Abstract
Cas12i is a recently identified type V CRISPR-Cas endonuclease that predominantly cleaves the non-target strand of a double stranded DNA substrate. The nicking activity of Cas12i can be potentially used for genome editing with high specificity. To elucidate its mechanisms for target recognition and cleavage, we determined the structures of Cas12i in multiple functional states by cryo-EM. Cas12i pre-orders a 7-nucleotide seed sequence of the crRNA for target recognition and undergoes a two-step activation by crRNA-DNA hybridization. Formation of 14 base pairs activates the nickase activity, and 28-bp hybridization promotes cleavage of the target strand. The atomic structures and mechanistic insights would facilitate the manipulation of Cas12i for genome editing applications.
CRISPR-Cas systems provide adaptive immunity in bacteria and archaea to resist invasion of mobile genetic elements (MGEs) via RNA-guided cleavage of target nucleic acids 1-6. The CRISPR-Cas acquired immunity occurs through three major steps: short fragments of the invasive nucleic acids are incorporated into CRISPR array as new spacers by Cas1-Cas2 integrase complex (1, acquisition or adaptation); transcription and maturation of crRNAs that assemble with Cas proteins into ribonucleoprotein (RNP) complex (2, expression step); the invasive nucleic acids are recognized and cleaved by the RNP complex (3, interference step) 7,8. Dynamics between the host and MGEs have resulted in diverse CRISPR-Cas systems, which are grouped into class 1 systems (types I, III and IV) and class 2 systems (types II, V and VI) 9-12. Class 2 systems utilize a single effector endonuclease and have been harnessed for genome editing and targeting tools 13-16, with Cas9 from the type II systems being the most widely used 13,14. In addition to Cas9, three type V CRISPR-Cas systems were successfully harnessed for genome editing in mammalian cells, including Cas12a (Cpf1) 17, Cas12b (C2c1) 18,19 and Cas12e (CasX) 20. Both type II and type V effectors adopt a similar bilobed architecture comprising of a recognition lobe (REC) and a nuclease lobe (NUC). Cas9 endonucleases employ both the RuvC and HNH domains for DNA cleavage; however, Cas12 endonucleases use a single RuvC domain for DNA cleavage.
Recently, a number of new type V systems were identified, revealing functional diversity in the type V systems 21 and providing new opportunities to improve and expand CRISPR-based applications. Among the newly identified type V endonucleases, Cas12i is a single RNA-guided nickase that predominantly cleaves the non-target strand of target DNA with a 5’-TTN-3’ protospacer adjacent motif (PAM). The nicking activity of Cas12i can be potentially used for genome editing with high specificity 21. Like Cas12a, Cas12i can process its own pre-crRNA into mature crRNA, providing a promising alternative platform for multiplexed genome editing 21. Intriguingly, the protein sequences of Cas12i effectors are evolutionarily diverged from other sub-types in the type V systems 21. The absence of an acquisition module further suggests the specialized features of the CRISPR-Cas12i system 21. Together, sequence and functional differences between Cas12i and other type V endonucleases indicate that Cas12i may have different molecular mechanisms in target recognition and cleavage.
Structure of Cas12i-crRNA-dsDNA
To reveal the mechanism of target recognition by Cas12i, we assembled a Cas12i-crRNA-DNA ternary complex by incubating a catalytically inactive Cas12i (E894A) with a 60-nucleotide (nt) precursor crRNA (pre-crRNA), a 36-nt target DNA strand, and a 10-nt non-target DNA strand containing a 5’-TTN-3’ PAM sequence, and then determined the structure of the ternary complex to an average resolution of 2.9 Å by cryo-EM (Extended Data Figs. 1 and 2, Table 1). The pre-crRNA was processed into a mature crRNA by cleavage of a segment at the 5’ end by the RNase activity of Cas12i (Extended Data Fig. 1b), consistent with a previous study 21.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| Cas12iE894A-crRNA- dsDNA (EMD-21541, PDB 6W5C) |
Cas12i-crRNA (EMD-21551, PDB 6W62) |
I1 (EMD-21552, PDB 6W64) |
|
|---|---|---|---|
| Data collection and processing | |||
| Magnification | 81,000 | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 54 | 36 | 36 |
| Defocus range (μm) | 1.5 – 2.5 | 1.5 – 2.5 | 1.5 – 2.5 |
| Pixel size (Å) | 1.05 | 1.05 | 1.05 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (no.) | 4,944,615 | 1,507,592 | 1,507,592 |
| Final particle images (no.) | 731,231 | 122,435 | 75,406 |
| Map resolution (Å) | 2.9 | 3.9 | 3.9 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 2.7 – ~20 | 3.6 – ~20 | 3.6 – ~20 |
| Refinement | |||
| Initial model used | PDB 5U30 | PDB 6W5C | PDB 6W5C |
| Model resolution (Å) | 3.1 | 4.2 | 4.1 |
| FSC threshold | 0.5 | 0.5 | 0.5 |
| Model resolution range (Å) | 2.7 – 50 | 3.8 – 50 | 3.8 – 50 |
| Map sharpening B factor (Å2) | −91 | −161 | −136 |
| Model composition | |||
| Non-hydrogen atoms | 10186 | 7390 | 8506 |
| Protein residues | 1023 | 910 | 928 |
| Nucleotides | 105 | 24 | 56 |
| B factors (Å2) | |||
| Protein | 37.44 | 30.00 | 25.49 |
| Nucleic acids | 47.28 | 39.80 | 28.77 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.004 | 0.003 | 0.003 |
| Bond angles (°) | 0.625 | 0.617 | 0.568 |
| Validation | |||
| MolProbity score | 1.90 | 2.26 | 2.03 |
| Clashscore | 8.04 | 16.14 | 9.32 |
| Poor rotamers (%) | 0.85 | 0.00 | 1.29 |
| Ramachandran plot | |||
| Favored (%) | 92.57% | 90.00% | 92.90% |
| Allowed (%) | 7.43% | 9.89% | 7.10% |
| Disallowed (%) | 0.00% | 0.11% | 0.00% |
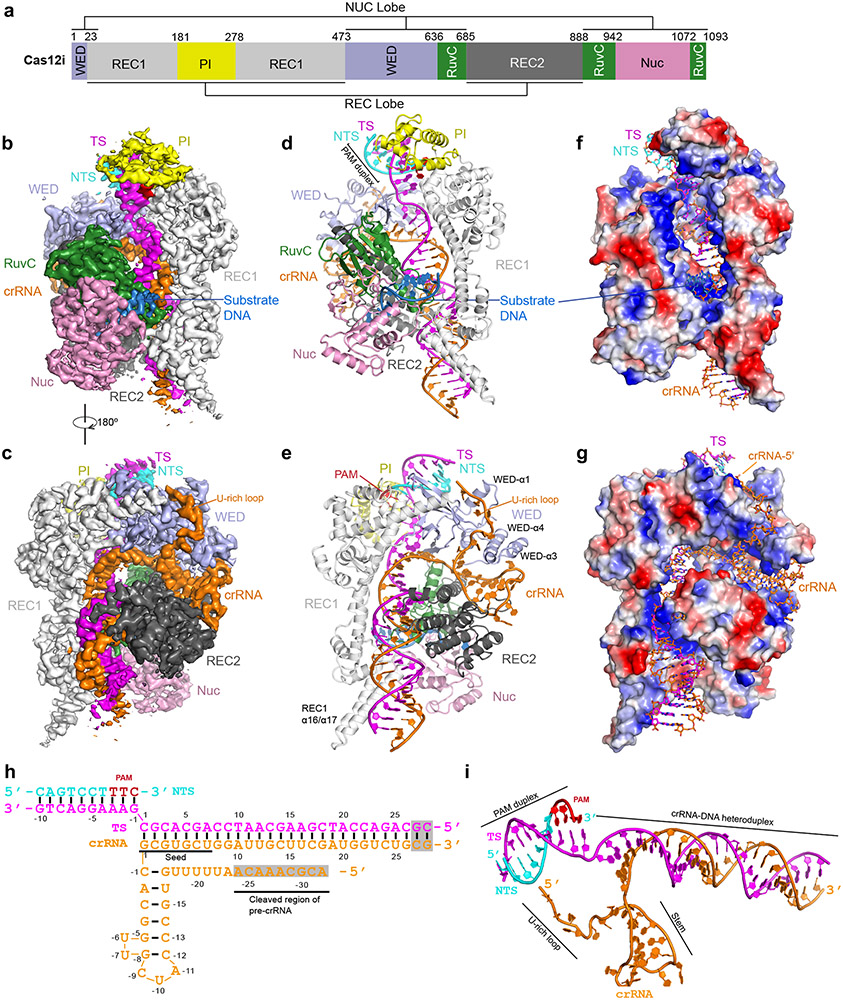
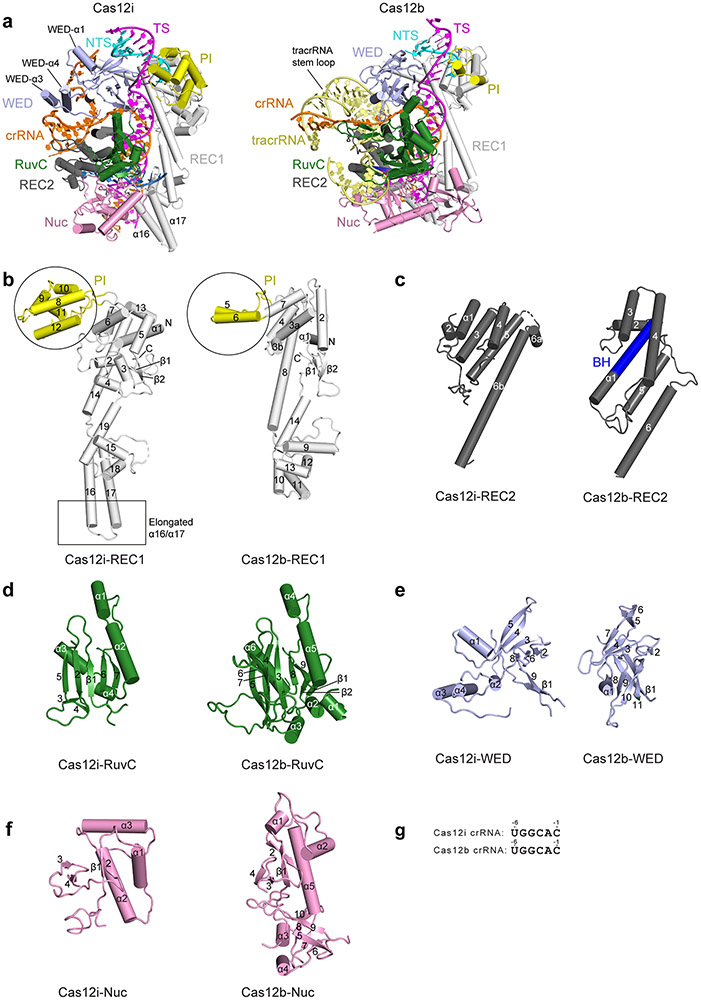
The overall structure of Cas12i displays a bilobed architecture encompassing a recognition lobe (REC) composed of the REC1, REC2, and PAM-interacting (PI) domains, and a nuclease lobe (NUC) composed of the RuvC, Wedge (WED), and Nuc domains (Fig. 1a-g). The crRNA contains a 28-nt spacer-derived guide segment (positions 1–28) at the 3’ end and a 23-nt repeat-derived sequence (positions −1 to −23) at the 5’ end, with the repeat-derived sequence forming a stem-loop structure (Fig. 1h,i). The stem-loop is sandwiched by the REC2 and WED domains, with an extended U-rich sequence preceding the stem-loop held by the WED domain (Fig. 1c,e). The spacer-derived guide segment of the crRNA and the protospacer-derived sequence of the target DNA strand form a 28-bp heteroduplex, which runs through a central channel surrounded by the WED, REC1, REC2, and RuvC domains (Fig. 1d-g). The 3’ end of target strand base pairs with the non-target strand, forming the PAM duplex inserted into a groove created by the PI, WED, and REC1 domains (Fig. 1d).
Figure 1. Structure of the Cas12i-crRNA-dsDNA ternary complex.
(a) Domain organization of Cas12i. (b, c) Cryo-EM map of the Cas12i-crRNA-dsDNA ternary complex in two views with each domain of Cas12i color-coded as in a. The crRNA, TS, NTS and substrate DNA are colored in orange, magenta, cyan, and sky blue, respectively. The PAM sequence is colored in red. (d,e) Atomic model of the Cas12i-crRNA-dsDNA ternary complex in two views shown in cartoon. (f,g) Surface potential of Cas12i with crRNA and DNA in stick presentation. (h) Schematic of the crRNA and target DNA. (i) Structure of the crRNA and target DNA in the Cas12i-crRNA-dsDNA ternary complex.
Despite low sequence identity (5.5%) and similarity (14.2%) (Supplementary Note), a DALI search 22 revealed Cas12b to be the closest structural homolog to Cas12i. Substantial differences in their overall architectures are observed (Extended Data Fig. 3a-f). First, an additional helix (α1) is present in the WED domain of Cas12i to accommodate the extended U-rich motif of crRNA (Fig. 1e, Extended Data Fig. 3a,e). Second, the tracrRNA stem-loop region of Cas12b is replaced by two additional helices (α3 and α4) in Cas12i, which are involved in the stabilization of the stem-loop of crRNA (Fig. 1e, Extended Data Fig. 3a,e). Moreover, the REC1 domain of Cas12i is longer than that of Cas12b by ~ 20 Å, featured by a long anti-parallel helix pair α16 and α17 that hold the crRNA-target DNA heteroduplex (Fig. 1e, Extended Data Fig. 3b). The PI domain of Cas12i contains three additional helices (α8, α9, and α12) relative to that of Cas12b, while the RuvC and Nuc domains of Cas12i are more compact than those of Cas12b (Extended Data Fig. 3b,d,f). These structural differences between Cas12i and Cas12b account for their distinct functions (to be discussed below).
Pre-crRNA processing
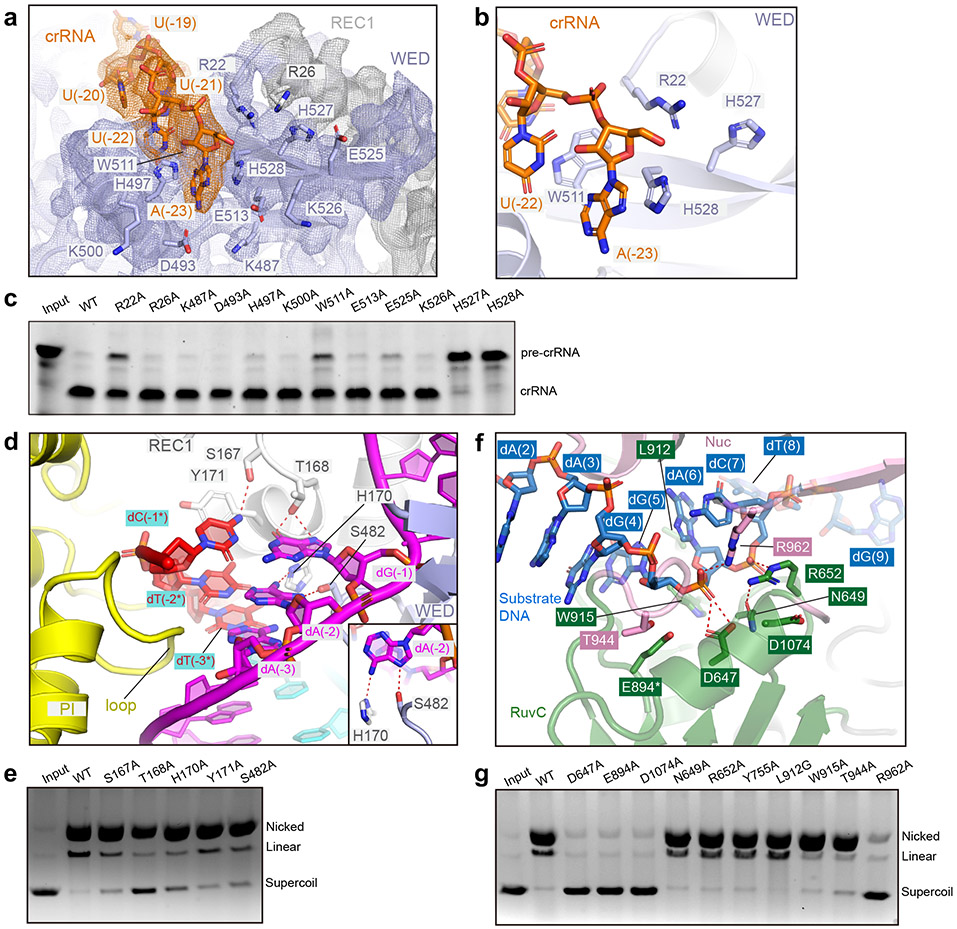
Similar to Cas12a, Cas12i is capable of processing its pre-crRNA 21. To understand the mechanism of pre-crRNA processing by Cas12i, we analyzed the structure around the 5’ end of the crRNA. The first observed nucleotide at the 5’ end of the crRNA in the EM map is A(−23), surrounded by H527, H528 and R22 (Fig. 2a,b). H528 is positioned close to the 5’ oxygen of A(−23), whereas H527 projects away from A(−23) and possibly lies in close proximity to A(−24). To test the role of these residues, we introduced alanine substitutions at each position. Pre-crRNA cleavage assays revealed that the single mutation of H527 or H528 almost completely abolishes while mutation of R22 reduces pre-crRNA cleavage (Fig. 2c). W511 forms pi-stacking interactions with U(−22) and A(−23), positioning the pre-crRNA for cleavage (Fig. 2b). Mutation of W511 also reduced pre-crRNA cleavage (Fig. 2c). Thus, H527, H528 and R22 of the WED domain likely constitute the RNase catalytic site for pre-crRNA maturation, with a cleavage site possibly located between A(−23) and A(−24) 21 (Fig. 2a, Extended Data Fig.1b). The structural and biochemical analysis suggest that Cas12i processes pre-crRNA by an acid-base catalytic mechanism similar to those of Cas12a 23 and Cas13a 24, with H527 and H528 being the acid-base catalyst pair, and R22 being the stabilizer. The three residues and W511 are not conserved in Cas12b (Supplementary Note), suggesting that Cas12b uses a distinct mechanism for pre-crRNA processing, for example via RNase III 25. Interestingly, Cas12i cleaves pre-crRNA in an extended loop preceding the stem-loop structure, different from Cas12a that cleaves pre-crRNA adjacent to the 5’ pseudoknot 23, and Cas6 that cleaves crRNA immediately downstream of the stem-loop structure 26, but similar to Cas13a which also cleaves a loop preceding the repeat-derived stem-loop 27.
Figure 2. Pre-crRNA processing, substrate recognition and cleavage by Cas12i.
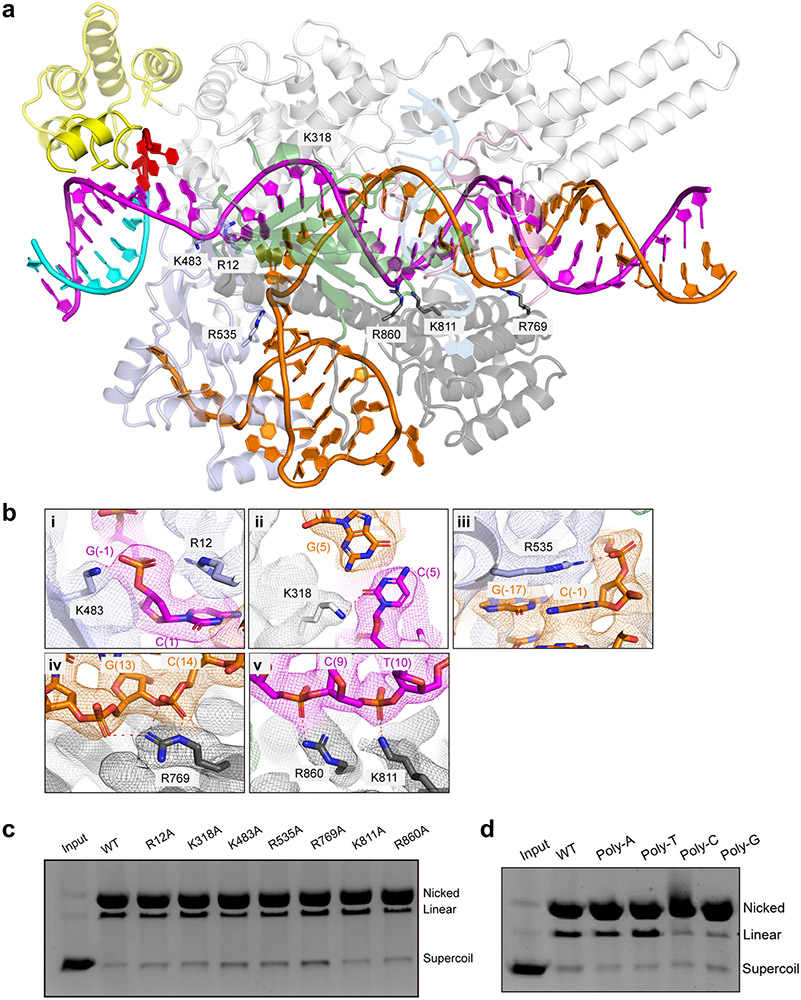
(a) Structure of the crRNA 5’ end and surrounding Cas12i protein in the Cas12i-crRNA-dsDNA ternary complex. Polar residues potentially involved in pre-crRNA processing (tested by mutagenesis) are shown in sticks with cryo-EM map shown in mesh. (b) Close-up view of the 5’ end of the crRNA with four key residues for pre-crRNA cleavage shown in sticks. (c) Pre-crRNA cleavage assay using wild-type Cas12i and alanine substitutions for potential residues involved in pre-crRNA cleavage. The results shown are representative of three experiments. (d) Detailed interactions between the PAM duplex and the REC1, WED, and PI domains. Inset shows the interactions between A(−2) of TS and Cas12i. (e) Cleavage assay using a supercoil plasmid with a 5’-TTN-3’ PAM sequence. Activities of wild-type Cas12i and alanine substitutions for residues involved in PAM recognition are tested. Nicking and double strand breaks in substrate are detected by agarose gel. The results shown are representative of three experiments. (f) Detailed interactions between the substrate DNA and the RuvC and Nuc domains. Note: * indicates E894 modeled in the structure is substituted by an alanine in the complex for cryoEM. (g) Substrate cleavage assay as shown in e using wild-type Cas12i and alanine substitutions for residues involved in substrate DNA recognition. The results shown are representative of three experiments. Uncropped images for panels c,e and g are available as source data online.
PAM recognition
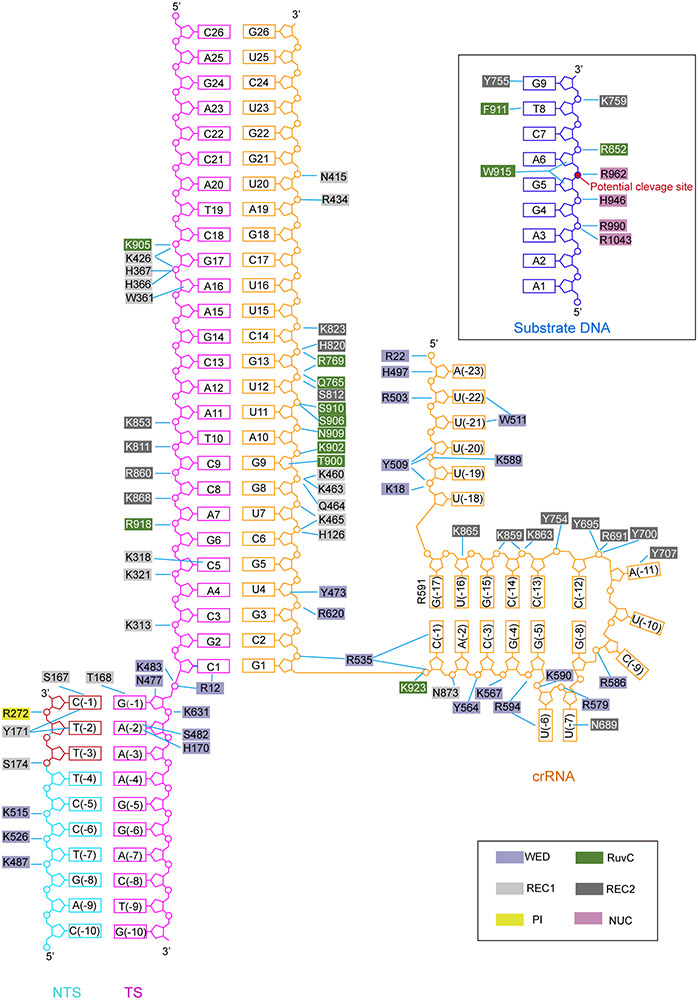
The PAM duplex is recognized by the PI, WED, and REC1 domains (Fig. 2d). A loop from the REC1 domain and a loop from the WED domain insert into the major groove of the PAM duplex and recognize the PAM sequence via hydrogen bonds (Fig. 2d). T168 interacts with dG (−1) while H170 and S482 interact with dA(−2) of the target strand. S167 forms a hydrogen bond with dC(−1*), and Y171 forms stacking interactions with dC(−1*) and dT(−2*) of the non-target strand. Alanine substitution of any of these residues resulted in reduced substrate cleavage activity (Fig. 2e). A loop in the PI domain is inserted into the minor groove of the PAM duplex and closely positioned to the thymine bases of dT(−2*) and dT(−3*) in the non-target strand (Fig. 2d); however, specific interactions were not identified due to poor density in the region. In addition to the sequence specific recognition, a number of positively charged residues form polar interactions with the phosphate groups of the PAM duplex (Extended Data Fig. 4).
crRNA-DNA heteroduplex recognition
The crRNA-DNA heteroduplex is clamped in the positively charged central channel of Cas12i. Both strands of 1-14 bp form extensive contacts with the WED, REC1, REC2, and RuvC domains, mainly through the charged interactions between basic residues and the phosphate groups of the duplex (Extended Data Fig. 4). However, only a few residues of Cas12i contact the 15-26 bp of the heteroduplex (Extended Data Fig. 4). Single alanine substitutions for the residues (such as K483, R535, and R769) involved in the recognition of the crRNA-DNA heteroduplex resulted in only modest reductions in the cleavage activity of Cas12i (Extended Data Fig. 5a-c).
Substrate cleavage
Density consistent with nucleic acid (named ‘substrate DNA’) around the RuvC active site is observed (Fig. 2f, Extended Data Fig. 2i), which is likely from excess DNA oligos used for Cas12i-crRNA-DNA complex assembly. Indeed, the sequence of the target strand can be modelled into the density, nevertheless unambiguous assignment was not feasible at the present resolution (Extended Data Fig. 2i). The presence of excess DNA in the catalytic pocket was also previously observed in Cas12b 28. Cas12i binds to substrate DNA mainly through residues surrounding the catalytic site in a sequence-independent manner (Extended Data Fig. 4). Consistently, Cas12i shows similar nickase activity for poly-A, -T, -C or -G substrates (Extended Data Fig. 5d). However, further studies will be required to investigate substrate preference of Cas12i.
Mutagenesis and substrate cleavage assays showed that three acidic residues (D647, E894, and D1074) in the RuvC domain constitute the catalytic center (Fig. 2g). A basic residue (R962) from the Nuc domain points towards the catalytic center and is also critical for substrate cleavage (Fig. 2f,g). The structure suggests a cleavage site between dG(5) and dA(6), as the scissile phosphate between the two nucleic acid is well positioned towards the catalytic center (Fig. 2f). Interestingly, when W915 from the RuvC domain or T944 from the Nuc domain are substituted by alanine, cleavage of the non-target strand is sustained but cleavage of the target strand is impaired, indicating that they play a role in target strand cleavage (Fig. 2g).
Cas12i activation
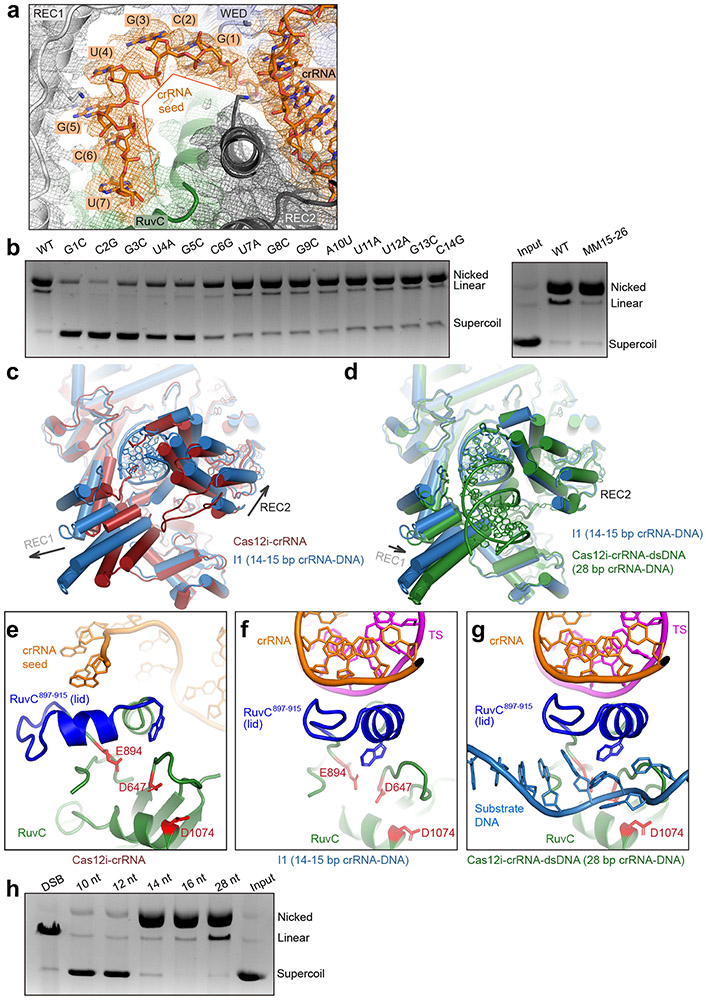
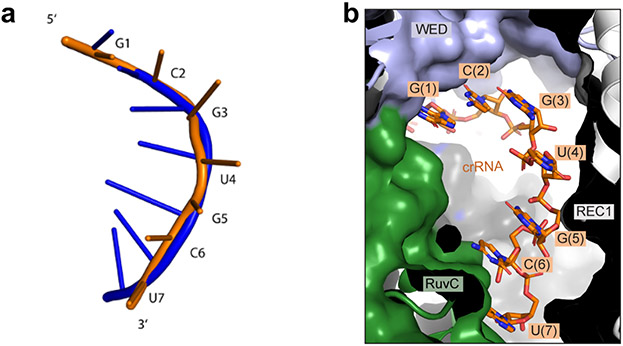
To understand the mechanism of activation, we analyzed the structure of wild-type Cas12i in complex with crRNA and target DNA (Extended Data Fig. 6). By 3D classification, we determined a 3.9 Å resolution cryo-EM structure of a Cas12i-crRNA binary complex (Extended Data Fig. 6c-f). A 7-nt guide segment (G1-U7) immediately after the crRNA hairpin is held by the WED and REC1 domains and ordered in a helical conformation (Fig. 3a, Extended Data Fig. 7a), reminiscent of the seed region observed in Cas12a and Cas12b 23,28,29. Furthermore, the 7-nt guide segment located in the central positively charged channel is exposed to solvent (Fig. 3a, Extended Data Fig. 7b), potentially facilitating the hybridization with the target stand of substrate. Single-nucleotide mismatch in this segment resulted in significant reduction of substrate cleavage activity (Fig. 3b), indicating substrate recognition is likely initiated from this segment. Together, the 7-nt segment is a plausible seed candidate for Cas12i.
Figure 3. Mechanism of Cas12i activation.
(a) 7-nt seed sequence in the Cas12i-crRNA binary complex with cryo-EM density shown in mesh. (b) Substrate cleavage assay using wild-type crRNA and crRNAs with mismatches. Right panel: Substrate cleavage assay using WT crRNA (WT15-26: AAGCTACCAGAC) and crRNA with mismatches in positions 15-26 (MM 15-26: UUCGAUGGUCUG). The results shown are representative of three experiments. (c) Superimposition of structures for the Cas12i-crRNA binary complex and the I1 state. (d) Superimposition of structures for the I1 state and the Cas12i-crRNA-DNA ternary complex. (e) Close-up view of the lid in the RuvC domain (RuvC897-915) in the Cas12i-crRNA binary complex. Catalytic residues are colored in red. (f) Close-up view of the lid in the RuvC domain (RuvC897-915) in the I1 state. (g) Close-up view of the lid in the RuvC domain (RuvC897-915) in the Cas12iE894A-crRNA-DNA ternary complex. (h) Substrate cleavage assay using crRNAs with increasing lengths (10-28 nt) of spacer-derived sequence. The first lane shows the supercoil plasmid substrate treated by a restriction enzyme to create double-strand breaks (DBS), resulting in complete linear product. The results shown are representative of three experiments. Uncropped images for panels b and h are available as source data online.
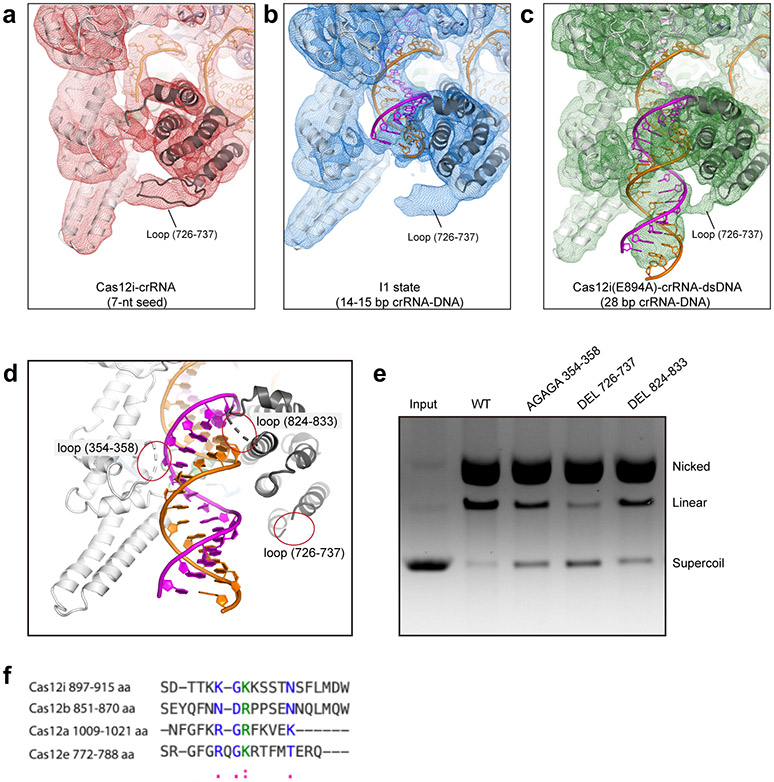
By 3D classification, we also observed an intermediate state (I1 state) with a crRNA-DNA heteroduplex of 14-15 bp, determined to 3.9 Å resolution (Extended Data Fig. 6). Structural comparison of the binary, I1 state, and Cas12iE894A-crRNA-dsDNA ternary complexes shows that formation of 14-15 bp crRNA-target DNA heteroduplex results in the outward movement of REC1 and REC2 (Fig. 3c); however, propagation of the heteroduplex to 28 bp induces an inward movement of REC1, establishing interactions between the 15-28 bp and the REC1 domain (Fig. 3d, Extended Data Fig. 4). Progression of the crRNA-DNA heteroduplex displaces a loop in the REC2 domain (aa 726-737) (Extended Data Fig. 8a-c). Deletion of this loop resulted in reduced cleavage activity (Extended Data Fig. 8d,e), possibly because the coupled conformational changes required for activation are interrupted.
Notably, a short helix (α1) and its preceding loop in the RuvC domain (RuvC897-915, the lid motif) undergo a 90° rotation upon crRNA-target DNA heteroduplex formation, and both are involved in recognition of the minor groove of the heteroduplex at positions 9-11 bp (Fig. 3e-g, Extended Data Fig. 4). Specifically, a number of polar residues contact the phosphate groups in the heteroduplex, assisting the stabilization of the heteroduplex (Extended Data Fig. 4). Rotation of the lid motif opens the substrate binding groove and thus enables substrate binding (Fig. 3e-g). Furthermore, the lid motif also stabilizes the substrate for cleavage such as W915 described above (Fig. 2g). Collectively, the lid motif may act as a key regulator for DNA cleavage by Cas12i.
To understand the biological significance of the different lengths of crRNA-target DNA heteroduplex observed in our structures, we designed crRNAs with different lengths and measured the cleavage activity. The results suggested that a 14-nt spacer-derived sequence is sufficient to activate the nickase activity of Cas12i (Fig. 3h). However, a 28-nt spacer-derived sequence is required for cleavage of the target strand, because deletion or mismatch of positions 15-28 showed no cleavage of the target strand (Fig. 3b,h). Taken together, Cas12i undergoes a two-step activation, wherein formation of 14-bp crRNA-DNA heteroduplex activates the nickase activity for cleavage of the non-target strand, and further base pairing to 28 bp is required for cleavage of the target strand.
Discussion
Type V CRISPR-Cas systems are featured by a single Cas12 effector protein with a RuvC endonuclease domain. To date, 11 sub-types are reported (Cas12a-k) 30-32, and structures for three sub-types were determined including Cas12a 23,33-41, Cas12b 28,29,42 and Cas12e 20. Most Cas12 proteins target dsDNA to create double strand breaks; however, Cas12i predominantly cleaves the non-target strand, creating a nick on target dsDNA 21.
Structure and function of Cas12i
Cas12i, as well as Cas12a, has several functional features distinct from Cas12b and Cas12e, including utilization of a single guide RNA and autonomous pre-crRNA processing. Cas12i also displays unique features in type V systems. First, Cas12i pre-orders a 7-nt seed sequence, in comparison to a 5-nt seed sequence in Cas12a 23,35 and Cas12b 28,29,42. Second, Cas12i is activated by a 14-15 bp crRNA-target DNA heteroduplex, but at least 16-17 bp are required for activation of Cas12a 38. Another significant difference between Cas12i and other Cas12 effectors is the length of crRNA-target DNA heteroduplex, wherein Cas12i accommodates 28 bp compared to 20 bp for Cas12a 39, Cas12b 28 and Cas12e 20, as well as for Cas9 43,44. The presence of more nucleotides for base paring indicates that Cas12i may impose stricter selection for a target sequence in order to create a double strand break 45. Interestingly, formation of a 28-bp heteroduplex is critical for cleavage of the target strand. The requirement of longer sequence complementation for double strand cleavage implies that Cas12i may exhibit higher fidelity for target selection, therefore reducing off-targeting in genome editing applications. It has been shown that rationally engineered Cas9 with longer spacer would increase the fidelity 46, the intrinsic preference for longer spacer base pairing length in Cas12i should motivate the exploration of more Cas12i orthologs with high efficiency and accuracy. It would therefore be interesting to investigate the genome editing activity of Cas12i in eukaryotic cells.
Conserved mechanisms for Cas12 endonucleases
Similar structural patterns (stem-loop or pseudoknot) are observed in the crRNA repeat region adjacent to the spacer-derived sequence in class 2 CRISPR-Cas systems, which not only stabilize the crRNA and allow for recognition by specific CRISPR effectors, but also likely facilitate the correct guide positioning and faithful target binding. Interestingly, six nucleotides immediately adjacent to the spacer-derived guide are identical between Cas12i and Cas12b (Extended Data Fig. 3g). Therefore, the crRNA motif adjacent to spacer-derived segment may be one signature for classification of CRISPR-Cas systems.
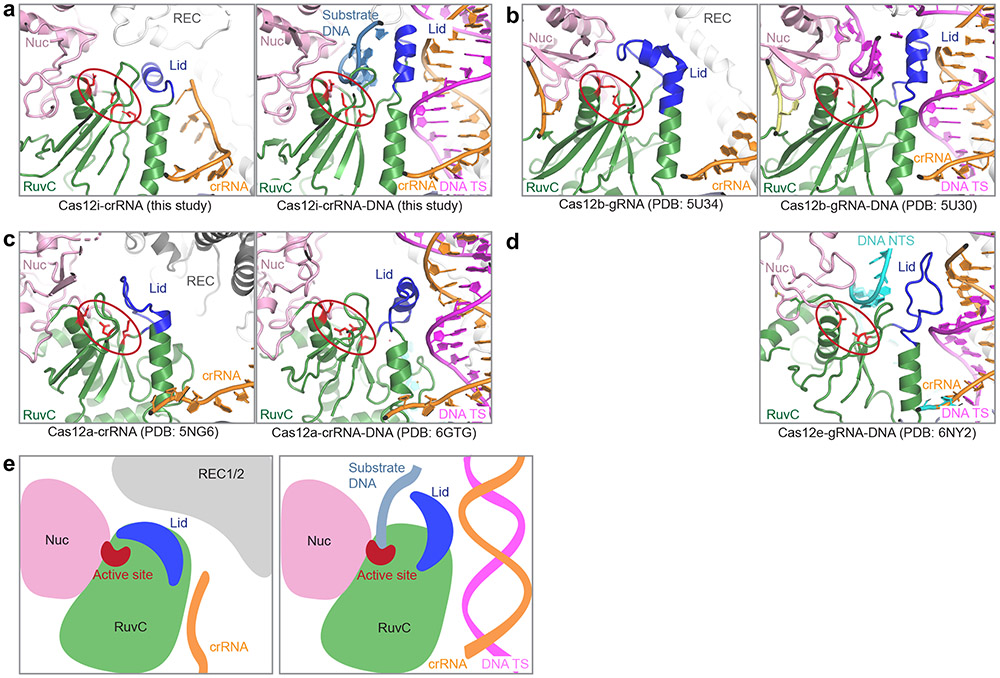
Structures of Cas12i reveal a lid (RuvC897-915) located between the last two β strands (β4 and β5) in the RuvC domain and following the second acidic residue of the conserved carboxylate triad, which undergoes a conformational change upon crRNA-DNA heteroduplex formation to facilitate substrate binding and cleavage (Fig. 3e-g). Interestingly, the structural element equivalent to lid in Cas12i is conserved in Cas12a, Cas12b, and Cas12e (Fig. 4a-d), although these Cas12 orthologs have divergent sequence of the lid region (Extended Data Fig. 8f). First, the lid engages the heteroduplex in a similar sequence-independent manner at positions 8-11. Second, conformational changes of the lid open the RuvC catalytic center. Third, the lid contributes to substrate binding, thereby facilitating DNA cleavage. The lid was demonstrated to be critical for RuvC activation 38, suggesting a conserved activation mechanism for Cas12 endonucleases (Fig. 4e).
Figure 4. The lid in Cas12 endonucleases.
(a-d) The lids (colored in blue) before and after DNA substrate recognition in Cas12i (a), Cas12b (b), Cas12a (c), and Cas12e (d) are shown in parallel. Left panels are the structures without substrate (structure for Cas12e is not available), while right panels are structures with substrate. The RuvC catalytic centers are indicated by red circles, with the three acidic amino acids shown in sticks. (e) Cartoon model showing that the movement of the lid is coupled with activation of Cas12 endonucleases.
In summary, our results shed light on the mechanism of Cas12i system in substrate recognition and activation, and the evolution of the type V endonuclease to acquire distinct functions. Atomic structures would facilitate the manipulation of Cas12i for genome editing with high specificity and diagnostic applications.
Methods
Protein expression and purification
The plasmid encoding full-length Cas12i (Cas12i1) is obtained from Addgene (Catalog # 120882) 21. Cas12i is fused with an N-terminal 6xHis tag. The mutations were generated by quickchange mutagenesis. These plasmids were transformed into BL21-CodonPlus (DE3)-RIL cells for overexpression in Terrifc Broth medium. The protein expression was induced by adding 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 18 °C overnight. The cells were collected and then disrupted by sonication in buffer A (25 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5% Glycerol, 1 mM PMSF and 5 mM β-mercaptoethanol). After centrifugation, the supernatants were incubated with the Ni-NTA resin and washed with buffer B (25 mM Tris–HCl, pH 8.0, 500 mM NaCl, 30 mM imidazole and 5 mM β-mercaptoethanol), and the His-tagged target proteins were eluted using buffer B supplemented with 300 mM imidazole. Subsequently, the target proteins were loaded onto the Heparin column (GE) and eluted with a linear sodium chloride gradient, followed by size-exclusion chromatography (Superdex 200, GE) in buffer C (25 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM DTT and 0.1 mM MgCl2). Peak fractions were concentrated and stocked in −80 °C.
To assemble the Cas12i-crRNA binary complex, Cas12i proteins were mixed with crRNA (synthesized from IDT) at a ratio of 1:1.3 on ice for 1 hours. To reconstitute the Cas12i-crRNA-target DNA ternary complexes, the Cas12i WT or E894A mutant proteins were incubated with crRNA for 30 mins, followed by adding the target DNA synthesized from IDT at a ratio of 1:1.3:2. The mixture was applied to a Superdex 200 (GE) column equilibrated with buffer C.
In vitro DNA cleavage assay
Target DNA containing the 5’-TTN-3’ PAM was synthesized from IDT and cloned into pET28-MHL vector using the InFusion™ cloning kit (ClonTech). Poly-A, -T, -C and -G substrates were generated by substitution at the target-strand of protospacer-derived sequence at positions 17-28. Cas12i proteins (250 ng) were mixed with crRNA at a ratio of 1:1.5 on ice for 30 mins in the cleavage buffer containing 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM MgCl2, 5 mM DTT and 5% glycerol. The purified plasmids (100 ng) were then added into the mixture for another 30 mins. The reactions were quenched by adding EDTA and Proteinase K. The cleavage products were resolved on 0.5% agarose gels and visualized by ethidium bromide staining.
Pre-crRNA cleavage Assays
The 60 nt pre-crRNA synthesized from IDT was incubated with 2 μg Cas12i proteins on ice at a ratio of 1:3 in the cleavage buffer. The reactions were stopped by adding TBE-Urea Sample Buffer (Thermo Fisher Scientific) and 66°C quenched for 10 mins. The products were separated on a 15% TBE-urea gel and stained using SYBR Gold (Invitrogen).
Electron Microscopy
Aliquots of 3 μL samples (Cas12i-crRNA, Cas12iWT-crRNA-dsDNA, and Cas12iE894A-crRNA-dsDNA) at 0.75 mg/ml were applied to glow-discharged Quantifoil holey carbon girds (Au, R1.2/1.3, 300 mesh). The grids were blotted for 4 s and plunged into liquid ethane using a Gatan Cryoplunge 3 plunger. Cryo-EM data were collected with a Titan Krios microscope (FEI) operated at 300 kV and images were collected using Leginon 50 at a nominal magnification of 81,000x (resulting in a calibrated physical pixel size of 1.05 Å/pixel) with a defocus range of 1.2 – 2.5 μm. The images were recorded on a K2 summit (for wild-type Cas12i-crRNA-dsDNA) or K3 (for Cas12iE894A-crRNA-dsDNA) electron direct detector in super-resolution mode at the end of a GIF-Quantum energy filter operated with a slit width of 20 eV. For data collection with a K2 detector, a dose rate of 5 e− per pixel per second and an exposure time of 8 seconds were used, generating 40 movie frames (200 milliseconds per frame) with a total dose of ~ 35 electrons per Å2. For data collection with a K3 detector, a dose rate of 20 electrons per pixel per second and an exposure time of 3.12 seconds were used, generating 40 movie frames (78 milliseconds per frame) with a total dose of ~ 54 electrons per Å2. Statistics for cryo-EM data are listed in Table 1.
Image Processing
The movie frames were imported to RELION-3 51. Movie frames were aligned using MotionCor2 52 with a binning factor of 2. Contrast transfer function (CTF) parameters were estimated using Gctf 53. A few thousand particles were auto-picked without template to generate 2D averages for subsequent template-based auto-picking.
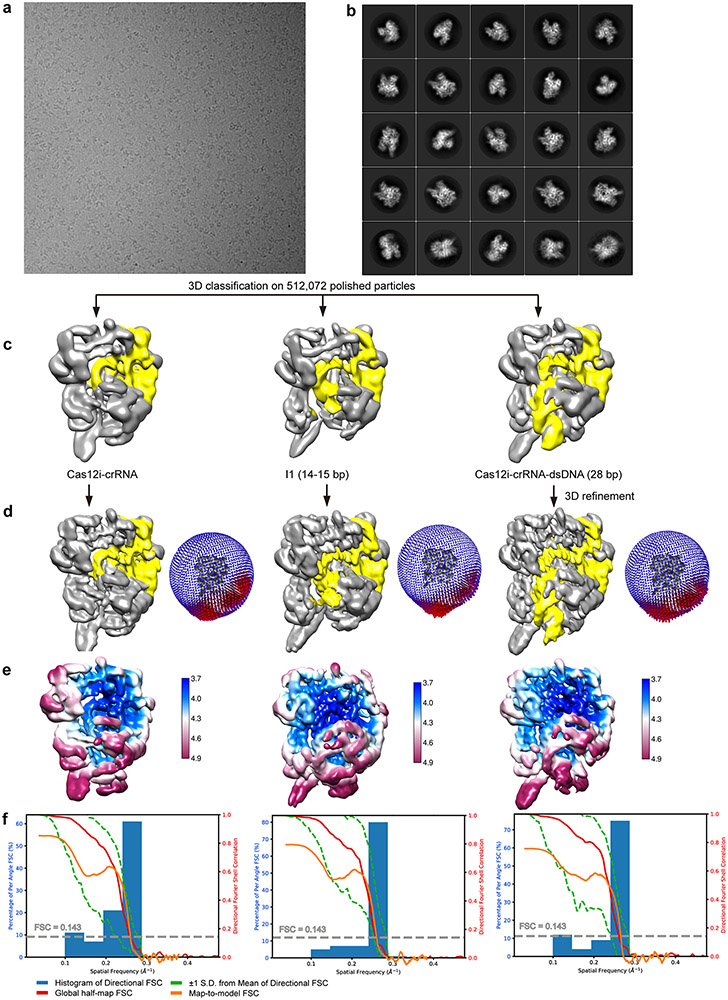
For Cas12iE894A-crRNA-dsDNA dataset, five million particles were auto-picked and extracted from the dose weighted micrographs. The dataset was split into batches for 2D classifications, which were used to exclude false and bad particles that fall into 2D averages with poor features. Particles from different views were used to generate initial model in cryoSPARC 54. The dataset was split into six batches for 3D refinement, each converging at resolution of around 3.3 Å. After Bayesian polishing, each part was classified into six classes in 3D classification. 3D classification shows that the complex adopts a single conformation. The classes representing bad particles from Bayesian polishing and the classes with less alignment accuracy were excluded. Remaining 731,231 particles were used for 3D refinement, CTF refinement and 3D refinement, which finally converged at 2.9 Å resolution. Focused 3D refinement, focused 3D classification and 3D refinement were performed sequentially around the PAM binding region to improve the local map quality.
For Cas12iWT-crRNA-dsDNA dataset, 1.5 million particles were auto-picked and extracted from the dose weighted micrographs. The dataset was split into batches for 2D classifications, which were used to exclude false and bad particles that fall into 2D averages with poor features. Particles from different views were used to generate initial model in cryoSPARC 54. The dataset was split into two batches for 3D refinement, each converging at a resolution of around 4 Å. After Bayesian polishing, each part was classified into six classes in 3D classification. 3D classification shows that the complex adopts different conformation. Cas12i accommodates crRNA-DNA duplex with different lengths, and different conformational states were distinguished accordingly. Dataset representing the Cas12i-crRNA binary complex contains 122,435 particles, converging at a resolution of 3.9 Å. Dataset representing the I1 state (14-15 bp crRNA-DNA heteroduplex) contains 75,406 particles, converging at a resolution of 3.9 Å. Dataset representing the Cas12i-crRNA-DNA ternary complex (26-28 bp crRNA-DNA heteroduplex) contains 99,516 particles, resulting in a 3.8 Å resolution map similar to the map of Cas12iE894A-crRNA-dsDNA.
3D FSC analysis for the presented maps were performed using the Remote 3DFSC Processing Server (https://3dfsc.salk.edu/upload/) 55. Map-to-model FSC analysis was performed using phenix.validation_cryoem tool in Phenix.
Model Building and refinement
De novo model building of the Cas12iE894A-crRNA-dsDNA structure was performed manually in COOT 56. Secondary structure predictions by PSIPRED 57 were used to assist manual building. Refinement of the structure models against corresponding maps were performed using phenix.real_space_refine tool in Phenix 58. For the Cas12i-crRNA binary complex and the I1 state, the Cas12iE894A-crRNA-dsDNA structure was firstly docked to the corresponding cryo-EM maps using UCSF Chimera 59, then manually adjusted in COOT, and finally refined in Phenix.
Visualization
Figures were generated using PyMOL and UCSF Chimera 59.
Reporting Summary statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability statement
Cryo-EM reconstructions of Cas12i(E894A)-crRNA-dsDNA, Cas12i-crRNA, and the I1 complexes have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-21541, EMD-21551 and EMD-21552, respectively. Coordinates for atomic models of Cas12i(E894A)-crRNA-dsDNA, Cas12i-crRNA, and the I1 complexes have been deposited in the Protein Data Bank under the accession numbers 6W5C, 6W62, and 6W64, respectively. Source data are available with the paper online.
Extended Data
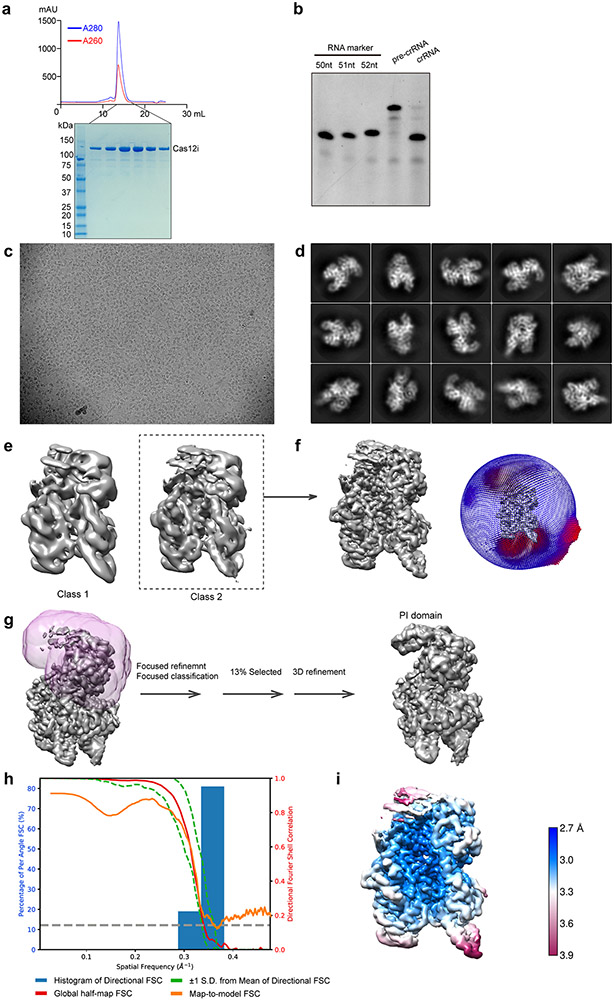
Extended Data Figure 1. Sample preparation and cryo-EM for the Cas12i(E894A)-crRNA-dsDNA complex.
(a) Purification of Cas12i. Upper: Size exclusion chromatography (SEC) profile of Cas12i. UV absorbance curves at 280 nm and 260 nm are shown in blue and red, respectively. Lower: SDS-PAGE analysis of the elution fractions from SEC as indicated. (b) Urea-PAGE analysis of pre-crRNA before and after processing by Cas12i. RNA markers indicate that the mature crRNA is 51nt. (c) A representative raw cryo-EM micrograph of the Cas12i-crRNA-dsDNA ternary complex. (d) Representative 2D class averages. (e) Two major classes from 3D classification. The two maps are similar, with class 2 at better quality. (f) 3D auto-refinement using particles from class 2. Angular distribution of particles is shown on the right. (g) Procedures used for improving the resolution of the PI domain. Focused refinement followed by focused 3D classification using a soft mask (in pink mesh) resulted in a 3D class from 13% particles with improved density in the PI domain. (h) Plot of the global half-map FSC (solid red line), map-to-model FSC (solid orange line), and spread of directional resolution values (±1σ from mean, green dotted lines; the blue bars indicate a histogram of 100 such values evenly sampled over the 3D FSC). (i) Local resolution map of the final reconstruction. Uncropped images for panels a and b are available as source data online.
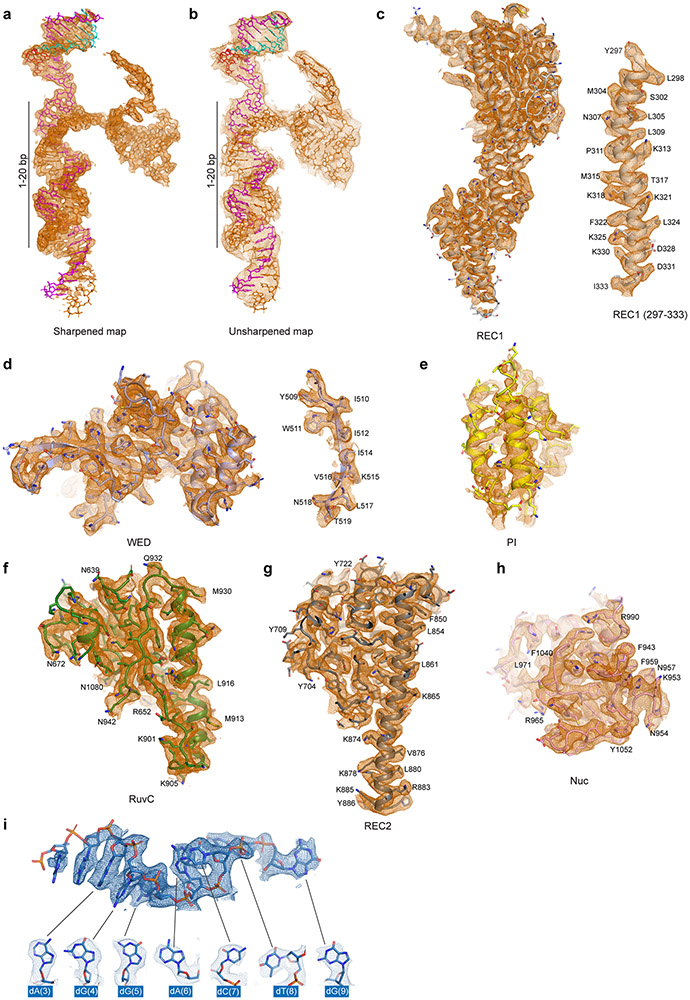
Extended Data Figure 2. Detailed cryo-EM density map of the Cas12i(E894A)-crRNA-dsDNA complex with final atomic model fitted in.
(a, b) Fitting of nucleic acids to the corresponding cryo-EM map. The atomic models are shown in stick with crRNA, the target strand and the non-target strand colored in orange, magenta and cyan, respectively. Cryo-EM density from the sharpened map (a) or the unsharpened map (b) is shown in mesh. (c) Fitting of the REC1 domain. A representative α-helix from the REC1 domain is shown in details on the right. (d) Fitting of the WED domain. A representative β-strand is shown in details on the right. (e) Fitting of the PI domain. (f) Fitting of the RuvC domain. (g) Fitting of the REC2 domain. (h) Fitting of the Nuc domain. (i) Fitting of the substrate DNA.
Extended Data Figure 3. Structural comparison of Cas12i and Cas12b.
(a) Overall structures of the Cas12i-crRNA-dsDNA and the Cas12b-gRNA-dsDNA complexes. The structures are aligned by secondary-structure matching (SSM) in COOT. (b-f) Structural comparison of each domain. Secondary structures in each domain are labeled. The PI domain and the extended α16 and α17 helix pair are indicated by circles and a box, respectively. (g) Sequence alignment of the crRNA repeat adjacent to the spacer-derived segment between Cas12i and Cas12b.
Extended Data Figure 4. Schematic of nucleic acid recognition in the Cas12i(E894A)-crRNA-dsDNA complex.
Intermolecular contacts between Cas12i and nucleic acids, including the target and non-target strands and the substrate DNA, are shown by solid lines. The Cas12i residues are colored based on the domain architecture.
Extended Data Figure 5. Structure and mutagenesis analysis for the recognition of the crRNA-target DNA heteroduplex.
(a) Structure of the Cas12i-crRNA-DNA complex with selected key residues involved in the recognition of the heteroduplex shown as sticks. (b) Close-up view of the interactions shown in a with cryo-EM map shown in mesh. (c) Substrate cleavage assay using wild-type Cas12i and Cas12i with single mutations on the key residues shown in a. The results shown are representative of three experiments. (d) Substrate cleavage assay using poly-A, -T, -C, and G as substrates. The results shown are representative of three experiments. Uncropped images for panels c and d are available as source data online.
Extended Data Figure 6. Cryo-EM data processing for the wild-type Cas12i-crRNA-dsDNA complex.
(a) A representative raw cryo-EM micrograph of the wild-type Cas12i-crRNA-DNA complex. (b) Representative 2D class averages. (c) 3D classification. Three major classes are observed, representing the Cas12i-crRNA binary complex, the intermediate state (I1 state, 14-15 bp heteroduplex), and the fully assembled Cas12i-crRNA-DNA complex (26-28 bp heteroduplex). Densities corresponding to the crRNA and target DNA are colored in yellow. The Cas12i-crRNA map is comparable to a reconstruction using biochemically purified Cas12i-crRNA complex (data not shown). (d) 3D auto-refinement for the three classes shown in c. Angular distribution of each reconstruction is shown below. (e) Local resolution map of the final reconstructions in d. (f) Plot of the global half-map FSC (solid red line), map-to-model FSC (solid orange line), and spread of directional resolution values (±1σ from mean, green dotted lines; the blue bars indicate a histogram of 100 such values evenly sampled over the 3D FSC).
Extended Data Figure 7. Structural analysis of the seed sequence.
(a) Superposition of the seed sequence of Cas12i (orange) with the same sequence simulated in an A-form geometry (blue). (b) Structure of the seed sequence with Cas12i domains shown in surface representation.
Extended Data Figure 8. Conformational changes of recognition loops in Cas12i upon heteroduplex formation.
(a) Structure of the Cas12i-crRNA binary complex with cryo-EM density shown in mesh. Loop 726-737 is indicated. (b) Structure of the I1 state with cryo-EM density shown in mesh. (c) Structure of the Cas12i(E894A)-crRNA-DNA ternary complex with cryo-EM density shown in mesh. (d) Structure showing three loops from the REC1 and REC2 domains of Cas12i that are likely involved in heteroduplex recognition. (e) Substrate cleavage assay using wild-type Cas12i and Cas12i with each of the three loops shown in d deleted or mutated. The results shown are representative of three experiments. (f) Sequence alignment of the lid region among Cas12 orthologs using ClustalW program 47. Uncropped image for panel e is available as source data online.
Supplementary Material
Structure-based sequence alignment between Cas12i and Cas12b. PROMALS3D 48 program was used to align the sequences based on structures. The alignment diagram was plotted using ESPript 49. Domains of Cas12i are indicated by background colors according to domain architecture in Fig. 1a. Secondary structures of each domain of Cas12i are labeled. The extended α16 and α17 helix pair in the REC1 domain, three additional helices in the WED domain, and residues involved in pre-crRNA processing are indicated by boxes.
Acknowledgments
We thank Thomas Klose and Valorie Bowman for help with cryo-EM, Steven Wilson for computation, and John Tesmer and Clinton Gabel for critical reading of the manuscript. This work was supported by the NIH grant R01GM138675 and a Showalter Trust Research Award to L.C.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Mohanraju P et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Wright AV, Nuñez JK & Doudna JA Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Sorek R, Lawrence CM & Wiedenheft B CRISPR-mediated adaptive immune systems in bacteria and archaea. Annual review of biochemistry 82, 237–266 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Marraffini LA CRISPR-Cas immunity in prokaryotes. Nature 526, 55 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Jiang F & Doudna JA CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys 46, 505–529, doi: 10.1146/annurev-biophys-062215-010822 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Hille F et al. The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259, doi: 10.1016/j.cell.2017.11.032 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Barrangou R & Marraffini LA CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell 54, 234–244, doi: 10.1016/j.molcel.2014.03.011 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Oost J, Jore MM, Westra ER, Lundgren M & Brouns SJ CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34, 401–407, doi: 10.1016/j.tibs.2009.05.002 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Shmakov S et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nature Reviews Microbiology 15, 169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS et al. An updated evolutionary classification of CRISPR–Cas systems. Nature Reviews Microbiology 13, 722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Molecular cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koonin EV, Makarova KS & Zhang F Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol 37, 67–78, doi: 10.1016/j.mib.2017.05.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrangou R & Doudna JA Applications of CRISPR technologies in research and beyond. Nature biotechnology 34, 933 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Komor AC, Badran AH & Liu DR CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi H et al. CRISPR Disruption of BmOvo Resulted in the Failure of Emergence and Affected the Wing and Gonad Development in the Silkworm Bombyx mori. Insects 10, doi: 10.3390/insects10080254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickar-Oliver A & Gersbach CA The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol 20, 490–507, doi: 10.1038/s41580-019-0131-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771, doi: 10.1016/j.cell.2015.09.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strecker J et al. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun 10, 212, doi: 10.1038/s41467-018-08224-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng F et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov 4, 63, doi: 10.1038/s41421-018-0069-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JJ et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223, doi: 10.1038/s41586-019-0908-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan WX et al. Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91, doi: 10.1126/science.aav7271 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Rosenstrom P & Schenkel A Searching protein structure databases with DaliLite v.3. Bioinformatics (Oxford, England) 24, 2780–2781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swarts DC, van der Oost J & Jinek M Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol Cell 66, 221–233 e224, doi: 10.1016/j.molcel.2017.03.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knott GJ et al. Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat Struct Mol Biol 24, 825–833, doi: 10.1038/nsmb.3466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murugan K, Babu K, Sundaresan R, Rajan R & Sashital DG The Revolution Continues: Newly Discovered Systems Expand the CRISPR-Cas Toolkit. Mol Cell 68, 15–25, doi: 10.1016/j.molcel.2017.09.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K & Doudna JA Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358, doi: 10.1126/science.1192272 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L et al. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 170, 714–726 e710, doi: 10.1016/j.cell.2017.06.050 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Yang H, Gao P, Rajashankar KR & Patel DJ PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell 167, 1814–1828 e1812, doi: 10.1016/j.cell.2016.11.053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L et al. C2c1-sgRNA Complex Structure Reveals RNA-Guided DNA Cleavage Mechanism. Mol Cell 65, 310–322, doi: 10.1016/j.molcel.2016.11.040 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Swarts DC Stirring Up the Type V Alphabet Soup. CRISPR J 2, 14–16, doi: 10.1089/crispr.2019.29044.dcs (2019). [DOI] [PubMed] [Google Scholar]
- 31.Strecker J et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53, doi: 10.1126/science.aax9181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Shayeb B et al. Clades of huge phages from across Earth's ecosystems. Nature 578, 425–431, doi: 10.1038/s41586-020-2007-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong D et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Gao P, Yang H, Rajashankar KR, Huang Z & Patel DJ Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell research 26, 901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamano T et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamano T et al. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Molecular cell 67, 633–645. e633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stella S, Alcón P & Montoya G Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546, 559 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Stella S et al. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell (2018). [DOI] [PubMed] [Google Scholar]
- 39.Nishimasu H et al. Structural basis for the altered PAM recognition by engineered CRISPR-Cpf1. Molecular cell 67, 139–147. e132 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swarts DC & Jinek M Mechanistic Insights into the cis-and trans-acting DNase activities of Cas12a. Molecular cell 73, 589–600. e584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H et al. Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins. Cell Host Microbe 25, 815–826 e814, doi: 10.1016/j.chom.2019.05.004 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Wu D, Guan X, Zhu Y, Ren K & Huang Z Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA. Cell Res 27, 705–708, doi: 10.1038/cr.2017.46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimasu H et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949, doi: 10.1016/j.cell.2014.02.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders C, Niewoehner O, Duerst A & Jinek M Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573, doi: 10.1038/nature13579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ran FA et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389, doi: 10.1016/j.cell.2013.08.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kulcsár PI et al. Blackjack mutations improve the on-target activities of increased fidelity variants of SpCas9 with 5′G-extended sgRNAs. Nature Communications, doi: 10.1038/s41467-020-15021-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson JD, Gibson TJ & Higgins DG Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2, Unit 2 3, doi: 10.1002/0471250953.bi0203s00 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Pei J, Kim BH & Grishin NV PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36, 2295–2300, doi: 10.1093/nar/gkn072 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert X & Gouet P Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324, doi: 10.1093/nar/gku316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suloway C et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60, doi: 10.1016/j.jsb.2005.03.010 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Tan YZ et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat Methods 14, 793–796, doi: 10.1038/nmeth.4347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones DT Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292, 195–202, doi: 10.1006/jmbi.1999.3091 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544, doi: 10.1107/S2059798318006551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure-based sequence alignment between Cas12i and Cas12b. PROMALS3D 48 program was used to align the sequences based on structures. The alignment diagram was plotted using ESPript 49. Domains of Cas12i are indicated by background colors according to domain architecture in Fig. 1a. Secondary structures of each domain of Cas12i are labeled. The extended α16 and α17 helix pair in the REC1 domain, three additional helices in the WED domain, and residues involved in pre-crRNA processing are indicated by boxes.
Data Availability Statement
Cryo-EM reconstructions of Cas12i(E894A)-crRNA-dsDNA, Cas12i-crRNA, and the I1 complexes have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-21541, EMD-21551 and EMD-21552, respectively. Coordinates for atomic models of Cas12i(E894A)-crRNA-dsDNA, Cas12i-crRNA, and the I1 complexes have been deposited in the Protein Data Bank under the accession numbers 6W5C, 6W62, and 6W64, respectively. Source data are available with the paper online.