Graphical abstract
Keywords: ACE2, SARS-CoV-2, Spike glycoprotein, Pomegranate, Punicalagin, Punicalin, Urolithin A
Abstract
The novel coronavirus disease (Covid-19) has become a major health threat globally. The interaction of SARS-CoV-2 spike (S) glycoprotein receptor-binding domain (RBD) with ACE2 receptor on host cells was recognized as the first step of virus infection and therefore as one of the primary targets for novel therapeutics. Pomegranate extracts are rich sources of bioactive polyphenols that were already recognized for their beneficial health effects. In this study, both in silico and in vitro methods were employed for evaluation of pomegranate peel extract (PoPEx), their major polyphenols, as well as their major metabolite urolithin A, to attenuate the contact of S-glycoprotein RBD and ACE2. Our results showed that PoPEx, punicalin, punicalagin and urolithin A exerted significant potential to block the S-glycoprotein-ACE2 contact. These in vitro results strongly confirm the in silico predictions and provide a valuable insight in the potential of pomegranate polyphenols for application in SARS-CoV-2 infection.
1. Introduction
Since December 2019 when the first media statement was released by the Wuhan Municipal Health Commission on cases of ‘viral pneumonia’ in China, the pandemic has spread over 200 countries with over 100 million confirmed cases (https://covid19.who.int/). The SARS-CoV-2, a newly discovered type of coronavirus, was identified as the main causative pathogen of disease named coronavirus disease (COVID-19). Coronaviruses (CoV) (Family: Coronaviridae) are viruses with a spiked envelope which is crucially involved in a process of viral infection with nonsegmented, positive‐stranded genomic RNA contained therein [1]. The SARS-CoV-2 contains 4 main structural proteins: Envelope (E), Membrane (M), Nucleocapsid (N) and Spike (S) transmembrane glycoprotein. While other proteins are included in different processes of virus life cycle, only S-glycoprotein is directly involved in virus attachment to specific receptors on the host cells and presents druggable target of SARS-CoV-2 with therapeutic perspective [2]. This process is essential for virus entry and the beginning of infection [3]. The S-glycoprotein is composed of two subunits, S1 and S2, with different functions. The S1 subunit contains a receptor-binding domain (RBD), which binds to the cell surface receptor Angiotensin-Converting Enzyme 2 (ACE2), causing conformational changes to the S2 subunit, which allows the membrane fusion process. In particular, SARS-CoV-2 has a higher affinity for the human ACE2 receptors than other coronaviruses and the described mechanism is considered as the primary mechanism of SARS-CoV-2 entry into the host cells. The increased transmission of SARS-CoV-2 in comparison with SARS and MERS viruses could be explained by the increased number of cellular ACE2 receptors that enable the SARS-CoV-2 to enter host cells [4]. Both processes depend on proteolytic activity of the enzymes present in the membrane of the host cell, such as transmembrane protease, serine 2 (TMPRSS2) and furin [5], [6]. Natural products that are used as food or herbal medicines have historically been used to prevent viral infections and are generally proved as beneficial and safe [7]. It was previously reported that several naturally occurring molecules exert significant antiviral activity against different strains of coronaviruses. Although these molecules represent different chemical classes, they all possess functional groups which can serve as hydrogen bonds acceptors or donors. Saikosaponins are triterpene glycosides that effectively disable virus attachment and penetration into the host cell. Other polyphenolic compounds such as amentoflavone, lycorine, myricetin and scutellarein have documented anti-SARS-CoV effects, as well [7]. Among the myriad of substances that are currently being studied for anti SARS-CoV-2 activity, natural products are of particular importance [8]. Computational methods were introduced to achieve fast screening of a large natural products databases [9]. This approach could easily identify herbal substances with the most promising characteristics. One of the chemical classes that have recently drawn significant attention for its anti-SARS-CoV-2 potential, are polyphenols from various natural sources [10], [11]. Frank et al. (2020) tested and compared virucidal activity of different food products as natural beverages. Results of their study showed a significant ability of pomegranate juice to inactivate SARS-CoV-2 particles in vitro. Considering that for certain pomegranate polyphenols, such as punicalin, the ability to inhibit influenza virus through interaction with its surface glycoproteins has already been shown, it was logical to assume that similar mechanism for SARS-CoV-2 inhibition is involved [12]. In our previous computational study, we confirmed that the most abundant polyphenols from pomegranate peel extract (PoPEx) formed stable interactions with S-glycoprotein residues from the predicted active site and therefore proved to be potential candidates to prevent the process of virus internalization into the host cells [13]. Some other studies have shown that certain hydrolysable tannins including punicalagin and punicalin had the potential not only to intervene in SARS-CoV-2 entry process but also to interfere with the viral replication process inside the host cell [14], [15], [16]. Pomegranate fruit peel is usually discarded as waste, but chemical analysis of peel extracts showed they are also a rich source of bioactive polyphenols [17], which affects the increase in their use as the ingredients in herbal products and dietary supplements. Moreover, the highest total phenolic content was detected in pomegranate peel in comparison with juice and seeds [18]. Although being promising candidates, the potential therapeutic application of pomegranate polyphenols is contingent upon their bioavailability. Several studies investigated ellagitannin biotransformation in rats and humans. Just in one study, punicalagin was detected in biologically relevant concentrations in rats [19]. However, it has generally been accepted that the majority of these compounds undergo intensive metabolism in humans, and one of the major metabolites was identified as urolithin A [20]. Urolithins are biologically active metabolites with confirmed anti-inflamatory activity through the mechanism of mitophags activation [21]. Recently reported results from in silico study showed that urolithin formed stable complexes with enzymes involved in the SARS-CoV-2 replication process. In our previous in silico study, we searched for the most druggable binding site on RBD of S-glycoprotein for testing the PoPEx potential for interaction. However, interactions with any of RBD amino-acid residues might interfere with S-glycoprotein and ACE2 receptor contacts [22]. Therefore, in this study, we expanded our docking simulation to the whole S-glycoprotein RBD region and introduced the molecular dynamic simulation to test the ligand–protein dynamics. Furthermore, along with the PoPEx polyphenolic constituents analysis, we also tested the urolithin A, for the same activity in vitro. According to the available literature, urolithin A as a major metabolite of ellagitannins from pomegranate and biologically active compound haven’t been previously evaluated for anti SARS-CoV-2 activity. The obtained in silico and in vitro results for all tested compounds in this study were compared with the results obtained for umifenovir as a positive control under the same experimental conditions.
2. Results
2.1. HPLC analysis
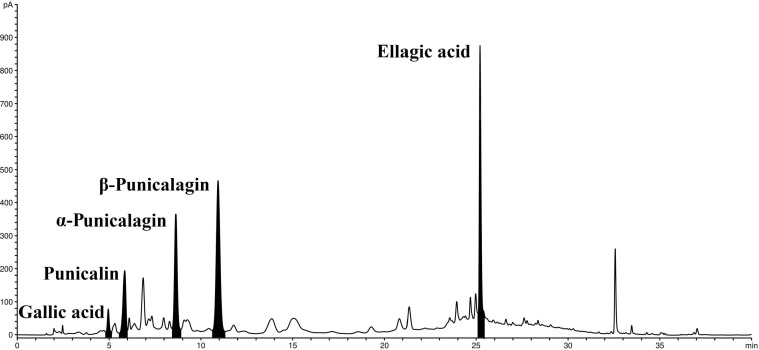
We studied anti-SARS-CoV-2 activity of PoPEx with main polyphenols quantified by HPLC (Table 1 , Fig. 1 ). The selected polyphenols eluted as sharp, well separated symmetrical peaks under analytical conditions. Moreover, α- and β-punicalagin were completely separated with retention times of 8.6 and 10.9 min, respectively.
Table 1.
The quantity of the most abundant pomegranate peel extract (PoPEx) polyphenols and their retention times determined by using HPLC method.
| Retention time (min) | Compound | mg/g DW* |
|---|---|---|
| 4.9 | Gallic acid | 7.74 ± 0.29 |
| 5.8 | Punicalin | 31.31 ± 0.71 |
| 8.6 | α-Punicalagin | 26.02 ± 6.77 |
| 10.9 | β-Punicalagin | 45.57 ± 4.62 |
| 25.2 | Ellagic acid | 22.82 ± 0.22 |
milligrams per gram of dry weight.
Fig. 1.
The HPLC chromatogram of pomegranate peel extract (PoPEx) major constituents.
The results showed that PoPEx phenols were principally composed of punicalagin isomers: α- and β-punicalagin (26.02 and 45.57 mg/g DW, respectively). The second most abundant ellagitannin was punicalin (31.31 mg/g DW), followed by ellagic (22.82 mg/g DW) and gallic acid (7.74 mg/g DW). Ellagic acid is commonly found in ellagitannin-rich fruits in free or bound forms from which it can spontaneously hydrolyze.
2.2. Molecular docking simulation
Molecular docking studies were employed for the prediction of the most stable binding pose of the investigated molecules in complex with S-glycoprotein. The data obtained from these protein–ligand interactions are summarized in Table 2 .
Table 2.
Molecular docking study results for the PoPEx most abundant compounds and their metabolite urolithin A.
| Molecule | Binding energy [kcal/mol] | H-bond receptor-ligand interactions |
|---|---|---|
| Ellagic acid | −7.56 | Asn 343, Ser 373, Asn 437, Asn 440, Arg 509 |
| Gallic acid | −6.29 | Asp 467, Ser 469, Glu 471 |
| Punicalin | −9.25 | Asp 428, Pro 463, Phe 515, Glu 516, Leu 517 |
| Punicalagin | −7.79 | Glu 340, Ala 348, Ala 352, Ser 399, Asn 450 |
| Urolithin A | −6.86 | Asn 343, Asp 364, Val 362 |
| Umifenovir (positive control) | −5.97 | Thr 430 |
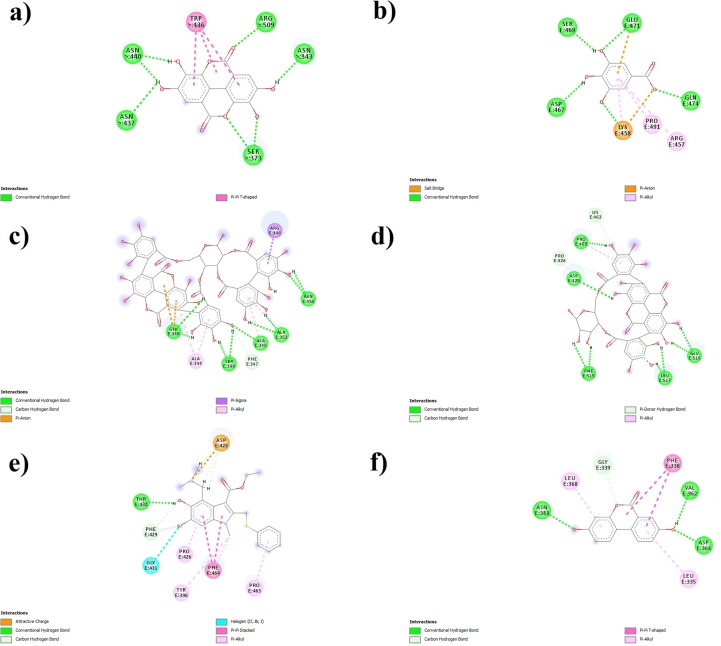
The molecular docking study revealed that punicalin had the highest docking score (−9.25 kcal/mol) compared to other tested compounds from PoPEx. Punicalagin interacted as the second most stable complex with target protein with the score of −7.79 kcal/mol. All other tested compounds could also form hydrogen bond complexes with S-glycoprotein amino acid residues but with higher binding energies. Some of the RBD amino acid residues which stabilize complexes through interactions with PoPEx polyphenols have been previously identified as key interaction residues with other potent phytochemicals (Fig. 2 ). Predicted active sites for all tested compounds except for ellagic acid and umifenovir contain aspartic and glutamic acid which are negatively charged at physiological pH. Considering that gallic acid according to its pKa value (4.4 at 25 ℃) is a negatively charged molecule at pH = 7.4 it is expected that presence of negatively charged residues in active site significantly influences binding affinity of this compound.
Fig. 2.
Molecular docking ligand S-glycoprotein interactions with: a) Ellagic acid, b) Gallic acid, c) Punicalagin, d) Punicalin, e) Umifenovir and f) Urolithin A.
2.3. Molecular dynamics simulation
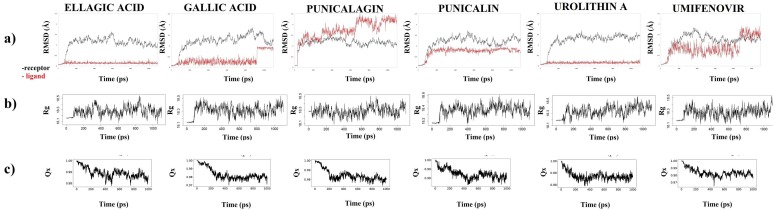
For the following set of experiments, we introduced the ligand–protein MD studies. The RMSD is the measure of the average distance between the atoms of superimposed structures. The RMSD results of ligand–protein MD simulation of 6 selected docked complexes showed that the interactions were stable over 1 ns simulation time step. The Rg parameter is very important for the evaluation of protein folding state. The detected values of Rg were in a range of 18.06 Å to 18.55 Å (interaction of 6M0J and ellagic acid), 18.11 Å to 18.54 Å (interaction of 6M0J and gallic acid), 18.05 Å to 18.50 Å (interaction of 6M0J and punicalagin), 18.15 Å to 18.60 Å (interaction of 6M0J and punicalin), 18.12 Å to 18.57 Å (interaction of 6M0J and urolithin A), as well as 18.11 Å to 18.52 Å (interaction of 6M0J and umifenovir). Another important parameter that changes with the unfolding of protein is the Qx. The MD simulation results with Qx values close to 1 indicate a native state of protein (Fig. 3 ).
Fig. 3.
The MD simulation results of a) RMSD, b) Radius of gyration (Rg) and c) Fraction of Native Contacts (Qx) of the most stable complexes ligand-receptor complexes from the docking studies.
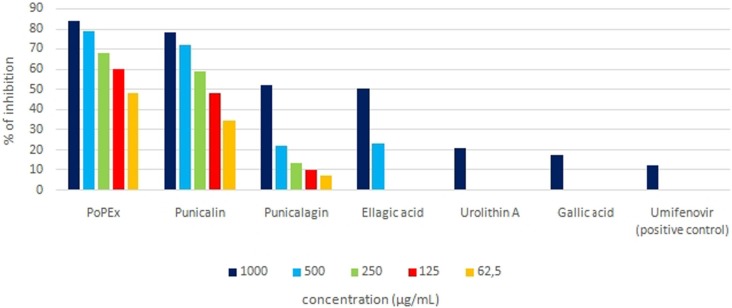
2.4. In vitro SARS-CoV-2 inhibition assay
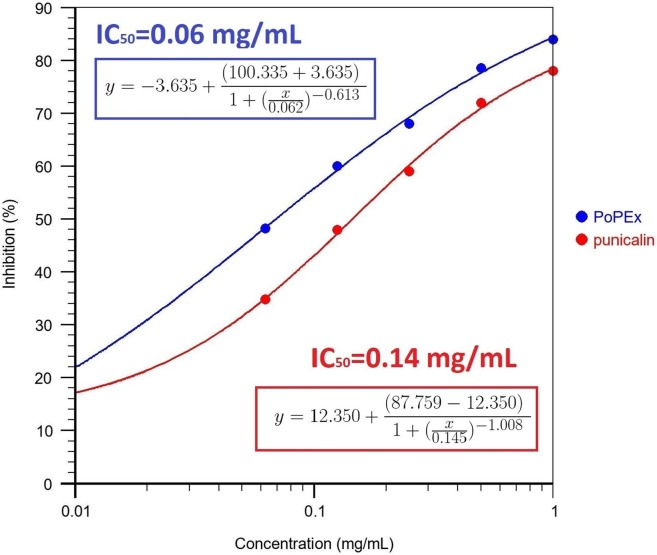
Different concentration ranges of the selected PoPEx polyphenols samples were evaluated in vitro for S-glycoprotein-ACE2 receptor contact inhibition. The results of SARS-CoV-2 inhibitory activity of tested samples and reference compounds at the concentration range of (62.5–1000 μg/mL) are presented in Fig. 4 . The PoPEx exhibited the highest inhibition against SARS-CoV-2 compared to the other tested samples. The SARS-CoV-2 inhibitory activity of all tested compounds was as follows: PoPEx > Punicalin > Punicalagin ≫ Ellagic acid > Urolithin A > Gallic acid > Umifenovir (positive control). The dose-dependent responses of PoPEx, punicalin, punicalagin and ellagic acid were observed in the tested concentration ranges. While punicalagin inhibited 50% of S-glycoprotein and ACE2 contact only in the sample of the highest concentration (1 mg/mL), PoPEx and punicalin attenuated this contact in different concentration ranges in a dose-dependent manner. The calculated IC50 values for PoPEx and punicalin were 0.06 and 0.14 mg/mL, respectively (Fig. 5 ).
Fig. 4.
The SARS-CoV-2 inhibition activity in vitro results.
Fig. 5.
The IC50 equations and concentration-inhibition response of PoPEx and punicalin against S-glycoprotein-ACE2 interaction.
In the tested concentration range the highest inhibitory activity for S-glycoprotein-ACE2 receptor-binding interaction was 83.25%, recorded for PoPEx sample (1 mg/mL). In the same concentration, punicalin inhibited 78% of the same binding activity. The lowest inhibition activity was observed for gallic acid which in maximally tested concentration inhibited the S-glycoprotein-ACE2 binding interaction for 17.42%. However, gallic acid was still more potent inhibitor than umifenovir with 12.68% inhibition activity in the same concentration. These in vitro results regarding the order of PoPEx polyphenols inhibiting potential for S-glycoprotein-ACE2 binding interaction are in exact accordance with our in silico docking study results.
3. Discussion
3.1. HPLC analysis
Pomegranate is a rich natural source of vitamins, polyphenols and dietary fibers, which makes it valuable and worldwide popular dietary fruit [23]. It is even considered as a superfood due to its high antioxidant potential and other beneficial effects on human health [24], [25]. The most abundant phenolic compounds found in pomegranate belong to the ellagitannins and anthocyanins. Interestingly, non edible parts of pomegranate such as peel and seeds are also a rich source of polyphenols, which are considered as principal contributors to pomegranate beneficial effects [24]. The selection of extraction method parameters such as solvent, temperature, solid–solvent ratio or particle size could significantly influence the total polyphenolic content [26]. Moreover, the ratio of ellagitannins and ellagic acid content was also influenced by the origin of pomegranate material [27]. Although certain variations in the amount of PoPEx ellagitannins detected in our sample were observed, the content of punicalagin remains inside a variation range reported in the literature [15], [17], [28].
3.2. Molecular docking simulation
The results obtained from the molecular docking simulation study showed that all the most abundant PoPEx polyphenols formed more stable complexes with amino-acid residues in receptor-binding site than positive control umifenovir (−5.97 kcal/mol), under the same experimental conditions. Umifenovir is a well known synthetic broad-spectrum antiviral agent, which mechanism of action is based on prevention of the S-glycoprotein contact with the host cell receptor [29]. Punicalin with the highest binding affinity was the most promising candidate for the inhibition of S-glycoprotein-ACE2 interaction, which is in accordance with the results from recent in silico studies [13], [30]. This complex of S-glycoprotein and punicalin is also stabilized with interactions of Pro 463 and Glu 516, which are previously marked as key interactions of ashwagandhanolide, a bioactive herbal ligand with the hydrogen bond interactions at the active site of SARS-CoV-2 S-glycoprotein [31]. Lower binding affinity of gallic acid compared to other tested compound could be explained by repulsive forces between negatively charged ligand and residues from the active site at pH = 7.4 [32]. However, other negatively charged ligands (ellagic acid and umifenovir) are not expected to be affected by these forces due to lack of negatively charged residues at the respective active sites. Urolithin A also formed a complex with RBD residues with higher binding affinity (−6.86 kcal/mol) than umifenovir, which indicates a significant potential for inhibitory activity.
3.3. Molecular dynamics simulation
RMSD plots from MD simulation revealed that ellagic acid, gallic acid, punicalin and urolithin were stable during simulation while a low fluctuation was recorded for punicalagin and umifenovir (positive control). Moreover, other MD simulation parameters confirmed that S-glycoprotein interacted in its native state that are only considered energetically favorable [33]. Equalized RMSD and Rg plots indicate the equilibrium of the system and minimal changes in the macromolecule folding state, respectively [34]. The detected Rg values range from 18.05 to 18.60 Å represents a characteristic of the compactness of protein structure and shows that globular portion of spike protein containing RBD region possesses structural characteristics of class A globular proteins [35].
3.4. In vitro SARS-CoV-2 inhibition assay
In silico simulation results obtained in this study were fully confirmed by the in vitro results. In vitro inhibition activities of studied polyphenols were in the exact order as predicted in molecular docking simulations. As expected, predicted repulsive forces between negatively charged gallic acid and negatively charged residues at the active site resulted in lower in vitro activity of this PoPEx constituent compared to the other tested compounds. Moreover, the in vitro results correspond with a recently reported study, which tested PoPEx potential on inhibition of S-glycoprotein-ACE2 interaction with the screening kit from another manufacturer [15]. They tested extracts in a concentration range from 40 to 1000 µg/mL and registered inhibition of up to 74%, which was dose-dependent, but lower than in this study. Additionally, the order of the exhibited activity for the individual PoPEx constituent which they considered relevant was similar to the present findings, except for punicalin, which was not tested [15]. However, in the present study, punicalin was the most potent polyphenol tested, both in silico and in vitro. Considering that none of the individually tested PoPEx polyphenols exceeded the inhibitory activity potential of the PoPEx, we assumed that some other constituents could contribute to the observed activity. It is also possible that PoPEx constituents possess a significant synergistic effect on the tested activity, similar to the higher efficiency of plant-based molecules and synthetic drugs combination in reducing the viral load showed in other studies [36]. Virucidal effects of pomegranate constituent have been confirmed in studies that evaluated pomegranate juice and lozenge potential for decreasing SARS-CoV-2 loads in oral cavity [11], [37]. This activity could be beneficial regarding prevention of the initial infection, viral dissemination into the lower parts of the respiratory system or transmission to the next individual [37]. Prevention of infection and viral dissemination are related to virus S-glycoprotein-ACE2 receptor interaction as the first step toward the virus internalization. In order to be able to prevent the interaction of SARS-CoV-2 with the host cell receptor, the small molecule must have appropriate bioavailability. The PoPEx polyphenols undergo intensive metabolism by human gut microbiota forming urolithins, which are primarily responsible for health beneficial effects [38]. Considering the aforementioned, PoPEx polyphenols could have the most significant contribution when applied locally for a viral load decrease in oral or nasal cavity which is important for transmission prevention. Nevertheless, other in vivo PoPEx effects should be considered through their main metabolites (urolithins) activity as well. To the best of the authors' knowledge, this is the first report about urolithin A (major ellagitannins metabolite) in silico and in vitro potential to attenuate the SARS-CoV-2 S-glycoprotein binding ability to ACE2 receptor.
4. Materials and methods
4.1. Chemicals
The chemical substances such as ellagic acid, gallic acid, punicalagin (mixture of anomers), punicalin, umifenovir, urolithin A were obtained from Sigma Chemical Co. (St. Louis, USA). Solvents used for dissolution of the tested compounds were dimethylsulfoxide (DMSO) and phosphate-buffered saline (PBS) which were also obtained from Sigma Chemical Co. (St. Louis, USA). DMSO didn’t exceed screening kit recommended concentration.
4.2. Plant material
Pomegranate fruits were collected during November 2019 in village Do, the Republic of Srpska, Bosnia and Herzegovina at natural locality. Manually separated peels were air-dried at room temperature for 4–6 days and grounded to provide particles of 0.75–2 mm size used further for extraction.
4.3. Pomegranate peel extract
Dry PoPEx was prepared from triple percolate (1:1) obtained with 70% ethanol as a solvent further evaporated to dryness in a vacuum evaporator. The most abundant polyphenols were quantified by the HPLC method.
4.4. HPLC analysis
The analyses were carried out on Agilent 1200 RR HPLC instrument (Agilent, Waldbronn, Germany), equipped with DAD detector, on a reverse phase Zorbax SB-C18 (Agilent) analytical column (150 mm × 4.6 mm i.d., 5 µm particle size), and the column temperature was maintained at 25 °C. The mobile phase consisted of solvent A (1% v/v solution of orthophosphoric acid in water) and mobile phase B (acetonitrile). The injection volume was 3 μL, the flow rate was adjusted to 1 mL/min, and detection wavelengths were set at 260 and 320 nm. Gradient elution was applied according to the following scheme: 0–5 min, 98–90% A; 5–15 min, 90% A; 15–20 min, 90–85% A; 20–25 min, 85–70% A; 25–30 min, 70–40% A; 30–34 min, 40–0% A, with post-time of 2 min. Quantification of ellagic acid, gallic acids, punicalagin and punicalin was done using calibration curves of authentic standards. The results are presented as milligrams per gram of dry weight (mg/g DW).
4.5. Datasets
The crystal structure of S-glycoprotein RBD in complex with human ACE2 was retrieved from the Protein Data Bank (PDB; www.pdb.org, PDB ID:6M0J). Protein structure was prepared for the docking analysis using Yasara Structure (v. 20.4.24) (http://www.yasara.org/). This procedure included deletion of solvents from the PDB files, adding hydrogens and charges to the structure, and process of energy minimization. The 3D molecular structures of molecules of ellagic acid, gallic acid, urolithin A and umifenovir were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) whereas Chemspider (http://www.chemspider.com/) was used to download the punicalagin and punicalin molecules. All ligands were further prepared for the docking study using the biocomputing software Yasara Structure energy minimization process at physiological pH (7.4).
4.6. Molecular docking simulation
The RBD residues (306–527) were selected as the binding site for the ligand molecules with the grid box generated around amino acid residues within a distance of 5 Å. Thereafter, a docking procedure was conducted through Yasara Structure software based on the AutoDockLGA algorithm and AMBER03 force field [39]. Output files of the most stable complexes were further analyzed with the visualization software (Discovery Studio Visualizer v.20.1.0.19295).
4.7. Molecular dynamics simulation
The molecular dynamics (MD) of six most stable docked complexes with: 6M0J and ellagic acid, gallic acid, punicalagin, punicalin, urolithin A and umifenovir were investigated on LARMD server (http://chemyang.ccnu.edu.cn/ccb/server/LARMD/) at Int mod parameter for 1 ns in an explicit water model [34]. LARMD provides an automatic protocol for conformational sampling and analysis. The root-mean-square deviation (RMSD), radius of gyration (Rg) and fraction of native contacts (Qx) were evaluated.
4.8. In vitro SARS-CoV-2 inhibition assay
To investigate the effects of PoPEx polyphenols on SARS-CoV-2 binding activity to ACE2 the MBS669459 screening kit (MyBiosource.com) was employed. This assay is based on a colorimetric ELISA kit that measures the binding of RBD of the S-glycoprotein from SARS-CoV-2 to its human receptor ACE2. All tested samples were dissolved in phosphate buffer solution or DMSO with a final concentration of ≤0.1%. Reagents preparation and assay procedure steps were conducted strictly following the provided protocol for default configuration (https://www.mybiosource.com/covid-19-assay-kits/covid-19-coronavirus/669459).
4.9. Statistical analysis of IC50
The IC50 values were calculated by The Quest Graph™ IC50 online calculator (https://www.aatbio.com/tools/ic50-calculator) [40].
5. Conclusions
In this study, we confirmed that PoPEx and its major polyphenols, such as punicalin and punicalagin, have a strong potential to attenuate the SARS-CoV-2 S-glycoprotein binding ability to ACE2 receptor using in silico and in vitro approaches. It was shown that pomegranate bioactivity could also be attributed to urolithin A, one of the major PoPEx metabolites in humans. The most pronounced in vitro activity was observed for PoPEx, suggesting a possible synergistic effect of pomegranate polyphenols. We have shown that pomegranate polyphenols have a potential to attenuate the SARS-CoV-2 S-glycoprotein binding ability to ACE2 receptor and therefore to be excellent candidates for possible therapeutic application.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2021.105145.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faheem, B.K. Kumar, K. Sekhar, S. Kunjiappan, J. Jamalis, R. Balana-Fouce, B.L. Tekwani, M. Sankaranarayanan, Druggable targets of SARS-CoV-2 and treatment opportunities for COVID-19, Bioorg Chem 104 (2020) 104269, 10.1016/j.bioorg.2020.104269. [DOI] [PMC free article] [PubMed]
- 3.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahmoud I.S., Jarrar Y.B., Alshaer W., Ismail S. SARS-CoV-2 entry in host cells-multiple targets for treatment and prevention. Biochimie. 2020;175:93–98. doi: 10.1016/j.biochi.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M., Hofmann-Winkler H., Pöhlmann S. In: Activation of Viruses by Host Proteases. Böttcher-Friebertshäuser E., Garten W., Dieter Klenk H., editors. Springer; Cham: 2018. Priming Time: How Cellular Proteases Arm Coronavirus Spike Proteins; pp. 71–98. [Google Scholar]
- 7.Lin L.T., Hsu W.C., Lin C.C. Antiviral natural products and herbal medicines. J Tradit Complement Med. 2014;4(1):24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahzad F., Anderson D., Najafzadeh M. The antiviral, anti-inflammatory effects of natural medicinal herbs and mushrooms and SARS-CoV-2 infection. Nutrients. 2020;12(9) doi: 10.3390/nu12092573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W., Jr. Computational methods in drug discovery. Pharmacol Rev. 2014;66(1):334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. Sharma, V. Kumar Bhardwaj, R. Singh, V. Rajendran, R. Purohit, S. Kumar, An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2, Food Chem 346 (2021) 128933, https://doi.org/10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed]
- 11.B. Frank, C. Conzelmann, T. Weil, R. Groß, P. Jungke, M. Eggers, J.A. Müller, J. Münch, U. Kessler, Antiviral activity of plant juices and green tea against SARS-CoV-2 and influenza virus in vitro, bioRxiv 2020.10.30.360545 (2020), https://doi.org/10.1101/2020.10.30.360545.
- 12.M. Haidari, M. Ali, S. Ward Casscells, 3rd, M. Madjid, Pomegranate (Punica granatum) purified polyphenol extract inhibits influenza virus and has a synergistic effect with oseltamivir, Phytomedicine 16(12) (2009) 1127-36, https://doi.org/10.1016/j.phymed.2009.06.002. [DOI] [PubMed]
- 13.Suručić R., Tubić B., Stojiljković M.P., Djuric D.M., Travar M., Grabež M., Šavikin K., Škrbić R. Computational study of pomegranate peel extract polyphenols as potential inhibitors of SARS-CoV-2 virus internalization. Mol. Cell Biochem. 2020 doi: 10.1007/s11010-020-03981-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puttaswamy H., Gowtham H.G., Ojha M.D., Yadav A., Choudhir G., Raguraman V., Kongkham B., Selvaraju K., Shareef S., Gehlot P., Ahamed F., Chauhan L. In silico studies evidenced the role of structurally diverse plant secondary metabolites in reducing SARS-CoV-2 pathogenesis. Sci. Rep. 2020;10(1):20584. doi: 10.1038/s41598-020-77602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A. Tito, A. Colantuono, L. Pirone, E. Pedone, D. Intartaglia, G. Giamundo, I. Conte, P. Vitaglione, F. Apone, A pomegranate peel extract as inhibitor of SARS-CoV-2 Spike binding to human ACE2 (in vitro): a promising source of novel antiviral drugs, bioRxiv 2020.12.01.406116 (2020), https://doi.org/10.1101/2020.12.01.406116. [DOI] [PMC free article] [PubMed]
- 16.Khalifa I., Zhu W., Mohammed H.H.H., Dutta K., Li C. Tannins inhibit SARS-CoV-2 through binding with catalytic dyad residues of 3CL(pro): an in silico approach with 19 structural different hydrolysable tannins. J. Food Biochem. 2020 doi: 10.1111/jfbc.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Šavikin K., Živković J., Alimpić A., Zdunić G., Janković T., Duletić-Laušević S., Menković N. Activity guided fractionation of pomegranate extract and its antioxidant, antidiabetic and antineurodegenerative properties. Ind. Crops Prod. 2018;113:142–149. doi: 10.1016/j.indcrop.2018.01.031. [DOI] [Google Scholar]
- 18.Gozlekci S., Saracoglu O., Onursal E., Ozgen M. Total phenolic distribution of juice, peel, and seed extracts of four pomegranate cultivars. Pharmacogn. Mag. 2011;7(26):161–164. doi: 10.4103/0973-1296.80681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerda B., Tomas-Barberan F.A., Espin J.C. Metabolism of antioxidant and chemopreventive ellagitannins from strawberries, raspberries, walnuts, and oak-aged wine in humans: identification of biomarkers and individual variability. J. Agric. Food Chem. 2005;53(2):227–235. doi: 10.1021/jf049144d. [DOI] [PubMed] [Google Scholar]
- 20.Espin J.C., Larrosa M., Garcia-Conesa M.T., Tomas-Barberan F. Biological significance of urolithins, the gut microbial ellagic acid-derived metabolites: the evidence so far. Evid. Based Complement Alternat. Med. 2013;2013 doi: 10.1155/2013/270418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. Ryu, L. Mouchiroud, P.A. Andreux, E. Katsyuba, N. Moullan, A.A. Nicolet-Dit-Felix, E.G. Williams, P. Jha, G. Lo Sasso, D. Huzard, P. Aebischer, C. Sandi, C. Rinsch, J. Auwerx, Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents, Nat Med 22(8) (2016) 879-88, 10.1038/nm.4132. [DOI] [PubMed]
- 22.Garcia-Iriepa C., Hognon C., Frances-Monerris A., Iriepa I., Miclot T., Barone G., Monari A., Marazzi M. Thermodynamics of the interaction between the spike protein of severe acute respiratory syndrome coronavirus-2 and the receptor of human angiotensin-converting enzyme 2. Effects of possible ligands. J. Phys. Chem. Lett. 2020;11(21):9272–9281. doi: 10.1021/acs.jpclett.0c02203. [DOI] [PubMed] [Google Scholar]
- 23.A.A. Asmah R, Evaluation of Total Phenolic Content, Total Antioxidant Activity, and Antioxidant Vitamin Composition of Pomegranate Seed and Juice, General Med 03(01) (2014), https://doi.org/10.4172/2327-5146.1000164.
- 24.Vučić V., Grabež M., Trchounian A., Arsić A. Composition and potential health benefits of pomegranate: a review. Curr. Pharm. Des. 2019;25(16):1817–1827. doi: 10.2174/1381612825666190708183941. [DOI] [PubMed] [Google Scholar]
- 25.Grabež M., Škrbić R., Stojiljković M., Rudić-Grujić V., Šavikin K., Menković N., Zdunić G., Vasiljević N. Beneficial effects of pomegranate peel extract treatment on anthropometry and body composition of overweight patients with diabetes mellitus type-2: a randomised clinical trial. Scr. Med. 2020;51(1):21–27. doi: 10.5937/scriptamed51-25763. [DOI] [Google Scholar]
- 26.Venkataramanamma D., Aruna P., Singh R.P. Standardization of the conditions for extraction of polyphenols from pomegranate peel. J. Food Sci. Technol. 2016;53(5):2497–2503. doi: 10.1007/s13197-016-2222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masci A., Coccia A., Lendaro E., Mosca L., Paolicelli P., Cesa S. Evaluation of different extraction methods from pomegranate whole fruit or peels and the antioxidant and antiproliferative activity of the polyphenolic fraction. Food Chem. 2016;202:59–69. doi: 10.1016/j.foodchem.2016.01.106. [DOI] [PubMed] [Google Scholar]
- 28.Lu J., Ding K., Yuan Q. Determination of punicalagin isomers in pomegranate husk. Chromatographia. 2008;68(3–4):303–306. doi: 10.1365/s10337-008-0699-y. [DOI] [Google Scholar]
- 29.Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int. J. Antimicrob. Agents. 2020;56(2) doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.I. Khalifa, W. Zhu, M.S. Nafie, K. Dutta, C. Li, (2020), 10.20944/preprints202003.0277.v1.
- 31.Chikhale R.V., Gurav S.S., Patil R.B., Sinha S.K., Prasad S.K., Shakya A., Shrivastava S.K., Gurav N.S., Prasad R.S. Sars-cov-2 host entry and replication inhibitors from Indian ginseng: an in-silico approach. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1778539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H.C., Briggs J.M. The association between a negatively charged ligand and the electronegative binding pocket of its receptor. Biopolymers. 2002;63(4):247–260. doi: 10.1002/bip.10050. [DOI] [PubMed] [Google Scholar]
- 33.Meshkin H., Zhu F. Thermodynamics of protein folding studied by umbrella sampling along a reaction coordinate of native contacts. J. Chem. Theory Comput. 2017;13(5):2086–2097. doi: 10.1021/acs.jctc.6b01171. [DOI] [PubMed] [Google Scholar]
- 34.Yang J.F., Wang F., Chen Y.Z., Hao G.F., Yang G.F. LARMD: integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinform. 2020;21(6):2206–2218. doi: 10.1093/bib/bbz141. [DOI] [PubMed] [Google Scholar]
- 35.Lobanov M.Y., Bogatyreva N.S., Galzitskaya O.V. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 2008;42(4):623–628. doi: 10.1134/s0026893308040195. [DOI] [PubMed] [Google Scholar]
- 36.Prasad A., Muthamilarasan M., Prasad M. Synergistic antiviral effects against SARS-CoV-2 by plant-based molecules. Plant Cell Rep. 2020;39(9):1109–1114. doi: 10.1007/s00299-020-02560-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belcaro G. Virucidals control the presence of covid in mouth/Saliva. Med. Clin. Res. 2020 [Google Scholar]
- 38.Piwowarski J.P., Granica S., Stefanska J., Kiss A.K. Differences in metabolism of ellagitannins by human gut microbiota ex vivo cultures. J. Nat. Prod. 2016;79(12):3022–3030. doi: 10.1021/acs.jnatprod.6b00602. [DOI] [PubMed] [Google Scholar]
- 39.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.I. AAT Bioquest, Quest Graph™ IC50 Calculator (v.1). 2021. (Accessed February 25 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.