Abstract
The variant B.1.1.7 of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the RNA virus causing the pandemic more than a year worldwide, was reported from United Kingdom (UK) in late December 2020. It was reported that mortality increases by 65% and transmissibility increases by 70%, which may result in an increase of reproduction number to 1.13-1.55 from 0.75-0.85. To analyze the global increasing trend of the variant B.1.1.7, we extracted results of B.1.1.7 from GISAID on May 11 and May 12, 2021, and conducted a dose-response regression. It took 47 days to reach 20% and 121 days to reach 50% among the sequence submitted from UK. In Korea, cases of B.1.1.7 have increased since the first report of three cases on December 28, 2020. Positive rate of B.1.1.7 in Korea was 21.6% in the week from May 9 to May 15, 2021. Detection rate of the variants is expected to increase further and new variants of SARS-CoV-2 are emerging, so a close monitoring and control would be maintained for months.
Keywords: COVID-19, Epidemics, Epidemiology, SARS-CoV-2, Variant
Introduction
The world has suffered extensively from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, with 167 million cases and 3.5 million deaths worldwide as of May 23, 2021 [1]. Since its introduction, several mutations have been reported, such as D614G in China and the 69/70 deletion in Denmark [2−4]. In mid-December 2020, the United Kingdom (UK) and South Africa reported locally emerged variants, followed by a report a few days later of a new variant in 4 people entering Japan from Brazil on January 10, 2021 [5]. The variant that emerged in England (a.k.a., the B.1.1.7 lineage, GR, VOC 202012/01, N501Y, 501Y.V1, or 20I/501Y.V1) was assessed by the UK’s advisory panel on December 18, 2020, and the Prime Minister Boris Johnson mentioned that the B.1.1.7 variant could increase the transmissibility of SARS-CoV-2 by 70% and/or the reproduction number (R) by 0.4 [6]. These possibilities pose global concerns for the impact of the B.1.1.7 variant on the SARS-CoV-2 pandemic. Here, we summarized the epidemiological findings of the UK variant and its impact on transmissibility based on investigations in the UK [7−12], and analyzed global trends in its spread.
Materials and Methods
A Short History of the Discovery of the UK Variant
The UK has a coronavirus disease 2019 (COVID-19) surveillance system including genomic profiling under the COVID-19 Genomics Consortium UK (CoG-UK). As part of an epidemiological investigation to investigate the increasing trend of COVID-19 cases starting in September 2020 in the UK, available genomic data from 255 cases in Kent, England (a county in the southeast region) were analyzed [8]. A large phylogenetic cluster of 117 genomically similar cases was discovered from the samples of 255 cases in the Kent data collected from November 10 to 18, which was very distinct from the rest of the UK dataset. In the UK dataset (available genomes of 915 cases on the analysis date of December 8, 2020), a distinct Kent cluster was observed mostly from November (828 cases, 90.5%) and to a lesser extent in October (79 cases, 8.6%), with the earliest records dating from September (4 cases, 0.4%).
Information regarding the B.1.1.7 variant was simultaneously or subsequently reported from the UK, either by the government or research institutes, starting on December 20, 2020. This paper summarizes the main results of recent reports, which are: (1) a paper regarding the genomic characteristics of B.1.1.7 reported by researchers from CoG-UK [11]; (2) 4 technical reports including epidemic and epidemiological characteristics reported by Public Health England (PHE) from December 20 to January 15 [7−10]; and (3) a mathematical modeling paper on the transmissibility and reproduction number of B.1.1.7 in the UK by researchers from Imperial College and CoG-UK [12].
Profile and Nomenclature of the B.1.1.7 Variant
The two earliest samples were collected from Kent on September 20 and from Greater London on September 21, 2020 [11], and samples continued to be detected through early December 2020. The new distinct variant belonged to the B.1.1.7 lineage and the GR clade. The variant involves non-synonymous mutations or deletions (e.g., N501Y, 69/70 deletion, P681H, and the ORF8 stop codon mutation) in the receptor binding domain (RBD) of the spike protein, which can induce an increased ACE2 receptor affinity of the spike protein of SARS-CoV-2 in the airway [13].
The variant has been named in several ways based on different nomenclature systems: (1) “VOC 202012/01,” designated as a variant of concern (VOC) followed by year, month, and number [8]; (2) the “B.1.1.7 lineage,” based on the dynamic lineage nomenclature system of SARS-CoV-2 suggested by Rambaut et al. (PANGO lineages, cov-lineages.org) [14]; (3) “20I/501Y.V1,” applying “20I” from the Nextstrain clade naming strategy combining the year and letter in a system developed 5 years ago for seasonal influenza and extended for SARS-CoV-2 since January 2020 (nextstrain.org) [15], and “501Y.V1” which denotes the replacement of asparagine (N) by tyrosine (Y) in the RBD of the spike protein at position 501. “V1” stands for the first variant examined of N501Y.
Analysis of Global Trends of B.1.1.7 Based on the Reports of GISAID
GISAID is a publicly accessible database launched in 2008 that contains genomic information on influenza viruses and SARS-CoV-2. Information on 6 SARS-CoV-2 variants (B.1.1.7, B.1.351, P.1, B.1.429 & B.1.427, B.1.525, and B.1.617) is provided through the web-based platform as of May 26, 2021 [16]. A weekly report of relative variant genome frequency per region provided as a graph from GISAID was extracted on May 11 and May 12, 2021 and analyzed using dose-response regression in MedCalc version 19.8 (MedCalc Software Ltd., Ostend, Belgium).
Results
Epidemic of the Variant in the UK
Through routine genomic surveillance of SARS-CoV-2 in the UK, a total of 6,008 cases of the B.1.1.7 variant were identified as of January 4, 2021 [9]. Geographically, the B.1.1.7 variant has largely been reported in the Southeast region and Greater London compared to other regions, although the samples are non-random and not representative of all COVID-19 cases in the country (Fig. 1A). Instances of local authorities identifying the B.1.1.7 variant increased steeply from late October to early December (Fig. 1C) [12].
Fig. 1.
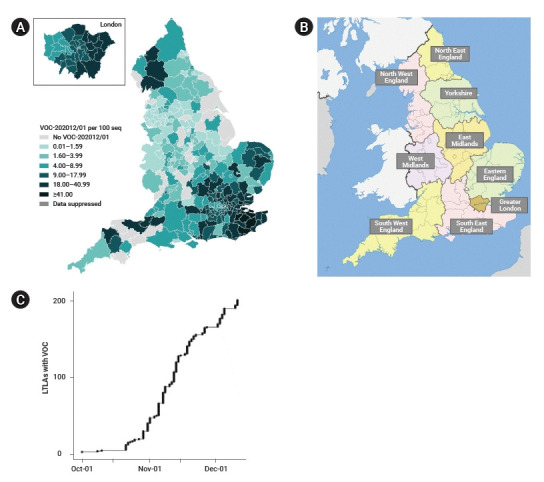
Geographic distribution and time trend of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) B.1.1.7 cases by whole genome sequencing in England. (A) SARS-CoV-2 B.1.1.7 cases per 100 sequences in England by local authorities from September 20, 2020 to January 4, 2021 (extracted from Chand et al. [9]). (B) Nine regions in England (map downloaded from Wikipedia). (C) Local authorities of identifying SARS-CoV-2 B.1.1.7 cases in England from October 1 to December 5, 2020 (extracted from Volz et al. [12]). LTLA, lower tier local authority; VOC, variant of concern.
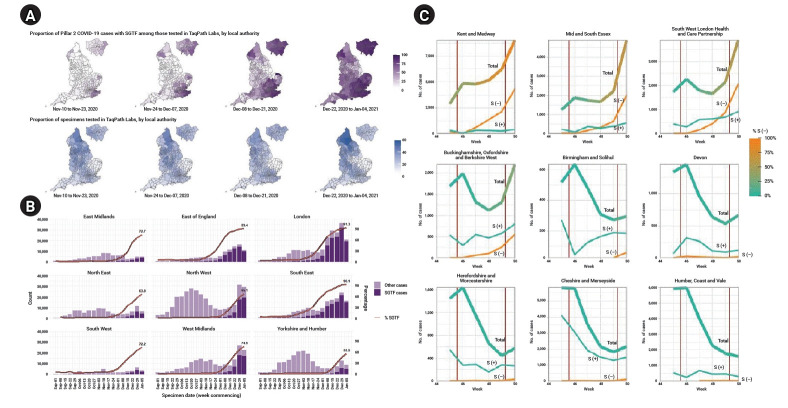
Since the genomic surveillance program includes only a small fraction of COVID-19 cases and it takes 2 weeks to analyze the results from testing using whole-genome scans, the UK researchers used a proxy to indicate the presence of the B.1.1.7 variant: S gene target failure (SGTF) [10,12]. The UK has a high-throughput national testing program including 4 types of tests known as pillars [8]. Pillar 2 is the UK government testing program, including lighthouse laboratories and specially arranged laboratories from the public, private, and academic sectors. SGTF was observed from the results using a Thermo Fisher (TaqPath) probe in the lighthouse laboratories. Coincidently, the B.1.1.7 variant includes the 69/70 deletion (Δ69−70 sequences) of the spike protein, which was found to be highly correlated with SGTF, as 99.5% of Δ69−70 sequences showed SGTF, whereas SGTF was found in only 0.05% of sequences without Δ69−70 sequences, based on an analysis of 14,950 tested samples from October 12 to December 27, 2020 that had both sequencing and SGTF results. In addition, the weekly proportion of the B.1.1.7 variant among all Δ69−70 sequences among all pillar 2 sequences increased from 3% in the week of October 12 to 88% in the week of November 16 and over 98% in the week of December 7, suggesting that the B.1.1.7 variant is correlated with SGTF [9].
Correlated with the spatial distribution of B.1.1.7 cases (Fig. 1A), the SGTF cases were predominantly distributed in the southeast region and Greater London, spreading out to other regions in later weeks (Fig. 2A) [9]. The proportion of SGTF cases increased every week in every region in England since September 1, and SGTF cases accounted for more than 50% of all tested cases in most of the regions except Yorkshire and Humber. In particular, it comprised over 75% of cases in the east of England, London, and southeast since December (Fig. 2B) [7]. Volz et al. [12] examined the time trend of cases in the SGTF group (S−) and non-SGTF group (S+) separately, and reported that the S− group consistently increased, while the incidence of the S+ group was controlled under the lockdown (Fig. 2C).
Fig. 2.
Weekly trend of S gene target failure (SGTF) cases in England. (A) Mapping of proportion of SGTF and specimens tested in TaqPath in England by weeks (extracted from Chand et al. [9]). (B) Proportion of SGTF cases tested in TaqPath Labs in England by region and week (extracted from Chand et al. [7]). (C) Weekly trend of number of cases by SGTF group (S−) and non-SGTF group (S+) in several areas (thick line, total cases; yellow line, S− cases; thin greenish line, S+ cases) (extracted from Volz et al. [12]).
The sex distribution was similar between the S− and S+ groups (Fig. 3A). The number of SGTF cases was largest in the age group of 25 to 59, while the proportion of the S− group similarly increased in recent weeks in every age group, reaching 76.2% in those aged under 25, 77.1% in those aged between 25 and 59, and 74.5% in those aged over 60 in the most recent week of January 5, 2021 (Fig. 3B) [7].
Fig. 3.
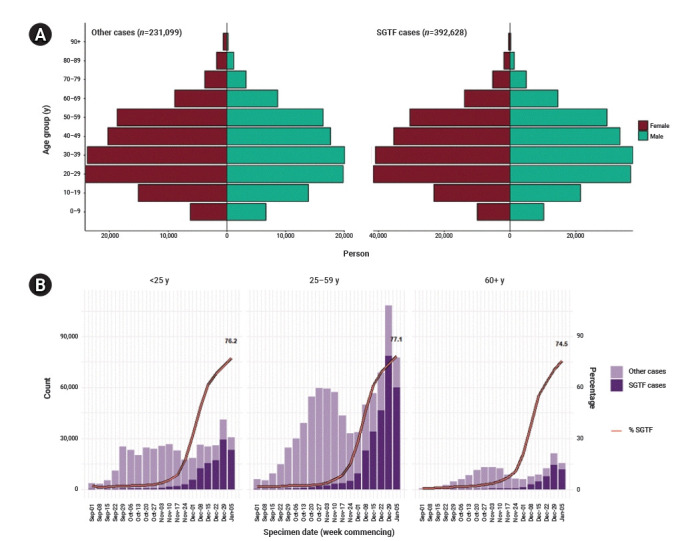
Sex and age distribution of S gene target failure (SGTF) in England. (A) Sex-age pyramid of non-SGTF group (left) and SGTF group (right) from December 1, 2020 to January 11, 2021 (extracted from Chand et al. [7]) (brown, female; green, male). (B) Weekly number and proportion of SGTF cases tested in TaqPath Labs in England by age groups (extracted from Chand et al. [7]).
Epidemiological Characteristics of B.1.1.7
The preliminary results of a case-case comparison study reported epidemiological characteristics including the hospitalization and fatality rates [10]. In total, 1,769 cases of the B.1.1.7 variant were identified from September 20 to December 15, 2020 based on the specimen date. The cases were 1:1 frequency-matched to wild-type (non-SGTF) cases by sex, age group, upper tier local authority of residence, and specimen date (2-week period).
The median age of patients was about 36 years old, with a sex distribution of 48.6% male and 51.4% female (Table 1). The proportion of Asians was slightly higher in the wild-type group. Compared to the wild-type group, the B.1.1.7 variant group showed a lower hospitalization rate (0.9% vs. 1.5%), a higher case fatality rate (0.89% vs. 0.73%; odds ratio, 1.21), and a lower re-infection rate (1.13 vs. 1.70 per 1,000), but all results were statistically non-significant [10]. However, from an updated result published on January 21, 2021 in a SAGE Meeting Paper, the relative risk of death within 28 days after having a polymerase chain reaction (PCR) test was 1.65 in SGTF cases versus non-SGTF cases (95% confidence interval [CI], 1.21−2.25) [17].
Table 1.
Epidemiologic characteristics of 3,538 VOC and non-VOC cases in England
| Characteristic | VOC group (n=1,769) | Wild type group (n=1,769) | p |
|---|---|---|---|
| Median age (y) | 36 | 35 | NR |
| Sex (%) | NR | ||
| Male | 48.6 | 48.6 | |
| Female | 51.4 | 51.4 | |
| Race (%) | NR | ||
| White | 75.2 | Proportion of White and Black similar to VOC group | |
| Black | 5.9 | ||
| Asian | 10.1 | 13.5 | |
| Residential typea) (%) | 63.5 | 56.1 | <0.01 |
| Hospitalization rate (case, %) | 16 (0.9) | 26 (1.5) | 0.16 |
| Case fatality rate (case, %)b) | 12/1,340 (0.89) | 10/1,360 (0.73) | 0.65 |
| Reinfection rate (/1,000)c) | 1.13 | 1.70 | 1.00 |
Organized from Chand et al. [10].
VOC, variant of concern; PHE, Public Health England; NR, not reported.
Resides in residential cluster.
Men and women who have passed 28 days since the last date of sample collection (n=2,700). In another report discussed at the SAGE meeting of PHE, United Kingdom on January 21, 2021 [16], it was updated to 104 deaths in non-S gene target failure (SGTF) cases (0.2%) and 65 deaths in SGTF cases (0.1%) with a risk ratio of 1.65 (95% confidence interval, 1.21−2.25).
Those with positive results of polymerase chain reaction (PCR) within 90 days of the last date of positive PCR.
The secondary attack rate was analyzed from the contact tracing data in the NHS Test and Trace in the UK [9]. In total, 956,519 contacts were reported to the NHS Test and Trace from November 30 to December 20, 2020. To examine the attack rate by B.1.1.7 status or SGTF positivity, the attack rate was calculated by test type (whole-genome scanning or PCR by TaqPath) (Table 2) [9]. The attack rate of the B.1.1.7 group was 14.7% and that of the SGTF group was 14.9%; these rates were higher than those of the non-B.1.1.7 group (11.0%) and the non-SGTF group (11.0%). The average attack rate of total cases and contacts was 12.7%.
Table 2.
Attack rate of severe acute respiratory syndrome coronavirus 2 in United Kingdom grouped by test type and the test results
| Test type | Contacts of the group | Contacts become cases | Attack rate (%) |
|---|---|---|---|
| Whole genome sequencing | 956,519 | 121,072 | 12.7 |
| VOC | 9,228 | 1,361 | 14.7 |
| Non-VOC | 11,269 | 1,244 | 11.0 |
| No sequence data | 936,022 | 118,467 | 12.7 |
| TaqPath assay | 956,519 | 12,1072 | 12.7 |
| SGTF (S−) | 262,769 | 39,277 | 14.9 |
| Non-SGTF (S+) | 262,232 | 28,770 | 11.0 |
| No SGTF data | 431,518 | 53,025 | 12.3 |
Organized from Chand et al. [9].
VOC, variant of concern; SGTF, S gene target failure.
Transmissibility of B.1.1.7
Volz et al. assessed the associations of the B.1.1.7 variant with the SARS-CoV-2 reproduction number [12,18]. To summarize the results from regression models (fixed effect, random effect, Bayesian), the additive effect size of time-varying reproduction number (Rt) of the B.1.1.7 variant was between 0.36 to 0.68 compared to the non-B.1.1.7 group. In detail, the Rt was estimated to be 0.48 to 0.68 higher in B.1.1.7 variant cases and 0.36 to 0.52 higher in SGTF cases compared to other variants. The mean Rt difference was 0.51 (95% CI, −0.09 to 1.10) and the mean ratio of the Rt of B.1.1.7 and non-B.1.1.7 groups was 1.56 (95% CI, 0.92−2.28), as estimated using SGTF frequencies in data from November 1 to December 12, 2020 [12].
Increasing Trend of B.1.1.7 by Continents
Since the first week when B.1.1.7 was reported in the UK and each continent, the days taken to reach a 20% positive rate among the sequences submitted to GISAID was 47 days in the UK (Fig. 4A), 59 days in Oceania, 83 days in Europe (excluding the UK), and 89 days in North America (Fig. 4B−D). The most rapid increase to 50% since the week of the first report was found in the UK, followed by Europe (excluding the UK), North America, and Oceania, where it took 84 days, 116 days, 125 days, and 141 days, respectively. The global average was 178 days for B.1.1.7 to reach 50% among the reported sequences to GISAID (Fig. 4F). In Asia, the growth rate of B.1.1.7 has been slower than the global average: it took 140 days to reach a 20% positive rate of B.1.1.7, and it has not yet reached 50% as of May 12, 2021 (Fig. 4E).
Fig. 4.
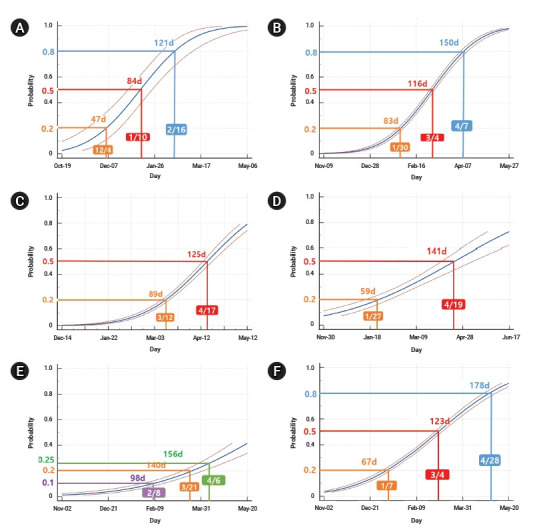
Dose-response plot of probability of B.1.1.7 positive rate in United Kingdom (UK) and in several continents from GISAID [16] (extracted on May 11 and May 12, 2021). (A) UK, (B) Europe except UK, (C) North America, (D) Oceania, (E) Asia, (F) global.
Discussion
It was found that B.1.1.7 emerged in September 2020 in southeast England, and its prevalence dramatically increased through December 2020. More than half of COVID-19 cases throughout England and more than 75% in east England and Greater London were estimated to be B.1.1.7 as of December 15, 2020. The reproduction number was estimated to be 1.57-fold higher in the B.1.1.7 group compared to the non-VOC group. Transmissibility and fatality rates were 70% [12] and 65% [17] higher in the B.1.1.7 group than in the non-B.1.1.7 group, respectively. The secondary attack rate of the B.1.1.7 variant was also higher (14.7%) than that of the non-B.1.1.7 group (11.0%) [9].
In the PHE reports, SGTF was used as a proxy for 501Y.V1. Since SGTF is a result of 69/70 deletion, which has arisen multiple times through the global SARS-CoV-2 pandemic, SGTF testing detects any variant containing the 69/70 deletion, including but not necessarily limited to the B.1.1.7 variant. The PHE recognized this concern and examined the weekly percentage of B.1.1.7 in all cases of 69/70 deletion detected through pillar 2, and found that it was 80% since November 9, over 90% since November 23, and over 98% since December 7, and was therefore confident in applying SGTF as a proxy for the B.1.1.7 variant [9].
SARS-CoV-2 is a rapidly-evolving RNA virus that is suggested to have 1 to 2 mutations per month [19]. The first widely distributed mutation was the spike mutation D614G, which first emerged from China in late January or early February 2020 and rapidly spread geographically and temporally, and became the dominant genotype globally in May 2020 [4]. The D614G mutation is expected not to be associated with a greater severity, but to be associated with a higher viral load and to affect younger people; thus, it may have contributed to the second wave in Korea, Europe, and worldwide. A distinct characteristic of B.1.1.7 compared to D614G is that the proportion of B.1.1.7 cases has been consistent across age groups, although the number of affected individuals was highest in the age group of 25 to 49 years old in the UK.
Just a few days after B.1.1.7 was reported, a similar variant (including N501Y, but not Δ69−70) was reported from South Africa in samples from October 2020 [20] (named the B.1.351 lineage, GH, or 20C/501Y.V2), followed by reports from Botswana and Zambia [21,22]. As of January 25, 2021, the World Health Organization (WHO) reported that B.1.1.7 had been detected in 70 countries in all 6 WHO regions and 501Y.V2 in 31 countries [23]. A few days later, another emerging variant from Brazil was reported from travelers entering Japan (named the P.1 variant, GR, B.1.1.28., 20B, or E484K) [5]. This variant includes mutations of N501Y, E484K, K417T, and a deletion in ORF1b (del11288-11296) in the spike protein. In addition to the P.1 variant, a distinct but similar variant was reported in Brazil (P.2, B.1.1.28., E484K mutation), expanding to 8 countries including Japan, Korea, and Singapore in Asia as of January 25, 2021 [23].
Distinct from B.1.1.7, the B.1.1.351 and P.1 variants can escape the neutralizing antibody responses of plasma antibody treatment, thereby reducing the efficacy of S-protein-based vaccine-induced antibodies, which is alarming [24]. The effectiveness of the Pfizer-BioNTech vaccine was 86.5% for B.1.1.7, and 75.0% for B.1.1.351 for those who received at least 1 dose of the vaccine in Qatar [25]. The efficiency of the AstraZeneca vaccine was 72.3% against all symptomatic cases, 70.4% against B.1.1.7, and 81.5% against other variants in a phase 2/3 vaccine efficacy study conducted in the UK [26]. As a summary of the current knowledge, the developed vaccines for COVID-19 are effective for reducing symptomatic cases of B.1.1.7. In contrast, vaccine efficiency has been reported to be much lower for B.1.351; the efficacy of the AstraZeneca vaccine was 21.9% in all cases but 10.4% in B.1.1351 cases in a phase 1b-2 clinical trial conducted in South Africa [27].
The first official report of the B.1.1.7 variant in Korea was 3 cases in a family, all of whom entered Korea from the UK on December 22 [28]. As of January 29, 2021, 23 cases of B.1.1.7, 6 cases of 501.V2, and 5 cases of the P.1 or P.2 variants had been reported in Korea. Among 23 cases of B.1.1.7, except 4 cases of family members who contacted the case at the airport or at home, all of the cases were people who entered from abroad. Cases of variants have increased since December 2020 in Korea, and cumulatively 904 cases of B.1.1.7, 111 cases of 501.V2, and 11 cases of P.1 or P.2 variants were reported among 9,977 genetically analyzed cases as of May 18, 2021 [29]. In the second week of May 2021 (from May 9 to May 15, 2021), the number of detected cases of B.1.1.7 was 199 among 921 sequence-analyzed cases (21.6%). Unlike the variant cases in January which were mostly detected at entry or during quarantine after entry to Korea from abroad, many of the recent B.1.1.7 cases involved transmission within Korea and surpassed cases from abroad: 592 cases (65.5%) were domestic and 312 (34.5%) cases were from abroad out of the 904 cases of B.1.1.7 as of May 18. Meanwhile, the reproduction number in Korea increased from 0.82−0.96 in January 2021, to 0.94−1.04 in May 2021 [30]. This increment is less than the expected increase suggested by Volz et al. [12,18], which can be attributed to the strengthened control during quarantine and within Korea, including identification of a broader range of contacts of variant cases compared to non-variant cases. However, the types and number of cases of SARS-CoV-2 variants are increasing in Korea, and close monitoring must be continued.
As of May 26, 2021, it is reported that B.1.1.7 has spread to 151 countries, B.1.1351 to 106 countries, and P.1 to 61 countries, including verified and unverified cases [31]. Ongoing research is exploring the transmissibility and epidemiological characteristics of SARS-CoV-2 variants, as well as their associations with vaccines and treatment. Even though the characteristics of SARS-CoV-2 variants are not clearly understood, it is well-known that non-pharmaceutical interventions such as social distancing and wearing masks are effective for reducing the transmissibility of SARS-CoV-2.
Footnotes
Ethics Approval
Not applicable.
Conflicts of Interest
The authors have no conflicts of interest to declare.
Funding
None.
Availability of Data
All data generated or analysed during this study are included or cited in this published article. For other data, these may be requested through the corresponding author.
Authors’ Contributions
Conceptualization: YK, DK, SWL; Data review and analysis: YK, DK, EJK; Writing–original draft: YK. Writing–review & editing: DK, SWL, EJK.
References
- 1. World Health Organization (WHO). WHO coronavirus (COVID-19) dashboard [Internet]. Geneva: WHO; 2020 [cited 2021 May 23]. Available from: https://covid19.who.int.
- 2.Baric RS. Emergence of a highly fit SARS-CoV-2 variant. N Engl J Med. 2020;383:2684–6. doi: 10.1056/NEJMcibr2032888. [DOI] [PubMed] [Google Scholar]
- 3.Kemp SA, Datir RP, Collier DA, et al. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70 [Preprint] doi: 10.1101/2020.12.14.422555. Posted 2020 Dec 21. bioRxiv 2020.12.14.422555. [DOI] [Google Scholar]
- 4.World Health Organization (WHO) Geneva: WHO; 2020. Disease outbreak news: SARS-CoV-2 variants [Internet] World Health Organization (WHO). Disease outbreak news: SARS-CoV-2 variants [Internet]. Geneva: WHO; 2020 [2020 Dec 31]. Available from: https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/ [Google Scholar]
- 5. Reuters. Japan finds new coronavirus variant in travellers from Brazil [Internet]. Reuters; 2021 Jan 10 [cited 2021 Jan 10]. Available from: https://www.reuters.com/article/us-health-coronavirus-japan-variant-idUSKBN29F08R.
- 6. Gallagher J. New coronavirus variant: what do we know? [Internet]. BBC; 2020 Dec 20 [cited 2020 Dec 20]. Available from: https://www.bbc.com/news/health-55388846.
- 7. Chand M, Hopkins S, Achison C, et al. Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01, technical briefing 4 [Internet]. London: Public Health England; 2021 [cited 2021 Jan 15]. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959359/Variant_of_Concern_VOC_202012_01_Technical_Briefing_4.pdf.
- 8. Chand M, Hopkins S, Dabrera G, et al. Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01, technical briefing 1 [Internet]. London: Public Health England; 2020 [cited 2021 Jan 5]. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdf.
- 9. Chand M, Hopkins S, Dabrera G, et al. Investigation of novel SARS-CoV-2 variant: variant of concern 202012/01, technical briefing 3 [Internet]. London: Public Health England; 2021 [cited 2021 Jan 15]. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959360/Variant_of_Concern_VOC_202012_01_Technical_Briefing_3.pdf.
- 10. Chand M, Hopkins S, Dabrera G, et al. Investigation of novel SARS-CoV-2 variant [Internet]. London: Public Health England; 2020 [cited 2021 Jan 5]. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959361/Technical_Briefing_VOC202012-2_Briefing_2.pdf.
- 11. Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations [Internet]. Virological; 2020 Dec 20 [cited 2021 Jan 5]. Available from: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 12.Volz E, Mishra S, Chand M, et al. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data [Preprint] doi: 10.1101/2020.12.30.20249034. Posted 2020 Dec 31. medRxiv 2020.12.30.20249034. [DOI] [Google Scholar]
- 13.Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rambaut A, Holmes EC, O'Toole A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–7. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. GISAID. Tracking of variants [Internet]. GISAID; 2020 [cited 2021 May 11]. Available from: https://www.gisaid.org/hcov19-variants/
- 17. Horby P, Huntley C, Davies N, et al. NERVTAG note on B. 1.1. 7 severity: presented to SAGE on 21/1/21 [Internet]. [cited 2021 Jan 21]. Available from: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/961037/NERVTAG_note_on_B.1.1.7_severity_for_SAGE_77__1_.pdf.
- 18.Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duchene S, Featherstone L, Haritopoulou-Sinanidou M, et al. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6:veaa061. doi: 10.1093/ve/veaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa [Preprint] doi: 10.1101/2020.12.21.20248640. Posted 2020 Dec 22. medRxiv 2020.12.21.20248640. [DOI] [Google Scholar]
- 21. World Health Organization (WHO). Seven things to know about COVID-19 variants in Africa [Internet]. Brazzaville: WHO Africa; 2021 Jan 22 [cited 2021 Jan 22]. Available from: https://www.afro.who.int/news/seven-things-know-about-covid-19-variants-africa.
- 22. Centers for Disease Control and Prevention (CDC). Science brief: emerging SARS-CoV-2 variants [Internet]. Atlanta: CDC; 2021 [cited 2021 Jan 28]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html. [PubMed]
- 23. World Health Organization (WHO). COVID-19 weekly epidemiological update, 27 January 2021 [Internet]. Geneva: WHO; 2021 [cited 2021 Jan 28]. Available from: https://apps.who.int/iris/handle/10665/339547.
- 24. Reuters. New COVID-19 variant defeats plasma treatment, may reduce vaccine efficacy [Internet]. Reuters; 2021 Jan 21 [cited 2021 Jan 28]. Available from: https://www.reuters.com/article/us-health-coronavirus-safrica-variant-idUSKBN29P2HJ. [Google Scholar]
- 25.Abu-Raddad LJ, Chemaitelly H, Butt AA, et al. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med. 2021 doi: 10.1056/NEJMc2104974. 2021 May 5 [Epub]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emary KR, Golubchik T, Aley PK, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–62. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhi SA, Baillie V, Cutland CL, et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1885–98. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korea Centers for Disease Control and Prevention (KCDC). COVID-19 in Korea: current status [Internet]. Cheongju (KR): KCDC; 2020 Dec 28 [cited 2021 May 24]. Available from: http://www.kdca.go.kr/board/board.es?mid=a20501020000&bid=0015&list_no=711602&cg_code=C01&act=view&nPage=42.
- 29. Korea Centers for Disease Control and Prevention (KCDC). COVID-19 in Korea: current status [Internet]. Cheongju (KR): KCDC; 2021 May 18 [cited 2021 May 24]. Available from: http://www.kdca.go.kr/board/board.es?mid=a20501020000&bid=0015&list_no=713349&cg_code=C01&act=view&nPage=3.
- 30. Korea Centers for Disease Control and Prevention (KCDC). COVID-19 in Korea: current status [Internet]. Cheongju (KR): KCDC; 2021 May 24 [cited 2021 May 24]. Available from: http://www.kdca.go.kr/board/board.es?mid=a20501020000&bid=0015&list_no=713398&cg_code=C01&act=view&nPage=1.
- 31. Centers for Disease Control and Prevention (CDC). Global variants report [Internet]. CDC; 2020 [cited 2021 May 24]. Available from: https://covid.cdc.gov/covid-data-tracker/#global-variant-report-map. [Google Scholar]