Abstract
The SARS-CoV-2 virus causes the coronavirus disease 19 emerged in 2020. The pandemic triggered a turmoil in public health and is having a tremendous social and economic impact around the globe. Upon entry into host cells, the SARS-CoV-2 virus hijacks cellular machineries to produce and maintain its own proteins, spreading the infection. Although the disease is known for prominent respiratory symptoms, accumulating evidence is also demonstrating the involvement of the central nervous system, with possible mid- and long-term neurological consequences. In this study, we conducted a detailed bioinformatic analysis of the SARS-CoV-2 proteome aggregation propensity by using several complementary computational tools. Our study identified 10 aggregation prone proteins in the reference SARS-CoV-2 strain: the non-structural proteins Nsp4, Nsp6 and Nsp7 as well as ORF3a, ORF6, ORF7a, ORF7b, ORF10, CovE and CovM. By searching for the available mutants of each protein, we have found that most proteins are conserved, while ORF3a and ORF7b are variable and characterized by the occurrence of a large number of mutants with increased aggregation propensity. The geographical distribution of the mutants revealed interesting differences in the localization of aggregation-prone mutants of each protein. Aggregation-prone mutants of ORF7b were found in 7 European countries, whereas those of ORF3a in only 2. Aggregation-prone sequences of ORF7b, but not of ORF3a, were identified in Australia, India, Nepal, China, and Thailand. Our results are important for future analysis of a possible correlation between higher transmissibility and infection, as well as the presence of neurological symptoms with aggregation propensity of SARS-CoV-2 proteins.
Keywords: SARS-CoV-2, Protein aggregation, Proteostasis, Bioinformatics
Abbreviations: APR, aggregation-prone regions; A3D, Aggrescan3D; CoVex, Coronavirus Explorer; COVID-19, coronavirus disease 19; NSP, non-structural proteins; ORF, open reading frame; PASTA, prediction of amyloid structural aggregation; PDB, Protein Data Bank; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense single-stranded RNA virus from the Coronaviridae family [1] that causes the coronavirus disease 2019 (COVID-19). In January 2020, human-to-human transmission was confirmed, with respiratory symptoms being the most prominent effect. [2] Other COVID-19 symptoms were also reported and included shortness of breath, sore throat, sneezing, fever, exhaustion and, in some cases, gastrointestinal and neurological effects. [3] In fact, around 80% of the patients develop some type of neurological manifestation throughout the infection, ranging from anosmia (11% of the cases) to encephalopathy (up to 32% of the cases). [[4], [5], [6]] Therefore, alongside with other coronaviruses, SARS-CoV-2 is also considered a neurotropic virus. [7,8] Concerningly, several reports link the infection with cognitive impairment and even with neurodegeneration. [9,10] Therefore, the long-term consequences of the infection for the central nervous system need to be further investigated, and targets for therapeutic intervention need to be defined. [11,12]
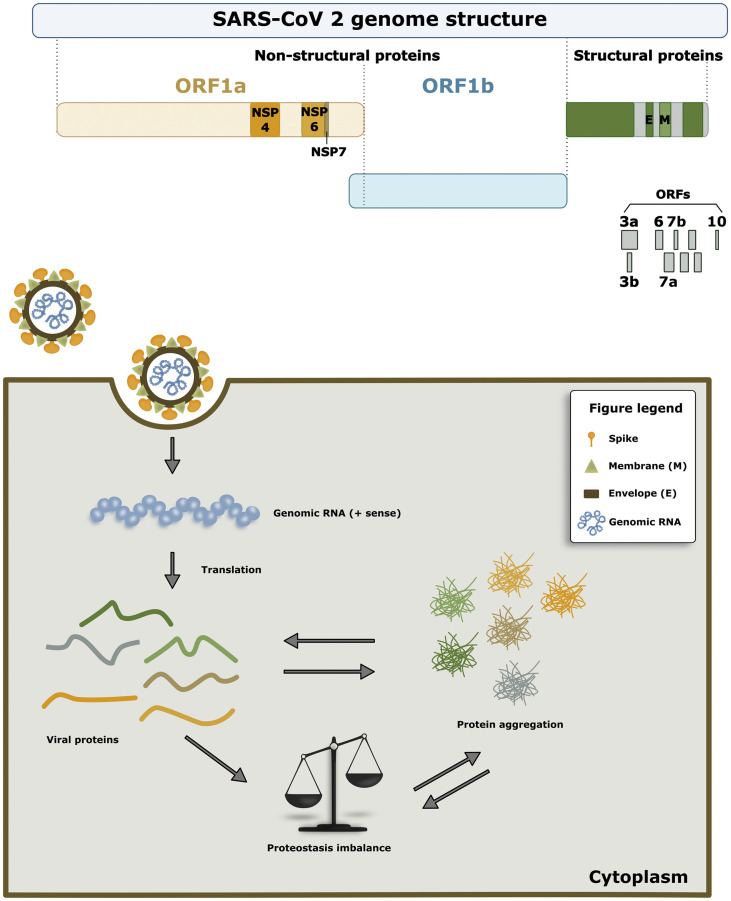
The SARS-CoV-2 genome comprises an open reading frame (ORF) 1a/b encoding for non-structural proteins (NSP), as well as for structural proteins (spike, envelope, membrane, and nucleocapsid proteins), and for accessory proteins such as ORF 3, 6, 7a, 7b, 8 and 9b. [13,14] The NSPs form the replicase-transcriptase complex, which include two viral proteases, Nsp3 (papain-like protease) and Nsp5 (chymotrypsin-like, 3C-like), the primase complex (NSP7-NSP8), the primary RNA-dependent RNA polymerase (Nsp12), an helicase (Nsp13), an exoribonuclease (Nsp14), an endonuclease (Nsp15) and N7- and 2′O-methyltransferases (Nsp10 and Nsp16). [15,16] The spike protein is one of the essential proteins for the infection success of the virus since it mediates the virus-host cell surface attachment. Once inside the host cell, the replication cycle is a regulated process where viral proteins are expressed, folded, and assembled, allowing the production of new viral particles. [17,18] As the virus hijacks the host-cell protein homeostasis (proteostasis) network to ensure the folding, assembly, and release of new viral particles, the cell becomes overloaded with both viral proteins and also endogenous proteins. At some point, the accumulation of proteins may exceed the capacity of cellular quality control systems, resulting in protein misfolding and aggregation, which may, in turn, exacerbate cellular pathologies.
For a long time, protein aggregation was considered an unspecific process. However, it is now known that it relies on a combination of physicochemical parameters within protein sequences and structure. [19] Therefore, several experimental-based models enabled the creation of databases and bioinformatic tools and algorithms to predict and identify aggregation-prone regions (APR) in proteins. These models explore the concept that aggregation is enhanced by the presence of amino acids with higher hydrophobicity and β-sheet propensity, and with lower net charge. [19,20] Therefore, an important aspect to consider in the SARS-CoV-2 proteome is the aggregation-potential of the viral proteins, as this may dramatically change due to mutations.
Here, we employed computational APR-prediction tools to assess the aggregation propensity of the SARS-CoV-2 proteins. We found that 10 proteins can be classified as aggregation-prone. While some of these proteins display low or no variability across geographical regions, others accommodate a large number of mutant sequences. For all the analyzed proteins, we identified mutations that increase the aggregation-propensity of the protein. Interestingly, aggregation-prone variants of ORF3a and ORF7b seem to display specific geographic distribution.
In total, our study provides the foundation for future experimental studies focusing on the analysis of cellular pathologies associated with the aggregation of selected candidates of the SARS-CoV-2 proteome and may enable the identification of novel targets for therapeutic intervention.
2. Methods
2.1. Generation of the SARS-CoV-2 protein sequence set
A full proteome of SARS-CoV-2 virus isolated in Wuhan, China and released on January 17th, 2020 was obtained from the NCBI Virus database. [15,[21], [22], [23]] These protein sequences were set as reference and were used for the prediction of aggregation propensity. As positive controls for aggregation-prone proteins, human aSyn and Aβ peptide 1–42 were used. Bovine serum albumin was used as a negative control as it is a soluble globular protein. Only complete sequences were included for each protein.
2.2. Prediction of aggregation propensity of individual SARS-CoV-2 proteins
The aggregation propensity of SARS-CoV-2 proteins was predicted using four different methods. The cutoffs for each method were determined based on the results obtained for the positive (strongly aggregation prone proteins) and negative (soluble globular protein) controls used in the study.
2.2.1. PASTA 2.0 (prediction of amyloid structural aggregation)
The full proteomes of SARS-CoV-2, with the controls, were analyzed using the PASTA 2.0 server. [24] As an output, the method provides the prediction of the aggregation prone proteins and protein fragments, as well as the analysis of protein disorder and secondary structure. For further analysis, we focused solely on the prediction of the aggregation-prone proteins, displayed as aggregation pairing energy, whereby the lowest aggregation pairing energy signifies the highest aggregation propensity. All proteins smaller than 50 residues with the aggregation pairing energy below -7 kJ, and proteins larger than 1000 residues with the aggregation pairing energy below −20 kJ were considered aggregation prone and were included into further analysis. For proteins of sizes between 100 and 1000 residues, the limit of aggregation pairing energy was −15 kJ.
2.2.2. AmyloGram
Briefly, this approach predicts amyloid proteins using random forests algorithm trained on the n-gram encoded peptides. [25] Using R package, [26] the set of SARS-CoV-2 proteins was analyzed by AmyloGram and all proteins with the score above 0.85 were considered aggregation-prone and included in further analysis.
2.2.3. CamSol
CamSol is a free online software that predicts the solubility of proteins. [27] The use of this method requires either the knowledge of the native structure of the target protein to calculate the structure-corrected solubility profile or the intrinsic solubility profile can be predicted based on the amino acid sequence.
The profiles consist of a score for each residue and it characterizes their impact on the overall solubility of the protein. This sequence-based method predicts protein solubility and generic aggregation propensity. The lower the score is, the lower the solubility of the protein, i.e. the higher the aggregation propensity. Proteins with the intrinsic solubility lower than −1.5 (a.u.) were deemed aggregation prone.
2.2.4. Tango
TANGO is a statistical mechanics algorithm that predicts protein aggregation based on the physico-chemical principles of β-sheet formation. [19,28,29] It predicts secondary structure content of analyzed proteins. A high propensity to form β-sheet structure and the burial of the residues of the β-region in the hydrophobic interior means that a particular amino acid sequence is aggregation-prone. The prediction of aggregation propensity is done in a sequence specific manner.
SARS-CoV-2 full protein sequences were imported to TANGO in FASTA format. For each aminoacid of each analyzed protein, the percentage of beta turn, beta strand, beta sheet and alpha helix aggregation were obtained and exported in a .csv file. Subsequently, for each protein the mean beta sheet aggregation propensity was calculated by normalization to the length of the protein. Any analyzed protein with an aggregation tendency above 10% over 5–6 residues was assigned as a potential aggregation prone protein. The graphs for each protein were generated and the data compared with controls. All the acquired data was statistically analyzed by GraphPad Prism 8 software.
2.3. Selection criteria
Proteins predicted as likely to aggregate with at least three of the methods applied were employed in further analysis. A second set of sequences was generated with a non-redundant list of all mutants of each SARS-CoV-2 protein from all available geographic locations until August 23rd, 2020 (NCBI Virus database). Starting with this set of sequences, from each occurring mutant, only a unique representative was included in the further analysis. The aggregation propensity was then computed with the methods mentioned above for the representative sequences of each mutant. All mutations that increased the aggregation propensity were included in further analysis.
To evaluate if a particular mutation increased the aggregation propensity of a protein variant, we established the following procedure: the output score of each method was normalized to the value of the same protein from the reference strain by dividing the values of each studied protein with the value of the same protein from the reference strain. Next, the log2 of the obtained number was calculated and all protein variants with log2 > 0 had increased aggregation propensity. A sum, mean and median of this score were calculated and each was used to perform the hierarchical clustering of sequences. All the protein variants with sum, mean and median larger than 0 were selected as those where the mutation triggered an increase in aggregation propensity. Finally, all geographical locations with mutations that increased the protein aggregation propensity were marked on maps using R software, package ‘maps’.
2.4. SARS-CoV-2 protein structural analysis
SARS-CoV-2 protein structures were obtained from the Protein Data Bank (PDB). [30] The curation of the redundant structures was performed taking into account the following parameters: when possible, monomers and/or non-conjugated or bound proteins were used, crystalized proteins with the highest resolution were included regardless of the method used as well as the structure with the most complete amino-acid sequence. More detailed characteristics are summarized in Table 1 .
Table 1.
PDB codes from the available SARS-CoV-2 proteins. The chain used for the structural analyses of the crystallized structure is shown along with the complex that the main structure was crystallized with.
| Code | Chain | Protein | Complex |
|---|---|---|---|
| 6YB7 | A | Main Protease (Active site) | None |
| 6 W75 | A | NSP16 | NSP10 |
| 6 W75 | B | NSP10 | NSP16 |
| 7BV1 | B | NSP8 | NSP7 - NSP12 |
| 7BV1 | C | NSP7 | NSP8 - NSP12 |
| 6WLC | A | NSP15 (Endoribonuclease) | UMP |
| 6WOJ | A | NSP3 (macro domain) | ADP ribose |
| 6WXD | A | NSP9 (RNA replicase) | None |
| 6WJI | A | Nucleocapsid phosphoprotein (C-terminal) | None |
| 6YI3 | A | Nucleocapsid phosphoprotein (N-Terminal) | None |
| 6 W37 | A | ORF7A | None |
| 6W9C | A | Papain-like protease | None |
| 6YYT | A | Polymerase | None |
| 6VSB | A | Spike glycoprotein | Prefusion with a single receptor-binding domain |
| 6XDC | A | ORF3a | None |
| 7JTL | A | ORF8 | None |
| 6ZLW | i | Nsp1 | Bound to human 40S ribosomal unit |
| 6ZSL | A | Helicase | None |
2.5. Aggrescan3D (A3D)
The Aggrescan3D 2.0 server was used to evaluate the aggregation propensity of the proteins according to their tertiary structure. [31] PDB codes (from the curation process) were submitted from the available crystalized proteins. The chain identifier used for each PDB entry is listed in Table 1.
Additional parameters that were selected as part of the analysis: Stability calculations - on, Dynamic mode - on, and a distance of Aggregation of 10 Å. Once the analysis was finalized, the .csv file that had the score of each amino acid was downloaded and processed in Microsoft Excel®. Using GraphPad Prism 8® the aggregation propensity graph was obtained, and it was analyzed to obtain the Area under the curve. Amino acids below an Aggregation score of 1 were excluded as well as peaks that contained less than 2 continuous amino-acids prone to aggregate.
To evaluate the mutant forms reported in the literature, the same tool was used to mutate the residue in each variant that the PDB sequence allowed us to work with. The same parameters and exclusion criteria were used as in the analysis of the wild type variant.
2.6. Coronavirus explorer (CoVex)
The Coronavirus Explorer Online tool was used to generate a string analysis of the human proteins that could interact with the SARS-CoV-2 proteins. [32,33] To generate the specific network, the desired SARS-CoV-2 protein was selected in the “Filter Viral Protein” section and “Brain - Substantia nigra” was selected in the “Tissue” section.
3. Results
3.1. Aggregation propensity of the reference SARS-CoV-2 proteome
In order to analyze the aggregation propensity of the full proteome of the SARS-CoV-2 virus, the protein sequences of the Wuhan reference strain were analyzed using four different bioinformatic tools: Pasta2.0, CamSol, Amylogram and Tango (Fig. 1 ). The proteins were then sorted according to their likelihood to aggregate (as likely or unlikely). At a false positive rate of <5%, Pasta2.0 has a sensitivity of 40%, and at a false positive rate of <10%, it has a sensitivity of 30% for predicting APRs, making it the most sensitive tool compared to the others. Therefore, to minimize false APR predictions, the results obtained with Pasta2.0 were selected as our reference predictions.
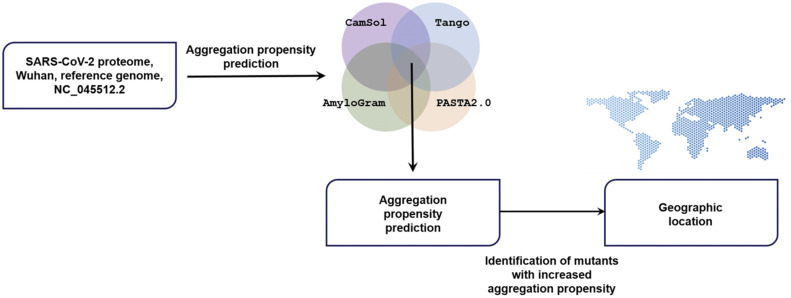
Fig. 1.
Workflow of the study. Aggregation prone proteins were predicted in the SARS-CoV-2 reference strain. Among all mutant sequences available in August 2020, mutations increasing the aggregation propensity were identified and mapped according to their geographical location.
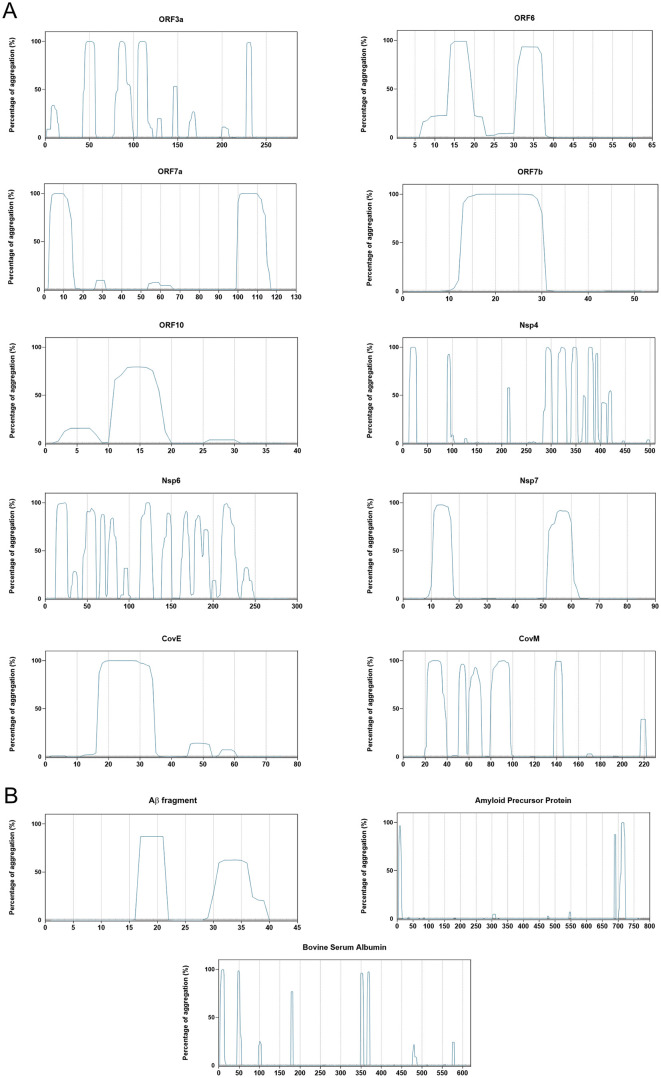
To further strengthen our aggregation propensity prediction, we then analyzed the selected proteins using the other 3 algorithms (Table 2 ). If the protein was predicted as aggregation-prone using at least 2 other algorithms, we classified it as aggregation-prone. The analyses showed that 10 proteins from the SARS-CoV-2 proteome were prone to aggregate. The non-structural proteins Nsp4, Nsp6 and Nsp7 as well as ORF3a, ORF6, ORF7a, ORF7b, ORF10, CovE and CovM were predicted as aggregation-prone. Thus, we continued our study with these 10 selected proteins.
Table 2.
Full set of values/scores of aggregation propensity predictions from all SARS-CoV-2 proteins using 4 different algorithms.
| Protein | CamSol Intrinsic solubility | Tango (beta aggregation propensity) | Amylogram | PASTA (Pairing energy, kJ) | Prone to aggregate |
|---|---|---|---|---|---|
| BSA | 0.479889 | 6.513342 | 0.67476910427 | −9.230629 | Negative control |
| Beta-Amyloid | 1.103345 | 22.4587 | 0.964286175387 | −20.171292 | Positive control |
| just Aβ fragment | 0.655945 | 3.635395 | 0.5936772918 | −6.888103 | Positive control |
| 3′-to-5′ exonuclease | −1.365414 | 3.540996 | 0.780836059694905 | −10.987355 | No |
| 2-O-Ribose Methyltransferase | −0.503960 | 7.303587 | 0.917586033222489 | −7.926963 | No |
| 3C Like Proteinase | −1.094241 | 5.254686 | 0.866995916960981 | −12.714527 | No |
| endoRNAse | −0.225170 | 5.126858 | 0.889817000611829 | −11.885505 | No |
| Envelope protein | −2.915438 | 25.39225 | 0.914774818 | −29.181144 | Yes |
| Helicase | −1.206905 | 2.752962 | 0.885824647794826 | −8.055405 | No |
| Leader protein | 1.394877 | 0.2026 | 0.674893206737557 | −5.359897 | No |
| Membrane glycoprotein | −2.887348 | 26.75686 | 0.862681744 | −34.513196 | Yes |
| Nsp2 | −0.623592 | 4.99456 | 0.810156395859043 | −12.65233 | No |
| Nsp3 | −3.745792 | 5.9662 | 0.888048791900172 | −8.066283 | No |
| Nsp4 | −4.727092 | 19.00763 | 0.855524114 | −32.389486 | Yes |
| Nsp6 | −6.013864 | 36.30086 | 0.887343189 | −32.815411 | Yes |
| Nsp7 | 0.231983 | 17.76802 | 0.889817001 | −7.767171 | Yes |
| Nsp8 | 0.816455 | 2.594571 | 0.672595099068358 | −6.916269 | No |
| Nsp9 | 1.063820 | 7.472212 | 0.691380284008589 | −5.15149 | No |
| Nsp10 | −0.040202 | 0.665628490992486 | −5.345617 | No | |
| Nsp11 | 0.964712 | 0.071538 | 0.52229465066422 | −2.681862 | No |
| Nucleocapsid phosphoprotein | 2.216086 | 4.942186 | 0.850755764881348 | −6.850497 | No |
| ORF3a | −3.400784 | 19.49843 | 0.888048792 | −26.32989 | Yes |
| ORF6 | −0.804210 | 23.35634 | 0.866119264 | −21.15957 | Yes |
| ORF7a | −1.501395 | 22.73566 | 0.816087315 | −16.067259 | Yes |
| ORF7b | −3.341397 | 0.858967046 | −21.061641 | Yes | |
| ORF8 | −1.072771 | 10.9687 | 0.83488134871516 | −10.071886 | No |
| ORF10 | −1.501395 | 18.52229 | 0.804993185 | −10.10498 | Yes |
| RNA-dependent RNA polymerase | −1.702360 | 0.6386355287 | −9.090557 | No | |
| Surface Glycoprotein | −3.379700 | 26.76 | 0.862681743572924 | −16.434159 | No |
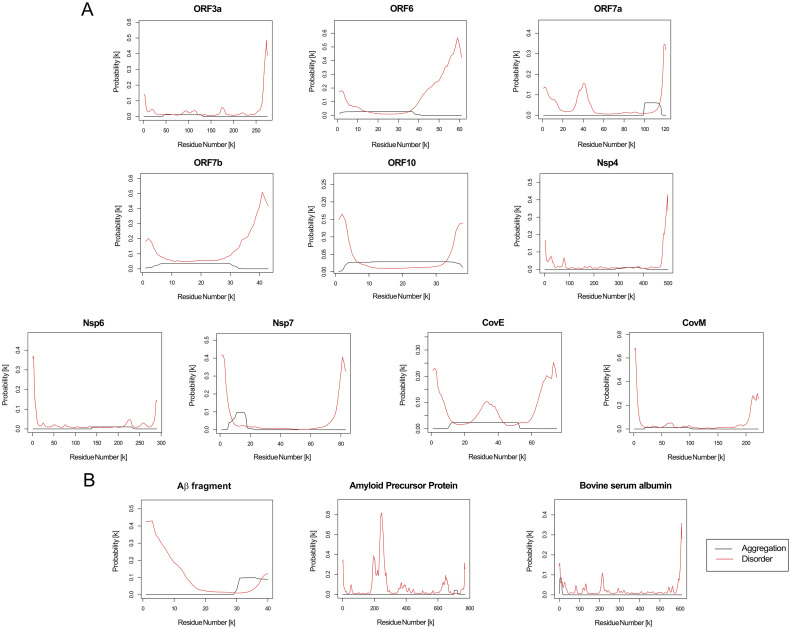
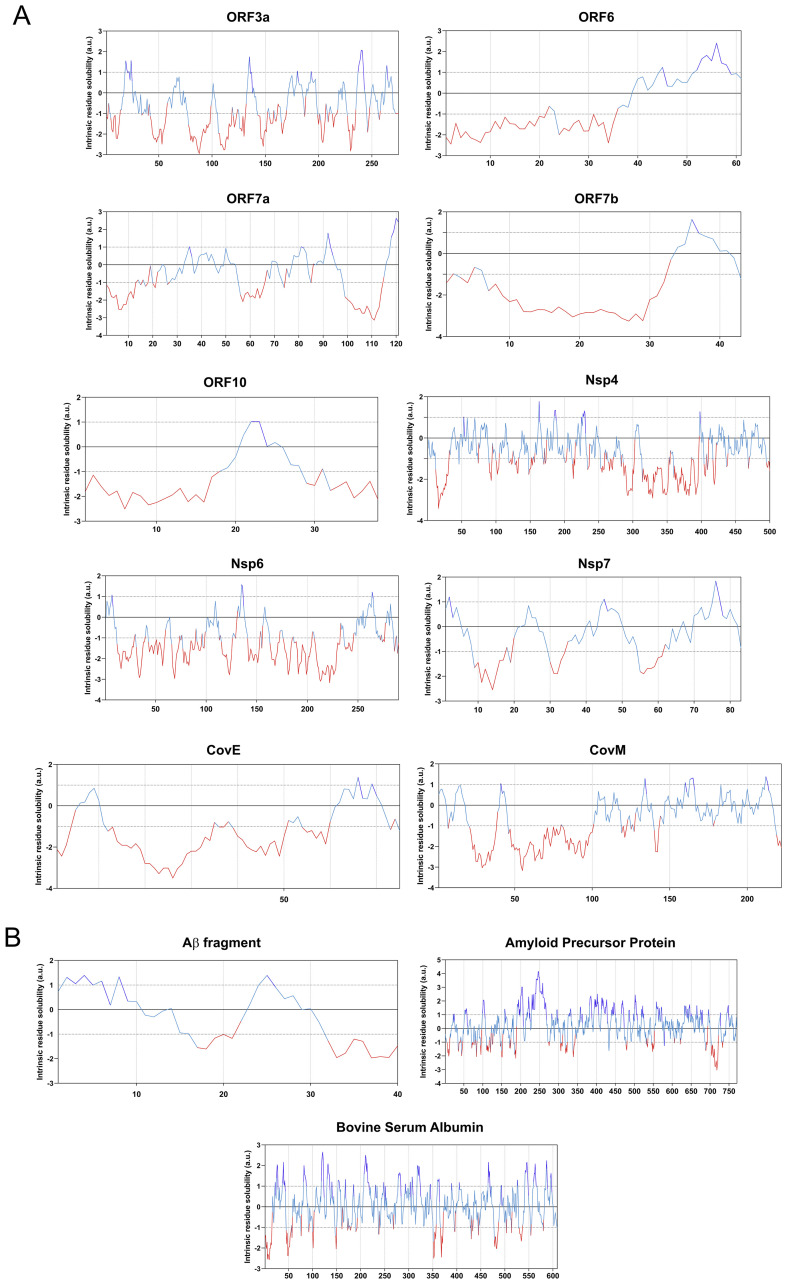
Using PASTA, we obtained the probability of the individual residues of each studied protein sequence to aggregate (black line) and to be disordered (red line), and found that proteins predicted to be prone to aggregate typically had APRs between 30 and 100 residues long (Fig. 2 ). Interestingly, these regions were also recognized as regions of low disorder. On the other hand, CamSol informed on the intrinsic residue solubility for each residue in the analyzed sequences. According to this parameter, the aggregation prone proteins were characterized according to large domains with low solubility (Fig. 3 ).
Fig. 2.
Aggregation probability calculated using PASTA2.0. The graphs displays the probability of each amino acid to be part of a prone to aggregate region (black curve) and to be a part of disordered regions for: a) each SARS-CoV-2 aggregation-prone protein; b) positive and negative controls proteins used in the study.
Fig. 3.
CamSol based analyses of protein solubility. Each graph displays the sequence based solubility profile for each query protein. The part of the curve corresponding to the most soluble protein fragments are shown in blue, and the regions of low solubility are shown in red for: a) SARS-CoV-2 aggregation prone protein solubility graphs; and b) positive and negative control proteins used in the study. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
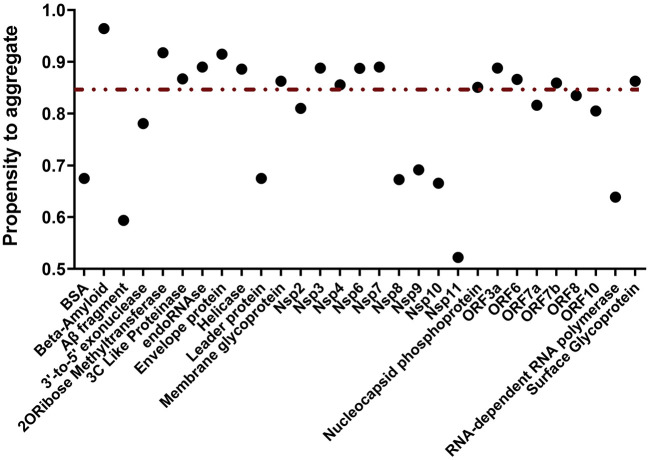
Using Amylogram we obtained the propensity for aggregation into amyloid structure of each protein from the reference SARS-CoV-2 strain. The results showed a wide range of aggregation propensities. However, only the proteins characterized by a score higher than 0.85 were considered prone to aggregate into amyloid structure (Fig. 4 ). Furthermore, TANGO provided an average aggregation score per residue for every analyzed sequence, and the output displayed the likelihood of each amino acid to adopt certain conformation. In our case, we focused on aggregation into amyloid-like structure (Fig. 5 ).
Fig. 4.
Amylogram based calculations of aggregation propensity. The complete set of the SARS-CoV-2 proteome and the positive and negative control proteins are displayed. The non-continuous line marks the threshold (>0.85) considered as “high propensity to aggregate”. The beta-amyloid peptide has the highest value, which relates to its propensity to aggregate.
Fig. 5.
Aggregation propensity calculated using TANGO. Graphs display the percentage of aggregation, signifying the probability of aggregation throughout the queried amino acid sequence for: a) SARS-CoV-2 proteins characterized by high aggregation propensity; b) positive and negative control proteins used in the study.
3.2. Effect of mutations on the aggregation of selected SARS-CoV-2 proteins
Since the sequence of the reference strain was reported, numerous mutants of SARS-CoV-2 virus were reported during 2020, from different geographical locations. Therefore, we collected and analyzed the mutant sequences for each protein, reported until August 2020, for their likelihood to aggregate. First, we curated the set of unique mutant sequences available per protein and kept only unique mutant sequences to avoid redundancy (Table 3 ). This showed that some proteins, like ORF7b or ORF10, appeared to be rather conserved, while others, like ORF3a or Nsp4, displayed a larger variability in their sequence, with numerous mutants occurring in different geographical regions. Next, using a set of unique sequences for each protein, we predicted the aggregation propensity for each mutant sequence using all 4 algorithms employed in our pipeline (full set of results are available in supplementary data, appendix A). In most cases, single point mutations did not alter the protein's susceptibility to aggregate. However, ~40% of the mutant sequences displayed an increased aggregation proneness score in at least one of the algorithms employed.
Table 3.
Number of unique mutant sequences reported per protein. Proteins shown are the ones analyzed according to the established aggregation-prone criteria.
| ORF3a | ORF6 | ORF7a | ORF7b | ORF10 | Nsp4 | Nsp6 | Nsp7 | E protein | M protein | |
|---|---|---|---|---|---|---|---|---|---|---|
| Total number of unique sequences | 102 | 21 | 32 | 6 | 12 | 52 | 47 | 24 | 12 | 25 |
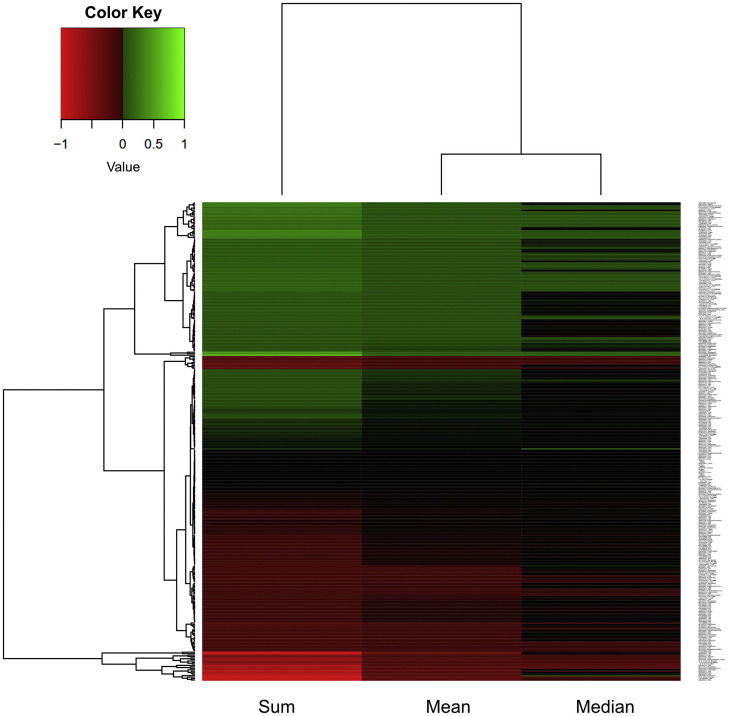
In order to reduce the possibility of false positive predictions, our selection criteria was to choose mutants with increased aggregation propensity when compared to the reference sequence. First, the output score of each method used for the predictions of the likelihood of the protein to aggregate was normalized to the score of the reference strain. Then, the log2 of each value was calculated and this number was termed AggreScore. AggreScore equaled to 0 for the variants with unchanged aggregation propensity compared to the reference strain. All sequences with the AggreScore greater than 0 were considered to be prone to aggregate. Next, we calculated the sum, mean and median of AggreScores for each sequence and performed hierarchical clustering (Fig. 6 ). The sum, mean and median were used to integrate the information in the AggreScore obtained by the four employed methods and build selection criteria stringent enough to avoid false positives. Therefore, we selected all sequences with sum, mean and median greater than 0 for subsequent analyses.
Fig. 6.
Hierarchical clustering heatmap of the sequence variants from different geographical locations based on aggregation propensity. The sum, mean and median value of the log2 of their normalized aggregation score was used to evaluate their aggregation propensity and to cluster them. Green labels an increase and red a decrease in aggregation propensity.
3.3. Geographical distribution of aggregation-prone variants of selected SARS-CoV-2 proteins
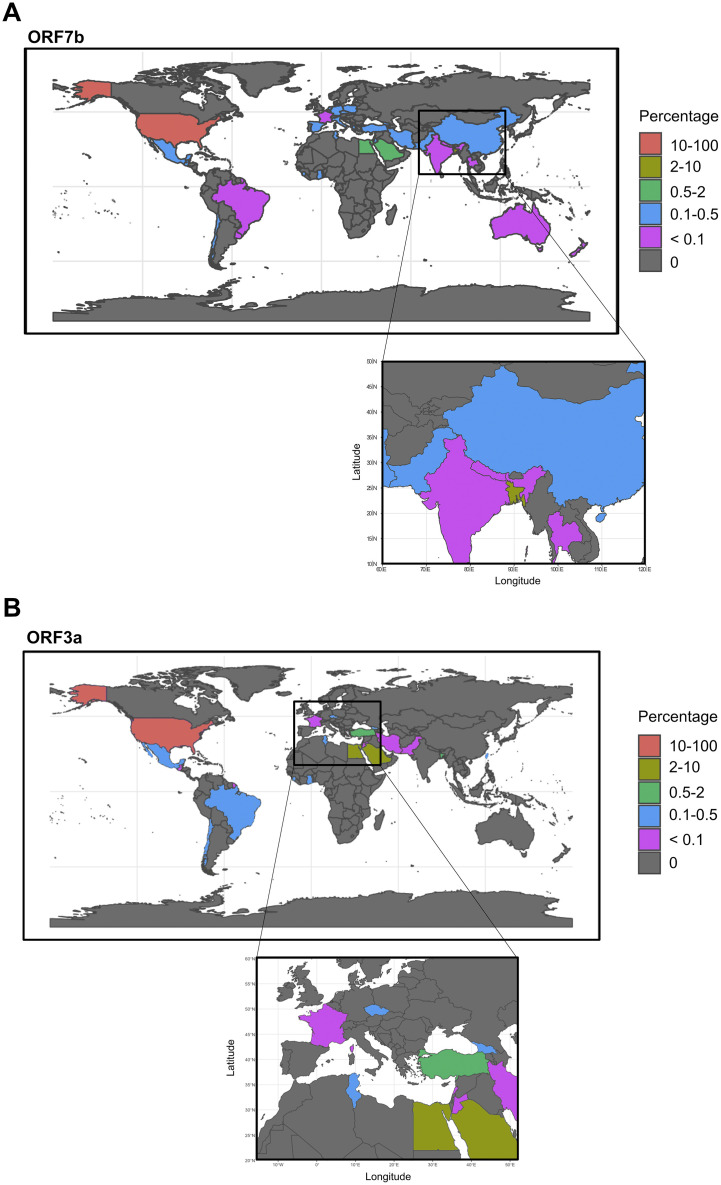
To assess a possible correlation between geographical distribution and SARS-CoV-2 mutations, we retrieved the available sequences of each mutant and plotted on the world map according to the countries where they emerged. 3948 sequences were available for ORF7b, and 1981 sequences for ORF3a. A significantly lower number of sequences was available for the other studied proteins (supplementary data, appendix A) and, therefore, we could not proceed with the analysis of their geographical distribution. The geographical distribution of aggregation-prone mutants of ORF7b and ORF3a is presented in Fig. 7 . For ORF7b (Fig. 7a) and ORF3a (Fig. 7b), over 80% of mutant sequences were found in the USA followed by Bangladesh, Egypt and Saudi Arabia. Interestingly, we detected several differences among less abundant mutant sequences: aggregation-prone mutants of ORF7b appeared in 7 different European countries, whereas those of ORF3a appeared in only 2. Moreover, aggregation-prone sequences of ORF7b, but not of ORF3a, were also identified in Australia. Aggregation-prone variants of ORF3a were absent in India, Nepal, China, and Thailand, regions where aggregation-prone ORF7b were detected. ORF3a variants were, however, present in Taiwan.
Fig. 7.
Geographical distribution of variants of (A) ORF7b and (B) ORF3a characterized by increased aggregation propensity. The location of the SARS-CoV-2 variants are shown as a percentage marked by different colors (key, right side) in each reported country.
3.4. Structural alterations in ORF3a mutants and possible biological effects
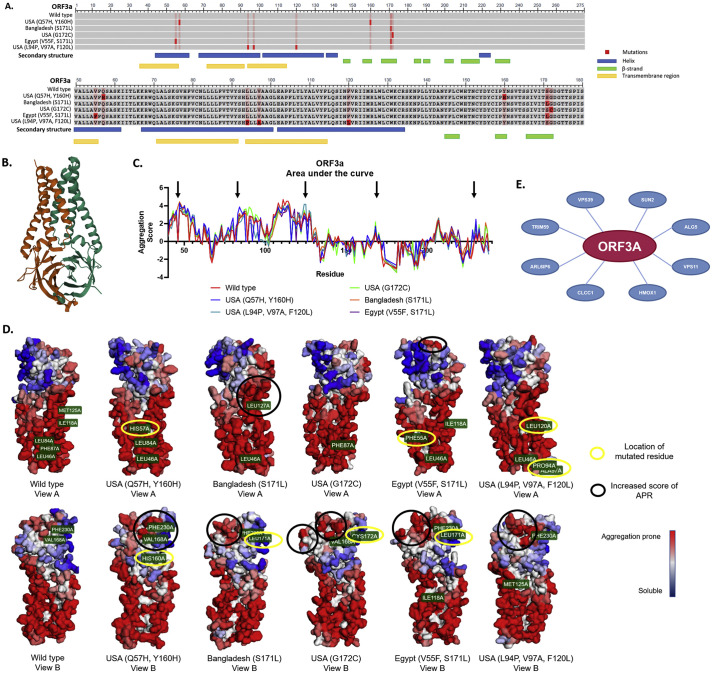
To assess the effects of mutations at the level of the tertiary structure of the protein, we combined data available in the PDB and the use of the online tool A3D to visualize the protein topology, APR locations, and how the mutations affected them. For this, we used two criteria: (i) that the PDB data was available and (ii) a high ΔG difference between the variants and the reference sequence (wild type) (predicted by A3D) (Table 4 ). PDB data was only available for ORF3a. From the 17 variants predicted by A3D to affect the structure, we selected the top 5 for subsequent analysis: USA (Q57H, Y160H), Bangladesh (S171L), USA (G172C), Egypt (V55F, S171L), and USA (L94P, V97A, F120L). ORF3a is a 275 amino acid transmembrane protein (Fig. 8a) with 5 helixes, 8 β-strands and 3 transmembrane domains (Fig. 8a & b). Most of the mutations found in these variants were located in structural domains of the ORF3a protein (Fig. 8a) and, therefore, the structural destabilization might be due to an increase in the aggregation score of the APR. To evaluate this, quantification of the area under the curve for the wild type and the variants with higher ΔG was performed (Fig. 8c) and Table 4). This curve showed an increased aggregation score for some amino acids (black arrows) when compared to the wild type protein. Nevertheless, the % of the peak area did not change drastically (Table 4). The 3D structural analysis of the APR showed that the regions that harbor the mutations did not change their propensity to aggregate (Fig. 8d), yellow circles), but that the greater change was in nearby regions. These regions (Fig. 8c) corresponded to the marked APR (Fig. 8d, black circles) in the protein structure. Interestingly, two of the variants with the highest ΔG (Egypt and Bangladesh) had a higher APR score on two different regions of the protein. This might be related to the predicted destabilization of the protein structure.
Table 4.
ORF3a mutants compared to the wild type. The percentage of the peak area of the APRs is shown. The ΔG difference is associated with the destabilization of the protein's structure. Underlined ΔG show the analyzed variants.
| ORF3a Variant | % Peak Area | Energy Difference (kcal/mol) |
|---|---|---|
| Wild type | 65.91 | 0.0 |
| QNG41554.1 USA (L53F) | 72.24 | 2.2044 |
| QNG41770.1 USA (L140F) | 66.80 | 2.1808 |
| QND77223.1 USA (Q57H, P104S) | 67.97 | 2.4939 |
| QNA38024.1 USA (Q57H, Y160H) | 69.82 | 5.0052 |
| QNA38792.1 USA (Q57H, I123V) | 70.09 | 1.5448 |
| QMU94777.1 Bangladesh (S171L) | 69.53 | 11.2476 |
| QMI98320.1 USA (Q57H, Q185H) | 69.55 | 1.3686 |
| QLM05656.1 USA (G172C) | 68.89 | 4.3654 |
| QLM05764.1 USA (L53H, Q57H) | 67.44 | 2.9557 |
| QLH56255.1 Saudi Arabia (Q57H, S216P) | 66.35 | 3.3689 |
| QKS66941.1 Egypt (V55F, S171L) | 69.13 | 9.9923 |
| QKN20824.1 USA (Q57H, A59V) | 65.81 | 1.0337 |
| QKG90399.1 USA (Q57H, R134C) | 71.74 | 1.5122 |
| QKG64052.1 USA (F56C) | 71.57 | 1.2334 |
| QKE44990.1 USA (L94P, V97A, F120L) | 67.98 | 5.4940 |
| QJD47849.1 Taiwan (Q57H, M125I) | 74.34 | 1.2187 |
| QJD23418.1 USA (Q57H, A99D) | 67.94 | 1.0058 |
Fig. 8.
ORF3a variant structural analysis. a) COBALT sequence alignment analysis of the mutant variants vs. wild type. The bright red boxes represent the mutation in each variant and the light red boxes represent the residue that was changed in another variant. Below the sequence, the important characteristics of the secondary structure are shown; the blue boxes are alpha-helixes, the green boxes represent the beta-strands and the yellow boxes the transmembrane regions. b) Secondary structure of the ORF3a protein wild type reported in the PDB (6XDC). c) Area under the curve analysis of the aggregation score of each amino acid from each ORF3a variant. Black arrows point towards the peaks that showed a higher increase in the aggregation score compared to the wild type. d) Aggrescan 3D 2.0 reconstruction of the variant structures. Yellow circles show where the mutated amino acid is located, black circles show the important changes in the aggregation prone regions. e) CoVex Protein-Protein interaction network analysis. Red circle is the viral protein, blue circles are the host proteins. The interaction prediction tool is based on the human cell line HEK-293 T.
Finally, in order to address the possible impact in the host cell biological functions, we used Coronavirus Explorer (CoVex) in order to identify human proteins that could possibly interact with ORF3a. This interaction network identified 8 proteins that can potentially interact with ORF3a (Fig. 8e). Among the predicted proteins, four (VPS39, VPS11, HMOX1 and CLCC1) have been associated with inflammatory processes, endoplasmic reticulum stress, protein misfolding/clearance and autophagy. [[34], [35], [36], [37]]
4. Discussion
Cellular proteostasis is maintained through a well-orchestrated network that ensures proteins are produced, folded, maintained, and degraded according to quality control standards that enable proper cellular function. External stimuli, such as temperature or chemical stress, mutations, or age-associated alterations can affect the activity of the various surveillance components of the proteostasis network, leading to protein misfolding and accumulation, and eventually to cellular pathologies.
Viruses are infectious agents that hijack different cellular components to produce and process their own proteins, often in high amounts, thereby affecting the overall cellular proteostasis. This can lead to the aggregation of endogenous cellular proteins, or to the aggregation of viral proteins, creating a vicious cycle that can further disrupt the proteostasis network and cause disease. [38]
In the context of the current SARS-CoV-2 pandemic, we reasoned that it would be important to assess whether certain viral proteins might be more aggregation-prone, thereby compromising the proteostasis of the cells they infect. In our study, we took advantage of several available algorithms (TANGO, CamSol and AmyloGram) to predict/assess the aggregation potential of the whole reference viral-proteome from the original Wuhan strain. The validity of the predicted APRs is based on the reliability of the employed tools, as well as of the controls used in the study. The use of different algorithms for aggregation propensity prediction is essential for increasing the confidence in the predictions, as the algorithms use slightly different parameters. We found that the aggregation-prone regions are usually between 30 and 100 residues long and are located in the low disorder regions of the proteins, which provides insight into the aggregation propensity propagation and stability of the proteins throughout the life cycle of the virus.
From the various algorithms employed, we used PASTA 2.0 for the first selection, as this seems the most accurate in the prediction of aggregation propensity. Our results showed that different types of proteins had the highest aggregation propensity scores: Nsp 4, Nsp 6, Nsp7, ORF 3a, ORF 6, ORF 7a, ORF 7b, ORF10, CoVE and CovM (Fig. 9a). Interestingly, NSPs synthesis requires 2/3 of the genome of the virus and these are of upmost importance for the viral replication cycle within the host cell. [39] Likewise, ORF proteins are essential for promoting the membrane rearrangement and cell death in the host cells, as well as other structural changes that lead to the propagation of the virus and the demise of the host cell. [40,41] The CovE and CovM proteins are known to play essential roles in virus morphogenesis and assembly through interactions with other viral proteins. [42,43] Therefore, understanding the propensity of these proteins for aggregation is of great importance for understanding the long-term effects in the host cells. The hijacking of the cellular machinery to produce viral proteins in high amounts may, on one hand, result in the aggregation of aggregation-prone proteins and, in parallel, overwhelm the proteostasis network, leading to additional protein aggregation (Fig. 9).
Fig. 9.
Proteostasis imbalance due to SARS-CoV-2 aggregation-prone proteins. a) In our study, we identified 10 proteins that showed an increase in the predicted aggregation propensity: non-structural proteins Nsp4, Nsp6 and Nsp7, as well as ORF3a, ORF6, ORF7a, ORF7b, ORF10, CovE and CovM. b) One of the common consequences of protein aggregation is, for example, impairment of the protein degradation systems. This extra burden on the degradation system can promote additional protein accumulation, ultimately causing a proteostasis imbalance. This may result in the accumulation of viral proteins that can engage in promiscuous interactions with other proteins, and may increase the virulence of the virus.
Accessory proteins are considered non-essential for the viral structure but, in some viral families, they are important for the virulence or evasion of the host immune responses. Among these, ORF3a, ORF7a, and ORF7b show a high propensity for aggregation. The molecular function of the accessory proteins, such as ORF3a and ORF7b, has been characterized in SARS-CoV and they seem to be important for the virulence and host interaction. Given that ORF3a and ORF7b share approximately 80% homology with those of SARS-CoV-2 [44], their function might be similar in this virus. ORF3a is an ion-channel viroporin through where the virus might be released. [45,46] In mice, deletion of ORF3a reduces viral replication [47] suggesting that this accessory protein plays an important role in the virus' life cycle. Less is known regarding the function of ORF7b. It has been characterized as an important multimeric structural component of the virions and seems to be necessary for the infection. [48,49] Thus, it is possible that mutations or conditions that alter the function of ORF3a or ORF7b may affect the spreading of infection by SARS-CoV-2.
Subsequently, after identifying the 10 proteins most prone to aggregate, we asked whether mutations (reported around the world until August of 2020) might preferentially affect their aggregation. We identified several mutations predicted to increase protein aggregation and, strikingly, we found a correlation with the localization in different geographical regions. Future studies of the symptomatology of neurological disorders may reveal whether a connection exists with the presence of aggregation prone SARS-CoV-2 variants. It should be noted that due to possible sequencing bias, these are the only conclusions that can reliably be drawn from the geographical spreading of aggregation prone SARS-CoV-2 variants.
Thus far, we have not analyzed the variants of the virus that emerged in Brazil, South Africa and UK in the late 2020, beginning of 2021, which showed to be more transmissible and infectious, leading to a higher death toll. Therefore, in a future study, it will be of great interest to assess these new mutations for their effect in the aggregation propensity of the proteins in which they occur, and to investigate a possible correlation between higher transmissibility (and/or infection) and aggregation propensity. [50]
Protein aggregation is a major pathological hallmark in many neurodegenerative disorders. [38,51] Interestingly, several studies have hinted at possible connections between viral infections and the onset of diseases such as Parkinson's disease [11,52] or Alzheimer's disease. [12,[53], [54], [55]] Therefore, it is of great interest to investigate whether this connection results from direct interactions between host and viral aggregation-prone proteins, or from a more general effect at the level of the proteostasis network. Consistently, SARS-CoV-2 mRNA is present in brain samples of COVID-19 patients, [56] suggesting that the virus may also directly infect the brain. Moreover, it was demonstrated that the virus can infect neurons through the ACE2 receptor and that the infection modifies the brain vascular topology, ultimately affecting the microenvironment in ways that may be hazardous for the neighboring cells. [57] Astrocytes can also be infected by SARS-CoV2, reducing their energy metabolism, and triggering an inflammatory response that reduces neuronal viability. [58] These reports support the idea that understanding the infection mechanisms and consequences for neuronal and glial cells will be important as they may inform on possible strategies for therapeutic intervention.
5. Conclusions
In the present study, we conducted a detailed bioinformatic analysis of the SARS-CoV-2 proteome aggregation propensity by using several complementary computational tools such as PASTA2.0, CamSol, AmyloGram and TANGO. To our knowledge, this is the first study to analyze and compare the SARS-CoV-2 protein variants according to their geographical location. First, we found that the aggregation-prone regions are usually between 30 and 100 residues long and are located in the low disorder regions of these proteins. Second, the analyses showed that 10 proteins were predicted as aggregation prone. Furthermore, the number of reported variants showed that some proteins, like ORF7b or ORF10, appeared to be rather conserved with a few reported variants, while others, like ORF3a or Nsp4, displayed a larger variability. From these mutants, ~40% displayed an increased aggregation propensity score in at least one of the algorithms employed. Additionally, the aggregation-prone mutants of ORF7b were identified in 7 different European countries, Australia, India, Nepal, China, and Thailand. Compared to this, the ORF3a variants were detected in 2 European countries and in Taiwan. Given that the immunological response and its components play an important role in the SARS-CoV-2 infection, this may vary depending on specific genetic backgrounds of different populations. Consistently, several studies have reported a link between the immunotype and the severity of the disease, mortality and/or prognosis. [[59], [60], [61], [62]] However, how the immunotype affects the cellular machinery, and which are the mechanisms involved, are still important and open questions that need to be addressed in the context of aggregation-prone proteins.
Finally, to shed light on the possible cellular mechanisms affected by these prone-aggregation proteins, like ORF3a, 8 proteins that can potentially interact with ORF3a were identified. These have been associated with inflammatory processes, endoplasmic reticulum stress, protein misfolding/clearance and autophagy. Taking all of these into account, this work provides new insights into the importance of understanding and studying the long-term effects of the SARS-CoV-2 infection. Additionally, it reinforces the emerging idea that it is important to address the effects of the infection on neuronal and glial cells in order to develop future therapeutic strategies.
Data availability
All data analyzed (accession numbers) during this study are included in this published article in the supplementary data, appendix B.
Funding
Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Acknowledgements
The authors acknowledge funding by the Sino-German Center for Research Promotion in Beijing (GZ C-0054). AK is funded by the Heisenberg program of the German Research Foundation (DFG). The authors are grateful to Dr. Nicolás Lemus Diaz for his help with data curation, analysis, and presentation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbapap.2021.140693.
Appendix A. Supplementary data
Supplementary material
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F.W., Yuan S., Kok K.H., K.K.W. To, Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S., Tsoi H.W., Lo S.K.F., Chan K.H., Poon V.K.M., Chan W.M., Ip J.D., Cai J.P., Cheng V.C.C., Chen H., Hui C.K.M., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machhi J., Herskovitz J., Senan A.M., Dutta D., Nath B., Oleynikov M.D., Blomberg W.R., Meigs D.D., Hasan M., Patel M., Kline P., Chang R.C.C., Chang L., Gendelman H.E., Kevadiya B.D. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. NeuroImmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liotta E.M., Batra A., Clark J.R., Shlobin N.A., Hoffman S.C., Orban Z.S., Koralnik I.J. Frequent neurologic manifestations and encephalopathy-associated morbidity in Covid-19 patients. Ann. Clin. Transl. Neurol. 2020;7:2221–2230. doi: 10.1002/acn3.51210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Román G.C., Spencer P.S., Reis J., Buguet A., Faris M.E.A., Katrak S.M., Láinez M., Medina M.T., Meshram C., Mizusawa H., Öztürk S., Wasay M. The neurology of COVID-19 revisited: a proposal from the environmental neurology specialty Group of the World Federation of neurology to implement international neurological registries. J. Neurol. Sci. 2020;414:116884. doi: 10.1016/j.jns.2020.116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nepal G., Rehrig J.H., Shrestha G.S., Shing Y.K., Yadav J.K., Ojha R., Pokhrel G., Tu Z.L., Huang D.Y. Neurological manifestations of COVID-19: A systematic review. Crit. Care. 2020;24 doi: 10.1186/s13054-020-03121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desforges M., Le Coupanec A., Brison É., Meessen-Pinard M., Talbot P.J. Neuroinvasive and neurotropic human respiratory coronaviruses: potential neurovirulent agents in humans. Adv. Exp. Med. Biol. 2014;807:75–96. doi: 10.1007/978-81-322-1777-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramani A., Müller L., Ostermann P.N., Gabriel E., Abida-Islam P., Müller-Schiffmann A., Mariappan A., Goureau O., Gruell H., Walker A., Andrée M., Hauka S., Houwaart T., Dilthey A., Wohlgemuth K., Omran H., Klein F., Wieczorek D., Adams O., Timm J., Korth C., Schaal H., Gopalakrishnan J. <scp>SARS</scp> -CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39 doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen M.E., Eichel R., Steiner-Birmanns B., Janah A., Ioshpa M., Bar-Shalom R., Paul J.J., Gaber H., Skrahina V., Bornstein N.M., Yahalom G. A case of probable Parkinson's disease after SARS-CoV-2 infection. Lancet Neurol. 2020;19:804–805. doi: 10.1016/S1474-4422(20)30305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adam H., Trender W., Chamberlain S.R., Jolly A., Grant J.E., Patrick F., Mazibuko N., Williams S., Barnby J.M., Hellyer P., Mehta M.A. Cognitive deficits in people who have recovered from COVID-19 relative to controls: An N=84,285 online study. MedRxiv. 2020 doi: 10.1101/2020.10.20.20215863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lippi A., Domingues R., Setz C., Outeiro T.F., Krisko A. SARS-CoV-2: at the crossroad between aging and Neurodegeneration. Mov. Disord. 2020;35:716–720. doi: 10.1002/mds.28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingues R., Lippi A., Setz C., Outeiro T.F., Krisko A. SARS-CoV-2, immunosenescence and inflammaging: Partners in the COVID-19 crime. Aging (Albany NY) 2020;12:18778–18789. doi: 10.18632/aging.103989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fehr A.R., Perlman S. Coronaviruses Methods Protoc. Springer; New York: 2015. Coronaviruses: An overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan J.F.W., Kok K.H., Zhu Z., Chu H., K.K.W. To, Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Y.A. Malik, Properties of Coronavirus and SARS-CoV-2, n.d. [PubMed]
- 17.Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS -CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39 doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee T.K., Malik P., Maitra R., Hoidal J.R. Ravaging SARS-CoV-2: rudimentary diagnosis and puzzling immunological responses. Curr. Med. Res. Opin. 2021;37:207–217. doi: 10.1080/03007995.2020.1862532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. A comparative study of the relationship between protein structure and β-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 20.Deforte S., Uversky V.N. Intrinsically disordered proteins in PubMed: what can the tip of the iceberg tell us about what lies below? RSC Adv. 2016;6:11513–11521. doi: 10.1039/c5ra24866c. [DOI] [Google Scholar]
- 21.Williams G.D., Chang R.-Y., Brian D.A. A Phylogenetically conserved hairpin-type 3′ Untranslated region Pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robertson M.P., Igel H., Baertsch R., Haussler D., Ares M., Scott W.G. The structure of a rigorously conserved RNA element within the SARS virus genome. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh I., Seno F., Tosatto S.C.E., Trovato A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burdukiewicz M., Sobczyk P., Rödiger S., Duda-Madej A., MacKiewicz P., Kotulska M. Amyloidogenic motifs revealed by n-gram analysis. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-13210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R: The R Project for Statistical Computing, (n.d.). https://www.r-project.org/ (accessed April 7, 2021).
- 27.Sormanni P., Aprile F.A., Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J. Mol. Biol. 2015;427:478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 29.Rousseau F., Schymkowitz J., Serrano L. Protein aggregation and amyloidosis: confusion of the kinds? Curr. Opin. Struct. Biol. 2006;16:118–126. doi: 10.1016/j.sbi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E., The Protein Data Bank Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuriata A., Iglesias V., Pujols J., Kurcinski M., Kmiecik S., Ventura S. Aggrescan3D (A3D) 2.0: prediction and engineering of protein solubility. Nucleic Acids Res. 2019;47:W300–W307. doi: 10.1093/nar/gkz321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadegh S., Matschinske J., Blumenthal D.B., Galindez G., Kacprowski T., List M., Nasirigerdeh R., Oubounyt M., Pichlmair A., Rose T.D., Salgado-Albarrán M., Späth J., Stukalov A., Wenke N.K., Yuan K., Pauling J.K., Baumbach J. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-17189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.CoVex, (n.d.). https://exbio.wzw.tum.de/covex/explorer (accessed April 7, 2021).
- 34.Zeng X., Carlin C.R. Host cell autophagy modulates early stages of adenovirus infections in airway epithelial cells. J. Virol. 2013;87:2307–2319. doi: 10.1128/jvi.02014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., Bi Y., Wang P., Zhang H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell. 2021;56 doi: 10.1016/j.devcel.2020.12.010. 427–442.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi M., Piagnerelli M., Van Meerhaeghe A., Zouaoui Boudjeltia K. Heme oxygenase-1 (HO-1) cytoprotective pathway: a potential treatment strategy against coronavirus disease 2019 (COVID-19)-induced cytokine storm syndrome. Med. Hypotheses. 2020;144:110242. doi: 10.1016/j.mehy.2020.110242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia Y., Jucius T.J., Cook S.A., Ackerman S.L. Loss of Clcc1 results in ER stress, misfolded protein accumulation, and neurodegeneration. J. Neurosci. 2015;35:3001–3009. doi: 10.1523/JNEUROSCI.3678-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marreiros R., Müller-Schiffmann A., Trossbach S.V., Prikulis I., Hänsch S., Weidtkamp-Peters S., Moreira A.R., Sahu S., Soloviev I., Selvarajah S., Lingappa V.R., Korth C. Disruption of cellular proteostasis by H1N1 influenza a virus causes α-synuclein aggregation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:6741–6751. doi: 10.1073/pnas.1906466117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 41.Jose D. SARS-Cov-2 systems biology. Ann. Syst. Biol. 2020:029–032. doi: 10.17352/asb.000009. [DOI] [Google Scholar]
- 42.Liu D.X., Yuan Q., Liao Y. Coronavirus envelope protein: a small membrane protein with multiple functions. Cell. Mol. Life Sci. 2007;64:2043–2048. doi: 10.1007/s00018-007-7103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G., Stamou D.G., Wilson I.A., Kuhn P., Buchmeier M.J. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariano G., Farthing R.J., Lale-Farjat S.L.M., Bergeron J.R.C. Structural characterization of SARS-CoV-2: where we are, and where we need to be. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu W., Zheng B.J., Xu K., Schwarz W., Du L., Wong C.K.L., Chen J., Duan S., Deubel V., Sun B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Issa E., Merhi G., Panossian B., Salloum T., Tokajian S. Vol. 5. 2020. SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis, MSystems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. MBio. 2018;9 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaecher S.R., Mackenzie J.M., Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007;81:718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fogeron M.-L., Montserret R., Zehnder J., Nguyen M.-H., Dujardin M., Brigandat L., Cole L., Ninot-Pedrosa M., Lecoq L., Böckmann A. SARS-CoV-2 ORF7b: is a bat virus protein homologue a major cause of COVID-19 symptoms? BioRxiv. 2021 doi: 10.1101/2021.02.05.428650. [DOI] [Google Scholar]
- 50.Toovey O.T.R., Harvey K.N., Bird P.W., Tang J.W.T. Introduction of Brazilian SARS-CoV-2 484K.V2 related variants into the UK. J. Inf. Secur. 2021 doi: 10.1016/j.jinf.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Müller-Schiffmann A., Trossbach S.V., Lingappa V.R., Korth C. Viruses as ‘Truffle Hounds’: molecular tools for untangling brain cellular pathology. Trends Neurosci. 2020 doi: 10.1016/j.tins.2020.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Yamada T. Viral etiology of Parkinson's disease: focus on influenza a virus. Parkinsonism Relat. Disord. 1996;2:113–121. doi: 10.1016/1353-8020(96)00006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cribbs D.H., Azizeh B.Y., Cotman C.W., LaFerla F.M. Fibril formation and neurotoxicity by a herpes simplex virus glycoprotein B fragment with homology to the Alzheimer's Aβ peptide. Biochemistry. 2000;39:5988–5994. doi: 10.1021/bi000029f. [DOI] [PubMed] [Google Scholar]
- 54.Eimer W.A., Vijaya Kumar D.K., Navalpur Shanmugam N.K., Rodriguez A.S., Mitchell T., Washicosky K.J., György B., Breakefield X.O., Tanzi R.E., Moir R.D. Alzheimer's disease-associated β-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron. 2018;99 doi: 10.1016/j.neuron.2018.06.030. 56–63.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sochocka M., Zwolińska K., Leszek J. The infectious etiology of Alzheimer's disease. Curr. Neuropharmacol. 2017;15 doi: 10.2174/1570159x15666170313122937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., Braun F., Lu S., Pfefferle S., Schröder A.S., Edler C., Gross O., Glatzel M., Wichmann D., Wiech T., Kluge S., Pueschel K., Aepfelbacher M., Huber T.B. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/nejmc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song E., Zhang C., Israelow B., Lu-Culligan A., Prado A.V., Skriabine S., Lu P., El Weizman O., Liu F., Dai Y., Szigeti-Buck K., Yasumoto Y., Wang G., Castaldi C., Heltke J., Ng E., Wheeler J., Alfajaro M.M., Levavasseur E., Fontes B., Ravindra N.G., van Dijk D., Mane S., Gunel M., Ring A., Kazmi S.A. Jaffar, Zhang K., Wilen C.B., Horvath T.L., Plu I., Haik S., Thomas J.L., Louvi A., Farhadian S.F., Huttner A., Seilhean D., Renier N., Bilguvar K., Iwasaki A. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021;218 doi: 10.1084/JEM.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crunfli F., Carregari V.C., Veras F.P., Vendramini P.H., Valença A.G. Fragnani, Antunes A.S.L. Marcelo, Brandão-Teles C., Zuccoli G. de S., De Oliveira G. Reis, Silva-Costa L.C., Saia-Cereda V.M., Codo A.C., Parise P.L., Teixeira D.A. Toledo, de Souza G.F., Muraro S.P., Melo B.M. Silva, Almeida G.M., Firmino E.M. Silva, Ludwig R.G., Ruiz G.P., Knittel T.L., Davanzo G.G., Gerhardt J.A., Rodrigues P.B., Forato J., Amorim M.R., Silva N.B., Martini M.C., Benatti M.N., Batah S., Siyuan L., Silva R.E.M. Pereira, João R.B., Silva L.S., Nogueira M.H., Aventurato Í.K., de Brito M.R., Alvim M.K. Machado, da Silva J.R., Damião L.L., Stefano M.E.D.P. Castilho, De Sousa I.M.P., da Rocha E.D., Gonçalves S.M., da Silva L.H.L., Bettini V., de Campos B.M., Ludwig G., Viana R.M. Mendes, Martins R., Vieira A.S., Alves-Filho J.C., Arruda E., Sebollela A.S., Cendes F., Cunha F.Q., Damásio A., Vinolo M.A. Ramirez, Munhoz C.D., Rehen S.K., Mauad T., Duarte-Neto A.N., da Silva L.F.F., Dolhnikoff M., Saldiva P., Fabro A.T., Farias A.S., Moraes-Vieira P.M.M., Módena J.L. Proença, Yasuda C.L., Mori M.A., Cunha T.M., De Souza D. Martins. SARS-CoV-2 infects brain astrocytes of COVID-19 patients and impairs neuronal viability. MedRxiv. 2020;16 doi: 10.1101/2020.10.09.20207464. [DOI] [Google Scholar]
- 59.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D., Cuapio A., Ask E.H., Hull R.M., Haroun-Izquierdo A., Schaffer M., Klingström J., Folkesson E., Buggert M., Sandberg J.K., Eriksson L.I., Rooyackers O., Ljunggren H.G., Malmberg K.J., Michaëlsson J., Marquardt N., Hammer Q., Strålin K., Björkström N.K. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5:6832. doi: 10.1126/SCIIMMUNOL.ABD6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D'Andrea K., Manne S., Chen Z., Huang Y.J., Reilly J.P., Weisman A.R., Ittner C.A.G., Kuthuru O., Dougherty J., Nzingha K., Han N., Kim J., Pattekar A., Goodwin E.C., Anderson E.M., Weirick M.E., Gouma S., Arevalo C.P., Bolton M.J., Chen F., Lacey S.F., Ramage H., Cherry S., Hensley S.E., Apostolidis S.A., Huang A.C., Vella L.A., Betts M.R., Meyer N.J., Wherry E.J., Alam Z., Addison M.M., Byrne K.T., Chandra A., Descamps H.C., Kaminskiy Y., Hamilton J.T., Noll J.H., Omran D.K., Perkey E., Prager E.M., Pueschl D., Shah J.B., Shilan J.S., Vanderbeck A.N. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science (80) 2020;369 doi: 10.1126/SCIENCE.ABC8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernal E., Gimeno L., Alcaraz M.J., Quadeer A.A., Moreno M., Martínez-Sánchez M.V., Campillo J.A., Gomez J.M., Pelaez A., García E., Herranz M., Hernández-Olivo M., Martínez-Alfaro E., Alcaraz A., Muñoz Á., Cano A., McKay M.R., Muro M., Minguela A. Activating killer-cell immunoglobulin-like receptors are associated with the severity of coronavirus disease 2019. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martín-Sánchez E., Garcés J.J., Maia C., Inogés S., López-Díaz de Cerio A., Carmona-Torre F., Marin-Oto M., Alegre F., Molano E., Fernandez-Alonso M., Perez C., Botta C., Zabaleta A., Alcaide A.B., Landecho M.F., Rua M., Pérez-Warnisher T., Blanco L., Sarvide S., Vilas-Zornoza A., Alignani D., Moreno C., Pineda I., Sogbe M., Argemi J., Paiva B., Yuste J.R. Immunological biomarkers of fatal COVID-19: a study of 868 patients. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.659018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data analyzed (accession numbers) during this study are included in this published article in the supplementary data, appendix B.