Abstract
Objectives
Growth hormone‐releasing hormone (GHRH) is a potent stimulator of growth hormone (GH) secretion from the pituitary gland. Although GHRH is essential for the growth of immune cells, the regulatory effects of its antagonist in granulomatous disease remain unknown.
Methods
Here, we report expression of GHRH receptor (R) in human tissue with sarcoidosis granuloma and demonstrate the anti‐inflammatory effects of MIA602 (a GHRH antagonist) in two in vitro human granuloma models and an in vivo granuloma model using different methods including ELISA, immunohistochemistry, RNA‐seq analysis and flow cytometry.
Results
MIA602 decreases the levels of IL‐2, IL‐2R, IL‐7, IL‐12, IL‐17A and TNF‐α in an in vitro granuloma model. Further, we show that the anti‐inflammatory effect of MIA602 appears to be mediated by a reduction in CD45+CD68+ cells in granulomatous tissue and upregulation in PD‐1 expression in macrophages. Analysis of the expression of proteins involved in the mitochondrial stage of apoptosis showed that MIA602 increases the levels of caspase‐3, BCL‐xL/BAK dimer and MCl‐1/Bak dimer in the granuloma. These findings indicate that MIA602 may not induce apoptosis.
Conclusions
Our findings further suggest that GHRH‐R is potentially a clinical target for the treatment of granulomatous disease and that MIA602 may be used as a novel therapeutic agent for sarcoidosis.
Keywords: GHRH antagonist, granuloma, MIA602, sarcoidosis
GHRH‐R is potentially a target for treatment of granulomatous diseases such as sarcoidosis, which could be blocked by growth hormone‐releasing hormone‐R antagonists such as MIA602 as novel therapeutic agents for sarcoidosis.
Introduction
Sarcoidosis is an inflammatory disease characterised pathologically by non‐caseating granulomas. 1 Sarcoidosis affects every race and ethnicity, but in the United States, it is more commonly seen in African Americans. 2 In addition to genetic predisposition, environmental factors such as dust, mould, mycobacteria and occupational exposures also trigger sarcoidosis. 3 , 4
No known medications specifically treat sarcoidosis, but corticosteroids such as prednisone and prednisolone have been used because of their anti‐inflammatory properties. 5 Despite corticosteroids showing some positive treatment results, many patients continue to experience significant problems because of the development of fibrosis from previously active or smouldering granulomatous inflammation. 5 , 6 In addition, many patients report significant steroid‐associated side effects. 7 , 8 Therefore, there is a great need for sarcoid‐specific medications to target inflammatory pathways and prevent sarcoidosis sequelae. If inflammation can be modulated, this would likely cause a decrease in symptoms associated with sarcoidosis, leading to disease remission.
Growth hormone‐releasing hormone (GHRH) is a hypothalamic 40–44 amino acid peptide hormone that specifically stimulates the synthesis and release of growth hormone (GH) from the pituitary gland. 9 GHs are active throughout the body and promote growth and development. The GHRH receptor (GHRH‐R) is a seven‐transmembrane subunit G protein‐linked receptor found predominantly in the pituitary gland. 10 GHRH‐R can be blocked by synthetic peptides known as GHRH‐R antagonists. These peptides include the MIA and AVR classes of antagonists. 11 MIA602, which is a GHRH‐R antagonist, has been shown to modulate lung inflammation and fibrosis. 12 GHRH‐R antagonists function in a cAMP‐dependent pathway and downregulate p21‐activated kinase 1 (PAK1)‐mediated signal transducer and activation of transcription factor 3 (STAT3)/nuclear factor‐kB (NF‐kB). 13 To date, however, none of GHRH‐R antagonists have known anti‐inflammatory properties in granulomatous diseases or sarcoidosis. 14
In this study, we investigated the anti‐inflammatory properties of MIA602 by measuring cytokines and gene expression in both in vitro and in vivo granuloma models. We developed the in vitro model by exposing human peripheral blood mononuclear cells (PBMCs) to microparticles generated from Mycobacterium abscessus (MAB) cell walls. 15 We exposed mouse lungs to the same microparticles to study lung granulomatous disease in vivo. 16
Results
MIA602 has anti‐inflammatory effects in an in vitro sarcoidosis‐like granuloma model
To determine whether MIA602 can reduce inflammation in granulomatous disease, we first asked whether GHRH‐R is expressed in sarcoidosis granuloma. To address this question, we stained 5 human lung samples with pathological confirmation of sarcoidosis for GHRH‐R using immunohistochemistry (IHC). Pathology blocks from 5 patients with confirmed sarcoidosis (treatment naïve) were recut and stained for GHRH‐R. Lung parenchymal biopsy of patient 1 with stage II pulmonary sarcoidosis showed 90% expression of GHRH‐R in cytoplasmic and cell membrane regions. The biopsy of the paratracheal lymph node at station 7 of both patients 2 and 3 with stage I pulmonary sarcoidosis showed 90% and 40% expression of GHRH‐R in granuloma, respectively. The biopsy of the paratracheal lymph node at station 4 of patient 4 with stage III pulmonary sarcoidosis showed 20% expression of GHRH‐R in the granuloma. The biopsy of the paratracheal lymph node at station 4 of patient 5 with stage I pulmonary sarcoidosis showed 10% expression of GHRH‐R in the granuloma. As shown in Figure 1, granulomata significantly expressed levels of GHRH‐R. Table 1 summarises the demographic data of the patients and their sarcoidosis stage.
Figure 1.
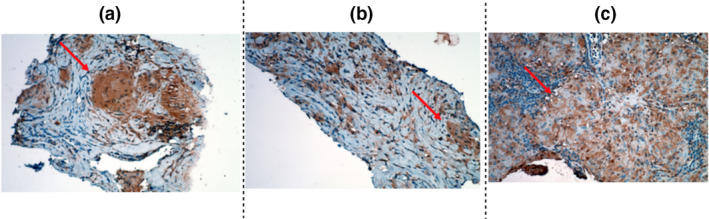
Lung histopathology. Lymph node tissue from three subjects, ( a ) patient 1; (b ) patient 2; and (c) patient 4, with confirmed sarcoidosis was recut. 5‐µm sections were stained for GHRH‐R. The arrow shows a granuloma. The brown colour in the tissue indicates the expression of GHRH‐R. Data are from one experiment that is representative of three separate experiments.
Table 1.
Demographic and sarcoidosis stage of five enrolled subjects with confirmed pulmonary sarcoidosis
| Patients IDs | Age | Gender | Ethnicity | Location of granuloma used for Tissue microarray | Sarcoidosis stage |
|---|---|---|---|---|---|
| 1 | 56 | Male | Hispanic | Lung parenchyma | 2 |
| 2 | 54 | Male | Hispanic | Station 7 lymph node | 1 |
| 3 | 65 | Female | Hispanic | Station 7 lymph node | 1 |
| 4 | 52 | Female | Hispanic | Neck/left lymph node | 3 |
| 5 | 51 | Female | Hispanic | Station 4 lymph node | 1 |
After confirming the presence of GHRH‐R in granuloma, we aimed to determine the anti‐inflammatory properties of the GHRH‐R antagonist (MIA602) in an in vitro sarcoidosis‐like granuloma system. To achieve this goal, we used PBMCs from five confirmed subjects with sarcoidosis who participated in an IRB‐approved sarcoidosis biobank. Samples of PBMCs were challenged with microparticles generated from M. abscessus cell wall (called MAB microparticles in this manuscript) and then treated with saline (granuloma group), 1 μm MIA602 (MIA602 group) or high dose (138 μm) of methylprednisolone (methylprednisolone group) compared with control PBMCs. PBMCs challenged with MAB microparticles all formed granulomas. The medium was removed 48 h after treatments, and cytokines were measured using a multiplex ELISA instrument. We found a significant difference in the expression of several cytokines in the granuloma group in comparison with the control. MIA602 significantly reduced the production of IL‐2, IL‐2R, IL‐7, IL‐12, IL‐17A and TNF‐α, as shown in Figure 2. In comparison with methylprednisolone, MIA602 showed a non‐inferior anti‐inflammatory effect in the in vitro model.
Figure 2.
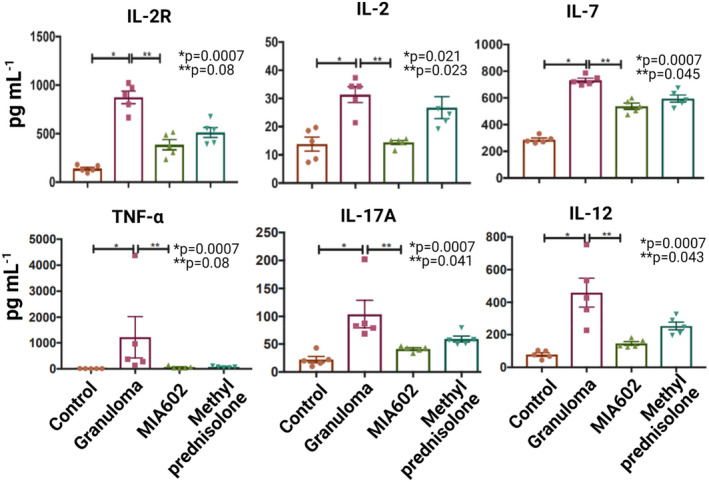
ELISA results for in vitro sarcoidosis‐like granulomas. Granulomas were developed using PBMCs from subjects with confirmed sarcoidosis. Four different groups were studied: control, PBMC from sarcoidosis patients (not challenged with microparticles); granuloma, PBMCs challenged with microparticles; MIA602, granuloma treated with MIA602; and methylprednisolone, granuloma treated with methylprednisolone. Data shown are mean cytokine concentrations ± SEM at 48 h after challenge with MAB microparticles. For each group, five replications were included. Data are from one experiment that is representative of three separate experiments.
Effect of MIA602 on apoptosis in an in vitro sarcoidosis‐like granuloma model
We tested the hypothesis that MIA602 may influence respiration and apoptosis in granuloma cells. The specific processes of apoptosis and its relation to mitochondrial dynamics in granulomas have not been fully elucidated. To test whether MIA602 has apoptotic effects, we measured the protein levels of pro‐apoptotic (active caspase‐3) and anti‐apoptotic (survivin, Bcl‐xL/Bak dimer and Mcl‐1/Bak dimer) factors in the in vitro granuloma model. PBMC samples from 5 confirmed sarcoidosis subjects were challenged with microparticles and treated with saline, 1 μm MIA602, methylprednisolone (138 μm) and control PBMCs. Medium samples were removed 48 h after treatments.
As shown in Figure 3, the level of Mcl‐1/Bak dimer was significantly decreased in granulomas but was restored by MIA602. Bcl‐xL/Bak dimer levels increased in granulomas treated with MIA602 in comparison with those treated with saline. Active caspase‐3 level also increased significantly in granulomas compared with PBMC, possibly because of early lymphocyte activation. MIA602 further increased the levels of active caspase‐3. Survivin levels showed no difference between groups.
Figure 3.
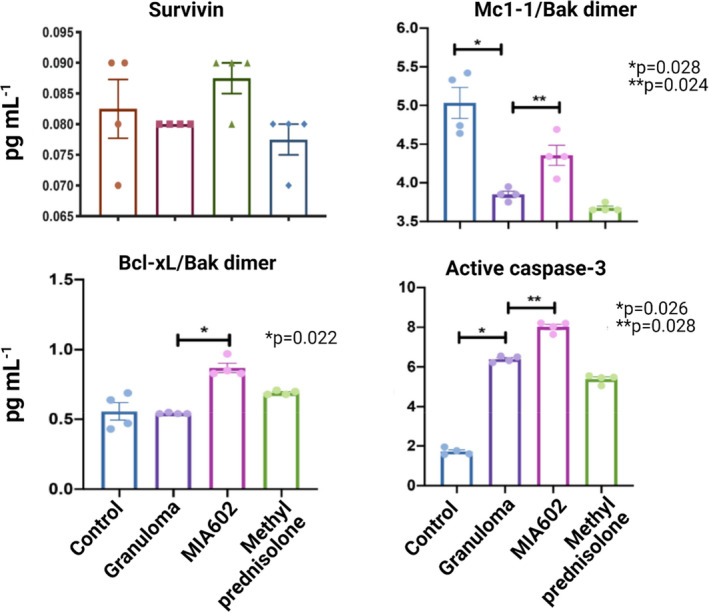
The protein levels of pro‐apoptotic (active caspase‐3) and anti‐apoptotic (survivin, Bcl‐xL/Bak dimer and Mcl‐1/Bak dimer) factors in the in vitro granuloma model measured by ELISA. We used four different groups: control, PBMCs of sarcoidosis subjects; granuloma, PBMCs of sarcoidosis subjects challenged with MAB microparticles; MIA602, granuloma treated with 1 μm MIA602; and methylprednisolone, granuloma treated with 138 μm methylprednisolone. Each group had the PBMCs from five sarcoidosis subjects. Data are from one experiment that is representative of three separate experiments.
MIA602 reduces inflammation in the lung of a sarcoidosis mouse model
We established a mouse pulmonary granuloma model from exposure to the microparticles and have used it to explore the role of type I IFN pathways in inflammation. This model is applicable to pulmonary sarcoidosis studies because of the similarity of granulomas to human sarcoidosis. We used C57BL/6 mice to develop the model as presented in detail elsewhere. 16 Granulomatous disease was induced by the addition of the MAB microparticles in the lower respiratory tract of mice, and we confirmed that the expression of GHRH‐R was significantly increased in the lungs with granulomata relative to control (Figure 4a). Subcutaneous injection of 5 µg MIA602 daily to mice resulted in a noticeable decrease in inflammation (Figure 4a). The granuloma and MIA602 models had higher expression of GHRH‐R (more green) relative to control. Scoring of the inflammation in our animal model revealed that GHRH‐R was significantly increased over granulomata, but treatment with MIA602 resulted in lower expression of GHRH‐R in granulomata (Figure 4b).
Figure 4.
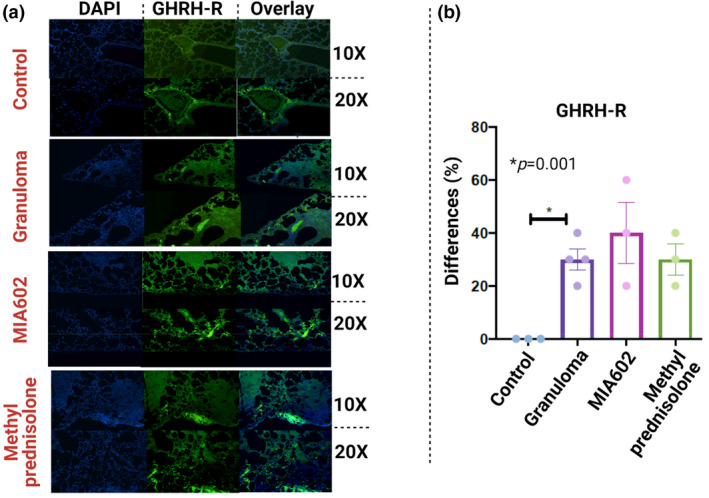
A representative image immunofluorescence microscopic detection of GHRH‐R in lungs of mice (right lower lobe). Magnifications are 10× and 20× for all representative images (a). P‐value shows percentage differences of lung‐stained cells between challenged mice and controls (N = 3 control, four challenged and three MIA602 mice) (b). Control mice were not challenged, while granuloma and MIA602 groups were challenged with MAB microparticles. The granuloma group received saline as treatment, while the MIA602 group received 5 μg MIA602 daily injection. Control also received saline. The staining intensity and percentage of area extent of the immunohistochemistry were evaluated by two lung pathologists blindly. Data are from one experiment that is representative of three separate experiments.
No significant variation was detected in the body weight of mouse models of MIA602 in comparison with control, showing that MIA602 did not interfere with normal growth (Supplementary figure 1). Lung samples were stained with H&E, CD68, PD‐1, PD‐L1 and CD30 and scored based on the percentage of cells expressing each marker (Figure 5a and b, Supplementary figure 2). CD30 was used as an indicator of lymphocyte activation over sarcoidosis inflammation. 17 The inflammation score of the lungs showed a higher percentage of inflammation in the granuloma group treated with saline. Lung inflammation was almost normalised in mice that received treatment with MIA602 in comparison with granuloma alone, and there was no significant variation between the anti‐inflammatory effect of MIA602 and methylprednisolone as shown in Figure 5b and Supplementary figure 2. CD30+ cells were significantly increased in the lungs of mice treated with MIA602, which also showed a reduced number and size of granuloma. The percentage of CD68+ cells in the lung increased in granuloma but decreased in MIA602 (not statistically significant). This same pattern can also be seen in PD‐1‐ and PD‐L1‐positive cells in granuloma (treated with saline) and MIA602 groups (Figure 5b).
Figure 5.
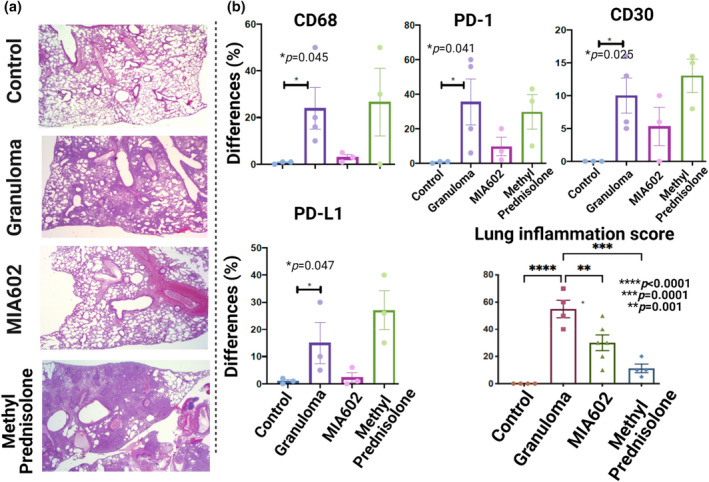
Representative images for H&E of lungs are shown for control, granuloma, granuloma treated with MIA602 and granuloma treated with methylprednisolone (a). Percentage differences of lung‐stained cells for different markers (CD30, CD68, PD‐1 and PD‐L1) between challenged mice and controls (b). Control mice (three mice) were not challenged, while granuloma (four mice) and MIA602 (three mice) groups were challenged with MAB microparticles. Mice in the MIA602 group received 5 μg MIA602 daily subcutaneous injection, while granuloma and control groups only received saline. The percentage of the involved area of the immunohistochemistry was evaluated by two lung pathologists separately. Data are from one experiment that is representative of three separate experiments.
Mice were challenged with the microparticles and treated with saline, MIA602 (5 μg) or methylprednisolone (100 μg) and were compared with control mice. After 3 weeks, lungs were harvested from all groups and single cells were generated for flow cytometric analysis. As shown in Figure 6, the population of CD45+CD68+ cells significantly increased in challenged mice. MIA602 treatment significantly reduced these cell populations. As it has been previously shown that PD‐1 and PD‐L1 play important roles in granuloma formation, 18 we hypothesised that MIA602 would restore the number of CD68+ cells that express PD‐1 and PD‐L1. Figure 6 shows that CD45+CD68+ cells that weakly expressed PD‐1 were indeed significantly reduced in mice challenged with granulomatous reaction. The percentage of cells significantly increased with MIA602 and reached the levels of the control group (Figure 6, Supplementary figure 3).
Figure 6.
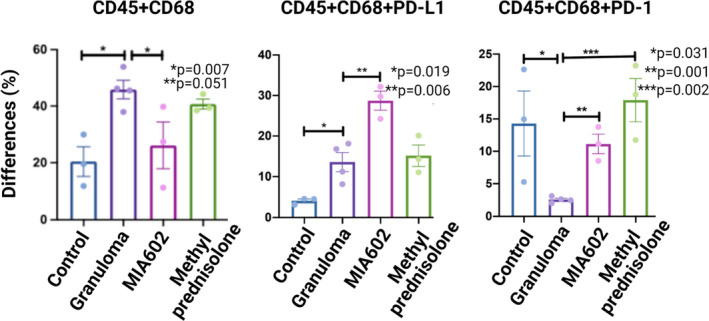
Flow cytometric analysis of single cells generated from mice lungs. The live leucocytes were defined as CD45+, and the CD45+CD68+ population was defined as macrophage. We used four different mice groups: control (three mice); granuloma (four mice), challenged with MAB microparticles; MIA602 (three mice), challenged with MAB microparticles and treated with MIA602; and methylprednisolone (three mice), challenged with MAB microparticles and treated with methylprednisolone. Significant variation (P‐value) is indicated for each plot. Data are from one experiment that is representative of three separate experiments.
To confirm that mouse lungs with granulomas showed a higher percentage of CD45+CD68+ cells that weakly expressed PD‐L1, we stained lung single cells in all experimental groups. As shown in Figure 6 and Supplementary figure 3, the population of CD45+CD68+PD‐L1 cells was significantly increased in granulomas and further increased after treatment with MIA602. Thus, we conclude that the anti‐inflammatory effects of MIA602 appear primarily through a reduction in CD68+ cells and upregulation of PD‐1 expression.
Inducible nitric oxide synthase and nitrotyrosine are increased in the lung of a sarcoidosis mouse model
Inducible nitric oxide synthase (iNOS) produces nitric oxide and plays a role in granuloma development as macrophages increase expression of iNOS after exposure to bacterial lipopolysaccharides and IFN‐γ. 19 We have previously shown that GHRH also increases the expression of iNOS. 20 While the effects of GHRH‐R antagonists in oxidative stress have been explored previously, 21 the effects of these peptides in nitrosative stress have not been well evaluated. To understand the effects of MIA602 on the nitric oxide response, we measured iNOS and nitrotyrosine (as an indicator of NO function) in mouse lungs that were challenged with microparticles (sarcoidosis model) and injected daily with saline and subcutaneous injections of 5 μg MIA602, and we compared them with the control group that were never challenged. Lung tissue was harvested after 3 weeks and stained with NOS2 and nitrotyrosine. As shown in Figure 7, expression of NOS2 was increased in sarcoidosis‐like granulomas and statistically unchanged in MIA602‐treated mice.
Figure 7.
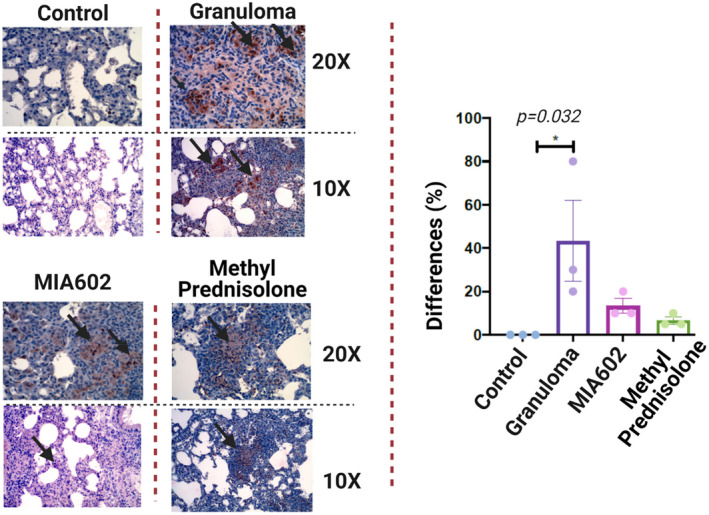
Representative image of immunohistochemical staining for iNOS2 staining in the lung of mice (control; granuloma, challenged with MAB microparticles; and MIA602, challenged with MAB microparticles and treated with MIA602). The black arrow shows granulomas with iNOS2 staining. Three mice were studied in each group. The brown colour in the tissue indicates the expression of iNOS2. Low magnification (10×) and high magnification (20×) are presented. Data are from one experiment that is representative of three separate experiments.
Nitrotyrosine expression was statistically significantly increased in the lungs of mice with sarcoidosis‐like granuloma as shown in Figure 8. Treatment with MIA602 decreased the nitrotyrosine levels, but the decrease was not statistically significant. This confirms that mouse lung after challenge with microparticles activates iNOS and increases nitrotyrosine. MIA602 reduced both iNOS and nitrotyrosine, but not to a statistically significant extent, in our experiments. This demonstrates a mild anti‐nitrosative effect of MIA602 in the sarcoidosis mouse model.
Figure 8.
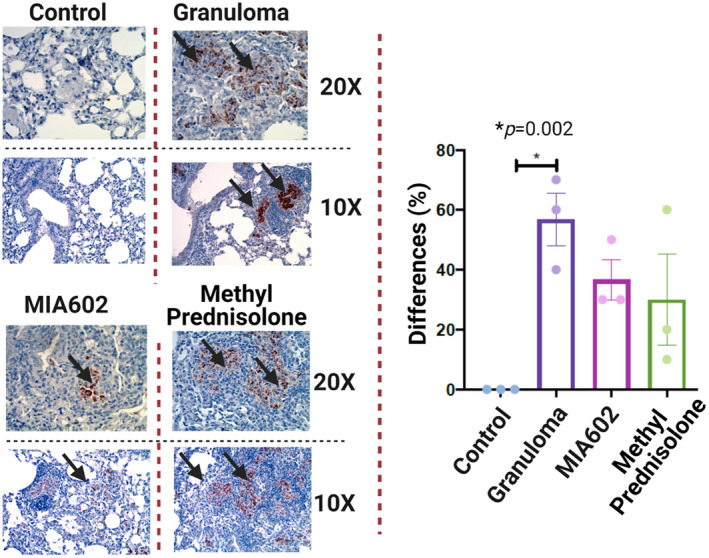
Representative immunohistochemical staining for nitrotyrosine in lung of mice (control; granuloma, challenged with MAB microparticles; and MIA602, challenged with MAB microparticles and treated with MIA602). The black arrow shows granulomas with nitrotyrosine staining. Three mice were studied in each group. The brown colour in the tissue indicates the expression of nitrotyrosine. Low magnification (10×) and high magnification (20×) are presented. Data are from one experiment that is representative of three separate experiments.
MIA602 significantly changed transcriptomics in the lung of a sarcoidosis mouse model
We extracted RNA from lungs challenged with the microparticles and treated daily with saline or 5 μg MIA602. RNA was isolated from lung tissue 3 weeks after the challenge, and RNA‐seq was performed.
Overall, we identified 1407 protein‐coding genes differentially expressed between granuloma and MIA602 with a false discovery rate P‐value < 0.05 (599 upregulated in MIA602 and 808 downregulated in MIA602). To identify the most robustly changing genes, we extracted only those with at least a twofold change and identified 489 genes (226 upregulated in MIA602 and 263 downregulated in MIA602). Supplementary table 1 shows the top 15s up‐ and downregulated coding genes after MIA602 treatment. Genes with a twofold change are visualised in a heatmap (Figure 9) showing consistent and robust differences between these two groups of samples.
Figure 9.
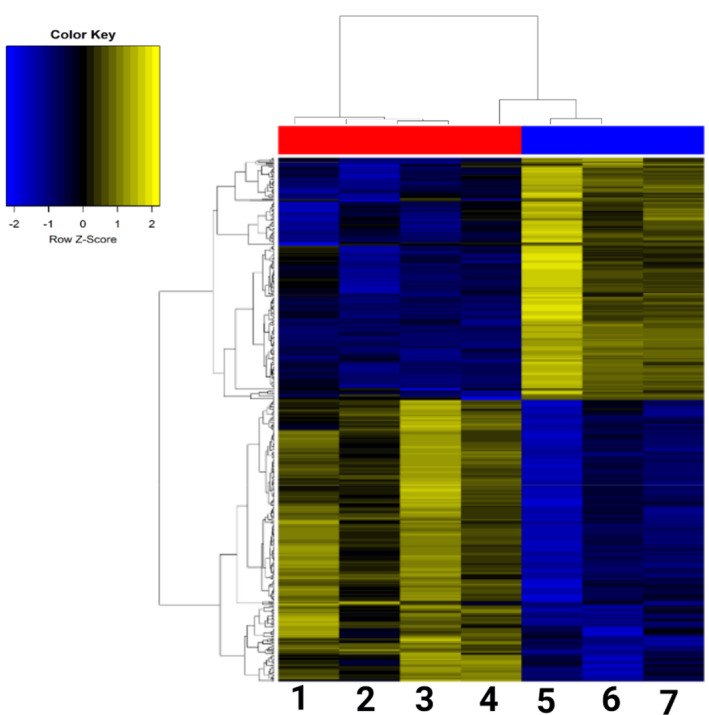
Heatmap of genes with FC > 2.5. Coding genes were illustrated in the heatmap between mouse lung with granuloma and treated with saline (four mice; codes 1, 2, 3 and 4) vs. mouse lung with granuloma and treated with MIA602 (three mice; codes 5, 6 and 7).
Discussion
MIA602 is a novel, potentially therapeutic agent for sarcoidosis. We found that MIA602 has an anti‐inflammatory effect in an in vitro sarcoidosis model, with a significant reduction in inflammatory cells. Furthermore, we show that the effect of MIA602 may not be because of apoptosis in granulomata. The anti‐inflammatory effect of MIA602 appears to be mediated by a reduction in CD45+CD68+ cells in granulomatous tissue and upregulation in PD‐1 expression in macrophages. We also showed significant changes in the gene profiles of mouse lungs treated with MIA602 vs. saline. For example, Serpina1d, which encodes alpha‐1‐antitrypsin 1–4 protein and is upregulated by MIA602, is an alpha‐1‐proteinase inhibitor (A1PI), which inhibits various proteases and protects tissues from enzymes of inflammatory cells. 22 More interestingly, the keratin gene family (Krt4, Krt5, Krt13, Krt14, Krt31 and Krt32) is upregulated by MIA602. These proteins are important for the mechanical stability and integrity of epithelial cells and tissues. In addition, they have regulatory functions and are involved in intracellular signalling pathways, for example protection from stress, wound healing and apoptosis. 23
The anti‐inflammatory effects of MIA602 in granulomas are a novel finding consistent with previous reports on anti‐inflammatory properties of GHRH‐R antagonists in pulmonary fibrosis, 12 ocular inflammation 24 and chronic prostatitis. 25
The current study suggests that MIA602 may alleviate inflammatory response by reduction of central cytokines in sarcoidosis, including IL‐17A. IL‐2, IL‐12, IL‐17 and TNF‐α that are also major cytokines in the pathogenesis of sarcoidosis. 26 Prasse et al. 27 demonstrated that bronchoalveolar lavage fluid of patients with sarcoidosis has high concentrations of IL‐2 and IFN‐γ. The roles of Th17 cells and IL‐17A in sarcoidosis are well known. 28
MIA602 restored the level of Mcl‐1/Bak dimer level in granulomas treated with MIA602 relative to those treated with saline, while we found a higher level of Bcl‐xL/Bak dimer and active caspase‐3 in the granuloma model treated with MIA602 than in the granuloma model treated with saline. There was also no variation in survivin levels after MIA602. Our data suggest that MIA602 may not induce apoptosis; however, further investigation is needed to characterise the effect of MIA602 on apoptosis based on labelling of DNA strand breaks (TUNEL) and DNA degradation assays. 29
The in vivo experiments in the sarcoidosis mouse model show that tissue inflammation scores were significantly reduced by MIA602 treatment, and this effect is mainly macrophage‐dependent. GH has an important role in the maturation and activation of macrophages. 30 While the role of the GHRH agonist on T cells was described previously by Khorram et al., 31 the current study suggests an anti‐inflammatory role of the GHRH‐R antagonist in a macrophage phenotype. Our IHC study shows that PD‐1 expression in lung macrophages was significantly reduced in the granuloma sarcoid model, and MIA602 restored PD‐1 expression. Thus, the GHRH‐R antagonist's anti‐inflammatory effects may be partially because of the expression of PD‐1+ macrophages. PD‐L1 is an activation marker of macrophages, M1 and M2 32 ; however, CD68 is a marker for the quantity of macrophages. 32 Our experiments show that the number of CD68+ cells significantly reduced with MIA602. We also found a weak activation of residential cells. Further investigation is needed to find out whether the residential cells are dominantly M1 or M2 phenotypes. We also found that healthy controls and methylprednisolone‐treated animals had a higher rate of CD45+CD68+PD‐1 compared with CD45+CD68+PD‐L1 cells, whereas MIA602‐treated animals had a higher rate of CD45+CD68+PD‐L1 compared with CD45+CD68+PD‐1. This could be because of a significant decrease in the number of CD68+ cells, but more activation of these cells in mice with MIA602 treatment. Further investigation is needed to explore more about the activation of M2 phenotype through MIA602 treatment.
Gordon et al. suggest that PD‐1+ macrophages may be associated with type 2 polarisation and suppression of phagocytosis. 33 The M2 phenotype of macrophages is known to have an immunosuppressive effect, with anti‐inflammatory cytokine secretion to allow wound healing. 34 PD‐L1+ macrophages increased in the lungs of mice with sarcoidosis‐like granulomas, and treatment with MIA602 induced higher numbers of PD‐L1 macrophages. The functionality of PD‐L1 in macrophages is not well understood. It has been suggested that macrophages are destroyed by T cells via PD‐L1. 35
Our study has several limitations. We did not incorporate a healthy control in this study. However, our previous study demonstrated that sarcoidosis models generated from PBMCs of healthy control and sarcoidosis subjects showed a similar pattern of proinflammatory cytokine response, but different concentrations in the majority of measured cytokines. 15 We were also unable to collect lung biopsies from normal healthy people, or inflamed lung tissue from patients without granulomata (pneumonia, bronchiolitis) or other granulomatous diseases such as anti‐neutrophil cytoplasmic autoantibody (ANCA) vasculitis because of the IRB limitations. Our results showed that MIA602 significantly decreases IL‐17A; however, we did not test the effect of MIA602 on other isoforms of IL‐17, which is another limitation of this study. We studied iNOS expression in this study by immunohistochemical staining, but it will be measured in response to MIA602 in the future.
The effect of MIA602 in neutrophils and lymphocytes was not evaluated in our study. Lymphocytes are important effectors in the granuloma. The role of a ratio between neutrophils and lymphocytes in sarcoidosis has been discussed elsewhere. 36 , 37 In addition, the effect of MIA602 in caspase‐3 needs further investigation. MIA602 significantly increased active caspase‐3 concentration. It may be associated with inducing apoptosis in granuloma or may be because of the early activation of stimulated T cells. 36 MIA602 reduced iNOS, but not to a statistically significant degree, in our experiment. This finding could be because of the limitations in our sample size.
The current study shows that MIA602 has important anti‐inflammatory properties through a reduction in IL‐2, IL‐2R, IL‐12, IL‐17A and TNF‐α release and reduction in CD68+ cells in the lung. These effects may be mediated through checkpoint receptors (PD‐1 and PD‐L1). Our results showed that MIA602 has non‐inferiority anti‐inflammatory effects in comparison with corticosteroid. Further investigation will lead to the understanding of the anti‐inflammatory effects of MIA602 in sarcoidosis and explore its potential therapeutic role.
Methods
Ethics
All human and animal methods were carried out following the relevant guidelines and regulations. The sarcoidosis biobank and animal studies were approved by the Miami VA Health Care System's Institutional Review Board and Institutional Animal Care and Use Committees.
MIA602 preparation
MIA602 was prepared by Dr Renzi Cai and Dr Andrew Schally. The chemical structure of MIA602 is [PhAc‐Ada0, D‐Arg2, Fpa56, Ala8, Har9, Tyr (Me)10, His11, Orn12, Abu15, His20, Orn21, Nle27, D‐Arg28, Har29] hGH‐RH (1–29) NH2. MIA602 was dissolved in 100% dimethyl sulphoxide (DMSO, ACS grade; Sigma, St. Louis, MO, USA) for stock and diluted at 1:1000 in the corresponding culture medium to a final concentration of 1 μm before use as previously presented. The control group in vitro and in vivo received a placebo with the same volume and concentration of DMSO.
Microparticle development
As previously presented in detail, 15 microparticles were generated from a rough colony of a clinical strain of M. abscessus by sonicating and heating live bacilli. High‐quality images of non‐infectious MAB particles were obtained by scanning electron microscope (SEM).
Human blood samples
Blood samples were collected from nine patients with confirmed pulmonary sarcoidosis randomly selected from the University of Miami Sarcoidosis Biobank and matched by age, sex and race with 10 healthy control by the University of Miami Institutional Review Board (approval number 20150612). None of the sarcoidosis or healthy controls subjects were on immunosuppressive therapy. To avoid the inconvenience and risks associated with additional venipunctures, a 10 mL blood specimen was collected during an already‐scheduled venipuncture. Patients who currently had an hgb < 7 mg dL−1 were excluded from participating in this study.
Maturing in vitro granuloma‐like formation
In vitro granulomas were developed from challenging PBMCs to microparticles as previously described. 15 Our IRB did not allow us to study PBMCs of the healthy controls for this study. However, our previous study demonstrated that sarcoidosis models generated from PBMCs of healthy control and sarcoidosis subjects show a similar pattern of proinflammatory cytokines but different concentrations in the majority of measured cytokines. 15
Mouse model exposure to MAB microparticles
The granuloma model was developed in the mouse lung as previously described. 16 After challenge and treatment (3 weeks), mice were sacrificed on day 14. We measured the weight of each mouse (whole‐body), lung, spleen, liver, and kidney. The left lungs were harvested for pathology after perfusion to remove blood. Lungs were filled with 10% buffered formalin and fixed in formalin for at least 72 h before IHC staining. H&E staining was used on the right lower lobe of the lung to determine inflammatory pathology. 16 Two pathologists with no previous information about the detail of the sample (blinded) analysed the inflammation. Lung inflammation was scored using the three fields with the highest infiltrate's intensity at 100X power magnification. The area of inflammation was measured and averaged for the three examined high‐power fields. 16
ELISA
As we previously presented, 15 PBMCs were lysed in lysis buffer (Cat #9803; Cell Signaling Technology, Beverly, MA, USA) with protease inhibitor cocktail (Cat#5871; Cell Signaling Technology) and sonicated three times for 2 s each with at least 1‐min rest on ice between each 2‐s pulse. Samples were centrifuged at 10 000 g for 5 min at 4°C, and the supernatant was collected. Protein concentration was determined by the BCA protein assay kit (Cat #7780; Cell Signaling Technology).
Thirty micrograms of total protein was mixed in a reducing sample buffer and used for mitochondrial apoptosis assay per the kit instruction. The assay was performed using a Bio‐Plex Pro™ RBM Apoptosis Panel 1 Kit (Cat#171WAR1CK; Life Science, San Francisco, CA, USA).
To measure the cytokines in media, supernatant aliquot samples were analysed, thawed and spun at 16 128 g for 10 min to separate the particulate material at the bottom. From each sample, 50 μL of undiluted media was plated onto a 96‐well V‐bottom plate (source plate) by manual pipetting according to the predefined maps. The aliquots were wrapped in parafilm and kept in a humid chamber at 4°C during the entire process, but not longer than 72 h. Growth factors and their receptors' capture antibodies were reconstituted and diluted according to the manufacturer's instructions, and 50 μL was plated into each well of respective 96‐well high‐binding half‐well plates, which were then sealed and incubated overnight at 4°C. The cytokine levels were measured using the Th1/Th2 Cytokine 11‐Plex Human ProcartaPlex Panel (Cat # EPX110‐10810‐901; Invitrogen, Suwanee, GA, USA).
Immunofluorescence confocal microscopy
Detailed methods have been discussed elsewhere. 15 In summary, mice were sacrificed on day 14, and the left lungs were harvested as presented earlier. Lungs were filled with 10% buffered formalin and fixed in formalin for at least 72 h before IHC staining. H&E staining was used to determine inflammatory pathology.
For immunofluorescence, paraffin‐embedded serial sections (5 μm) first underwent standard deparaffinisation and rehydration procedures and were then probed with GHRH‐R (Cat# TA311715; OriGene Technologies, Inc., Rockville, MD, USA) as primary antibody and anti‐rabbit antibody from Sigma (Cat# F‐9887; Sigma‐Aldrich) as secondary antibody. Nuclei were counterstained with DAPI (Cat# 28718‐90‐3; Sigma‐Aldrich). Tissue sections were analysed using fluorescence microscopy and ImageJ software (version 6.0; NIH, Bethesda, MD, USA) to quantitate fluorescence intensity. In trichrome‐stained slides, blue stain (collagen content) was also quantitatively analysed using ImageJ.
Confocal immunofluorescence images were acquired using a Leica DM6000 microscope with a SP5 confocal module at the University of Miami McKnight Analytical Imaging Core Facility. Captured images were processed using Velocity Software version 6.1.1 software (PerkinElmer, Waltham, MA, USA).
For IHC, 5‐μm paraffin sections were processed by deparaffinisation and rehydration followed by endogenous peroxidase blocking (1% H2O2 in methanol for 20 min) and antigen retrieval (boiled in 10 mm citrate buffer for 30 min). Tissue sections were blocked with 2% goat or horse serum (Vector Laboratories, Burlingame, CA, USA) and incubated with antibody CD68 (Cat# 25747‐1‐AP; ProteinTech Group, Inc., Rosemont, IL, USA), PD‐1 (Cat # 84651; Cell Signaling Technology, Beverly, MA, USA), PD‐L1 (Cat# 17952‐1‐AP; ProteinTech Group, Inc., Rosemont, IL, USA), CD30 (Cat# LS‐c162069; LSBio, Seattle, WA, USA), CD3 (Cat# 99940; Cell Signaling Technology, Beverly, MA, USA), iNOS (Cat #PAI‐036; Invitrogen, USA) and nitrotyrosine (Cat# NBP2‐54606; Novus Biologicals, LLC, Littleton, CO, USA) overnight at 4°C and washed with TBST five times. Then, secondary antibodies were added (Cat# PI‐2000; Vector Laboratories, Inc.). Immunoreactivity was detected using the ABC Elite Kit (Cat# PK‐6200; Vector Laboratories, Inc.). We used DAB as the final chromogen and haematoxylin as the nuclear counterstain. Negative controls for all antibodies were made by replacing the primary antibody with non‐immunogen IgG. Lung inflammation was scored using the three fields with the highest infiltrate intensity at 100× power magnification as previously presented by our team. 16 The area of inflammation was measured and averaged for the three examined high‐power fields.
RNA isolation and analysis
RNA from mouse lungs was extracted using the RNA Miniprep Plus Kit (Cat# R2050; Zymo Research, Inc, Irvine, CA, USA). Briefly, the whole lung was homogenised in TRI, and total RNA extraction was performed following the instructions provided by the manufacturer with additional DNase treatment. The quantity and quality of the samples were determined by NanoDrop Spectrophotometer and Agilent Bioanalyzer 2100, respectively (Agilent, Santa Clara, CA, USA).
Preparation and sequencing of RNA libraries was performed at the John P Hussman Institute for Human Genomics Center for Genome Technology. Briefly, total RNA quantity and quality were determined using the Agilent Bioanalyzer. At least 300 ng of total RNA was used as input for the Nugen Universal Plus mRNA‐Seq Library Preparation Kit (Cat# 0520‐24; Tecan Genomics, Inc., Redwood, CA, USA) according to the manufacturer's protocol to create polyA‐enriched mRNA‐sequencing libraries. Sequencing was performed on the Illumina NovaSeq 6000, generating at least 40 million paired‐end 100 base reads per sample.
Sequencing data were processed with a bioinformatics pipeline including quality control, alignment to the mm10 mouse reference genome and gene quantification against the Ensembl build 91 mouse gene set. Count data were input into edgeR software for differential expression analysis. Counts were normalised using the trimmed mean of M‐values (TMM) method to account for the compositional difference between the libraries. Differential expression analysis between groups was performed using the quasi‐likelihood F‐test implemented in edgeR. Genes were considered to be statistically different with a false discovery rate P‐value (FDR) ≤ 0.05. The raw RNA‐seq data are available as part of GEO SuperSeries GSE140439.
Flow cytometry
Mice were sacrificed on day 14, and the left lungs were harvested for pathology after perfusion of the right ventricle with 10 mL of PBS.
The upper half of the left lung tissue (without trachea, main bronchus or branches) was removed and rinsed by PBS to clean off the blood. The tissue was minced and dispersed with scissors, to increase the total surface area. To develop single‐cell suspension, we used the rubber end of a 5‐mL plastic syringe to mesh cells through a 100‐µm cell strainer with continuous rinse of the cell strainer with ice‐cold RPMI 1640. We performed meshed cell suspension again through a 70‐µm cell strainer and rinsed thoroughly with 3 mL of washing buffer containing DNAse followed by 15 mL of DNAse‐free washing buffer. We then used centrifugation at 286 g and 18°C for 5 min and discarded the supernatant. 38
The cells (106 cells mL−1) were resuspended in 100 μL protein‐blocking solution with 5 μL fluorescent‐conjugated antibodies: CD8 (Cat# 100714), CD45 (Cat# 103130), CD68 (Cat# 137004), PD‐1 (Cat# 135219), PD‐L1 (Cat# 124308), CD4 (Cat# 100510), CD11b (Cat# 101243), CD11c (Cat# 117318), F4/80 (Cat# 123146) and IFNg (Cat# 505836). All antibodies were purchased from BioLegend, Inc., San Diego, CA, USA. Samples were analysed on a BD LSR II flow cytometer using BD FACSDiva software, and data analysis was performed using FlowJo software (TreeStar, Ashland, OR, USA). Cell populations were identified using a sequential gating strategy. Viable cells were sorted using CD45 expression. 39 We used CD68 expression as described previously to study the leucocyte population in lung samples. 39 CD68 expression has been widely applied as a pan‐macrophage marker and used to classify leucocyte subpopulations in the lungs of adult mice into three groups: CD68 negative (CD68−), CD68low and CD68high. 39
Statistical analysis
Cytokine analysis was performed with one‐way analysis of variance (ANOVA) and non‐parametric analysis (Friedman and Dunn's multiple comparison tests). One‐way ANOVA unpaired analysis was used to compare the values of means of all experiments for mouse groups using GraphPad Prism 8 software (GraphPad Software, Inc., San Diego, CA, USA). Data represent mean ± SEM, and results with P‐value (two‐sided) < 0.05 were defined as statistically significant.
Study approval
Blood samples for this study were randomly selected from the University of Miami Sarcoidosis Biobank per the University of Miami Institutional Review Board, approval number 20150612. Written informed consent was received from participants before inclusion in the study.
The animal study was reviewed and approved by the Miami VA Healthcare System's Animal Care and Use Committee (IACUC).
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
Chongxu Zhang: Formal analysis; Methodology. Runxia Tian: Data curation; Formal analysis. Emilee M Dreifus: Data curation; Formal analysis. Abdolrazagh Hashemi Shahraki: Visualization; Writing‐review & editing. Greg Holt: Writing‐original draft; Writing‐review & editing. Renzhi Cai: Data curation; Formal analysis. Anthony Griswold: Software; Visualization; Writing‐original draft; Writing‐review & editing. Pablo Bejarano: Data curation; Formal analysis; Writing‐review & editing. Robert Jackson: Writing‐review & editing. Andrew V Schally: Writing‐review & editing.
Supporting information
Acknowledgments
The authors acknowledge the Sylvester Comprehensive Cancer Center Flow Cytometry. The research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA240139. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This study was funded by the VA Distinguished Scientist Award, VA (Grant No. 1 I01 BX004371), and VA Research Service.
References
- 1. Costabel U, Hunninghake GW. ATS/ERS/WASOG statement on sarcoidosis. Sarcoidosis Statement Committee. American Thoracic Society. European Respiratory Society. World Association for Sarcoidosis and Other Granulomatous Disorders. Eur Respir J 1999; 14: 735–737. [DOI] [PubMed] [Google Scholar]
- 2. Mirsaeidi M, Machado RF, Schraufnagel D, Sweiss NJ, Baughman RP. Racial difference in sarcoidosis mortality in the United States. Chest 2015; 147: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen ES, Moller DR. Etiologies of sarcoidosis. Clin Rev Allergy Immunol 2015; 49: 6–18. [DOI] [PubMed] [Google Scholar]
- 4. Chen ES, Moller DR. Sarcoidosis—scientific progress and clinical challenges. Nat Rev Rheumatol 2011; 7: 457–467. [DOI] [PubMed] [Google Scholar]
- 5. Ungprasert P, Ryu JH, Matteson EL. Clinical manifestations, diagnosis, and treatment of sarcoidosis. Mayo Clin Proc Innov Qual Outcomes 2019; 3: 358–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. James WE, Baughman R. Treatment of sarcoidosis: grading the evidence. Expert Rev Clin Pharmacol 2018; 11: 677–687. [DOI] [PubMed] [Google Scholar]
- 7. Ramachandraiah V, Aronow W, Chandy D. Pulmonary sarcoidosis: an update. Postgrad Med 2017; 129: 149–158. [DOI] [PubMed] [Google Scholar]
- 8. Drent M, Cremers JP, Jansen TL, Baughman RP. Practical eminence and experience‐based recommendations for use of TNF‐α inhibitors in sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis 2014; 31: 91–107. [PubMed] [Google Scholar]
- 9. Gaylinn BD. Molecular and cell biology of the growth hormone‐releasing hormone receptor. Growth Horm IGF Res 1999; 9: 37–44. [DOI] [PubMed] [Google Scholar]
- 10. Schally AV, Varga JL, Engel JB. Antagonists of growth‐hormone‐releasing hormone: an emerging new therapy for cancer. Nat Clin Pract Endocrinol Metab 2008; 4: 33–43. [DOI] [PubMed] [Google Scholar]
- 11. Schally AV, Zhang X, Cai R, Hare JM, Granata R, Bartoli M. Actions and potential therapeutic applications of growth hormone‐releasing hormone agonists. Endocrinology 2019; 160: 1600–1612. [DOI] [PubMed] [Google Scholar]
- 12. Zhang C, Cai R, Lazerson A et al. Growth hormone‐releasing hormone receptor antagonist modulates lung inflammation and fibrosis due to bleomycin. Lung 2019; 197: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gan J, Ke X, Jiang J et al. Growth hormone‐releasing hormone receptor antagonists inhibit human gastric cancer through downregulation of PAK1‐STAT3/NF‐κB signaling. Proc Natl Acad Sci USA 2016; 113: 14745–14750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Recinella L, Chiavaroli A, Orlando G et al. Antinflammatory, antioxidant, and behavioral effects induced by administration of growth hormone‐releasing hormone analogs in mice. Sci Rep 2020; 10: 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang C, Chery S, Lazerson A et al. Anti‐inflammatory effects of α‐MSH through p‐CREB expression in sarcoidosis like granuloma model. Sci Rep 2020; 10: 7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C, Asif H, Holt GE et al. Mycobacterium abscessus‐bronchial epithelial cells cross‐talk through type I interferon signaling. Front Immunol 2019; 10: 2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ahmad S, Azid NA, Boer JC et al. The key role of TNF‐TNFR2 interactions in the modulation of allergic inflammation: a review. Front Immunol 2018; 9: 2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Braun NA, Celada LJ, Herazo‐Maya JD et al. Blockade of the programmed death‐1 pathway restores sarcoidosis CD4+ T‐cell proliferative capacity. Am J Respir Crit Care Med 2014; 190: 560–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Facchetti F, Vermi W, Fiorentini S et al. Expression of inducible nitric oxide synthase in human granulomas and histiocytic reactions. Am J Pathol 1999; 154: 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barabutis N, Siejka A, Schally AV. Growth hormone releasing hormone induces the expression of nitric oxide synthase. J Cell Mol Med 2011; 15: 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barabutis N, Schally AV. Antioxidant activity of growth hormone‐releasing hormone antagonists in LNCaP human prostate cancer line. Proc Natl Acad Sci USA 2008; 105: 20470–20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mackiewicz A, Kushner I, Baumann H. Acute Phase Proteins Molecular Biology, Biochemistry, and Clinical Applications. Boca Raton, FL: CRC Press, 1993. [Google Scholar]
- 23. Moll R, Divo M, Langbein L. The human keratins: biology and pathology. Histochem Cell Biol 2008; 129: 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin YJ, Chan SO, Chong KK et al. Antagonist of GH‐releasing hormone receptors alleviates experimental ocular inflammation. Proc Natl Acad Sci USA 2014; 111: 18303–18308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Popovics P, Cai R, Sha W, Rick FG, Schally AV. Growth hormone‐releasing hormone antagonists reduce prostatic enlargement and inflammation in carrageenan‐induced chronic prostatitis. Prostate 2018; 78: 970–980. [DOI] [PubMed] [Google Scholar]
- 26. Ringkowski S, Thomas PS, Herbert C. Interleukin‐12 family cytokines and sarcoidosis. Front Pharmacol 2014; 5: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prasse A, Georges CG, Biller H et al. Th1 cytokine pattern in sarcoidosis is expressed by bronchoalveolar CD4+ and CD8+ T cells. Clin Exp Immunol 2000; 122: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramstein J, Broos CE, Simpson LJ et al. IFN‐γ‐producing T‐helper 17.1 cells are increased in sarcoidosis and are more prevalent than T‐helper type 1 cells. Am J Respir Crit Care Med 2016; 193: 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou J, Wang Y, Liu Y, Zeng H, Xu H, Lian F. Adipose derived mesenchymal stem cells alleviated osteoarthritis and chondrocyte apoptosis through autophagy inducing. J Cell Biochem 2019; 120: 2198–2212. [DOI] [PubMed] [Google Scholar]
- 30. Schneider A, Wood HN, Geden S et al. Growth hormone‐mediated reprogramming of macrophage transcriptome and effector functions. Sci Rep 2019; 9: 19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khorram O, Yeung M, Vu L, Yen SS. Effects of [norleucine27]growth hormone‐releasing hormone (GHRH) (1–29)‐NH2 administration on the immune system of aging men and women. J Clin Endocrinol Metab 1997; 82: 3590–3596. [DOI] [PubMed] [Google Scholar]
- 32. Francisco LM, Sage PT, Sharpe AH. The PD‐1 pathway in tolerance and autoimmunity. Immunol Rev 2010; 236: 219–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gordon SR, Maute RL, Dulken BW et al. PD‐1 expression by tumour‐associated macrophages inhibits phagocytosis and tumour immunity. Nat 2017; 545: 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fang P, Li X, Dai J et al. Immune cell subset differentiation and tissue inflammation. J Hematol Oncol 2018; 11: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singhal S, Stadanlick J, Annunziata MJ et al. Human tumor‐associated monocytes/macrophages and their regulation of T cell responses in early‐stage lung cancer. Sci Transl Med 2019; 11: eaat1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alam A, Cohen LY, Aouad S, Sekaly RP. Early activation of caspases during T lymphocyte stimulation results in selective substrate cleavage in nonapoptotic cells. J Exp Med 1999; 190: 1879–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mirsaeidi M, Mortaz E, Omar HR, Camporesi EM, Sweiss N. Association of neutrophil to lymphocyte ratio and pulmonary hypertension in sarcoidosis patients. Tanaffos 2016; 15: 44–47. [PMC free article] [PubMed] [Google Scholar]
- 38. Posel C, Moller K, Boltze J, Wagner DC, Weise G. Isolation and flow cytometric analysis of immune cells from the ischemic mouse brain. J Vis Exp 2016; 12: 53658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zaynagetdinov R, Sherrill TP, Kendall PL et al. Identification of myeloid cell subsets in murine lungs using flow cytometry. Am J Respir Cell Mol Biol 2013; 49: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


