Abstract
The spindle assembly checkpoint (SAC) is critical for sensing defective microtubule–kinetochore attachments and tension across the kinetochore and functions to arrest cells in prometaphase to allow time to repair any errors before proceeding into anaphase. Dysregulation of the SAC leads to chromosome segregation errors that have been linked to human diseases like cancer. Although much has been learned about the composition of the SAC and the factors that regulate its activity, the proximity associations of core SAC components have not been explored in a systematic manner. Here, we have taken a BioID2-proximity-labeling proteomic approach to define the proximity protein environment for each of the five core SAC proteins BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1 in mitotic-enriched populations of cells where the SAC is active. These five protein association maps were integrated to generate a SAC proximity protein network that contains multiple layers of information related to core SAC protein complexes, protein–protein interactions, and proximity associations. Our analysis validated many known SAC complexes and protein–protein interactions. Additionally, it uncovered new protein associations, including the ELYS–MAD1L1 interaction that we have validated, which lend insight into the functioning of core SAC proteins and highlight future areas of investigation to better understand the SAC.
Keywords: spindle assembly checkpoint (SAC), BioID2, proximity labeling, protein associations, protein networks, cell division
Introduction
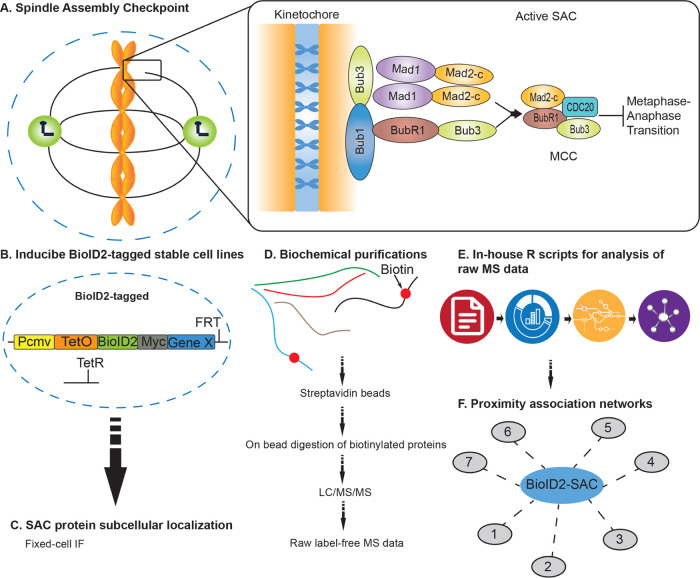
Human cell division is a highly coordinated set of events that ensure the proper transmission of genetic material from one mother cell to two newly formed daughter cells. Chromosome missegregation during cell division can lead to aneuploidy, an aberrant chromosomal number, which is a hallmark of many types of cancers and has been proposed to promote tumorigenesis.1 However, there is currently no consensus as to the pathways and factors that are deregulated to induce aneuploidy, why it is prevalent in cancer and how it contributes to tumorigenesis. Pivotal to cell division is the metaphase to anaphase transition, which is a particularly regulated process involving a multitude of protein–protein interactions that rely heavily on posttranslational modifications like phosphorylation and ubiquitination that function as switches to activate or inactivate protein function.2,3 For example, the multicomponent spindle assembly checkpoint (SAC) is activated when unattached kinetochores or nonproductive (monotelic, syntelic, and merotelic) attachments are sensed and functions to arrest cells in metaphase to give time to correct these deficiencies and generate proper microtubule–kinetochore attachments2 (Figure 1A). This ensures proper sister chromatid separation and minimizes segregation errors that lead to chromosomal instability, aneuploidy, and tumorigenesis.1 Core components of the SAC include BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1.4 Critical to the SAC is the mitotic checkpoint complex (MCC, composed of MAD2L1, BUBR1, BUB3, and CDC20) that maintains the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase substrate adaptor protein CDC20 sequestered and thereby inactivates the APC/C.5,6 Upon proper microtubule–kinetochore attachment, the SAC is satisfied and the inhibitory effect of the MCC on the APC/C is relieved2 (Figure 1A). Active APC/C then ubiquitinates and targets securin for degradation,2 which activates separase, the protease that cleaves RAD21, a component of the cohesin complex that holds sister chromatids together.7 This releases sister chromatid cohesion and chromatids are pulled to opposing poles of the cell by spindle microtubules, marking the entry into anaphase.
Figure 1.
Overview of the approach to generate core SAC protein BioID2-proximity association networks. (A) Schematic of the core spindle assembly checkpoint (SAC) components BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1 that localize to the kinetochore region during early mitosis. MCC denotes a mitotic checkpoint complex. (B) Generation of inducible BioID2-tagged stable cell lines for each core SAC protein. (C) Fixed-cell immunofluorescence microscopy to analyze BioID2-tagged SAC protein subcellular localization in time and space. (D) Biochemical purifications; affinity purification of biotinylated proteins; and identification of proteins by liquid chromatography with tandem mass spectrometry (LC/MS/MS). (E) Computational analysis of raw mass spectrometry data using in-house R scripts. (F) Generation of high-confidence SAC protein proximity association networks.
Because understanding the SAC is critical to understanding tumorigenesis and the response of tumor cells to antimitotic drugs that activate the SAC and trigger apoptotic cell death, it has become an intensive area of research.8,9 Although decades of research have shed light on the SAC, we are far from elucidating the full complement of regulatory factors involved in this complex pathway and from understanding how misregulation of this pathway can lead to tumorigenesis and resistance to chemotherapeutic drugs like antimitotics.10 Furthermore, models of proximity associations of the core SAC proteins with themselves and with structural and signaling components that mediate the establishment and silencing of the SAC are still being defined.11−13 Recently, proximity-labeling approaches like BioID and APEX have been used effectively to determine association networks among proteins and for defining the architecture of the centrosome, centrosome–cilia interface, and other organelles within the cell.14−19 However, proximity labeling has not been applied to the SAC in a systematic fashion, which could help to interrogate current models of core SAC protein associations and regulation.
Here, we have engineered vectors for establishing inducible BioID2-tagged protein stable cell lines. This system was used to establish stable cell lines with inducible BioID2-tagged core SAC protein (BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1) expression. These cell lines were utilized in BioID2-proximity biotin labeling studies, which were coupled to biotin biochemical purifications and mass spectrometry analyses to map the associations among core SAC proteins and other proteins in close proximity. These analyses yielded a wealth of information with regard to the protein environment of core SAC proteins in mitotic-enriched populations of cells where the SAC is active. In addition to validating well-established SAC protein complexes and protein–protein interactions, we defined new protein associations that warrant further investigation, including the ELYS–MAD1L1 interaction, to advance our understanding of SAC protein function and regulation.
Experimental Procedures
Cell Culture and Cell Cycle Synchronization
All media and chemicals were purchased from ThermoFisher Scientific (Waltham, MA) unless otherwise noted. HeLa Flp-In T-REx BioID2-tagged stable cell lines and retinal pigment epithelium (RPE) cells were grown in F12/Dulbecco’s modified Eagle’s medium (DMEM) 50:50 medium with 10% fetal bovine serum (FBS), 2 mM l-glutamine, in 5% CO2 at 37 °C. Cells were induced to express the indicated BioID2-tagged proteins by the addition of 0.2 μg/mL doxycycline (Sigma-Aldrich, St. Louis, MO) for 16 h. For synchronization of cells in mitosis, cells were treated with 100 nM Taxol (Sigma-Aldrich) for 16 h. A list of all reagents used is provided in Table S1.
Cell siRNA and Chemical Treatments
HeLa cell siRNA treatments were performed as described previously,20 with control siRNA (siControl, D-001810-10) or BUB1-targeting siRNA (siBUB1, L-004102-00) from Dharmacon (Lafayette, CO) for 48 h. For chemical treatments, RPE or HeLa cells were treated with a control dimethyl sulfoxide (DMSO) vehicle or the BUB1 inhibitor BAY 1816032 (HY-103020)21 from MedChemExpress (Monmouth Junction, NJ) at 10 μM for 5 h.
Generation of Inducible BioID2-tagging Vectors and Stable Cell Lines
For generating pGBioID2-27 or pGBioID2-47 vectors, the EGFP-S-tag was removed from pGLAP122 by digestion with BstBI and AflII. BioID2-Myc-27 (27 amino acid linker) or BioID2-Myc-47 (47 amino acid linker) was polymerase chain reaction (PCR) amplified, digested with NheI and XhoI and cloned into BstBI- and AflII-digested pGLAP1 to generate pGBioID2-27 or pGBioID2-47 (Figure S1A). For full-length human SAC core gene hBUB1, hBUB3, hBUBR1, hMAD1L1, and hMAD2L1 expression, cDNA corresponding to the full-length open reading frame of each gene was cloned into pDONR221 as described previously22,23 (Figure S1B). SAC core genes were then transferred from pDONR221 to pGBioID2-47 using the Gateway cloning system (Invitrogen, Carlsbad, CA) as described previously22,23 (Figure S1B). pGBioID2-47-SAC protein vectors were then used to generate doxycycline-inducible HeLa Flp-In T-REx BioID2 stable cell lines that expressed fusion proteins from a specific single locus within the genome as described previously22,23 (Figure S1C,D). All primers were purchased from ThermoFisher Scientific. A list of primers used is provided in Table S2. For a list of vectors generated in this study see Table S3. pGBioID2-27 and pGBioID2-47 vectors have been deposited at Addgene (Addgene IDs: 140276 and 140277, respectively) and are available to the scientific community.
Biotin Affinity Purifications
All media, chemicals, and beads were purchased from ThermoFisher Scientific unless otherwise noted. Biotin affinity purifications were conducted using previously described protocols with modifications.18,19 Briefly, 10% FBS was treated with 1 mL of MyOne streptavidin C1 Dynabeads overnight and passed through a 0.22 μm filter. BioID2-BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1, and BioID2 alone inducible stable cell lines were plated on six 150 mm tissue culture dishes, 24 h postplating; the cells were washed three times with phosphate-buffered saline (PBS) and once with DMEM without FBS, and shifted to the streptavidin Dynabead-treated 10% FBS DMEM. The cells were induced with 0.2 μg/mL Dox and treated with 100 nM Taxol and 50 μM Biotin for 16 h. Mitotic cells were collected by shake-off and centrifuged at 1500 rpm for 5 min and washed twice with PBS. The pellet was lysed with 3 mL of lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid (EGTA), 1% Triton-X-100, 0.1% sodium dodecyl sulfate (SDS), Halt Protease and Phosphatase Inhibitor Cocktail) and incubated with gentle rotation for 1 h at 4 °C, then centrifuged at 15 000 rpm for 15 min and transferred to a new 15 mL conical tube. The lysate was transferred to a TLA-100.3 tube (Beckman Coulter, Indianapolis, IN) and centrifuged at 45 000 rpm for 1 h at 4 °C. The lysate was then transferred to a new 15 mL conical tube and incubated with 300 μL of equilibrated streptavidin Dynabeads overnight with gentle rotation at 4 °C. The beads were separated with a magnetic stand and washed twice with 2% SDS, followed by a wash with WB1 (0.1% sodium deoxycholate, 1% Triton-X-100, 500 mM NaCl, 1 mM EDTA, 50 mM N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES)), a wash with WB2 (250 mM LiCl, 0.5% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0), and a final wash with 50 mM Tris-HCl pH 7.5. The beads were then resuspended in 50 mM triethylammonium bicarbonate (TEAB), 12 mM sodium lauroyl sarcosine, and 0.5% sodium deoxycholate. Ten percent of the beads were boiled with sample buffer and used for immunoblot analysis.
In Solution Tryptic Digestion
Streptavidin Dynabeads in 50 mM triethylammonium bicarbonate (TEAB), 12 mM sodium lauroyl sarcosine, and 0.5% sodium deoxycholate were heated to 95 °C for 10 min and then sonicated for 10 min to denature proteins. Protein disulfide bonds were reduced by treatment with 5 mM tris(2-carboxyethyl) phosphine (final concentration) for 30 min at 37 °C. Protein alkylation was performed with 10 mM chloroacetamide (final concentration) and incubation in the dark for 30 min at room temperature. The protein solutions were diluted 5-fold with 50 mM TEAB. Trypsin was prepared in 50 mM TEAB and added 1:100 (mass/mass) ratio to target proteins followed by a 4 h incubation at 37 °C. Trypsin was again prepared in 50 mM TEAB and added 1:100 (mass/mass) ratio to target proteins followed by overnight incubation at 37 °C. A 1:1 (volume/volume) ratio of ethyl acetate plus 1% trifluoroacetic acid (TFA) was added to the samples and samples were vortexed for 5 min. Samples were centrifuged at 16 000g for 5 min at room temperature and the supernatant was discarded. Samples were then lyophilized by SpeedVac (ThermoFisher Scientific) and desalted on C18 StageTips (ThermoFisher Scientific) as described previously.24
Nanoliquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) Analysis
Nano-LC-MS/MS with collision-induced dissociation was performed on a Q Exactive Plus Orbitrap (ThermoFisher Scientific) integrated with an Eksigent 2D nano-LC instrument. A laser-pulled reverse-phase column, 75 μm × 200 mm, containing 5 μm C18 resin with 300 Å pores (ThermoFisher Scientific) was used for online peptide chromatography. Electrospray ionization conditions using the nanospray source (ThermoFisher Scientific) for the Orbitrap were set as follows: capillary temperature at 200 °C, tube lens at 110 V, and spray voltage at 2.3 kV. The flow rate for reverse-phase chromatography was 500 nL/min for loading and analytical separation (buffer A, 0.1% formic acid and 2% acetonitrile; buffer B, 0.1% formic acid and 98% acetonitrile). Peptides were loaded onto the column for 30 min and resolved by a gradient of 0–80% buffer B over 174 min. The Q Exactive Plus Orbitrap was operated in data-dependent mode with a full precursor scan time at 180 min at high resolution (70 000 at m/z 400) from 350 to 1700 m/z and 10 MS/MS fragmentation scans at low resolution in the linear trap using charge-state screening excluding both unassigned and +1 charge ions. For collision-induced dissociation, the intensity threshold was set to 500 counts, and a collision energy of 40% was applied. Dynamic exclusion was set with a repeat count of 1 and exclusion duration of 15 s.
Experimental Design and Statistical Rationale
To enhance confidence in identifying core SAC protein proximity associations, we performed control and experimental purifications in biological replicates (three biological purifications for each core SAC protein, except for BUB3 where two biological purifications were performed, and two technical replicates were performed for each biological purification). This approach allowed for downstream comparison of control and experimental purifications, where proteins identified in the control BirA only (empty vector) were deemed potential nonspecific associations. For experimental mass spectrometry data acquisition and analysis workflow, see Figure S2. Database searches of the acquired spectra were analyzed with Mascot (v2.4; Matrix Science, Boston, MA) as described previously.25 The UniProt human database (October 10, 2018) was used with the following search parameters: trypsin digestion allowing up to two missed cleavages, carbamidomethyl on cysteine as a fixed modification, oxidation of methionine as a variable modification, 10 ppm peptide mass tolerance, and 0.02 Da fragment mass tolerance. With these parameters, an overall 5% peptide false discovery rate, which accounts for total false positives and false negatives, was obtained using the reverse UniProt human database as the decoy database. Peptides that surpassed an expectation cut-off score of 20 were accepted. For a list of all identified peptides, see Table S4, and for a list of all identified proteins, see Table S5. A list of all peptides that were used to identify proteins with one peptide sequence is provided in Table S6. All raw mass spectrometry files can be accessed at the UCSD Center for Computational Mass Spectrometry MassIVE datasets ftp://MSV000084975@massive.ucsd.edu. Peptides meeting the above criteria with information about their corresponding identified protein were further analyzed using in-house R scripts. All R scripts used in this study are freely available at GitHub https://github.com/uclatorreslab/MassSpecAnalysis. To increase precision and reduce error, a pseudo qualitative/quantitative approach was taken. Proteins identified in both the control and test purifications were assayed for significance, whereas proteins identified in test purifications but not present in control purifications were further considered. To handle proteins shared between test and control purifications, but only identified in less frequency, we measured the relative fold change or mean difference in a quantitative manner. To compare quantification between purifications, we used the Exponentially Modified Protein Abundance Index (emPAI).26 emPAI offers approximate relative quantitation of the proteins in a mixture based on protein coverage by the peptide matches in a database search result and can be calculated using the following equation26
where NObserved is the number of experimentally observed peptides and NObservable is the calculated number of observable peptides for each protein.26 To compare proteins across multiple replicates/baits, each emPAI score was normalized to pyruvate carboxylase, a protein that readily binds to biotin,27 and was found in high abundance in all purifications. Using a normalized emPAI (NemPAI) as a relative quantification score, we calculated the mean difference (the mean NemPAI for a certain protein across test replicates minus the mean NemPAI of the same protein across control replicates). Resampling involved recreating or estimating the normal distribution around a test statistic, in this case the mean difference, by calculating that statistic many times under rearrangement of labels. We performed 10 000 simulations per test statistic, resulting in normal distributions of the mean difference between values of proteins identified in the experimental and control purifications. Using this distribution, we related each individual mean difference to the mean difference observed in the overall population to get a relative idea of what might be significantly higher in value compared to the control, when taking what is observed in the entire population. Values that lied outside of the 95% confidence interval of the mean difference and showed a higher value in the experimental purification compared to the control were then considered for further analysis (see Table S7).
Protein Proximity Network Visualization and Integration of Systems Biology Databases
Visual renderings relating protein–protein interactions/associations were carried out using custom scripts in R. To incorporate protein complex information, we integrated the Comprehensive Resource of Mammalian Protein Complexes (CORUM v. 3.0).28 Protein–protein interaction information was derived and integrated from the Biological General Repository for Interaction Datasets (BioGRID v. 3.5).29 To create relational networks that associated proteins based on cellular mechanisms, Gene Ontology (GO) terms were incorporated into the search (Gene Ontology release June 2019).30 For a list of GO terms used, see Table S8. Pathway information was derived from Reactome, an open source and peer-reviewed pathway database.31 All databases were individually curated into an in-house system biology relational database using custom R scripts. Final visuals relating to protein associations were constructed using RCytoscapeJS, a developmental tool used to develop Cytoscape renderings in an R and JavaScript environment.32,33
Immunoprecipitations
For cell lysate immunoprecipitations (IPs), BioID2 (empty vector, EV), BioID2-MAD1L1, or BioID2-MAD2L1 HeLa stable cell lines were induced with 0.2 μg/mL Dox and treated with 100 nM Taxol for 16 h to arrest cells in mitosis. Cells were collected by shake-off and lysed with lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton-X-100, 0.1% SDS, Halt Protease and Phosphatase Inhibitor Cocktail) and incubated with gentle rotation for 1 h at 4 °C, then centrifuged at 15 000 rpm for 30 min and the supernatant was transferred to a microcentrifuge tube. Myc magnetic beads were equilibrated and incubated with mitotic cell extracts for 5 h at 4 °C with gentle rotation. The beads were then washed five times with wash buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM dithiothreitol (DTT), and Halt Protease and Phosphatase Inhibitor Cocktail) for 5 min each and bound proteins were eluted with 50 μL of 2× Laemmli SDS sample buffer. Ten percent of the sample inputs and the entire eluates from the immunoprecipitations were used for immunoblot analysis.
In Vitro Binding Assays
For in vitro binding assays, Myc or FLAG-tagged GFP, MAD1L1, MAD2L1, or ELYS (N-terminal fragment) were in vitro transcribed and translated (IVT) using TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI) in 10 μL reactions. Myc beads (MBL, Sunnyvale, CA) were washed three times and equilibrated with wash buffer (50 mM Tris pH 7.4, 200 mM KCl, 1 mM DTT, 0.5% NP-40, and Halt Protease and Phosphatase Inhibitor Cocktail). IVT reactions were added to equilibrated Myc beads and incubated for 1.5 h at 30 °C with gentle shaking, and after binding, beads were washed three times with wash buffer and eluted by boiling for 10 min with 2× Laemmli SDS sample buffer. The samples were then resolved using a 4–20% gradient Tris gel with Tris-Glycine SDS running buffer, transferred to an Immobilon poly(vinylidene difluoride) (PVDF) membrane (EMD Millipore, Burlington, MA), and the membranes were analyzed using a PharosFX Plus molecular imaging system (Bio-Rad, Hercules, CA).
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as described previously34 with modifications described in ref (25). Briefly, HeLa-inducible BioID2-tagged BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1 stable cell lines were treated with 0.2 μg/mL doxycycline for 16 h, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton-X-100/PBS, and costained with 0.5 μg/mL Hoechst 33342 and the indicated antibodies. Imaging of mitotic cells was carried out with a Leica DMI6000 microscope (Leica DFC360 FX Camera, 63×/1.40–0.60 NA oil objective, Leica AF6000 software, Buffalo Grove, IL) at room temperature. Images were subjected to Leica Application Suite 3D Deconvolution software and exported as TIFF files. The quantification of immunofluorescence microscopy images from BUB1 RNAi and BUB1 inhibitor-treated cells was performed by capturing intensity profiles in ImageJ for both a kinetochore section and a background section adjacent to the kinetochore. Each intensity value was normalized by the area of the captured image and the background signal was subtracted. The values were compared using a Student’s t-test. The number of samples used varied by experiment; knock-down experiments: BUB1 (n = 19), SGO2 (n = 50), and PLK1 (n = 13); inhibitor treatments: BUB1 (n = 20), SGO2 (n = 17), and PLK1 (n = 17). All calculations were performed in R.
Antibodies
Immunofluorescence microscopy and immunoblotting were performed using the following antibodies: BioID2 (BioFront Technologies, Tallahassee, FL), GAPDH (Preoteintech, Rosemont, IL), α-tubulin (Serotec, Raleigh, NC), anticentromere antibody (ACA, Cortex Biochem, Concord, MA), SGO2 (Bethyl, Montgomery, TX), PLK1, BUB1, and ELYS (Abcam, Cambridge, MA). Affinipure secondary antibodies labeled with FITC, Cy3, and Cy5 were purchased from Jackson Immuno Research (West Grove, PA). IRDye 680RD streptavidin was purchased from LI-COR Biosciences (Lincoln, NE). Immunoblot analyses were carried out using secondary antibodies conjugated to IRDye 680 and IRDye 800 from LI-COR Biosciences (Lincoln, NE) and blots were scanned using a LI-COR Odyssey infrared imager.
Results and Discussion
Generation of Inducible BioID2-Tagged SAC Protein Stable Cell Lines
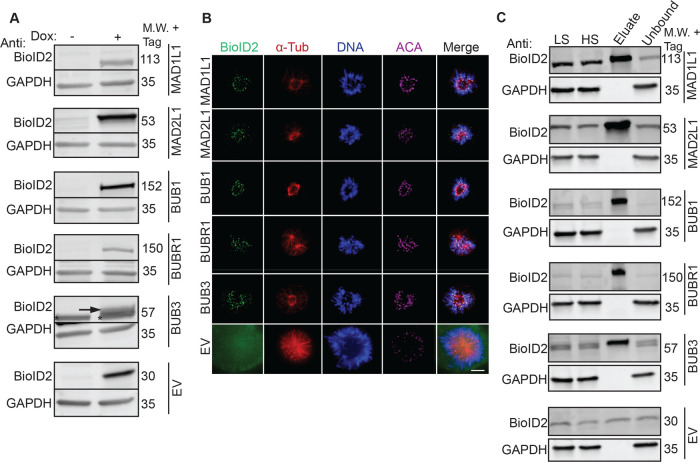
The spindle assembly checkpoint is essential for ensuring the fidelity of chromosome segregation during cell division35 (Figure 1A). To better understand how the SAC functions and is regulated, we sought to map the protein associations of the core SAC proteins BUB1, BUB3, BUBR1 (BUB1B), MAD1L1, and MAD2L1 using a BioID2-proximity-labeling proteomic approach18 (Figure 1B–F). The overexpression of critical cell division proteins often leads to cell division defects that can preclude the generation of epitope-tagged stable cell lines. Therefore, we first sought to generate BioID2 Gateway-compatible vectors with a doxycycline (Dox)-inducible expression functionality. To do this, we amplified BirA-Myc with linkers coding for 27 or 47 amino acid residues downstream of Myc (BirA-Myc-27/47) (Figure S1A, Table S2). These amplification products were cloned into the pGLAP1 vector,22 which had been previously modified by removal of its LAP-tag (EGFP-Tev-S-protein), to generate pGBioID2-27 and pGBioID2-47 vectors (Figure S1A). Full-length human open reading frames encoding for BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1 were cloned into the pGBioID2-47 vector. pGBioID2-47-SAC protein vectors (Figure S1B, Table S3) were cotransfected with a vector expressing the Flp recombinase (pOG44) into HeLa Flp-In T-REx cells (Figure S1C). Hygromycin-resistant clones were then selected (Figure S1D) and grown in the presence or absence of Dox for 16 h. The Dox-induced expression of each BioID2-47-SAC protein was then assessed by immunoblot analysis (Figure 2A). All of the BioID2-tagged core SAC proteins were expressed only in the presence of Dox (Figure 2A), indicating the successful establishment of inducible BioID2-tagged core SAC protein stable cell lines. Additionally, these BioID2-tagged core SAC proteins were expressed at lower levels than the untagged endogenous proteins (Figure S3A)
Figure 2.
Establishment of inducible BioID2-tagged SAC protein (BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1) stable cell lines and biochemical purifications. (A) Immunoblot analysis of extracts from doxycycline (Dox)-inducible BioID2-tag alone (EV, empty vector) or BioID2-tagged SAC protein (BUB1, BUB3, BUBR1, MAD1L1, MAD2L1) expression cell lines in the absence (−) or presence (+) of Dox for 16 h. For each cell line, blots were probed with anti-BioID2 (to visualize the indicated BioID2-tagged SAC protein) and anti-GAPDH as a loading control. M.W. indicates molecular weight. Note that BioID2-tagged SAC proteins are only expressed in the presence of Dox. The arrow points to the induced BioID2-BUB3 protein band and the asterisk denotes a nonspecific band recognized by the anti-BioID2 antibody. (B) Fixed-cell immunofluorescence microscopy of the BioID2-tag alone (EV) or the indicated BioID2-tagged SAC proteins during prometaphase, a time when the SAC is active. HeLa BioID2-tagged protein expression cell lines were induced with Dox for 16 h, fixed and stained with Hoechst 33342 DNA dye and anti-BioID2, anti-α-Tubulin, and anticentromere antibodies (ACA). Bar indicates 5 μm. Note that all BioID2-tagged SAC proteins localize to the kinetochore region (overlapping with the ACA signal), whereas the BioID2-tag alone (EV) was absent from kinetochores. (C) Immunoblot analysis of BioID2 biochemical purifications from cells expressing the indicated BioID2-tagged SAC proteins or the BioID2-tag alone (EV). For each cell line, blots were probed with anti-BioID2 (to visualize the indicated BioID2-tagged SAC protein) and anti-GAPDH as a loading control. M.W. indicates molecular weight, LS indicates low-speed supernatant, and HS indicates high-speed supernatant. Uncropped immunoblots are provided in Figures S17 and S18.
BioID2-SAC Proteins Localize Properly to Kinetochores during Prometaphase
Then, the ability of BioID2-SAC proteins to properly localize to kinetochores during prometaphase, a time when the SAC is active and core SAC proteins localize to the kinetochore region, was analyzed by immunofluorescence microscopy. BioID2-SAC protein HeLa-inducible stable cell lines were treated with Dox for 16 h, fixed, and stained with Hoechst 33342 DNA dye and anti-BioID2, anti-α-Tubulin, and anticentromere antibodies (ACA). The localization of BioID2-SAC proteins in prometaphase cells was then monitored by immunofluorescence microscopy. BioID2-tagged BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1 localized to kinetochores, overlapping fluorescence signal with anticentromere antibodies (ACA) during prometaphase (Figure 2B). In contrast, BioID2-tag alone showed no specific localization (Figure 2B). These results indicated that BioID2-tag was not perturbing the ability of SAC proteins to localize to kinetochores during the time when the SAC was active. Further, the addition of biotin did not perturb the localization of BioID2-SAC proteins to kinetochores (Figure S3B).
BioID2-SAC Protein Proximity Labeling, Purifications, and Peptide Identification
To define the protein proximity networks of core SAC proteins, inducible BioID2-SAC protein HeLa stable cell lines were used to perform BioID2-dependent proximity biotin labeling and biotinylated proteins were purified with a streptavidin resin (Figure 1D,C). Briefly, inducible BioID2-SAC protein HeLa stable cell lines were treated with 0.2 μg/mL Dox, 100 nM Taxol, and 50 μM Biotin for 16 h to induce the expression of BioID2-SAC proteins and to activate the SAC and arrest cells in prometaphase. Mitotic cells were collected by shake-off, lysed, and the cleared lysates were bound to streptavidin beads. Bound biotinylated proteins were trypsinized on the beads and the peptides were analyzed by 2D-LC-MS/MS (for details, see the Experimental Procedures section). A diagnostic immunoblot analysis of each purification, using anti-BioID2 antibodies, showed that BioID2-tagged BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1 were present in the extracts and were purified with the streptavidin beads, indicating that they had been biotinylated (Figure 2C). Additionally, western blots of each purification were probed with streptavidin, which showed that biotinylated proteins were present and efficiently captured in each purification (Figure S4A). In-house R scripts were then used to analyze the mass spectrometry results (for details, see the Experimental Procedures section), to draw significance between peptides shared between the experimental and control purifications, we estimated the distribution of the mean difference of normalized emPAI scores across proteins and selected proteins with a significantly higher difference (for details, see the Experimental Procedures section). Proteins that showed significantly higher values in test purifications compared to controls (values that lied outside of 95% confidence interval of the population mean difference) were considered hits and further analyzed (Table S7).
Analysis of the Core SAC Protein Proximity Association Network
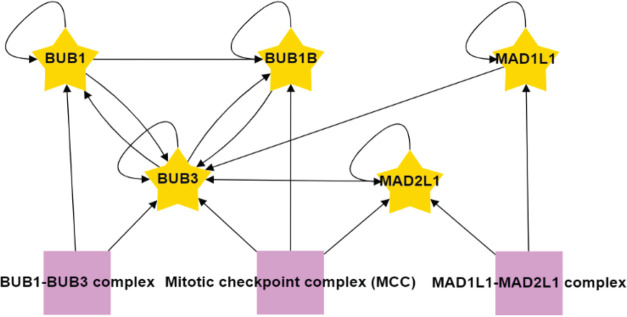
In-house R scripts were then used to integrate identified proteins from the mass spectrometry analysis with the data visualization application RCytoscapeJS32 to generate protein proximity association maps for each of the core SAC proteins (BUB1, BUB3, BUBR1, MAD1L1, MAD2L1) (Figure S5). These five maps were compiled to generate the SAC protein proximity network (Figure S6). To begin to digest the wealth of information within the SAC protein proximity network, we first analyzed the network with the CORUM database28 and examined the proximal associations between each of the core SAC proteins. This analysis revealed many of the previously characterized core SAC component protein–protein interactions and the BUB1-BUB3, BUBR1-BUB3, BUBR1-BUB3-CDC20 (BBC subcomplex of the MCC), and MAD2L1-BUBR1-BUB3-CDC20 (MCC) complexes (Figures 3 and S6).6,36−38 These SAC complexes are critical to the establishment and maintenance of the SAC39 and their identification was an indication that our proximity-based labeling approach was robust. Of interest, BUB3 was present in all of the purifications, consistent with its central role in recruiting other SAC proteins to the kinetochore and coordinating the formation of SAC subcomplexes (Figure 3).12 Although MAD1L1 and MAD2L1 had been previously determined to bind directly,40 our approach was unable to detect this association. However, previous proteomic analyses with N- or C-terminal BioID-tagged MAD1L1 were also unable to detect an association with MAD2L1, which was attributed to a low number of lysines on the surface of MAD2L1 that likely affected the efficiency of biotin labeling.41
Figure 3.
Associations among the core SAC proteins identified in the proximity protein network. The associations between each of the core SAC proteins (BUB1, BUB3, BUBR1, MAD1L1, MAD2L1) were isolated from the unified core SAC protein proximity association network (Figure S6). Purple boxes highlight protein complexes known to assemble with core SAC proteins as annotated by the CORUM database. Arrows indicate the direction of the detected associations.
Analysis of Core SAC Protein-Kinetochore Protein Proximity Associations
To specifically analyze kinetochore proteins identified in core SAC protein proximity networks, we applied a kinetochore-related Gene Ontology (GO) annotation analysis to the data sets. Briefly, R scripts were used to integrate the identified proteins with the bioinformatic databases CORUM,28 Gene Ontology,30 BioGRID,29 and Reactome31 using kinetochore-related GO terms (see Table S8 for a list of Kinetochore GO IDs) to reveal kinetochore-associated proteins. RCytoscapeJS32 was then used to generate GO, BioGRID, and Reactome kinetochore protein proximity association maps for each of the core SAC proteins (BUB1, BUB3, BUBR1, MAD1L1, MAD2L1) (Figures S7–S11). The five kinetochore GO maps (one for each core SAC protein) were compiled to generate one core SAC protein kinetochore GO network that visualized the proteins within the network that were active at the kinetochore (Figure S12A). A similar process was repeated to generate one core SAC protein BioGRID network that displayed the verified associations between the proteins that were active at the kinetochore (Figure S12B) and one core SAC protein Reactome network that highlighted the cellular pathways that proteins in the SAC proximity association network have been linked to (Figure S12C). Additionally, we generated core SAC protein GO, BioGRID, and Reactome networks using mitotic spindle-related GO annotations (Figure S13A–C) and centromere-related GO annotations (Figure S14A–C), see Table S8 for a list of GO IDs. Finally, we generated core SAC protein GO, BioGRID, and Reactome networks using the kinetochore, mitotic spindle, and centromere-related GO annotations (Figure 4A–C). Interestingly, of the proteins identified in the purifications, kinetochore-associated proteins were enriched in comparison to mitochondrial proteins (Figure S15). Together, these networks not only visualized the associations of each core SAC protein with kinetochore components and more broadly proteins implicated in mitotic spindle assembly, but they also provided a holistic view of their interconnectedness (i.e., associations among core SAC proteins and subcomplex and complex formation).
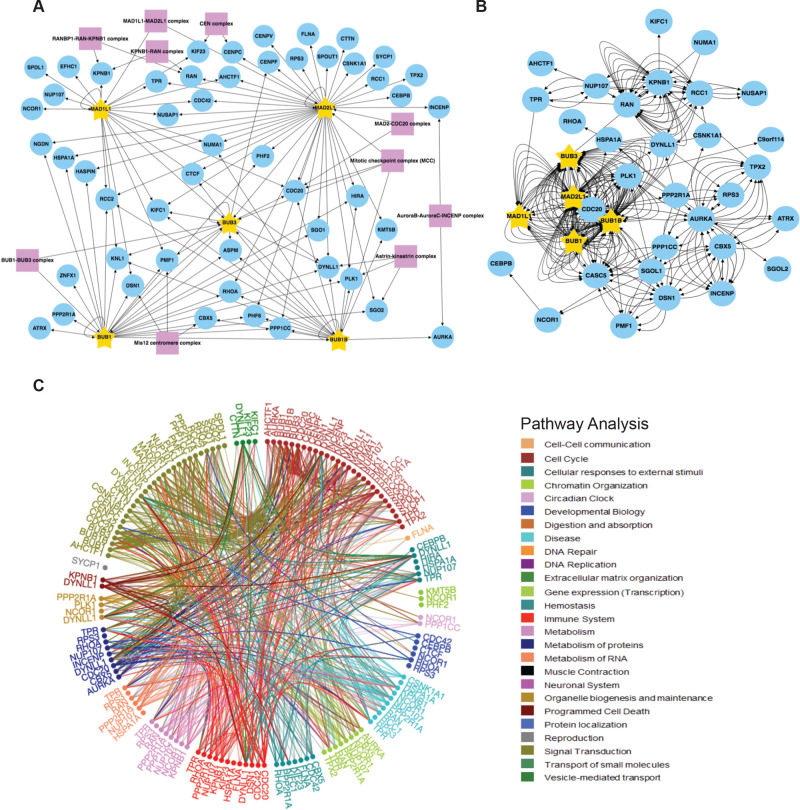
Figure 4.
SAC protein BioID2 kinetochore/mitotic spindle assembly/centromere proximity association network. (A) Individual core SAC protein (BUB1, BUB3, BUBR1, MAD1L1, MAD2L1) proximity protein maps were compiled and subjected to kinetochore, mitotic spindle assembly, and centromere GO annotation analysis along with a COURM complex annotation analysis to generate a core SAC protein kinetochore/mitotic spindle assembly/centromere proximity association network. Purple boxes highlight kinetochore, mitotic spindle assembly, and centromere-associated protein complexes present in the network. Arrows indicate the direction of the detected interactions. For a list of GO terms used, see Table S8. (B) Core SAC protein kinetochore/mitotic spindle assembly/centromere proximity association network was analyzed with BioGRID to reveal previously verified protein associations. Each arrow indicates an experimentally annotated interaction curated in the BioGRID database. The direction of arrows indicates an annotated interaction from a bait protein to the prey. (C) Reactome pathway analysis of the core SAC protein kinetochore/mitotic spindle assembly/centromere proximity association network. The Reactome circular interaction plot depicts the associations between the identified proteins within the SAC protein kinetochore/mitotic spindle assembly/centromere proximity association network and the corresponding pathways in which they function. Legend presents the color-coded pathways that correspond to the circular interaction plots.
Numerous insights were derived from these networks and we highlight four here. First, we identified the Mis12 centromere complex components DSN1 and PMF1 in the BUB1 and MAD1L1 purifications (Figures 4A, S7A, and S10A). The Mis12 complex is comprised of PMF1, MIS12, DSN1, and NSL142−44 and genetic and biochemical studies have shown that it coordinates communication from the outer kinetochore to the centromeric DNA in the inner kinetochore.44−46 PMF1 was also identified in the BUB3 purification (Figures 4A and S8A). To our knowledge, there have been no previous reports of a direct association between BUB3 and the Mis12 complex. Therefore, this BUB3-PMF1 proximity association could indicate a novel direct interaction or simply that these proteins reside within close proximity at the kinetochore. Of interest, the Mis12 complex recruits KNL1 to the kinetochore, which functions as a scaffold for the recruitment of BUB3 that subsequently recruits additional SAC components.4,38,47 Consistently, we observed the association of KNL1 with BUB1, BUB3, BUBR1, and MAD1L1 (Figure 4A). These associations were previously reported, as summarized in the Figure 4B BioGRID network, and had been established to have a role in checkpoint activation41,48−50 (reviewed in ref (5)). Additionally, MAD2L1 was not found to associate with KNL1, and to our knowledge, a KNL1–MAD2L1 interaction has not been reported.
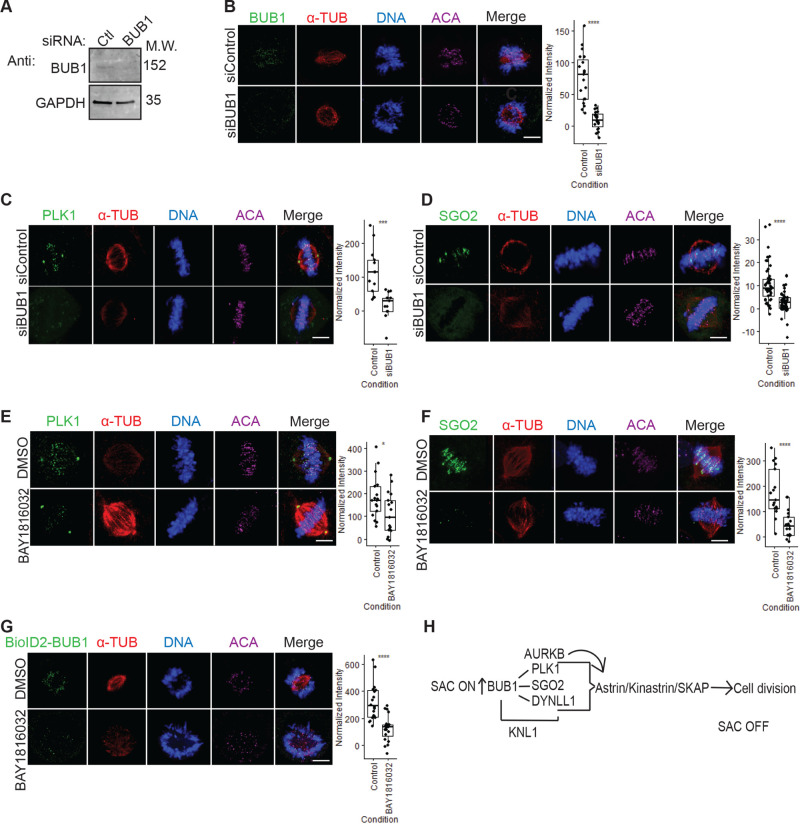
Second, minor components of the Astrin–Kinastrin complex (PLK1, DYNLL1, and SGO2)51 were found to associate with all of the core SAC proteins (Figures 4A, S7A, S8A, S9A, S10A, and S11A). The Astrin–Kinastrin complex is important for aligning and attaching microtubules to kinetochores.51−53 Previous studies showed that depletion of BUB1 led to the delocalization of PLK1 and SGO2 from the kinetochores during prometaphase.54,55 Additionally, the BUB1 kinase activity was shown to be important for SGO2 kinetochore localization56 and for the proper localization of BUB1 to the kinetochore55 and pharmacological inhibition of the BUB1 kinase activity led to delocalization of SGO2 away from kinetochores.57 However, whether the BUB1 kinase activity was required for PLK1 kinetochore localization remained unknown. To address this, we first sought to confirm that PLK1 and SGO2 were mislocalized in BUB1-depleted cells. HeLa cells were treated with control siRNA (siControl) or BUB1-targeting siRNA (siBUB1) capable of depleting BUB1 protein levels (Figure 5A). Immunofluorescence microscopy of these cells showed that BUB1 was absent from kinetochores in siBUB1-treated cells (Figure 5B). Additionally, the siBUB1 treatment reduced the levels of kinetochore-localized PLK1 and SGO2 (Figure 5C,D). Then, we asked if the BUB1 kinase activity was required for PLK1 and SGO2 kinetochore localization. RPE cells were treated with a control DMSO vehicle or the recently developed BUB1 kinase selective inhibitor BAY 1816032,21 and the localization of PLK1 and SGO2 was assessed in mitotic cells. In comparison to the control DMSO treatment, treatment with BAY 1816032 led to a reduction in the levels of kinetochore-localized PLK1 and SGO2 (Figure 5E,F). Additionally, treatment of BioID2-BUB1-expressing HeLa cells with BAY 1816032 also led to a reduction in the levels of kinetochore-localized BioID2-BUB1 (Figure 5G). This data indicated that the BUB1 kinase activity was important for its proper localization to kinetochores and for the localization of the Astrin–Kinastrin minor complex components PLK1 and SGO2 to the kinetochore.
Figure 5.
BUB1 as a hub for organizing the metaphase to anaphase transition. (A) Immunoblot analysis of protein extracts isolated from HeLa cells treated with control (Ctl) or BUB1 siRNA. GAPDH was used as a loading control. (B–D) Fixed-cell immunofluorescence microscopy of mitotic HeLa cells treated with control siRNA (siControl) or siRNA-targeting BUB1 (siBUB1). Cells were fixed and stained with Hoechst 33342 DNA dye and anti-BUB1 (B), anti-PLK1 (C), or anti-SGO2 (D) antibodies, along with anti-α-Tubulin and anticentromere antibodies (ACA). Bars indicate 5 μm. Box plots on the right of each panel show the quantification of the normalized fluorescence intensity for kinetochore-localized BUB1 (B), PLK1 (C), or SGO2 (D) and **** denotes P < 0.001. (E, F) Same as in (A), except that RPE cells were used and treated with control DMSO vehicle or BUB1 kinase inhibitor BAY 1816032. Note that the levels of kinetochore-localized PLK1 (E) and SGO2 (F) decrease in BAY 1816032-treated cells. Bars indicate 5 μm. Box plots on the right of each panel show the quantification of the normalized fluorescence intensity for kinetochore-localized PLK1 (E, * indicates P = 0.027) or SGO2 (F, **** indicates P < 0.001). (G) Same as in (E,F), except that a HeLa BioID2-BUB1-expressing cell line was used. Bar indicates 5 μm. The box plot shows the quantification of the normalized fluorescence intensity for kinetochore-localized BioID2-BUB1, **** indicates P < 0.001. (H) Model of BUB1 as an organizer of the metaphase to anaphase transition. BUB1 is critical for SAC protein binding to KNL1 to establish the SAC response and is also critical for the recruitment of the Astrin–Kinastrin minor complex, which is essential for the metaphase to anaphase transition.
Third, we identified CENPV as a MAD2L1-associating protein (Figure 4A). CENPV was identified in a proteomic screen for novel components of mitotic chromosomes58 and was later shown to localize to kinetochores early in mitosis and to have a major role in directing chromosomal passenger complex (CPC) subunits Aurora B and INCENP to the kinetochore.50,59 Although BUB1 has been shown to be important for the recruitment of the CPC to kinetochores,60 we are unaware of any reports of MAD2L1 being involved in this process. Interestingly, MAD2L1 has been shown to regulate the relocation of the CPC from centromeres through its inhibition of MKLP2, which is essential for proper cytokinesis.61 Thus, it is possible MAD2L1 could also be regulating CPC localization to kinetochores through its association with CENPV.
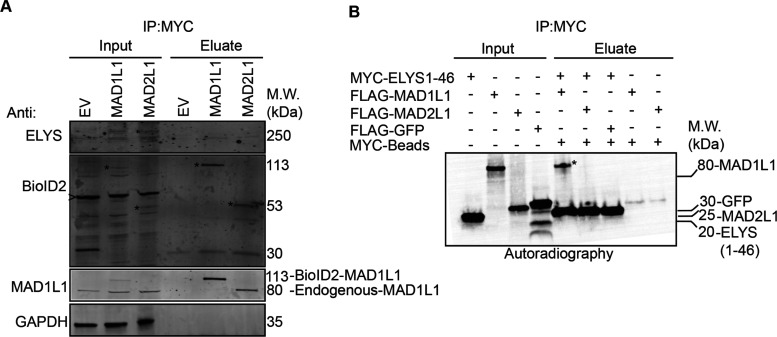
Fourth, components of the nuclear pore complex were found to associate with MAD1L1 and MAD2L1 (Figure S5). To better visualize these nuclear pore-associated proteins, we performed a proximity protein mapping analysis for each of the core SAC proteins using nuclear pore-related GO annotations (see Table S8 for a list of nuclear pore-related GO IDs) (Figure S16). This analysis revealed that MAD1L1 had associations with nuclear pore basket components including TPR, NUP153, NUP50, and other components of the nuclear pore that are in close proximity to the nuclear basket like ELYS/AHCTF1 (also known as MEL-28 in Caenorhabditis elegans) and NUP107 (Figure S16A). These data support previous studies in humans and other organisms that have shown that MAD1L1 associates with TPR, NUP153, ELYS, and NUP107 and is important for generating the MAD1L1–MAD2L1 complex in early mitosis to establish the SAC.62−68 Similarly, MAD2L1 was found to associate with TPR (previously verified in ref (63)), NUP50, Nup153, NUP210, and ELYS (Figure S16A). Of interest, we did not detect associations between other core SAC proteins (BUB1, BUB3, BUBR1) and nuclear pore basket proteins. These data are consistent with a model where MAD1L1 makes multiple direct contacts with the nuclear pore basket complex subunits and MAD2L1 is in close proximity to NUP153 and NUP50 due to its binding to MAD1L1. We note that ELYS was found in both the MAD1L1 and MAD2L1 proximity maps (Figure S16A). ELYS was discovered in a proteomic screen for NUP107-160 complex binding partners and was shown to localize to nuclear pores in the nuclear lamina during interphase and to kinetochores during early mitosis, similar to the NUP107-160 complex.69 More recently, ELYS was shown to function as a scaffold for the recruitment of Protein Phosphatase 1 (PP1) to the kinetochore during M-phase exit, which was required for proper cell division.70,71 Due to ELYS’s roles at the kinetochore and an identified yeast two-hybrid interaction between C. elegans MEL-28 (ELYS in humans) and MDF-1 (MAD1L1 in humans),65 we sought to determine if MAD1L1 and MAD2L1 were binding directly to ELYS. First, we performed MYC immunoprecipitations from mitotic protein extracts prepared from BioID2, BioID2-MAD1L1, and BioID2-MAD2L1 expressing cell lines that had been arrested in mitosis. Indeed, ELYS immunoprecipitated with both BioID2-MAD1L1 and BioID2-MAD2L1, albeit weakly, in these mitotic extracts (Figure 6A). Then, we sought to assess these interactions in a cell-free in vitro expression system. Although a validated full-length ELYS cDNA vector was not available and could not be generated, we were able to generate a MYC-tagged ELYS N-terminal fragment vector that expressed the first 46 amino acids of ELYS. This ELYS N-terminal fragment bound to FLAG-MAD1L1 (indicated by an asterisk in the eluate), but not FLAG-MAD2L1 (Figure 6B). Together, these data indicated that ELYS associated with MAD1L1 and MAD2L1 in mitotic cell extracts and that MAD1L1 bound to the ELYS N-terminal fragment in vitro.
Figure 6.
ELYS binds to MAD1L1 and MAD2L1 in mitotic cell lysates and to MAD1L1 in vitro. (A) BioID2-Myc (empty vector, EV), BioID2-Myc-MAD1L1, or BioID2-Myc-MAD2L1-inducible HeLa stable cell lines were induced with Dox and treated with 100 nM Taxol to arrest cells in mitosis. Mitotic cell lysates were then used for Myc immunoprecipitations and subjected to immunoblot analysis with the indicated antibodies. Note that endogenous ELYS immunoprecipitates with BioID2-Myc-tagged MAD1L1 and MAD2L1. Asterisks indicate BioID2-Myc-MAD1L1 or BioID2-Myc-MAD2L1 in the inputs or eluates. The arrowhead indicates a nonspecific background band recognized by the anti-BioID2 antibody. (B)35S-radiolabeled Myc-ELYS N-terminal fragment (ELYS1-46, first 46 amino acids); FLAG-MAD1L1, FLAG-MAD2L1, and FLAG-GFP (control) were used in in vitro binding assays. Myc immunoprecipitations were resolved by western blotting and the blots were analyzed by autoradiography. Note that the ELYS N-terminal fragment binds to MAD1L1 (indicated by the asterisk in the eluate) and not MAD2L1.
Core SAC Proteins in Cellular Homeostasis
It is important to note that most of the core SAC proteins have been shown to have roles in cellular homeostasis independent of their role in the SAC, which are predominantly mediated through protein–protein interactions with nonkinetochore proteins. Many of these associations were present in individual core SAC protein proximity maps where GO annotations were not applied (Figure S5). Consistently, Reactome pathway analysis of the core SAC protein proximity protein network showed that many of the SAC-associated proteins had roles in numerous pathways important for cellular homeostasis including the cell cycle, DNA repair, and gene expression (Figure 4C). We encourage researchers interested in nonmitotic SAC protein functions to explore SAC protein proximity association networks to gain further insights into these pathways.
Conclusions
The SAC is an important signaling pathway that is critical for proper cell division, which functions with great precision in a highly orchestrated manner.2 Due to the dynamic nature of the associations between core SAC proteins and the complexes and subcomplexes that they form, it has been difficult to generate a proteomic network view of the proteins that are in close proximity and that interact with core SAC proteins. Here, we have established an inducible BioID2-tagging system that allowed for the transient expression of BioID2-tagged core SAC proteins (BUB1, BUB3, BUBR1, MAD1L1, and MAD2L1), which bypasses issues associated with long-term overexpression of key cell division proteins that can compromise cellular homeostasis. We coupled this system to a proximity-labeling proteomic approach to systematically define a proximity protein association map for each of the core SAC proteins. These proximity maps were integrated to generate a core SAC protein proximity protein network. The coupling of the proximity maps/network with curated functional databases like CORUM, GeneOntology, BioGRID, and Reactome allowed for a system-level bioinformatic analysis of the associations within these maps/network. To our knowledge, this is the first systematic characterization of core SAC proteins by proximity-based proteomics.
Our analysis recapitulated many of the core SAC protein–protein interactions, subcomplexes, and complexes that had been previously described. Importantly, it also identified numerous novel associations that warrant further examination. Among these is ELYS, which is associated with MAD1L1 and MAD2L1. Although an interpretation of these associations could be that MAD1L1 and MAD2L1 associate with ELYS at the nuclear pore in preparation for mitotic entry and SAC activation, we favor a model where ELYS may be important for the recruitment of SAC proteins to the kinetochore and/or for checkpoint activation. Future studies aimed at addressing these models should bring clarity to the potential role of ELYS in SAC functioning and cell division. Of interest, previous studies had shown the importance of BUB1 for the localization of the Astrin–Kinastrin minor complex proteins to the kinetochore51−54 and our analysis further determined that the BUB1 kinase activity was important for this function. Together, these data indicate that BUB1 may have a central organizing role not only in SAC activation and function but in SAC silencing and mediating the transition from metaphase to anaphase through its association with the Astrin–Kinastrin minor complex (Figure 5H).
We note that there are limitations to the BioID2 approach (for review, see ref (72)). Although our analysis was conducted from mitotic-enriched populations of cells to enrich for mitotic protein associations, the biotinylation process is conducted over the time frame of hours and some identified associations could represent associations that take place outside of mitosis. These associations could inform on the nonmitotic functions of core SAC proteins, which is a rapidly growing field. Moving forward recent developments in BioID2 technology such as the mini-turboID system should help to resolve proximity associations in a time-dependent manner, as labeling occurs within minutes.73 Our analysis also employed N-terminal BioID2-tagging, and a similar approach using C-terminal tagging of core SAC proteins could lead to different results. Additionally, it is important to note that BioID systems do not identify all known interactions of any specific bait protein. For example, we did not identify the MAD1L1–MAD2L1 interaction in our BioID2 analysis, which is consistent with a previous BioID analysis of MAD1L1.41 Interestingly, we were able to detect the MAD1L1–MAD2L1 interaction when we performed immunoprecipitations with BioID2-MAD1L1 and BioID2-MAD2L1 and immunoblotted for endogenous MAD1L1 or MAD2L1 (Figures S4B and 6A). This indicates that BioID2-MAD1L1 is capable of binding to MAD2L1, but is not able to biotinylate it efficiently. There are many reasons why similar phenomena may occur with other protein pairs and these include a low abundance of surface-exposed lysines on prey proteins (whether absent from the protein surface or buried within a protein complex) and the orientation of the protein interaction could preclude access to lysines on prey proteins.72 Nonetheless, BioID systems have been invaluable to understanding the cellular process and the architecture of cellular structures.14,19,74−76
To facilitate the use and interrogation of the core SAC protein proximity maps/network generated in this study, all mass spectrometry data and R scripts used to analyze the data have been deposited in open-access databases that are freely available to the scientific community (see the Experimental Procedures section). These tools will enable researchers to define novel associations and to generate testable hypotheses to further advance the current understanding of SAC protein function and regulation.
Acknowledgments
This material is based upon the work supported by the National Institutes of Health NIGMS grant numbers R35GM139539 and R01GM117475 to J.Z.T.; any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health NIGMS. Y.A.G. and K.M.C. were supported by the UCLA Tumor Cell Biology Training Program (USHHS Ruth L. Kirschstein Institutional National Research Service Award # T32CA009056). This work was supported in part by a grant to The University of California, Los Angeles, from the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study Program (E.F.V.), by a UCLA Molecular Biology Institute Whitcome Fellowship (E.F.V.) and an NIH P30DK063491 grant (J.P.W.).
Glossary
Abbreviations
- BioID2
biotin identification 2
- SAC
spindle assembly checkpoint
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00941.
Generation of BioID2-tagged SAC protein-inducible stable cell lines (Figure S1); workflow of mass spectrometry data acquisition and analysis (Figure S2); establishment of inducible BioID2-tagged SAC protein (BUB1, BUB3, BUBR1, MAD1L1 and MAD2L1) stable cell lines (Figure S3); BioID2 biochemical purifications (Figure S4); core SAC protein proximity association maps (Figure S5); core SAC protein proximity network (Figure S6); BioID2-BUB1 proximity protein association map (Figure S7); BioID2-BUB3 proximity protein association map (Figure S8); BioID2-BUBR1/BUB1B proximity protein association map (Figure S9); BioID2-MAD1L1 proximity protein association map (Figure S10); BioID2-MAD2L1 proximity protein association map (Figure S11); core SAC protein proximity association network using kinetochore Gene Ontology annotations (Figure S12); core SAC protein proximity association network using mitotic spindle-related Gene Ontology annotations (Figure S13); core SAC protein proximity association network using centromere-related Gene Ontology annotations (Figure S14); kinetochore protein enrichment analysis (Figure S15); core SAC protein proximity association network using nuclear pore-related Gene Ontology annotations (Figure S16); uncropped immunoblots and autoradiography blots for all figures (Figure S17); and uncropped immunoblots for all supplemental figures (Figure S18) (PDF)
List of reagents used (Table S1) (XLSX)
List of primers used (Table S2) (XLSX)
List of vectors generated (Table S3) (XLSX)
Summary of all identified peptides from all BioID2 purifications (Table S4) (XLSX)
Summary of all identified proteins from all BioID2 purifications (Table S5) (XLSX)
Summary of peptides for all proteins that were identified with one peptide sequence (Table S6) (XLSX)
Summary of significant SAC protein proximity associated proteins (Table S7) (XLSX)
List of Gene Ontology (GO) annotations used in the core SAC protein proximity association network analyses (Table S8) (XLSX)
The authors declare no competing financial interest.
Supplementary Material
References
- Holland A. J.; Cleveland D. W. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 478–487. 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A.; Salmon E. D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Gelens L.; Qian J.; Bollen M.; Saurin A. T. The Importance of Kinase-Phosphatase Integration: Lessons from Mitosis. Trends Cell Biol. 2018, 28, 6–21. 10.1016/j.tcb.2017.09.005. [DOI] [PubMed] [Google Scholar]
- Foley E. A.; Kapoor T. M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. T.; Zhang H. The mitotic checkpoint complex (MCC): looking back and forth after 15 years. AIMS Mol. Sci. 2016, 3, 597–634. 10.3934/molsci.2016.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V.; Chan G. K.; Yen T. J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001, 154, 925–936. 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O.; Zou H.; Gerber S. A.; Gygi S. P.; Kirschner M. W. Dual inhibition of sister chromatid separation at metaphase. Cell 2001, 107, 715–726. 10.1016/S0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- Matson D. R.; Stukenberg P. T. Spindle poisons and cell fate: a tale of two pathways. Mol. Interventions 2011, 11, 141–150. 10.1124/mi.11.2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R. M.; van Deursen J. M. Correction of microtubule-kinetochore attachment errors: mechanisms and role in tumor suppression. Semin. Cell Dev. Biol. 2011, 22, 559–565. 10.1016/j.semcdb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S. The spindle assembly checkpoint: progress and persistent puzzles. Biochem. Soc. Trans. 2013, 41, 1755–1760. 10.1042/BST20130240. [DOI] [PubMed] [Google Scholar]
- Corbett K. D. Molecular Mechanisms of Spindle Assembly Checkpoint Activation and Silencing. Prog. Mol. Subcell. Biol. 2017, 56, 429–455. 10.1007/978-3-319-58592-5_18. [DOI] [PubMed] [Google Scholar]
- Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Dou Z.; Prifti D. K.; Gui P.; Liu X.; Elowe S.; Yao X. Recent Progress on the Localization of the Spindle Assembly Checkpoint Machinery to Kinetochores. Cells 2019, 8, 278 10.3390/cells8030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go C. D.; Knight J. D. R.; Rajasekharan A.; Rathod B.; Hesketh G. G.; Abe K. T.; Youn J.-Y.; Samavarchi-Tehrani P.; Zhang H.; Zhu L. Y.; Popiel E.; Lambert J.-P.; Coyaud É; Cheung S. W. T.; Rajendran D.; Wong C. J.; Antonicka H.; Pelletier L.; Raught B.; Palazzo A. F.; Shoubridge E. A.; Gingras A.-C. A proximity biotinylation map of a human cell. bioRxiv 2019, 796391 10.1101/796391. [DOI] [Google Scholar]
- Firat-Karalar E. N.; Stearns T. Probing mammalian centrosome structure using BioID proximity-dependent biotinylation. Methods Cell Biol. 2015, 129, 153–170. 10.1016/bs.mcb.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K. J.; Kim D. I.; Burke B. BioID: a screen for protein-protein interactions. Curr. Protoc. Protein Sci. 2013, 91, 19.23.1–19.23.15. 10.1002/cpps.51.. [DOI] [PubMed] [Google Scholar]
- Mehus A. A.; Anderson R. H.; Roux K. J. BioID Identification of Lamin-Associated Proteins. Methods Enzymol. 2016, 569, 3–22. 10.1016/bs.mie.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. I.; Jensen S. C.; Noble K. A.; Kc B.; Roux K. H.; Motamedchaboki K.; Roux K. J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell 2016, 27, 1188–1196. 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G. D.; Coyaud E.; Goncalves J.; Mojarad B. A.; Liu Y.; Wu Q.; Gheiratmand L.; Comartin D.; Tkach J. M.; Cheung S. W.; Bashkurov M.; Hasegan M.; Knight J. D.; Lin Z. Y.; Schueler M.; Hildebrandt F.; Moffat J.; Gingras A. C.; Raught B.; Pelletier L. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 2015, 163, 1484–1499. 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senese S.; Cheung K.; Lo Y. C.; Gholkar A. A.; Xia X.; Wohlschlegel J. A.; Torres J. Z. A unique insertion in STARD9ʼs motor domain regulates its stability. Mol. Biol. Cell 2015, 26, 440–452. 10.1091/mbc.E14-03-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemeister G.; Mengel A.; Fernandez-Montalvan A. E.; Bone W.; Schroder J.; Zitzmann-Kolbe S.; Briem H.; Prechtl S.; Holton S. J.; Monning U.; von Ahsen O.; Johanssen S.; Cleve A.; Putter V.; Hitchcock M.; von Nussbaum F.; Brands M.; Ziegelbauer K.; Mumberg D. Inhibition of BUB1 Kinase by BAY 1816032 Sensitizes Tumor Cells toward Taxanes, ATR, and PARP Inhibitors In Vitro and In Vivo. Clin. Cancer Res. 2019, 25, 1404–1414. 10.1158/1078-0432.CCR-18-0628. [DOI] [PubMed] [Google Scholar]
- Torres J. Z.; Miller J. J.; Jackson P. K. High-throughput generation of tagged stable cell lines for proteomic analysis. Proteomics 2009, 9, 2888–2891. 10.1002/pmic.200800873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley M.; Ramirez I.; Cheung K.; Gholkar A. A.; Torres J. Z. Inducible LAP-tagged Stable Cell Lines for Investigating Protein Function, Spatiotemporal Localization and Protein Interaction Networks. J. Visualized Exp. 2016, 118, 54870 10.3791/54870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J.; Mann M.; Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Cheung K.; Senese S.; Kuang J.; Bui N.; Ongpipattanakul C.; Gholkar A.; Cohn W.; Capri J.; Whitelegge J. P.; Torres J. Z. Proteomic Analysis of the Mammalian Katanin Family of Microtubule-severing Enzymes Defines Katanin p80 subunit B-like 1 (KATNBL1) as a Regulator of Mammalian Katanin Microtubule-severing. Mol. Cell. Proteomics 2016, 15, 1658–1669. 10.1074/mcp.M115.056465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y.; Oda Y.; Tabata T.; Sato T.; Nagasu T.; Rappsilber J.; Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 2005, 4, 1265–1272. 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Báez-Saldaña A.; Zendejas-Ruiz I.; Revilla-Monsalve C.; Islas-Andrade S.; Cardenas A.; Rojas-Ochoa A.; Vilches A.; Fernandez-Mejia C. Effects of biotin on pyruvate carboxylase, acetyl-CoA carboxylase, propionyl-CoA carboxylase, and markers for glucose and lipid homeostasis in type 2 diabetic patients and nondiabetic subjects. Am. J. Clin. Nutr. 2004, 79, 238–243. 10.1093/ajcn/79.2.238. [DOI] [PubMed] [Google Scholar]
- Giurgiu M.; Reinhard J.; Brauner B.; Dunger-Kaltenbach I.; Fobo G.; Frishman G.; Montrone C.; Ruepp A. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019, 47, D559–D563. 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C.; Breitkreutz B.-J.; Reguly T.; Boucher L.; Breitkreutz A.; Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–9. 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M.; Ball C. A.; Blake J. A.; Botstein D.; Butler H.; Cherry J. M.; Davis A. P.; Dolinski K.; Dwight S. S.; Eppig J. T.; Harris M. A.; Hill D. P.; Issel-Tarver L.; Kasarskis A.; Lewis S.; Matese J. C.; Richardson J. E.; Ringwald M.; Rubin G. M.; Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B.; Matthews L.; Viteri G.; Gong C.; Lorente P.; Fabregat A.; Sidiropoulos K.; Cook J.; Gillespie M.; Haw R.; Loney F.; May B.; Milacic M.; Rothfels K.; Sevilla C.; Shamovsky V.; Shorser S.; Varusai T.; Weiser J.; Wu G.; Stein L.; Hermjakob H.; D’Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M.; Lopes C. T.; Huck G.; Dong Y.; Sumer O.; Bader G. D. Cytoscape.js: a graph theory library for visualisation and analysis. Bioinformatics 2016, 32, 309–311. 10.1093/bioinformatics/btv557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X.; Gholkar A.; Senese S.; Torres J. Z. A LCMT1-PME-1 methylation equilibrium controls mitotic spindle size. Cell Cycle 2015, 14, 1938–1947. 10.1080/15384101.2015.1026487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A. P. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology 2016, 5, 44 10.3390/biology5040044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.; Mendez B. L.; Sedgwick G. G.; Nilsson J. Two functionally distinct kinetochore pools of BubR1 ensure accurate chromosome segregation. Nat. Commun. 2016, 7, 12256 10.1038/ncomms12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A.; Han J. S.; Cleveland D. W. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell 2009, 16, 105–117. 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S.; Ha E.; McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 1998, 142, 1–11. 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overlack K.; Primorac I.; Vleugel M.; Krenn V.; Maffini S.; Hoffmann I.; Kops G. J.; Musacchio A. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. eLife 2015, 4, e05269 10.7554/eLife.05269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antoni A.; Pearson C. G.; Cimini D.; Canman J. C.; Sala V.; Nezi L.; Mapelli M.; Sironi L.; Faretta M.; Salmon E. D.; Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005, 15, 214–225. 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Zhang G.; Kruse T.; Lopez-Mendez B.; Sylvestersen K. B.; Garvanska D. H.; Schopper S.; Nielsen M. L.; Nilsson J. Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat. Commun. 2017, 8, 15822 10.1038/ncomms15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskell D. P.; Hu X. W.; Singleton M. R. Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 2010, 190, 823–834. 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A.; Pasqualato S.; Dube P.; Krenn V.; Santaguida S.; Cittaro D.; Monzani S.; Massimiliano L.; Keller J.; Tarricone A.; Maiolica A.; Stark H.; Musacchio A. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 2010, 190, 835–852. 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S. L.; Cheeseman I. M.; Hori T.; Fukagawa T.; Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 2006, 173, 9–17. 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E.; De Antoni A.; Alushin G. M.; Petrovic A.; Melis T.; Nogales E.; Musacchio A. Direct Binding of Cenp-C to the Mis12 Complex Joins the Inner and Outer Kinetochore. Curr. Biol. 2011, 21, 391–398. 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obuse C.; Iwasaki O.; Kiyomitsu T.; Goshima G.; Toyoda Y.; Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004, 6, 1135–1141. 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- Ghongane P.; Kapanidou M.; Asghar A.; Elowe S.; Bolanos-Garcia V. M. The dynamic protein Knl1 - a kinetochore rendezvous. J. Cell Sci. 2014, 127, 3415–3423. 10.1242/jcs.149922. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez J. A.; Lewis C.; McKinley K. L.; Sikirzhytski V.; Corona J.; Maciejowski J.; Khodjakov A.; Cheeseman I. M.; Jallepalli P. V. Distinct Roles of RZZ and Bub1-KNL1 in Mitotic Checkpoint Signaling and Kinetochore Expansion. Curr. Biol. 2018, 28, 3422–3429. 10.1016/j.cub.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; e5
- Krenn V.; Wehenkel A.; Li X.; Santaguida S.; Musacchio A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 2012, 196, 451–467. 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Santos M. D.; Hervas-Aguilar A.; Sewart K.; Lancaster T. C.; Meadows J. C.; Millar J. B. Bub3-Bub1 Binding to Spc7/KNL1 Toggles the Spindle Checkpoint Switch by Licensing the Interaction of Bub1 with Mad1-Mad2. Curr. Biol. 2016, 26, 2642–2650. 10.1016/j.cub.2016.07.040. [DOI] [PubMed] [Google Scholar]
- Dunsch A. K.; Linnane E.; Barr F. A.; Gruneberg U. The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol. 2011, 192, 959–968. 10.1083/jcb.201008023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein K. H.; Kleylein-Sohn J.; Nigg E. A.; Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 2007, 178, 345–354. 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D. M.; Monda J. K.; Su K. C.; Wilson-Kubalek E. M.; Cheeseman I. M. Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. eLife 2017, 6, e26866 10.7554/eLife.26866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M.; Tanaka K. Plk1 bound to Bub1 contributes to spindle assembly checkpoint activity during mitosis. Sci. Rep. 2017, 7, 8794 10.1038/s41598-017-09114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar A.; Lajeunesse A.; Dulla K.; Combes G.; Thebault P.; Nigg E. A.; Elowe S. Bub1 autophosphorylation feeds back to regulate kinetochore docking and promote localized substrate phosphorylation. Nat. Commun. 2015, 6, 8364 10.1038/ncomms9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yakoubi W.; Buffin E.; Cladiere D.; Gryaznova Y.; Berenguer I.; Touati S. A.; Gomez R.; Suja J. A.; van Deursen J. M.; Wassmann K. Mps1 kinase-dependent Sgo2 centromere localisation mediates cohesin protection in mouse oocyte meiosis I. Nat. Commun. 2017, 8, 694 10.1038/s41467-017-00774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron A. P.; von Schubert C.; Cubizolles F.; Siemeister G.; Hitchcock M.; Mengel A.; Schroder J.; Fernandez-Montalvan A.; von Nussbaum F.; Mumberg D.; Nigg E. A. Probing the catalytic functions of Bub1 kinase using the small molecule inhibitors BAY-320 and BAY-524. eLife 2016, 5, 253 10.7554/eLife.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R.; Henzing A. J.; Earnshaw W. C. Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma 2005, 113, 385–397. 10.1007/s00412-004-0326-0. [DOI] [PubMed] [Google Scholar]
- Tadeu A. M.; Ribeiro S.; Johnston J.; Goldberg I.; Gerloff D.; Earnshaw W. C. CENP-V is required for centromere organization, chromosome alignment and cytokinesis. EMBO J. 2008, 27, 2510–2522. 10.1038/emboj.2008.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y.; Salic A.; Dasso M.; Arnaoutov A. Bub1 is essential for assembly of the functional inner centromere. J. Cell Biol. 2007, 176, 919–928. 10.1083/jcb.200609044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; McCormick F.; Saya H. Mad2 inhibits the mitotic kinesin MKlp2. J. Cell Biol. 2010, 191, 1069–1077. 10.1083/jcb.201003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R. J.; Lusk C. P.; Dilworth D. J.; Aitchison J. D.; Wozniak R. W. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 4362–4374. 10.1091/mbc.e05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; Sterling H.; Burlingame A.; McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008, 22, 2926–2931. 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussi Y. C.; Shumaker D. K.; Shimi T.; Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus 2010, 1, 71–84. 10.4161/nucl.1.1.10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ródenas E.; Gonzalez-Aguilera C.; Ayuso C.; Askjaer P. Dissection of the NUP107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein MAD1 in Caenorhabditis elegans. Mol. Biol. Cell 2012, 23, 930–944. 10.1091/mbc.e11-11-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V.; Maciejowski J.; Corona J.; Buch H. K.; Collin P.; Kanemaki M. T.; Shah J. V.; Jallepalli P. V. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 2014, 156, 1017–1031. 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilera C.; Askjaer P. Dissecting the NUP107 complex: multiple components and even more functions. Nucleus 2012, 3, 340–348. 10.4161/nucl.21135. [DOI] [PubMed] [Google Scholar]
- Mossaid I.; Fahrenkrog B. Complex Commingling: Nucleoporins and the Spindle Assembly Checkpoint. Cells 2015, 4, 706–725. 10.3390/cells4040706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A.; Orjalo A. V.; Shen Z.; Briggs S.; Forbes D. J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 17801–17806. 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N.; Cheerambathur D.; Moyle M.; Stefanutti M.; Richardson A.; Lee K. Y.; Dumont J.; Oegema K.; Desai A. A Nucleoporin Docks Protein Phosphatase 1 to Direct Meiotic Chromosome Segregation and Nuclear Assembly. Dev. Cell 2016, 38, 463–477. 10.1016/j.devcel.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley N.; Desai A. The nucleoporin MEL-28/ELYS: A PP1 scaffold during M-phase exit. Cell Cycle 2017, 16, 489–490. 10.1080/15384101.2017.1278929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P.; Samson R.; Gingras A. C. Proximity Dependent Biotinylation: Key Enzymes and Adaptation to Proteomics Approaches. Mol Cell Proteomics 2020, 19, 757–773. 10.1074/mcp.R120.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon T. C.; Bosch J. A.; Sanchez A. D.; Udeshi N. D.; Svinkina T.; Carr S. A.; Feldman J. L.; Perrimon N.; Ting A. Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J. Y.; Dunham W. H.; Hong S. J.; Knight J. D. R.; Bashkurov M.; Chen G. I.; Bagci H.; Rathod B.; MacLeod G.; Eng S. W. M.; Angers S.; Morris Q.; Fabian M.; Cote J. F.; Gingras A. C. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517.e11–532.e11. 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Coyaud E.; Mis M.; Laurent E. M.; Dunham W. H.; Couzens A. L.; Robitaille M.; Gingras A. C.; Angers S.; Raught B. BioID-based Identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 Ligase Substrates. Mol. Cell. Proteomics 2015, 14, 1781–1795. 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remnant L.; Booth D. G.; Vargiu G.; Spanos C.; Kerr A. R. W.; Earnshaw W. C. In vitro BioID: mapping the CENP-A microenvironment with high temporal and spatial resolution. Mol. Biol. Cell 2019, 30, 1314–1325. 10.1091/mbc.E18-12-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.