Abstract
Cas12g, the type V-G CRISPR-Cas effector, is an RNA-guided ribonuclease that targets single-stranded RNA substrate. CRISPR-Cas12g system offers a potential platform for transcriptome engineering and diagnostic applications. We determined the structures of Cas12g-guide RNA complexes in the absence and presence of target RNA by cryo-EM to 3.1 Å and 4.8 Å, respectively. Cas12g adopts a bilobed structure with miniature REC2 and Nuc domains, whereas the guide RNAs fold into a flipped “F” shape which is primarily recognized by the REC lobe. Target RNA and the crRNA guide form a duplex that inserts into the central cavity between the REC and NUC lobes, inducing conformational changes in both lobes to activate Cas12g. The structural insights would facilitate the development of Cas12g-based applications.
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) systems endow the prokaryotes with immunity against mobile genetic elements 1-4. CRISPR systems are grouped into two classes comprising six types 5-7. The class 1 system (types I, III and IV) employs multi-protein effector complexes to cleave foreign nucleic acid, while the class 2 system (types II, V and VI) utilizes a single multi-domain effector endonuclease. The specific DNA targeting and cleavage activities of CRISPR-Cas systems have inspired their successful applications in genome editing, with CRISPR-Cas9 from the type II system most widely used 8,9. However, RNA targeting CRISPR-Cas effectors have been developed into various tools for manipulating specific RNA molecules, such as site-specific RNA editing 10, RNA knockdown 11 and molecular diagnostic detection of nucleic acids 10. RNA editing offers a tool for temporary and reversible alteration in gene function, thus avoiding permanent changes to the genome as in the case of DNA editing, where off-targeting is a big concern when used for clinical treatment. RNA targeting endonucleases have been found in type II, III and VI CRISPR-Cas systems 12-16. Nevertheless, current RNA targeting applications are mostly based on the RNA-dedicated type VI systems (Cas13a-d), which exclusively catalyzes RNA cleavage with HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domains 17-19.
Cas12g is a recently identified type V RNA-guided endonuclease through mining the metagenomic database 20. Cas12g specifically recognizes RNA substrates, distinguishing it from other Cas12 proteins discovered to date, including the well-studied Cas12a (Cpf1), Cas12b (C2C1) and Cas12e (CasX) that target DNA and have been successfully harnessed for genome editing in mammalian cells 21-24. Cas12g employs two guide RNAs, a crRNA and a tracrRNA, to target single-stranded RNA (ssRNA) for cleavage without the requirement of a protospacer adjacent motif (PAM) sequence 20. After activation by target RNA, Cas12g displays collateral RNase and single-strand DNase activities 20, similar to the collateral activity of Cas12a and Cas13 proteins 25-28. Distinct from Cas12a and Cas13, in vitro experiments showed that Cas12g is incapable of processing pre-crRNA 20.
Originating from a hot spring metagenome, Cas12g is thermostable while harboring comparable RNA detection sensitivity compared to the best-performing Cas13 effectors 20. Another advantage of Cas12g is its compact size (only 767 aa), compared to Cas13 proteins (averages sizes for Cas13a-d are 1250 aa, 1150 aa, 1120 aa and 930 aa, respectively 29). The small size potentially facilitates flexible packaging into size-constrained therapeutic viral vectors such as adeno-associated virus (AAV) 30. Thus, Cas12g has the potential to enhance RNA detection and other in vivo transcriptome engineering applications.
Understanding the molecular basis underlying target RNA recognition and cleavage by Cas12g is critical for the development of Cas12g-based tools. Structural studies of type V effectors, including Cas12a 28,31-39, Cas12b 40-42, Cas12e 24, and Cas12i 43, showed that they display a canonical bilobed architecture encompassing a recognition lobe (REC) and a nuclease lobe (NUC). The REC lobe contains the REC1 and REC2 domains, which are responsible for substrate recognition, whereas the NUC lobe is composed of the RuvC, Wedge (WED), PAM-interacting (PI), and Nuc domains, with the nuclease site located in the RuvC domain. However, low sequence similarity between Cas12g and other Cas12 effectors and RNA targeting function of Cas12g indicates differences in its mechanisms for target recognition and cleavage, motivating us to investigate the structure of Cas12g.
Results
Overall structure of Cas12g-sgRNA
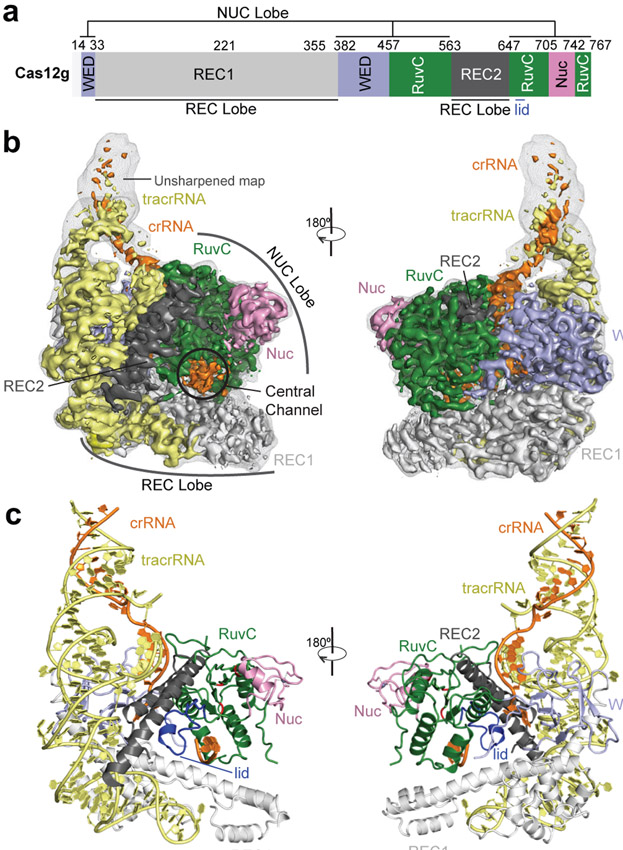
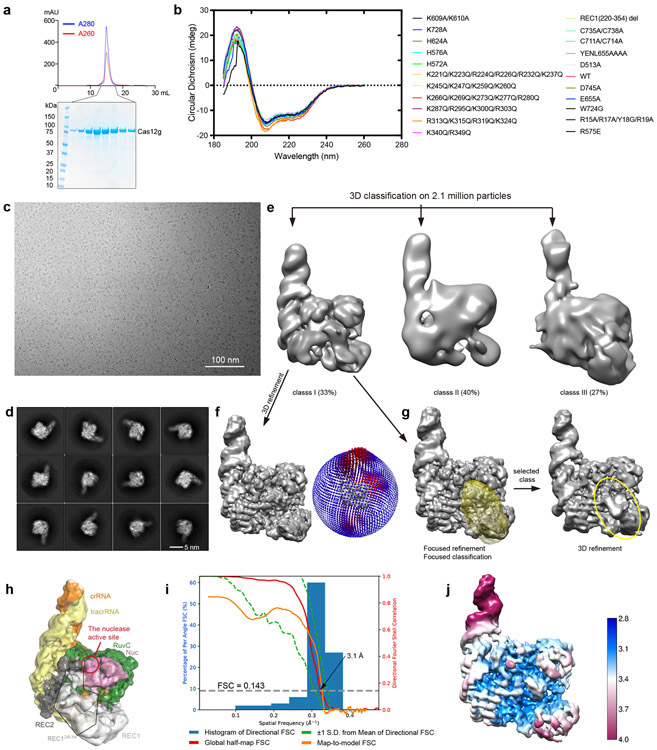
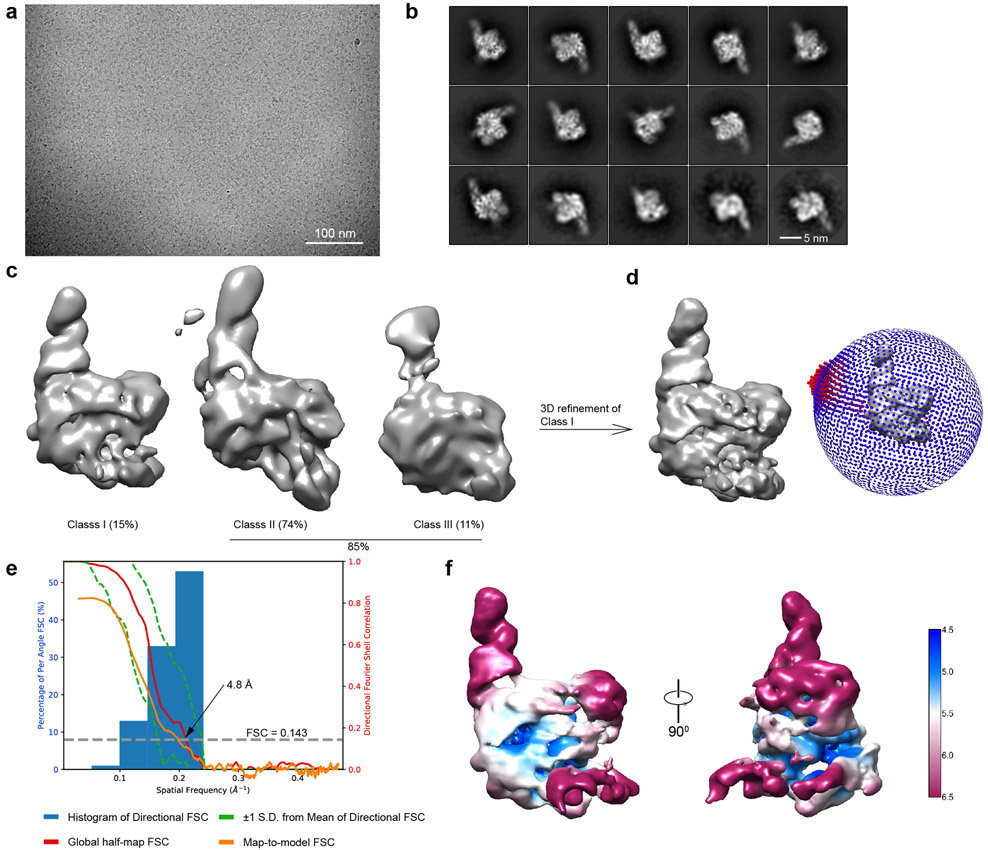
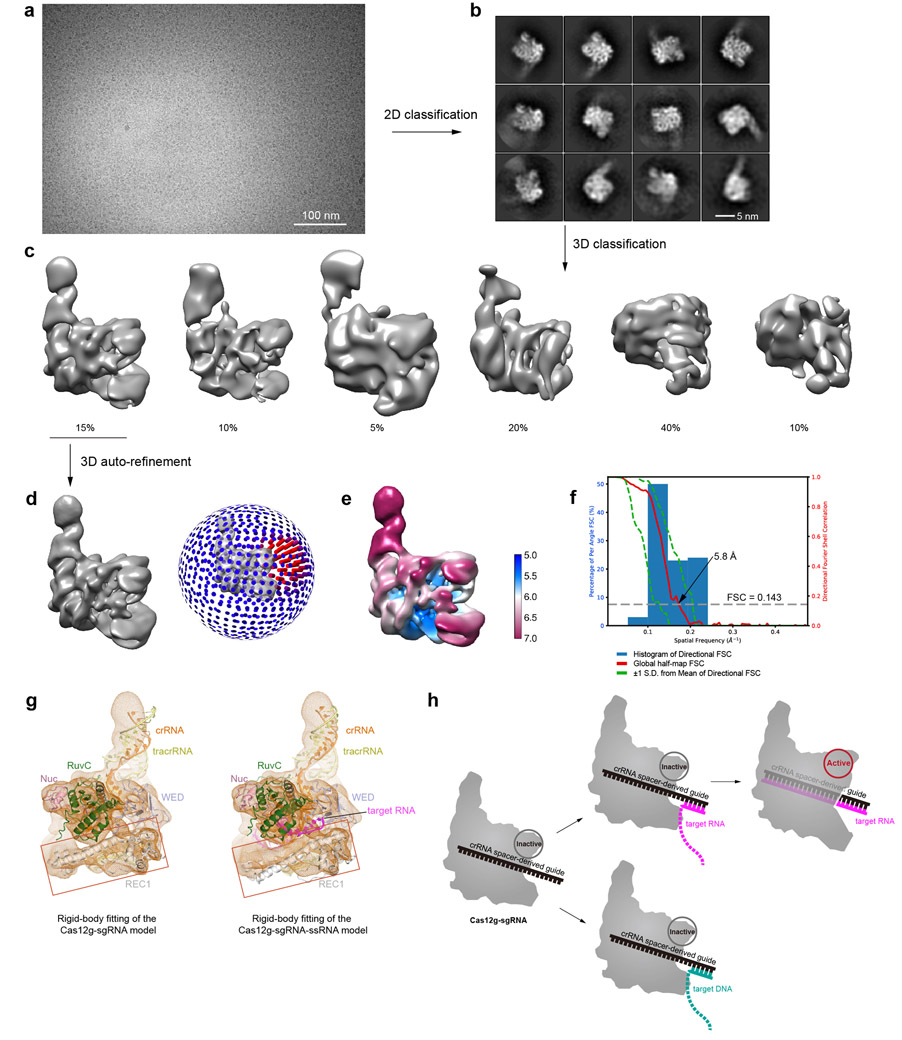
We assembled a binary complex of wild-type Cas12g and a 136-nt single-guide RNA (sgRNA, fusion of tracrRNA and crRNA) and determined the structure of this complex at an average resolution of 3.1 Å by cryo-EM (Extended Data Fig. 1 and Supplementary Table 1). The resultant map allowed ab initio model building of most residues and nucleotides of the complex, with the exception of several segments at the periphery of the complex (Extended Data Fig. 2 and Supplementary Fig. 1).
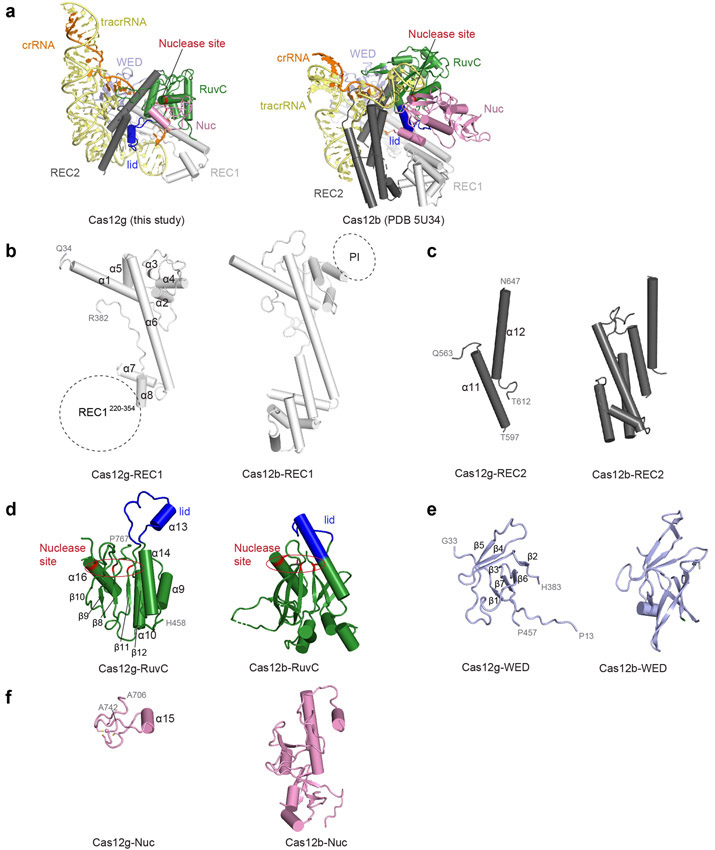
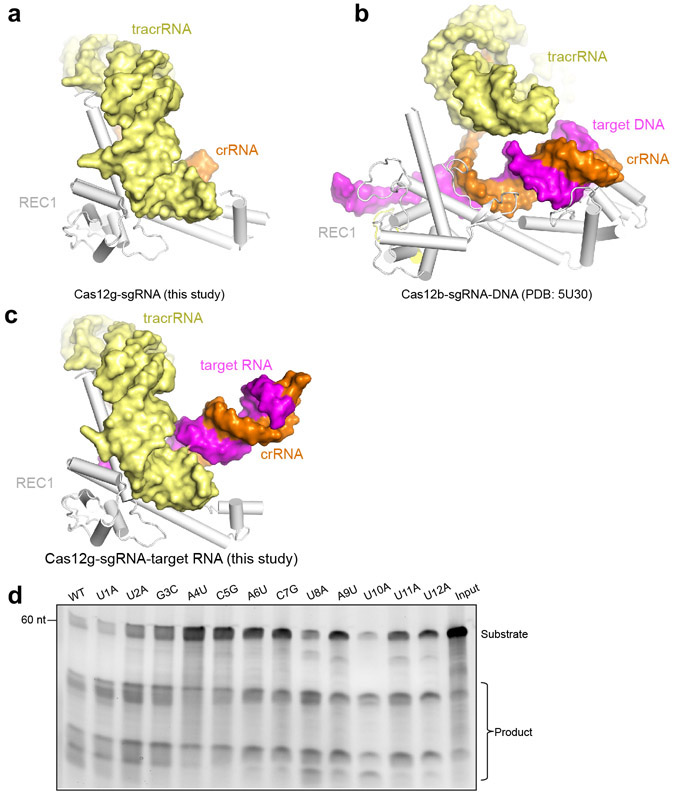
The overall structure of Cas12g displays a bilobed architecture containing the REC and NUC lobes, enclosing a positively charged central channel between the two lobes (Fig. 1a-c). sgRNA primarily engages Cas12g by contacting the REC lobe, with additional interactions with the WED domain from the NUC lobe (Fig. 1c). A DALI search revealed Cas12b as the closest structural match to Cas12g (Z scores: ~ 14; root-mean-square deviation: 5.8 Å over 466 amino acids) (Extended Data Fig. 3 and Supplementary Fig. 1), whereas Cas12a and Cas12e are also close matches (Z scores: 9-10).
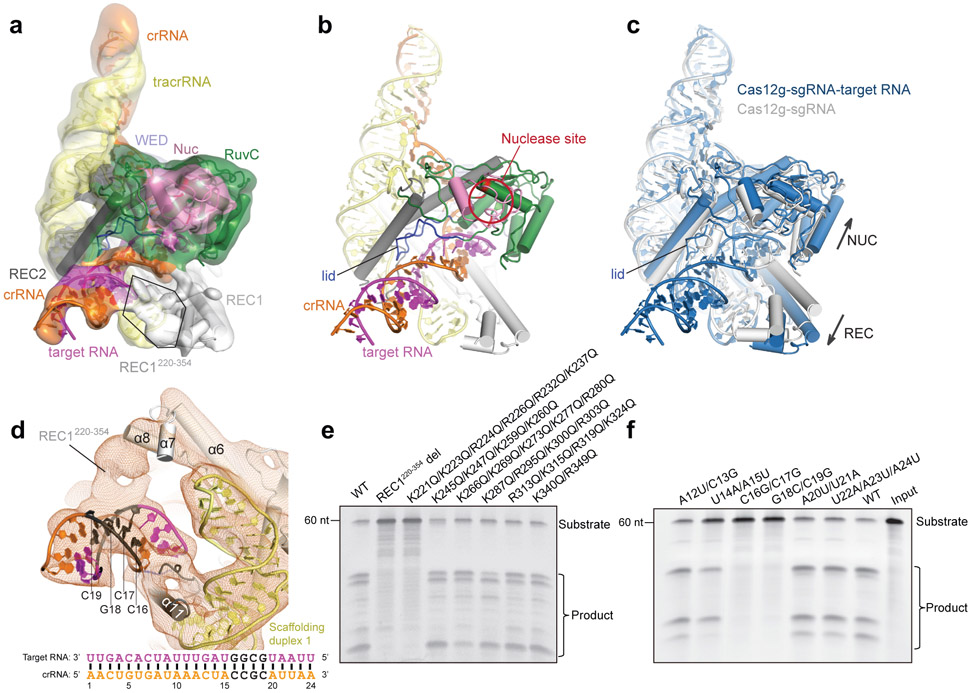
Figure 1. Overall structure of the Cas12g-sgRNA binary complex.
a, Domain organization of Cas12g. b, Cryo-EM map of the Cas12g-sgRNA complex in two views with each domain of Cas12g color-coded as in a. The crRNA and tracrRNA regions of sgRNA are colored in orange and light yellow, respectively. c, Atomic model of the Cas12g-sgRNA complex shown in cartoon.
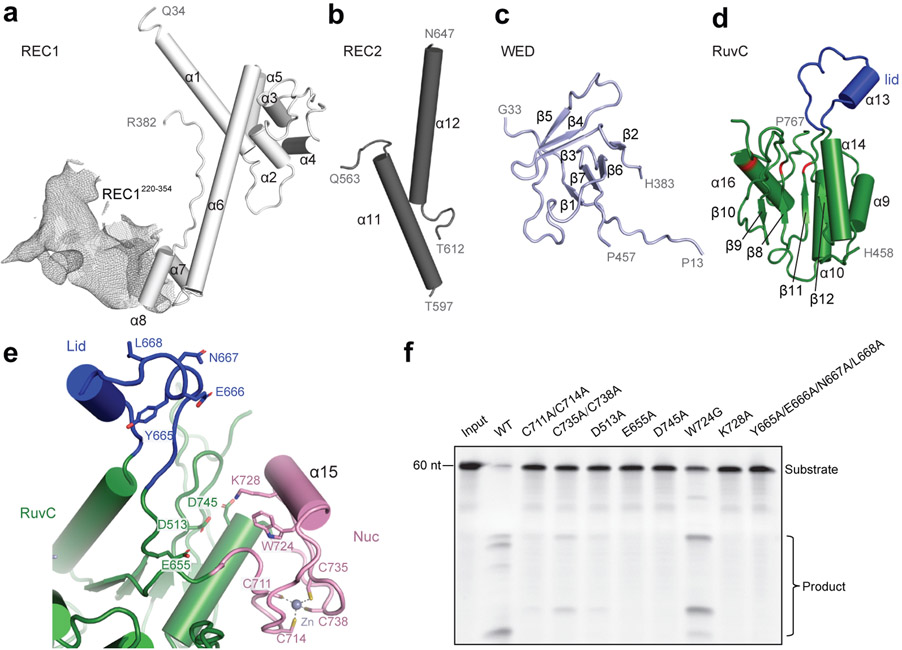
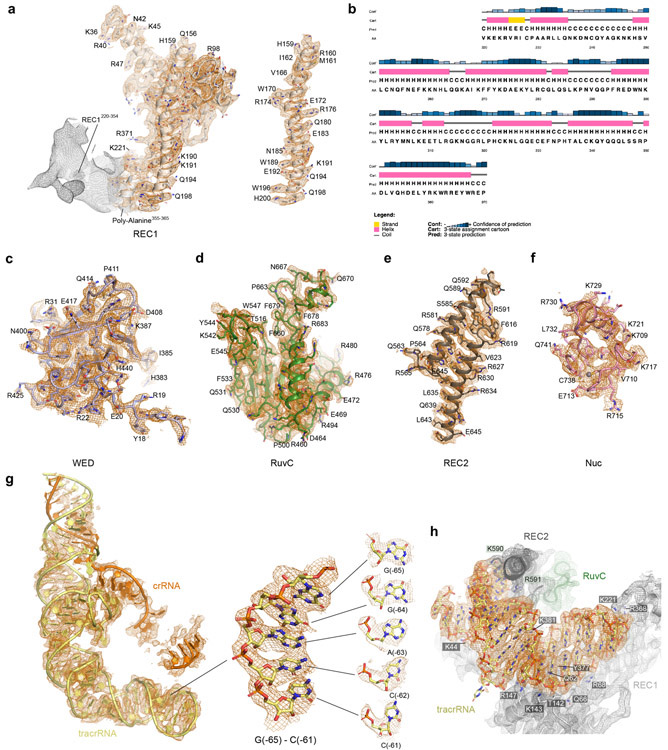
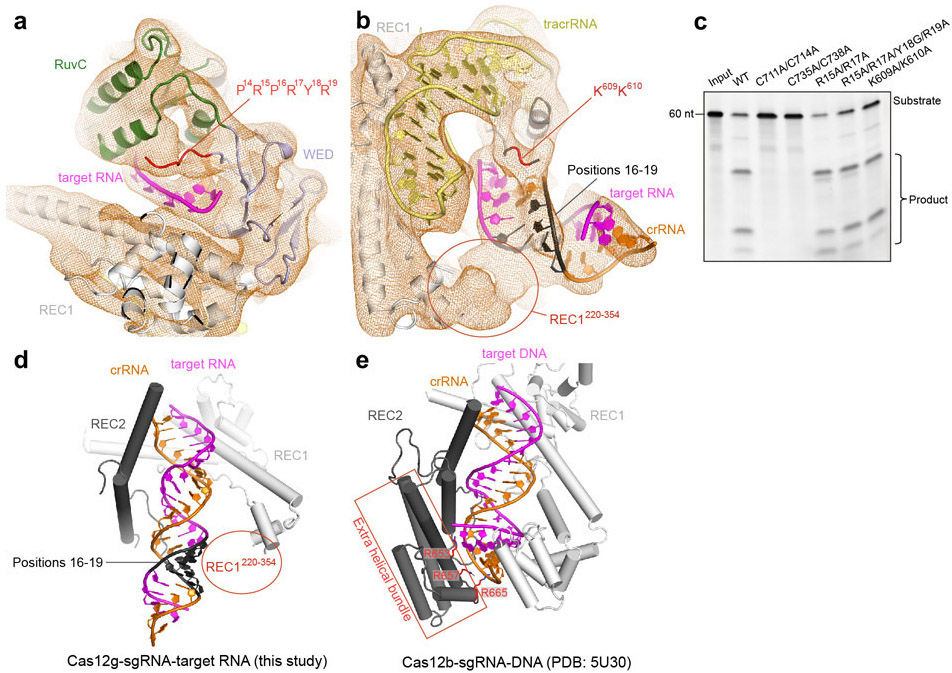
The dumbbell-shaped REC1 domain contains an N-terminal helical subdomain (α1-5) and two helices at the C-terminus (α7-8), connected by a featured long helix (α6) (Fig. 2a). A large portion of the C-terminus between α7 and α8 (a.a. 220-354, REC1220-354) is barely visible from the map (Figs. 1b and 2a), indicating that this region is flexible. Three-dimensional classification of cryo-EM images focusing on this region resulted in a major class with this region determined at a lower resolution (Extended Data Fig. 1g), showing that REC1220-354 is located on the exit of the central channel (Extended Data Fig. 1h). Secondary structure prediction suggests that this region is mainly composed of helices, similar to the structure of this region in Cas12b (Extended Data Figs. 2b and 3b). The REC2 domain is simplified into two helices, lacking a four-helix bundle as in Cas12b that is involved in the recognition of the crRNA-target DNA heteroduplex 40 (Fig. 2b and Extended Data Fig. 3c).
Figure 2. Domain structure and active site of Cas12g.
a-d, Cartoon presentation of the REC1 domain(a), REC2 domain(b), WED domain(c), and RuvC domain(d). Density of REC1220-354 observed by 3D focused classification is shown in mesh. The triplet of acidic residues in the active site and the lid motif colored in red and blue, respectively. e, The endonuclease center in the RuvC domain. The acidic residues D513, E655, and D745 are labeled. The Nuc domain is attached to the active center, with K728 and W724 pointing to the active site. A zinc ion chelated by four cysteines (C711, C714, C735 and C738) is present in the Nuc domain. f, Substrate RNA cleavage assay using wild-type Cas12g and Cas12g with mutations as indicated. The results shown are representative of three experiments.
The NUC lobe contains the WED, RuvC, and Nuc domains, but lacks the PI domain, consistent with the previous report that PAM is not required in substrate recognition of Cas12g 20. The WED domain consists of seven β-strands, folding into two β-sheet layers (Fig. 2c). The conserved RuvC domain is composed a five-β-strand (β8-12) core flanked by five α-helices, with α13 and surrounding loops (a.a. 659-679) equivalent to the lid motif of other Cas12 nucleases 36,43 (Fig. 2d). The lid motif in Cas12a is involved in substrate recognition, and conformational changes of the lid motif act as molecular checkpoints to activate catalysis 36,43. Attached to the RuvC domain is the Nuc domain, which is only 37 amino acids in length and contains two CXXC zinc finger motifs, 711-CAEC-714 and 735-CEGC-738 (Fig. 2e). Alanine substitutions of these cysteine residues abolished substrate RNA cleavage, suggesting the zinc finger motifs are critical for the nuclease activity of Cas12g (Fig. 2f). The zinc finger motifs were also observed in the Nuc domain of Cas12e 24, but not in Cas12a 28,31-39, Cas12b 40-42, and Cas12i 43 (Supplementary Table 2).
Nuclease site of Cas12g
Located in the interface between the RuvC and Nuc domains are a conserved triplet of acidic residues (D513, E655, and D745) from the RuvC domain and two bulky residues (W724 and K728) from the Nuc domain (Fig. 2e). Alanine substitutions of each of the triplet (D513A, E655A, and D745A) and K728 abolished substrate cleavage activity of Cas12g, while mutation of W724 (W724G) showed compromised nuclease activity (Fig. 2f). The lid motif in Cas12g lies above the active site (Fig. 2e). Substitution of key residues within the lid motif (665-YENL-668) with alanine abolished the substrate cleavage activity (Fig. 2f) as well as collateral cleavage activity towards ssDNA and ssRNA (Extended Data Fig. 4a,b), indicating that the lid motif plays a critical role in the activation of Cas12g, as observed in Cas12a 36. These results suggest that Cas12g utilizes a nuclease site at the interface between the RuvC and Nuc domains to perform multiple-turnover cleavage of RNA substrate (Extended Data Fig. 4c).
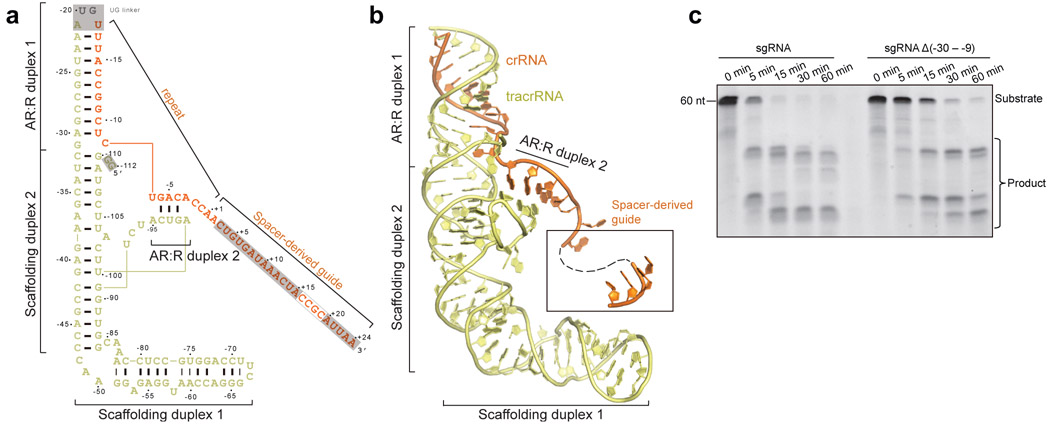
Structure of sgRNA
The sgRNA of Cas12g contains a 92-nt tracrRNA at the 5′ end, and a 42-nt crRNA at the 3′ end (18-nt repeat and 24-nt spacer-derived sequence) (Fig. 3a). The sgRNA folds into a flipped “F” shaped structure consisting of tracrRNA scaffolding duplexes 1-2 and anti-repeat:repeat (AR:R) duplexes 1-2 (Fig. 3a,b and Extended Data Fig. 2g). AR:R duplex 1 is extended from scaffolding duplex 2 with no observable contacts to Cas12g, consistent with its flexibility and compromised resolution in the cryo-EM map (Fig. 1b and Extended Data Fig. 1j). Lack of direct contact with Cas12g implies that the potential function of AR:R duplex 1 is to tether the tracrRNA and crRNA, rather than to directly contribute to the nuclease activity. Consistent with this hypothesis, truncation of the AR:R duplex 1 in the sgRNA presents comparable nuclease activity with the full-length sgRNA (Fig. 3c).
Figure 3. Overall structure of sgRNA.
a, Schematic of the sgRNA. Unmodeled regions are indicated by grey background. b, Structure of the sgRNA in the Cas12g-sgRNA complex in cartoon presentation. c, RNA substrate cleavage assay using full-length sgRNA and sgRNA with AR:R duplex 1 deleted. The results shown are representative of three experiments.
Interactions between sgRNA and Cas12g
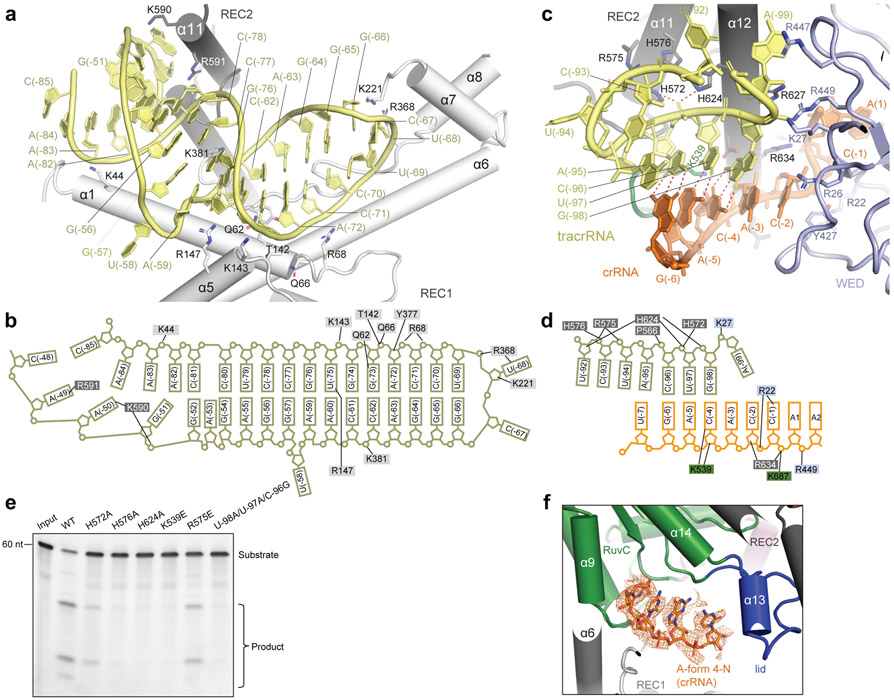
Scaffolding duplex 1, formed by tracrRNA (−82 to −51), is recognized by the concave surface of the REC1 domain and the REC2 domain mainly through non-sequence-specific interactions (Fig. 4a,b and Extended Data Fig. 2h), occupying a binding surface in REC1 that is used for recognition of the crRNA-target DNA duplex in other Cas12 effectors, such as Cas12b (Extended Data Fig. 5a-c). Scaffolding duplex 2, almost perpendicular to scaffolding duplex 1, is bracketed by the REC1, REC2 and WED domains (Fig. 1c).
Figure 4. Interactions between Cas12g and sgRNA.
a-b, Detailed interactions (a) between scaffolding duplex 1 and the REC lobe of Cas12g, and the schematic of the interactions (b). c-d, Detailed interactions (c) between the AR:R duplex 2 of sgRNA and Cas12g, and the schematic of the interactions (d). e, Substrate RNA cleavage assay using wild-type Cas12g and Cas12g with mutations in residues involved in interactions with the AR:R duplex 2, as well as mismatches in the AR:R duplex 2. The results shown are representative of three experiments. f, A four-nucleotide segment from the crRNA is pre-ordered in an A-form configuration, attached to the RuvC domain. EM density corresponding to the four-nucleotide segment is shown in mesh.
A stretch of tracrRNA (−99 to −92) protruding from the scaffolding duplex 2 base pairs with the crRNA repeat region (−6 to −4), forming the AR:R duplex 2 (Fig. 4c,d), which establishes extensive interactions with the REC2, RuvC and WED domains of Cas12g through charged interactions. For example, H572, R575, H576 and H624 of the REC2 domain and K539 of the RuvC domain establish charged interactions with multiple phosphate groups of the loop (Fig. 4c,d), stabilizing the AR:R duplex 2 and thereby facilitating the correct positioning of the crRNA guide sequence. Mutations of each of these residues (H572A, R575E, H576A, H624A and K539E), or introducing mismatches in the AR:R duplex 2 (U-98A/U-97A/C-96G) severely reduced the nuclease activity of Cas12g (Fig. 4e).
The spacer-derived guide inserts into the positively charged channel of Cas12g (Fig. 1b,c). However, the corresponding densities are very weak, indicating their flexibility (Extended Data Fig. 2g), with the exception of a four-nucleotide segment bound to the RuvC domain (Fig. 4f). This segment forms an A-form helical conformation at the exit of the central channel, reminiscent of Cas13a which is reported to adopt a “central seed” to initiate target recognition 44; however, the resolution of this region did not allow for unambiguous assignment of the crRNA sequence.
Structure of Cas12g-sgRNA-target RNA
To gain insights into target RNA recognition, we assembled a Cas12g-sgRNA-target RNA ternary complex by incubating a catalytically inactive Cas12g (E655A) with the sgRNA and a 24-nt target RNA. We determined the structure at 4.8 Å by cryo-EM (Fig. 5a,b, Extended Data Fig. 6 and Supplementary Table 1). Unambiguous secondary structure feature of the map allows fitting of individual domain structure into the map (cross correlation coefficient, 0.92). An A-form duplex is observed in the central channel of Cas12g, which is interpreted as the crRNA-target RNA heteroduplex (Fig. 5a,b). Modelling of the duplex showed a length of about 24 bp, consistent with the length of target RNA used for complex assembly. The crRNA-target RNA duplex is primarily attached to the RuvC, REC1 and REC2 domains (Fig. 5b).
Figure 5. Target RNA recognition of Cas12g.
a, Cryo-EM map and model of the Cas12g-sgRNA-target RNA complex with each domain of Cas12g color-coded as in Fig. 1a. The crRNA, tracrRNA, and target RNA are colored in orange, light yellow, and magenta, respectively. b, Cartoon presentation of the Cas12g-sgRNA-target RNA complex. c, Structural superimposition of Cas12g-sgRNA (grey) and Cas12g-sgRNA-target RNA (sky blue) complexes. Conformational changes in Cas12g domains and sgRNA after recognition of target RNA are indicated by arrows. d, Contact between REC1220-354 and positions 16-19 of the crRNA-target RNA duplex. Schematic of the crRNA spacer-derived guide and target RNA is shown below. e, Substrate RNA cleavage assay using wild-type Cas12g and Cas12g with REC1220-354 deleted or with group mutations within REC1220-354. f, Substrate RNA cleavage assay using wild-type and mutant target RNAs. The results in e and f are representatives of three experiments.
Structural comparison between Cas12g-sgRNA binary complex and Cas12g-sgRNA-target RNA ternary complex reveals significant conformational changes. The REC and NUC lobes move away from the central channel by approximately 10 Å and 3 Å, respectively, opening the central channel to accommodate the crRNA-target RNA duplex (Fig. 5c). The crRNA-target RNA duplex induces movement in the lid motif, which is presumably involved in the interactions with the duplex (Fig. 5c). Together with the mutagenesis study mentioned above (Fig. 2f), these evidences indicate that the lid motif plays a critical role in the activation of Cas12g. Different from the binary complex in which the REC1220-354 is located at the exit of the central channel, the REC1220-354 in the ternary complex is attached to the crRNA-RNA duplex (Fig. 5d and Extended Data Fig. 7a,b). Deletion of REC1220-354 abolished the nuclease activity, indicating an important role in substrate cleavage (Fig. 5e). The REC1220-354 region is rich in positively charged residues, which are likely involved in the recognition of the crRNA-target RNA duplex. Consistent with this hypothesis, mutation of six positively charged residues within the first 20 amino acids of this region (K221, K223, R224, R226, R232, and K237) showed similar effect in RNA cleavage efficiency as deletion of REC1220-354 (Fig. 5e).
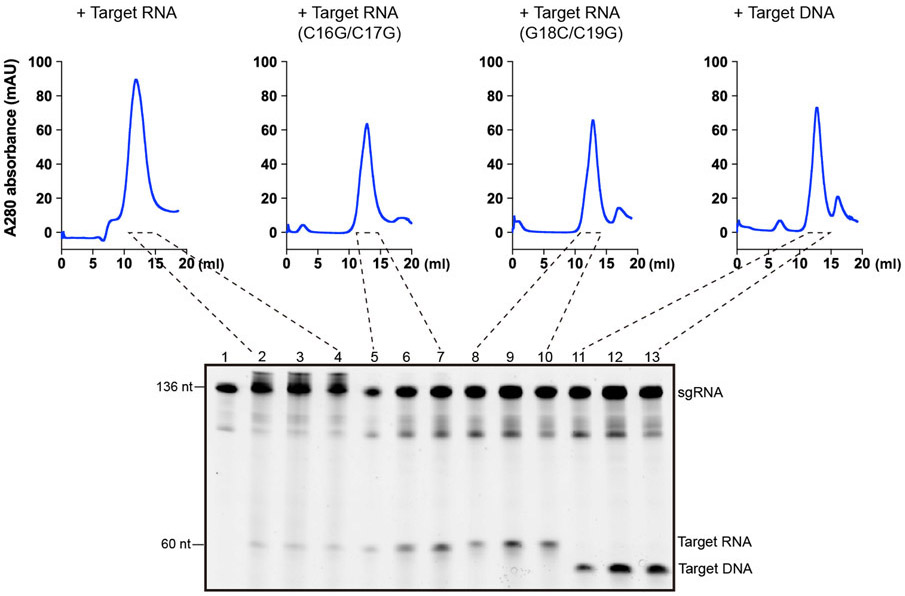
REC1220-354 engages the crRNA-target RNA duplex at positions 16-19. RNA substrate cleavage assay showed that mismatches between the crRNA and RNA substrate at positions 16-19 almost abolished substrate cleavage activity, while mismatches at flanking positions had little effects (Fig. 5f and Extended Data Fig. 5d), suggesting that recognition of the crRNA-target RNA duplex by REC1220-354 is critical for Cas12g activation. Mismatches at positions 16-19 also severely reduced the collateral cleavage activity towards ssDNA and ssRNA (Extended Data Fig. 4a,b). Substrate RNAs with mismatches at positions 16-19 are capable of binding to Cas12g-sgRNA (Extended Data Fig. 8), indicating that the base-pairing and stabilization of this region is vital for activation of Cas12g. Interestingly, target ssDNA is also able to bind to Cas12g-sgRNA (Extended Data Fig. 8). However, cryo-EM analysis suggests that target ssDNA failed to induce conformational changes in Cas12g as observed by target ssRNA (Extended Data Fig. 9), which might explain why ssDNA is incapable of activating Cas12g (discussed below). Collectively, the results provide mechanistic insights into target RNA recognition and activation of Cas12g.
Discussion
To date, 11 sub-types are reported (Cas12a-k) 45,46 within type V CRISPR-Cas systems. Those effectors are classified into three major branches based on phylogenetic analysis 20. Branches 1 (e.g. Cas12b 40-42) and 2 (e.g. Cas12a 31-36,38,39 and Cas12e 24) have been structurally studied previously. However, to our knowledge, the Cas12g structure reported here represents the first structure of branch 3 effector, shedding light on other members in this branch such as Cas12f (Cas14) 20.
The structures reported here suggest that Cas12g share similar domain architecture with several specialized features (Supplementary Table 2). The concave surface of the REC1 domain used for recognition of the crRNA-target DNA duplex in other Cas12 effectors is repurposed for recognition of tracrRNA scaffolding duplex 1. The C-terminal region of REC1, REC1220-354, is structurally flexible, in contrast to ordered structure of this region in other Cas12 effectors 24,31-36,38-42. However, this region plays critical regulatory role in the recognition of target RNA and activation of Cas12g.
Target recognition of type V and II systems are generally initiated by interactions between effectors and the PAM duplex of substrates. Recognition of PAM duplex triggers unwinding of DNA substrate, preparing the target strand for hybridization with the crRNA 36,47. Because Cas12g specifically recognizes ssRNA, it is not surprising that PI domain is not required. Target recognition in type VI effectors is initiated by protein-PFS (protospacer flanking sequence) interactions 44. However, bioinformatics analysis of target-flanking sequences of Cas12g revealed no such requirements for interference 20. No restrictions of PAM or PFS is an advantage of Cas12g when used for RNA-targeting applications.
In Cas12a, Cas12b and Cas12i, a short region (5-7 nt) at the 5′ end of the spacer-derived guide is pre-ordered in a helical conformation, serving as a seed for substrate recognition 37,40,41,43 (Supplementary Table 2). The seed sequence initiates substrate recognition and is sensitive to mismatches with the substrate. However, such pre-ordered segment is not observed at the 5′ end of the spacer-derived guide in the Cas12g-sgRNA complex. Instead, a four-nucleotide segment in an A-form configuration is observed at the exit of the central channel (Fig. 4e).
Most Cas12 nucleases, such as Cas12a, Cas12b, Cas12e, and Cas12i, specifically recognize target DNA. PAM recognition initiates unwinding of dsDNA and allow hybridization between the target strand and crRNA from the 5’ end of the crRNA guide. Activation of Cas12 nucleases requires the formation of the crRNA-target DNA heteroduplex at a certain length. For instance, activation of Cas12a requires the heteroduplex at least 16-17 bp 36,47, whereas activation of Cas12i requires at least 14-15 bp 43. Some anti-CRISPR proteins inhibit Cas12 nuclease by suppressing the progression of crRNA-target DNA heteroduplex, such as AcrVA4 which inhibits Cas12a by blocking the heteroduplex formation at 8 bp 39.
Cas12g specifically recognizes ssRNA. ssDNA with the sequence complementary to the crRNA is incapable of activating Cas12g 20 (Extended Data Fig. 4a,b). In the Cas12g-sgRNA complex, the 5’ end of the crRNA guide is embedded, whereas the 3’ end of the crRNA guide is exposed to solvent and a likely initiation site for hybridization between crRNA and target RNA (Fig. 1b,c). Cryo-EM analysis of the Cas12g-sgRNA-target DNA complex shows that the complex is in an inactivated conformation similar to the Cas12g-sgRNA binary complex (Extended Data Fig. 9g). A possible explanation is that hybridization between ssDNA and the 3’ end of the crRNA guide (presumably in a flexible conformation) failed to progress to a required length for activation of Cas12g (Extended Data Fig. 9h), although the possibility of ssDNA disassociation on cryo-EM grids cannot be completely ruled out. REC1220-354 and simplified REC2 domain are possible structural features associated with the RNA specificity of Cas12g (Extended Data Fig. 7).
It was recently shown that type V nucleases including Cas12a, Cas12b, Cas12e and Cas12i share a conserved activation mechanism 43. Before target DNA binding, the lid motif covers the RuvC active site, preventing access to the substrate. Formation of the crRNA-target DNA heteroduplex is accompanied with displacement of the lid motif, which senses formation of the crRNA-target DNA duplex. Displacement of the lid motif opens the RuvC active site and contributes to recruitment of substrate 43. Conformational changes observed in Cas12g upon target RNA binding and the critical role of the lid motif indicate that Cas12g may adopt a similar activation mechanism. Once activated by target RNA, both RNA and DNA substrates may be loaded to the RuvC active site in a favored geometry for catalysis. Future studies will be required to understand the catalytic mechanism of Cas12g towards both RNA and DNA substrates.
Methods
Protein expression and purification
The plasmid used for expressing full-length Cas12g with an N-terminal 6x His-tag was obtained from Addgene 20. The plasmid was transformed into BL21-CodonPlus (DE3)-RIL cells for protein expression in Terrifc Broth medium. The protein expression was induced by adding 0.5 mM IPTG (isopropyl β-D-thiogalactopyranoside) at 18 °C overnight. The cells were collected and then lysed by sonication in buffer A (25 mM Tris–HCl, 500 mM NaCl, 5% Glycerol, 1 mM PMSF and 5 mM β-mercaptoethanol, pH 8.0). After centrifugation, the supernatants were incubated with the Ni-NTA resin and washed extensively with buffer B (25 mM Tris–HCl, 500 mM NaCl, 30 mM imidazole and 5 mM β-mercaptoethanol, pH 8.0), and the His-tagged target proteins were eluted with buffer B supplemented with 300 mM imidazole. Subsequently, the target proteins were loaded into the Heparin column (GE) and eluted with a linear NaCl gradient, followed by size exclusion chromatography (Superdex 200, GE) in buffer C (25 mM Tris–HCl, 150 mM NaCl, 2 mM DTT and 0.1 mM MgCl2, pH 8.0). Peak fractions were pooled, concentrated and stocked at −80 °C.
To assemble the Cas12g-sgRNA complex, the sgRNA was incubated with Cas12g proteins at a ratio of 1.3:1 at 37°C for 30 mins. To reconstitute the Cas12g-sgRNA-target RNA complex, the Cas12g E655A mutant protein was incubated with sgRNA and target RNA or target DNA at a ratio of 1:1.3:5 at 37°C for 60 mins. These complexes were loaded onto a Superdex 200 (GE) column equilibrated with buffer C for further purification.
Mutagenesis
Single site mutations were introduced by QuikChange site-directed mutagenesis method. Group mutations containing multiple sites were introduced by ligating inverse PCR-amplified backbone with mutations-bearing DNA oligo via In-Fusion Cloning Kit. Large fragment deletion (REC1 220-354 deletion) was introduced by ligating inverse PCR-amplified backbone with DNA oligo encoding a flexible linker (“SGASGGAGS”). All mutants were confirmed by DNA sequencing. The mutant proteins were expressed and purified as described for wild-type.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectra of wild-type and mutants of Cas12g were measured at room temperature with a Chirascan (Applied Photophysics) spectro-polarimeter using a quartz cuvette with a 1 mm path length. The samples (1.3 mg/ml) were measured in low-salt buffer (20 mM Tris-HCl, pH 8.0, 50 mM NaCl). The measurements were recorded from 260 – 185 nm wavelengths (0.2 nm/step) with a total of three scans for each. The spectra were corrected by subtracting the spectrum from the buffer background.
In vitro transcription and purification of RNA
The DNA template was synthesized from IDT and amplified by PCR. In vitro transcription was carried out using the HiScribe T7 High Yield RNA synthesis kit (NEB) according to manufacturer's protocol. The transcribed RNA was loaded onto to the Resource-Q column (GE) and eluted with a linear sodium chloride gradient (50 mM-1000 mM) in 25 mM Tris–HCl, pH 8.0. The eluted RNA was concentrated and stocked at −80 °C.
In vitro RNA cleavage assay
Cas12g proteins (290 ng) purified from gel-filtration were pre-mixed with sgRNA at 37°C for 30 mins at a molar ratio of 4:1 in buffer C. Target RNA (160 ng; sequence: CUCAUGUUUGACAGCUUAUCAUCGAUAAGCUUUAAUGCGGUAGUUUAUCACAGUUAAAUU) synthesized from IDT was then added into the mixture for another 30 minutes unless otherwise specified. The reaction was quenched by adding EDTA and proteinase K. The products were denatured with TBE-Urea loading buffer (Thermo Fisher Scientific) at 66 °C for 10 mins, and analyzed on a 15% TBE-urea gel and stained using SYBR Gold (Invitrogen).
For stoichiometric titration assay for the turnover studies, the Cas12g-sgRNA complex were incubated with target RNA at different molar ratio at 37°C for 30 minutes. The experiment was done in triplicates. The resulting gels were scanned in grey mode and band intensities were measured by ImageJ 48.
Collateral cleavage assay
Collateral cleavage reactions were initiated by mixing a 267nt collateral ssRNA (215 ng. Sequence: GGAUAUUAAUAGCGCCGCAAUUCAUGCUGCUUGCAGCCUCUGAAUUUUGUUAAAUGAGGGUUAGUUUGACUGUAUAAAUACAGUCUUGCUUUCUGACCCUGGUAGCUGCUCACCCUGAUGCUGCUGUCAAUAGACAGGAUAGGUGCGCUCCCAGCAAUAAGGGCGCGGAUGUACUGCUGUAGUGGCUACUGAAUCACCCCCGAUCAAGGGGGAACCCUCCAAAAGGUGGGUUGAAAGGAGAAGUCAUUUAAUAAGGCCACUGUUAAA) or M13 ssDNA (315 ng, New England Biolabs) and different targets with the binary complex (wild-type or lid mutant) at 42°C for 60 minutes in buffer C. The reaction was quenched by adding EDTA and proteinase K. The collateral products were resolved on Urea-PAGE (15%) or 0.7% agarose gel and were visualized by SYBR® Gold staining (Invitrogen)
Size Exclusion Chromatography
The pre-assembled binary Cas12g-sgRNA complex was incubated with target DNA, target RNA and target RNA with mismatches with molar ratios of 1:1.15. The resulting mixture was incubated at 37°C for 30 minutes before applied to a Superdex 200 (GE) column which was equilibrated with buffer C. Urea-PAGE (15%) was used to analyze and monitor the complex formation.
Electron Microscopy
Aliquots of 3 μL samples (Cas12g-sgRNA and Cas12gE655A-sgRNA-target RNA) at different concentrations (0.3 mg/ml, and 0.5 mg/ml, respectively) were applied to glow-discharged Quantifoil holey carbon girds (Cu, R1.2/1.3, 400 mesh). The grids were blotted for 4 secs and plunged into liquid ethane with a Gatan Cryoplunge 3 plunger. Cryo-EM data were collected with a Titan Krios microscope (FEI) running at 300 kV and images were collected using Leginon 49 at a nominal magnification of 81,000x (with a calibrated pixel size of 1.05 Å/pixel) with a defocus range from 1.2 μm to 2.5 μm. The images were collected with a K3 summit electron direct detector in super-resolution mode together with a GIF-Quantum energy filter working at a slit width of 20 eV. A dose rate of 20 electrons per pixel per second and an exposure time of 3.12 seconds were used, generating 40 movie frames with a total dose of ~ 54 electrons per Å2. Appion 50 was used to do quick motion correction and CTF (contrast transfer function) estimation to monitor the data quality during data collection. A total of 2520 and 987 movie stacks were collected for Cas12g-sgRNA and Cas12gE655A-sgRNA-target RNA, respectively (Supplementary Table 1).
Image Processing
The movie frames were imported to RELION-3 51, and then aligned using MotionCor2 52 with a binning factor of 2. Contrast transfer function (CTF) parameters of the motion-corrected images were estimated using Gctf 53. For particle picking, 2D averages were first generated based on a few thousand particles picked without template, and then used for subsequent template-based auto-picking.
For Cas12g-sgRNA complex, particles were firstly auto-picked and extracted from the dose weighted micrographs. The dataset was split into batches for 2D classifications, which were used to remove false and bad particles that were assigned into 2D averages with poor features. Particles from different 2D averages were used to create initial model in cryoSPARC 54. The dataset was split into four parts for 3D classification. From 3D classification, three major classes can be discerned. Class I adopts a rigid structure with clear structural features, while classes II and III are with poor structural features (Extended Data Fig. 1e). Particles in Class I were selected for 3D refinement, converging at 3.4 Å resolution. After Bayesian polishing, another round of 3D classification was performed to exclude bad particles. Resulting dataset with 469,157 particles were used for CTF refinement and 3D refinement, converging at resolution of 3.1 Å. Focused 3D classification around REC1 domain was further performed without alignment resulted in a class with this region observed from ~ 10% of particles.
For Cas12gE655A-sgRNA-target RNA dataset, similar strategy was applied. 3D refinement was performed against the final dataset containing 38,238 particles, converging at 4.8 Å resolution (Extended Data Fig. 6). Cryo-EM image processing is summarized in Supplementary Table 1. For Cas12gE655A-sgRNA-target DNA dataset, 3D refinement was performed against 53,612 particles, converging at 5.8 Å resolution (Extended Data Fig. 9).
Model building, refinement and validation
De novo model building of the Cas12g-sgRNA structure were performed manually in COOT 55. Secondary structure predictions by PSIPRED 56 and the structure of Cas12b (PDB: 5U34) were used to assist manual building. Refinement of the structure models against corresponding maps were performed using phenix.real_space_refine tool in Phenix 57. For the Cas12gE655A-sgRNA-target RNA complex, the structure model of the Cas12g-sgRNA binary complex was fitted into the cryo-EM map, and each domain was manually adjusted in COOT, and the crRNA-target RNA duplex was built as a standard A-form double helix. The resultant model was refined against the corresponding cryo-EM map using phenix.real_space_refine tool in Phenix. 3D FSC analysis for the presented maps were performed using the Remote 3DFSC Processing Server (https://3dfsc.salk.edu/upload/). Map-to-model FSC analysis was performed using phenix.validation_cryoem tool in Phenix.
Sequence alignment and structural visualization
PROMALS3D 58 program was used to align the Cas12g and Cas12b sequences based on structures. Figures were prepared using PyMOL and UCSF Chimera 59.
Data availability
Cryo-EM reconstructions of Cas12g-sgRNA and Cas12g-sgRNA-target RNA complexes have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-22257 and EMD-22258, respectively. Coordinates for atomic models of Cas12g-sgRNA and Cas12g-sgRNA-target RNA complexes have been deposited in the Protein Data Bank under the accession numbers 6XMF and 6XMG, respectively. Source data are provided with this paper.
Extended Data
Extended Data Fig.1. Sample preparation and cryo-EM of the Cas12g-sgRNA binary complex.
a, Purification of Cas12g. Upper: Size exclusion chromatography (SEC) profile. UV absorbance curves at 280 nm and 260 nm are shown in blue and red, respectively. Lower: SDS-PAGE analysis. b, Circular dichroism (CD) spectra of wild-type and mutants of Cas12g used in this study. c, A representative raw cryo-EM micrograph of the Cas12g-sgRNA complex from a total of 2520 micrographs. d, Representative 2D class averages from a total of 100 images. e, Three major classes from 3D classification. f, 3D refinement for Class I with angular distribution of particles. g, Focused classification around the flexible region of the REC1 domain (REC1220-354). h, Cryo-EM map of class I with each domain of Cas12g and sgRNA colored coded. i, Plot of the global half-map FSC (solid red line), map-to-model FSC (solid orange line), and spread of directional resolution values (±1σ from mean, green dotted lines; the blue bars indicate a histogram of 100 such values evenly sampled over the 3D FSC). FSC plot for the reconstruction suggests an average resolution of 3.1 Å. j, Local resolution map for the reconstruction in f.
Extended Data Fig. 2. Detailed cryo-EM density map of the Cas12g-sgRNA complex with atomic model fitted in.
a, Fitting of the REC1 domain, with density of REC1220-354 from focused classification shown in grey mesh. A representative α-helix from the REC1 domain is shown in detail on the right. b, Secondary structure prediction of REC1220-354 using PSIPRED. c, Fitting of the WED domain. d, Fitting of the RuvC domain. e, Fitting of the REC2 domain. f, Fitting of the Nuc domain. g, Fitting of nucleic acids to the corresponding cryo-EM map, with details of a segment shown as indicated on the right. The atomic models are shown in stick with crRNA and tracrRNA regions colored in orange and yellow, respectively. h, Detailed interactions between scaffolding duplex 1 and Cas12g. Cryo-EM density map is shown in mesh.
Extended Data Fig. 3. Structural comparison between Cas12g and Cas12b.
a, Overall structures of the Cas12g-sgRNA and the Cas12b-sgRNA complexes. b-f, Structural comparison of each domain. The structures are aligned by secondary-structure matching (SSM) in COOT.
Extended Data Fig. 4. Collateral cleavage and stoichiometric titration assays for Cas12g.
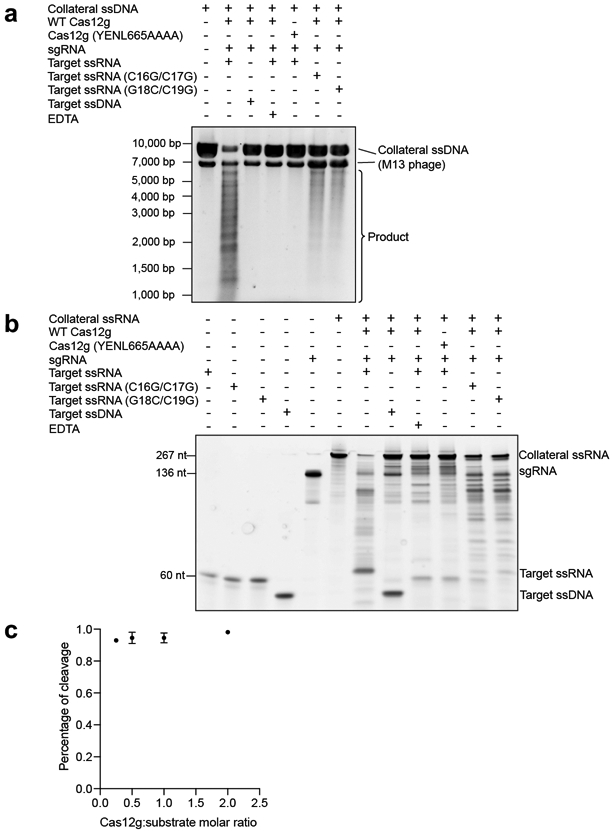
a, Collateral cleavage of unrelated ssDNA by Cas12g. The results shown are representative of three experiments. b, Collateral cleavage of unrelated ssRNA by Cas12g. The results shown are representative of three experiments. c, Stoichiometric titration assay indicates that Cas12g is a multiple turnover enzyme towards RNA substrate. Each point represents the cleavage ratio after 30 minutes at 37°C. Error bars represent mean ± SD, where n = 3 replicates.
Extended Data Fig. 5. Comparison of the REC1 domain in Cas12g and Cas12b, and crRNA guide mismatch assay for Cas12g.
a, The tracrRNA scaffolding duplex 1 is bound to the concave surface of the REC1 domain in the Cas12g-sgRNA binary complex. b, The crRNA-target DNA duplex is bound to the concave surface of the REC1 domain in the Cas12b-sgRNA-DNA complex. c, The tracrRNA scaffolding duplex 1 is bound to the concave surface of the REC1 domain in the Cas12g-sgRNA-target RNA complex. d, RNA cleavage assay using wild-type and mutant target RNAs. The results shown are representative of three experiments.
Extended Data Fig. 6. Cryo-EM data processing for the Cas12g-sgRNA-target RNA complex.
a, A representative raw cryo-EM micrograph of the Cas12g-sgRNA-target RNA complex from a total of 987 micrographs. b, Representative 2D class averages from a total of 100 images. c, 3D classification. Three major classes were observed. Classes II and III did not result in good reconstructions and likely represent damaged particles. d, 3D refinement for Class I. Angular distribution is shown on the right. e, Plot of the global half-map FSC (solid red line), map-to-model FSC (solid orange line), and spread of directional resolution values (±1σ from mean, green dotted lines; the blue bars indicate a histogram of 100 such values evenly sampled over the 3D FSC). FSC plot for the reconstruction suggests an average resolution of 4.8 Å. f, Local resolution map in two views.
Extended Data Fig. 7. Structural and biochemical analysis of the Cas12g-sgRNA-target RNA complex.
a-b, Structural elements involved in crRNA-target RNA duplex recognition. Three such structural elements are indicated by red lines, including 1) an N-terminal extension enriched in arginines within the WED domain, 2) a loop within the REC2 domain (608-AKKATLQP-615), and 3) REC1220-354. c, Substrate RNA cleavage assay using wild-type Cas12g and Cas12g with mutations in two loops involved in the recognition of the crRNA-target RNA duplex. The results shown are representative of three experiments. d-e, Comparison of the REC1 and REC2 domains for substrate recognition between Cas12g and Cas12b.
Extended Data Fig. 8. Binding assay between the Cas12g-sgRNA complex and target nucleic acids.
Upper: SEC profiles for the Cas12g-sgRNA complex incubated with different targets including target RNA, target RNA with mismatches at positions 16 and 17, target RNA with mismatches at positions 18 and 19, and target ssDNA. UV absorbance curves at 280 nm are shown. Lower: Urea-PAGE (15%) analysis of the elution fractions from SEC as indicated. The results shown are representative of three experiments.
Extended Data Fig. 9. Cryo-EM analysis for the Cas12g-sgRNA-target DNA complex.
a, A representative raw cryo-EM micrograph of the Cas12g-sgRNA-target DNA complex from a total of 1083 micrographs. b, Representative 2D class averages from a total of 100 images. c, 3D classification. Classes 2-6 did not result in good reconstructions and likely represent damaged particles. d, 3D refinement for Class 1 with angular distribution. e, Plot of the global half-map FSC (solid red line), map-to-model FSC (solid orange line), and spread of directional resolution values (±1σ from mean, green dotted lines; the blue bars indicate a histogram of 100 such values evenly sampled over the 3D FSC). FSC plot for the reconstruction suggests an average resolution of 5.8 Å. f, Local resolution map. g, Comparison of the rigid-body fitting between the Cas12g-sgRNA complex and the Cas12g-sgRNA-target RNA complex. Red squares indicate the fitting of the REC1 domain, which shows that the model of the Cas12g-sgRNA complex fits better to the map (left panel). h, Schematic model for Cas12g activation. Target DNA fails to form fully assembled duplex required for Cas12g activation.
Supplementary Material
Acknowledgments
We thank Thomas Klose and Valorie Bowman for help with cryo-EM, and Steven Wilson for computation. This work made use of the Purdue Cryo-EM Facility. This work was supported by the NIH grant R01GM138675 and a Showalter Trust Research Award to L.C.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Sorek R, Lawrence CM & Wiedenheft B CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem 82, 237–266 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Marraffini LA CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Jiang F & Doudna JA CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys 46, 505–529 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hille F et al. The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Shmakov S et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarova KS et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol 13, 722–736 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koonin EV, Makarova KS & Zhang F Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol 37, 67–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrangou R & Doudna JA Applications of CRISPR technologies in research and beyond. Nat. Biotechnol 34, 933–941 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Komor AC, Badran AH & Liu DR CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox DBT et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abudayyeh OO et al. RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terns MP CRISPR-Based Technologies: Impact of RNA-Targeting Systems. Mol. Cell 72, 404–412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dugar G et al. CRISPR RNA-Dependent Binding and Cleavage of Endogenous RNAs by the Campylobacter jejuni Cas9. Mol. Cell 69, 893–905 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strutt SC, Torrez RM, Kaya E, Negrete OA & Doudna JA RNA-dependent RNA targeting by CRISPR-Cas9. eLife 7, e32724 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rousseau BA, Hou Z, Gramelspacher MJ & Zhang Y Programmable RNA Cleavage and Recognition by a Natural CRISPR-Cas9 System from Neisseria meningitidis. Mol. Cell 69, 906–914 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connell MR et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan WX et al. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 70, 327–339 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smargon AA et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 65, 618–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abudayyeh OO et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan WX et al. Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zetsche B et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strecker J et al. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun 10, 212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng F et al. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 4, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JJ et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JS et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S-Y et al. CRISPR-Cas12a has both cis-and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gootenberg JS et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swarts DC & Jinek M Mechanistic Insights into the cis- and trans-Acting DNase Activities of Cas12a. Mol. Cell 73, 589–600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connell MR Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J. Mol. Biol 431, 66–87 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Konermann S et al. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173, 665–676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong D et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature 532, 522–526 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Gao P, Yang H, Rajashankar KR, Huang Z & Patel DJ Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 26, 901–913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamano T et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 165, 949–962 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamano T et al. Structural basis for the canonical and non-canonical PAM recognition by CRISPR-Cpf1. Mol. Cell 67, 633–645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stella S, Alcón P & Montoya G Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage. Nature 546, 559–563 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Stella S et al. Conformational Activation Promotes CRISPR-Cas12a Catalysis and Resetting of the Endonuclease Activity. Cell 175, 1856–1871 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Swarts DC, van der Oost J & Jinek M Structural Basis for Guide RNA Processing and Seed-Dependent DNA Targeting by CRISPR-Cas12a. Mol. Cell 66, 221–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimasu H et al. Structural basis for the altered PAM recognition by engineered CRISPR-Cpf1. Mol. Cell 67, 139–147 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H et al. Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins. Cell Host Microbe 25, 815–826 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Yang H, Gao P, Rajashankar KR & Patel DJ PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease. Cell 167, 1814–1828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L et al. C2c1-sgRNA Complex Structure Reveals RNA-Guided DNA Cleavage Mechanism. Mol. Cell 65, 310–322 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Wu D, Guan X, Zhu Y, Ren K & Huang Z Structural basis of stringent PAM recognition by CRISPR-C2c1 in complex with sgRNA. Cell Res. 27, 705–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Li Z, Xiao R & Chang L Mechanisms for target recognition and cleavage by the Cas12i RNA-guided endonuclease. Nat. Struct. Mol. Biol 27, 1069–1076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu L et al. The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a. Cell 170, 714–726 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Swarts DC Stirring Up the Type V Alphabet Soup. CRISPR J 2, 14–16 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Strecker J et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeon Y et al. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun 9, 2777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suloway C et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Lander GC et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol 166, 95–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buchan DWA & Jones DT The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. Struct. Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pei J, Kim BH & Grishin NV PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM reconstructions of Cas12g-sgRNA and Cas12g-sgRNA-target RNA complexes have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-22257 and EMD-22258, respectively. Coordinates for atomic models of Cas12g-sgRNA and Cas12g-sgRNA-target RNA complexes have been deposited in the Protein Data Bank under the accession numbers 6XMF and 6XMG, respectively. Source data are provided with this paper.