Abstract
Non‐small‐cell lung cancer (NSCLC) remains a huge obstacle to human health. Certain circular RNAs endow with crucial regulatory roles in NSCLC progression. Here, we investigated the functional effects of circ_0001821 on cellular behaviors of NSCLC cells and explored the possible mechanism. The expression of circ_0001821, microRNA (miR)‐526b‐5p, and G protein‐coupled receptor kinase 5 (GRK5) was determined by quantitative real‐time polymerase chain reaction or Western blot assay. Clonogenicity in NSCLC cells was detected via colony formation assay. Cell migration and invasion were monitored by Transwell assay. Cell sensitivity to paclitaxel (TAX) evaluated by Cell Counting Kit‐8 assay. Cell apoptosis was assessed by flow cytometry, caspase‐3 activity, and caspase‐9 activity. The targeted relationship between miR‐526b‐5p and circ_0001821 or GRK5 was confirmed by dual‐luciferase reporter or RNA pull‐down assay. Moreover, the role of circ_0001821 in vivo was examined by xenograft model assay. The results presented that the expression of circ_0001821 and GRK5 was increased, while miR‐526b‐5p expression was decreased in NSCLC tissues and cells. Circ_0001821 knockdown reduced colony formation ability and metastasis ability but enhanced TAX sensibility and apoptosis of NSCLC cells, which was attenuated by miR‐526b‐5p inhibition or GRK5 overexpression. Circ_0001821 targeted miR‐526b‐5p, and miR‐526b‐5p targeted GRK5. Circ_0001821 could upregulate GRK5 expression by sponging miR‐526b‐5p. Depletion of circ_0001821 also blocked tumor growth in vivo. In conclusion, the depletion of circ_0001821 inhibited NSCLC progression, at least in part, by modulating the miR‐526b‐5p/GRK5 axis.
Keywords: circ_0001821, GRK5, miR‐526b‐5p, NSCLC, TAX resistance, TAX sensibility
A diagram illustrated the summary of all findings in this study.

Abbreviations
- CCK‐8
Cell Counting Kit‐8
- circRNAs
certain circular RNAs
- GRK5
G protein‐coupled receptor kinase 5
- miR
microRNA
- NSCLC
non‐small‐cell lung cancer
- qRT‐PCR
quantitative real‐time polymerase chain reaction
1. INTRODUCTION
As one of the most frequently diagnosed malignancies and primary causes of cancer‐related death, non‐small‐cell lung cancer (NSCLC) consists of lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), and large‐cell lung carcinoma (LULC) and occupies about 85% of lung cancers all over the world. 1 During the past two decades, targeted therapies and immunotherapies for NSCLC have developed a lot and benefited much to patients. However, overall survival rate of advanced or metastatic NSCLC remains not optimistic. 2 Paclitaxel (TAX) is an efficient antitumor drug and affects cell cycle and autophagy of NSCLC cells. 3 , 4 In addition, the occurrence of chemoresistance is a critical barrier undermining the curative effects of NSCLC patients. 5 Hence, clarifying the mechanisms underlying NSCLC metastasis and TAX resistance is urgently needed.
Circular RNAs (circRNAs) are a group of noncoding RNA molecules, harboring special covalently closed continuous structure and numerous potential functions in human diseases. 6 Mounting evidence suggests that circRNAs have close association with tumor development and serve as diagnostic markers and therapeutic targets. 7 For example, circ‐ZKSCAN1 (hsa_circ_0001727) was reported to be an important positive regulator of NSCLC development. 8 Enforced expression of circ_0002483 could suppress proliferation and invasion but promote TAX sensibility of NSCLC cells. 9 High expression of circ_0074027 or circRNA ARHGAP10 indicates an unfavorable prognosis of NSCLC patients. 10 , 11 Located at chr8:128902834–128903244, hsa_circ_0001821 (also named as hsa_circ_000006) was reported to be upregulated in both LUAD and LUSC tissues. 12 However, the role of circ_0001821 in NSCLC progression is still uncertain.
MicroRNAs (miRNAs) are short noncoding RNAs, with only about 22 nucleotides in length, and play vital roles in regulating gene expression posttranscriptionally. 13 Functional miRNAs are generally dysregulated in NSCLC, acting tumor‐suppressive or tumor‐promotive roles, thus affecting NSCLC development. 14 MiRNAs are also involved in regulating drug resistance in NSCLC. 15 Previous research reported the tumor‐suppressive property of miR‐526b in hepatocellular carcinoma. 16 The suppressive effects of miR‐526b on breast cancer cell growth and metastasis were detected, highlighting its potential to be a therapeutic target of breast cancer. 17 In NSCLC, miR‐526b was manifested to suppress cell proliferation and tumor growth. 18 Here, we found that circ_0001821 harbor binding sites with miR‐526b‐5p, predicted by circinteractome. Whether miR‐526b‐5p involved in circ_0001821‐mediated NSCLC development needs to be investigated.
G protein‐coupled receptor kinase 5 (GRK5), a kind of serine/threonine kinase, could respond to intracellular diverse stimuli and regulate signaling pathways. Interestingly, it has dual roles in tumor growth. 19 In thyroid cancer, GRK5 inactivated G protein‐coupled receptors (GPCRs) to block tumor growth. 20 On the contrary, GRK5 promoted the progression of prostate cancer and renal cell carcinoma. 21 , 22 Additionally, the oncogenic role of GRK5 was also disclosed in NSCLC. 23 Apart from this, deficiency of GRK5 expression was reported to increase the sensibility of cervical cancer cells and breast cancer cells TAX. 24 starBase 3.0 predicted that GRK5 was a possible target of miR‐526b‐5p. We then explored the association of GRK5 with circ_0001821‐mediated NSCLC progression.
In the present study, the upregulation of circ_0001821 was identified in NSCLC tissues and cells. Its role in NSCLC development in vitro and in vivo was investigated, and the functional mechanism of circ_0001821 was further addressed to understand the development of NSCLC.
2. MATERIALS AND METHODS
2.1. Collection of clinical specimens
Prior to sample collection, we obtained the permission from the Ethics Committee of the Jingmen No. 2 People's Hospital. Sixty‐five NSCLC patients enrolled at Jingmen No. 2 People's Hospital from 2014 to 2015 were subjected to resection of tumor tissues and adjacent normal tissues after their submission of written informed consents. Later, these 65 patients were followed up from date of sample collection until death or ending date of this study.
2.2. Cell culture and transient transfection
Human bronchial epithelial cells 16HBE (CL‐0249; Procell) and NSCLC cells, including A549 (ATCC® CCL‐185; American Type Culture Collection), H3122 (CBP60133; Cobioer), H1975 (ATCC® CRL‐5908), and H2342 (ATCC® CRL‐5941) cells, were maintained in Dulbecco's Modified Eagle Medium (HyClone) containing 10% (v/v) fetal bovine serum (Beyotime) and 1% penicillin/streptomycin (Transgen) at 37℃ in a humidified atmosphere including 5% CO2 and 95% air.
Short hairpin RNA (shRNA) specially targeting circ_0001821 (sh‐circ) was introduced into A549 and H3122 cells to silence circ_0001821, with sh‐NC as negative control. To upregulate or downregulate miR‐526b‐5p, miR‐526b‐5p mimic (miR‐526b‐5p) or miR‐526b‐5p inhibitor (anti‐miR‐526b‐5p) was transfected into A549 and H3122 cells, with miR‐NC or anti‐NC as negative control. To overexpress GRK5, its overexpression plasmid (GRK5) was introduced into A549 and H3122 cells, with pcDNA as negative control. Above reagents were supplied by RIBOBIO Co. Ltd. Transfection assay was conducted by employing Lipofectamine 3000 (Invitrogen) for 48 h.
2.3. qRT‐PCR assay
To isolate total RNA, clinical specimens or cells were mixed with TRIzol Reagent (Invitrogen), followed by RNA detection with NanoDrop ND‐1000 spectrophotometer (Thermo Fisher Scientific). Then, RNA was subjected to reverse transcription into complementary DNA (cDNA) using RevertAid RT Reverse Transcription kit (Thermo Fisher Scientific) or TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) and quantitative real‐time polymerase chain reaction (qRT‐PCR) assay with GoScript Reverse Transcription System (Promega) or TaqMan MicroRNA Assays (Thermo Fisher Scientific). Relative expression of circ_0001821, miR‐526b‐5p, and GRK5 was assessed by 2−ΔΔCt method, and β‐actin (for circ_0001821 and GRK5) or U6 (for miR‐526b‐5p) served as internal control. All primers involved in qRT‐PCR assay were listed in Table 1.
TABLE 1.
Primers used for qPCR assay
| Gene | Forward primer (5′ →3′) | Reverse primer (5′ →3′) |
|---|---|---|
| circ_0001821 | CCCCGACTCTTCCTGGTGAA | CAGGCACAGCCATCTTGAGG |
| miR‐526b‐5p | CTCTTGAGGGAAGCACT | GAACATGTCTGCGTATCTC |
| GRK5 | GCCACATTAGGATCTCAGACCTG | GTACCTCTGGTTGTTCAGGACC |
| β‐Actin | CAAGAGATGGCCACGGCTGCT | TCCTTCTGCATCCTGTCGGCA |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
2.4. Colony formation assay
After transfection, about 500 A549 or H3122 cells were plated into six‐well plates and maintained at routine condition for 15 days. After discarding medium, generated cell colonies were rinsed with PBS, fixed with 4% paraformaldehyde, stained with crystal violet reagent (Beyotime), and counted using Image J software (NIH).
2.5. Transwell assay
The current assay was employed to evaluate tumor cell metastasis. For migration analysis, transfected A549 and H3122 cells (1 × 105) were harvested and resuspended into the top chambers (8‐μm pores; BD Biosciences) containing medium without serum. Meanwhile, the lower ones were added with complete medium. At 24 h post routine culture, cells migrated through the polycarbonic membrane were fixed with paraformaldehyde and stained with crystal violet solution, then counted under an inverted microscope in five randomly chosen fields. As for invasion detection, transfected NSCLC cells (4 × 105) were inoculated into the top chambers enveloped with Matrigel (BD Biosciences), the rest procedures were same to migration analysis.
2.6. CCK‐8 assay
To assess the sensibility of NSCLC cells to paclitaxel (TAX), Cell Counting Kit‐8 (CCK‐8) assay was performed. In brief, transfected A549 and H3122 cells were seeded into 96‐well plates and administered with TAX at different concentrations (0, 2.5, 5, 10, 20, 40, and 80 nM). After 48‐h treatment, 10‐μl CCK‐8 solution (Beyotime) was added and incubated for 2 h. Subsequently, a microplate reader (Bio‐Rad) was utilized to measure the absorbance at 450 nm to draw survival curves, followed by the computation of 50% inhibitory concentration (IC50).
2.7. Determination of cell apoptosis
After transfection, A549 and H3122 cells were disposed by TAX at different concentrations for 48 h (9.37 nM for transfected A549 cells and 7.44 nM for transfected H3122 cells). Flow cytometry assay was applied to monitor cell apoptosis exploiting an Annexin V‐fluorescein isothiocyanate (FITC) Apoptosis Detection kit (BD Biosciences). Treated cells were harvested and double stained with Annexin V‐FITC and propidium iodide (PI) in the absence of light. Apoptotic A549 and H3122 cells were determined using a flow cytometer (BD Biosciences) and analyzed Cell Quest 6.0 software. Apoptotic rate indicates the percentage of cells at quadrant Annexin V‐FITC + and PI − or +.
The activity of caspase‐3 and caspase‐9 was measured utilizing a commercial kit (ab219915; Abcam). According to the protocol booklet supplied by the manufacturer, treated A549 and H3122 cells inoculated into 96‐well plates were incubated with 10× test compounds in PBS for 4 h; then, loading solution (mixture of Assay Buffer and caspase‐3 substrate or caspase‐9 substrate) was added to react for 1 h in the dark. Then, the caspase activity was evaluated by monitoring the fluorescence increase (Ex/Em =535/620 nm [red] for caspase‐3 and Ex/Em =370/450 nm [blue] for caspase‐9) using a fluorescence microplate reader.
2.8. Targeted relationship prediction and validation
The potential target genes of circ_0001821 and miR‐526b‐5p were forecasted by circinteractome (https://circinteractome.nia.nih.gov), circBank (http://www.circbank.cn/), and starBase 3.0 (http://starbase.sysu.edu.cn/), respectively.
For dual‐luciferase reporter assay, fragmentary sequence of circ_0001821 or GRK5 3′‐untranslated region (3′UTR) that harboring the binding position with miR‐526b‐5p was inserted into the psiCHECK‐2 vector (Promega Corp.) to synthesize wide‐type luciferase reporter plasmids, circ_0001821 WT and GRK5 WT. Meanwhile, mutant‐type ones (circ_0001821 MUT and GRK5 MUT) were established after mutating the binding sits to the complementary sequences using GENEART Site‐Directed Mutagenesis System (Thermo Fisher Scientific). Generated luciferase reporter plasmid was cotransfected into A549 and H3122 cells with miR‐NC or miR‐526b‐5p using Lipofectamine 3000. Forty‐eight hours later, luciferase density was examined by Dual‐Luciferase Reporter detection System (Promega Corp.).
RNA pull‐down assay was also employed to verify the targeted relationship between circ_0001821 and miR‐526b‐5p. Initially, A549 cells were transfected with Biotin‐labeled miRNAs (50 nM, RIBOBIO Co. Ltd.) using Lipofectamine 3000, including biotinylated miR‐526b‐5p (Bio‐miR‐526b‐5p‐WT), its mutant (Bio‐miR‐526b‐5p‐MUT, binding sites were mutated to the complementary sequences), and negative control (Bio‐miR‐NC). Cells were collected after 48 h and lysed in lysis buffer (Ambion). Generated cell lysate was subjected to incubation with M‐280 streptavidin magnetic beads (Invitrogen) that pre‐enveloped with RNase‐free bovine serum albumin and yeast tRNA. At last, bound RNA was purified exploiting TRIzol Reagent for qRT‐PCR assay.
2.9. Western blot assay
The present assay was used to detect the protein level of GRK5, with β‐actin as loading control. 16HBE, H1975, or H2342 cells were lysed in RIPA buffer (Beyotime) to extract protein samples, followed by quantification utilizing a bicinchoninic acid assay (BCA) kit. Then, 40‐μg samples were subjected to separation through sodium dodecyl sulfate (SDS)‐polyacrylamide gels (PAGE) and transfer onto polyvinylidene difluoride membranes (Thermo Fisher Scientific). The membranes were then blocked in skim milk, incubated with primary antibody against GRK5 (ab64943; Abcam) or β‐actin (ab8227; Abcam) then incubated with secondary antibody (ab205718; Abcam). Finally, protein signals were activated by feat of a chemiluminescence assay kit (Thermo Fisher Scientific) and analyzed exploiting Quantity One software.
2.10. Xenograft mode in nude mice
Experiments conducted in animals were approved by the Ethics Committee of the Jingmen No. 2 People's Hospital. BALB/c nude mice (4–5 weeks old, male) purchased from Shanghai SLAC Laboratory Animal Co., Ltd. were subcutaneously inoculated with A549 cells (2 × 106) stably transfected with sh‐NC or sh‐circ, which were raised at specific pathogen‐free (SPF) condition supplemented with enough food and water. Additionally, the mice were intraperitoneally injected with DMSO (5 mg/kg) or TAX on at 1, 8, 15, and 22 days post inoculation (n = 5). Seven days after inoculation, size of formed tumors was recorded once a week and calculated using the formula: 5 × length × width2. Twenty‐eight days later, all mice were killed to resect tumors, for weight, qRT‐PCR assay and Western blot analysis. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
2.11. Statistical analysis
All data were generated form triplicate experiments, processed using GraphPad Prism 7 (GraphPad Inc.) and exhibited as mean ± standard deviation (SD). Overall survival rate of NSCLC patients was analyzed exploiting the Kaplan–Meier method with log‐rank test. Difference analyses were implemented by Student's t test or one‐way analysis of variance followed by Tukey's test. A p value less than .05 was identified to be significantly different.
3. RESULTS
3.1. Circ_0001821 was upregulated in NSCLC tissues and cells
In order to explore the functional role of circ_0001821 in NSCLC progression, we firstly detected the expression of circ_0001821 in NSCLC tissues and matched normal tissues. Data from qRT‐PCR assay showed that circ_0001821 expression in NSCLC tissues (n = 65) was higher than that in normal tissues (n = 65) (Figure 1A). After sample collection, all of these 65 NSCLC patients were followed up. According to the median expression value of circ_0001821 in NSCLC tissues, 65 NSCLC patients were divided into circ_0001821 high group (n = 32) and circ_0001821 low (n = 33) group. Results revealed that patients in circ_0001821 high group exhibited lower overall survival rate (p = .02) (Figure 1B). As expect, circ_0001821 was highly expressed in NSCLC cells (A549, H3122, H1975, and H2342 cells) relative to 16HBE cells (Figure 1C). Taken together, circ_0001821 was upregulated in NSCLC tissues and cells.
FIGURE 1.

Circ_0001821 was upregulated in non‐small‐cell lung cancer (NSCLC) tissues and cells. (A) Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay for the relative expression of circ_0001821 in NSCLC tissues (n = 65) and normal tissues (n = 65). (B) Overall survival rate of NSCLC patients with high or low circ_0001821 expression. (C) qRT‐PCR assay for the relative expression of circ_0001821 in 16HBE, A549, H3122, H1975, and H2342 cells. **p < .01
3.2. Circ_0001821 knockdown inhibited growth and metastasis but facilitated TAX sensibility and apoptosis of NSCLC cells
In view of the dysregulation of circ_0001821 in NSCLC tissues and cells, we performed loss‐of‐function assay to assess the influence of circ_0001821 on the behaviors of NSCLC cells. As illustrated in Figure 2A, circ_0001821 expression in A549 and H3122 cells exhibited a remarkable reduction due to transfection with sh‐circ, sh‐NC serving as negative control. Colony formation assay disclosed that depletion of circ_0001821 efficiently inhibited clonogenicity in NSCLC cells (Figure 2B). Circ_0001821 knockdown also repressed cell migrated and invasive abilities of NSCLC cells, as demonstrated by Transwell assay (Figure 2C and D). Following CCK‐8 assay was conducted to examine the TAX sensibility of NSCLC cells. The data suggested that circ_0001821 knockdown significantly reduced the IC50 of A549 cells to TAX (16.88) in contrast to cells transfected with sh‐NC (9.37). Likewise, IC50 of H3122 cells with circ_0001821 knockdown (12.24) was lower than that of cells transfected with sh‐NC (7.44). In other words, A549 and H3122 cells with circ_0001821 knockdown had an elevated TAX sensibility compared with cells transfected with sh‐NC (Figure 2E and F). Flow cytometry witnessed the circ_0001821 knockdown‐induced enhanced apoptosis in NSCLC cells (Figure 2G). Caspase family is the key regulator of apoptosis, including extracellular effector caspase‐3 and intracellular initiator caspase‐9. 25 Here, commercial kits were used to measure the activities of caspase‐3 and caspase‐9. As shown in Figure 2H, deficiency of circ_0001821 expression apparently improved caspase‐3 activity and caspase‐9 activity in NSCLC cells. Above results indicated that circ_0001821 knockdown inhibited NSCLC cell growth and metastasis, while facilitated TAX sensibility and apoptosis.
FIGURE 2.

Circ_0001821 knockdown inhibited growth and metastasis, while facilitated TAX sensibility and apoptosis of non‐small‐cell lung cancer (NSCLC) cells. A549 and H3122 cells were transfected with sh‐NC or sh‐circ. (A) Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay for the relative expression of circ_0001821 in transfected cells. (B) Colony formation assay for the clonogenicity in transfected cells. (C and D) Transwell assay for the migrated and invasive abilities of transfected cells. (E and F) CCK‐8 assay for the cell viability of A549 cells and H3122 cells treated with 0, 2.5, 5, 10, 20, 40, and 80 nM TAX. (G and H) Transfected A549 cells were disposed with 9.37 nM TAX for 48 h, and transfected H3122 cells were disposed with 7.44 nM for 48 h. (G) Flow cytometry for the apoptotic rate of treated cells. (H) Measurement of caspase‐3 activity and caspase‐9 activity in treated cells. **p < .01
3.3. Circ_0001821 acted as a sponge of miR‐526b‐5p
Here, circinteractome, starBase, and circBank were utilized to search the possible miRNA directly interacted with circ_0001821. Venn diagram showed that only miR‐526b‐5p was predicted by all tools (Figure S1A). MiR‐526b‐5p was thus used as a candidate in this study, and the binding site between circ_0001821 and miR‐526b‐5p was shown in Figure 3A. A549 and H3122 cells with miR‐526b‐5p overexpression were successfully constructed, with miR‐NC as control (Figure 3B). To validate the prediction, dual‐luciferase reporter assay and RNA pull‐down assay were performed. As exhibited, gain of miR‐526b‐5p triggered about 60% reduction in the luciferase activity of A549 and H3122 cells cotransfected with circ_0001821 WT, while it had no significant effect on that of cells cotransfected with circ_0001821 MUT (Figure 3C and D). Results from RNA pull‐down assay indicated that circ_0001821 was pulled down by biotinylated miR‐526b‐5p (Bio‐miR‐526b‐5p‐WT), rather than the mutated oligo (Figure 3E). Then, qRT‐PCR assay was implemented to evaluate the expression of miR‐526b‐5p in NSCLC cells. We found that miR‐526b‐5p expression was declined in A549 and H3122 cells when compared with 16HBE cells (Figure 3F). And circ_0001821 knockdown remarkably increased miR‐526b‐5p expression in A549 and H3122 cells (Figure 3G). To sum up, circ_0001821 could target miR‐526b‐5p in NSCLC cells.
FIGURE 3.

Circ_0001821 acted as a sponge of miR‐526b‐5p. (A) The binding sites between circ_0001821 and miR‐526b‐5p predicted by circinteractome. (B) Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay for the relative expression of miR‐526b‐5p in A549 and H3122 cells transfected with miR‐NC or miR‐526b‐5p. (C and D) Dual‐luciferase reporter assay for the luciferase activity of A549 and H3122 cells cotransfected with miR‐NC or miR‐526b‐5p and circ_0001821 WT or circ_0001821 MUT. (E) qRT‐PCR and RNA pull‐down assays for the enrichment of circ_0001821 in A549 cells were transfected with Bio‐miR‐NC, Bio‐miR‐526b‐5p‐WT, or Bio‐miR‐526b‐5p‐MUT. (F) qRT‐PCR assay for the relative expression of miR‐526b‐5p in 16HBE, A549, and H3122 cells. (G) qRT‐PCR assay for the relative expression of miR‐526b‐5p in A549 and H3122 cells transfected with sh‐NC or sh‐circ. **p < .01
3.4. Inhibition of miR‐526b‐5p could attenuate the effects of circ_0001821 knockdown on NSCLC cells
Having known that circ_0001821 could target miR‐526b‐5p in NSCLC cells, we then explored the role of miR‐526b‐5p in circ_0001821 knockdown‐induced NSCLC cells. qRT‐PCR assay manifested that transfection with anti‐miR‐526b‐5p prominently decreased expression of miR‐526b‐5p in A549 and H3122 cells, with anti‐NC as control (Figure 4A). What is more, circ_0001821 knockdown induced the repressed clonogenicity (Figure 4B), cell migration, and invasion in NSCLC cells (Figure 4C and D), as well as elevated TAX sensibility (Figure 4E), apoptosis (Figure 4F), caspase‐3 activity (Figure 4G), and caspase‐9 activity (Figure 4H) in NSCLC cells were all attenuated by miR‐526b‐5p downregulation. Taken together, circ_0001821 knockdown inhibited growth and metastasis, while facilitated TAX sensibility and apoptosis of NSCLC cells by sponging miR‐526b‐5p.
FIGURE 4.

Interference of miR‐526b‐5p could attenuate the effects of circ_0001821 knockdown on non‐small‐cell lung cancer (NSCLC) cells. (A) Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay for the relative expression of miR‐526b‐5p in A549 and H3122 cells transfected with anti‐NC or anti‐miR‐526b‐5p. (B–H) A549 and H3122 cells were transfected with sh‐NC, sh‐circ, sh‐circ + anti‐NC, or sh‐circ + anti‐miR‐526b‐5p. (B) Colony formation assay for the clonogenicity in transfected cells. (C and D) Transwell assay for the migration and invasion of transfected cells. (E) CCK‐8 assay for the IC50 of transfected cells to TAX. (F–H) Transfected A549 cells were disposed with 9.37 nM TAX for 48 h, and transfected H3122 cells were disposed with 7.44 nM for 48 h. (F) Flow cytometry for the apoptotic rate of treated cells. (G and H) Measurement of caspase‐3 activity and caspase‐9 activity in treated cells. **p < .01
3.5. MiR‐526b‐5p targeted GRK5 in NSCLC cells
Subsequently, we further explored the mechanism by which miR‐526b‐5p working through in NSCLC cells. starBase was utilized to find the target genes of miR‐526b‐5p, and numerous genes were obtained. Then, we examined the expression of these genes in A549 and H3122 cells with miR‐526b‐5p enrichment, and we found that GRK5 expression was significantly depleted by miR‐526b‐5p enrichment (Figure S1B and C). The binding site between GRK5 3′UTR and miR‐526b‐5p was shown in Figure 5A. To verify above prediction, dual‐luciferase reporter assay was conducted. As shown in Figure 5B and C, luciferase activity of A549 and H3122 cells cotransfected with miR‐526b‐5p and GRK5 WT was obviously declined compared with cells cotransfected with miR‐NC and GRK5 WT, while luciferase activity of cells in GRK5 MUT group was changeless. The expression of GRK5 mRNA was markedly increased in tumor tissues compared with that in normal tissues (Figure S2A). Besides, high GRK5 expression was related with relatively higher overall survival of NSCLC patients within 5 years (Figure S2B). qRT‐PCR and Western blot assays uncovered the upregulation of GRK5 in NSCLC cells relative to 16HBE cells at mRNA (Figure 5D) and protein (Figure 5E) levels, respectively. We also observed that the interference of miR‐526b‐5p could upregulate GRK5 expression in NSCLC cells, while overexpressed miR‐526b‐5p evidently downregulated GRK5 expression (Figure 5F and G). In addition, circ_0001821 knockdown induced the downregulation of GRK5 expression in A549 and H3122 cells, which was almost reversed by the interference of miR‐526b‐5p (Figure 5H and I). Therefore, miR‐526b‐5p could target GRK5 in NSCLC cells.
FIGURE 5.
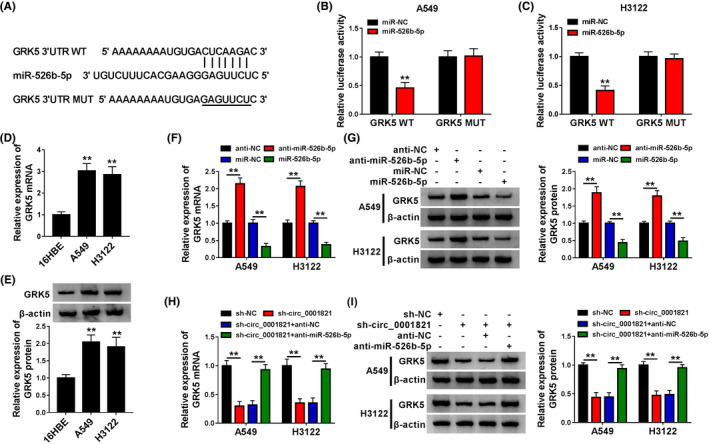
MiR‐526b‐5p targeted GRK5 in non‐small‐cell lung cancer (NSCLC) cells. (A) The binding sites between miR‐526b‐5p and GRK5 predicted by starBase. (B and C) Dual‐luciferase reporter assay for the luciferase activity of A549 and H3122 cells cotransfected with miR‐NC or miR‐526b‐5p and GRK5 WT or GRK5 MUT. (D and E) Quantitative real‐time polymerase chain reaction (qRT‐PCR) and Western blot assay for the relative expression of (D) GRK5 mRNA and (E) GRK5 protein in 16HBE, A549, and H3122 cells. (F and G) qRT‐PCR and Western blot assays for the relative expression of (F) GRK5 mRNA and (G) GRK5 protein in A549 and H3122 cells transfected with anti‐NC, anti‐miR‐526b‐5p, miR‐NC, or miR‐526b‐5p. (H and I) qRT‐PCR and Western blot assays for the relative expression of (H) GRK5 mRNA and (I) GRK5 protein in A549 and H3122 cells transfected with sh‐NC, sh‐circ, sh‐circ + anti‐NC, or sh‐circ + anti‐miR‐526b‐5p. **p < .01
3.6. Reintroduction of GRK5 attenuated the effects of circ_0001821 knockdown on NSCLC cells
Above findings, circ_0001821 could target miR‐526b‐5p and miR‐526b‐5p could target GRK5 in NSCLC cells, we further investigated the impact of GRK5 on circ_0001821 knockdown‐induced NSCLC cells. First, A549 and H3122 cells with GRK5 overexpression were established by transfection with GRK5, with pcDNA as control, which was demonstrated by qRT‐PCR and Western blot assays (Figure 6A and B). Western blot showed that the expression of GRK5 was depleted in A549 and H3122 cells transfected with sh‐circ_0001821 but partially repressed in cells transfected with sh‐circ_001821 + GRK5 (Figure S3). Enforced expression of GRK5 efficiently recovered the circ_0001821 knockdown‐induced the inhibited clonogenicity (Figure 6C), reduced migrated and invasive abilities (Figure 6D and E) of A549 and H3122 cells, as well as increased TAX sensibility (Figure 6F), apoptosis (Figure 6G), caspase‐3 activity (Figure 6H), and caspase‐9 activity (Figure 6I) in NSCLC cells. Collectively, circ_0001821 knockdown inhibited growth and metastasis, while facilitated TAX sensibility and apoptosis of NSCLC cells by downregulating GRK5 expression.
FIGURE 6.
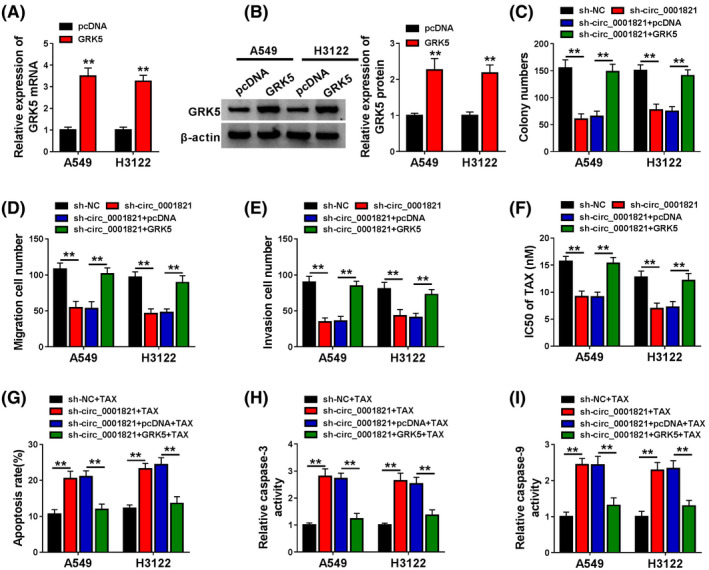
Introduction of GRK5 could also relieve the effects of circ_0001821 knockdown on non‐small‐cell lung cancer (NSCLC) cells. (A and B) Quantitative real‐time polymerase chain reaction (qRT‐PCR) and Western blot assays for the relative expression of (A) GRK5 mRNA and (B) GRK5 protein in A549 and H3122 cells transfected with pcDNA or GRK5. (C–I) A549 and H3122 cells were transfected with sh‐NC, sh‐circ, sh‐circ + pcDNA, or sh‐circ + GRK5. (C) Colony formation assay for the clonogenicity in transfected cells. (D and E) Transwell assay for the migrated and invasive abilities of transfected cells. (F) CCK‐8 assay for the IC50 of transfected cells to TAX. (G–I) Transfected A549 cells were disposed with 9.37 nM TAX for 48 h, and transfected H3122 cells were disposed with 7.44 nM for 48 h. (G) Flow cytometry for the apoptotic rate of treated cells. (H and I) Measurement of caspase‐3 activity and caspase‐9 activity in treated cells. **p < .01
3.7. Depletion of circ_0001821 suppressed tumor growth in vivo
At last, the role of circ_0001821 in vivo was studied through xenograft assay. A549 cells stably expressing sh‐NC or sh‐circ were subcutaneously injected into nude mice, followed by inoculation with DMSO or TAX. As shown in Figure 7A–C, TAX treatment or circ_0001821 knockdown significantly inhibited the size and weight of tumors formed in nude mice when compared with the corresponding control group. In addition, circ_0001821 knockdown aggravated the inhibitory effect of TAX. Moreover, qRT‐PCR and Western blot assays proved that the TAX treatment or circ_0001821 knockdown induced the downregulation of circ_0001821 (Figure 7D) and GRK5 (Figure 7F and G) and the upregulation of miR‐526b‐5p (Figure 7E) in tumors generated in nude mice in contrast to the control group. In summary, circ_0001821 knockdown blocked tumor growth in vivo.
FIGURE 7.

Depletion of circ_0001821 suppressed tumor growth in vivo. A549 cells stably expressing sh‐NC or sh‐circ were subcutaneously injected in to nude mice, followed by inoculation with DMSO or TAX (n = 5). (A) Volume of generated tumors measured once a week. (B) Images of generated tumors. (C) Weight of generated tumors. (D–F) Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay for the relative expression of (D) circ_0001821, (E) miR‐526b‐5p, and (F) GRK5 mRNA in generated tumors. (G) Western blot assay for the relative expression of GRK5 protein in generated tumors. **p < .01
4. DISCUSSION
Dysregulated circRNAs are closely associated with the diverse malignant behaviors of NSCLC cells and have potential to be biomarkers for NSCLC diagnosis, prognosis, and therapy. 26 Here, we investigated the function of a dysregulated circRNA, circ_0001821, in NSCLC development. Deficiency of circ_0001821 expression resulted in inhibitory effects on growth, metastasis, TAX resistance, and tumorigenesis of NSCLC in vitro and in vivo.
Kong et al. alleged that circ_0001821 was downregulated in gastric cancer tissues and whole‐blood samples, and its expression was inversely correlated with tumor depth and lymph node metastasis (LNM) of patients. 27 Moreover, they found that circ_0001821 was significantly upregulated in colorectal cancer (CRC) tissues but not upregulated in breast cancer or lung cancer tissues compared with matched normal tissues, suggesting the organ specificity of circ_0001821. 27 Furthermore, Wang et al. reported the upregulation of circ_0001821 in both LUAD and LUSC tissues, and circ_0001821 had potential to be diagnostic biomarker of NSCLC patients. 12 Likewise, we detected the upregulation of circ_0001821 in both NSCLC tissues and cells, and higher expression of circ_0001821 indicated lower overall survival rate of NSCLC patients, implying its potential function in NSCLC progression. We could conclude from our data that circ_0001821 knockdown repressed colony formation ability, metastasis ability, and TAX resistance of NSCLC cells in vitro, as well as blocked tumor growth in vivo, indicating the tumor‐promoting role of circ_0001821 in NSCLC.
Functionally, endogenous circRNAs could act as sponges of miRNAs, repressing their function and activity, so as to regulate multiple biological processes. 28 A previous study disclosed that circ_0001821 (circPVT1) acted as a sponge of miR‐125 family to promote the proliferation of gastric cancer cells, 29 suggesting that circ_0001821 played functions partially through the circRNA/miRNA pathway. Hence, circinteractome was used to seek the miRNAs directly interacted with circ_0001821, and miR‐526b‐5p was found to have binding region with circ_0001821. Following dual‐luciferase reporter and RNA pull‐down assays validated the target relationship between miR‐526b‐5p and circ_0001821. MiR‐526b‐5p was previously reported to be an antitumor factor in hepatocellular carcinoma, breast cancer, osteosarcoma, and cervical cancer, 16 , 17 , 30 , 31 as well as in NSCLC. 18 Zhang et al. demonstrated the downregulation of miR‐526b in NSCLC tissues, and its expression was correlated with poor outcomes of NSCLC patients; gain of miR‐526b significantly inhibited NSCLC cell proliferation and tumor growth. 18 Here, we also detected the downregulation of miR‐526b‐5p in NSCLC cells. And silencing of miR‐526b‐5p efficiently attenuated the circ_0001821 knockdown‐induced repressed impact on NSCLC cell growth, metastasis ability, and TAX resistance.
MiRNAs were said to perform crucial roles in NSCLC by targeting multiple tumor‐related effector genes. 32 In this project, we used starBase 3.0 to predict the potential target genes of miR‐526b‐5p, and 3′UTR of GRK5 could bind to miR‐526b‐5p, followed by confirmation via dual‐luciferase reporter assay. GRK5 participated in pathologic conditions of human cancers and exerted dual role. 19 In NSCLC, GRK5 was highly expressed in NSCLC tissues, and its high expression was correlated with bad prognosis of NSCLC patients. Besides, GRK5 could positively regulate NSCLC cell proliferation, migration, and tumorigenesis. 23 Also, we observed the upregulation of GRK5 in NSCLC cells. Exogenous introduction of GRK5 largely weakened the inhibitory effects of circ_0001821 knockdown on NSCLC cell growth, metastasis ability, and TAX resistance. Above results suggested the role of circ_0001821/miR‐526b‐5p/GRK5 in NSCLC progression.
In conclusion, circ_0001821 was significantly upregulated in NSCLC tissues and cells. Depletion of circ_0001821 could suppress growth and metastasis but promote TAX sensibility and apoptosis of NSCLC cells in vitro, as well as block tumor growth in vivo. Additionally, we testified that oncogenic role of circ_0001821 was attributed to its regulation on miR‐526b‐5p/GRK5 axis, at least in part (Figure 8). Our findings might provide a potential target for NSCLC therapy.
FIGURE 8.

A diagram illustrated the summary of all findings in this study
DISCLOSURE
The authors declare that they have no financial conflicts of interest.
AUTHORS’ CONTRIBUTIONS
YL and CL designed and performed the research; HL, JW analyzed the data; YL and CL wrote the manuscript. All authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Written informed consents were obtained from all participants and this study was permitted by the Ethics Committee of Jingmen NO.2 People's Hospital.
Supporting information
Figure S1
Figure S2
Figure S3
ACKNOWLEDGEMENT
None.
Liu Y, Li C, Liu H, Wang J. Circ_0001821 knockdown suppresses growth, metastasis, and TAX resistance of non‐small‐cell lung cancer cells by regulating the miR‐526b‐5p/GRK5 axis. Pharmacol Res Perspect. 2021;9:e00812. 10.1002/prp2.812
Ying Liu and Changchao Li contributed equally to this work.
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553(7689):446‐454. 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 3. Yang CH, Taxol HSB. The first microtubule stabilizing agent. Int J Mol Sci. 2017;18(8):1733. 10.3390/ijms18081733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhan Y, Wang K, Li Q, et al. The novel autophagy inhibitor alpha‐Hederin promoted paclitaxel cytotoxicity by increasing reactive oxygen species accumulation in non‐small cell lung cancer cells. Int J Mol Sci. 2018;19(10):3221. 10.3390/ijms19103221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garbuzenko OB, Kuzmov A, Taratula O, Pine SR, Minko T. Strategy to enhance lung cancer treatment by five essential elements: inhalation delivery, nanotechnology, tumor‐receptor targeting, chemo‐ and gene therapy. Theranostics. 2019;9(26):8362‐8376. 10.7150/thno.39816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greene J, Baird A‐M, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. 10.3389/fmolb.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hou LD, Zhang J. Circular RNAs: an emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30(1):1‐6. 10.1177/0394632016686985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Xu R, Zhang D, et al. Circ‐ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR‐330‐5p to promote non‐small cell lung cancer progression. Transl Lung Cancer Res. 2019;8(6):862‐875. 10.21037/tlcr.2019.11.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li X, Yang BO, Ren H, et al. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non‐small cell lung cancer by targeting miR‐182‐5p. Cell Death Dis. 2019;10(12):953. 10.1038/s41419-019-2180-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao P, Wang Z, Hu Z, Jiao X, Yao Y. Circular RNA circ_0074027 indicates a poor prognosis for NSCLC patients and modulates cell proliferation, apoptosis, and invasion via miR‐185‐3p mediated BRD4/MADD activation. J Cell Biochem. 2020;121(3):2632‐2642. 10.1002/jcb.29484 [DOI] [PubMed] [Google Scholar]
- 11. Jin M, Shi C, Yang C, Liu J, Huang G. Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR‐150‐5p/GLUT‐1 axis. Mol Ther Nucleic Acids. 2019;18:219‐231. 10.1016/j.omtn.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang C, Tan S, Liu W‐R, et al. RNA‐Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134. 10.1186/s12943-019-1061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harrandah AM, Mora RA, Chan EKL. Emerging microRNAs in cancer diagnosis, progression, and immune surveillance. Cancer Lett. 2018;438:126‐132. 10.1016/j.canlet.2018.09.019 [DOI] [PubMed] [Google Scholar]
- 14. Petrek H, Yu AM. MicroRNAs in non‐small cell lung cancer: gene regulation, impact on cancer cellular processes, and therapeutic potential. Pharmacol Res Perspect. 2019;7(6):e00528. 10.1002/prp2.528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wei X, Shen X, Ren Y, Hu W. The roles of microRNAs in regulating chemotherapy resistance of non‐small cell lung cancer. Curr Pharm Des. 2018;23(39):5983‐5988. 10.2174/1381612823666171018105207 [DOI] [PubMed] [Google Scholar]
- 16. Liu X, Yang L, Tu J, et al. microRNA‐526b servers as a prognostic factor and exhibits tumor suppressive property by targeting Sirtuin 7 in hepatocellular carcinoma. Oncotarget. 2017;8(50):87737‐87749. 10.18632/oncotarget.21209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu YQ, Cong YZ, Jiang J, et al. MiR‐526b suppresses cell proliferation, cell invasion and epithelial‐mesenchymal transition in breast cancer by targeting Twist1. Eur Rev Med Pharmacol Sci. 2020;24(6):3113‐3121. 10.26355/eurrev_202003_20678 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Z‐Y, Fu S‐L, Xu S‐Q, et al. By downregulating Ku80, hsa‐miR‐526b suppresses non‐small cell lung cancer. Oncotarget. 2015;6(3):1462‐1477. 10.18632/oncotarget.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gambardella J, Franco A, Giudice CD, et al. Dual role of GRK5 in cancer development and progression. Transl Med UniSa. 2016;14:28‐37. [PMC free article] [PubMed] [Google Scholar]
- 20. Métayé T, Menet E, Guilhot J, Kraimps JL. Expression and activity of g protein‐coupled receptor kinases in differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2002;87(7):3279‐3286. 10.1210/jcem.87.7.8618 [DOI] [PubMed] [Google Scholar]
- 21. Kim JI, Chakraborty P, Wang Z, Daaka Y. G‐protein coupled receptor kinase 5 regulates prostate tumor growth. J Urol. 2012;187(1):322‐329. 10.1016/j.juro.2011.09.049 [DOI] [PubMed] [Google Scholar]
- 22. Zhao TL, Gan XX, Bao Y, Wang WP, Liu B, Wang LH. GRK5 promotes tumor progression in renal cell carcinoma. Neoplasma. 2019;66(2):261‐270. 10.4149/neo_2018_180621N409 [DOI] [PubMed] [Google Scholar]
- 23. Jiang L‐P, Fan S‐Q, Xiong Q‐X, et al. GRK5 functions as an oncogenic factor in non‐small‐cell lung cancer. Cell Death Dis. 2018;9(3):295. 10.1038/s41419-018-0299-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lagman J, Sayegh P, Lee CS, et al. G protein‐coupled receptor kinase 5 modifies cancer cell resistance to paclitaxel. Mol Cell Biochem. 2019;461(1‐2):103‐118. 10.1007/s11010-019-03594-9 [DOI] [PubMed] [Google Scholar]
- 25. Gao J, Tian X, Yan X, et al. Selenium exerts protective effects against fluoride‐induced apoptosis and oxidative stress and altered the expression of Bcl‐2/Caspase family. Biol Trace Elem Res. 2021;199(2):682‐692. 10.1007/s12011-020-02185-w [DOI] [PubMed] [Google Scholar]
- 26. Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non‐small cell lung cancer. J Cancer Res Clin Oncol. 2019;145(12):2875‐2889. 10.1007/s00432-019-03045-4 [DOI] [PubMed] [Google Scholar]
- 27. Kong S, Yang Q, Tang C, Wang T, Shen X, Ju S. Identification of hsa_circ_0001821 as a novel diagnostic biomarker in gastric cancer via comprehensive circular RNA profiling. Front Genet. 2019;10:878. 10.3389/fgene.2019.00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42‐51. 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 29. Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208‐219. 10.1016/j.canlet.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 30. Yan M, Gao H, Lv Z, et al. Circular RNA PVT1 promotes metastasis via regulating of miR‐526b/FOXC2 signals in OS cells. J Cell Mol Med. 2020;24(10):5593‐5604. 10.1111/jcmm.15215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Wang J, Xu F, et al. By downregulating PBX3, miR‐526b suppresses the epithelial‐mesenchymal transition process in cervical cancer cells. Future Oncol. 2019;15(14):1577‐1591. 10.2217/fon-2018-0575 [DOI] [PubMed] [Google Scholar]
- 32. Gong F‐Y, Bai T, Zhang D‐Y, et al. Regulation mechanism of microRNAs in non‐small cell lung cancer. Curr Pharm Des. 2018;23(39):5973‐5982. 10.2174/1381612823666170714153424 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Data Availability Statement
Not applicable.


