Abstract
Objective
The NADPH oxidase Nox4 is an important source of H2O2. Nox4-derived H2O2 limits vascular inflammation and promotes smooth muscle differentiation. On this basis, the role of Nox4 for restenosis development was determined in the mouse carotid artery injury model.
Methods and results
Genetic deletion of Nox4 by a tamoxifen-activated Cre-Lox-system did not impact on neointima formation in the carotid artery wire injury model. To understand this unexpected finding, time-resolved single-cell RNA-sequencing (scRNAseq) from injured carotid arteries of control mice and massive-analysis-of-cDNA-ends (MACE)-RNAseq from the neointima harvested by laser capture microdissection of control and Nox4 knockout mice was performed. This revealed that resting smooth muscle cells (SMCs) and fibroblasts exhibit high Nox4 expression, but that the proliferating de-differentiated SMCs, which give rise to the neointima, have low Nox4 expression. In line with this, the first weeks after injury, gene expression was unchanged between the carotid artery neointimas of control and Nox4 knockout mice.
Conclusion
Upon vascular injury, Nox4 expression is transiently lost in the cells which comprise the neointima. NADPH oxidase 4 therefore does not interfere with restenosis development after wire-induced vascular injury.
Keywords: NADPH oxidase, Nox4, Restenosis, Reactive oxygen species, Carotid injury, Single-cell RNA sequencing, Inflammation
Abbreviations: CTL, Control; MACE, massive-analysis-of-cDNA-ends; ROS, Reactive oxygen species; scRNAseq, Single cell RNA sequencing; SMC, smooth muscle cells
Graphical abstract
1. Introduction
Reactive oxygen species (ROS) are important signaling molecules, with both protective and detrimental functions in the vascular system [[1], [2], [3]]. NADPH oxidases of the Nox family are important sources of ROS in the vascular system [4,5]. In vascular inflammatory diseases, like atherosclerosis and hypertension, ROS generated by the NADPH oxidase homologues Nox1, Nox2 and Nox5 have been suggested to promote disease [[5], [6], [7]]. For instance, deletion or inhibition of Nox1 reduces atherosclerosis in diabetic conditions and deletion of p47phox, a subunit of the Nox2 system, decreases atherosclerotic plaque formation [8].
The NADPH oxidase Nox4 differs from the above mentioned Nox homologues in two important aspects: 1) It is constitutively active; thus its ROS-production depends on the abundance of the enzyme [9]. 2) It directly produces H2O2 rather than O2− and cannot scavenge NO [10]. H2O2, different to O2−, activates protein kinase G signaling [11] and Nox4 has been suggested to in part act through this system [12,13].
Conflicting results have been published regarding the pathophysiological relevance of Nox4. We and others, using a knockout model, have suggested that Nox4 is protective [14,15] to limit angiotensin II-induced hypertension [16] and atherosclerosis development [1,17,18]. Others report with the aid of a dominant negative transgenic mouse rather a detrimental function of Nox4 [19,20]. In addition to its potential function in cardiovascular diseases, Nox4 has been linked to smooth muscle cell differentiation [21] through a p38 MAPK and SRF-dependent control of gene expression [22,23]. As cellular differentiation limits proliferation, it is interesting to speculate that Nox4 limits smooth muscle-dependent neointima formation and thus contributes to clinically relevant processes like restenosis. Interestingly, Nox4 was reported to be upregulated after vascular injury in mice [24] and Nox4 overexpression limited adventitial myofibroblast migration [25].
Based on these considerations, we hypothesized that endogenous Nox4 limits neointima development after vascular injury and studied this aspect in the carotid artery wire injury model of WT and Nox4 knockout mice.
2. Methods
2.1. Mouse studies
All animal experiments were performed in accordance with the National Institutes of Health Guidelines on the use of laboratory animals. The University Animal Care Committee and the Federal Authorities for Animal Research (Darmstadt, Germany) approved the study protocol (approval number: FU1185). Mice were housed in a specified pathogen-free facility with 12/12 h day and night cycle and free access to water and chow at all times.
Tamoxifen-inducible Nox4−/− mice were generated by crossing Nox4flox/flox with ERT2-Cre0/+ transgenic mice [16]. Activation of Cre-ERT2 was achieved by oral tamoxifen administration with the chow ad libitum for 10 days. In all experiments, only male animals were used. Cre-positive and Cre-negative littermates received tamoxifen to exclude direct effects of the anti-estrogen. According to the expression of Cre-ERT2, Nox4 carrying Nox4-flox-CMV-Cre-ERT20/0 mice are denoted as WT, whereas Nox4-deleted Nox4-flox-CMV-Cre-ERT2+/0 are denoted as Nox4−/− throughout the study.
2.2. Carotid injury model
Wire-induced injury surgery was applied to male mice at an age of 11 weeks, after Tamoxifen intake and washout. For wire-induced injury blood flow was stopped during the procedure by loops around the A. carotis interna (ICA), A. carotis communis (CCA) and A. carotis externa (ECA). An incision hole was placed using surgical scissors to introduce a 0.35 mm diameter flexible bended wire into the A. carotis interna. The bended wire was guided through the A. carotis interna into the A. carotis communis. After passing the vessel three times with rotation, the wire was removed and a suture around the A. carotis interna was placed to permanently stop blood flow in the A. carotis interna. The remaining loops were removed to resume blood flow via A. carotis communis and A. carotis externa (Supp. Fig. 1). Wire-injury was carried out only on the left carotid of the animal, the right carotid artery served as control [[26], [27], [28]]. The wire-induced injury model leads to endothelial disruption and damage of medial smooth muscle cells (SMCs) [27]. As typical for the model of severe deep injury, the variability between animals was fairly high, moreover, some vessels clotted post intervention. Vessels with signs of clots were removed from the analysis and the subsequent groups were designed to include at least 9 animals per group.
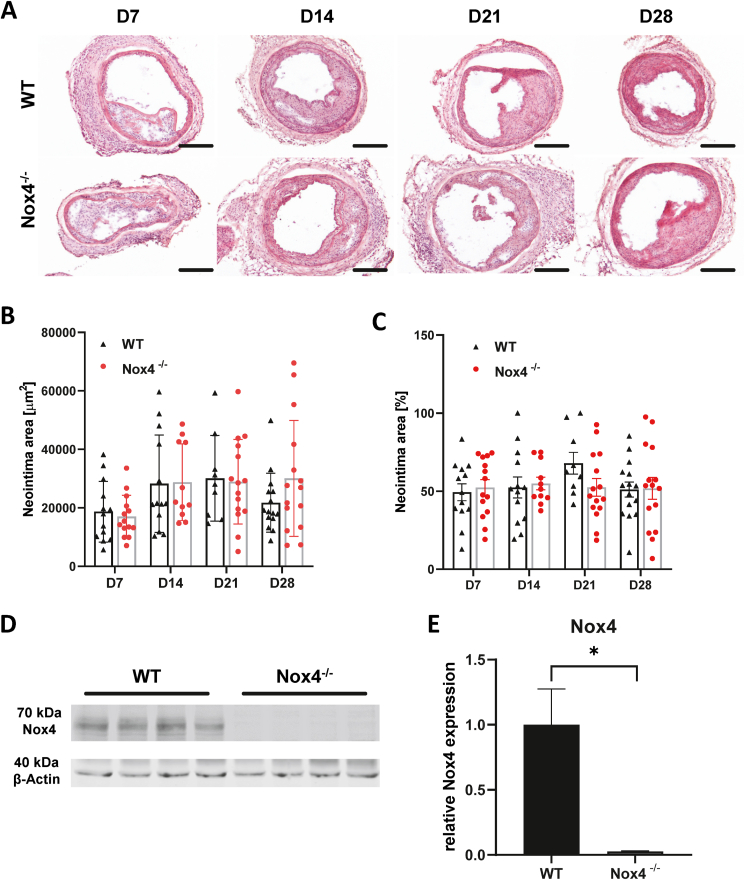
Fig. 1.
Nox4 does not limit neointima formation: A) Hematoxylin and eosin staining of injured carotids of WT and Nox4−/− (KO) mice D7, D14, D21 & D28 after injury; scale bar: 200 μm. B) Quantification of average neointima area [μm2] (C) and highest neointima thickening [%]; n = 9–15; GM 95% CI. D) and E) Western blot analysis showing the expression of Nox4 in aorta tissue samples obtained from WT and Nox4−/− animals after tamoxifen treatment; n = 4; t-test; *p < 0.05.
2.3. Immunoblotting
Pulverized aorta tissue samples were lysed using following lysis buffer (pH 7.4): Tris-HCl (50 mmol/L), NaCl (150 mmol/L), sodium pyrophosphate (10 mmol/L), sodium fluoride (20 mmol/L), nonidet P40 (1%), sodium desoxycholate (0.5%), proteinase inhibitor mix, phenylmethylsulfonyl fluoride (1 mmol/L), orthovanadate (2 mmol/L), okadaic acid (0.00001 mmol/L). For Nox4 detection, the disulfide bonds were cleaved with TCEP (Thermo Scientific) and samples were not boiled. Western blot analyses were performed with an infrared-based detection system (Odyssey, Licor). Nox4 primary antibody was provided by one of the co-authors (Ajay M. Shah) and anti-β-actin antibody was from Sigma (1:1000, #A1978, Sigma). Conjugated secondary antibodies were obtained from Licor (1:10,000, #926–68073/#926–32212).
2.4. Isolation of mRNA and qRT-PCR
Carotids were removed, snap frozen in liquid nitrogen and pulverized. Total mRNA was isolated from the upper half of left or right carotid arteries with the Bio&Sell RNA-Minikit (Bio&Sell) according to the manufacturer's instructions. cDNA synthesis was carried out using SuperScript III Reverse Transcriptase (Invitrogen) random hexamer primers with oligo (dT) primers, real-time PCR was performed with Eva Green Master Mix and ROX as reference dye (Thermo Scientific) in an Mx4000 cycler (Stratagene). Relative expressions of target genes were normalized to eukaryotic translation elongation factor 2 (EEF2), analyzed by delta-delta-CT method. The following primers were used: ACTA2-F 5′-ACAGAGGCACCACTGAACCCTAAG-3′; ACTA2-R 5′-ACAATCTCACGCTCGGCAGTAGTC-3′; CALD1-F 5′-AGAAGGAGTTTGATCCGACCA-3′; CALD1-R 5′-CATTTTCGGCGGAGTCATTTTG-3′; CNN1–F 5′-TCTGCACATTTTAACCGAGGTC-3′; CNN1-R 5′-GCCAGCTTGTTCTTTACTTCAGC-3′; EEF2-F 5′-GACATCACCAAGGGTGTGCAG-3′; EEF2-R 5′- GCGGTCAGCACACTGGCATA-3′; MYH11-F 5′-GTGTGGTGGTCAACCCCTAC-3′; MYH11-R 5′-GATGTGAGGCGGCATCTCAT-3′; NOX2-F 5′-GGGAACTGGGCTGTGAATGA-3′; NOX2-R 5′-CAGTGCTGACCCAAGGAGTT-3′; NOX4-F 5′-TGTTGGGCCTAGGATTGTGTT-3′; NOX4-R 5′-TGTTGGGCCTAGGATTGTGTT-3′; PECAM1-F 5′-GGACAGACCCTTCCACCAAG-3′; PECAM1-R 5′-CTGTTTGGCCTTGGCTTTCC-3′; SMTN-F 5′- GCCCTCAGATACCTTGGACTC-3′; SMTN-R 5′- GGCAGGATTTCGTTTCAGACG-3′.
2.5. Histology
Carotid tissue samples frozen in OCT compound were prepared as 10 μm serial sections. Sections were stained with Hematoxylin and Eosin staining solutions. The neointima area was analyzed by planimetry with ImageJ.
2.6. Single cell RNA sequencing
Carotids were minced in small pieces and digested for 1 h at 37 °C with an enzyme mix containing collagenase typ XI (125 U/ml), collagenase typ I (450 U/ml), hyaluronidase (60 U/ml) and deoxyribonuclease I (60 U/ml). Digestion was stopped with stopping solution (PBS; 2% FCS; 1 mM EDTA) and cell suspension was strained. Dead cells were removed by MACS Dead cell removal kit (Miltenyi, Germany). Cells were washed and resuspended in PBS and used for droplet scRNA-seq. Cellular suspensions were loaded on a 10x Chromium Controller (10x Genomics) according to manufacturer's protocol. All scRNA-seq libraries were prepared using Chromium Single Cell 30 v2 Reagent Kit (10x Genomics) according to manufacturer's protocol. In brief, the initial step consisted in generating an emulsion where individual cells were isolated into droplets together with gel beads coated with unique primers bearing 10x cell barcodes, UMI (unique molecular identifiers) and poly (dT) sequences. Reverse transcription reactions were engaged to generate barcoded full-length cDNA followed by the disruption of emulsions using the recovery agent and cDNA clean up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific, Germany). Bulk cDNA was amplified using a Biometra Thermocycler TProfessional Basic Gradient with 96-Well Sample Block (98 °C for 3min; cycled 14x: 98 °C for 15 s, 67 °C for 20 s, and 72 °C for 1min; 72 °C for 1min; held at 4 °C). Amplified cDNA product was cleaned with the SPRIselect Reagent Kit (Beckman Coulter, USA). Indexed sequencing libraries were constructed using the reagents from the Chromium Single Cell 30 v2 Reagent Kit, as follows: fragmentation, end repair and A-tailing; size selection with SPRIselect; adaptor ligation; post-ligation cleanup with SPRIselect; sample index PCR and cleanup with SPRI select beads. Library quantification and quality assessment were performed using Bioanalyzer Agilent 2100 using a High Sensitivity DNA chip (Agilent Genomics, USA). Indexed libraries were equimolarly pooled and sequenced on two Illumina HiSeq 4000 using paired-end 26 × 98 bp as sequencing mode by GenomeScan (Leiden, Netherlands).
Single-cell expression data were processed using the Cell Ranger Single Cell Software Suite (v2.1.1) to perform quality control, sample de-multiplexing, barcode processing, and single-cell gene counting. Sequencing reads were aligned to the mouse reference genome GRCm38 using the Cell Ranger suite with default parameters. A total of 5523 single cells were analyzed. UMAP (Uniform Manifold Approximation and Projection) cluster analysis and was performed by using Seurat (v2.3). The gene-cell-barcode matrix of the samples was log-transformed and normalized by the number of UMI's per cell. In brief, differential expression of genes was used utilizing the ‘FindMarkers’ function in the Seurat package for focused analyses. Genes with adjusted p-values <0.05 were considered as differentially expressed genes. Pre-annotation was conducted using scCATCH and further annotation was performed according to top 50 differentially expressed genes.
2.7. Laser capture microdissection
Frozen tissue sections were mounted on special LCM membrane slides and stained immunohistochemically with Hematoxylin and Eosin to detect carotids and elastic laminas. Carotid sections containing outer elastic lamina and neointima were then cut and catapulted into the lid of a collection tube by the laser. After collection of up to 70 sections, total RNA was extracted using a RNeasy Micro Kit (Qiagen, Hilden, Germany). NanoMACEseq was performed for further analysis.
2.8. nanoMACEseq and bioinformatics
The library preparation was conducted by GenXPro. 3′mRNA libraries were prepared using GenXPros “Rapid MACE-Seq Kit” for low-input mRNA according to the manual of the manufacturers. Briefly, RNA was fragmented to an average size of 350 bps, followed by poly-A specific cDNA synthesis, pooling and amplification by PCR using the minimum number of cycles as described in the manual. The PCR product was purified by SPRI purification. The final product of 200–400 bps was quality controlled on a PerkinElmer LabChip GXII, the concentration was measured on Qbit. Sequencing was performed on an Illumina NextSeq 500 machine with 1 × 75 bps. The MACE-reads were demultiplexed according to the sample IDs, PCR duplicates were removed with the help of the “TrueQuant” UMI barcodes. Low-quality and adapter-containing reads were cropped. The reads were mapped to the mouse genome (GRCm38.p6) and genes were quantified for the individual libraries for gene expression. Differential gene expression of the different pairwise comparisons was analyzed using DE-Seq2. The normalized gene counts, p-values and log2fold-change values of all pairwise comparisons were combined in a single table. Differentially expressed genes were used to calculate GO enrichment using hypergeometric distribution measurements using the GenXPro analyses pipeline and web-service.
2.9. Statistics
Unless otherwise indicated, data are given as means ± standard error of mean (SEM). N indicates the number of individual experiments or animals. Calculations were performed with Prism 8.0. The latter was also used to test for normal distribution and similarity of variance. Individual statistics of unpaired samples was performed by t-test Mann-Whitney test or ANOVA followed by post-hoc testing, according to normal distribution and group number. A p-value of <0.05 was considered as significant. For the analysis of RNAseq data, p-values were adjusted by Benjamini-Hochberg for multiple testing (FDR/False discovery rate).
3. Results
3.1. Deletion of Nox4 does not affect neointima formation
In order to determine the role of Nox4 in neointima development in vivo, the carotid artery wire injury model for severe injury was set up. Carotid artery injury resulted in neointima formation as documented by an increase in the neointima area and the percentage of neointima compared to total vessel size (Fig. 1A–C). In order to study the role of Nox4 a conditional knockout approach was chosen. To avoid compensation, the enzyme was acutely knocked out by a tamoxifen-activated Cre-recombinase 2 weeks prior to vascular injury and successful knockout was confirmed by Western blot from the aorta (Fig. 1D&E). Unexpectedly, deletion of Nox4 at the time points studied (7, 14, 21 and 28 days after injury) had no impact on neointima formation (Fig. 1A–C). Thus, despite the published induction of Nox4 in response to vascular injury [5,24], the enzyme appears to be functionally irrelevant for neointima formation.
3.2. Nox4 and Nox2 show cell-specific expression patterns
Vascular injury is a complex, highly dynamic process with profound changes in the cellular composition of the tissue and the individual cellular phenotype [29]. Nox4 expression has been reported for many cells of the vascular system including endothelial cells, SMCs and fibroblasts [5], and the latter cells massively expand in the adventitia upon vascular injury.
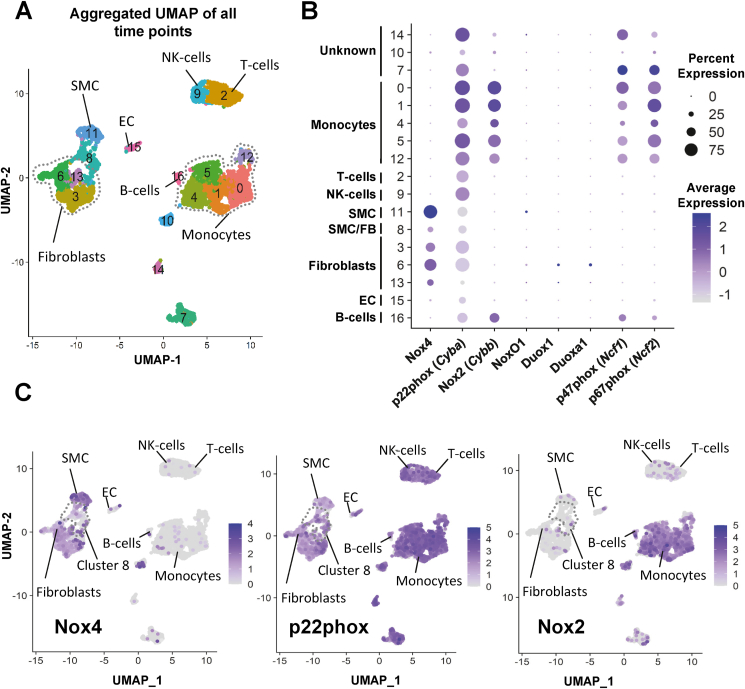
In order to determine the cell-specific expression of the different NADPH oxidase homologues after wire injury, single-cell RNA-sequencing (scRNAseq) was performed at different time points (3, 7 and 14 days) after wire injury and without injury (CTL). Aggregated analysis of the cells of all time points and k-means clustering revealed 17 distinct cell clusters (Fig. 2A, see Supplemental Table 1 for maker genes). Whereas scRNAseq is particularly well suited for cell identification, the sequencing depth is low. Therefore, only abundantly expressed genes are detected. In the aggregated data therefore Nox1 and Nox3 were not detected (Fig. 2B). Nox2 expression (Cybb) was particularly high in myeloid cells with some expression seen also in fibroblasts and endothelial cells (Fig. 2C). P47phox and p67phox (Ncf1 and Ncf2) expression resembled that of Nox2 (Fig. 2B). In contrast to this, Nox4 abundance was restricted to fibroblasts and SMCs but absent in inflammatory cells (Fig. 2C). P22phox (Cyba) was expressed in all cell types (Fig. 2C). In addition to Nox enzymes, also the expression of other ROS generators and antioxidant genes was different between the cell clusters (Supp. Fig. 2). Expression of these genes in endothelial cells, T-cellls, B-cells and NK-cells was somewhat similar, with the exception of Nos3, which was selectively expressed in the endothelium. Other uniquely detected genes were Gpx3, MaoA/B and SOD3 in fibroblasts. Hemoxygenase 1 and 2 were particularly well detected in monocytes, as was Glutaredoxin 1. These differences suggest that intense redox-crosstalk exists between the different cell types of the vascular compartment, which requires further investigation.
Fig. 2.
Nox-enyzmes exhibit a cell specific expression pattern after vascular injury: A) Aggregated UMAP plot and cluster annotation of single cell sequencing experiments performed in C57BL/6 mice without (CTL) and 3, 7 and 14 days after wire injury (aggregated time points; n = 6 animals per time point). B) Dot plot showing average and percentage expression of Nox enzymes. Not detected: Nox1, Noxa1, Nox3, Duox2, Duoxa2. C) Aggregated UMAP plot highlighting expression levels of Nox4, p22phox (Cyba) and Nox2 (Cybb).
3.3. The modified neointima SMCs are located in cluster 8
For neointima formation, SMCs have to acquire a pro-migratory, pro-proliferative phenotype. Neointimal SMCs are therefore characterized by a proliferative, intermediate phenotype sharing aspects of fibroblasts and differentiated SMCs [29]. Of the 5 clusters assigned to fibroblasts and SMCs, particularly cluster 8 exhibited these features. Cluster 8 also was the only cluster with proliferative activity as determined by CellCycleScoring (%G0 phase: cluster 8 46%, all other fibroblast & SMC clusters: >98%). This suggests that cluster 8 contains the modified SMC of the neointima. Other cells which may also contribute to cluster 8 are myofibroblasts and transformed cells of the media, but without lineage tracing and site-specific labelling it is not possible to deconvolute the cluster further. With respect to the expression of redox-modulating genes, cluster 8 grouped well with fibroblasts and smooth muscle cells, although it appears that most redox-genes are expressed slightly less, with the exception of peroxiredoxin 1 (Supp. Fig. 2).
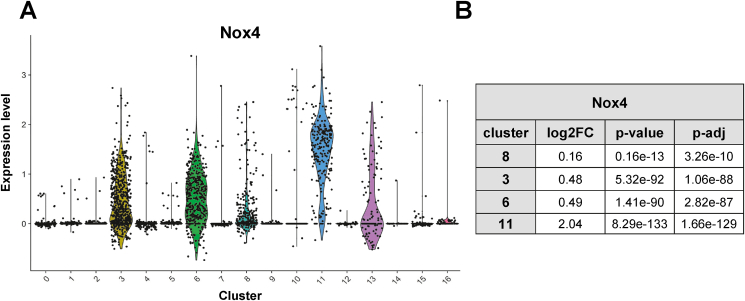
3.4. Modified SMCs express little Nox4
With the identification of the modified SMCs, it became possible to specifically determine Nox4 expression in this subset. As compared to the differentiated SMCs and the fibroblast clusters, the modified SMCs expressed very little Nox4 (Fig. 3A and B). Importantly, these data suggest that particularly those cells, which contribute to neointima formation lose most of their Nox4 expression and this observation could explain why Nox4 did not impact on the process of neointima formation in response to wire injury.
Fig. 3.
Nox4 expression of cell clusters: Single cell sequencing experiment performed in C57BL/6 mice without (CTL) and 3, 7 and 14 days after wire injury (aggregated time points; n = 6 animals per time point). A) and B) Nox4 expression levels of clusters shown by violin plot of scRNA-seq data. The Seurat tool ‘FindMarkers’ function was used to determine significance using Wilcoxen-test.
The neointima contains inflammatory activated, dedifferentiated smooth muscle cells.
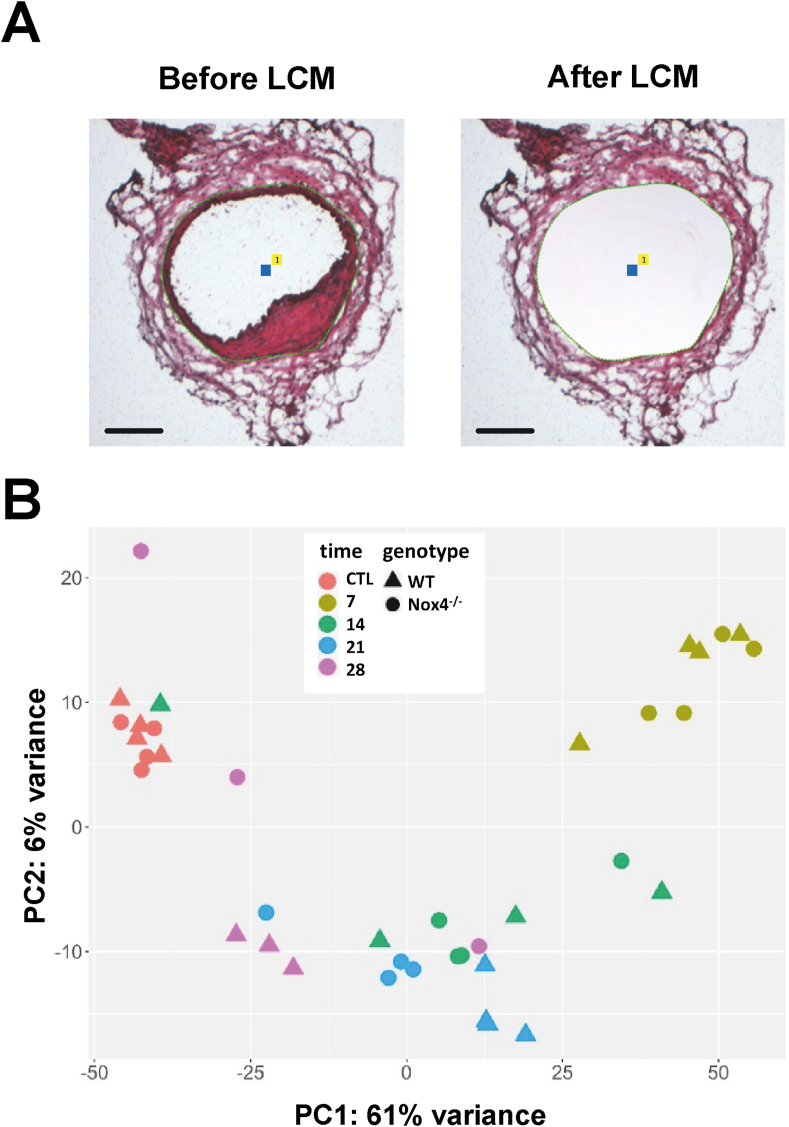
A disadvantage of scRNAseq is that the tissue integrity is lost during tissue digestion and that it is therefore not possible to specifically determine the gene expression of the cells of the neointima. To overcome this limitation, laser capture microdissection of the neointima of the injured side as well as of the control side was performed from vessels of WT and conditional Nox4−/− mice 7, 14, 21 and 28 days after wire-injury (Fig. 4A). RNA was extracted and subjected to massive-analysis-of-cDNA-ends-(MACE)-RNAseq to determine time- and Nox4-dependent neointimal mRNA expression. For this approach 70 microdissected sections of neointima of one animal were pooled per sequencing and 3–4 sequencings per time point and genotype were performed.
Fig. 4.
Principle component analysis from laser capture microdissections: A) Exemplary section before and after laser capture microdissection of the neointima. B) Principle component analysis of MACE-RNAseq of laser capture microdissection of the neointima and the control side from WT and Nox4−/− mice subjected to wire injury for 7, 14, 21 and 28 days. CTL indicates the non-operated control side (n = 3–4 per group).
Aggregated principal component analysis revealed that the injury itself has strong effects on gene expression, which was most evident between the control condition and day 7 after surgery as indicated by the highest deviation in PC1. Subsequently, the gene expression in the neointima started to approximate back to the control state (Fig. 4B). The time point 28 days after surgery was closest to the control condition indicating that the neointima almost completely consolidated. This development was also reflected by the gene ontology (GO) term analysis (Supp. Fig 3) with the highest p-value differences between control and day 7 after surgery and almost no GO terms remaining 28 days after injury. In keeping with the concepts of neointima formation, the GO terms analysis indicated, that the cells of the neointima exhibit a transient loss of the differentiated and contractile smooth muscle phenotype. In contrast, they express cytokines, adhesion molecules and markers for immune cell invasion. Interestingly, at day 28 after injury, particularly markers for resolution of inflammation (Col2A1, Col8a1, Mmp12, Comp, Il10ra) were increased.
3.5. Vascular injury decreases Nox4 expression
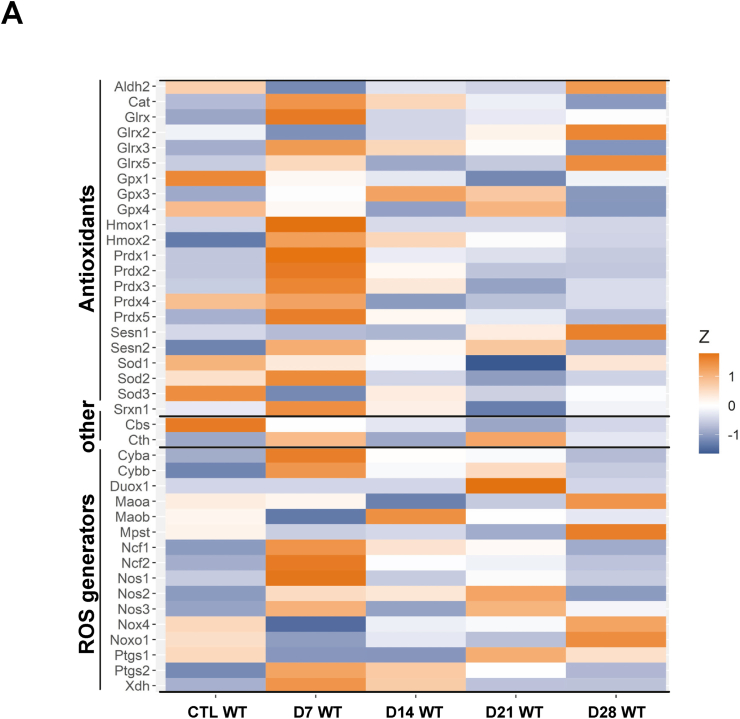
In keeping with the data obtained by MACEseq analysis, Nox4 expression was significantly reduced in the neointima early after wire injury, as compared to the non-operated condition (Fig. 5A). Gradually, Nox4 expression subsequently increased. 28 days after injury, no difference in Nox4 expression to the situation before the surgery remained. As compared to Nox4, the leukocyte NADPH Oxidase Nox2 (Cybb) together with its subunits p22phox (Cyba) and p47phox (Ncf1) exhibited an inverse expression profile, with a marked increase early after operation (Fig. 5; Supp table 3). This is certainly a reflection of the inflammation-induced invasion of leukocytes in response to the injury. As another sign of inflammation, expression of inducible NO-synthase was also increased (Nos2), as was xanthine dehydrogenase (Xdh) and Hemoxygenase (Hmox1, Hmox2). As a sign of early endothelial regrowth, endothelial NO synthase (Nos3) was already increased 7 days after injury (Fig. 5, Supp table 3). Collectively, these data illustrate, that the neointima compartment is initially characterized by inflammation which subsequently consolidates. Early in the processes, markers of muscle cell differentiation as well as Nox4 are low and with the consolidation of the injury they are re-expressed.
Fig. 5.
ROS generator and antioxidant expression in course of inflammation in neointima: A) MACEseq dataset of wildtype neointima samples without (CTL), D7, D14, D21 and D28 after wire-induced injury. Heatmap of ROS generator and antioxidant gene expression (Z-score of normalized mean counts).
3.6. Nox4 does not affect neointimal gene expression early after injury
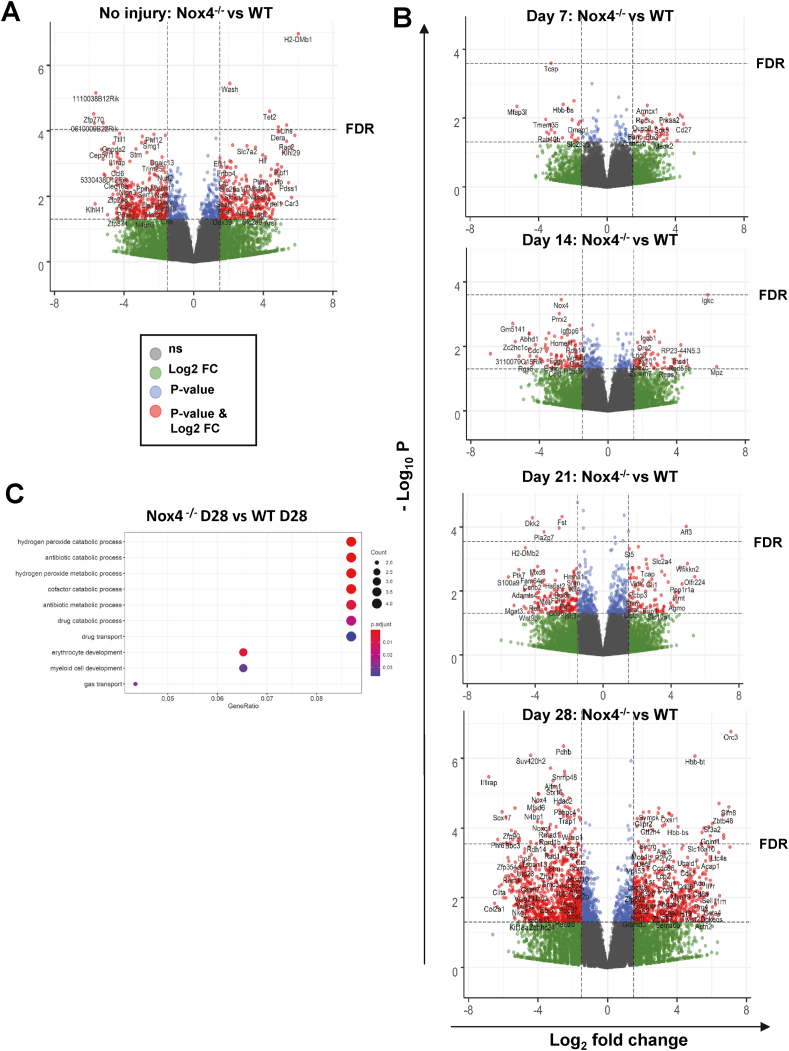
In order to determine the impact of Nox4 on gene expression, MACEseq data under basal conditions and of the neointima were compared between WT and conditional Nox4−/− mice. Before the surgery, several differentially expressed genes were detected, albeit without an obvious connection to redox-regulation (Fig. 6A–B). These differences were lost and in the early neointima, 7 days after surgery, not a single Nox4-differentially expressed gene (padj<0.05) remained. Subsequently, differences increased and at day 28 after injury, more differentially expressed genes were observed than under resting conditions. Interestingly, GO-term analysis at day 28 returned “hydrogen peroxide catalytic processes” (Fig. 6C), which was primarily the consequence of an increased expression of genes of the haemoglobin B chain. The abundance of the other detected Nox genes was not changed on mRNA level upon loss of Nox4, therefore compensatory effects of other Nox family members are negligible.
Fig. 6.
Role of Nox4 for neointimal gene expression: A) and B) Volcano plot on neointima gene expression differences between Nox4−/− and WT mice CTL (A), 7, 14, 21 and 28 days after wire-induced injury (B); n = 3–4. C) GO term analysis of D28 Nox4−/− vs WT significant upregulated genes; p < 0.05.
4. Discussion
In this manuscript we set out to determine the role of Nox4 after deep carotid artery wire injury in mice. Unexpectedly, we observed that Nox4 did not influence the process of neointima formation. Mechanistically, Nox4 is down-regulated after vascular injury and is re-expressed during the consolidation phase.
Nox4 was discovered more than 20 years ago [30] and despite more than 3000 publications covering this protein, it is hardly possible to name it's true physiological function. This somewhat surprising statement has several causes. Unspecific inhibitors, transgenic model systems with unknown specificity like dominant-negative constructs, correlative study design, publication bias and analysis performed in dedifferentiated cells all resulted in a colorful picture even attributing function to Nox4 in cells which clearly do not express the protein.
Interestingly, it is not well understood what governs Nox4 expression. The highest expression of Nox4 is found in cells of the proximal renal tubule followed by stromal cells like fibroblasts [31,32], but the underlying transcription factor network needs to be better defined. The fairly global transcription factors Sp3 [33] and E2F1 [34] as well as STAT3 [35] contribute to the basal transcriptional activity for Nox4. Interestingly, several pathways activated by injury, inflammation and oxidative stress have been shown to downregulate Nox4: In EA.hy926 cells, activation of PKC results in an initial PKC ε-dependent downregulation of Nox4 followed by a later PKC α-dependent induction of the enzyme [36]. In a study on cyclic strain, NO downregulated Nox4 [37] and oxidative stress as present in high shear models through Nrf 2 and an atypical ARE-like/Oct-1 binding site also reduced Nox4 expression [38]. Vascular injury is a situation of oxidative stress with strong Nox2 accumulation and iNOS induction, as also observed in the MACEseq analysis of neointima in the present study. The loss of Nox4 expression would have therefore been expected, if there wasn't a second highly important mechanism of Nox4 expression control through transforming growth factor β1 (TGFβ1) and SMAD [[39], [40], [41]]. Indeed, numerous studies have demonstrated that TGFβ1 is a strong inducer of Nox4, particularly in fibroblasts and myofibroblasts [42], and especially the healing phase after vascular injury is a situation of increased TGFβ1 signaling. The cytokine is produced by M2 macrophages, and promotes matrix formation and limits neointima development [43,44]. This may explain why Nox4 increases late after vascular injury as observed in rats [24] as well as in the present study.
The physiological importance of TGFβ1-induced Nox4 expression is unclear but it is obvious to assume a link to H2O2 formation by the enzyme. The link to TGFβ1 sets to context for matrix formation. In C. elegans, NADPH oxidase-derived H2O2 facilitates oxidative matrix crosslink on tyrosine [45]. Matrix synthesis also requires oxidative protein crosslinking in the ER as well as in the extracellular spaces [46,47]. Particularly for the latter situation, catalyzed by vascular peroxidase-1, H2O2 is needed. Although ex vivo it is possible to demonstrate that TGFβ promotes protein crosslinking [48], several studies failed to detect a function of Nox4 in oxidative protein folding in physiological situations [49,50]. It is not known, whether this is due to redundancy in the system or because the current paradigms in the field are simply wrong. In addition to this aspect, several studies support the concept that Nox4 tunes the antioxidant response and therefore loss of Nox4 had negative effects in these studies. The fact that after vascular injury, Nox4 expression is drastically decreased explains why in contrast to those publications, we failed to observe a function of Nox4 in the present study.
Collectively, the present work supports the concept that Nox4 in vivo is a marker of differentiated cells. Upon injury, Nox4 expression is lost and therefore the enzyme turned out to be unimportant for neointima formation after wire injury.
Source of funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft (GRK 2336, SFB815 (TPA01) and SFB834 (TPA02), Excellence Cluster EXS2026 CPI - Cardiopulmonary Institute). AMS is supported by the British Heart Foundation (CH/1999001/11735).
Declaration of competing interest
The authors declare that they have no relevant financial, personal or professional relationships to disclose which could be perceived as a conflict of interest or as potentially influencing or biasing the authors’ work.
Acknowledgement
We are grateful for the excellent technical assistance of Susanne Wienstroer, Katalin Palfi and Tanja Lüneburg.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102050.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schürmann C., Rezende F., Kruse C., Yasar Y., Löwe O., Fork C., van de Sluis B., Bremer R., Weissmann N., Shah A.M., Jo H., Brandes R.P., Schröder K. The NADPH oxidase Nox4 has anti-atherosclerotic functions. Eur. Heart J. 2015;36:3447–3456. doi: 10.1093/eurheartj/ehv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray S.P., Di Marco E., Kennedy K., Chew P., Okabe J., El-Osta A., Calkin A.C., Biessen E.A.L., Touyz R.M., Cooper M.E., Schmidt H.H.H.W., Jandeleit-Dahm K.A.M. Reactive oxygen species can provide atheroprotection via NOX4-dependent inhibition of inflammation and vascular remodeling. Arterioscler. Thromb. Vasc. Biol. 2016;36:295–307. doi: 10.1161/ATVBAHA.115.307012. [DOI] [PubMed] [Google Scholar]
- 3.Wind S., Beuerlein K., Armitage M.E., Taye A., Kumar A.H.S., Janowitz D., Neff C., Shah A.M., Wingler K., Schmidt H.H.H.W. Oxidative stress and endothelial dysfunction in aortas of aged spontaneously hypertensive rats by NOX1/2 is reversed by NADPH oxidase inhibition. Hypertension. 2010;56:490–497. doi: 10.1161/HYPERTENSIONAHA.109.149187. [DOI] [PubMed] [Google Scholar]
- 4.Brandes R.P., Weissmann N., Schröder K. NADPH oxidases in cardiovascular disease. Free Radic. Biol. Med. 2010;49:687–706. doi: 10.1016/j.freeradbiomed.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Knock G.A. NADPH oxidase in the vasculature: expression, regulation and signalling pathways; role in normal cardiovascular physiology and its dysregulation in hypertension. Free Radic. Biol. Med. 2019;145:385–427. doi: 10.1016/j.freeradbiomed.2019.09.029. [DOI] [PubMed] [Google Scholar]
- 6.Gray S.P., Jandeleit-Dahm K.A.M. The role of NADPH oxidase in vascular disease--hypertension, atherosclerosis & stroke. Curr. Pharmaceut. Des. 2015;21:5933–5944. doi: 10.2174/1381612821666151029112302. [DOI] [PubMed] [Google Scholar]
- 7.Gray S.P., Di Marco E., Okabe J., Szyndralewiez C., Heitz F., Montezano A.C., de Haan J.B., Koulis C., El-Osta A., Andrews K.L., Chin-Dusting J.P.F., Touyz R.M., Wingler K., Cooper M.E., Schmidt H.H.H.W., Jandeleit-Dahm K.A. NADPH oxidase 1 plays a key role in diabetes mellitus-accelerated atherosclerosis. Circulation. 2013;127:1888–1902. doi: 10.1161/CIRCULATIONAHA.112.132159. [DOI] [PubMed] [Google Scholar]
- 8.Sobey C.G., Judkins C.P., Rivera J., Lewis C.V., Diep H., Lee H.W., Kemp-Harper B.K., Broughton B.R.S., Selemidis S., Gaspari T.A., Samuel C.S., Drummond G.R. NOX1 deficiency in apolipoprotein E-knockout mice is associated with elevated plasma lipids and enhanced atherosclerosis. Free Radic. Res. 2015;49:186–198. doi: 10.3109/10715762.2014.992893. [DOI] [PubMed] [Google Scholar]
- 9.Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K.-H. NOX4 activity is determined by mRNA levels and reveals a unique pattern of ROS generation. Biochem. J. 2007;406:105–114. doi: 10.1042/BJ20061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takac I., Schröder K., Zhang L., Lardy B., Anilkumar N., Lambeth J.D., Shah A.M., Morel F., Brandes R.P. The E-loop is involved in hydrogen peroxide formation by the NADPH oxidase Nox4. J. Biol. Chem. 2011;286:13304–13313. doi: 10.1074/jbc.M110.192138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burgoyne J.R., Madhani M., Cuello F., Charles R.L., Brennan J.P., Schröder E., Browning D.D., Eaton P. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science. 2007;317:1393–1397. doi: 10.1126/science.1144318. [DOI] [PubMed] [Google Scholar]
- 12.Ray R., Murdoch C.E., Wang M., Santos C.X., Zhang M., Alom-Ruiz S., Anilkumar N., Ouattara A., Cave A.C., Walker S.J., Grieve D.J., Charles R.L., Eaton P., Brewer A.C., Shah A.M. Endothelial Nox4 NADPH oxidase enhances vasodilatation and reduces blood pressure in vivo. Arterioscler. Thromb. Vasc. Biol. 2011;31:1368–1376. doi: 10.1161/ATVBAHA.110.219238. [DOI] [PubMed] [Google Scholar]
- 13.Burgoyne J.R., Rudyk O., Cho H.-J., Prysyazhna O., Hathaway N., Weeks A., Evans R., Ng T., Schröder K., Brandes R.P., Shah A.M., Eaton P. Deficient angiogenesis in redox-dead Cys17Ser PKARIα knock-in mice. Nat. Commun. 2015;6:7920. doi: 10.1038/ncomms8920. [DOI] [PubMed] [Google Scholar]
- 14.Zhang M., Mongue-Din H., Martin D., Catibog N., Smyrnias I., Zhang X., Yu B., Wang M., Brandes R.P., Schröder K., Shah A.M. Both cardiomyocyte and endothelial cell Nox4 mediate protection against hemodynamic overload-induced remodelling. Cardiovasc. Res. 2018;114:401–408. doi: 10.1093/cvr/cvx204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M., Brewer A.C., Schröder K., Santos C.X.C., Grieve D.J., Wang M., Anilkumar N., Yu B., Dong X., Walker S.J., Brandes R.P., Shah A.M. NADPH oxidase-4 mediates protection against chronic load-induced stress in mouse hearts by enhancing angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18121–18126. doi: 10.1073/pnas.1009700107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schröder K., Zhang M., Benkhoff S., Mieth A., Pliquett R., Kosowski J., Kruse C., Luedike P., Michaelis U.R., Weissmann N., Dimmeler S., Shah A.M., Brandes R.P. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circ. Res. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 17.Craige S.M., Chen K., Pei Y., Li C., Huang X., Chen C., Shibata R., Sato K., Walsh K., Keaney J.F. NADPH oxidase 4 promotes endothelial angiogenesis through endothelial nitric oxide synthase activation. Circulation. 2011;124:731–740. doi: 10.1161/CIRCULATIONAHA.111.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morawietz H. Cardiovascular protection by Nox4. Cardiovasc. Res. 2018;114:353–355. doi: 10.1093/cvr/cvx252. [DOI] [PubMed] [Google Scholar]
- 19.Tong X., Khandelwal A.R., Wu X., Xu Z., Yu W., Chen C., Zhao W., Yang J., Qin Z., Weisbrod R.M., Seta F., Ago T., Lee K.S.S., Hammock B.D., Sadoshima J., Cohen R.A., Zeng C. Pro-atherogenic role of smooth muscle Nox4-based NADPH oxidase. J. Mol. Cell. Cardiol. 2016;92:30–40. doi: 10.1016/j.yjmcc.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong X., Khandelwal A.R., Qin Z., Wu X., Chen L., Ago T., Sadoshima J., Cohen R.A. Role of smooth muscle Nox4-based NADPH oxidase in neointimal hyperplasia. J. Mol. Cell. Cardiol. 2015;89:185–194. doi: 10.1016/j.yjmcc.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Clempus R.E., Sorescu D., Dikalova A.E., Pounkova L., Jo P., Sorescu G.P., Schmidt H.H.H., Lassègue B., Griendling K.K. Nox4 is required for maintenance of the differentiated vascular smooth muscle cell phenotype. Arterioscler. Thromb. Vasc. Biol. 2007;27:42–48. doi: 10.1161/01.ATV.0000251500.94478.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Garrido A., Brown D.I., Lyle A.N., Dikalova A., Seidel-Rogol B., Lassègue B., San Martín A., Griendling K.K. NADPH oxidase 4 mediates TGF-β-induced smooth muscle α-actin via p38MAPK and serum response factor. Free Radic. Biol. Med. 2011;50:354–362. doi: 10.1016/j.freeradbiomed.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee M., San Martín A., Valdivia A., Martin-Garrido A., Griendling K.K. Redox-sensitive regulation of myocardin-related transcription factor (MRTF-A) phosphorylation via palladin in vascular smooth muscle cell differentiation marker gene expression. PloS One. 2016;11 doi: 10.1371/journal.pone.0153199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szöcs K., Lassègue B., Sorescu D., Hilenski L.L., Valppu L., Couse T.L., Wilcox J.N., Quinn M.T., Lambeth J.D., Griendling K.K. Upregulation of Nox-based NAD(P)H oxidases in restenosis after carotid injury. Arterioscler. Thromb. Vasc. Biol. 2002;22:21–27. doi: 10.1161/hq0102.102189. [DOI] [PubMed] [Google Scholar]
- 25.Haurani M.J., Cifuentes M.E., Shepard A.D., Pagano P.J. Nox4 oxidase overexpression specifically decreases endogenous Nox4 mRNA and inhibits angiotensin II-induced adventitial myofibroblast migration. Hypertension. 2008;52:143–149. doi: 10.1161/HYPERTENSIONAHA.107.101667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bigalke B., Pohlmeyer I., Schönberger T., Griessinger C.M., Ungerer M., Botnar R.M., Pichler B.J., Gawaz M. Imaging of injured and atherosclerotic arteries in mice using fluorescence-labeled glycoprotein VI-Fc. Eur. J. Radiol. 2011;79:e63–e69. doi: 10.1016/j.ejrad.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 27.Xu Q. Mouse models of arteriosclerosis. Am. J. Pathol. 2004;165:1–10. doi: 10.1016/S0002-9440(10)63270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindner V., Fingerle J., Reidy M.A. Mouse model of arterial injury. Circ. Res. 1993;73:792–796. doi: 10.1161/01.res.73.5.792. [DOI] [PubMed] [Google Scholar]
- 29.Roostalu U., Aldeiri B., Albertini A., Humphreys N., Simonsen-Jackson M., Wong J.K.F., Cossu G. Distinct cellular mechanisms underlie smooth muscle turnover in vascular development and repair. Circ. Res. 2018;122:267–281. doi: 10.1161/CIRCRESAHA.117.312111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiszt M., Kopp J.B., Várnai P., Leto T.L. Identification of renox, an NAD(P)H oxidase in kidney. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8010–8014. doi: 10.1073/pnas.130135897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsuyama M., Hirai H., Iwata K., Ibi M., Matsuno K., Matsumoto M., Yabe-Nishimura C. Sp3 transcription factor is crucial for transcriptional activation of the human NOX4 gene. FEBS J. 2011;278:964–972. doi: 10.1111/j.1742-4658.2011.08018.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L., Sheppard O.R., Shah A.M., Brewer A.C. Positive regulation of the NADPH oxidase NOX4 promoter in vascular smooth muscle cells by E2F. Free Radic. Biol. Med. 2008;45:679–685. doi: 10.1016/j.freeradbiomed.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Guo S., Chen X. The human Nox4: gene, structure, physiological function and pathological significance. J. Drug Target. 2015;23:888–896. doi: 10.3109/1061186X.2015.1036276. [DOI] [PubMed] [Google Scholar]
- 36.Xu H., Goettsch C., Xia N., Horke S., Morawietz H., Förstermann U., Li H. Differential roles of PKCalpha and PKCepsilon in controlling the gene expression of Nox4 in human endothelial cells. Free Radic. Biol. Med. 2008;44:1656–1667. doi: 10.1016/j.freeradbiomed.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 37.Goettsch C., Goettsch W., Arsov A., Hofbauer L.C., Bornstein S.R., Morawietz H. Long-term cyclic strain downregulates endothelial Nox4, Antioxid. Redox Signal. 2009;11:2385–2397. doi: 10.1089/ars.2009.2561. [DOI] [PubMed] [Google Scholar]
- 38.Goettsch C., Goettsch W., Brux M., Haschke C., Brunssen C., Muller G., Bornstein S.R., Duerrschmidt N., Wagner A.H., Morawietz H. Arterial flow reduces oxidative stress via an antioxidant response element and Oct-1 binding site within the NADPH oxidase 4 promoter in endothelial cells. Basic Res. Cardiol. 2011;106:551–561. doi: 10.1007/s00395-011-0170-3. [DOI] [PubMed] [Google Scholar]
- 39.Cucoranu I., Clempus R., Dikalova A., Phelan P.J., Ariyan S., Dikalov S., Sorescu D. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ. Res. 2005;97:900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- 40.Sturrock A., Cahill B., Norman K., Huecksteadt T.P., Hill K., Sanders K., Karwande S.V., Stringham J.C., Bull D.A., Gleich M., Kennedy T.P., Hoidal J.R. Transforming growth factor-beta1 induces Nox4 NAD(P)H oxidase and reactive oxygen species-dependent proliferation in human pulmonary artery smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290:L661–L673. doi: 10.1152/ajplung.00269.2005. [DOI] [PubMed] [Google Scholar]
- 41.Bai G., Hock T.D., Logsdon N., Zhou Y., Thannickal V.J. A far-upstream AP-1/Smad binding box regulates human NOX4 promoter activation by transforming growth factor-β. Gene. 2014;540:62–67. doi: 10.1016/j.gene.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Touyz R.M., Montezano A.C. Vascular Nox4: a multifarious NADPH oxidase. Circ. Res. 2012;110:1159–1161. doi: 10.1161/CIRCRESAHA.112.269068. [DOI] [PubMed] [Google Scholar]
- 43.Yokote K., Kobayashi K., Saito Y. The role of Smad3-dependent TGF-beta signal in vascular response to injury. Trends Cardiovasc. Med. 2006;16:240–245. doi: 10.1016/j.tcm.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Khan R., Agrotis A., Bobik A. Understanding the role of transforming growth factor-beta1 in intimal thickening after vascular injury. Cardiovasc. Res. 2007;74:223–234. doi: 10.1016/j.cardiores.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Aguirre J., Lambeth J.D. Nox enzymes from fungus to fly to fish and what they tell us about Nox function in mammals. Free Radic. Biol. Med. 2010;49:1342–1353. doi: 10.1016/j.freeradbiomed.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhave G., Cummings C.F., Vanacore R.M., Kumagai-Cresse C., Ero-Tolliver I.A., Rafi M., Kang J.-S., Pedchenko V., Fessler L.I., Fessler J.H., Hudson B.G. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat. Chem. Biol. 2012;8:784–790. doi: 10.1038/nchembio.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulleid N.J. Disulfide bond formation in the mammalian endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a013219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thannickal V.J., Fanburg B.L. Activation of an H2O2-generating NADH oxidase in human lung fibroblasts by transforming growth factor beta 1. J. Biol. Chem. 1995;270:30334–30338. doi: 10.1074/jbc.270.51.30334. [DOI] [PubMed] [Google Scholar]
- 49.Sirokmány G., Kovács H.A., Lázár E., Kónya K., Donkó Á., Enyedi B., Grasberger H., Geiszt M. Peroxidasin-mediated crosslinking of collagen IV is independent of NADPH oxidases. Redox Biol. 2018;16:314–321. doi: 10.1016/j.redox.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tavender T.J., Springate J.J., Bulleid N.J. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 2010;29:4185–4197. doi: 10.1038/emboj.2010.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.