 10.1101/2020.04.08.20057679v2
- 143.Ye Y, Hou S, Fan Y, Qian Y, Zhang Y, Sun S, Peng Q, Laparo K (2020) -satellite: an AI-driven system and benchmark datasets for hierarchical community-level risk assessment to help combat COVID-19, preprint on webpage at arXiv:2003.12232 [DOI] [PMC free article] [PubMed]
- 144.Vasileios L, Majumder MS, Elad Y-T, Edelstein M, Moura S, Yohhei H, Rangaka MX, McKendry RA, Cox IJ. Tracking COVID-19 using online search. NPJ Digit Med. 2021;4(1):1–11. doi: 10.1038/s41746-021-00384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Heldt FS, Vizcaychipi MP, Peacock S, Cinelli M, McLachlan L, Andreotti F, Jovanović S, Dürichen R, Lipunova N, Fletcher RA, et al. Early risk assessment for COVID-19 patients from emergency department data using machine learning. Sci Rep. 2021;11(1):1–13. doi: 10.1038/s41598-021-83784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wong ZS, Zhou J, Zhang Q. Artificial intelligence for infectious disease big data analytics. Infect Dis Health. 2019;24(1):44–48. doi: 10.1016/j.idh.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 147.Gozes O, Frid-Adar M, Greenspan H, Browning PD, Zhang H, Ji W, Bernheim A, Siegel E (2020) Rapid AI development cycle for the coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis, preprint on webpage at arXiv:2003.05037
- 148.Pirouz B, ShaffieeHaghshenas S, ShaffieeHaghshenas S, Piro P. Investigating a serious challenge in the sustainable development process: analysis of confirmed cases of COVID-19 (newtype of coronavirus) through a binary classification using artificial intelligence and regression analysis. Sustainability. 2020;12(6):2427. doi: 10.3390/su12062427. [DOI] [Google Scholar]
- 149.Smeulders A, Van Ginneken A. An analysis of pathology knowledge and decision making for the development of artificial intelligence-based consulting systems. Anal Quant Cytol Histol. 1989;11(3):154–165. [PubMed] [Google Scholar]
- 150.McCall B. COVID-19 and artificial intelligence: protecting health-care workers and curbing the spread. Lancet Digit Health. 2020;2(4):e166–e167. doi: 10.1016/S2589-7500(20)30054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Whitelaw S, Mamas MA, Topol E, Van Spall HGC. Applications of digital technology in COVID-19 pandemic planning and response. Lancet Digit Health. 2020;2(8):e435–e440. doi: 10.1016/S2589-7500(20)30142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Booth AL, Abels E, McCaffrey P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod Pathol. 2021;34(3):522–531. doi: 10.1038/s41379-020-00700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chun A (2020) In a time of coronavirus, chinasinvestment in AI is payingoff in a bigway. https://www.scmp.com/comment/opinion/article/3075553/time-coronavirus-chinas-investment-ai-paying-big-way. Accessed 17 July 2021
- 154.Dickson B (2020) Why AI might be the most effective weapon we have to fight COVID-19. https://bit.ly/3qd0KDB. Accessed 17 July 2021
- 155.Rivas A (2020) Drones and artificial intelligence to enforce social isolation during COVID-19 outbreak. https://linkmn.gr/dOoW5O. Accessed 17 July 2021
- 156.(2020) How AI, big data and machine learning can be used against the corona virus. https://ars.electronica.art/aeblog/en/2020/03/19/ki-corona-part1/. Accessed 15 Jan 2021
- 157.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MU, Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J Travel Med. 2020;27(2):taaa008. doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.BlueDot: Outbreak risk software (2020) https://bluedot.global/. Accessed 6 June 2020
- 159.HealthMap (2020) http://www.diseasedaily.org/. Accessed 22 Aug 2020
- 160.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks, preprint on webpage at arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 161.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of xception and resnet50v2. Inform Med Unlocked. 2020;19:100360100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang P, Zheng X, Li J, Zhu B. Prediction of epidemic trends in COVID-19 with logistic model and machine learning technics. Chaos Solitons Fractals. 2020;139:110058. doi: 10.1016/j.chaos.2020.110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Panwar H, Gupta P, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using ncovnet. Chaos Solitons Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J, Cai M, Yang J, Li Y, Meng X et al (2020) A deep learning algorithm using CT images to screen for corona virus disease (COVID-19), preprint on webpage at 10.1101/2020.02.14.20023028v5 [DOI] [PMC free article] [PubMed]
- 166.Bai X, Fang C, Zhou Y, Bai S, Liu Z, Xia L, Chen Q, Xu Y, Xia T, Gong S et al (2020) Predicting COVID-19 malignant progression with AI techniques, preprint on webpage at 10.1101/2020.03.20.20037325v2
- 167.Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R et al (2020) Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images, preprint on webpage at 10.1101/2020.02.23.20026930v1 [DOI] [PMC free article] [PubMed]
- 168.Zheng C, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W, Wang X (2020) Deep learning-based detection for COVID-19 from chest CT using weak label, preprint on webpage at 10.1101/2020.03.12.20027185v2
- 169.Liu B, Liu P, Dai L, Yang Y, Xie P, Tan Y, Du J, Shan W, Zhao C, Zhong Q, et al. Assisting scalable diagnosis automatically via CT images in the combat against COVID-19. Sci Rep. 2021;11(1):1–8. doi: 10.1038/s41598-021-83424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, Chen Q, Huang S, Yang M, Yang X, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ucar F, Korkmaz D. Covidiagnosis-net: Deep bayes-squeezenet based diagnostic of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Programs Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rahaman MM, Li C, Yao Y, Kulwa F, Rahman MA, Wang Q, Qi S, Kong F, Zhu X, Zhao X. Identification of COVID-19 samples from chest X-ray images using deep learning: a comparison of transfer learning approaches. J X Ray Sci Technol. 2020;28(5):1–19. doi: 10.3233/XST-200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rajpal S, Kumar N, Rajpal A (2020) Cov-elm classifier: an extreme learning machine based identification of COVID-19 using chest-ray images, preprint on webpage at arXiv:2007.08637
- 175.Sarkar J, Chakrabarti P (2020) A machine learning model reveals older age and delayed hospitalization as predictors of mortality in patients with COVID-19, preprint on webpage at 10.1101/2020.03.25.20043331v1
- 176.Du S, Wang J, Zhang H, Cui W, Kang Z, Yang T, Lou B, Chi Y, Long H, Ma M, et al. Predicting COVID-19 using hybrid AI model. Lancet. 2020;50(7):2891–2904. doi: 10.1109/TCYB.2020.2990162. [DOI] [PubMed] [Google Scholar]
- 177.Assaf D, Gutman Y, Neuman Y, Segal G, Amit S, Gefen-Halevi S, Shilo N, Epstein A, Mor-Cohen R, Biber A, et al. Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern Emerg Med. 2020;15:1–9. doi: 10.1007/s11739-020-02475-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hofmarcher M, Mayr A, Rumetshofer E, Ruch P, Renz P, Schimunek J, Seidl P, Vall A, Widrich M, Hochreiter S et al (2020) Large-scale ligand-based virtual screening for SARS-COV-2 inhibitors using deep neural networks, preprint on webpage at arXiv:2004.00979
- 179.Beck BR, Shin B, Choi Y, Park S, Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-COV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shin B, Park S, Kang K, Ho JC (2019) Self-attention based molecule representation for predicting drug-target interaction. In: Machine learning for healthcare conference. PMLR, pp 230–248
- 181.Moskal M, Beker W, Roszak R, Gajewska EP, Wołos A, Molga K, Szymkuć S, Grzybowski BA (2020) Suggestions for second-pass anti-COVID-19 drugs based on the artificial intelligence measures of molecular similarity, shape and pharmacophore distribution, preprint on webpage at https://chemrxiv.org/ndownloader/files/22217781
- 182.Hu F, Jiang J, Yin P (2020) Prediction of potential commercially inhibitors against SARS-COV-2 by multi-task deep model, preprint on webpage at arXiv:2003.00728 [DOI] [PMC free article] [PubMed]
- 183.Kadioglu O, Saeed M, Johannes Greten H, Efferth T. Identification of novel compounds against three targets of SARS COV-2 coronavirus by combined virtual screening and supervised machine learning. Bull World Health Organ. 2021;133:104359. doi: 10.1016/j.compbiomed.2021.104359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhavoronkov A, Aladinskiy V, Zhebrak A, Zagribelnyy B, Terentiev V, Bezrukov D, Polykovskiy D, Shayakhmetov R, Filimonov A, Orekhov P et al (2020) Potential covid-2019 3c-like protease inhibitors designed using generative deep learning approaches. 2020. chemrxiv, preprint on webpage at 10.26434/chemrxiv.12301457.v1
- 185.McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-COV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pan X, Dong L, Yang N, Chen D, Peng C. Potential drugs for the treatment of the novel coronavirus pneumonia (COVID-19) in China. Virus Res. 2020;286:198057. doi: 10.1016/j.virusres.2020.198057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jin Z, Liu J-Y, Feng R, Ji L, Jin Z-L, Li H-B. Drug treatment of coronavirus disease 2019 (COVID-19) in China. Eur J Pharmacol. 2020;883:173326. doi: 10.1016/j.ejphar.2020.173326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lu H. Drug treatment options for the 2019-new coronavirus (2019-ncov) Biosci Trends. 2020;14(1):69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 189.Yang X, Liu Y, Liu Y, Yang Q, Wu X, Huang X, Liu H, Cai W, Ma G. Medication therapy strategies for the coronavirus disease 2019 (COVID-19): recent progress and challenges. Expert Rev Clin Pharmacol. 2020;13(9):957–975. doi: 10.1080/17512433.2020.1805315. [DOI] [PubMed] [Google Scholar]
- 190.Grippo A, Assenza G, Scarpino M, Broglia L, Cilea R, Galimberti CA, Lanzo G, Michelucci R, Tassi L, Vergari M, et al. Electroencephalography during SARS-COV-2 outbreak: practical recommendations from the task force of the Italian society of neurophysiology (sinc), the Italian league against epilepsy (lice), and the Italian association of neurophysiology technologists (aitn) Neurol Sci. 2020;41(9):2345–2351. doi: 10.1007/s10072-020-04585-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hazafa A, Ur-Rahman K, Haq I-U, Jahan N, Mumtaz M, Farman M, Naeem H, Abbas F, Naeem M, Sadiqa S, et al. The broad-spectrum antiviral recommendations for drug discovery against COVID-19. Drug Metab Rev. 2020;52(3):408–424. doi: 10.1080/03602532.2020.1770782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Siddiqui AJ, Jahan S, Ashraf SA, Alreshidi M, Ashraf M, Patel M, Snoussi M, Singh R, Adnan M. Current status and strategic possibilities on potential use of combinational drug therapy against COVID-19 caused by SARS-COV-2. J Biomol Struct Dyn. 2020;40:1–14. doi: 10.1080/07391102.2020.1802345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Khuroo MS, Khuroo M, Khuroo MS, Sofi AA, Khuroo NS. COVID-19 vaccines: a race against time in the middle of death and devastation! J Clin Exp Hepatol. 2020;10:610–621. doi: 10.1016/j.jceh.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen J, Li K, Zhang Z, Li K, Yu PS (2020) A survey on applications of artificial intelligence in fighting against COVID-19, preprint on webpage at arXiv:2007.02202
- 195.Qiao R, Tran NH, Shan B, Ghodsi A, Li M (2020) Personalized workflow to identify optimal t-cell epitopes for peptide-based vaccines against COVID-19, preprint on webpage at arXiv:2003.10650
- 196.Herst CV, Burkholz S, Sidney J, Sette A, Harris PE, Massey S, Brasel T, Cunha-Neto E, Rosa DS, Chao WCH, et al. An effective ctl peptide vaccine for ebola zaire based on survivors’ cd8+ targeting of a particular nucleocapsid protein epitope with potential implications for COVID-19 vaccine design. Vaccine. 2020;38:4464–4475. doi: 10.1016/j.vaccine.2020.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ward D, Higgins M, Phelan J, Hibberd ML, Campino S, Clark TG (2021) An integrated in silico immuno-genetic analytical platform provides insights into COVID-19 serological and vaccine targets. bioRxiv 13(1):4 [DOI] [PMC free article] [PubMed]
- 198.Sarkar B, Ullah MA, Johora FT, Taniya MA, Araf Y (2020) The essential facts of Wuhan novel corona virus outbreak in China and epitope-based vaccine designing against 2019-ncov, preprint on webpage at 10.1101/2020.02.05.935072v2
- 199.Rahman MS, Hoque MN, Islam MR, Akter S, Rubayet-Ul-Alam A, Siddique MA, Saha O, Rahaman MM, Sultana M, Crandall KA, et al. Epitope-based chimeric peptide vaccine design against s, m and e proteins of SARS-COV-2 etiologic agent of global pandemic COVID-19: an in silico approach. PeerJ. 2020;8:e9572. doi: 10.7717/peerj.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Prachar M, Justesen S, Steen-Jensen DB, Thorgrimsen SP, Jurgons E, Winther O, Bagger F (2020) COVID-19 vaccine candidates: prediction and validation of 174 SARS-COV-2 epitopes, preprint on webpage at 10.1101/2020.03.20.000794v4 [DOI] [PMC free article] [PubMed]
- 201.Nguyen DD, Gao K, Wang R, Wei G (2020) Machine intelligence design of 2019-ncov drugs, preprint on webpage at 10.1101/2020.01.30.927889v1
- 202.Singh R, Singh R, Bhatia A. Sentiment analysis using machine learning technique to predict outbreaks and epidemics. Int J Adv Sci Res. 2018;3(2):19–24. [Google Scholar]
- 203.Rekha Hanumanthu S (2020) Role of intelligent computing in COVID-19 prognosis: a state-of-the-art review. Chaos Solitons Fractals 138:109947 [DOI] [PMC free article] [PubMed]
- 204.Patel BN, Rosenberg L, Willcox G, Baltaxe D, Lyons M, Irvin J, Rajpurkar P, Amrhein T, Gupta R, Halabi S, et al. Human–machine partnership with artificial intelligence for chest radiograph diagnosis. NPJ Digit Med. 2019;2(1):1–10. doi: 10.1038/s41746-019-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rajendrakumar AL, Nair ATN, Nangia C, Chourasia PK, Chourasia MK, Syed MG, Nair AS, Nair AB, Koya MSF. Epidemic landscape and forecasting of SARS-COV-2 in India. J Epidemiol Glob Health. 2020;11(1):55–59. doi: 10.2991/jegh.k.200823.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mondal S, Ghosh S (2020) Fear of exponential growth in covid19 data of India and future sketching, preprint on webpage at 10.1101/2020.04.09.20058933v1
- 207.Chatterjee K, Chatterjee K, Kumar A, Shankar S. Healthcare impact of COVID-19 epidemic in India: a stochastic mathematical model. Med J Armed Forces India. 2020;76(2):147–155. doi: 10.1016/j.mjafi.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ghosal S, Sengupta S, Majumder M, Sinha B. Linear regression analysis to predict the number of deaths in India due to SARS-COV-2 at 6 weeks from day 0 (100 cases-march 14th 2020) Diabetes Metab Syndr Clin Res Rev. 2020;14(4):311–315. doi: 10.1016/j.dsx.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sujatha R, Chatterjee J et al (2020) A machine learning methodology for forecasting of the COVID-19 cases in India, preprint on webpage at 10.36227/techrxiv.12143685.v1
- 210.Singh R, Adhikari R (2020) Age-structured impact of social distancing on the COVID-19 epidemic in India, preprint on webpage at arXiv:2003.12055
- 211.Virk JS, Ali SA, Kaur G (2020) Recent update on COVID-19 in India: is locking down the country enough? Preprint on webpage at 10.1101/2020.04.06.20053124v2
- 212.Ranjan R (2020) Predictions for COVID-19 outbreak in India using epidemiological models, preprint on webpage at 10.1101/2020.04.02.20051466v1
- 213.Biswas S, Mukherjee M (2020) Risk assessment of ncovid-19 pandemic in India: a mathematical model and simulation, preprint on webpage at 10.1101/2020.04.10.20060830v1
- 214.DAS A, Mishra S, Gopalan SS (2020) Predicting community mortality risk due to COVID-19 using machine learning and development of a prediction tool, preprint on webpage at 10.1101/2020.04.27.20081794v2
- 215.Chakraborty T, Ghosh I. Real-time forecasts and risk assessment of novel coronavirus (COVID-19) cases: a data-driven analysis. Chaos Solitons Fractals. 2020;135:109850. doi: 10.1016/j.chaos.2020.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gupta R, Pal SK, Pandey G (2020) A comprehensive analysis of COVID-19 outbreak situation in India, preprint on webpage at 10.1101/2020.04.08.20058347v2
- 217.Ray D, Salvatore M, Bhattacharyya R, Wang L, Du J, Mohammed S, Purkayastha S, Halder A, Rix A, Barker D, et al. Predictions, role of interventions and effects of a historic national lockdown in India’s response to the COVID-19 pandemic: data science call to arms. Harv Data Sci Rev. 2020;2020(Suppl 1):1–45. doi: 10.1162/99608f92.60e08ed5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bhardwaj R. A predictive model for the evolution of COVID-19. Trans Indian Natl Acad Eng. 2020;5:133–140. doi: 10.1007/s41403-020-00130-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Singh S, Parmar KS, Makkhan SJS, Kaur J, Peshoria S, Kumar J. Study of arima and least square support vector machine (LS-SVM) models for the prediction of SARS-COV-2 confirmed cases in the most affected countries. Chaos Solitons Fractals. 2020;139:110086. doi: 10.1016/j.chaos.2020.110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tuan NH, Mohammadi H, Rezapour S. A mathematical model for COVID-19 transmission by using the caputo fractional derivative. Chaos Solitons Fractals. 2020;140:110107. doi: 10.1016/j.chaos.2020.110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Arti M, Bhatnagar K (2020) Modeling and predictions for COVID 19 spread in India, preprint on webpage at 10.13140/RG.2.2.11427.81444
- 222.Rai B, Shukla A, Dwivedi LK (2020) COVID-19 in India: predictions, reproduction number and public health preparedness, preprint on webpage at 10.1101/2020.04.09.20059261v1
- 223.Croccolo F, Roman HE. Spreading of infections on random graphs: a percolation-type model for COVID-19. Chaos Solitons Fractals. 2020;139:110077. doi: 10.1016/j.chaos.2020.110077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Shuja J, Alanazi E, Alasmary W, Alashaikh A. COVID-19 open source data sets: a comprehensive survey. Appl Intell. 2020;51:1296–1325. doi: 10.1007/s10489-020-01862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Xu B, Kraemer MU, Gutierrez B, Mekaru S, Sewalk K, Loskill A, Wang L, Cohn E, Hill S, Zarebski A, et al. Open access epidemiological data from the COVID-19 outbreak. Lancet Infect Dis. 2020;20(5):534. doi: 10.1016/S1473-3099(20)30119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Frazer JS, Shard A, Herdman J. Involvement of the open-source community in combating the worldwide COVID-19 pandemic: a review. J Med Eng Technol. 2020;44(4):169–176. doi: 10.1080/03091902.2020.1757772. [DOI] [PubMed] [Google Scholar]
- 227.Alimadadi A, Aryal S, Manandhar I, Munroe PB, Joe B, Cheng X. Artificial intelligence and machine learning to fight COVID-19. Physiol Genom. 2020;54(4):200–202. doi: 10.1152/physiolgenomics.00029.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pham Q-V, Nguyen DC, Hwang W-J, Pathirana PN et al (2020) Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: a survey on the state-of-the-arts, preprint on webpage at https://www.preprints.org/manuscript/202004.0383/v1 [DOI] [PMC free article] [PubMed]
- 229.Textual Data Set T1 (2020) https://datahub.io/core/covid-19. Accessed 15 Aug 2020
- 230.Medical Data Set M1 (2020) https://ai.nscc-tj.cn/thai/deploy/public/pneumonia_ct. Accessed 15 Aug 2020
- 231.Textual Data Set T2 (2020) https://github.com/CSSEGISandData/COVID-19. Accessed 15 Aug 2020
- 232.Medical Data Set M2 (2020) https://zenodo.org/record/3757476. Accessed 15 Aug 2020
- 233.Textual Data Set T3 (2020) https://ncov.dxy.cn/ncovh5/view/pneumonia. Accessed 15 Aug 2020
- 234.Medical Data Set M3 (2020) https://gitee.com/junma11/COVID-19-CT-Seg-Benchmark. Accessed 15 Aug 2020
- 235.Textual Data Set T4 (2020) https://www.arcgis.com/apps/opsdashboard/index.html. Accessed 15 Aug 2020
- 236.Medical Data Set M4 (2020) http://medicalsegmentation.com/covid19/. Accessed 15 Aug 2020
- 237.Textual Data Set T5 (2020) https://www.kaggle.com/covid-19-contributions. Accessed 15 Aug 2020
- 238.Medical Data Set M5 (2020) https://www.sirm.org/en/category/articles/covid-19-database/. Accessed 15 Aug 2020
- 239.Textual Data Set T6 (2020) https://github.com/WeileiZeng/Open-Source-COVID-19. Accessed 15 Aug 2020
- 240.Medical Data Set M6 (2020) https://coronacases.org/. Accessed 15 Aug 2020
- 241.Textual Data Set T7 (2020) https://dataverse.harvard.edu/dataverse/2019ncov. Accessed 15 Aug 2020
- 242.Medical Data Set M7 (2020) https://www.bsti.org.uk/training-and-education/covid-19-bsti-imaging-database/. Accessed 15 Aug 2020
- 243.Textual Data Set T8 (2020) https://www.kaggle.com/lachmann12/world-population-demographics-by-age-2019. Accessed 15 Aug 2020
- 244.Medical Data Set M8 (2020) https://www.sirm.org/en/category/articles/covid-19-database/. Accessed 15 Aug 2020
- 245.Textual Data Set T9 (2020) https://github.com/Emergent-Epidemics/covid19_npi_china. Accessed 15 Aug 2020
- 246.Medical Data Set M9 (2020) https://radiopaedia.org/articles/covid-19-3. Accessed 15 Aug 2020
- 247.Textual Data Set T10 (2020) https://www.ecdc.europa.eu/en/covid-19-pandemic. Accessed 15 Aug 2020
- 248.Textual Data Set T11 (2020) https://github.com/BayesForDays/coronada. Accessed 15 Aug 2020
- 249.Textual Data Set T12 (2020) https://www.kaggle.com/smid80/coronavirus-covid19-tweets. Accessed 15 Aug 2020
- 250.Speech Data Set S1 (2020) https://coswara.iisc.ac.in/. Accessed 15 Aug 2020
- 251.Textual Data Set T13 (2020) https://covidscholar.org. Accessed 15 Aug 2020
- 252.Speech Data Set S2 (2020) https://github.com/iiscleap/Coswara-Data. Accessed 15 Aug 2020
- 253.Textual Data Set T14 (2020) https://www.kaggle.com/chaibapat/google-mobility. Accessed 15 Aug 2020
- 254.Speech Data Set S3 (2020) https://www.covid-19-sounds.org/en/. Accessed 15 Aug 2020
- 255.Textual Data Set T15 (2020) https://www.apple.com/covid19/mobility. Accessed 15 Aug 2020
- 256.Speech Data Set S4 (2020) https://cvd.lti.cmu.edu/. Accessed 15 Aug 2020
- 257.Textual Data Set T16 (2020) https://geods.geography.wisc.edu/covid19/physical-distancing/. Accessed 15 Aug 2020
- 258.Speech Data Set S5 (2020) https://coughvid.epfl.ch/. Accessed 15 Aug 2020
- 259.Textual Data Set T17 (2020) http://qianxi.baidu.com/. Accessed 15 Aug 2020
- 260.Speech Data Set S6 (2020) http://virufy.org/. Accessed 15 Aug 2020
- 261.Textual Data Set T18 (2020) https://www.google.com/covid19/mobility/. Accessed 15 Aug 2020
- 262.Speech Data Set S7 (2020) https://github.com/virufy/covid. Accessed 15 Aug 2020
- 263.Xu B, Gutierrez B, Mekaru S, Sewalk K, Goodwin L, Loskill A, Cohn EL, Hswen Y, Hill SC, Cobo MM, et al. Epidemiological data from the COVID-19 outbreak, real-time case information. Sci Data. 2020;7(1):1–6. doi: 10.1038/s41597-020-0448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kucharski AJ, Russell TW, Diamond C, Liu Y, Edmunds J, Funk S, Eggo RM, Sun F, Jit M, Munday JD, et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis. 2020;20(5):553–558. doi: 10.1016/S1473-3099(20)30144-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Benvenuto D, Giovanetti M, Vassallo L, Angeletti S, Ciccozzi M. Application of the arima model on the COVID-2019 epidemic dataset. Data Brief. 2020;29:105340. doi: 10.1016/j.dib.2020.105340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lachmann A (2020) Correcting under-reported COVID-19 case numbers: estimating the true scale of the pandemic, preprint on webpage at 10.1101/2020.03.14.20036178v2
- 267.Obeid JS, Davis M, Turner M, Meystre SM, Heider PM, O’Bryan EC, Lenert LA. An artificial intelligence approach to COVID-19 infection risk assessment in virtual visits: a case report. J Am Med Inform Assoc. 2020;27(8):1321–1325. doi: 10.1093/jamia/ocaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Kasilingam D, Prabhakaran SS, Dinesh Kumar R, Rajagopal V, Santhosh Kumar T, Soundararaj A. Exploring the growth of COVID-19 cases using exponential modelling across 42 countries and predicting signs of early containment using machine learning. Transbound Emerg Dis. 2020;68(3):1001–1018. doi: 10.1111/tbed.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zheng N, Du S, Wang J, Zhang H, Cui W, Kang Z, Yang T, Lou B, Chi Y, Long H, et al. Predicting COVID-19 in china using hybrid AI model. IEEE Trans Cybern. 2020;50(7):2891–2894. doi: 10.1109/TCYB.2020.2990162. [DOI] [PubMed] [Google Scholar]
- 270.Kraemer MU, Yang C-H, Gutierrez B, Wu C-H, Klein B, Pigott DM, Du Plessis L, Faria NR, Li R, Hanage WP, et al. The effect of human mobility and control measures on the COVID-19 epidemic in China. Science. 2020;368(6490):493–497. doi: 10.1126/science.abb4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Anzai A, Kobayashi T, Linton NM, Kinoshita R, Hayashi K, Suzuki A, Yang Y, Jung S-M, Miyama T, Akhmetzhanov AR, et al. Assessing the impact of reduced travel on export at ion dynamics of novel coronavirus infection (COVID-19) J Clin Med. 2020;9(2):601. doi: 10.3390/jcm9020601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lai S, Ruktanonchai NW, Zhou L, Prosper O, Luo W, Floyd JR, Wesolowski A, Santillana M, Zhang C, Du X, et al. Effect of non-pharmaceutical interventions for containing the COVID-19 outbreak in China. medRxiv. 2020;585(7825):410–413. doi: 10.1038/s41586-020-2293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wells CR, Sah P, Moghadas SM, Pandey A, Shoukat A, Wang Y, Wang Z, Meyers LA, Singer BH, Galvani AP. Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. Proc Natl Acad Sci. 2020;117(13):7504–7509. doi: 10.1073/pnas.2002616117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Tian H, Liu Y, Li Y, Wu C-H, Chen B, Kraemer MU, Li B, Cai J, Xu B, Yang Q, et al. An investigation of transmission control measures during the first 50 days of the COVID-19 epidemic in china. Science. 2020;368(6491):638–642. doi: 10.1126/science.abb6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kleinberg B, van der Vegt I, Mozes M (2020) Measuring emotions in the COVID-19 real world worry dataset, preprint on webpage at arXiv:2004.04225
- 276.Banda JM, Tekumalla R, Wang G, Yu J, Liu T, Ding Y, Artemova K, Tutubalina E, Chowell G (2020) A large-scale COVID-19 twitter chatter dataset for open scientific research—an international collaboration, preprint on webpage at arXiv:2004.03688 [DOI] [PMC free article] [PubMed]
- 277.Covid-19: The first public coronavirus twitter dataset (2020) https://github.com/echen102/COVID-19-TweetIDs. Accessed 08 Jan 2021
- 278.Alqurashi S, Alhindi A, Alanazi E (2020) Large arabic twitter dataset on COVID-19, preprint on webpage at arXiv:2004.04315
- 279.Yu J (2020) Open access institutional and news media tweet dataset for COVID-19 social science research, preprint on webpage at arXiv:2004.01791
- 280.Zarei K, Farahbakhsh R, Crespi N, Tyson G (2020) A first instagram dataset on COVID-19, preprint on webpage at arXiv:2004.12226
- 281.Sarker A, Lakamana S, Hogg-Bremer W, Xie A, Al-Garadi MA, Yang Y-C. Self-reported COVID-19 symptoms on twitter: an analysis and a research resource. J Am Med Inform Assoc. 2020;27(8):1310–1315. doi: 10.1093/jamia/ocaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ahamed S, Samad M (2020) Information mining for COVID-19 research from a large volume of scientific literature, preprint on webpage at arXiv:2004.02085
- 283.Fister I Jr, Fister K, Fister I (2020) Discovering associations in COVID-19 related research papers, preprint on webpage at arXiv:2004.03397
- 284.Adhikari SP, Meng S, Wu Y-J, Mao Y-P, Ye R-X, Wang Q-Z, Sun C, Sylvia S, Rozelle S, Raat H, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Arksey H, O’Malley L. Scoping studies: towards a methodological framework. Int J Soc Res Methodol. 2005;8(1):19–32. doi: 10.1080/1364557032000119616. [DOI] [Google Scholar]
- 286.Moons KG, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, Reitsma JB, Kleijnen J, Mallett S. Probast: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1–W33. doi: 10.7326/M18-1377. [DOI] [PubMed] [Google Scholar]
- 287.Chen E, Lerman K, Ferrara E. Tracking social media discourse about the COVID-19 pandemic: development of a public coronavirus twitter data set. JMIR Public Health Surveill. 2020;6(2):e19273. doi: 10.2196/19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Alamo T, Reina DG, Mammarella M, Abella A (2020) Open data resources for fighting COVID-19, preprint on webpage at arXiv:2004.06111
- 289.Cohen JP, Bertin P, Frappier V (2019) Chester: a web delivered locally computed chest X-ray disease prediction system, preprint on webpage at arXiv:1901.11210
- 290.Zhao J, Zhang Y, He X, Xie P (2020) COVID-CT-dataset: a CT scan dataset about COVID-19, preprint on webpage at https://covid-19.conacyt.mx/jspui/handle/1000/4157
- 291.Khan SH, Sohail A, Zafar M, Khan A (2020) Coronavirus disease analysis using chest X-ray images and a novel deep convolutional neural network, preprint on webpage at 10.13140/RG.2.2.35868.64646 [DOI] [PMC free article] [PubMed]
- 292.Savadjiev P, Chong J, Dohan A, Vakalopoulou M, Reinhold C, Paragios N, Gallix B. Demystification of AI-driven medical image interpretation: past, present and future. Eur Radiol. 2019;29(3):1616–1624. doi: 10.1007/s00330-018-5674-x. [DOI] [PubMed] [Google Scholar]
- 293.Shan F, Gao Y, Wang J, Shi W, Shi N, Han M, Xue Z, Shi Y (2020) Lung infection quantification of COVID-19 in CT images with deep learning, preprint on webpage at arXiv:2003.04655
- 294.Jun M, Cheng G, Yixin W, Xingle A, Jiantao G, Ziqi Y, Minqing Z, Xin L, Xueyuan D, Shucheng C, et al. (2020) COVID-19 CT lung and infection segmentation dataset. 10.5281/zenodo.3757476
- 295.Ma J, Wang Y, An X, Ge C, Yu Z, Chen J, Zhu Q, Dong G, He J, He Z, Cao T, Zhu Y, Nie Z, Yang X. Toward data-efficient learning: a benchmark for COVID-19 CT lung and infection segmentation. Med Phys. 2021;48(3):1197–1210. doi: 10.1002/mp.14676. [DOI] [PubMed] [Google Scholar]
- 296.Rajinikanth V, Dey N, Raj ANJ, Hassanien A, Santosh K, Raja N (2020) Harmony-search and otsu based system for coronavirus disease (COVID-19) detection using lung CT scan images, preprint on webpage at arXiv:2004.03431
- 297.Apostolopoulos ID, Aznaouridis SI, Tzani MA. Extracting possibly representative COVID-19 biomarkers from x-ray images with deep learning approach and image data related to pulmonary diseases. J Med Biol Eng. 2020;40:462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Lin ZQ, Shafiee M, Bochkarev S, Jules MS, Wang X, Wong A (2019) Explaining with impact: a machine-centric strategy to quantify the performance of explain ability algorithms, preprint on webpage at 10.1101/2020.05.10.20097063v1
- 299.Wang L, Lin ZQ, Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 301.Born J, Brändle G, Cossio M, Disdier M, Goulet J, Roulin J, Wiedemann N (2020) Pocovid-net: automatic detection of COVID-19 from a new lung ultrasound imaging dataset (pocus), preprint on webpage at arXiv:2004.12084
- 302.Sharma A, Rani S, Gupta D. Artificial intelligence-based classification of chest X-ray images into COVID-19 and other infectious diseases. Int J Biomed Imaging. 2020;2020:1–10. doi: 10.1155/2020/8889023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Imran A, Posokhova I, Qureshi HN, Masood U, Riaz S, Ali K, John CN, Hussain I, Nabeel M. Ai4covid-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform Med Unlocked. 2020;20:100378. doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Brown C, Chauhan J, Grammenos A, Han J, Hasthanasombat A, Spathis D, Xia T, Cicuta P, Mascolo C (2020) Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data, preprint on webpage at arXiv:2006.05919
- 305.Sharma N, Krishnan P, Kumar R, Ramoji S, Chetupalli SR, Ghosh PK, Ganapathy S et al (2020) Coswara—a database of breathing, cough, and voice sounds for COVID-19 diagnosis, preprint on webpage at arXiv:2005.10548
- 306.Greenhalgh T, Koh GCH, Car J. COVID-19: a remote assessment in primary care. BMJ. 2020;368:m1182. doi: 10.1136/bmj.m1182. [DOI] [PubMed] [Google Scholar]
- 307.Faezipour M, Abuzneid A. Smartphone-based self-testing of COVID-19 using breathing sounds. Telemed e-Health. 2020;26(10):1202–1205. doi: 10.1089/tmj.2020.0114. [DOI] [PubMed] [Google Scholar]
- 308.Trivedy S, Goyal M, Mohapatra PR, Mukherjee A. Design and development of smartphone-enabled spirometer with a disease classification system using convolutional neural network. IEEE Trans Instrum Meas. 2020;69(9):7125–7135. doi: 10.1109/TIM.2020.2977793. [DOI] [Google Scholar]
- 309.Han J, Qian K, Song M, Yang Z, Ren Z, Liu S, Liu J, Zheng H, Ji W, Koike T et al (2020) An early study on intelligent analysis of speech under COVID-19: Severity, sleep quality, fatigue, and anxiety, preprint on webpage at arXiv:2005.00096
- 310.Yao H, Zhang N, Zhang R, Duan M, Xie T, Pan J, Peng E, Huang J, Zhang Y, Xu X, et al. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. Front Cell Dev Biol. 2020;8:683. doi: 10.3389/fcell.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Kim AW, Adam EK, Bechayda SA, Kuzawa CW. Early life stress and HPA axis function independently predict adult depressive symptoms in metropolitan Cebu, Philippines. Am J Phys Anthropol. 2020;173(3):448–462. doi: 10.1002/ajpa.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Kim AW, Nyengerai T, Mendenhall E (2020) Evaluating the mental health impacts of the COVID-19 pandemic in urban South Africa: perceived risk of COVID-19 infection and childhood trauma predict adult depressive symptoms, preprint on webpage at 10.1101/2020.06.13.20130120v1 [DOI] [PMC free article] [PubMed]
- 313.Nour M, Cömert Z, Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl Soft Comput. 2020;97:106580. doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Heidari M, Mirniaharikandehei S, Khuzani AZ, Danala G, Qiu Y, Zheng B. Improving performance of CNN to predict likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int J Med Inform. 2020;144:104284. doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Farid AA, Selim GI, Awad H, Khater A. A novel approach of CT images feature analysis and prediction to screen for corona virus disease (COVID-19) Int J Sci Eng Res. 2020;11(3):1–9. [Google Scholar]
- 316.Mbuvha R, Marwala T. Bayesian inference of COVID-19 spreading rates in South Africa. medRxiv. 2020;15(8):e0237126. doi: 10.1371/journal.pone.0237126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Lai C-C, Hsu C-Y, Jen H-H, Yen M-F, Chan C-C, Chen H-H (2020) Bayesian approach for modelling the dynamic of COVID-19 outbreak on the diamond princess cruise ship, preprint on webpage at 10.1101/2020.06.21.20136465v1
- 318.Karmakar S, Das S (2020) Evaluating the impact of COVID-19 on cyberbullying through Bayesian trend analysis. In: Proceedings of the European interdisciplinary cybersecurity conference (EICC) co-located with European Cyber Week. pp 1–6
- 319.Campbell F, Cori A, Ferguson N, Jombart T. Bayesian inference of transmission chains using timing of symptoms, pathogen genomes and contact data. PLoS Comput Biol. 2019;15(3):e1006930. doi: 10.1371/journal.pcbi.1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Jewell CP, Kypraios T, Neal P, Roberts GO, et al. Bayesian analysis for emerging infectious diseases. Bayesian Anal. 2009;4(3):465–496. doi: 10.1214/09-BA417. [DOI] [Google Scholar]
- 321.Franco-Villoria M, Ventrucci M, Rue H, et al. A unified view on Bayesian varying coefficient models. Electron J Stat. 2019;13(2):5334–5359. doi: 10.1214/19-EJS1653. [DOI] [Google Scholar]
- 322.Albahri AS, Hamid RA et al (2020) Role of biological data mining and machine learning techniques in detecting and diagnosing the novel coronavirus (COVID-19): a systematic review. J Med Syst 44(7):122 [DOI] [PMC free article] [PubMed]
- 323.Medel-Ramírez C, Medel-Lopez H (2020) Data mining for the study of the epidemic (SARS-COV-2) COVID-19: algorithm for the identification of patients (SARS-COV-2) COVID 19 in Mexico. Available at SSRN 3619549Preprint on webpage at 10.2139/ssrn.3619549
- 324.Kumar S. Monitoring novel corona virus (COVID-19) infections in India by cluster analysis. Ann Data Sci. 2020;7(3):417–425. doi: 10.1007/s40745-020-00289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Ding Z, Qin Z, Qin Z. Frequent symptom sets identification from uncertain medical data in differentially private way. Sci Program. 2017;2017:1–10. [Google Scholar]
- 326.Gurwitz D. Repurposing current therapeutics for treating COVID-19: a vital role of prescription records data mining. Drug Dev Res. 2020;81:777–781. doi: 10.1002/ddr.21689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wahbeh A, Nasralah T, Al-Ramahi M, El-Gayar O. Mining physicians’ opinions on social media to obtain insights into COVID-19: mixed methods analysis. JMIR Public Health Surveill. 2020;6(2):e19276. doi: 10.2196/19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Liu J, Zhou J, Yao J, Zhang X, Li L, Xu X, He X, Wang B, Fu S, Niu T, et al. Impact of meteorological factors on the COVID-19 transmission: a multi-city study in China. Sci Total Environ. 2020;726:138513. doi: 10.1016/j.scitotenv.2020.138513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Wang J, Tang K, Feng K, Lv Wf (2020) Impact of temperature and relative humidity on the transmission of COVID-19: A modeling study in china and the united states, preprint on webpage at 10.2139/ssrn.3551767 [DOI] [PMC free article] [PubMed]
- 330.Fang Y, Nie Y, Penny M. Transmission dynamics of the COVID-19 outbreak and effectiveness of government interventions: a data-driven analysis. J Med Virol. 2020;92(6):645–659. doi: 10.1002/jmv.25750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Rodriguez-Diaz CE, Guilamo-Ramos V, Mena L, Hall E, Honermann B, Crowley JS, Baral S, Prado GJ, Marzan-Rodriguez M, Beyrer C, et al. Risk for COVID-19 infection and death among latinos in the united states: examining heterogeneity in transmission dynamics. Ann Epidemiol. 2020;52:46–53. doi: 10.1016/j.annepidem.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M. Classification of the COVID-19 infected patients using densenet201 based deep transfer learning. J Biomol Struct Dyn. 2020;40:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 333.Tindale L, Coombe M, Stockdale JE, Garlock E, Lau WYV, Saraswat M, Lee Y-HB, Zhang L, Chen D, Wallinga J et al (2020) Transmission interval estimates suggest pre-symptomatic spread of COVID-19, preprint on webpage at 10.1101/2020.03.03.20029983v1
- 334.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis. 2020;93:284–286. doi: 10.1016/j.ijid.2020.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Lopez CE, Vasu M, Gallemore C (2020) Understanding the perception of COVID-19 policies by mining a multilanguage twitter dataset, preprint on webpage at arXiv:2003.10359