Abstract
The testis transcriptome is highly complex and includes RNAs that potentially hybridize to form double-stranded RNA (dsRNA). We isolated dsRNA using the monoclonal J2 antibody and deep-sequenced the enriched samples from testes of juvenile Dicer1 knockout mice, age-matched controls, and adult animals. Comparison of our data set with recently published data from mouse liver revealed that the dsRNA transcriptome in testis is markedly different from liver: In testis, dsRNA-forming transcripts derive from mRNAs including promoters and immediate downstream regions, whereas in somatic cells they originate more often from introns and intergenic transcription. The genes that generate dsRNA are significantly expressed in isolated male germ cells with particular enrichment in pachytene spermatocytes. dsRNA formation is lower on the sex (X and Y) chromosomes. The dsRNA transcriptome is significantly less complex in juvenile mice as compared to adult controls and, possibly as a consequence, the knockout of Dicer1 has only a minor effect on the total number of transcript peaks associated with dsRNA. The comparison between dsRNA-associated genes in testis and liver with a reported set of genes that produce endogenous siRNAs reveals a significant overlap in testis but not in liver. Testis dsRNAs also significantly associate with natural antisense genes—again, this feature is not observed in liver. These findings point to a testis-specific mechanism involving natural antisense transcripts and the formation of dsRNAs that feed into the RNA interference pathway, possibly to mitigate the mutagenic impacts of recombination and transposon mobilization.
The mammalian testis displays the highest transcriptome diversity of all organs, even exceeding that of brain (Werner et al. 2007; Soumillon et al. 2013). All transcript categories including protein-coding and noncoding RNAs as well as antisense transcripts are overrepresented in testis. Many of even highly expressed protein-coding transcripts are not translated (Wang et al. 2019), suggesting that the RNA itself or the process of transcription fulfills a biological role.
A family of RNAs that is prominently expressed in the testis are natural antisense transcripts (NATs). NATs are fully processed RNAs, that is, spliced, polyadenylated, and capped, that are produced from the opposite strand of protein-coding genes and share sequence complementarity with the related sense transcript (Faghihi and Wahlestedt 2009; Zinad et al. 2017). Depending on sequencing depth and data processing, 40% or more of all protein-coding genes in humans produce NATs; a further hallmark of which is the significant underrepresentation of NATs on mammalian X Chromosomes. The sequence complementarity between sense and antisense transcripts and the prevalence of NATs suggest that double-stranded RNA (dsRNA) formation may be common, especially in testis with its complex transcriptome (Kiyosawa et al. 2003; Chen et al. 2004).
dsRNA formation is, in theory, a frequent event in mammalian cells, not only as a result of widespread NAT expression but also of pervasive transcription from repetitive regions of the genome and bidirectional transcription of mitochondrial DNA (Kim et al. 2019). The formation of endogenous dsRNA, however, conflicts with the cell's defense system against viruses, which is geared to recognize dsRNA. The dilemma appears to be solved, at least in somatic cells, by tight containment of dsRNA within mitochondria as well as efficient transcriptional repression of repetitive elements and retention of spurious transcripts in the nucleus (Dhir et al. 2018; Kim et al. 2018). A breakdown of these defense mechanisms, for example, by an increased transcription of nuclear elements with sequence complementarity or reduced removal of mitochondrial dsRNA, has potentially catastrophic consequences for a cell and possibly the entire organism. A cytosolic accumulation of dsRNA is typical of viral infection and triggers a strong inflammatory response. Accordingly, stimulation of dsRNA sensor proteins, such as interferon induced with helicase C domain 1 (IFIH1, also known as MDA5), eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2, also known as PKR), adenosine deaminase RNA specific (ADAR), DExD/H-box helicase 58 (DDX58, also known as RIG-I) has been reported in response to various stressors that affect mitochondrial integrity or reduce transcriptional repression by DNA methylation (Hur 2019).
Because testis represents an immunologically privileged environment, the effects of dsRNA formation may be different in male germ cells as compared to somatic tissues (Zhao et al. 2014). Hence high levels of dsRNA formation as a result of pervasive transcription may be tolerated in male germ cells and have a specific purpose there. We have previously suggested an evolutionary mechanism involving antisense transcription, dsRNA formation, and endogenous siRNA (endo-siRNA) production to parse a viable sperm population from those that suffered catastrophic mutagenic events during recombination and transposon mobilization (Werner et al. 2009, 2015). The hypothesis is supported by the recent finding that the complex transcriptome in testis results in lower mutation rates in genes that are expressed during spermatogenesis (Xia et al. 2020).
Male germ cells display a highly complex transcriptome that potentially produces intermolecular dsRNA structures. Here, we enriched and sequenced dsRNA from mouse testis and compared it to a similar data set from somatic liver cells. We aimed to establish potential differences between the dsRNA transcriptome of male germ cells versus somatic (liver) cells. We then sought to establish whether dsRNA is a substrate for DICER1 to generate endo-siRNAs (endo-siRNAs) and if natural antisense transcripts coexpressed with the cognate sense transcripts contribute form detectable levels of dsRNA.
Results
dsRNAs are enriched in mouse testis
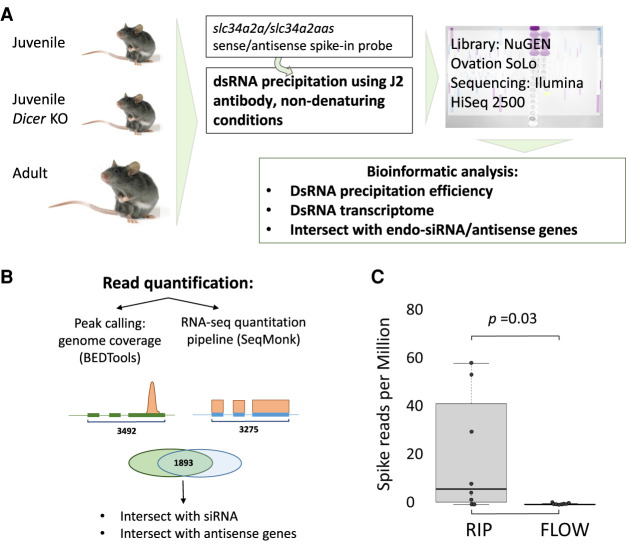
Testes show the most complex transcriptome of all organs in mammals including a comprehensive array of natural antisense transcripts. We hypothesized that such transcriptional complexity may result in substantial formation of dsRNA that can be isolated using the dsRNA-specific antibody J2 (Dhir et al. 2018). Because RNA extraction methods introduce, depending on the particular methodology, either a significant positive or negative bias toward dsRNA, antibody pull-down of dsRNA was performed without prior RNA extraction. A hybridized sense–antisense transcript pair (slc34a2a/slc34a2aas) from zebrafish (Nalbant et al. 1999) was added to the tissue lysate as a spike-in probe (Fig. 1A).
Figure 1.
Experimental strategy to characterize the dsRNA transcriptome of mouse testis. (A) Testes from three groups of animals were collected, including 18-d-old wild-type and Dicer1 knockout mice as well as adult mice (4–6 mo old). The dsRNA was immune purified using the specific J2 antibody and both bound and unbound RNA samples were sequenced. (B) Two quantification methods were applied to call expressed genes; (1) peak calling using BEDTools (Quinlan and Hall 2010) followed by quantification using DESeq2, and (2) the RNA-seq pipeline in SeqMonk (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) that quantifies exon mapping reads and returns RPKM values. Both pipelines yield lists of gene coordinates in BED/bedGraph format. (C) Enrichment of the spike-in probe in the J2 treated samples. Reads per million of the spike-in probe in the J2 immuno-enriched samples (RIP) versus the flowthrough samples (FLOW). The dots represent results from the eight different samples. t-test confirmed a significant accumulation of dsRNA in the J2-enriched samples.
Testes of eight mice were used in the study: three juvenile (18 d old) wild-type (WT) mice, three age-matched male germ cell–specific Dicer1 knockout animals (Zimmermann et al. 2014), and two adult WT control mice. After incubation with the J2 antibody, both bound dsRNA and the flowthrough were sequenced. A total of 18.44 million reads were obtained from the antibody-bound samples (RIP) and 73.30 million reads from the flowthrough (FLOW); of these, 10.16 and 54.52 million reads, respectively, mapped uniquely to the mouse reference genome (Supplemental Table S1). The reads derived from the spike-in probe were almost entirely found in the RIP samples, although in some samples in very low numbers, indicating a comparably shallow read depth (Fig. 1C). Nevertheless, the significant enrichment of the dsRNA probe in the RIP samples confirmed the validity of the protocol and the specificity of the antibody.
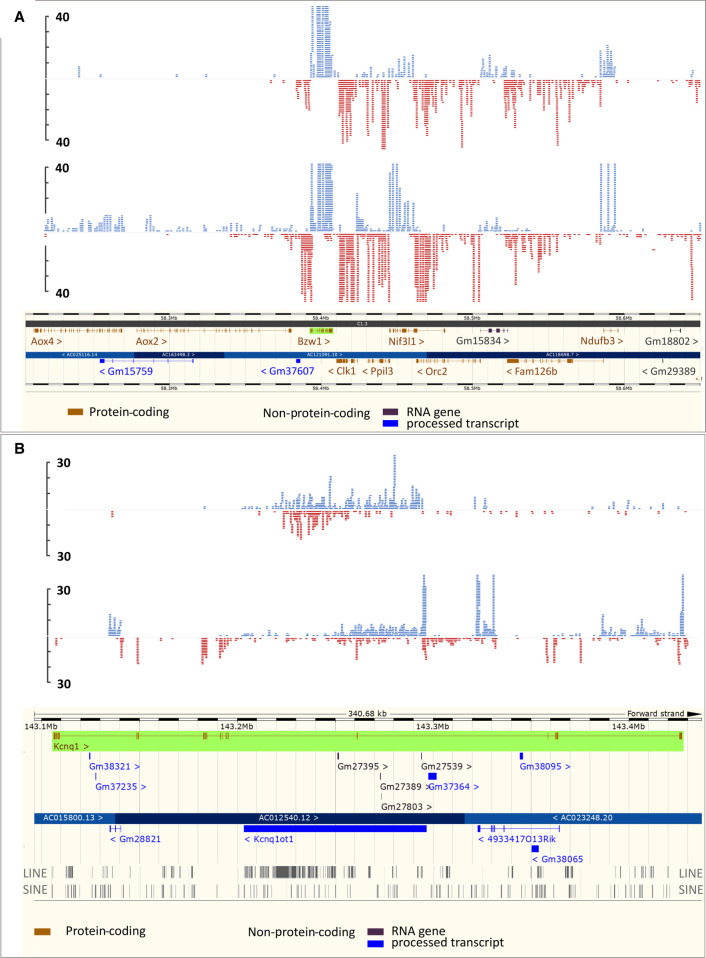
Figure 2 includes two examples of aligned reads from a J2-enriched sample (R2) and a total RNA-seq testis experiment (Pervouchine et al. 2015), showing a cluster including the genes Gm37600, Bzw1, Clk1, Ppil3, Nif3l1, Orc2, Gm15834, Fam126b, and Ndufb3 (194 kbp) that potentially form dsRNA on several occasions (Fig. 2A). The pattern is reflected in the total RNA-seq testis sample. The reads are not enriched in double-stranded regions, in line with the experimental procedure that recovers the entire transcripts that form hybrids and not only the complementary regions. An adjacent cluster (Aox4, Gm15759, Aox2) with potentially overlapping transcripts is not expressed, hence no dsRNA was detectable. On the other hand, Kcnq1ot1 shows reads on both strands with minimal expression of Kcnq1, possibly due to the clustering of LINE and SINE elements in Kcnq1ot1 that can form intramolecular dsRNA (Fig. 2B). Moreover, the read pattern is different between the J2-enriched and the total RNA-seq samples.
Figure 2.
Genome browser snapshots (SeqMonk) of representative examples of dsRNA-associated gene clusters. Two individual data sets with high read coverage from adult murine kidney (R2 and GSM900193) (Pervouchine et al. 2015) are shown. The upper panels represent the dsRNA-enriched sample (R2), the lower panels represent testis RNA (GSM900193). (A) The cluster encompassing the genes Bzw1, Clk1, Ppil3, Nif3l1, Orc2, Gm15834, Fam126b, and Ndufb3 shows reads mapping to both strands in exons of the related genes, although at low levels. There are hardly any regions that contain reads in both orientations. The adjacent cluster, also containing genes with complementary exons is not expressed. (B) Snapshot of the parentally imprinted Kcnq1 gene. The protein-coding sense transcript is not expressed in males; however, the related antisense transcript is expressed, and reads in both orientations are detected. This could be the result of intramolecular dsRNA formation by SINE and LINE elements enriched in this region. The blue bars represent the + and the red bars the – strand.
dsRNAs derive from testis- and liver-specific genic regions on autosomes
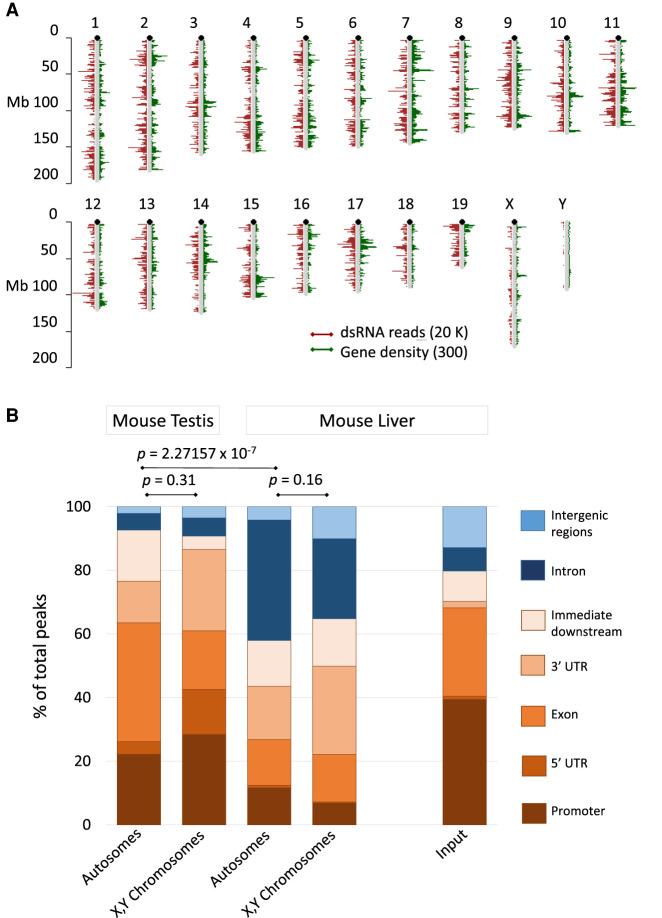
To characterize a comprehensive dsRNA transcriptome we pursued two strategies, one focusing on read peaks and the other on gene expression (Fig. 1B). The first one involved peak calling using genome coverage (genomecov, BEDTools) (Quinlan and Hall 2010), and calls from all samples were combined followed by the identification of genes associated with peaks. Combined reads from all eight samples were visualized per chromosome in relation to gene density (Fig. 3A). The dsRNA read depth follows the pattern of protein-coding gene density indicating that transcripts from these genes give rise to dsRNAs (Fig. 3A). Peaks that reached a threshold of five times more than background or higher were then annotated using ChIPpeakAnno (Zhu et al. 2010), and the genomic coordinates were compiled (Fig. 3B). Peaks were predominantly associated with protein-coding sequences, including regulatory features such as promoters, exons, and flanking regions confirming the matching appearance of read and gene densities in Figure 3A. The proportion between peaks in exons versus introns was 7.2 to 1. A total of 3328 peaks were detected on autosomes, 128 on the X, and 13 on the Y Chromosome, respectively, indicating a bias against the sex chromosomes. The vast majority of these peaks (97.2%) were associated with protein-coding and a few noncoding genes, whereas only 2.8% of peaks were found in intergenic regions. The sex chromosomes showed a comparable trend (97.6 vs. 2.4% on the X Chromosome and 84.6 vs. 15.4% on the Y Chromosome) (Fig. 3; Supplemental Table S2).
Figure 3.
dsRNA transcriptome of combined mouse testis samples and liver. (A) Chromosomal distribution of dsRNA reads from testis. The dsRNA reads are in red; annotated protein-coding and noncoding genes in green are for comparison. dsRNA reads and genes show a comparable pattern. Sex chromosomes display less dsRNA compared to autosomes. The bars represent dsRNA read- or gene density per million bases. (B) J2 antibody enriched reads from mouse testis and liver were aligned and quantified; the input from mouse liver served as a control. The resulting peaks were then annotated to biotypes (color-coded as indicated) using ChIPpeakAnno. Features related to the regulation or the structure of annotated transcripts (promoters, exons, UTRs, and immediate downstream regions; GENCODE.vM20.annotation.gff3) are shaded in brown. Introns and intergenic regions are in shades of blue. Promoters encompass 1000 bp upstream of the transcription start; “immediate downstream” includes 1000 bp downstream from the transcript end. Autosomes and sex chromosomes are displayed in separate columns because sex chromosomes are enriched in repetitive elements and depleted in natural antisense transcripts, both of which are associated with dsRNA formation. The pipeline is described in the Methods and graphically outlined in Supplemental Figure S1.
To investigate a potential difference between male germ cells and somatic cells, we used our pipeline to analyze a published data set from mouse fetal liver including J2-enriched samples and an input control (Gao et al. 2020). The experiments revealed a significantly different distribution of loci associated with dsRNA, particularly in peak numbers associated with exons versus introns. In testis the proportion was 37.3% to 5.2% compared to liver with 14.4% and 37.8% of total peaks, respectively (Fig. 3B). The overall difference between the two samples is highly significant (χ2, 89.92, P-value = 3.13 × 10−17). An additional difference between male germ cells and somatic cells concerns the 5′ flanking regions, which are associated with 4% of dsRNA peaks in testis but <1% in mouse liver, respectively. The sex chromosomes are distinct from autosomes in two features relevant to dsRNA formation, specifically repetitive elements are enriched, whereas antisense transcripts are depleted, and both features are associated with dsRNA formation. However, neither in mouse testis nor liver was a significant difference in peak-associated biotypes between sex chromosomes and autosomes observed. On the other hand, both J2-purified samples showed a distinct biotype distribution when compared to mouse liver input control (P < 0.001). Promoter-associated peaks are enriched in the input sample (39.4% vs. 11.6% liver autosomes or 22.2% testis autosomes in J2-purified samples) possibly related to shorter, promoter-associated transcripts that do not form dsRNA (Preker et al. 2008; Djebali et al. 2012). These results confirm that male germ cells and somatic cells produce a notably different set of transcripts that form dsRNA: In male germ cells, the clear majority of reads are associated with exons or regulatory sequences of protein-coding genes. Conversely, in somatic cells, introns and intergenic regions of the genome contribute to ∼50% or more to the dsRNA transcriptome.
dsRNAs are highly abundant in pachytene spermatocytes
Because the majority of dsRNA-related read peaks in testis associate with annotated genes, we generated a list of the expressed transcripts using the RNA-seq quantitation pipeline in SeqMonk with a cutoff of eight times more than background and a “present” call in at least six of the eight samples. This approach produced a set of 3275 genes that form dsRNA in testis. Again, genes on the sex chromosomes are significantly underrepresented in this list (42 on the X Chromosome and one on the Y Chromosome vs. 3232 on autosomes) (Supplemental Table S2).
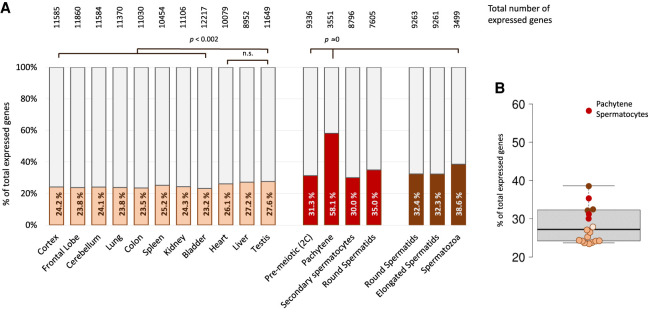
We used this list to obtain an impression of whether the genes associated with dsRNA formation show tissue-specific expression using publicly available transcriptome data (brain cortex, frontal lobe, cerebellum, lung, colon, spleen, kidney, bladder, heart, liver, and testis) (Pervouchine et al. 2015) as well as from staged developing male germ cells (premeiotic, pachytene, secondary spermatocytes, round spermatids) (da Cruz et al. 2016); round spermatids, elongated spermatids, and spermatozoa) (Zuo et al. 2016). Among the compilation of different tissues, testis expressed the highest proportion of dsRNA-forming genes with 27.6% versus 23.5–27.2% in other tissues (Fig. 4A). The percentage of expressed dsRNA-forming genes was elevated in isolated, developing male germ cells (33.3 ± 3.1%) and particularly in pachytene spermatocytes with a proportion of 58.1% (P = close to 0) (Fig. 4B). Of note, the number of genes with positive calls is clearly reduced in pachytene spermatocytes and spermatozoa as compared to other stages. Despite the fact that the pipeline may only identify one transcript of a pair that forms dsRNA, the significant accumulation of mRNAs that form dsRNA (with an unidentified complementary transcript) in pachytene spermatocytes supports our focus on testis and suggests a biological role for dsRNA in this particular cell type.
Figure 4.
Expression of potentially dsRNA-forming genes in various mouse tissues and staged male germ cells. (A) Bar graph indicating the total number of expressed genes (100%, indicated above the bars) and the percentage of dsRNA-forming genes within the colored, lower part of the bars. Brown and red areas reflect the proportion of genes that form dsRNA in testis (dsRNA-associated genes), and the light gray areas are expressed genes without evidence of dsRNA. The different colors represent the data sets from Pervouchine (light brown), da Cruz (red), and Zuo (brown) (Pervouchine et al. 2015; da Cruz et al. 2016; Zuo et al. 2016). (B) Compilation of all the values presented in A. Box plot indicating the median (solid black line) and 25th and 75th percentiles as box limits. Whiskers show 1.5 times the interquartile range; pachytene spermatocytes represent a clear outlier. χ2 test was performed, and related P-values are indicated.
We tested four loci with established complementary sense and antisense transcripts by RT-qPCR. Loci that contained a protein-coding gene and also produced a spliced, lowly expressed antisense transcript that shares complementarity in exons with the sense gene were selected. We designed four primer pairs for each sense–antisense pair that amplify fragments from noncomplementary and complementary regions of the mRNAs (Supplemental Fig. S2). Bidirectional transcription of the four loci was confirmed with a general trend that protein-coding sense transcripts are expressed at higher levels. Expression levels determined by RNA-seq and RT-qPCR did not correspond well, likely because RT-qPCR focuses on specific small regions, whereas RNA-seq quantification integrates the entire transcript.
The analysis of the dsRNA transcriptome has so far established that dsRNA is significantly more prevalent in testis, particularly in pachytene spermatocytes, than in somatic cells. Moreover, dsRNA in testis derives from annotated genic regions rather than intergenic sequences and introns.
dsRNAs are associated with endo-siRNAs and antisense transcripts
Significant levels of endogenous siRNA have been reported in testis, suggesting that the dsRNA may be processed by Dicer1 into endo-siRNAs (Song et al. 2011). To test this hypothesis, we first compared a published endo-siRNA data set from mouse testis (Hilz et al. 2017) with our dsRNA data. Moreover, we assessed the dsRNA transcriptome from juvenile, male, germ cell–specific Dicer1 knockout (KO) mice and age-matched wild-type littermate controls on the assumption that DICER1 may be involved in dsRNA processing. Reads were aligned and expression quantified using the pipeline for peak calling as applied previously for the testis and liver data sets. To determine overlapping regions, the coordinates of genes associated with dsRNA and with endo-siRNA were intersected using the online tool BedSect (https://imgsb.org/bedsect/) (Mishra et al. 2020). We compared the dsRNA-associated genes from mouse testis (3492 entries) and liver (RIP, 4900 entries, and input control, 4061) to the published data set of endo-siRNAs (3712 entries) in testis (Hilz et al. 2017) as well as to a list of annotated natural antisense genes (2991 entries, Ensembl BioMart).
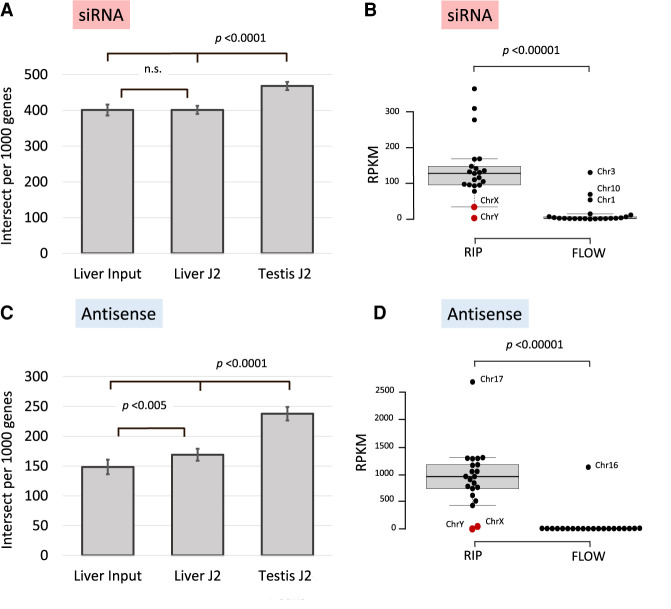
The coordinates of 1000 randomly selected dsRNA genes from mouse testis and liver were intersected with endo-siRNA-associated genes. The number of genes with dsRNA and endo-siRNA formation was comparable in mouse liver J2-enriched samples and input control (401.2 ± 15.2 vs. 401.4 ± 11.4 per 1000 genes) but significantly different from a set of genes associated with dsRNA reads in testis (468.3 ± 11.2; P < 0.0001) (Fig. 5A). This result was confirmed by comparing normalized reads in mouse testis RIP and FLOW samples. The RIP samples displayed about 10 times as many normalized reads that intersected with siRNAs signals as counted with the FLOW samples (140.04 vs. 15.37 RPKM, P < 0.0001, respectively) (Fig. 5B). Three chromosomes showed higher counts in the FLOW samples (Chr 1, 3, and 10) caused by highly expressed peaks that skew the otherwise “normal” proportion (Supplemental Fig. S3). Of note, X and Y Chromosomes display a significantly lower number of dsRNA reads that correlate with endo-siRNAs (Fig. 5B).
Figure 5.
Reads in J2-enriched samples that intersect with endo-siRNAs (Hilz et al. 2017) and antisense transcripts. Samples from testis and liver as well as the flowthrough (FLOW) were analyzed, the upper panel (A,B) showing the overlap with siRNA forming genomic regions, the lower panel (C,D) with antisense genes. (A) Number of intersected genes per 1000 genes, dsRNA samples from testis show significantly more genes that generate endo-siRNAs than both J2-enriched samples and input control from liver. (B) Normalized number of fragments on individual chromosomes per megabase in combined RIP samples from mouse testis as compared to FLOW samples. Each dot represents a specific chromosome; the box plot indicates the median, 25th, and 75th percentiles as box limits and 1.5 times the interquartile range (whiskers); Chromosomes 1, 3, and 10 are clear outliers in the FLOW samples (Supplemental Fig. S3). The sex chromosomes tend to show a lesser overlap between dsRNA and endo-siRNA formation even if the lower number of dsRNA peaks is considered. (C) Number of dsRNA-forming genes intersected with antisense genes. The coordinates of 2991 antisense genes were retrieved and lists of 1000 randomly selected genes were intersected with lists from testis (1000 of 3492) and liver (1000 of 4900 J2 and 1000 of 4061 input control). The dsRNA-associated genes in both testis and liver are significantly associated with antisense genes (P < 0.0001 and P < 0.005, respectively). (D) Total number of reads on individual chromosomes per megabase in combined RIP and FLOW samples that intersect with antisense transcripts. The box plot gives the median, 25th, and 75th percentiles as box limits and 1.5 times the interquartile range (whiskers); Chromosome 16 represents an outlier in the FLOW samples.
We then interrogated the two dsRNA-enriched samples from mouse testis and liver (same data sets as for the endo-siRNA analysis) for a potential association with natural antisense transcripts. The coordinates of antisense genes (2991) were intersected with 1000 randomly selected dsRNA-associated genes from mouse testis and liver (3492 and 4900 genes, respectively). Again, the input sample from mouse liver was used as a control. As shown in Figure 5C, both dsRNA-enriched samples (testis and liver) are associated with antisense transcripts, although in testis the link is clearly more pronounced (input control 148.3 ± 12.3 vs. 168.9 ± 10.2 and 237.8 ± 11.4, liver and testis, respectively). Moreover, the association between dsRNA and antisense transcripts was significantly higher in the J2-enriched as compared to the FLOW samples (P < 0.0001) (Fig. 5D).
The second strategy to investigate a potential link between dsRNA and endo-siRNA focused on the dsRNA transcriptome from juvenile, male germ cell–specific Dicer1 knockout (KO) mice and age-matched wild-type littermate controls. The late phases of spermatogenesis are severely disrupted in Dicer1 KO mice (Korhonen et al. 2011); therefore, we used testes from 18-d-old juvenile mice before the onset of spermatogenic defects.
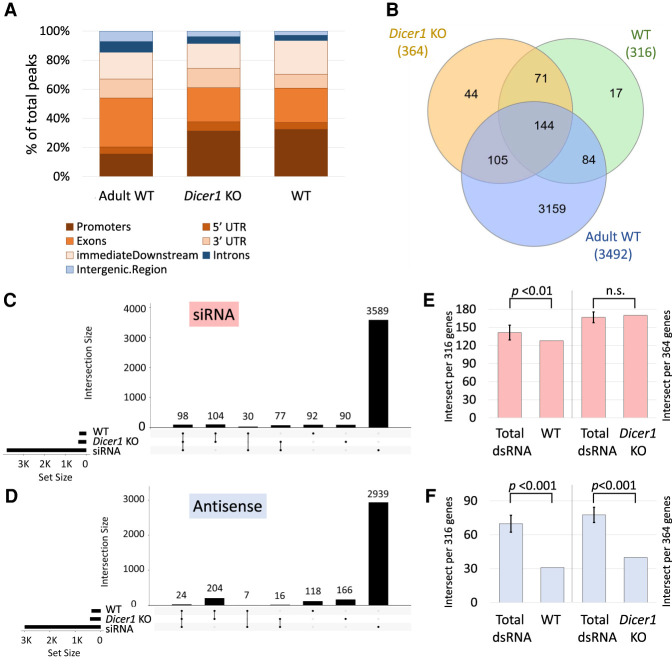
In general, the dsRNA transcriptome was comparable between juvenile WT and Dicer1 KO mice (316 vs. 364 peaks) but significantly less complex than in adult WT mice (3461 peaks). Again, the peaks are predominantly associated with protein-coding sequences, with <10% mapping to introns or intergenic regions (Fig. 6A). However, almost half of the genes (148) were differentially expressed: 95 were significantly overexpressed (P ≤ 0.05), and 53 were suppressed in Dicer1 KO animals (Supplemental Table S3). Of note, none of the 148 genes mapped to the mitochondrial genome, which produces the highest density of dsRNA. The peak finding pipeline applied to the WT and Dicer1 KO samples failed to establish significant differences in dsRNA peak occurrence (Fig. 6A). We next overlapped the genomic regions of J2-bound peaks in the three data sets (juvenile WT, Dicer1 KO, and adult WT mice). We found 144 genes to be present in all three data sets and only 44 and 17 peak-associated genes solely present in Dicer1 KO or juvenile WT controls, respectively (Fig. 6B). To test whether the data sets from juvenile mice were associated with either siRNAs or antisense genes, we intersected the list of coordinates with the previously described set of siRNA and antisense genes. As shown by the upset plots there are more dsRNA peak-associated genes in both the siRNA and antisense list for the samples from Dicer1 KO mice versus WT (77 vs. 30 siRNA and 16 vs. 7 antisense, respectively); notably, the numbers are very small (Fig. 6C,D). To assess whether the juvenile animals show differences in the association between dsRNA and endo-siRNA/antisense seen for the total dsRNA data set, we intersected Dicer1 KO and WT samples with the endo-siRNA and antisense gene coordinates and compared it to size-matched control sets of random dsRNA-associated genes. Figure 6, E and F, indicate that the overlap between dsRNA genes and siRNA-related genes is comparable between adult and juvenile animals, whereas the juvenile data sets contain clearly less antisense-associated peaks (P < 0.001). Of note, these experiments involved juvenile mice, and the complexity of the dsRNA transcriptome is expected to increase with age as may the contribution of DICER1 in processing dsRNA (Björkgren and Sipilä 2015).
Figure 6.
dsRNA formation in juvenile Dicer1 KO mice and age-matched wild-type controls. (A) dsRNA peaks in the three samples (Adult WT, Dicer1 KO, and WT) were annotated to biotypes using ChIPpeakAnno: Promoter, 5′ UTR, exon, 3′ UTR, immediate downstream (1000 bp) are in shades of brown, and introns and intergenic regions are in shades of blue. (B) Venn diagram depicting the genes associated with dsRNA peaks in Dicer1 KO, WT, and adult mouse testis and the overlaps between the samples. (C) The coordinates of genes associated with peaks in Dicer1 KO samples and WT controls were intersected with the coordinates of siRNA-associated genes (in red) (Hilz et al. 2017) and antisense genes (in blue) in D, respectively, and visualized in upset plots. In both cases, the Dicer1 KO specific genes show greater association with siRNA-associated and antisense genes (siRNA 77 vs. 30, antisense 16 vs. 7). (E) Number of dsRNA-associated genes in Dicer1 KO and WT controls that intersect with siRNA-associated genes, compared to an average of 10 size-matched random samples from the total dsRNA gene set. (F) Number of dsRNA-associated genes in Dicer1 KO and WT controls that intersect with antisense transcripts compared to the control total dsRNA gene set. Significance was determined by one-sample t-test.
dsRNA formation is essential for male germ cell development
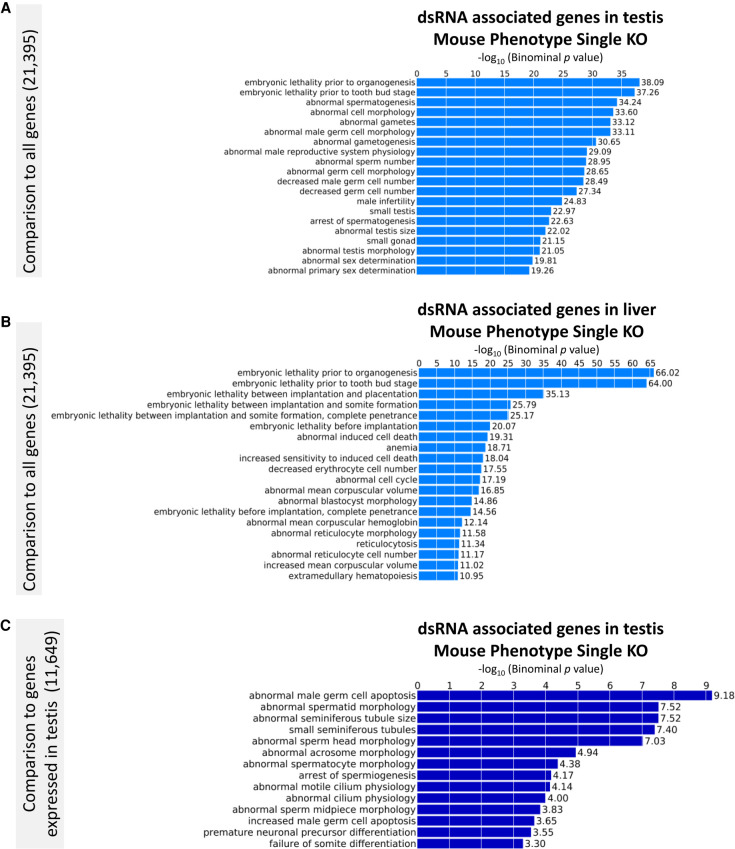
Finally, we generated a parsed list of dsRNA genes by intersecting the two dsRNA lists from testis (1893) (Fig. 1B; Supplemental Table S2). We used this list of dsRNA-related genes to assess phenotypic consequences of a single gene knockout (Fig. 7). The same approach was performed with the dsRNA-associated genes in liver as a comparison. We also examined the extent to which dsRNA-forming genes contributed to specific phenotypes on the background of genes expressed in testis (Pervouchine et al. 2015) using Genomic Regions Enrichment of Annotations Tool (GREAT) analysis (McLean et al. 2010).
Figure 7.
Consequences of dsRNA-associated gene deletion on mouse phenotypes using GREAT (McLean et al. 2010). Only the Gene Ontology “Mouse Phenotype Single knockout” is shown. The list of the top 20 enriched terms of the other gene ontologies is given in Supplemental Table S4. (A) The parsed list of 1893 genes that form dsRNA was tested for enrichment against all protein-coding mouse genes (21,395). The mouse phenotype single KO database contains 9170 entries that cover 9466 or 44% of all genes. (B) Phenotypes enriched after knockout of dsRNA genes expressed in mouse liver on the background of all genes. (C) Phenotypes enriched after knockout of dsRNA genes expressed in mouse testis (1888 genes or 16%) on a background of genes that are expressed in testis (11,649).
In general, the knockout of dsRNA genes had a strong influence on cellular and embryonic development related to the investigated tissue, that is, sperm development in testis (Fig. 7A,B; Supplemental Table S4). The knockout of genes present in both data sets (1759) was predominantly associated with defects in embryonic development (Supplemental Fig. S4). The same approach with the liver input control, again, showed an enrichment of developmental phenotypes, reflecting a tightly regulated transcriptional program rather than a particular role of dsRNA formation (Supplemental Table S4). When the dsRNA-generating genes in testis were assessed for enrichment on a testis transcriptome background, traits related to sperm morphology, male germ cell apoptosis, and developmental arrest constituted the first 12 entries of a list of 14 terms that showed significant enrichment (Fig. 7C). These findings suggest that dsRNA formation may constitute an essential checkpoint for male germ cell development.
We made a considerable effort to visualize dsRNA in adult mouse tissue by fluorescence immunohistochemistry using the J2 antibody. However, the signal did not reach the detection limit. As a positive control, we used cultured cell lines (A375 and CCD1106) and treated the cells either with the dsRNA analog poly I:C or stressed them with azacytidine to provoke endogenous dsRNA production. Both procedures resulted in clearly enhanced staining with the J2 antibody, confirming on the one hand the specificity of the antibody and on the other suggesting that the level of endogenous dsRNA is low and spatially dispersed (Supplemental Fig. S5). An additional limitation of this study is the different mouse strains used for J2 enrichment of dsRNA. The processing of dsRNA and its role in innate immunity are well-established and generally conserved in vertebrates. Therefore, it is unlikely that strain-specific differences account for the reported differences between the analyzed data sets.
To conclude, we have shown that the dsRNA transcriptome in testis is fundamentally different from that in liver. Moreover, our evidence suggests that the dsRNA structures are formed between natural sense–antisense transcripts, recognized by DICER1, and processed into endo-siRNAs. These findings corroborate a testis-specific biological role of dsRNA for which the dsRNA structure of the molecule is the key determinant for function rather than the protein-coding potential of the particular genes.
Discussion
The highly complex transcriptome of testis appears to greatly exceed the demands of functioning simply as a (reproductive) organ (Soumillon et al. 2013). Various explanations have been offered, ranging from transcriptional fallout after DNA demethylation to RNA- or transcription-dependent genomic quality control (Werner et al. 2015; Xia et al. 2020). The recent findings that genes transcribed during sperm development show lower mutation rates than silent loci strongly support the latter hypothesis and emphasize the role of transcription and transcription-related repair (Xia et al. 2020). However, no mechanistic insights to underpin such biological role have been reported so far. Our investigations here suggest a mechanism during sperm development that involves dsRNA formation from genic regions including natural antisense transcripts followed by processing into endo-siRNAs. Failure of the mechanism appears to interfere with sperm development and promote apoptosis of male germ cells.
Analysis of the dsRNA transcriptome of fetal mouse testis and liver indicates that mitochondria contribute substantially to the dsRNA transcriptome (Dhir et al. 2018; Kim et al. 2018); conversely, nuclear transcripts that form dsRNA show significant differences regarding their origin (Fig. 3B). In testis, >90% of dsRNA-forming transcripts are associated with mRNA features, including promoters as compared to 58% in liver. The difference suggests that fully processed mRNAs significantly contribute to the formation of dsRNA in testis. Our analysis indicated that dsRNA formation in testis occurs in pachytene spermatocytes, whereas the cellular origin of dsRNA in liver is less clear. The observation that the knockout of dsRNA-forming genes in liver is associated with developmental defects indicates that dsRNA formation in undifferentiated cells may play a biological function. Accordingly, stem cells tolerate dsRNA in the cytoplasm without triggering an immune response (Wang et al. 2013).
A recent study has compared single-cell sequencing from mouse and human male germ cells and found comparably high levels of genic transcription in both species, predominantly in spermatocytes and round spermatids (Xia et al. 2020). Of note, these are also the stages identified in this study that express the highest proportion of dsRNA-forming genes. The particular and prominent expression of dsRNA in male germ cells raises the question whether oocytes show a comparable expression pattern. Studies using genetically modified mice with mutations in retrotransposon defense mechanisms have identified three specific pathways that protect the oocyte genome, including piRNAs, RNA interference, and transcriptional silencing mechanisms controlling LINE-1 elements (Taborska et al. 2019). The dsRNA feeding into piRNA and endo-siRNA pathways in oocytes derives from repetitive elements; whether genic dsRNA formation as observed in male germ cells occurs also in oocytes remains to be established.
The formation of endogenous dsRNA comes with a significant risk for mammalian cells. RNA duplexes of 30 bp and longer are reminiscent of dsRNA viruses and recognized by sensor proteins that trigger a strong innate immune response (Wang and Carmichael 2004). The discoveries of RNA interference and the widespread antisense transcription in mammalian genomes have indicated that, despite the danger of eliciting an unwanted immune reaction, endogenous dsRNA formation occurs (Carlile et al. 2008; Ghildiyal et al. 2008; Okamura and Lai 2008; Watanabe et al. 2008). However, the nature of the dsRNA transcriptome established here suggests that RNA duplexes have distinctly different biological roles in sperm and somatic cells. In the latter, dsRNA is contained in mitochondria and the nucleus and only leaks out when the barriers break down or production is increased owing to pathologies or drugs (Tarallo et al. 2012; Tsai et al. 2012). Accordingly, the inactivation of polynucleotide phosphorylase (PNPase), which breaks down mitochondrial dsRNA, leads to IFIH1 activation and ultimately triggers an interferon-mediated innate immune response (Dhir et al. 2018). Moreover, drugs that reduce DNA methylation and increase spurious transcription of nuclear Alu elements (e.g., azacytidine) induce a dsRNA response (Ahmad et al. 2018). The transcripts generated from repetitive elements are generally not processed and remain in the nucleus and are thus segregated from the dsRNA sensors in the cytoplasm (Kiyosawa et al. 2005; Elbarbary et al. 2016).
The situation in testis presents differently. Here, transcripts that form dsRNA derive from genic regions and are spliced, hence they are more likely to reach the cytoplasm. The mouse gene expression database (http://www.informatics.jax.org/expression.shtml) and the Human Protein Atlas (https://www.proteinatlas.org/) indicate that IFIH1 and EIF2AK2 (also known as PKR) are expressed at a low to medium level in developing male germ cells, whereas DICER1 and ADAR in the nucleus are more abundant. Accordingly, spermatocytes show a reduced response to poly I:C, a synthetic dsRNA analog widely used to experimentally trigger an antiviral response (Li et al. 2012). Male germ cells may therefore constitute a cellular environment that is more tolerant against cytoplasmic dsRNA essential for a posttranscriptional regulatory role of dsRNA.
The connection between dsRNA and a processing by DICER1 into endo-siRNAs is well-established in Caenorhabditis elegans (Duchaine et al. 2006; Vasale et al. 2010) and Drosophila (Czech et al. 2008; Ghildiyal et al. 2008; Lucchetta et al. 2009), less so in vertebrates (Watanabe et al. 2006, 2008; Carlile et al. 2009). Nevertheless, endo-siRNAs in mouse testis have been characterized previously, and a regulatory role reminiscent of miRNA function has been proposed (Song et al. 2011). Limited endo-siRNAs have also been found in a human cell line (HEK293), potentially linked to sense/antisense transcript pairs, but with unknown cellular function (Werner et al. 2014).
Our results confirm the link between dsRNA and the formation of endo-siRNAs, although the depletion of DICER1 in the testes of young mice only marginally affected the levels of dsRNA, which would be expected if DICER1 is efficiently processing RNA hybrids. The key concern relates to the young age of the Dicer1 KO animals and the limited complexity of the dsRNA transcriptome at this stage, which makes a potential impact of the knockout difficult to monitor. On the other hand, the minor bias toward overexpression of a heterogeneous (small) group of genes in KO animals is consistent with a role for DICER1 in processing dsRNA. Of note, DICER1 is localized to the chromatoid body in spermatocytes and round spermatids where also most of the polyadenylated RNA is accumulated (Kotaja et al. 2006; Jiang et al. 2020). A related observation was also made by Zimmermann and coworkers who reported a small up-regulation of protein-coding genes in Dicer1 KO animals (Zimmermann et al. 2014). Of importance, however, DICER1 is essential for miRNA processing and a general stimulation could also be the effect of decreased levels of miRNAs (Korhonen et al. 2011).
A recurrent and striking feature in the analysis of dsRNA is the distinct underrepresentation of dsRNA peaks, dsRNA-associated genes, as well as endo-siRNAs mapping to the X and Y Chromosomes. A similar bias against the X Chromosome has also been observed in the context of antisense transcripts generated from the mouse (and human) genome (Kiyosawa et al. 2003; Chen et al. 2004). The X Chromosome bias is not observed in sense–antisense transcript pairs with complementarity restricted to introns. Our findings that introns contribute only marginally to dsRNA structures in testis (Fig. 3B) and that dsRNA is associated with antisense transcription (Fig. 6C) are in line with the early observations by Kiyosawa et al. (2003) and Chen et al. (2004). Accordingly, we could corroborate the link between natural antisense transcripts and dsRNA formation in testis with a less pronounced association between dsRNA-associated genes and antisense transcripts in liver (Fig. 6). This observation contrasts with the accumulation of repetitive elements on the X Chromosome (Komissarov et al. 2011). The significant contribution of Alu and LINE-1 elements to dsRNA formation in somatic cells (Sadeq et al. 2021) may again point to different biological roles of dsRNA formation in germ and somatic cells.
The analysis of mouse phenotypes with deletions of dsRNA-associated genes suggests an essential role of these genes and potentially dsRNA formation in developmental processes. In testis, knockouts affected disproportionally sperm morphology and caused male germ cell apoptosis. These observations concur with a model in which genic transcription followed by dsRNA formation and siRNA production enables a control mechanism to mitigate DNA damage from recombination and transposon mobilization (Werner et al. 2015).
Visual scrutiny of dsRNA-related peaks indicated that very often the genes were indeed transcribed in both directions, but the reads did not necessarily map to the complementary parts of the transcripts (Fig. 2). This was also observed with the spike-in reads that mapped to single-stranded rather than complementary regions, suggesting a bias against sequencing double-stranded structures. The sequencing bias against RNA hybrids makes is impossible to unambiguously match the interacting sense–antisense transcripts. This also highlights a major experimental challenge when investigating dsRNA structures. Experimental strategies and procedures such as RNA extraction and reverse transcription are generally optimized for single-stranded molecules, and dsRNA may show a different behavior. For example, guanidinium salts as used in TRIzol strongly promotes double-strand formation (Mölder and Speek 2016), whereas standard reverse transcription and library synthesis as used here are inhibited by long dsRNA stretches. These considerations suggest that occurrence of dsRNA-forming sense–antisense transcript pairs may be underestimated here because only one part of the complementary gene pair was used for the intersections.
To conclude, we have shown that the dsRNA transcriptome in testis is fundamentally different from somatic liver cells. Moreover, our evidence suggests that the dsRNA structures involve pairs of natural sense–antisense transcripts. The RNA hybrids are processed into endo-siRNAs, most likely by DICER1. These findings are in line with the highly complex transcriptome and suggest a testis-specific biological role of dsRNA in which the double-strand structure is the key determinant rather than the protein-coding potential of the particular genes.
Methods
Animals
The mice used in this study were three Dicer1 KO mice (18 d old) (for details, see Korhonen et al. 2011), three age-matched wild-type juvenile mice (18 d old), and two adult control mice (BALB/c, ca. 4-6 mo old). Mice were housed under a controlled environment (12 h light cycle, temperature 22°C, humidity 55% ± 15%, specific pathogen free) at the Central Animal Laboratory of the University of Turku. Standard pellet chow and reverse osmosis water were available ad libitum. Male germ cell–specific Dicer1 knockout mice were generated as previously described by crossing mice with floxed Dicer1 alleles with mice expressing transgenic Cre under the neurogenin 3 (Neurog3) promoter (Korhonen et al. 2011). Dicer1 (fx/wt) littermates without Cre expression were used as controls. The mice were of mixed genetic background (C57BL/6J and SV129). All procedures were performed in accordance with Finnish laws and the Guide for Care and Use of Laboratory Animals (National Academy of Science, License number: 2009-1206-Kotaja).
Spike-in probe
Plasmids encoding the natural sense–antisense transcript pair (slc34a2a and slc34a2aas from zebrafish) (Nalbant et al. 1999) were linearized with XbaI and transcribed in vitro using the MEGAscript T7 Transcription Kit (Invitrogen). The transcripts are 2607 bases (sense, NM_131624) and 1371 bases (antisense, NR_002876.2) long and share 563 bp of complementarity over two exons. The resulting RNA was quantified and mixed in equimolar concentrations to a total concentration of 0.4 μg/μL. One microliter was diluted 500× with 0.1 M NaCl, heated to 70°C, and gradually cooled to hybridize the two strands. One microliter of the spike-in probe was added (0.8 ng) to the testis homogenate before J2 binding (see below).
Double-stranded RNA immunopurification
The protocol published by Dhir and coworkers was followed with slight modifications (Dhir et al. 2018). Both testes of the juvenile mice and half of a testis of adult mice were homogenized in 220 µL of NP-40 lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5% Na-deoxycholine, 220 units RNasin) using a disposable pestle followed by DNA shearing with a 25G needle. Cell debris was removed by centrifugation, and the volume was increased to 1 mL per sample with NET2+DOC buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5% Na-deoxycholine, 0.2 units/µL DNase I) plus 5 µg of J2 antibody (Scicons 10010500). At this point, the spike-in probe was added. Samples were rotated for 3 h at 4°C, then 100 µL of µMACS Protein G MicroBeads (Miltenyi Biotec) per sample was added followed by a 1-h incubation. The samples were then loaded onto µMACS columns equilibrated with NP-40 buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% NP-40), and the flowthrough (FLOW) was collected. Columns were washed with 300 µL of NP-40 and 3× 250 µL of wash buffer (50 mM Tris pH 7.5, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.5% Na-deoxycholine, and 0.1% SDS). The columns were removed, and beads with dsRNA were washed off the column with water. Beads and dsRNA were mixed with TRIzol and purified according to established protocols. In parallel, the RNA from 200 µL of the FLOW were purified with TRIzol and used as a background.
Sequencing
RNA samples were quantified, and the integrity was tested using a Bioanalyzer (Agilent). Strand-specific RNA-seq libraries were prepared using the NuGEN Ovation SoLo kit (Ovation SoLo RNA-seq System, Human, Tecan) without size selection or fragmentation of RNA. The supplier's guidelines were closely followed with the exception that the antibody-bound samples were excluded from rRNA depletion, whereas the flowthrough samples were rRNA depleted. The stranded library contained inserts of 300–350 bp. Paired-end sequencing was performed on an Ilumina HiSeq 2500 platform at the Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research (Darlinghurst, Australia) (sequencing read length 42–117 bases).
Data analysis
The quality of reads was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and adaptors were trimmed using Trimmomatic (version was 0.3.6) (Bolger et al. 2014). The spike probe reads in the different samples were aligned to a fragment of zebrafish Chromosome 1 (Chr 1: 14,432,434–14,454,662) that contains the slc34a2a gene and the related natural antisense transcript (slc34a2aas) originating from the bidirectional Rbpja promoter using STAR version 2.5.2b (Dobin et al. 2013). All data sets were then quantified using Salmon (Patro et al. 2017), and expression differences between KO mice, juvenile wild-type mice, and adult controls were established using DESeq2 (Love et al. 2014).
To establish the dsRNA transcriptome, reads from all samples were mapped to the reference genome (GRCm38.p5) using STAR. Then, dsRNA-derived reads were assessed in two different ways, one focusing on read peaks, the other using the RNA-seq quantitation pipeline in SeqMonk (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). Peaks and coverage were established with BEDTools genomecov (-bg -ibam) and multicov (BEDTools suite version 2.28.0) (Supplemental Fig. S1; Quinlan and Hall 2010). Regions with a coverage greater than or equal to five times more than background were annotated using ChIPpeakAnno (version 3.19.4) (Zhu et al. 2010). Conversely, a gene was considered expressed when at least eight times more than background and positive in six out of eight samples as established using the RNA-seq quantitation pipeline in SeqMonk. If the data sets contained fewer samples, the proportions were adjusted to four of six data samples, three of four samples, one of two samples, or one of one sample as appropriate. The list for dsRNA-forming genes was then intersected with the expressed genes in different tissues and male germ cells that were aligned and quantitated using the STAR/SeqMonk pipeline. Pearson's χ2 tests with Yates’ continuity correction and two-sample tests for equality proportions with continuity correction were performed in R (version3.4.3) (R Core 2020). For all data, a P-value ≤ 0.05 was determined to be statistically significant. To quantify regions that expressed both dsRNA and endo-siRNAs, genome coverage (BEDTools, genomecov and multiconv) was determined and replicates were merged (BEDTools, unionbedg with default settings). BED files representing the coverage of combined samples were intersected using the BEDTools intersect (intersect –wb –a) (Quinlan and Hall 2010). The resulting peak list was then used to retrieve gene annotations (GENCODE.vM20.annotation.gff3) and a minimal overlap of 0.5 times peak width with the feature was set. The primary BAM files were then used to quantify the chromosome coverage and distribution for the individual samples. Flowcharts summarizing the different pipelines are provided in Supplemental Figure S1. Murine tissue expression data from Pervouchine et al. (2015) was accessed from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE36025 (deposited under NCBI BioProject [https://www.ncbi.nlm.nih.gov/bioproject/] study PRJNA66167). Sequencing data from isolated, staged male germ cells were accessed from the NCBI BioProject study PRJNA317251 (da Cruz et al. 2016) and the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP078798 (Zuo et al. 2016). Short RNA reads from mouse testis were accessed from GEO accession number GSE83264 (Hilz et al. 2017). Gene Ontology and Mouse Phenotype Single KO analysis were performed using GREAT (McLean et al. 2010). A minimal enrichment of twofold was set with a cutoff of 20 terms. As background for enrichment the 21,395 protein-coding mouse genes (NCBI build 38; UCSC mm10, December 2011) were used for testis and liver samples. Alternatively, the testis sample was also tested for enrichment against the protein-coding genes expressed in testis as determined using the SeqMonk pipeline (11,622). False discovery rates as well as P-values were determined.
RT-qPCR
For each gene, four primer pairs were designed to analyze the expression of (1) the sense gene with at least one exon overlap with the antisense gene, (2) the sense gene with no exon overlap with the antisense gene, (3) the antisense gene with at least one exon overlap with the sense gene, and (4) the antisense gene with no exon overlap with the sense gene (Supplemental Fig. S2; Supplemental Table S5). Actin, beta (Actb) was used as a reference gene. First, a DNA digestion step was implemented (DNase I, Promega) followed by SigmaSpin Sequencing Reaction Clean-Up (Sigma-Aldrich). RT-qPCR was performed using the Luna Universal One-Step RT-qPCR Kit (NEB) according to the manufacturer's instructions, in a LightCycler 480 System (Roche) with the following parameters: Reverse transcription for 10 min at 55°C; initial denaturation for 1 min at 95°C; denaturation for 10 sec at 95°C and an extension for 30 sec at 60°C for 45 cycles; melting curve at 95°C. The RNA used was for RT-qPCR was from an 18-d-old mouse testis sample.
Immunofluorescence
For immunohistochemistry, 5-µm sections of adult mouse testis fixed in 4% paraformaldehyde/tris-buffered saline (TBS) were permeabilized with either 100% ice cold methanol (10 min) or 0.1% Triton X-100 in 5% normal serum in TBS (30 min). Unspecific binding was blocked with 5% bovine serum in TBS (30 min). The primary J2 antibody (Scicons 10010500) was diluted 1:200 (1 µg/µL stock), the secondary antibody, Alexa Fluor 488 (A-21131) or Alexa Fluor 594-coupled goat anti-mouse IgG2a (Thermo Fisher Scientific A-11032), was used at 1:1000. The cell nuclei were counterstained using DAPI (4′,6-diamidino-2-phenylindole). Sections were washed with TBS, mounted using VECTASHIELD (Vector Laboratories) and imaged using a Unit Zeiss AxioImager1 fluorescence microscope.
Immunohistochemistry was performed to test the specificity of the J2 antibody and to examine the expression of dsRNA-binding proteins. CCD1106 keratinocytes or A375 cells were treated with poly I:C (0.5 µg/mL) or azacytidine (500 nM) for 24 h. Cells were washed with PBS and fixed with 4% paraformaldehyde in PBS (Affymetrix) for 10 min. Cells were permeabilized in 0.25% Triton X-100 followed by blocking with 3% BSA in PBS (Albumin fraction V, USB) for 1 h. The antibodies used were J2 (Sicons 10010500, 1:200), PKR (Abcam ab32052, 1:1000), IFIH1 (Abcam ab126630, 1:1000) and RIG-1 (Abcam EPR18629, 1:1000), incubation was for 1 h at room temperature. Cells were washed with PBS-Tween and then incubated with the secondary antibodies Alexa Fluor 488 goat anti-mouse IgG2a (Thermo Fisher Scientific A-21131) or Alexa Fluor 594 goat anti-rabbit IgG H + L (Thermo Fisher Scientific A-11032) and analyzed as above.
Data access
Raw and processed sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA630221. Codes generated for this work are available as Supplemental Code and at GitHub (https://github.com/James-E-Clark/Masters-dsRNA-Project and https://github.com/jwcasement/dsRNA-seq-project).
Supplementary Material
Acknowledgments
We thank Eva Maria Novoa Pardo and David Burns for technical help and fruitful discussions. This work was partly funded by The Northern Counties Kidney Research Fund, Grant 18.011 (A.W.), The Iraqi Ministry of Higher Education (S.S. and S.A.-H.), The Higher Committee for Education Development in Iraq and the University of Baghdad (H.S.Z.). This research made use of the Rocket High Performance Computing service at Newcastle University.
Author contributions: A.W., N.K., M.S., and J.S.M. conceived the project and wrote the manuscript. A.W., J.E.C., C.S., J.C., H.S.Z., S.S., S.A.-H., and N.K. performed experiments and analyzed the data. M.S. and J.S.M. also contributed to data analysis and interpretation.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.265603.120.
Competing interest statement
The authors declare no competing interests.
References
- Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang CZ, Hur S. 2018. Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172: 797–810.e13. 10.1016/j.cell.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkgren I, Sipilä P. 2015. The role of Dicer1 in the male reproductive tract. Asian J Androl 17: 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile M, Nalbant P, Preston-Fayers K, McHaffie GS, Werner A. 2008. Processing of naturally occurring sense/antisense transcripts of the vertebrate Slc34a gene into short RNAs. Physiol Genomics 34: 95–100. 10.1152/physiolgenomics.00004.2008 [DOI] [PubMed] [Google Scholar]
- Carlile M, Swan D, Jackson K, Preston-Fayers K, Ballester B, Flicek P, Werner A. 2009. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res 37: 2274–2282. 10.1093/nar/gkp088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. 2004. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820. 10.1093/nar/gkh818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. 10.1038/nature07007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz I, Rodríguez-Casuriaga R, Santiñaque FF, Farías J, Curti G, Capoano CA, Folle GA, Benavente R, Sotelo-Silveira JR, Geisinger A. 2016. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic- to postmeiotic-related processes at pachytene stage. BMC Genomics 17: 294. 10.1186/s12864-016-2618-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rötig A, Crow YJ, Rice GI, Duffy D, Tamby C, et al. 2018. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560: 238–242. 10.1038/s41586-018-0363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354. 10.1016/j.cell.2005.11.036 [DOI] [PubMed] [Google Scholar]
- Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351: aac7247. 10.1126/science.aac7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643. 10.1038/nrm2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Vasic R, Song Y, Teng R, Liu C, Gbyli R, Biancon G, Nelakanti R, Lobben K, Kudo E, et al. 2020. m6A modification prevents formation of endogenous double-stranded RNAs and deleterious innate immune responses during hematopoietic development. Immunity 52: 1007–1021.e8. 10.1016/j.immuni.2020.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081. 10.1126/science.1157396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilz S, Fogarty EA, Modzelewski AJ, Cohen PE, Grimson A. 2017. Transcriptome profiling of the developing male germ line identifies the miR-29 family as a global regulator during meiosis. RNA Biol 14: 219–235. 10.1080/15476286.2016.1270002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. 2019. Double-stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37: 349–375. 10.1146/annurev-immunol-042718-041356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Soboleva TA, Tremethick DJ. 2020. Short histone H2A variants: small in stature but not in function. Cells 9: 867. 10.3390/cells9040867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Park J, Kim S, Kim M, Kang MG, Kwak C, Kang M, Kim B, Rhee HW, Kim VN. 2018. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol Cell 71: 1051–1063.e6. 10.1016/j.molcel.2018.07.029 [DOI] [PubMed] [Google Scholar]
- Kim S, Ku Y, Ku J, Kim Y. 2019. Evidence of aberrant immune response by endogenous double-stranded RNAs: attack from within. Bioessays 41: e1900023. 10.1002/bies.201900023 [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. 2003. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res 13: 1324–1334. 10.1101/gr.982903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. 2005. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res 15: 463–474. 10.1101/gr.3155905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarov AS, Gavrilova EV, Demin SJ, Ishov AM, Podgornaya OI. 2011. Tandemly repeated DNA families in the mouse genome. BMC Genomics 12: 531. 10.1186/1471-2164-12-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen HM, Meikar O, Yadav RP, Papaioannou MD, Romero Y, Da Ros M, Herrera PL, Toppari J, Nef S, Kotaja N. 2011. Dicer is required for haploid male germ cell differentiation in mice. PLoS One 6: e24821. 10.1371/journal.pone.0024821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. 2006. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci 103: 2647–2652. 10.1073/pnas.0509333103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wang T, Han D. 2012. Structural, cellular and molecular aspects of immune privilege in the testis. Front Immunol 3: 152. 10.3389/fimmu.2012.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Carthew RW, Ismagilov RF. 2009. The endo-siRNA pathway is essential for robust development of the Drosophila embryo. PLoS One 4: e7576. 10.1371/journal.pone.0007576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GP, Ghosh A, Jha A, Raghav SK. 2020. BedSect: an integrated web server application to perform intersection, visualization, and functional annotation of genomic regions from multiple data sets. Front Genet 11: 3. 10.3389/fgene.2020.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mölder T, Speek M. 2016. [Letter to the editor] Accelerated RNA–RNA hybridization by concentrated guanidinium thiocyanate solution in single-step RNA isolation. BioTechniques 61: 61–65. 10.2144/000114441 [DOI] [PubMed] [Google Scholar]
- Nalbant P, Boehmer C, Dehmelt L, Wehner F, Werner A. 1999. Functional characterization of a Na+-phosphate cotransporter (NaPi-II) from zebrafish and identification of related transcripts. J Physiol 520 Pt 1: 79–89. 10.1111/j.1469-7793.1999.00079.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. 2008. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 9: 673–678. 10.1038/nrm2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervouchine DD, Djebali S, Breschi A, Davis CA, Barja PP, Dobin A, Tanzer A, Lagarde J, Zaleski C, See LH, et al. 2015. Enhanced transcriptome maps from multiple mouse tissues reveal evolutionary constraint in gene expression. Nat Commun 6: 5903. 10.1038/ncomms6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. 10.1126/science.1164096 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Sadeq S, Al-Hashimi S, Cusack CM, Werner A. 2021. Endogenous double-stranded RNA. Noncoding RNA 7: 15. 10.3390/ncrna7010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Hennig GW, Wu Q, Jose C, Zheng H, Yan W. 2011. Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci 108: 13159–13164. 10.1073/pnas.1108567108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthes P, Kokkinaki M, Nef S, Gnirke A, et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3: 2179–2190. 10.1016/j.celrep.2013.05.031 [DOI] [PubMed] [Google Scholar]
- Taborska E, Pasulka J, Malik R, Horvat F, Jenickova I, Jelić Matošević Z, Svoboda P. 2019. Restricted and non-essential redundancy of RNAi and piRNA pathways in mouse oocytes. PLoS Genet 15: e1008261. 10.1371/journal.pgen.1008261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. 2012. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell 149: 847–859. 10.1016/j.cell.2012.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. 2012. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 21: 430–446. 10.1016/j.ccr.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D Jr. 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/argonaute pathway. Proc Natl Acad Sci 107: 3582–3587. 10.1073/pnas.0911908107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carmichael GG. 2004. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev 68: 432–452. 10.1128/MMBR.68.3.432-452.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wang J, Paul AM, Acharya D, Bai F, Huang F, Guo YL. 2013. Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem 288: 15926–15936. 10.1074/jbc.M112.421438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Eraslan B, Wieland T, Hallström B, Hopf T, Zolg DP, Zecha J, Asplund A, Li LH, Meng C, et al. 2019. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Syst Biol 15: e8503. 10.15252/msb.20188503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. 2006. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20: 1732–1743. 10.1101/gad.1425706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543. 10.1038/nature06908 [DOI] [PubMed] [Google Scholar]
- Werner A, Schmutzler G, Carlile M, Miles CG, Peters H. 2007. Expression profiling of antisense transcripts on DNA arrays. Physiol Genomics 28: 294–300. 10.1152/physiolgenomics.00127.2006 [DOI] [PubMed] [Google Scholar]
- Werner A, Carlile M, Swan D. 2009. What do natural antisense transcripts regulate? RNA Biol 6: 43–48. 10.4161/rna.6.1.7568 [DOI] [PubMed] [Google Scholar]
- Werner A, Cockell S, Falconer J, Carlile M, Alnumeir S, Robinson J. 2014. Contribution of natural antisense transcription to an endogenous siRNA signature in human cells. BMC Genomics 15: 19. 10.1186/1471-2164-15-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Piatek MJ, Mattick JS. 2015. Transpositional shuffling and quality control in male germ cells to enhance evolution of complex organisms. Ann N Y Acad Sci 1341: 156–163. 10.1111/nyas.12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Yan Y, Baron M, Wagner F, Barkley D, Chiodin M, Kim SY, Keefe DL, Alukal JP, Boeke JD, et al. 2020. Widespread transcriptional scanning in the testis modulates gene evolution rates. Cell 180: 248–262.e21. 10.1016/j.cell.2019.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhu W, Xue S, Han D. 2014. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol 11: 428–437. 10.1038/cmi.2014.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C, Romero Y, Warnefors M, Bilican A, Borel C, Smith LB, Kotaja N, Kaessmann H, Nef S. 2014. Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS One 9: e107023. 10.1371/journal.pone.0107023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinad HS, Natasya I, Werner A. 2017. Natural antisense transcripts at the interface between host genome and mobile genetic elements. Front Microbiol 8: 2292. 10.3389/fmicb.2017.02292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo H, Zhang J, Zhang L, Ren X, Chen X, Hao H, Zhao X, Wang D. 2016. Transcriptomic variation during spermiogenesis in mouse germ cells. PLoS One 11: e0164874. 10.1371/journal.pone.0164874 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.