Abstract
Poly(ethylene terephthalate) (PET) is an abundant and extremely useful material, with widespread applications across society. However, there is an urgent need to develop technologies to valorise post-consumer PET waste to tackle plastic pollution and move towards a circular economy. Whilst PET degradation and recycling technologies have been reported, examples focus on repurposing the resultant monomers to produce more PET or other second-generation materials. Herein, we report a novel pathway in engineered Escherichia coli for the direct upcycling of PET derived monomer terephthalic acid into the value-added small molecule vanillin, a flavour compound ubiquitous in the food and cosmetic industries, and an important bulk chemical. After process optimisation, 79% conversion to vanillin from TA was achieved, a 157-fold improvement over our initial conditions. Parameters such as temperature, cell permeabilisation and in situ product removal were key to maximising vanillin titres. Finally, we demonstrate the conversion of post-consumer PET from a plastic bottle into vanillin by coupling the pathway with enzyme-catalysed PET hydrolysis. This work demonstrates the first biological upcycling of post-consumer plastic waste into vanillin using an engineered microorganism.
An engineered biosynthetic pathway in Escherichia coli enables the one-pot upcycling of post-consumer plastic waste into vanillin.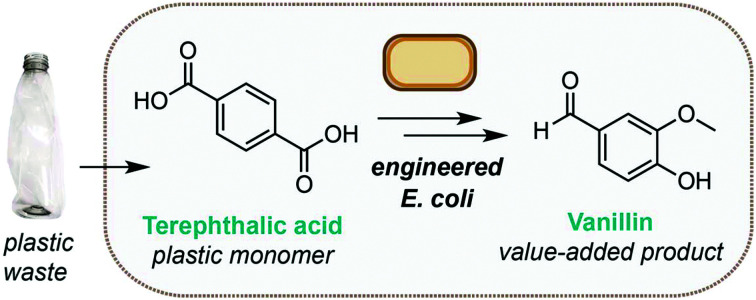
Plastics have widespread and important applications across society, yet poor management of these fossil fuel derived resources is causing widespread pollution. The global plastic waste crisis is now recognised as one of the most pressing environmental issues facing our planet, prompting urgent calls for new technologies to enable a circular plastics economy.1–3 There is also a strong economic incentive, with plastics losing 95% of their material value after a single use, leading to an estimated $110 bn loss to the global economy per annum.4 Therefore, new methods to degrade and valorise plastic waste would have considerable economic as well as environmental impact.
As such, the emerging field of plastic degradation and recycling has attracted significant attention across the fields of microbiology,5–7 biocatalysis and directed evolution,8 synthetic biology1 and non-enzymatic catalysis.9–13 In particular, there have been significant developments in enzymatic PET degradation. The most promising enzymes discovered to date are variants of PETase from Ideonella sakaiensis14 and leaf-branch compost cutinase (LCC).15 Whilst PETase and its engineered variants16–20 operate at ambient temperature (30–37 °C) to release bis- and mono-(2-hydroxyethyl)terephthalate, the LCC derived enzyme is thermostable with maximum productivity at 72 °C and exhibits the highest PET to terephthalate (TA) degradation productivity reported to date (up to 16.7 g L−1 h−1).15
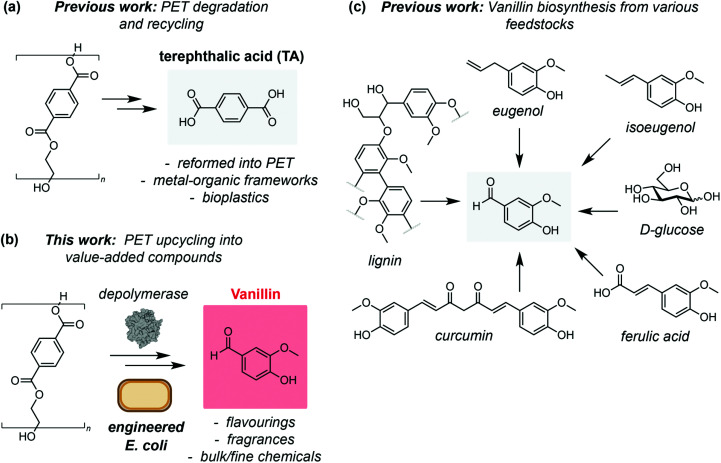
As increasingly efficient PET degradation methods are developed,16,17,21–23 the recycling of PET monomers TA and ethylene glycol has also been demonstrated. For example, TA from LCC catalysed PET degradation has been used as a feedstock for production of second generation PET,15 and as a building block for synthesis of value-added metal organic frameworks.24 Post-consumer PET has also been pyrolyzed and the TA produced used as a feedstock for bacterial production of polyhydroxyalkanoates (PHAs),25 a promising class of bioplastics with a superior biodegradability profile to PET26,27 (Fig. 1a). Chemically hydrolysed PET has also been used as a feedstock for protocatechuate (PC) derived small molecules using microbial co-cultures.28 However, to date there have been no examples of interfacing enzyme-catalysed PET degradation with biological upcycling pathways. To this end, we hypothesised that TA derived from PET waste could be valorised into the value-added product vanillin via a 5-step enzymatic pathway (Fig. 1b). Vanillin is the primary component of the extract of vanilla beans and is responsible for the characteristic taste and smell of vanilla. Consequently, vanillin is widely used across the food and cosmetics industries, as well as in pharmaceutical synthesis and formulation, herbicides, antifoaming agents and cleaning products, leading to a global demand in excess of 37 000 tonnes in 2018.29 The demand for vanillin is growing rapidly and is projected to exceed 59 000 tonnes with a revenue forecast of $734 000 000 by 2025.29
Fig. 1. (a) Chemical and enzymatic degradation of PET has been used to recycle the monomer terephthalic acid for use in second-generation PET and as a building block for value-added products such as metal–organic frameworks (MOFs), and as a feedstock for bioplastics production. (b) A novel biosynthetic route to vanillin from plastic (PET) waste using engineered E. coli cells. (c) Biotechnological routes to vanillin from renewable or waste feedstocks.
As the demand for vanillin far exceeds the supply from vanilla beans (‘natural vanillin’), methods for its synthesis (‘synthetic vanillin’) have been widely investigated. Biotechnological approaches are particularly attractive from a sustainability perspective owing to the inherently mild reaction conditions and use of renewable feedstocks.30 The most widely employed of these is the production of vanillin from lignin biomass,31 whilst methods for fermentation of vanillin using microbial and fungal hosts from ferulic acid,32,33 glucose, glycerol, l-tyrosine, xylose,34 curcumin,35 eugenol33 and isoeugenol36 have also been demonstrated (Fig. 1c). However, although conversion of TA to vanillic acid is known,28 direct access to the high value aldehyde vanillin from PET has not been reported. Herein, we demonstrate a novel enzymatic pathway using engineered Escherichia coli cells to synthesise vanillin from TA. This process falls within the ‘biotechnological vanillin’ sub-category of ‘synthetic vanillin’,30 namely vanillin produced by microbial fermentation via a non-natural, engineered pathway. After process optimisation, we obtain conversions in excess of 79%. Finally, we show that the pathway can be combined with LCC catalysed PET degradation to obtain vanillin directly from post-consumer plastic waste at ambient temperature and in aqueous conditions.
Results and discussion
Construction of a novel enzymatic pathway for vanillin production from TA
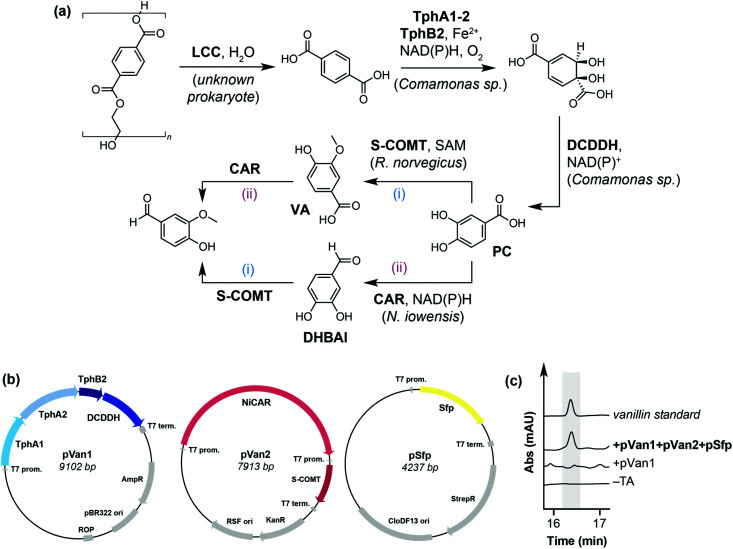
Our initial focus was on developing a novel in vivo enzymatic pathway for the conversion of TA to the target compound vanillin. Due to the inherent ability of E. coli to reduce aldehydes to the corresponding alcohol,37 we chose E. coli MG1655 RARE (reduced aromatic aldehyde reduction) as the host microorganism, which has previously been used for the biosynthesis of vanillin from glucose.38 Inspired by the biodegradation and metabolism of PET via PC by Ideonella sakaiensis,14 we hypothesised that TA could be converted into vanillin by a de novo pathway comprising: (i) terephthalate 1,2-dioxygenase (TPADO), (ii) dihydroxy-3,5-cyclohexadiene-1,4-dicarboxylic acid dehydrogenase (DCDDH), (iii) carboxylic acid reductase (CAR) and (iv), catechol O-methyltransferase (COMT) (Fig. 2a). It is noteworthy that CARs39 and O-MTs40 have both been reported to accept PC, dihydroxybenzaldehyde (DHBAl) and vanillic acid (VA) as substrates, such that the pathway may proceed by two possible intermediates (VA or DHBAl) to produce vanillin.
Fig. 2. (a) Proposed enzymatic pathway for the conversion of PET to value added product vanillin. LCC: Leaf-branch compost cutinase; TPADO: terephthalate 1,2-dioxygenase; DCDDH: 1,4-dicarboxylic acid dehydrogenase; O-MT: O-methyltransferase; CAR: carboxylic acid reductase; FAD: flavin adenine dinucleotide; NAD(P)H: reduced nicotinamide adenine dinucleotide (phosphate); SAM: S-adenosyl-l-methionine (b) design of pathway enzyme expression constructs pVan1 and pVan2. (c) HPLC traces showing proof of concept for the pathway. Vanillin was only detected when TA was added to cells expressing pVan1, pVan2 and pSfp.
The pathway enzymes were assembled onto two plasmids, named pVan1 and pVan2. pVan1 encodes TPADO from Comamonas sp., a heterotrimer comprising subunits TphA1, TphA2 and TphB2, and DCDDH, also from Comamonas sp., which together catalyse the conversion of TA to PC using atmospheric oxygen as the terminal oxidant. Both enzymes have previously been shown to express in E. coli and show activity towards TA and DCD, respectively.41–44 pVan2 encodes a carboxylic acid reductase from Nocardia iowensis (NiCAR) and a single point mutant of the soluble form of catechol O-methyltransferase (S-COMT Y200L), from Rattus norvegicus (Fig. 2b and Fig. S1†).39,40 This mutant was chosen for its high stereoselectivity for methylation at the meta-position of PC and DHBAl.40 NiCAR was selected for the reduction step as it has previously been demonstrated to efficiently reduce both PC and VA to the corresponding aldehydes.39 Additionally, cells were co-transformed with a third plasmid encoding the phosphopantetheinyl transferase (pSfp) from Bacillus subtilis, which is necessary for post-translational modification of NiCAR.45 For proof of concept, resuspended E. coli RARE cells expressing pVan1, pVan2 and pSfp (E. coli RARE_pVanX) were used in a screening scale biotransformation reaction for the conversion of TA to vanillin. Whilst no vanillin was detected from cells expressing only pVan1 or experiments lacking TA substrate, cells expressing all three plasmids with added TA (5 mM) gave a detectable amount of the target compound vanillin (5 μM, <1% conversion) (Fig. 2c and Fig. S3–S5†), with intermediates PC, DHBAl and vanillic acid also being detected at concentrations of 18 μM, 10 μM and 2 μM, respectively (Fig. S5†).
Process optimisation for maximum vanillin titres
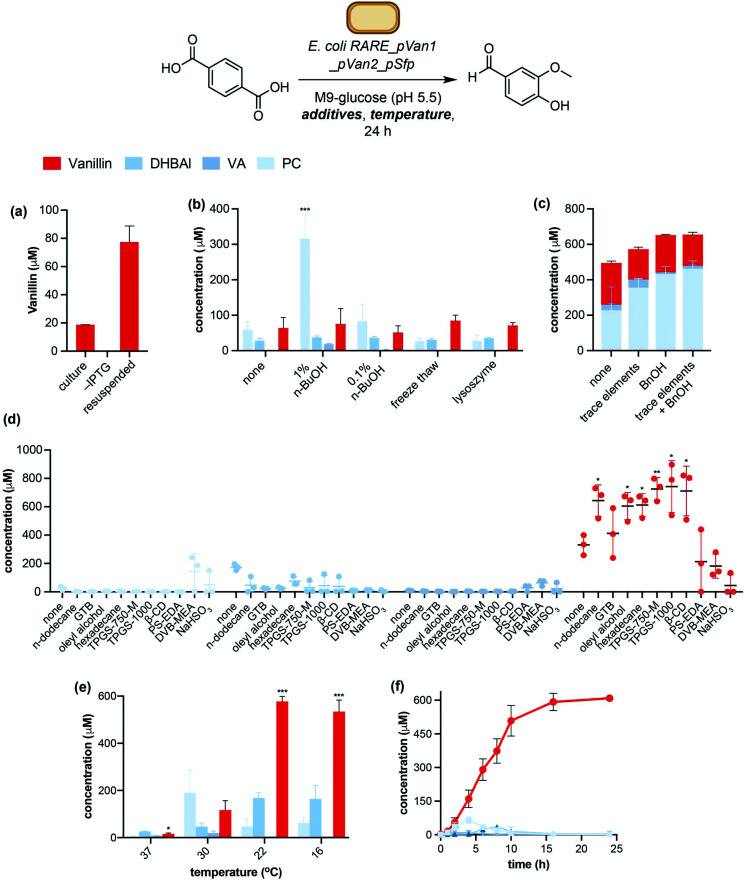
Encouraged by these initial data, we sought to maximise vanillin yields by optimisation of protein expression and whole cell reaction conditions. A screen of protein expression media showed M9 minimal media supplemented with casamino acids (M9-CA) to give the highest combined levels of conversion to vanillin and key intermediate PC (Fig. S5†). Resuspending whole cells in fresh M9 media proved superior to adding TA to expression cultures during the exponential growth phase, giving a 4-fold increase in vanillin titres (77 μM ± 11 μM, Fig. 3a). This was hypothesised to be due to the higher cell density and therefore biocatalyst loading of resuspended cells (OD600(ferm.) = 0.6–2; OD600(resusp.) = 20). Examination of standard expression parameters such as induction OD, inducer concentration, time and temperature did not lead to significant improvement in vanillin yields. Likewise, sulfur and iron containing expression medium additives such as cysteine, ferrous sulfate and ferric ammonium citrate, which were hypothesised to increase expression levels of [2Fe–2S] containing TPADO, did not give a significant increase in TA conversion levels (Fig. S6†).43,46 However, the addition of trace elements to the growth media led to a 1.5-fold increase in PC titres (Fig. 3c). This was hypothesised to improve TPADO and DCDDH activity, which are Fe2+ and Zn2+ dependent, respectively.41,43,44 Interestingly, the addition of benzyl alcohol (BnOH) to the expression culture prior to induction led to a 2-fold increase in PC levels even in the absence of trace elements. Benzyl alcohol is an osmolyte that has been reported to induce expression of endogenous chaperones in E. coli, leading to increased soluble expression of heterologous genes.47 Accordingly, analysis of protein expression by SDS-PAGE showed more soluble protein expression in the presence of BnOH or trace metals, whilst adding both BnOH and trace metals to the expression culture did not give further increase in either soluble protein expression or vanillin yields (Fig. 3c and Fig. S7†).
Fig. 3. Optimisation of vanillin pathway expression and biotransformation conditions. (a) Comparison between fermentation and resuspended whole cells (conditions: 5 mM TA added to 10 mL fermentation reactions at time of induction, incubated at 30 °C for 24 hours). (b) Effect of cell membrane permeabilisation (conditions: 5 mM TA, 30 °C, 24 hours). (c) Effect of addition of trace elements and benzyl alcohol (BnOH) to expression medium (conditions: 5 mM TA, 30 °C, 24 hours). (e) Effect of in situ product removal (ISPR) (conditions: 1 mM TA, 22 °C, 24 hours). (e) Effect of biotransformation temperature (conditions: 5 mM TA, 24 hours). (f) Time course of TA conversion under optimised conditions (1 mM TA, 22 °C, 20%v/v oleyl alcohol (OA)). GTB: Glycerol tributyrate; TPGS: dl-α-tocopherol methoxypolyethylene glycol succinate; β-CD: β-cyclodextrin; PS-EDA: ethylenediamine, polymer bound, 1% cross-linked (1%w/v loading); DVB-MEA: mercaptoethylamine, polymer bound, 1% cross-linked with divinylbenzene (1%w/v loading). *P < 0.05, **P < 0.005, ***P < 0.0005 (Welch's T-test).
Next, we investigated the effect of the whole cell biotransformation conditions on conversion of TA to vanillin and pathway intermediates. As TPADO is O2 dependant, we hypothesised that increasing reaction headspace would increase conversion of TA to PC. This was confirmed by observing a 65-fold improvement in vanillin titres (5 μM ± 3 μM to 327 μM ± 15 μM) when the headspace to reaction volume ratio was increased from 1 : 5 to 1 : 99 (Fig. S8†).
Further improvement in TA conversion was accomplished by increasing E. coli cell membrane permeability to TA. Whilst an influx transporter for TA in Pseudomonas sp. has been reported,48E. coli is not known to express a transporter capable of importing TA to the cytosol. Therefore, cell membrane permeabilization strategies were investigated. In particular, n-butanol (n-BuOH) has been shown to improve cell membrane permeability towards small molecules49,50 and was therefore hypothesised to improve TA conversion by increasing substrate concentration in the cell interior. Whilst 0.1%v/v had little effect on conversion, the addition of 1%v/v n-BuOH to the biotransformation buffer resulted in a 3-fold increase in cumulative conversion of TA to vanillin and pathway intermediates (Fig. 3b). Conversely, attempts to improve membrane permeability through freeze/thaw cycles or the addition of lysozyme to the reaction buffer did not lead to increased vanillin concentrations. Experiments using lysed E. coli RARE_pVanX cells and experiments using co-cultures of E. coli RARE_pVan1 and E. coli RARE_pVan2_pSfp gave negligible (<1%) levels of vanillin, indicating that co-localisation of the pathway enzymes within a single cell is advantageous in this system. Finally, the addition of l-Met (10 mM) resulted in a 2-fold increase in vanillin titres (Fig. S9†). This aligns with previous reports of in vivo pathways employing S-adenosylmethionine (SAM) dependant methyltransferases (MTs), which note supplementation of reaction buffer with l-Met as being vital to achieve maximum product titres.51
The doubly charged nature of terephthalate at neutral pH was also theorised to impede diffusion of TA across the cell membrane and hence the effect of biotransformation buffer pH was investigated. As predicted, a sharp decrease in TA to PC conversion from pH6 to 8 was observed. Upon screening conditions <pH7, pH5.5 was found to be optimum to balance maximum TA diffusion into the cell whilst minimising acid induced stress to the cell (Fig. S10†).
Cell density of resuspended E. coli RARE_pVanX was also studied. Whilst there was minimal effect on product titres and distribution between OD600 = 10–60, a significant increase in vanillin concentration (350 μM ± 62 μM to 480 μM ± 36 μM) was obtained at OD600 = 80, with a switch from PC to vanillin being the predominant product (Fig. S11†).
Biotransformation temperature also had a dramatic effect on vanillin yields (Fig. 3e). Decreasing the reaction temperature from 30 °C to 22 °C gave a 5-fold improvement in vanillin yields (577 μM ± 22 μM compared to 117 μM ± 40 μM at 30 °C). Interestingly, the ratio of pathway intermediates formed also changed upon lowering the reaction temperature. Vanillic acid (VA) was produced at 30 °C (20 μM ± 8 μM), whereas VA titres from lower temperature reactions were negligible. This indicates that at biotransformation temperatures of 22 °C and lower, the pathway proceeds via NiCAR mediated reduction of PC followed by S-COMT mediated methylation to give vanillin, whereas at 30 °C the alternative pathway via vanillic acid also operates. Decreasing the temperature to 16 °C gave no further improvement in product conversion.
Finally, we hypothesised that in situ product removal (ISPR) could improve vanillin yields through mitigating the toxicity of vanillin to E. coli (Fig. S12†) and increasing flux towards vanillin, the most hydrophobic molecule in the pathway (log Pvanillin = 1.2).52 Three ISPR strategies were investigated: (i) organic solvent overlays,53 (ii) product entrapment in biocompatible micelles54 or β-cyclodextrin55 and (iii) product trapping via reversible nucleophilic addition to the aldehyde moiety of DHBAl and vanillin.56,57 Whilst the product trapping reagents did not improve product titres, all the solvent overlays and product entrapment ISPR reagents increased vanillin yields relative to the control experiment (Fig. 3d). Out of these, oleyl alcohol (OA) and vitamin E derived biocompatible micelles TPGS-750-M were selected for further study based on initial data showing the highest vanillin concentrations and lowest levels of the intermediate DHBAl.
The biotransformation of TA to vanillin in the presence of no ISPR reagent, 20%v/v OA, or 2%w/v TPGS-750-M was carried out with varying TA concentrations to investigate whether ISPR enabled higher vanillin titres. These data showed OA to be the superior ISPR reagent, giving maximum vanillin titres of 744 μM ± 100 μM from 1 mM TA. The presence of OA also led to increased conversion of PC to DHBAl, however there was no significant increase in vanillin titres with increased TA concentration, indicating the bottleneck in the pathway to be the final COMT mediated methylation step (Fig. S13†). Strategies to alleviate this bottleneck, such as screening a library of alternative O-MTs and upregulating expression of SAH degradation enzymes,58,59 will be the focus of further study and development of this pathway. Finally, a time course experiment under the optimised conditions with an oleyl alcohol overlay showed maximum vanillin titres to be obtained after 16 hours, with sequential formation and consumption of PC and DHBAl also being observed (Fig. 3f). This led to the final optimised biotransformation conditions for TA to vanillin conversion: namely E. coli RARE_pVanX cells resuspended in M9-glucose supplemented with l-Met and nBuOH, pH 5.5, incubated with TA for 24 hours at room temperature with an oleyl alcohol overlay. Product formation under the optimised conditions was confirmed upon scaling up the biotransformation to 40 mL scale and analysing the reaction products by NMR spectroscopy (Fig. S14†).
Upcycling of post-consumer PET waste into vanillin
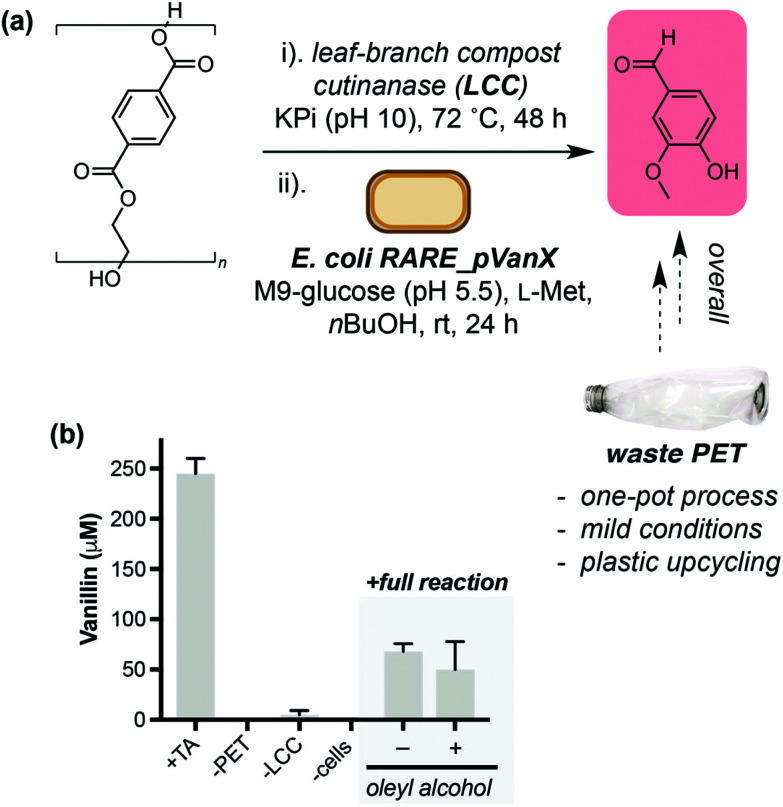
Finally, we set out to demonstrate the application of the TA to vanillin pathway in valorisation of post-consumer plastic waste. We selected the thermostable enzyme LCC WCCG15 (hereafter referred to as LCC) as a biocatalyst to aid hydrolysis of PET into TA. Unlike PETase from Ideonella sakaiensis, LCC releases TA directly and does not require an additional enzyme to hydrolyse mono-2-hydroxyethyl terephthalate (MHET) for release of TA. PET from a post-consumer plastic bottle was treated with semi-purified LCC at 72 °C (Fig. 4a). The reaction was cooled to room temperature and freshly prepared E. coli RARE_pVanX and a biotransformation buffer concentrate were added, and reactions were analysed after 24 hours. Gratifyingly, without any process optimisation, vanillin was detected when all pathway components were present (68 μM, Fig. 4b). Vanillin was not detected in control experiments lacking PET or cells expressing the pathway enzymes. Low levels of vanillin were detected in experiments lacking LCC, which is hypothesised to be due to background PET hydrolysis in LCC reaction buffer (pH10) in the absence of LCC (Fig. S15†). The addition of an oleyl alcohol overlay did not lead to a significant increase in vanillin titres, which was hypothesised to be due to the lower TA concentrations from PET degradation (300–400 μM).
Fig. 4. Conversion of a post-consumer PET bottle into vanillin. (a) Overview of the one-pot, two-step process to convert PET into value-added product vanillin. E. coli RARE_pVanX refers to E. coli RARE expressing plasmids pVan1, pVan2 and pSfp. (b) Data showing production of vanillin only in the presence of cells expressing the vanillin pathway enzymes and the feedstock PET.
Conclusions
Low cost and low intensity technologies to valorise post-consumer plastic waste are urgently required to tackle the plastic waste crisis and enable a circular economy. We have addressed this by engineering a novel biosynthetic pathway in the laboratory bacterium E. coli to convert plastic derived monomer terephthalic acid directly into the value-added molecule vanillin using a single engineered microorganism. The reaction is mild, uses a whole-cell catalyst produced from renewable feedstocks and occurs under ambient conditions (room temperature, pH5.5–7), in aqueous media, requires no additional cofactors or reagents and generates no hazardous waste. Maximum vanillin titres of 785 μM (119 mg L−1, 79% conversion) were achieved after extensive process optimisation studies, a 157-fold improvement over titres from pre-optimisation experiments. Key to this high conversion was biotransformation temperature, cell permeabilisation and use of ISPR to increase flux towards vanillin. Moreover, we have demonstrated that this pathway can utilise TA derived directly from post-consumer plastic waste in a one-pot bioprocess – the first example of biological upcycling of waste PET into a single value-added small molecule. Future studies will focus on intensifying this process through further strain engineering, process optimisation and extension of the pathway to other metabolites. Fundamentally, this work substantiates the philosophy that post-consumer plastic may be viewed not as a waste product, but rather as a carbon resource and feedstock to produce high value and industrially relevant materials and small molecules.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
J. C. S. acknowledges a Discovery Fellowship from BBSRC (BB/S010629/1) and S. W. acknowledges a Future Leaders Fellowship from UKRI (MR/S033882/1).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1gc00931a
References
- Wei R. et al., Possibilities and limitations of biotechnological plastic degradation and recycling. Nat. Catal. 2020;3:867–871. doi: 10.1038/s41929-020-00521-w. [DOI] [Google Scholar]
- Blank L. M. Narancic T. Mampel J. Tiso T. O'Connor K. Biotechnological upcycling of plastic waste and other non-conventional feedstocks in a circular economy. Curr. Opin. Biotechnol. 2020;62:212–219. doi: 10.1016/j.copbio.2019.11.011. [DOI] [PubMed] [Google Scholar]
- Payne J. McKeown P. Jones M. D. A circular economy approach to plastic waste. Polym. Degrad. Stab. 2019;165:170–181. doi: 10.1016/j.polymdegradstab.2019.05.014. [DOI] [Google Scholar]
- The New Plastics Economy: Rethinking the future of plastics | World Economic Forum. https://www.weforum.org/reports/the-new-plastics-economy-rethinking-the-future-of-plastics
- Danso D. Chow J. Streita W. R. Plastics: Environmental and biotechnological perspectives on microbial degradation. Appl. Environ. Microbiol. 2019;85:e01095–e01019. doi: 10.1128/AEM.01095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador M. et al., Microbial Genes for a Circular and Sustainable Bio-PET Economy. Genes. 2019;10:373. doi: 10.3390/genes10050373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes R. A. Aristilde L. Degradation and metabolism of synthetic plastics and associated products by Pseudomonas sp.: capabilities and challenges. J. Appl. Microbiol. 2017;123:582–593. doi: 10.1111/jam.13472. [DOI] [PubMed] [Google Scholar]
- Wei R. Zimmermann W. Biocatalysis as a green route for recycling the recalcitrant plastic polyethylene terephthalate. Microb. Biotechnol. 2017;10:1302–1307. doi: 10.1111/1751-7915.12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jehanno C. Pérez-Madrigal M. M. Demarteau J. Sardon H. Dove A. P. Organocatalysis for depolymerisation. Polym. Chem. 2019;10:172–186. doi: 10.1039/C8PY01284A. [DOI] [Google Scholar]
- Celik G. et al., Upcycling Single-Use Polyethylene into High-Quality Liquid Products. ACS Cent. Sci. 2019;5:1795–1803. doi: 10.1021/acscentsci.9b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y. et al., Towards the circular economy: converting aromatic plastic waste back to arenes over Ru/Nb2O5 catalyst. Angew. Chem., Int. Ed. 2021:202011063. doi: 10.1002/ange.202011063. [DOI] [PubMed] [Google Scholar]
- Jie X. et al., Microwave-initiated catalytic deconstruction of plastic waste into hydrogen and high-value carbons. Nat. Catal. 2020;3:902–912. doi: 10.1038/s41929-020-00518-5. [DOI] [Google Scholar]
- Tennakoon A. et al., Catalytic upcycling of high-density polyethylene via a processive mechanism. Nat. Catal. 2020;3:893–901. doi: 10.1038/s41929-020-00519-4. [DOI] [Google Scholar]
- Yoshida S. et al., A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351:1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- Tournier V. et al., An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580:216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Austin H. P. et al., Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4350–E4357. doi: 10.1073/pnas.1718804115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott B. C. et al., Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc. Natl. Acad. Sci. U. S. A. 2020;117:25476–25485. doi: 10.1073/pnas.2006753117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H. F. et al., Rational Protein Engineering of Thermo-Stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019;9:3519–3526. doi: 10.1021/acscatal.9b00568. [DOI] [Google Scholar]
- Chen Z. et al., Efficient biodegradation of highly crystallized polyethylene terephthalate through cell surface display of bacterial PETase. Sci. Total Environ. 2020;709:136138. doi: 10.1016/j.scitotenv.2019.136138. [DOI] [PubMed] [Google Scholar]
- Puspitasari N. Tsai S. L. Lee C. K. Fungal Hydrophobin RolA Enhanced PETase Hydrolysis of Polyethylene Terephthalate. Appl. Biochem. Biotechnol. 2020:1–12. doi: 10.1007/s12010-020-03358-y. [DOI] [PubMed] [Google Scholar]
- Yan F. Wei R. Cui Q. Bornscheuer U. T. Liu Y. J. Thermophilic whole-cell degradation of polyethylene terephthalate using engineered Clostridium thermocellum. Microb. Biotechnol. 2021;14:374–385. doi: 10.1111/1751-7915.13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K. et al., Organocatalytic depolymerization of poly(ethylene terephthalate) J. Polym. Sci., Part A: Polym. Chem. 2011;49:1273–1281. doi: 10.1002/pola.24551. [DOI] [Google Scholar]
- Sun J. et al., Solubilization and Upgrading of High Polyethylene Terephthalate Loadings in a Low-Costing Bifunctional Ionic Liquid. ChemSusChem. 2018;11:781–792. doi: 10.1002/cssc.201701798. [DOI] [PubMed] [Google Scholar]
- Srivastava M. Kumar Roy P. Ramanan A. Rajagopal C. Post consumer PET waste as potential feedstock for metal organic frameworks. Mater. Lett. 2013;106:390–392. doi: 10.1016/j.matlet.2013.05.058. [DOI] [Google Scholar]
- Kenny S. T. et al., Up-Cycling of PET (Polyethylene Terephthalate) to the Biodegradable Plastic PHA (Polyhydroxyalkanoate) Environ. Sci. Technol. 2008;42:7696–7701. doi: 10.1021/es801010e. [DOI] [PubMed] [Google Scholar]
- Emadian S. M. Onay T. T. Demirel B. Biodegradation of bioplastics in natural environments. Waste Manage. 2017;59:526–536. doi: 10.1016/j.wasman.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Suzuki M. Tachibana Y. Kasuya K. Biodegradability of poly(3-hydroxyalkanoate) and poly(ε-caprolactone) via biological carbon cycles in marine environments. Polym. J. 2021;53:47–66. doi: 10.1038/s41428-020-00396-5. [DOI] [Google Scholar]
- Kim H. T. et al., Biological Valorization of Poly(ethylene terephthalate) Monomers for Upcycling Waste PET. ACS Sustainable Chem. Eng. 2019;7:19396–19406. doi: 10.1021/acssuschemeng.9b03908. [DOI] [Google Scholar]
- Vanillin Market Size, Share| Global Industry Report, 2018-2025. https://www.grandviewresearch.com/industry-analysis/vanillin-market
- Banerjee G. Chattopadhyay P. Vanillin biotechnology: the perspectives and future. J. Sci. Food Agric. 2019;99:499–506. doi: 10.1002/jsfa.9303. [DOI] [PubMed] [Google Scholar]
- Fache M. Boutevin B. Caillol S. Vanillin Production from Lignin and Its Use as a Renewable Chemical. ACS Sustainable Chem. Eng. 2016;4:35–46. doi: 10.1021/acssuschemeng.5b01344. [DOI] [Google Scholar]
- Luziatelli F. Brunetti L. Ficca A. G. Ruzzi M. Maximizing the Efficiency of Vanillin Production by Biocatalyst Enhancement and Process Optimization. Front. Bioeng. Biotechnol. 2019;7:279. doi: 10.3389/fbioe.2019.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J. Steinbüchel A. Priefert H. Highly efficient biotransformation of eugenol to ferulic acid and further conversion to vanillin in recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 2003;69:6569–6576. doi: 10.1128/AEM.69.11.6569-6576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J. Tao F. Du H. Xu P. Mimicking a natural pathway for de novo biosynthesis: natural vanillin production from accessible carbon sources. Sci. Rep. 2015;5:13670. doi: 10.1038/srep13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparan V. Krings U. Struch M. Berger R. G. A three-enzyme-system to degrade curcumin to natural vanillin. Molecules. 2015;20:6640–6653. doi: 10.3390/molecules20046640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M. Okada Y. Yoshida T. Nagasawa T. Biotransformation of isoeugenol to vanillin by Pseudomonas putida IE27 cells. Appl. Microbiol. Biotechnol. 2007;73:1025–1030. doi: 10.1007/s00253-006-0569-1. [DOI] [PubMed] [Google Scholar]
- Li T. Rosazza J. P. N. Biocatalytic synthesis of vanillin. Appl. Environ. Microbiol. 2000;66:684–687. doi: 10.1128/AEM.66.2.684-687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjapur A. M. Tarasova Y. Prather K. L. J. Synthesis and accumulation of aromatic aldehydes in an engineered strain of escherichia coli. J. Am. Chem. Soc. 2014;136:11644–11654. doi: 10.1021/ja506664a. [DOI] [PubMed] [Google Scholar]
- Finnigan W. et al., Characterization of Carboxylic Acid Reductases as Enzymes in the Toolbox for Synthetic Chemistry. ChemCatChem. 2017;9:1005–1017. doi: 10.1002/cctc.201601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law B. J. C. et al., Effects of active-site modification and quaternary structure on the regioselectivity of catechol-O-methyltransferase. Angew. Chem., Int. Ed. 2016;55:2683–2687. doi: 10.1002/anie.201508287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains J. Wulff J. E. Boulanger M. J. Investigating terephthalate biodegradation: Structural characterization of a putative decarboxylating cis-dihydrodiol dehydrogenase. J. Mol. Biol. 2012;423:284–293. doi: 10.1016/j.jmb.2012.07.022. [DOI] [PubMed] [Google Scholar]
- Schlafli H. R. Weiss M. A. Leisinger T. Cook A. M. Terephthalate 1,2-dioxygenase system from Comamonas testosteroni T-2: Purification and some properties of the oxygenase component. J. Bacteriol. 1994;176:6644–6652. doi: 10.1128/jb.176.21.6644-6652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara Y. Kasai D. Katayama Y. Fukuda M. Masai E. Enzymatic properties of terephthalate 1,2-dioxygenase of Comamonas sp. strain E6. Biosci. Biotechnol. Biochem. 2008;72:2335–2341. doi: 10.1271/bbb.80236. [DOI] [PubMed] [Google Scholar]
- Sasoh M. et al., Characterization of the terephthalate degradation genes of Comamonas sp. strain E6. Appl. Environ. Microbiol. 2006;72:1825–1832. doi: 10.1128/AEM.72.3.1825-1832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tramontina R. et al., Consolidated production of coniferol and other high-value aromatic alcohols directly from lignocellulosic biomass. Green Chem. 2020;22:144–152. doi: 10.1039/C9GC02359C. [DOI] [Google Scholar]
- Jaganaman S. Pinto A. Tarasev M. Ballou D. P. High levels of expression of the iron-sulfur proteins phthalate dioxygenase and phthalate dioxygenase reductase in Escherichia coli. Protein Expression Purif. 2007;52:273–279. doi: 10.1016/j.pep.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco A. Vigh L. Diamant S. Goloubinoff P. Native folding of aggregation-prone recombinant proteins in Escherichia coli by osmolytes, plasmid- or benzyl alcohol-overexpressed molecular chaperones. Cell Stress Chaperones. 2005;10:329–339. doi: 10.1379/CSC-139R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosaka M. et al., Novel Tripartite Aromatic Acid Transporter Essential for Terephthalate Uptake in Comamonas sp. Strain E6. Appl. Environ. Microbiol. 2013;79:6148–6155. doi: 10.1128/AEM.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. C. Currin A. Kell D. B. Ultra-high throughput functional enrichment of large monoamine oxidase (MAO-N) libraries by fluorescence activated cell sorting. Analyst. 2018;143:4747–4755. doi: 10.1039/C8AN00851E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher E. Pilizota T. Davies P. R. McVey A. French C. E. Characterization of the effects of n-butanol on the cell envelope of E. coli. Appl. Microbiol. Biotechnol. 2016;100:9653–9659. doi: 10.1007/s00253-016-7771-6. [DOI] [PubMed] [Google Scholar]
- Kunjapur A. M. Hyun J. C. Prather K. L. J. Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell Fact. 2016;15:61. doi: 10.1186/s12934-016-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noubigh A. Abderrabba M. Provost E. Salt addition effect on partition coefficient of some phenolic compounds constituents of olive mill wastewater in 1-octanol-water system at 298.15 K. J. Iran. Chem. Soc. 2009;6:168–176. doi: 10.1007/BF03246517. [DOI] [Google Scholar]
- Dafoe J. T. Daugulis A. J. In situ product removal in fermentation systems: improved process performance and rational extractant selection. Biotechnol. Lett. 2014;36:443–460. doi: 10.1007/s10529-013-1380-6. [DOI] [PubMed] [Google Scholar]
- Wallace S. Balskus E. P. Designer Micelles Accelerate Flux Through Engineered Metabolism in E. coli and Support Biocompatible Chemistry. Angew. Chem., Int. Ed. 2016;55:6023–6027. doi: 10.1002/anie.201600966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayaci F. Uyar T. Solid inclusion complexes of vanillin with cyclodextrins: Their formation, characterization, and high-temperature stability. J. Agric. Food Chem. 2011;59:11772–11778. doi: 10.1021/jf202915c. [DOI] [PubMed] [Google Scholar]
- Larock R. Leung W. Stolz-Dunn S. Ehrlich J. Bogart M. T. Hamon D. P. G. J. A Novel, Nonaqueous Method for Regeneration of Aldehydes from Bisulfite Adducts. J. Org. Chem. 1992;35:4168. doi: 10.1021/jo990543v. [DOI] [PubMed] [Google Scholar]
- Inoue K. Fukuda K. Yoshimura T. Kusano K. Comparison of the Reactivity of Trapping Reagents toward Electrophiles: Cysteine Derivatives Can Be Bifunctional Trapping Reagents. Chem. Res. Toxicol. 2015;28:1546–1555. doi: 10.1021/acs.chemrestox.5b00129. [DOI] [PubMed] [Google Scholar]
- Della Ragione F. Porcelli M. Cartenì-Farina M. Zappia V. Pegg A. E. Escherichia coli S-adenosylhomocysteine/5′-methylthioadenosine nucleosidase. Purification, substrate specificity and mechanism of action. Biochem. J. 1985;232:335–341. doi: 10.1042/bj2320335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler J. C. Humphreys L. D. Snajdrova R. Burley G. A. A Tandem Enzymatic sp 2 -C-Methylation Process: Coupling in Situ S-Adenosyl- l -Methionine Formation with Methyl Transfer. ChemBioChem. 2017;18:992–995. doi: 10.1002/cbic.201700115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.