Abstract
Gamma-aminobutyric acid (GABA) is the main inhibitory neurotransmitter in the human brain and plays a key role in several brain functions and neuropsychiatric disorders such as anxiety, epilepsy, and depression. For decades, several in vivo and ex vivo techniques have been used to highlight the mechanisms of the GABA system, however, no studies have currently combined the techniques to create a high-resolution multimodal view of the GABA system.
Here, we present a quantitative high-resolution in vivo atlas of the human brain benzodiazepine receptor sites (BZR) located on postsynaptic ionotropic GABAA receptors (GABAA Rs), generated on the basis of in vivo [11C]flumazenil Positron Emission Tomography (PET) data. Next, based on ex vivo autoradiography data, we transform the PET-generated atlas from binding values into BZR protein density. Finally, we examine the brain regional association between BZR protein density and ex vivo mRNA expression for the 19 subunits in the GABAA R, including an estimation of the minimally required expression of mRNA levels for each subunit to translate into BZR protein.
This represents the first publicly available quantitative high-resolution in vivo atlas of the spatial distribution of BZR densities in the healthy human brain. The atlas provides a unique neuroscientific tool as well as novel insights into the association between mRNA expression for individual subunits in the GABAA R and the BZR density at each location in the brain.
Keywords: GABA, PET, Atlas, Autoradiography, mRNA, Benzodiazepine binding site
1. Introduction
Gamma-aminobutyric acid (GABA) is the main inhibitory neurotransmitter in the human brain and is a key component in several brain functions and in neuropsychiatric disorders, including anxiety, epilepsy, and depression (Sequiera et al. 2019; Zhu et al., 2018). The GABAA receptor (GABAA R) is a pentameric complex that functions as a ligand-gated chloride ion channel and it is the target of several pharmacological compounds such as anesthetics, antiepileptics, and hypnotics.
Benzodiazepines are well-known for their sedative, anticonvulsive and anxiolytic effects. They act as positive allosteric modulators via the benzodiazepine binding sites (BZR) which are located between the α1,2,3,5 and γ1–3 subunits in the pentameric constellation of the postsynaptic ionotropic GABAA R (Fig. 1). In total, 19 subunits (α1–6, β1–3, γ1–3, ρ1–3, δ, ε, θ, π) have currently been identified, and the constellation of subunits to form GABAA Rs, and consequently the expression of and affinity to the BZR site, has been shown to differ between individuals, between brain regions, and across the life span (Sequiera et al., 2019; Mulligan et al., 2012).
Fig. 1.
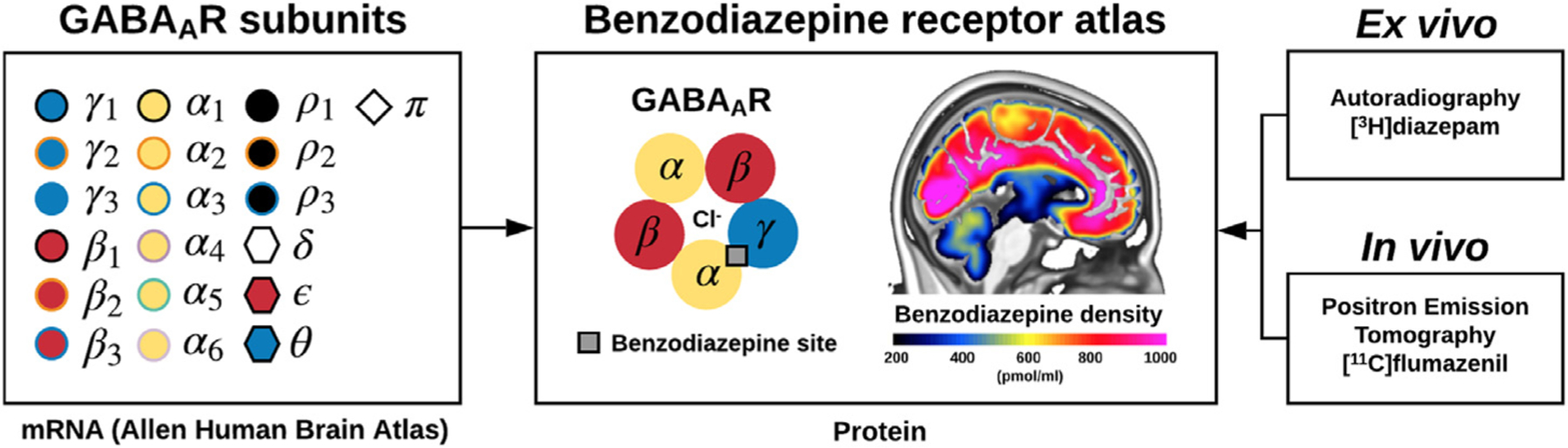
Overview of the construction of the high-resolution benzodiazepine receptor atlas using (1) mRNA data of the 19 subunits of the GABAA receptor (α1–6, β1–3, γ1–3, ρ1–3, δ, ε, θ, π) obtained from the Allen Human Brain Atlas (left), (2) ex vivo autoradiography data of the protein density of benzodiazepine receptors obtained from Braestrup et al., 1977 using the radioligand [3H]diazepam (right, upper), and (3) in vivo Positron Emission Tomography data of the protein density of benzodiazepine receptors obtained using the radioligand [11C]flumazenil (right, lower).
Several in vivo and ex vivo neuroimaging techniques have been developed over time to capture the complex mechanisms of the GABA system, all with benefits and drawbacks. The technique MR spectroscopy (MRS) has been used to quantitatively measure the concentration of GABA in vivo by direct detection of endogenous metabolites (Puts et al., 2012). The GABA signal from the MRS spectrum can then be isolated, to estimate the concentration of GABA at millimolar levels. However, MRS is often only used for a single brain region or voxel (single-voxel MRS), providing only partial coverage of the concentration of GABA within the brain. For an excellent review on MRS to measure GABA we refer to Puts et al. 2012.
In contrast, the technique Positron Emission Tomography (PET) has been used to quantitatively measure the BZR availability in vivo using the radioligand [11C]flumazenil, indirectly reflecting the density of GABAA Rs containing the BZR site. The radioligand [11C]flumazenil is a reversible BZR antagonist, and PET is used to detect the spatial location in 3D of the radioactive decay, providing full coverage of the BZR density within the brain, and with high-resolution PET scanners providing a resolution up to 1–2 mm (Feng et al., 2016; Price et al., 1991; Yamauchi, 2005). A third technique, autoradiography, has been used as a postmortem ex vivo technique to quantitatively measure BZR availability (pmol/g tissue) in several brain regions, however, similar to MRS, this technique often only provides partial coverage of the brain (Zilles et al., 2002; Braestrup et al., 1977). Other novel techniques to understand the GABA system include the Allen Human Brain Atlas (AHBA) that was developed from ex vivo postmortem brains, providing mRNA levels from complete microarray gene expression analyses for the 19 subunits of the GABAA R measured throughout the entire brain (Hawrylycz et al., 2012). Together, these techniques provide us with different views of the GABA system ranging from the concentration of GABA (MRS), density of GABA receptors (PET, autoradiography), and mRNA expression (microarray gene expression).
In recent years, high-resolution quantitative human brain atlases have emerged to represent a highly valuable reference tool in neuroimaging research to inform about the in vivo 3D distribution and density of specific targets. On the basis of high-resolution PET neuroimaging data from 210 healthy individuals, Beliveau et al., 2017 created a quantitative high-resolution atlas of the serotonin system (four receptors, and the serotonin transporter), providing a valuable reference for researchers studying human brain disorders, or effects of pharmacological interventions on the serotonin system that may subsequently down-or upregulate the receptors. Because the functional organization of receptor systems may be different than commonly defined structural organizations such as Brodmann areas (Amunts and Zilles, 2015), it is important to define and use the spatial distribution of the receptor systems modulating brain function not only to capture a more refined view of the brain (Beliveau et al., 2020), but also to provide insights into novel brain parcellations. Here, we present the first quantitative PET-based high-resolution in vivo 3D human brain atlas of the distribution of BZRs; an atlas to serve as a reference in future studies. Based on autoradiography data, we transform the PET generated atlas into brain regional protein densities of BZR and compare it to the mRNA expression for individual subunits in the GABAA R, using the Allen Human Brain Atlas (Hawrylycz et al., 2012) (Fig. 1).
2. Materials and methods
2.1. Study participants and neuroimaging
Sixteen healthy participants (9 females; mean age ± SD: 26.6 ± 8 years, range 19–46 years) were included in the study (Knudsen et al., 2016). The study was approved by the Regional Ethics Committee (KF 01280377), and all subjects provided written informed consent prior to participation, in accordance with The Declaration of Helsinki II. Data from 10 individuals have entered a previous paper (Feng et al., 2016) where more detailed accounts of the methods can be found. The participants were scanned between 1 and 3 times with the High-Resolution Research Tomograph (HRRT, CTI/Siemens) PET scanner at Rigshospitalet (Copenhagen, Denmark) with the radioligand [11C]flumazenil (26 unique PET scans in total). After an initial transmission scan (Keller et al., 2013), the radioligand was given either as an intra-venous bolus injection or as a bolus-infusion, and a PET emission scan was conducted over 90 min, starting at the time of injection. PET data was reconstructed into 35 frames (6 × 5, 10 × 15, 4 × 30, 5 × 120, 5 × 300, 5 × 600 s) with isotropic voxels of 1.2 mm using a 3D-ordered subset expectation maximum and point spread function modeling (3D-OSEM-PSF) (16 subsets, 10 iterations) with [137 Cs]transmission scan-based attenuation correction and no postfil-tering (Sureau et al., 2008; Hong et al., 2007; Mazoyer et al., 1991). Arterial sampling was also performed and the arterial input function was corrected for radiometabolites (Feng et al., 2016). The reconstructed PET data were motion corrected using the reconcile procedure in AIR (v. 5.2.5, http://loni.usc.edu/Software/AIR), and our criterion for acceptable motion was a median movement less than 3 mm across frames (Nørgaard et al., 2019). All participants had acceptable median motion below 3 mm. The PET tracer injection protocols carried out for each subject are listed in Table S1 in the supplementary.
An isotropic T1-weigthed MP-RAGE was acquired for all participants (matrix size = 256 × 256 × 192; voxel size = 1 mm; TR/TE/TI = 1550/3.04/800 ms; flip angle = 9°) using either a Magne-tom Trio 3T or a 3T Verio MR scanner (both Siemens Inc.). Furthermore, an isotropic T2-weighted sequence (matrix size 256 × 256 × 176; voxel size = 1 mm; TR/TE = 3200/409 ms; flip angle = 120°) was acquired for all participants. All acquired MRI’s were corrected for gradient non-linearities (Jovicich et al., 2006) and examined to ensure the absence of structural abnormalities.
The MR data was processed using FreeSurfer (v.6.0) (Fischl et al. 2012), and co-registered to the PET data using a rigid transformation and a normalized mutual information cost function.
Regional time-activity curves (TACs) of the target concentration, CT, were obtained using PETsurfer (Greve et al., 2014) defined by the Desikan-Killiany atlas. Voxel-wise TACs in common volume space (MNI152) were obtained using the Combined Volumetric and Surface (CVS) registration, and volume smoothed with a 5 mm FWHM Gaussian filter. Surface-based TACs in common space (fsaverage) were obtained using the cortical-surface registration provided by FreeSurfer and surface smoothed by a Gaussian filter with 10 mm FWHM (Fischl et al. 2012; Greve et al., 2014).
2.2. Quantification of BZR availability
The PET data were quantified to estimate total distribution volumes (VT) for each region using steady-state analysis for the bolus-infusion experiments and Logan analysis (Logan et al., 1996) for the bolus injections, both using the metabolite-corrected plasma curve as input function (Figure S1) (Feng et al., 2016).
2.2.1. Blood acquisition and analysis
For the bolus-infusion experiments, blood sampling was carried out by cannula insertion into one of the cubital veins, while another cannula was inserted to the other cubital vein for radiotracer administration.
For the bolus experiments, arterial blood sampling was carried out by cannula insertion into the radial artery of the non-dominant arm. During the first 10 min of the PET scan, blood measurements were counted continuously in whole-blood using an ABSS autosampler (Allogg Technology), and three samples were drawn manually to calibrate the autosamples.
The plasma to whole-blood ratio was obtained by manually drawing blood samples at 18 time points, measured using a well counter (Cobra 5003; Packard Instruments), and decay-corrected to the time of injection of the radiotracer. For correction of radiometabolites, nine blood samples were drawn during the PET scan and analyzed for metabolites using radio-high performance liquid chromatography (HPLC). The metabolite-corrected plasma input function, CP, was estimated as the product of the parent compound fitted using a biexponential function and the total plasma concentration.
2.2.2. Data analysis
An overview of the complete analysis workflow can be found in Figure S1 in the supplementary. The metabolite-corrected blood data curves were interpolated to the time points of the PET scan using a cubic spline. All resulting blood curves used for kinetic modeling can be found in the supplementary material.
For bolus-infusion experiments, the plateau for steady-state was used to calculate VT as an average of the ratio between CT and CP from 35 to 80 min after the beginning of tracer administration (Feng et al., 2016). For the bolus injections, the invasive Logan was used to estimate VT using the metabolite-corrected input function with t*= 35 min for all regions and subjects (Magata et al., 2003). Since the non-displaceable distribution volume, VND, is small but not negligible for [11C]flumazenil (Magata et al., 2003), the outcome of these analyses, VT, reflects the sum of specific (i.e., BZR) bound distribution volume, VS, and VND for all regions, including surface- and voxel maps for all scans. All kinetic models applied in this paper were implemented in MATLAB v. 2016b.
2.3. In vivo binding and autoradiography
The conversion from 3D VT images with the unit ml/cm 3 to BZR density (pmol per gram protein) was done by normalizing regional VT with the corresponding regional postmortem human brain [3H]diazepam autoradiography data from Braestrup et al., 1977. This included average estimates and standard deviations across 4 humans (two males, two females, aged 18, 32, 54 and 78y) in 11 brain regions; inferior frontal cortex (860 ± 130 pmol per g tissue, N = 3), inferior central cortex (760 ± 90 pmol per g tissue, N = 4), occipital cortex (840 ± 110 pmol per g tissue, N = 3), temporal cortex (670 ± 150 pmol per g tissue, N = 3), cerebellum (580 ± 40 pmol per g tissue, N = 3), hippocampus (670 ± 120 pmol per g tissue, N = 3), amygdala (510 pmol per g tissue, N = 1), thalamus (330 ± 55 pmol per g tissue, N = 3), caudate (440 ± 90 pmol per g tissue, N = 3), putamen (360 ± 60 pmol per g tissue, N = 3) and pons (160 ± 20 pmol per g tissue, N = 3). As a linear relationship between the regional average VT’s across subjects and the corresponding BZR densities could be established, VND could be estimated from the intercept (Fig. 2 B). To account for noise in the autoradiography estimates (y-axis) due to low sample size and age differences, the standard deviation reported in Braestrup et al., 1977 was used to draw random samples from a normal distribution for each region, specified by the region-specific mean and standard deviation. This procedure was repeated 1000 times, and parameter estimates were fitted for each iteration to obtain an intercept and a slope using a linear model. A subject-specific mean estimate of the slope and intercept was obtained by averaging over the 1000 estimates (Table S2). Finally, average VT estimates were obtained by averaging across subjects, and fitted using a linear model to obtain a groupwise association between autoradiography and VT (Fig. 2 B). This model was used to convert all surface- and voxel maps into densities of BZR availability (Fig. 2 A & Figure S1). Units of pmol/g tissue were converted to pmol/ml using a gray matter density of 1.045 g/mL, as done in Beliveau et al., 2017 (DiResta et al., 1991).
Fig. 2.
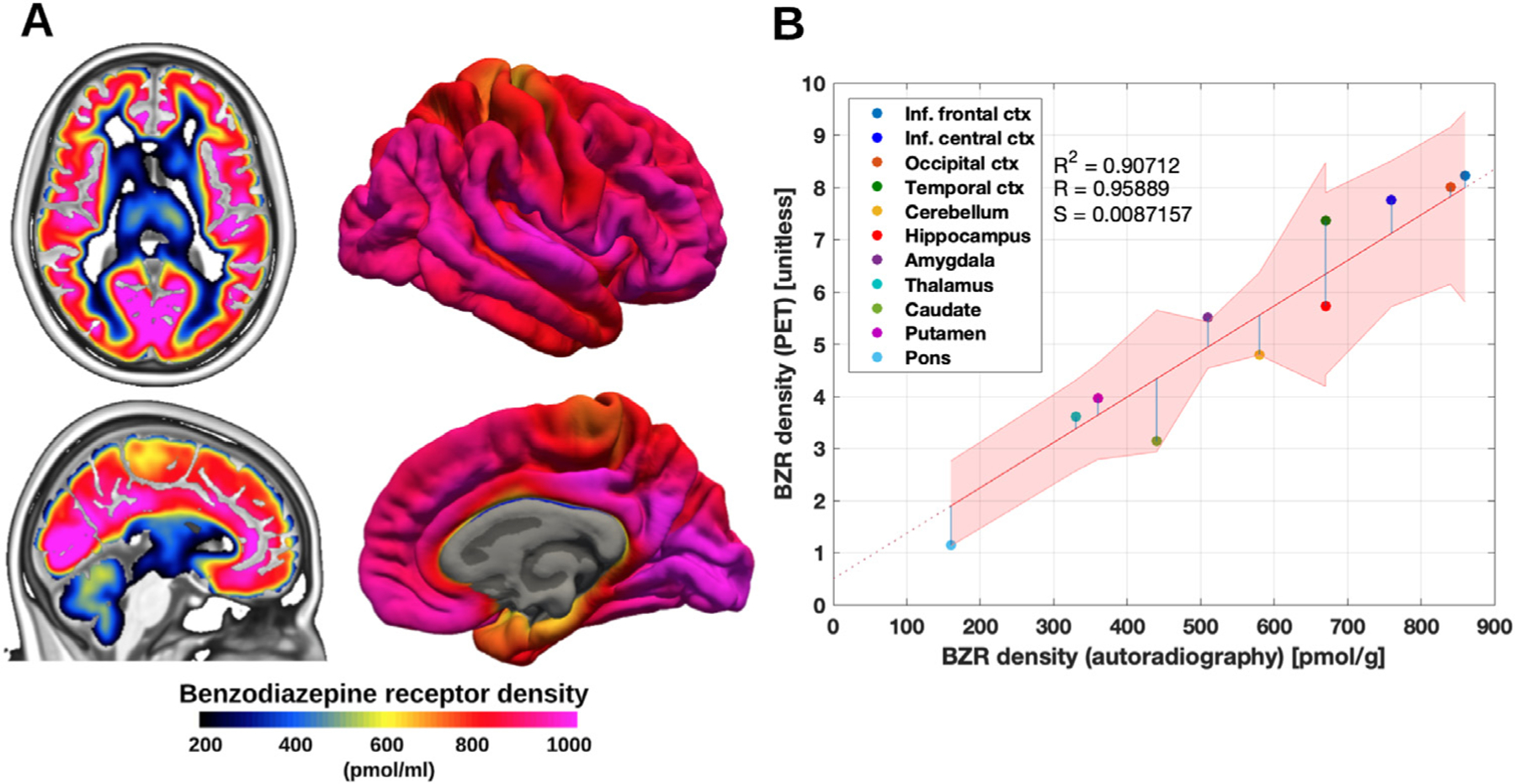
(A) High resolution human brain atlas of GABAA R density (pmol/mL) in MNI152 space (left) and on the surface (right) (B) Average regional benzodiazepine receptor density (from PET) and benzodiazepine receptor density (from mRNA, unit pmol/g). The regression is shown as the black line, and the intercept is the non-specific binding of the PET radioligand. The shaded area is the 95% confidence interval.
2.4. In vivo binding and mRNA expression
Regional VT estimates were compared to mRNA expression values of the 19 subunits of the GABAA R obtained using the Allen Human Brain Atlas (AHBA) (Hawrylycz et al., 2012; French and Paus 2015). The AHBA contains probe information from 6 human postmortem brains (five males, aged 24–57y), where each probe provides mRNA levels (log2 intensity) for all genes and coordinates in MNI152 space. The values from each probe were matched to a brain region defined by the Desikan-Killiany atlas (48 brain regions in total), and averaged within regions to provide a regional estimate of mRNA expression for each of the 19 subunits. Further information can be found in Beliveau et al., 2017.
A linear model between the BZR density and the mRNA expression for each of the 19 subunits was used to examine the correspondence between regions. The intercept (I) was estimated to represent the minimally required expression of mRNA levels for each subunit to translate into BZR protein.
2.5. Statistical analysis
A linear mixed-effects model was used to examine the effects of age, sex, administration form (bolus or bolus-infusion) and multiple scan sessions for a single subject, on the regional estimates of VT. The model included age, sex, and the interaction between age and sex as fixed effects. The administration form (bolus or bolus-infusion) and subjects (i.e. multiple scan sessions for a single subject) were modeled as random effects. The P-values obtained from each regional analysis (N = 11) were adjusted for multiple comparisons using false-discovery rate (FDR = 0.05).
A principal component analysis (PCA) was carried out using the mRNA expression for the 19 subunits and the BZR density for all subcortical and cortical regions (N = 48). Prior to running the PCA, the data was standardized (mean subtracted and normalized by the standard deviation). All the data is available in the supplementary material.
3. Results
The association between BZR availability measured with PET and the postmortem human brain autoradiography data revealed a strong correlation (Pearson’s R = 0.96), across all individuals (Fig. 2 B). Using a linear mixed-effects model, we found no age or gender related effects on regional estimates of VT (Table 1), nor an effect of tracer administration or multiple scan sessions for a single subject (P > 0.05, corrected). All estimates of VT across subjects and scans, including estimates for within- and between-subject variability, for all brain regions are available in Table S2.
Table 1.
Linear mixed-effects model for VT ~ age + gender + age * gender + (1|administration) + (1|scan_number). The P-values were obtained using ANOVA with Kenward-Roger’s method for estimating the degrees of freedom. All P-values are displayed without correction for multiple comparisons using False-Discovery Rate (FDR). With correction, no P-values survive the 0.05 threshold, with the smallest P-values = 0.33 (range: 0.33–0.94).
| Variable/Region | Fron | Cent | Occ | Temp | Cereb | Hippo | Amyg | Thal | Cau | Put | Pons |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 0.88 | 0.66 | 0.93 | 0.94 | 0.82 | 0.61 | 0.55 | 0.55 | 0.84 | 0.84 | 0.49 |
| Gender | 0.06 | 0.09 | 0.04 | 0.03 | 0.06 | 0.12 | 0.11 | 0.24 | 0.45 | 0.27 | 0.62 |
| Age × Gender | 0.08 | 0.11 | 0.07 | 0.05 | 0.12 | 0.16 | 0.14 | 0.31 | 0.51 | 0.3 | 0.54 |
The established association between the average VT estimates across subjects from PET and autoradiography (Fig. 2 B) was used to construct the high-resolution atlas of the spatial distribution of BZR density (Fig. 2 A). The BZR densities were high in cortical areas (600–1000 pmol/mL), medium in subcortical regions (300–600 pmol/mL), and low in the brainstem (< 200 pmol/mL).
The association between BZR densities and mRNA expression showed a medium to high correlation for each of the subunits included in the most common pentameric GABAA R, i.e., α1β2γ2 (48% mRNA expression in the brain Sequeira et al., 2019, Table 2) with Pearson’s R > 0.62 (range: 0.62–0.88, P < 0.0001) for all 3 subunits (Fig. 3 A–C). The association between BZR and mRNA was established for 48 brain regions (available in the supplementary file “Bmax_vs_mRNA.xlsx ”).
Table 2.
Association between mRNA subunit expression (log2 units) and benzodiazepine receptor density (pmol/ml) for 14 of the available 19 subunits of the GABAA receptor, explaining 98.4% of the total variance across brain regions. The associations for the individual subunits are summarized by the Pearson correlation coefficient (R), the uncorrected P-value (P), the intercept (I), and the variance explained (V). The variance explained was obtained from Sequiera et al. 2019.
| α1 | α2 | α3 | α4 | α5 | α6 | β1 | β2 | β3 | γ1 | γ2 | γ3 | θ | δ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | 0.67 | 0.28 | 0.39 | 0.33 | 0.15 | 0.36 | 0.03 | 0.88 | 0.26 | −0.37 | 0.62 | 0.92 | −0.33 | 0.71 |
| P | <0.01 | 0.05 | < 0.01 | 0.02 | 0.31 | 0.01 | 0.84 | <0.01 | 0.07 | 0.01 | <0.01 | <0.01 | 0.02 | < 0.01 |
| I | 5.8 | 7.2 | 5.1 | 4.9 | 6.9 | 1.9 | 8.5 | 6.5 | 7.4 | 8.2 | 9.1 | 1.5 | 3.2 | 4.5 |
| V | 16 | 11 | 2 | 2 | 5 | 3 | 5 | 12 | 7 | 7 | 20 | 0.2 | 0.2 | 8 |
Fig. 3.
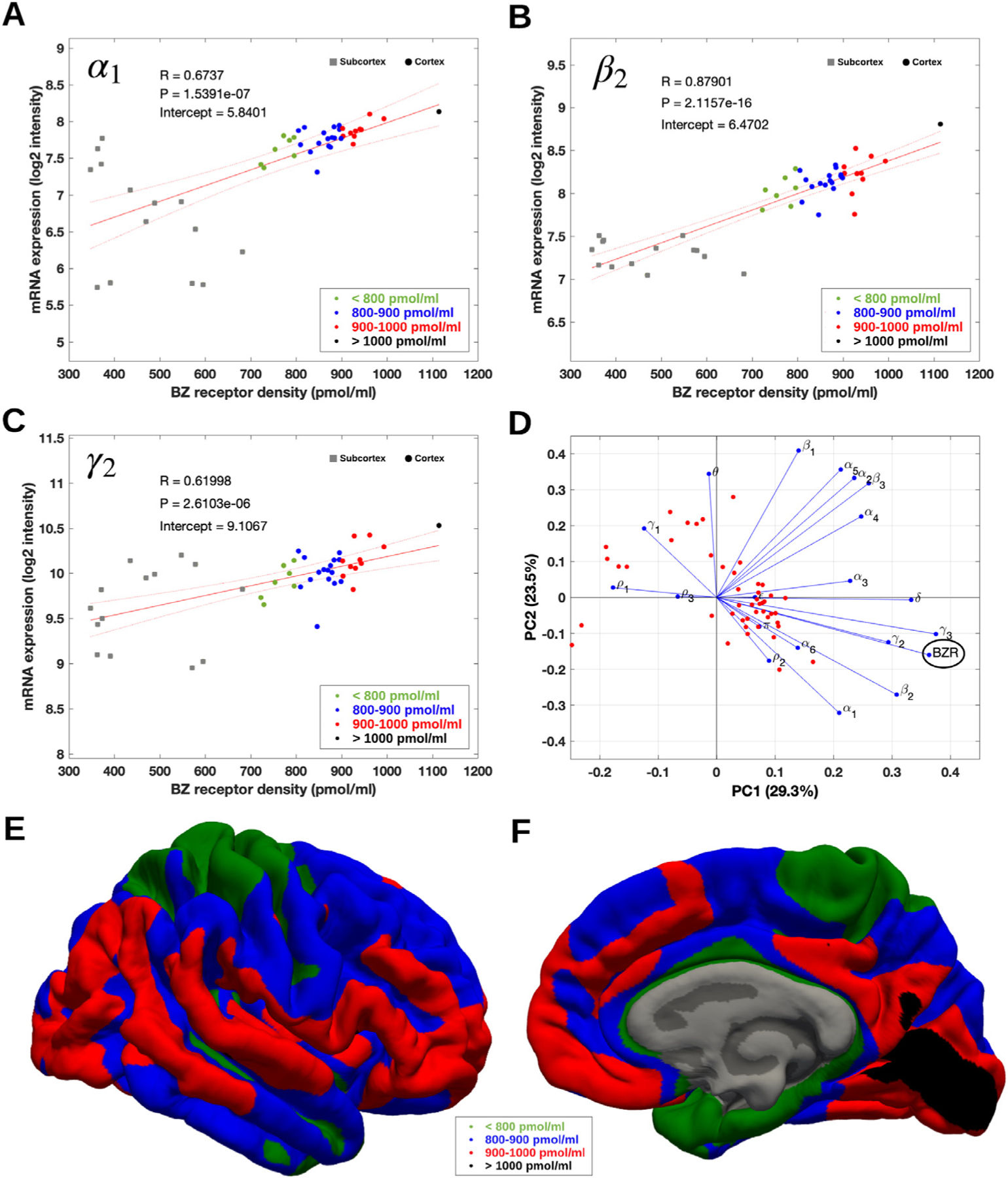
(A–C) Association between mRNA expression (log2 intensity) and BZR density across 48 brain regions for the subunits of the GABAA R most commonly represented in the BZR, α1 in (A), β2 in (B), and γ2 in (C). The points are regional estimates for subcortex (squares) and cortex (round dots), and are color coded according to the density, green (< 800 pmol/mL), blue (800–900 pmol/mL), red (900–1.000 pmol/mL), and black (> 1.000 pmol/mL). (D) Biplot of the (scaled) first two principal components (% variance explained) of a PCA of the 19 subunits and BZR. (E and F) The spatial distribution of BZR density according to the specified color coding shown on the lateral and medial surface of the brain.
A principal component analysis (PCA) of the mRNA and BZR density showed that 6 components explained >90% of the total variance, with the first and second components explaining 29.3% and 23.5%, respectively (Fig. 3 D). The biplot in Fig. 3 D shows the contribution of each subunit to the BZR density, indicating a high correlation between the BZR density and the subunits α1, β2, γ2, γ3, whereas the subunits γ1, ρ1 and ρ3 were negatively correlated with the BZR density.
Table 2 shows the linear association (Pearson’s R) between mRNA expression of 14 of the 19 available subunits of GABAA R and BZR density, ranging between −0.37 (γ1) and 0.92 (γ3). Six of the subunits (α1, α3, β2, γ2, γ3 and δ) showed a significant correlation (corrected for multiple comparisons using Bonferroni, P < 0.01). The intercept (I) for each of the subunits is the minimal mRNA expression (log2 intensity unit) required for translation into BZR protein and ranged from 1.5 (γ3) to 9.1 (γ2). Only the six subunits with a significant association (P < 0.01) between mRNA expression and BZR density (α1, α3, β2, γ2, γ3 and δ) were considered to have valid intercepts due to the strong correlation.
The variance explained (V) for each of the subunits was obtained from Sequiera et al. 2019 and shows the proportional contribution (%) from each subunit on the total mRNA expression in the brain across all subunits. Most of the variance was explained by the subunits α1, α2, β2, γ2, and δ, contributing with 67% of the variance (Table 2).
4. Discussion
Here, we present the first quantitative high-resolution in vivo human brain atlas of BZR protein density, freely available at https://nru.dk/BZR-atlas. All the data and supplementary information is also publicly available.
4.1. Association between BZR availability and autoradiography
First, we validated the association between the in vivo BZR availability from PET and protein density from autoradiography. This validation showed a very strong relationship between the BZR availability from PET and the autoradiography data from Braestrup et al., 1977, supporting the use of [11C]flumazenil-PET as a reliable putative marker for BZR availability in the living human brain.
Autoradiography is the gold standard for estimating the density of specific receptors in the brain (Braestrup et al., 1977). Braestrup et al., 1977 used the agonist tracer [3H]diazepam to estimate the density of available BZRs, but despite that [3H]diazepam (Videæk et al., 1993) and the antagonist tracer [11C]flumazenil used in PET (Whitwam et al. 1995; Lassen et al., 1995) have different pharmacological properties, there is no evidence for differences in their ability to bind to other receptors, i.e. they both display low non-specific binding. However, while the non-specific component is often neglected in [11C]flumazenil-PET studies, we observed a minor non-specific component across all subjects and scans that will potentially bias the subject-specific estimate of BZR availability if not taken into account. [11C]flumazenil-PET studies commonly use reference tissue modeling and the pons as reference region for estimation of BZR availability (Klumpers et al., 2012). For all subjects and scans in this study, the VT in pons was higher than the VND, suggesting that this region has specific BZR binding. Therefore, reference tissue modeling of the PET data using pons as a reference region is not entirely valid and may be associated with an underestimation of BZR availability (Klumpers et al., 2012; Price et al., 1991).
4.2. Association between BZR density and mRNA expression
The second validation showed a significant association across brain regions between the expression of mRNA levels for the subunits α1, α3, β2, γ2, γ3 and δ, and the BZR protein density. Together, these six subunits explain 67% of the total mRNA expression across the brain. The results confirm previous evidence on the existence of a BZR binding site located between the α1 and γ2 subunits in the commonly expressed pentameric GABAA R subtype α1β2γ2 (Zhu et al., 2018). We provide novel evidence of the minimally required expression of mRNA for the translation of BZR proteins for the α1 (= 5.8 log2 intensity), β2 (= 6.5 log2 intensity) and γ2 (= 9.1 log2 intensity) subunits. Previous work by Mulligan et al., 2012 also supports similar magnitudes of differences in mRNA expression between subunits. Future studies focusing on evaluating a transcription threshold, or whether altered transcription initiation or a dysregulation of the mRNA transcription leads to altered distribution of BZRs are highly encouraged. This is important since it may have consequences in neuropsychiatric disorders, with the inability to produce BZR proteins due to altered expression of mRNA (Mulligan et al., 2012). In general, the association between mRNA subunit expression and BZR density was particularly high throughout the neocortex (Fig. 3), whereas the associations varied more in the subcortical regions (BZR density < 700 pmol/ml). The brain region with the highest BZR density was the occipital cortex, which has been documented to be particularly rich in both α1 and β2 subunits (Sequiera et al., 2019). The remaining subunits showed a more variable association between transcription and translation, which might be caused by regional variations, due to either 1) transport of the subunit protein away from the site of production(e.g. long-range GABAergic cells, Caputi et al., 2013), 2) effects such as post-translational protein degradation and mRNA decay, meaning that a certain amount of mRNA is not translated into (functional) proteins, or 3) due to non-linear relationships between expression and translation (Mulligan et al., 2012). A non-linear relationship between transcription and translation is not uncommon and has also been observed for other proteins, e.g., the serotonin transporter and the 5-HT2A receptor (Beliveau et al., 2017). Subcortical regions such as the striatum have a high expression of α4 subunits, and the amygdala has a high expression of the α2, β1 and γ1 subunits (Sequiera et al., 2019). In our work, the association between mRNA expression of the γ1 subunit and BZR density was found to be negative, consistent with existing evidence that the expression of β1 is negatively correlated with the expression of β2 (Sequiera et al., 2019). Together, this evidence points toward a reduced BZR density in these regions, likely caused by the stoichiometry of the subunits to form the GABAA R. For an in-depth analysis on the subunits only we refer the reader to Sequiera et al., 2019.
4.3. Representativeness of the atlas
GABAA R subunit mRNA expression varies considerably, both across the human brain and between individuals (Sequiera et al., 2019), with the α1 subunit mRNA showing variations up to a 100-fold range across neocortex, and for subunits γ2 and β1 a 10–20 fold range (Mulligan et al., 2012). The high variability of subunit mRNA does not, however, reflect phenotypic variation (Mulligan et al., 2012) and the variability in BZR protein levels across brain regions and across individuals is much less, approximately 15–20% (Braestrup et al., 1977). Consistent with this observation, we find a similar variability between individuals using high-resolution PET (Table S2). We included all [11C]flumazenil PET scanned individuals from our database (Knudsen et al., 2016), consisting of mainly younger individuals and both men and women. Since we did not find any significant sex- or age-differences in the cerebral GABAA R binding (Kim et al., 2011), we consider our atlas to be representative for both men and women, at least in the age range from 19–49 years.
4.4. Methodological considerations and their impact on the atlas
Several methodological choices may have had an impact on our ability to correctly measure BZR availability with PET and create a valuable atlas, such as motion artifacts, resolution of the scanner and the delineation of brain regions (Nørgaard et al., 2019). For all individuals, within-scan motion was low (< 3 mm) and is therefore not expected to have a major impact on the resolution of the PET images, even after motion correction. The alignment between PET and MRI, and the parcellation from FreeSurfer to define the brain regions, was also precise due to the high resolution of both PET and MRI (Greve et al., 2014), adequately capturing the cortical boundaries of gray matter (Fig. 4).
Fig. 4.
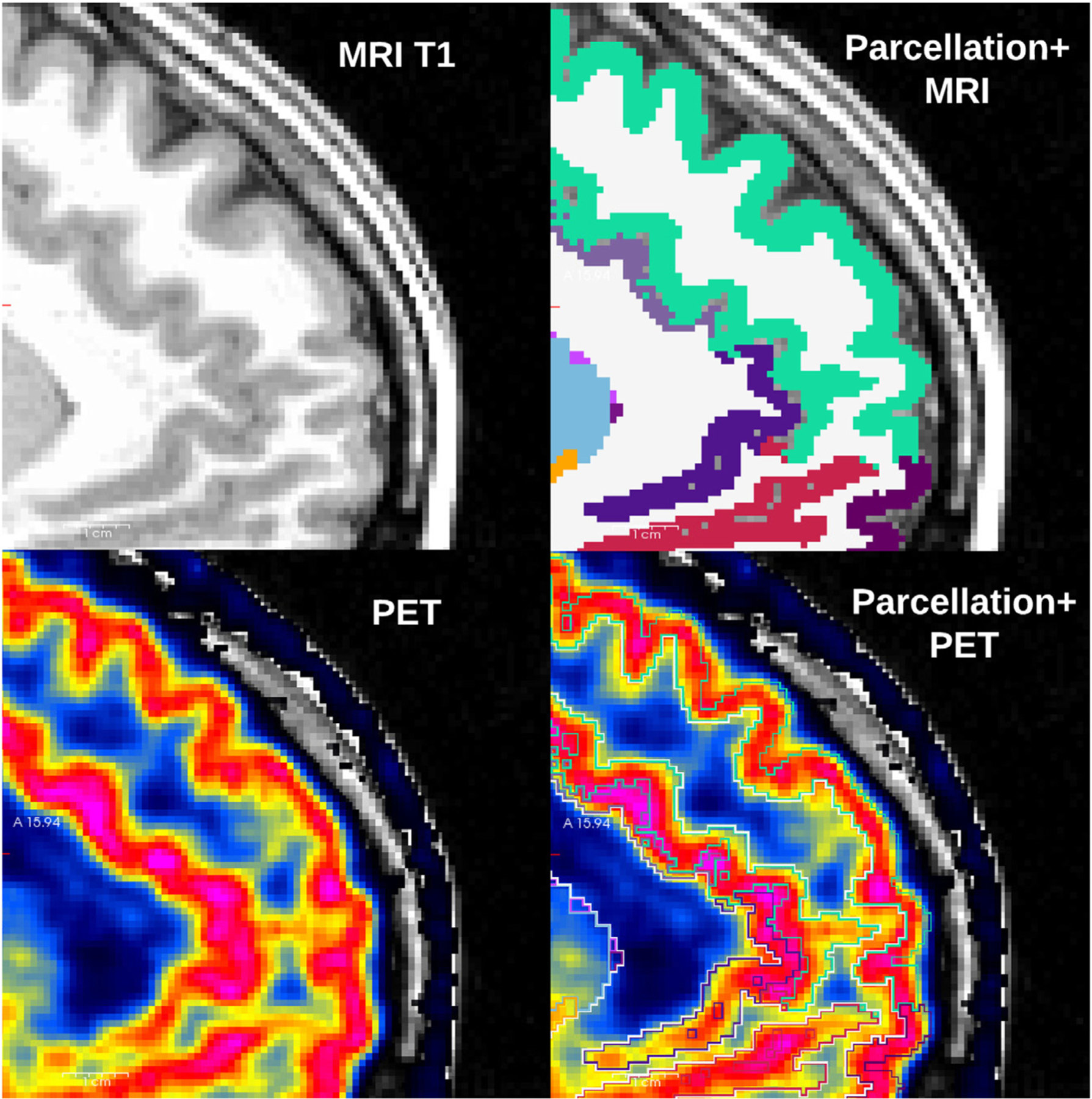
Unique T1-weighted image (upper left), and the corresponding parcellation obtained from Freesurfer (upper right). The MRI and PET were spatially aligned (lower left), and the parcellation was overlayed (lower right) and validated to correctly capture the uptake of [11C]flumazenil in cortical gray matter.
The resolution of the scanner and the PET data reconstruction will also have an impact on the ability to correctly estimate the BZR density. For example, the PSF modeling step may create ringing artefacts at the GM/WM boundary, potentially causing a misestimation of the true underlying BZR density. However, the PSF modeling in the PET data reconstruction on the HRRT scanner was implemented conservatively (Comtat et al., 2008; Sureau et al., 2008) without any variation across the FOV in part to avoid ringing artifacts. Previous work by Angelis et al., 2015 investigated the effect of applying a spatially variant PSF, showing some ringing using a sphere phantom, and minimal ringing for the conservative standard PSF, but at the level of or just above the general image noise. For studies using a PET scanner with lower resolution, partial volume effects may be a problem particularly for small brain regions, and partial volume correction may be necessary to correctly estimate the BZR density (Greve et al., 2014).
Overall, since all these methodological steps ranging from reconstruction to preprocessing were carefully analyzed and validated throughout the creation of the atlas, we believe that most of the remaining variability originates from true underlying biological variation as supported by the literature (Braestrup et al., 1977). However, because of the noticeable variability between individuals, the atlas must necessarily represent an average of BZR density in the healthy brain but it also provides measures of the interindividual variability.
4.5. Limitations
Our study is not without limitations. The association between mRNA and BZR density only allows for a direct comparison between the individual subunit and BZR density, and not their interaction with other subunits. In addition, it cannot be excluded that different BZR binding pentamers have different affinity for [11C]flumazenil. For example, it has been shown that flumazenil has a 200-fold higher affinity to receptors with the configuration α1, β2 and γ2, than the affinity to receptors containing the α6 subunit [19]. However, as PET acquisitions are carried out using tracer doses this should not impact the estimation of the specific binding of BZRs. In the future, the development of GABAA R subunit-specific radioligands would be helpful to examine the GABAergic system in more detail. With the addition of such tools, one could gain even more insight into if pentamer subunit composition affects the affinity to pharmacological targets and if certain brain disorders are associated with abnormalities in subunit composition of the GABAA R.
5. Conclusions
This high-resolution in vivo atlas of the spatial distribution of BZR densities in the healthy human brain provides a highly valuable tool for investigation of the GABA system, e.g., as a reference for patient groups with alterations in the cerebral GABA system. The findings provide additional insights into the association between mRNA expression for individual subunits in the GABAA R and the BZR density at each location in the brain and may be used to evaluate the efficacy of pharmacological targets acting on the BZR.
Supplementary Material
Acknowledgments
We wish to thank all the participants for kindly joining the research project. We thank the John and Birthe Meyer Foundation for the donation of the cyclotron and HRRT scanner used in this study. Nic Gillings, Bente Dall, and Ling Feng are also gratefully acknowledged for their expert assistance. We also thank the VU University Medical centre, Am-sterdam, Netherlands, for sharing their knowledge and expertise on [11C]flumazenil.
MN was supported by the Lundbeck Foundation (Grant R90-A7722), and the Independent Research Fund Denmark (DFF-133100109 & DFF-418300627). DNG was supported by NIH grant R01EB023281 and R01NS105820. MGB was funded by Elsass Fonden (18-3-0147).
Footnotes
Declaration of Competing Interest
GMK has received honoria as an expert advisor for Sage Therapeutics and as a speaker for Jannsen.
Data and Code availability statement
Researchers who wish to get access to the data used in the manuscript (stored in the Cimbi database, (Knudsen et al., 2016)) can apply through a standardized database application form (downloadable at http://www.cimbi.dk/db).
The code used to preprocess and create the results used in this manuscript mostly consists of open-source software (FreeSurfer and AIR). Code for running the pipeline and obtain the atlas can be found at www.nru.dk/BZR-atlas.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2021.117878.
References
- Angelis GI, Kotasidis FA, Matthews JC, Markiewicz PJ, Lionheart WR, Reader AJ, 2015. Full field spatially-variant image-based resolution modelling reconstruction for the HRRT. Phys. Med 31 (2), 137–145 March. [DOI] [PubMed] [Google Scholar]
- Amunts K, Zilles K, 2015. Architectonic mapping of the human brain beyond brodmann. Neuron 88 (6), 1086–1107. [DOI] [PubMed] [Google Scholar]
- Beliveau V, Ganz M, Feng L, Svarer C, Knudsen GM, Fisher PM, Ozenne B, Højgaard L, … Knudsen GM, 2017. A high-resolution in vivo atlas of the human brain’s serotonin system. J. Neurosci 37 (1), 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau V, Ozenne B, Strother S, Greve DN, Svarer C, Knudsen GM, Ganz M, 2020. The structure of the serotonin system: a PET imaging study. Neuroimage 205 (April 2019), 116240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braestrup C, Albrechtsen R, Squires R, 1977. High densities of benzodiazepine receptors in human cortical areas. Nature 269 (October), 702–704. [DOI] [PubMed] [Google Scholar]
- Caputi A, Melzer S, Michael M, Monyer H, 2013. The long and short of GABAergic neurons. Curr. Opin. Neurobiol 23 (2), 179–186 April. [DOI] [PubMed] [Google Scholar]
- Comtat C, Sureau FC, Sibomana M, et al. , 2008. Image based resolution modeling for the HRRT OSEM reconstructions software. In: 2008 IEEE Nuclear Science Symposium Conference Record, Dresden, Germany, pp. 4120–4123. [Google Scholar]
- DiResta GR, Lee J, Arbit E, 1991. Measurement of brain tissue specific gravity using pycnometry. J. Neurosci. Methods 39, 245–251. [DOI] [PubMed] [Google Scholar]
- Feng L, Svarer C, Madsen K, Ziebell M, Dyssegaard A, Ettrup A, … Pinborg LH, 2016. Design of Infusion Schemes for Neuroreceptor Imaging: application to [11C]Flumazenil-PET Steady-State Study. Biomed. Res. Int 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, 2012. FreeSurfer. Neuroimage 62 (2), 774–781 August 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French L, Paus T, 2015. A free surfer view of the cortical transcriptome generated from the Allen Human Brain Atlas. Front. Neurosci 9, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greve DN, Svarer C, Fisher PM, Feng L, Hansen AE, Baare W, … Knudsen GM, 2014. Cortical surface-based analysis reduces bias and variance in kinetic modeling of brain PET data. Neuroimage 92, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, … Jones AR, 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489 (7416), 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong IK, Chung ST, Kim HK, et al. , 2007. Ultra fast symmetry and SIMD-based projection-backprojection (SSP) algorithm for 3-D PET image reconstruction. IEEE Trans. Med. Imaging 26, 789–803. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve DN, Haley E, van der Kouwe A, Gollub R, Kennedy D, et al. , 2006. Reliability in multi–site structural MRI studies: effects of gradient non–linearity correction on phantom and human data. Neuroimage 30 (2), 436–443. [DOI] [PubMed] [Google Scholar]
- Keller SH, Svarer C, Sibomana M, 2013. Attenuation correction for the HRRT PET-s-canner using transmission scatter correction and total variation regularization. IEEE Trans. Med. Imaging 32, 1611–1621. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim YK, Yoon EJ, et al. , May 2011. Age-related changes in the availability of GABAa receptors in healthy subjects: imaging study with F-18 flumazenil PET. J. Nucl. Med 52 (supplement 1) 2009. [Google Scholar]
- Klumpers UMH, Boellard R, Veltman DJ, Kloet RW, Hoogendijk JG, Lammertsma AA, 2012. Parametric [11C]flumazenil images. Nucl. Med. Commun 33, 422–430. [DOI] [PubMed] [Google Scholar]
- Knudsen GM, Jensen PS, Erritzoe D, Baaré WFC, Ettrup A, Fisher PM, Frokjaer VG, 2016. The Center for Integrated Molecular Brain Imaging (Cimbi) database. Neuroimage 124, 1213–1219. [DOI] [PubMed] [Google Scholar]
- Lassen NA, Bartenstein PA, Lammertsma AA, Prevett MC, Turton DR, Luthra SK, … Vanggaard Andersen J, 1995. Benzodiazepine receptor quantification in vivo in humans using [11C]flumazenil and PET: application of the steady-state principle. J. Cerebr. Blood Flow Metab 15 (1), 152–165. [DOI] [PubMed] [Google Scholar]
- Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, 1996. Alexoff DL: distribution volume ratios without blood sampling from graphical analysis of PET data. J. Cereb. Blood Flow Metab 16 (5), 834–840. [DOI] [PubMed] [Google Scholar]
- Magata Y, Mukai T, Ihara M, Nishizawa S, Kitano H, Ishizu K, Saji H, Konishi J, 2003. Simple analytic method of 11C-flumazenil metabolite in blood. J. Nucl. Med 44, 417–421. [PubMed] [Google Scholar]
- Mazoyer B, Trebossen R, Deutch R, Casey M, Blohm K, 1991. Physical characteristics of the ECAT 953B/31: a new high resolution brain positron tomograph. IEEE Trans. Med. Imaging 10 (4), 499–504. [DOI] [PubMed] [Google Scholar]
- Mulligan MK, Wang X, Adler AL, Mozhui K, Lu L, Williams RW, 2012. Complex control of GABA(A) receptor subunit mRNA expression: variation, covariation, and genetic regulation. PLoS ONE 7 (4), e34586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørgaard M, Ganz M, Svarer C, Frokjaer VG, Greve DN, Strother SC, Knudsen GM, 2019. Optimization of preprocessing strategies in Positron Emission Tomography (PET) neuroimaging: a [11C]DASB PET study. Neuroimage 199, 466–479 October 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Mayberg HS, Dannals RF, Wilson AA, Ravert HT, Frost JJ, 1991. Estimation of benzodiazepine receptor bind ing parameters using llC-flumazenii and PET: equilibrium and kinetic methods. J. Cereb. Blood Flow Metab Ll (Suppl 2), S612. [Google Scholar]
- Puts NA, Edden RA, 2012. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog. Nucl. Magn. Reson. Spectrosc 60, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Shen K, Gottlieb A, Limon A, 2019. Human brain transcriptome analysis finds region- and subject-specific expression signatures of GABAAR subunits. Commun. Biol 2 (1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau FC, Reader AJ, Comtat C, et al. , 2008. Impact of image-space resolution modeling for studies with the high-resolution research tomograph. J. Nucl. Med 49, 1000–1008. [DOI] [PubMed] [Google Scholar]
- Videæk C, Friberg L, Holm S, Wammen S, Foged C, Andersen JV, … Lassen NA, 1993. Benzodiazepine receptor equilibrium constants for flumazenil and midazolam determined in humans with the single photon emission computer tomography tracer [123I]iomazenil. Eur. J. Pharmacol 249 (1–2), 43–51. [DOI] [PubMed] [Google Scholar]
- Yamauchi Hiroshi, et al. , 2005. Selective neuronal damage and borderzone infarction in carotid artery occlusive disease: a 11C-flumazenil PET study. Journal of Nuclear Medicine 46 (12), 1973–1979. [PubMed] [Google Scholar]
- Zhu S, Noviello CM, Teng J, Walsh RM, Kim JJ, Hibbs RE, 2018. Structure of a human synaptic GABAA receptor. Nature 559 (7712), 67–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Grefkes C, et al. , 2002. Architectonics of the human cerebral cortex and transmitter receptor fingerprints: reconciling functional neuroanatomy and neurochemistry. Eur. Neuropsychopharmacol 12 (6), 587–599. [DOI] [PubMed] [Google Scholar]
- Whitwam JG, Amrein R, 1995. Pharmacology of flumazenil. Acta Anaesthesiol. Scand 39 (10), 3–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


