Abstract
Ras gene family members play a relevant role in cancer, especially when they are mutated. However, they may play additional roles in other conditions beside cancer. We performed gene expression analysis using the NanoString PanCancer IO 360 panel in the peripheral blood mononuclear cell (PBMC) of six COVID-19 patients and we found that H-Ras gene was significantly upregulated, while both K-Ras and N-Ras genes were downregulated. In particular, H-Ras gene upregulation was more evident in COVID-19 patients with a more severe disease. We compared our results with those obtained by analyzing two different and independent datasets, including a total of 53 COVID-19 patients, in which the gene expression analysis was performed using the Immunology_V2 panel. Comparative analysis of the H-Ras gene expression in these patients confirmed our preliminary results. In both of them, in fact, we were able to confirm the upregulation of the expression of the H-Ras gene. The exact role of this specific upregulation of the H-Ras gene in response to SARS-CoV-2 infection and its possible role in cancer still remains to be elucidated. In conclusion, H-Ras gene participates to the host immune response to SARS-CoV-2 virus infection, especially in patients affected by the most severe form of the COVID-19.
Subject terms: Viral infection, Predictive markers
Introduction
Growth factor receptor (GFR) signaling besides playing an important role in cancer pathogenesis is also crucial for some virus infections [1–3]. GFRs activate various intracellular signaling pathways, including the one mediated by RAS and RAF. The Ras gene family members encode for four different, highly related protein isoforms (HRAS, N-RAS, K-RAS4A, and K-RAS4B) that are almost ubiquitously expressed in all cell lineages and organs, with some quantitative and qualitative differences of their expression. The Ras gene products are small GTP-binding proteins that play a critical role in the control of basic eukaryotic cell functions, such as proliferation, survival, and differentiation. In addition, they might play specific or overlapping functional roles in many physiological and pathological processes beside cancer, including inflammation.
It has been recently reported that SARS-CoV-2 infection is able to induce a strong up- and downregulation of components of many cellular signaling pathways involved in cancer, including the RAS-RAF/MEK/ERK signaling pathway [4]. Therefore, it appears likely that the activation of this signaling pathway could be involved in the SARS-CoV-2 virus infection and survival too. In line with this, inhibition of Ras signaling pathway by drugs has been suggested as potential antiviral treatment of COVID-19 [5].
Moreover, recent reported cases suggest that COVID-19 may accelerate the course of malignant hematological diseases and induce or exacerbate autoreactive hematopoietic disease [6]. In particular, the temporal relationship of the events may suggest a potential causal relationship between the immunological host response to SARS-CoV-2 infection and the hematopoietic disorder.
No data has been reported in the literature so far regarding the possible role of Ras family genes deregulation, and specifically, of H-Ras gene in COVID-19. In the present study, we analyzed the Ras family gene expression levels in peripheral blood mononuclear cell (PBMC) of COVID-19 patients.
Methods
Total RNA was extracted and purified from PBMC isolated from eight blood samples, six from of COVID-19 patients, subdivided in mild/moderate (n = 2) and severe/critical (n = 4), according to WHO criteria and two from healthy controls (HC). Gene expression was measured with the NanoString PanCancer IO 360 panel using as input 30–100 ng total RNA of each sample, as previously reported [7, 8] and according to the manufacturer’s instructions. After the Codeset hybridization the samples were analysed with the nCounter Digital Analyzer. RCC files were analyzed using nSolver analysis software (Version 4.0) following the manufacturer’s protocols. Negative and positive controls were included in probe sets and used for background thresholding and for normalization. Normalized counts were further analyzed by MATLAB R2020b. Gene expression differences were evaluated by a Wilcoxon rank sum test for two groups and Kruskal-Wallis test for more than two groups. Same statistical tests were used to asses gene expression differences in two validation cohorts, analyzed using the NanoString Immunology_V2 panel [9, 10] (Table 1).
Table 1.
Comparative Ras genes expression in three independent dataset of COVID-19 patients analyzed by NanoString nCounter technology.
| NanoString nCounter human gene expression panels | Genes included | COVID-19 patients | Healthy controls | Reference | ||
|---|---|---|---|---|---|---|
| Mild to moderatea | Severe to criticala | Total | ||||
| PanCancer IO 360 | 770 | 2 | 4 | 6 | 2 | Present study |
| Immunology_V2 | 594 | 10 | 21 | 31 | 13 | Hadjadj et al. [9] |
| Immunology_V2 | 594 | N/A | N/A | 22 | 10 | Vastrad et al. [10] |
| Total | 12 | 25 | 59 | 25 | ||
aWorld Health Organization (WHO) COVID-19 severity criteria.
Results
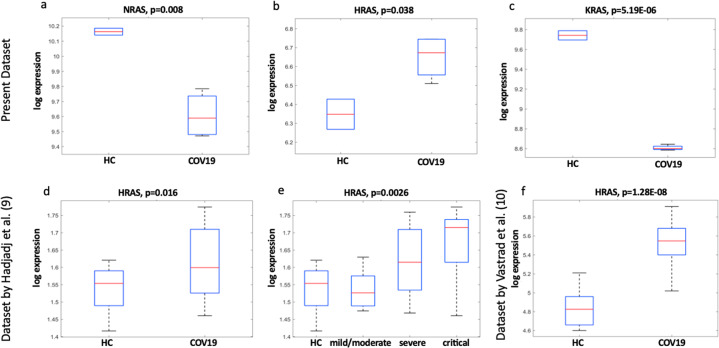
Dysregulated Ras family gene expressions represent a common event in our samples obtained from COVID-19 patients, compared to HC. This feature is even more evident when we consider severe/critical disease. In particular, COVID-19 patients with a more severe disease showed a significant upregulation of H-Ras and a coexistent downregulation of both K-Ras and N-Ras gene expressions when compared to control cases (Fig. 1a–c). In order to verify these results, we retrieved and analyzed data of gene expression signatures from two large cohorts of COVID-19 blood samples. In particular, by reanalyzing one published independent large dataset [9] we confirmed that H-Ras was indeed expressed at a higher level in COVID-19 patients compared to control ones and that its expression was correlated also with the disease severity in a statistically significant manner (Fig. 1d, e). To obtain another confirmation of such results, we analyzed another large microarray dataset of COVID-19 patients [10]. A significant H-Ras upregulation compared to control ones was, again, observed also in this study (Fig. 1f). Taken together, results of our study and the re-analysis of data from two large and independent case series, indicates that H-Ras gene expression is significantly increased in PBMC isolated from blood of COVID-19 patients and its increased expression correlates with the disease severity.
Fig. 1. Ras genes expression in COVID-19 patients.
The expression of H-Ras genes is reported in log-expression units for the three different dataset (b, d, e, f). Data regarding N- and K-Ras are available only for our dataset and were not present in the immunology panels used for the analysis of the other datasets (a, c). Statistically significant differences in the expression of Ras genes between healthy controls (HC) and COVID-19 patients (COV19) are reported.
Discussion
Dysregulation of RAS cycling can promote human disease conditions, with somatic mutations in the Ras genes being prominent drivers of cancerogenesis and Ras germline mutations contributing to a group of related developmental disorders known as the RASopathies [11]. Despite being one of the most frequently mutated signaling pathways in human cancer, various aspects of RAS biology are still poorly understood. In particular, dysregulated RAS signaling affects directly not only the development of various pathological condition in infected cells but also in host physiological processes, such as immunological response to viremic condition and inflammation [12]. Our preliminary observation, confirmed by bioinformatics analysis of two large independent datasets, indicates that H-Ras overexpression in PBMCs is part of the immune response to COVID-19, especially in patients with severe disease conditions. On the contrary, K-Ras and N-Ras, showed an opposite behavior and are both downregulated. It is attractive to speculate that H-Ras overexpression, triggered by viral infection, might act as a stimulator of precancerous conditions or of cancer stem cells compartment and that could represent one of the links connecting viral infection to cancer. A crescent number of studies reported that patients with malignant hematological diseases are at increased risk of complications from SARS-CoV-2 not only due to immune compromise related to the malignancy but also due to the host immunologic and cytokine response to SARS-CoV-2 infection [6]. We hypothesize that, due to the specific interaction between SARS-CoV-2 virus and infected patients, a cascade of out-of-control events can ensue determining some unexpected pathologic conditions such as the progression, reactivation of hematological malignancies or the development of proliferative disorders. It would be interesting to elucidate whether H-Ras gene overexpression that we observed in the PBMC of COVID-19 patients is part of a specific response to SARS-CoV-2 infection or it occurs also with other viruses.
In conclusion, we report here that the H-Ras gene the is overexpressed in the PBMC of COVID-19 patients, especially in those presenting a more severe condition. Such overexpression represents a molecular event activated by the immune cells upon SARS-CoV-2 infection. Our observation paves the way to a new area of research focused the role of Ras family genes in the host immune response to COVID-19 as well as to other viral infectious diseases.
Acknowledgements
We are deeply grateful to Benjamin Terrier for giving us the opportunity to use his dataset for our comparative analysis.
Funding
This work was supported by: (1) Italian Association for Cancer Research (AIRC) grants IG15216 to GC and IG24451 to RM; (2) the LazioInnova grant 2018 n.85-2017-13750 to RM; (3) PRIN Bando 2017 (Prot. 2017HWTP2K) to GC and RM; (4) Soka Gakkai grant 8 × 1000 2020-2016 RIC2 to GC and RM.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by I. Amelio
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Rita Mancini, Carlo Capalbo.
References
- 1.Beerli C, Yakimovich A, Kilcher S, Reynoso GV, Flaschner G, Muller DJ, et al. Vaccinia virus hijacks EGFR signalling to enhance virus spread through rapid and directed infected cell motility. Nat Microbiol. 2019;4:216–25. doi: 10.1038/s41564-018-0288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kung CP, Meckes DG, Raab-Traub N. Epstein-Barr virus LMP1 activates EGFR, STAT3, and ERK through effects on PKCd. J Virol. 2011;85:4399–408. doi: 10.1128/JVI.01703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu L, Lee PK, Lee WM, Zhao Y, Yu D, Chen Y. Rhinovirus induced major airway mucin production involves a novel TLR3-EGFRdependent pathway. Am J Respir Cell Mol Biol. 2009;40:610–9. doi: 10.1165/rcmb.2008-0223OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghasemnejad-Berenji M, Pashapour S. SARS-CoV-2 and the possible role of Raf/MEK/ERK pathway in viral survival: is this a potential therapeutic strategy for COVID-19? Pharmacology. 2021;106:119–22. doi: 10.1159/000511280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemmat N, Asadzadeh Z, Ahangar NK, Alemohammad H, Najafzadeh B, Derakhshani A, et al. The roles of signaling pathways in SARS-CoV-2 infection; lessons learned from SARS-CoV and MERS-CoV. Arch Virol. 2021;166:675–96. doi: 10.1007/s00705-021-04958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taliani G, Follini E, Guglielmetti L, Bernuzzi P, Faggi A, Ferrante P, et al. Case report: B lymphocyte disorders under COVID-19 inflammatory pressure. Front Oncol. 2020;10:582–901. doi: 10.3389/fonc.2020.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajeevan MS, Patel S, Li T, Unger ER. Nanostring technology for human papillomavirus typing. Viruses. 2021;13:188. doi: 10.3390/v13020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sciacchitano S, De Vitis C, D’Ascanio M, Giovagnoli S, De Dominicis C, Laghi A, et al. Gene signature and immune cell profiling by high-dimensional, single-cell analysis in COVID-19 patients, presenting Low T3 syndrome and coexistent hematological malignancies. J Trans Med. 2021;19:139. doi: 10.1186/s12967-021-02805-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–24. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vastrad B, Vastrad C, Tengli A. Identification of potential mRNA panels for severe acute respiratory syndrome coronavirus 2 (COVID19) diagnosis and treatment using microarray dataset and bioinformatics methods. 3 Biotech. 2020;10:422. doi: 10.1007/s13205-020-02406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki Y, Niihori T, Inoue S, Matsubara Y. Recent advances in RASopathies. J Hum Genet. 2016;61:33–9. doi: 10.1038/jhg.2015.114. [DOI] [PubMed] [Google Scholar]
- 12.Johnson DS, Chen YH. Ras family of small GTPases in immunity and inflammation. Curr Opin Pharmacol. 2012;12:458–63. doi: 10.1016/j.coph.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]