Abstract
Background: Due to the migratory flow of infected people with severe acute respiratory syndrome virus (SARS COV-2), the number of confirmed cases of coronavirus disease 2019 (COVID-19) is accelerating globally; preclinical evidence of antiviral agents that can combat this pandemic is still elusive. We identified published articles on SARS-COV efficacy experiments in which some selected compounds were used to test the reduction of the virus load in mice.
Methods: A systematic search of articles was conducted in PubMed, Web of Science, and Scopus. We then developed a combined model based on a systematic review, meta-analyses, and molecular docking studies to evaluate the effect size of preclinical studies of compounds that have been tested against SARS-COV. Because substantial heterogeneity was expected, random effect model meta-analyses were carried out to estimate the overall pooled disease’s prevalence. All meta-analyses were performed with Stata version 15.0. Subgroup analyses on therapies were conducted as well. Molecular docking studies of the inhibitors in the active pocket of COVID-19 protease were also performed.
Results: From all screened articles, six studies were appropriate for ultimate meta-analysis and systematic review. The residual amount of heterogeneity was high (τ2 =0.02; heterogeneity I2 =85.5 % with heterogeneity chi-square =103.57, a degree of freedom =15, and p <0.001). The overall random pooled prevalence of infected mice treated with the selected compounds was 78.1 % [95 % Confidence Interval (CI): 14.7-17.0 %]. Prophylactic has a significantly higher pooled prevalence than therapeutic, with 21.8 % (95 % CI: 16.4 % to 28.8 %). Our results indicated that most of the SARS-COV inhibitors analyzed were less effective in reducing the lung virus titer of SARS-COV infection in animal models. The findings from molecular docking studies also identified COVID-19 inhibitors that are good for optimization and drug development to fight against COVID-19 infection.
Conclusions: Findings from the review showed that studies on the preclinical compounds targeting SARS-COV and COVID-19 are limited. Furthermore, molecular docking studies and meta-analysis results substantiated three compounds, i.e., EIDD-2801, GS-5734, and amodiaquine. HIPPOKRATIA 2020, 24(3): 99-106.
Keywords: Coronavirus disease 2019, COVID-19, systematic review, meta-analysis, molecular docking, preclinical, severe acute respiratory syndrome coronavirus 2, SARS-COV-2
Introduction
The coronavirus disease originated in 2019 (also referred to as COVID-19) is an emerging infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). The global pandemic disease which sprung forth from Wuhan City, in the Hubei Province of China, was first reported by the World Health Organization (WHO) on December 31, 2019. Globally, the estimate of SARS-COV-2 on April 7, 2020, indicated that ~1.4 million people were infected, and 75,933 deaths had been recorded. Meanwhile, the migratory flow of infected people has accelerated the spread of the virus, especially in the United States of America, Spain, Italy and United Kingdom which led to mandatory isolation quarantine across the globe. Moreover, the outbreak of COVID-19 infection is not effectively controlled due to its strong capacity of transmission: i) human to human, i.e., proximity with infected people via respiratory droplets from cough or sneezing, ii) touching the surface or object contaminated with the virus and later on touching their mouth, nose, or eyes, and iii) aerosol transmission1-7. Consequently, the aftermath could potentially threaten worldwide health systems and negatively influence the global economy8-11. Heshui et al noted that older people and those with an underlying illness such as chronic pulmonary disease and diabetes are more susceptible to COVID 19 infection5. In another study, Ying and co-workers reported that many patients with underlying cardiovascular diseases (CVDs) might have an increased risk of death12. Additionally, the expert consensus statement released by Yen et al reviewed that no positive cases of newborns have been reported yet, and infected children show symptoms such as; fever, dry cough, and fatigue, and few have upper respiratory tract infection13. Due to a rapid increase in confirmed cases, COVID-19 has been considered a menace to humankind. Hence, an urgent need for drastic measures is required to design and develop an antiviral agent that can target COVID-19 so that treatment can be timely and expeditious while reducing the mortality rate12.
Severe acute respiratory syndrome virus (SARS-COV) and Middle East respiratory syndrome virus (MERS-COV) are in the same class of betacoronavirus alongside SARS-COV-2. SARS-COV main protease, 3CLpro, also referred to as Mpro, is a homodimer with three structural domains: domain I, II, and III and highly conserved among SARS-COV and MERS-COV, displaying 40-44 % sequence homology12,14-15. Whereas 3CLpro protease of SARS-COV-2 share over 95 % of sequence similarity with those of the SARS-COV. These two viruses demonstrated only 79 % sequence similarity at the genome level and similar active sites8,16-19. Also, 3CLpro plays a crucial role as functional intermediates involved in the replication and transcription of viral RNA, which makes it turn-out as the most potential attractive anti-coronavirus target20-22.
Unfortunately, at the time this review was conducted, there is only one approved vaccine to combat COVID-19. This has caused a global race for researchers and scientists to develop a useful, bioavailable antiviral agent with minimal side effects to fight COVID-19. To support the ongoing research and development efforts to discover effective agents that can target coronavirus. We used a combined method, including systematic review, meta-analyses, and molecular docking studies of selected preclinical (in vivo) compounds (Figure 1) targeting SARS-COV to identify active compounds with possibilities of combating COVID-19 infection8. Meta-analyses were used to calculate the effect size by exploring the experimental models’ main characteristics, virus strains, and the lung virus titer since clinical decisions should be based on the best evidence’s totality results of individual studies. Combining the in silico method, we also explored to what level the effect size attained for each compound could be explained by disparities in chemical structures, molecular interactions, and the rate of SARS-COV inhibition. Hence, meta-analyses on the preclinical studies and molecular docking studies were accomplished to analyze and obtain SARS-COV inhibitors that can be optimized to combat COVID-19 infection.
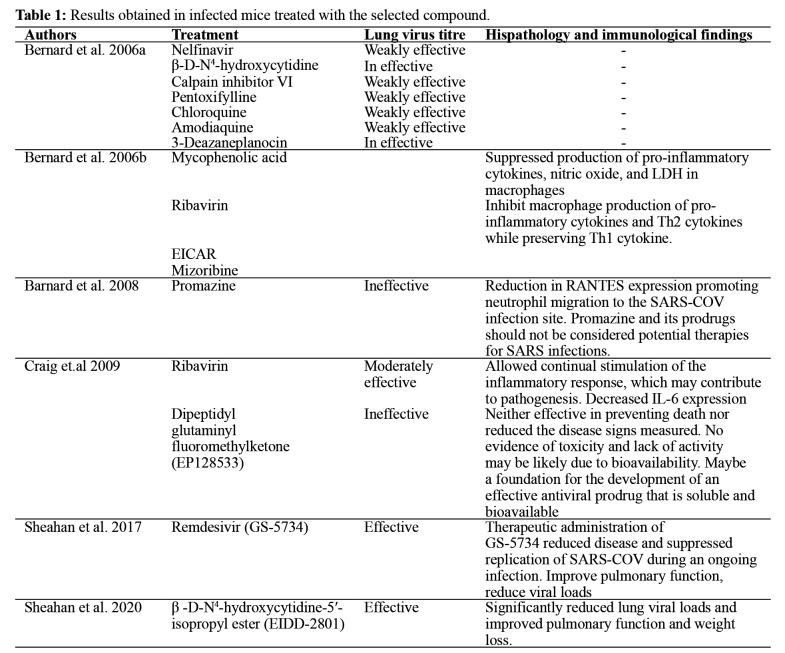
Figure 1. Structures of compounds evaluated for efficacy of SARS-COV in the animal models.
Materials and methods
Search Strategies
The search strategy was based on the methodological framework suggested by Moher and colleagues, namely, Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA)23. The meta-analysis of preclinical evidence of SAR-COV inhibitors was initiated with focus incline keywords using search words: ‘SARS-COV’, ‘SARS’, ‘COVID-19 OR COVID-19 infection’, ‘coronavirus’, ‘severe acute respiratory syndrome virus’, OR ‘in vivo’, ‘SARS-COV main protease’, ‘protease’, ‘preclinical evidence’, ‘animal model’, mice, ‘SARS-COV inhibitors’, ‘mouse’, and ‘BALB/c’. The search was carried out in three electronic databases: PubMed, Web of Science, and Scopus. The entire databases were searched until April 5, 2020, and all relevant studies were recovered and included in the systematic review and meta-analysis. The criteria ‘filtered results’ was applied to exclude conference proceedings, review articles, and book chapters.
Study Selection and Eligibility Criteria
In the systematic review and meta-analysis, we included published studies that are preclinical (in vivo) evidence of SARS-COV agents, regardless of the country. The included studies were published in an English language reputable journal. Since the number of articles on preclinical (in vivo) evidence on the SARS-COV agent is relatively small, no restrictions were made for publication dates. The exclusion criteria include i) no full-text available, ii) secondary studies (i.e., editorials, commentaries, letters to the editor and articles blog), iii) databased and epidemiological reports, iv) studies without control groups, v) experimental studies investigating in vitro and human systems, and vi) studies without SARS-COV strains.
Selections of the Studies
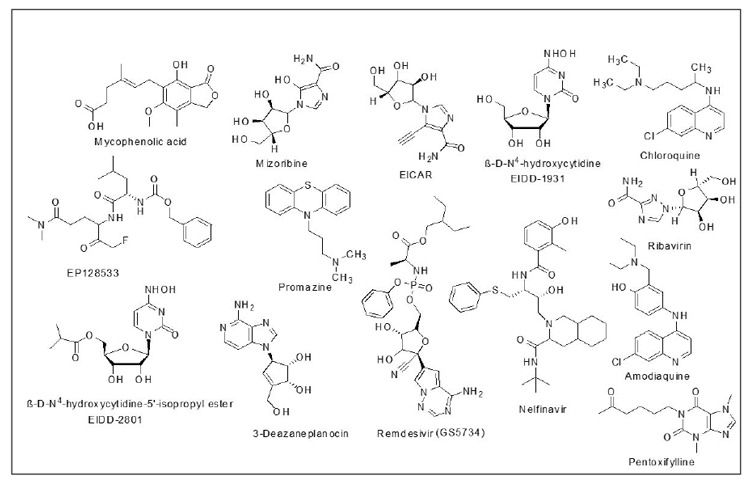
The study selection process is shown in Figure 2. All citations (n =225) identified through our search strategy were imported into EndNote version X7 (Clarivate Analytics, Philadelphia, PA, United States) reference management software and used the automated “Find Duplicates” function to exclude duplicates. The title and abstracts of the 225 articles were assessed by two reviewers (OE & REO).
Figure 2. Flow diagram of the database searches and inclusion/exclusion process.
Data Extraction
A standardized and pre-piloted checklist was used to extract the required information. Data were extracted on i) publication characteristics, ii) experimental design (animal models and disease models): species, linage, sex, age, and weight, virus species and strain, route of inhibitors administration, and iii) research outcomes (virus titer, histopathological, immunological maker, findings and mortality). Only the outcomes of the final tests were included in case they were performed at different points. Only the data of the highest dose of the inhibitors in the experimental group of animals were included. The mean and standard deviation (mean ± SD) were directly extracted when available in the tables or text, and values reported in graphics were extracted with an image analysis software program (Image-Pro Plus 4.5, Media Cybernetics, Bethesda, Maryland, USA).
Data Analysis and Synthesis
The data extracted were captured into an excel spreadsheet and later imported to Stata/IC software, version 15.0 (StataCorp LLC, College Station, Texas, USA) for further analysis. An essential statistical issue in meta-analysis is the handling of heterogeneity among studies. We evaluated heterogeneity with DerSimonian and Laird I2 statistics for each analysis category24. Funnel plots assessed the visual assessment of publication bias. A Begg-adjusted rank correlation test was also used to check for publication bias. If these tests showed non-significant heterogeneity, we used a fixed-effects model; otherwise, a more conservative random-effects model was used25. Because substantial heterogeneity was expected, a random-effects meta-analysis was done to estimate heterogeneity being taken from an inverse-variance model. Between studies, heterogeneity was assessed using I2, which estimated the percentage of total variation across studies and Cochran’s Q method. I2 values of 25 %, 50 %, and 75 % were considered low, moderate, and high heterogeneity, respectively24,26. According to the heterogeneity of studies, subgroup analysis was conducted and stratified the studies according to prophylactic and therapeutic therapies.
Molecular Docking
We performed molecular docking analysis to explore the SARS-COV agents’ binding pattern into the binding site of COVID-19 main protease. We retrieved the X-ray crystal structure of COVID-19 main protease (PDB ID: 6lu7) from RCSB Protein Data Bank27. We prepared the ligand and receptor complexes for docking using Autodock vina. We used the Chem3D Ultra to construct a three-dimensional structure for all the study compounds, followed by structure optimization. We used AutoDock Tools (V 1.5.6) to establish grid box parameters; we merged all non-polar hydrogens, added Gasteiger partial atomic charges, assigned and rotatable bonds28. The grid box of the complex had a grid spacing of 0.375 Å. The grid center and grid size were set: X =40/-20, Y =31/13, Z =65/47 Å. Ligand-receptor interaction of the ligands was after that simulated into the binding site of COVID-19 main protease. The resulting docking studies were validated by redocking the native ligand of the protein crystal structures into the binding site. Analysis and visualization of binding interactions in the protein-ligand complex were achieved using Discovery Studio Visualizer software developed by Accelrys (Dassault Systemes, BIOVIA Corp., San Diego, CA, USA).
Results
The literature search strategy included 225 articles. After removing the duplicates, 190 were excluded just by reading their titles. Of the remaining studies, four were excluded based on the abstract and the outcome variable. Hence, the remained six studies were included in the final analysis (Figure 2).
Characteristics features of the included studies
A total of six studies were included in systematic review and meta-analysis29-34. All studies originated from the United States of America (USA) and utilized mice and marmosets, but only the analyses with mice are considered. Besides, none of the studies reported the approach used to determine the sample size calculation utilized and the order in which the groups were treated. For the allocation of the administration usage: intraperitoneal (n =2, 33.3 %); intraperitoneal and intranasal (n =2, 33.3 %); oral (n =1, 16.6 %), and subcutaneous (n =1, 16.7 %).
Features of the animal model
As reported above, all the studies utilized mice as the animal model. The linage is distributed as follows: BALB/C (n =4, 66.7 %); C57B46J (n =1, 16.7 %), and both C57B46J and Ces/-/- (n =1, 16.7 %). Four of the studies (n =4) used female animals, and it represents 66.7 % of the entire studies while 33.3 % (n =2) of the studies used animals of both sexes. None of the studies utilized exclusively male animals. The animals’ age was reported in weeks, of which each category represented about 17 %. Three of the studies did not report the age of their animal. Meanwhile, the animal weight ranged from 11 to 18 g; animals with weight 11-16 g had the highest proportion (n =2, 33.3 %), while two of the studies did not report the weight of the mice. Urbani (n =3, 50.0 %); v2163 (n =1, 16.7 %), and MA15 (n =2, 33.3 %) were the SARS-COV strains finally used for the in vivo experiment.
Meta-analysis result
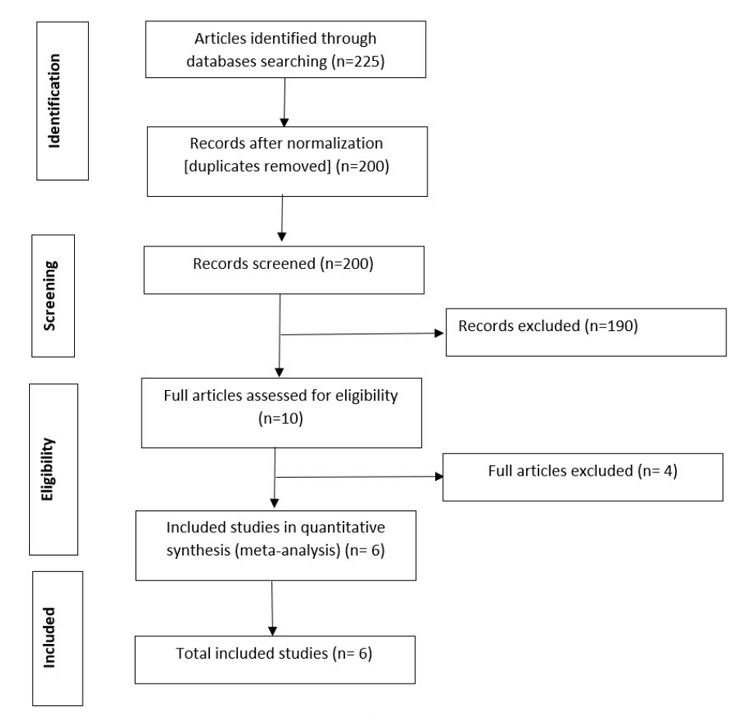
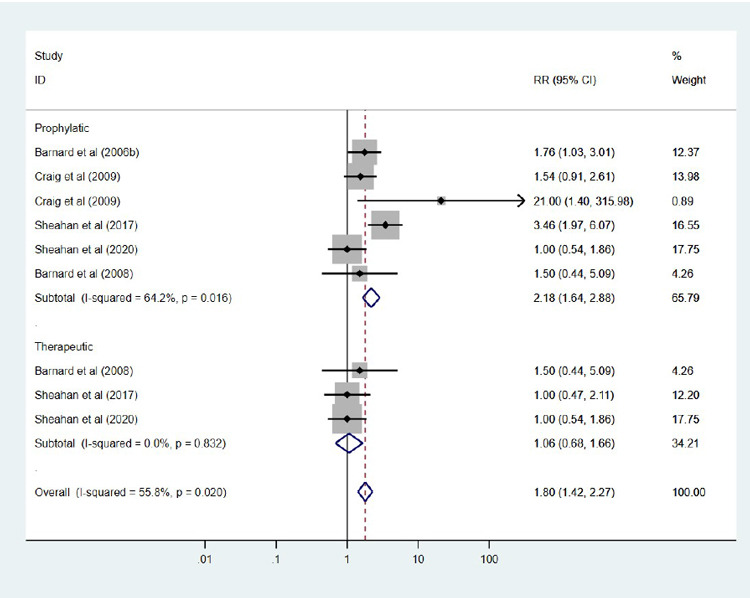
We use the standardized mean difference (SMD) for this meta-analysis. An SMD is adopted regularly in clinical meta-analyses due to its ability to express the difference between the groups relative to the standard deviation. It can be seen from the result that the test of heterogeneity yields Q =103.57, degree of freedom (df) =15, resulting in a p-value <0.0001. This indicates statistically significant heterogeneity among the sixteen compounds, which leads to the random-effects model. Using the random-effects model, the combined SMD =0.781 (95 % CI: 0.147, 1.595) with p-value <0.001, indicating a statistically significant difference between the compounds and control (placebo or vehicle) in terms of virus titer. Eleven out of the fifteen compounds showed a significant reduction in virus titer, indicating an effect in favor of treated animals compared to placebo (Figure 3). Subgroup analysis by study compounds was conducted to assess the potential heterogeneity between studies. Out of the two therapies, the highest estimated of mortality was found in studies with prophylactic [21.8 % (95 % CI: 16.4 % to 28.8 %), I2=64.2 %], followed by studies conducted with therapeutic, was 10.6 % (95 % CI: 14.2 %, 22.7 %), I2 =0.0 % (Figure 4). Hence, the overall mortality was reduced in mice treated with the compounds [RR: 1.80 (95 % CI: 1.42, 2.27), p =0.020].
Figure 3. Forest plot obtained from meta-analysis comparing the mean difference of virus titer in infected mice treated with the selected compounds and those untreated (control).
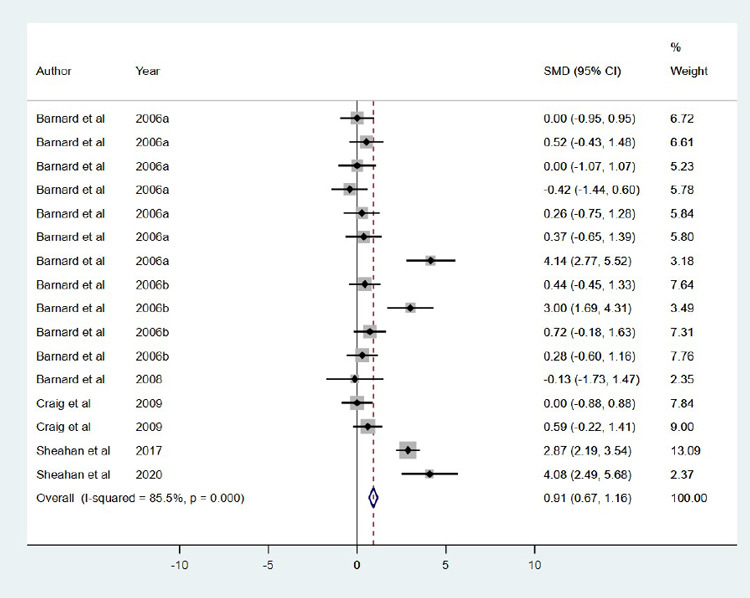
Figure 4. Forest plot obtained from meta-analysis comparing the mortality in infected mice treated with the selected compounds.
Discussion
SARS-COV emerged from a zoonotic reservoir and couple with cytokine, chemokine, and Interferon Stimulated Gene (ISG) responses in patients provided evidence that SARS-COV pathogenesis is partially controlled by innate immune signaling35-37. Dysregulation of inflammatory cytokines such as tumor necrosis factor (TNF)-α, interleukin (IL)-1β, IL-6, IL-10, polymorphonuclear neutrophil (PMN), and chemokine IL-8 are responsible for the indication of viral infection. These may be due to activation of transcription factor nuclear factor (NF)-κB, activator protein (AP)-1, and activating factor-2 (ATF-2)38-41. These can also be observed in SARS-COV-2. Thus, identifying specific molecules to destroy the immune evasion of SARS-COV-2 is very crucial. Systematic or scoping reviews of preclinical have served as a robust form of knowledge synthesis to evaluate transparently experimental therapies for more than a decade42.
Interestingly, this is the first paper using systematic review, meta-analyses, and molecular docking studies to identify SARS-COV preclinical (in vivo) compounds targeting COVID-19 main protease to the best of our ability. Further, the current meta-analysis aimed to estimate the pooled effect of compounds used to treat infected mice with SARS-COV. There was scarce data (in vivo) on infected mice with SARS-COV treated with active compounds. The results presented in this manuscript were from the analysis of data obtained through a systematic review of scientific publications of the efficacy of compounds used to treat infected mice with SARS-COV until April 5, 2020. The final quantitative and meta-analysis were done only on six articles that met the inclusion criteria. Our findings from the meta-analysis indicated the impacts of the selected compounds in reducing the virus titer. The treatment efficacy is represented by the midpoint and each segment’s length in the forest plot, while the diamond shape showed the combined effect (Figure 3). The random-effect meta-analysis result showed high variability with Higgin’s I2, which indicates that the variability between studies was not by chance alone. Because of the considerable variability between studies, the random effects meta-analysis weight of studies was nearly equal. This study’s findings affirmed a higher, statistically significant pooled effectiveness of compounds used to treat infected mice with SARS-COV. The effectiveness rate of infected mice was estimated to be 78.1 %. A possible explanation for this might be the differences attributed to the sample size, experimental models’ main characteristics, and the mice’s virus strains. The articles’ analysis in this domain showed that the effectiveness rate of infected mice treated with the selected compounds was higher in prophylactic than the therapeutic. This inference is reasonable since prophylactic is a preventive measure used to prevent the occurrence of disease. Nevertheless, further population-based research would help confirm the difference in the effectiveness of infected mice treated with the selected compounds against other infected mice treated based on different compounds examined in this study and appraise the factors that might lead to such differences. The histopathology and immunological outcomes of the selected compounds against the SARS-COV are detailed in Table 1 below.
Table 1. Results obtained in infected mice treated with the selected compound.
Understanding the similarity between the 3CLpro main protease of SARS-COV and COVID-19, which is very important in the viral life cycle, has highlighted the protease as an attractive potential target for anti-coronavirus drug development43-45. Besides, they both have catalytic dyad composed of conserved residues His41 and Cys145. Considering the explored compounds’ in vivo activity, their molecular affinity in the COVID-19 main protease’s binding pocket was evaluated. Only seven out of the compounds showed good binding energy. They bind with COVID-19 protease via multiple hydrogen bonding and hydrophobic contacts. Residues Phe140, 185 Leu141, His163, Met165, Glu166, and His172 form hydrophobic interactions with the ligands. While Asn142, Gly143, and Cys145 form intermolecular H-bond with the ligands. The binding mode indicates useful clues to the possible molecular basis of these compounds.
The oxygen atom of the primary hydroxyl functional group of nucleoside unit β-D-N4-hydroxycytidine (EIDD-1931; Binding Energy (B.E) = ‒6.5 kcal/mol) is H-bonded to Cys145 (bond distance =2.59 Å; bond angle 127.7 and 100.5o, respectively) (relevant Figure 1S can be found in the supplementary material: see Acknowledgement). Meanwhile, the hydrogen atom is H-bonded Ser144 and Le141 (bond distance (b.d) =2.73 and 2.68 Å; bond angle =129.1, 114.7, 127.7, and 100.5o, respectively). The conserved amino acid His41 is sandwiched between the pyrimidinone ring and the hydroxy amino group via H-bonding and hydrophobic interaction. The interactions of 3-Deazaneplanocin (B.E = ‒6.5 kcal/mol) with the COVID-19 main protease is described in detail in the supplementary material (Figure S2: see Acknowledgement). Interestingly, the cyclopentene unit of this compound interacts with Cys145 via H-bonding. In contrast, an oxygen atom of the primary hydroxy functional group on the cyclopentene ring formed three outstanding hydrogen bonds with a backbone amino acid residue Gly143 and 145 having the bond distance of 2.59 and 2.25 Å, respectively. The hydrogen atom formed H-bond interactions with Ser144 and Leu 141; these interactions were also observed in the docked complex of EP128533. Meanwhile, the amino acid Thr26 is interpolated between NH2 and NH functional groups of imidazo[4,5-c]pyridine moiety (supplementary material). The oxygen atoms on the carbonyl group of fluoromethyl ketone and dimethylamine (EP128533, B.E = -6.9 kcal/mol), dimethylamine forms an H-bond with the side-chain nitrogen of Gly143 and Thr26, the F atom formed interaction with Cys145 and Gly143 via H-bonding. The benzyl group in the unit formed interaction with the sulfur of Met165. Also, the carbamate group fits into a small hydrophobic pocket, which is favorable for π-σ and alkyl interaction between the methylene as well as methyl group and amino acid His41 and Met49, respectively, whereas the amine group H-bonded with Gln189. The increase in the inhibitor’s interaction in the active pocket of COVID-19 protease can be rationalized to be the reason for the increase in the inhibitor’s binding affinity (Figure S3: see Acknowledgement). The amodiaquine docked complex (Figure 5) was similar to that of EIDD-1931. Chloroquinoline group was inserted in the hydrophobic pocket consisting of Met165 and His41. The amino acid Cys145 was H-bonded to the benzyl ring and the tertiary hydroxyl functional group’s oxygen atom in the unit. The hydrogen atom formed H-bond, as shown in Figure 5: the first H-bond is from Leu141 (b.d =1.90 Å), the second is from Ser144 (b.d =2.53 Å). These two interactions in the binding pocket help in positioning and stabilizing the docked complex of amodiaquine. Furthermore, the interactions of nelfinavir (B.E = -8.2 kcal/mol) in the binding pocket of COVID-19 protease are shown in the supplementary material (Figure S4: see Acknowledgement). The phenylthiol was deeply inserted within the hydrophobic pocket and interacted to formed: π-π-T stacking contact with amino acid His41, π-alkyl with Met49, and π-sulfur with Met165. Moreover, the -CH3 group on hydroxymethylbenzamide interact with His41 and Leu27 via hydrophobic. In contrast, the carbonyl oxygen atom is involved in the H-bond network with Gly143. Moreover, Asn142 formed H-bonding with an amine and hydroxyl functional group of isoquinoline carboxamide. It was noted that nelfinavir has the highest binding affinity (-8.2 kcal/mol), which correlates with its highest interaction in the binding pocket of COVID-19 protease, hence confirm its highest stabilization compared to other inhibitors. Interestingly, the predicted binding mode of remdesivir (B.E = -7.5 kcal/mol) in Figure 6A showed interaction with the catalytic dyad. The phenyl moiety formed π-sulfur interaction with catalytic amino acid His41 and π-cation with Met49. All the amines group formed H-bond with amino acid Thr45, Thr25, and Gln149 (b.d =1.84, 2.34, and 2.53 Å, respectively), hydroxyl functional group which is near Cys145 accept H-bond from Thr26 (2.38 Å). Besides, the methyl group of methyl pentyl acetate formed hydrophobic interaction with imidazoles of His163 and His172. Remdesivir reproduces Adenosine Triphosphate (ATP) structure and shams as a nucleoside during viral RNA replication on entry into the cell and further joins to a different RNA molecule; thus, terminate the replication of viral RNA; this process is known as chain termination46. On February 21, 2020, the National Institutes of Health declared the Adaptive COVID-19 Treatment Trial (ACTT - NCT04280705), a double-blind, randomized, placebo-controlled trial. The optimistic outcomes from remdesivir make the U.S. Food and Drug released a EUA for the emergency approval of remdesivir to treat COVID-19 patients in the hospital47,48.
Figure 5. Schematic representation of COVID-19 main protease (PDB: 6LU7) interactions with amodiaquine. Blue: protein, Grey: ligand, Yellow: hydrophobic interaction, Green: hydrogen bond.
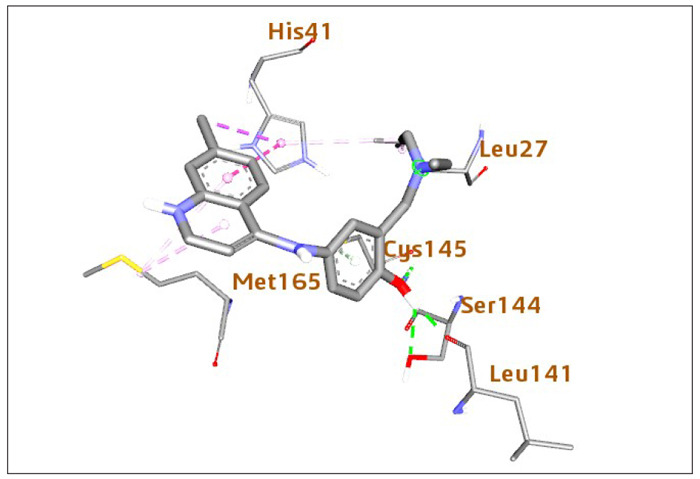
Figure 6. Schematic representation of COVID-19 main protease (PDB: 6LU7) interactions with compound (A) remdesivir and (B) EIDD-2801. Blue: protein, Grey: ligand, Yellow: hydrophobic interaction, Green: hydrogen bond.
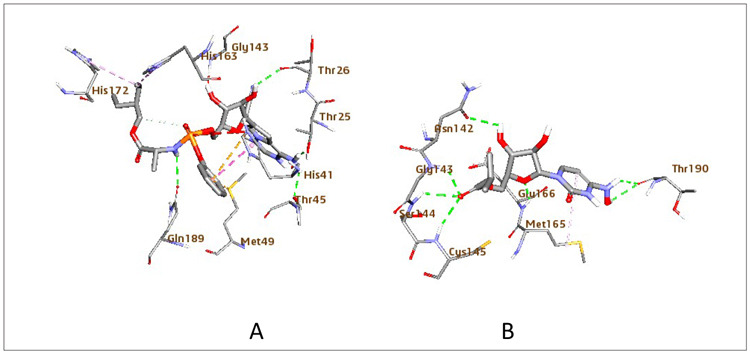
β-D-N4-hydroxycytidine-5′-isopropyl ester (EIDD-2801), which is also a nucleoside analog, not only showed high binding but also better interaction compared to β-D-N4-hydroxycytidine. Interestingly, the carbonyl oxygen of EIDD-2801 (B.E = -7.2 kcal/mol) accepts H-bond from the backbone of amino acids Gly143, Cys145, and Ser144. The presence of ester in this compound rationalized the increased inhibitory potency of this compound against SARS-COV-2. The amino acid, Thr190, was H-bonded with the hydroxylamine group. Meanwhile, the pyrimidinone ring formed hydrophobic interaction with Met165. Further, the oxygen atom in the nucleoside ring and the primary hydroxy functional group interacts with Glu166 and Asn142 via H-bonding (Figure 6B). The preclinical study detailed a reduction in lung injury and weight loss in mice 12 or 24 hours after the virus infection. Even so, the therapeutic gap of prospect for treatment in mice was relatively narrow. This treatment window will likely be extended in humans because it takes longer for the disease to improve in the human system than in mice. EIDD-1931 is orally bioavailable, compared to remdesivir. Hence, EIDD-2801 serves as a promising antiviral agent for treating COVID-19 infection in the future compared to remdesivir.
A meta-analysis of the preclinical studies and molecular docking studies results corroborate three compounds: EIDD-2801, GS-5734, and amodiaquine. This combined method would help develop primary deterrence policies in the health care providers to lessen the death rate and combat this pandemic to save lives using limited resources. Additionally, some limitations must be considered when interpreting this meta-analysis’s findings due to the limited information. The implication could lead to the likelihood of inaccuracies in reporting the effectiveness of therapies. Using the secondary data of a study assessing a different primary outcome, measurement bias, and residual confounding will inevitably influence this meta-analysis. Besides, due to the small sample size of the studies included and the samples of most of the studies not being population-based, the effectiveness estimate is disposed to be unstable. The other constraint observed was that published articles in English only were included in the meta-analysis. However, we only performed subgroup analysis on therapies; subgroup analyses on other modifiable risk factors could not be performed due to the included studies’ limited information. Another curb is that this meta-analysis included only published articles; thus, publication bias may exist. Like other systematic reviews, a comprehensive review of available literature provides a systematic identification of gaps and limitations, leading to improved future research designs.
Conclusion
Systematic reviews and meta-analyses are essential to accurately summarize evidence relative to drug development, efficacy, and healthcare interventions safety. This manuscript used systematic review, meta-analysis, and molecular docking studies to gather and analyze the preclinical evidence on SARS-COV inhibitors’ effect in coronavirus infections. The result indicated that the identified studies’ significant methodological limitations were centered on scarce data from the experimental designs as well as the outcome measures. Using more thorough and controlled studies appears possible. Our meta-analysis showed the overall effect in the reduction of virus titer and mortality in SARS-COV infected animals. Ribavirin was the most studied compound but was less active compared to EIDD-2801, GS-5734, and amodiaquine.
Further, optimization of the docked compounds as a molecular scaffold to synthesis more active structures using molecular synthesis or library screening of these compounds could be a reasonable strategy or approach in the development of new antiviral agents that targets COVID-19 infection. A meta-analysis of the preclinical studies and molecular docking studies results corroborate three compounds: EIDD-2801, GS-5734, and amodiaquine. This combined method would help develop primary deterrence policies in the health care providers to lessen the death rate and combat this pandemic to protect the survival of the human race using limited resources.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgment
In the electronic version of the paper on the journals’ website, supplementary material is provided, which includes supplementary Figure S1 (https://www.hippokratia.gr/images/PDF/24-3/FigureS1.pdf) , Figure S2 (https://www.hippokratia.gr/images/PDF/24-3/FigureS2.pdf), Figure S3 (https://www.hippokratia.gr/images/PDF/24-3/FigureS3.pdf), Figure S4 (https://www.hippokratia.gr/images/PDF/24-3/FigureS4.pdf). The authors are thankful to the Center for High-performance Computing (CHPC), Cape Town, South Africa, for providing the necessary facilities to carry out the present research work. The authors would like to thank Dr. Nkululeko Damoyi for his contribution to the development of this manuscript.
References
- 1.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9:29. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Transmission of SARS-CoV-2: implications for infection prevention precautions. Available at: https://www.who.int/publications-detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations. date accessed: 05/04/2020.
- 4.Meng L, Hua F, Bian Z. Coronavirus Disease 2019 (COVID-19): Emerging and Future Challenges for Dental and Oral Medicine. J Den Res. 2020;99:481–487. doi: 10.1177/0022034520914246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morawska L, Tang JW, Bahnfleth W, Bluyssen PM, Boerstra A, Buonanno G, et al. How can airborne transmission of COVID-19 indoors be minimised? Environ Int. 2020;142:105832. doi: 10.1016/j.envint.2020.105832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, et al. Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases. ACS Cent Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pak A, Adegboye OA, Adekunle AI, Rahman KM, McBryde ES, Eisen DP. Economic Consequences of the COVID-19 Outbreak: the Need for Epidemic Preparedness. Front Public Health. 2020;8 doi: 10.3389/fpubh.2020.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldekop JA, Horner R, Hulme D, Adhikari R, Agarwal B, Alford M, et al. COVID-19 and the case for global development. World Dev. 2020;134:105044. doi: 10.1016/j.worlddev.2020.105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akseer N, Kandru G, Keats EC, Bhutta ZA. COVID-19 pandemic and mitigation strategies: implications for maternal and child health and nutrition. Am J Clin Nutr. 2020;112:251–256. doi: 10.1093/ajcn/nqaa171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng YY, Ma YT, Zhang JY, Xie X. COVID-19 and the cardiovascular system. Nat Rev Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen K, Yang Y, Wang T, Zhao D, Jiang Y, Jin R, et al. Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: experts’ consensus statement. World J Pediatr. 2020;16:223–231. doi: 10.1007/s12519-020-00343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn. 2010:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 15.Le Bert N, Tan AT, Kunasegaran K, Tham CYL, Hafezi M, Chia A, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 16.Ton AT, Gentile F, Hsing M, Ban F, Cherkasov A. Rapid Identification of Potential Inhibitors of SARS‐CoV‐2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol Inform. 2020;39:e2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar Y, Singh H, Patel CN. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J Infect Public Health. 2020;13:1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2020:1–14. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nukoolkarn V, Lee VS, Malaisree M, Aruksakulwong O, Hannongbua S. Molecular dynamic simulations analysis of ritronavir and lopinavir as SARS-CoV 3CL(pro) inhibitors. J Theor Biol. 2008;254:861–867. doi: 10.1016/j.jtbi.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang SG, Shen H, Wang J, Tay FP, Liu DX. Proteolytic processing of polyproteins 1a and 1ab between non-structural proteins 10 and 11/12 of Coronavirus infectious bronchitis virus is dispensable for viral replication in cultured cells. Virology. 2008;379:175–180. doi: 10.1016/j.virol.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snijder EJ, Decroly E, Ziebuhr J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8:336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 25.George BJ, Aban IB. An application of meta-analysis based on DerSimonian and Laird method. J Nucl Cardiol. 2016;23:690–692. doi: 10.1007/s12350-015-0249-6. [DOI] [PubMed] [Google Scholar]
- 26.IntHout J, Ioannidis JP, Borm GF. The Hartung-Knapp-Sidik-Jonkman method for random effects meta-analysis is straightforward and considerably outperforms the standard DerSimonian-Laird method. BMC Med Res Methodol. 2014;14:25. doi: 10.1186/1471-2288-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Protein Data Bank. 6LU7: The crystal structure of COVID-19 main protease in complex with an inhibitor N3. Available at: https://www.rcsb.org/structure/6LU7. date accessed: 05/03/2020.
- 28.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnard DL, Day CW, Bailey K, Heiner M, Montgomery R, Lauridsen L, et al. Is the anti-psychotic, 10-(3-(dimethylamino)propyl)phenothiazine (promazine), a potential drug with which to treat SARS infections? Lack of efficacy of promazine on SARS-CoV replication in a mouse model. Antiviral Res. 2008;79:105–113. doi: 10.1016/j.antiviral.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Day CW, Baric R, Cai SX, Frieman M, Kumaki Y, Morrey JD, et al. A new mouse-adapted strain of SARS-CoV as a lethal model for evaluating antiviral agents in vitro and in vivo. Virology. 2009;395:210–222. doi: 10.1016/j.virol.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnard DL, Day CW, Bailey K, Heiner M, Montgomery R, Lauridsen L, et al. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS- coV replication in BALB/c mice. Antivir Chem Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 32.Barnard DL, Day CW, Bailey K, Heiner M, Montgomery R, Lauridsen L, et al. Enhancement of the infectivity of SARS-CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antiviral Res. 2006;71:53–63. doi: 10.1016/j.antiviral.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Totura AL, Baric RS. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol. 2012;2:264–275. doi: 10.1016/j.coviro.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hui KPY, Cheung MC, Perera RAPM, Ng KC, Bui CHT, Ho JCW, et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sallenave JM, Guillot L. Innate Immune Signaling and Proteolytic Pathways in the Resolution or Exacerbation of SARS-CoV-2 in Covid-19: Key Therapeutic Targets? Front Immunol. 2020;11:1229. doi: 10.3389/fimmu.2020.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Reeth K, Van Gucht S, Pensaert M. In vivo studies on cytokine involvement during acute viral respiratory disease of swine: troublesome but rewarding. Vet immunol Immunopathol. 2002;87:161–168. doi: 10.1016/S0165-2427(02)00047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung CY, Poon LL, Lau AS, Luk W, Lau YL, Shortridge KF, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 40.Mogensen TH, Paludan SR. Molecular pathways in virus-induced cytokine production. Microbiol Mol Biol Rev. 2001;65:131–150. doi: 10.1128/MMBR.65.1.131-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lalu MM, Fergusson DA, Cheng W, Avey MT, Corbett D, Dowlatshahi D, et al. Identifying stroke therapeutics from preclinical models: A protocol for a novel application of network meta-analysis. F1000Res. 2019;8:11. doi: 10.12688/f1000research.15869.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia B, Kang X. Activation and maturation of SARS-CoV main protease. Protein Cell. 2011;2:282–290. doi: 10.1007/s13238-011-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu IL, Mahindroo N, Liang PH, Peng YH, Kuo CJ, Tsai KC, et al. Structure-based drug design and structural biology study of novel nonpeptide inhibitors of severe acute respiratory syndrome coronavirus main protease. J Med Chem. 2006;49:5154–5161. doi: 10.1021/jm060207o. [DOI] [PubMed] [Google Scholar]
- 45.Kumar Y, Singh H, Patel CN. In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J Infect Public Health. 2020;13:1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha A, Sharma AR, Bhattacharya M, Sharma G, Lee SS, Chakraborty C. Probable Molecular Mechanism of Remdesivir for the Treatment of COVID-19: Need to Know More. Arch Med Res. 2020;51:585–586. doi: 10.1016/j.arcmed.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eastman RT, Roth JS, Brimacombe KR, Simeonov A, Shen M, Patnaik S, et al. Remdesivir: A Review of Its Discovery and Development Leading to Emergency Use Authorization for Treatment of COVID-19. ACS Cent Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abrams-Downey A, Saabiye J, Vidaurrazaga M. Investigational Therapies for the Treatment of COVID-19: Updates from Ongoing Clinical Trials. Eur Urol Focus. 2020;6:1028–1031. doi: 10.1016/j.euf.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]