Abstract
Approximately 1% of the human genome has the ability to fold into G-quadruplexes (G4s)—noncanonical strand-specific DNA structures forming at G-rich motifs. G4s regulate several key cellular processes (e.g., transcription) and have been hypothesized to participate in others (e.g., firing of replication origins). Moreover, G4s differ in their thermostability, and this may affect their function. Yet, G4s may also hinder replication, transcription, and translation and may increase genome instability and mutation rates. Therefore, depending on their genomic location, thermostability, and functionality, G4 loci might evolve under different selective pressures, which has never been investigated. Here we conducted the first genome-wide analysis of G4 distribution, thermostability, and selection. We found an overrepresentation, high thermostability, and purifying selection for G4s within genic components in which they are expected to be functional—promoters, CpG islands, and 5′ and 3′ UTRs. A similar pattern was observed for G4s within replication origins, enhancers, eQTLs, and TAD boundary regions, strongly suggesting their functionality. In contrast, G4s on the nontranscribed strand of exons were underrepresented, were unstable, and evolved neutrally. In general, G4s on the nontranscribed strand of genic components had lower density and were less stable than those on the transcribed strand, suggesting that the former are avoided at the RNA level. Across the genome, purifying selection was stronger at stable G4s. Our results suggest that purifying selection preserves the sequences of functional G4s, whereas nonfunctional G4s are too costly to be tolerated in the genome. Thus, G4s are emerging as fundamental, functional genomic elements.
The three-dimensional conformation of DNA at certain motifs may deviate from the canonical B-DNA (Watson and Crick 1953). According to a recent estimate, as much as 13% of the human genome has the potential to fold into non-B DNA structures (Guiblet et al. 2018), which include Z-DNA, H-DNA, A-phased bends, cruciforms, slipped-strand structures, and G-quadruplexes (G4s) (Zhao et al. 2010). Among non-B DNA structures, G4s have been investigated the most. Over the last several years, G4 structure formation was unequivocally shown in the native chromatin environment in human cells (Biffi et al. 2013; Hänsel-Hertsch et al. 2017). G4 formation is intermittent and may be sensitive to environment and to temporal signals associated with cell cycle (Hänsel-Hertsch et al. 2017) and development (Maizels 2015). Because of this intermittent formation and their ability to form in both DNA and RNA, G4 structures are emerging as key regulators of fundamental cellular processes (for review, see Varshney et al. 2020) and have been implicated in multiple human diseases, including neurological disorders (Maizels 2015; Simone et al. 2015) and cancer (Chambers et al. 2015; Hänsel-Hertsch et al. 2016).
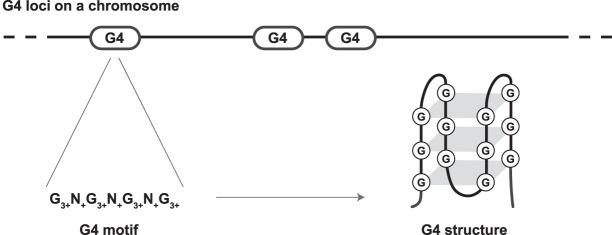
A G4 motif is composed of at least four stems, each including three or more guanines, and stems are separated by loops including one to 12 unspecified nucleotides (Fig. 1), leading to the consensus motif sequence of G3+N1−12G3+N1−12G3+N1−12G3+ (Huppert and Balasubramanian 2005). A G4 structure (Fig. 1) is formed on the G-rich DNA strand of the motif because of stacking of guanine stems in quartets (or quadruplexes) held together by Hoogsten hydrogen bonds (Sen and Gilbert 1988). Usually, at least four stretches of three guanines in tandem are required for G4 structure formation, with substitutions of the middle guanine in a stretch having particularly detrimental effects on structure formation (Lee and Kim 2009). G4 structures are stabilized by potassium ions (Pinnavaia et al. 1978; Sen and Gilbert 1990). The opposite C-rich strand can form a less stable i-motif structure (Takahashi et al. 2017). More than 670,000 loci in the human genome possess G4 motifs (Sahakyan et al. 2017a). With a mean motif length of 36 bp, the loci capable of forming G4 structures constitute ∼1% of the human genome (for a comparison, protein-coding sequences constitute ∼1.5% of the genome) (Chiaromonte et al. 2003).
Figure 1.
The schematic presentation of the G4 consensus motif and structure.
The initial clues about the potential functionality of G4 loci came from studies showing their enrichment in several genic components and nongenic functional regions of the genome. For instance, it was shown that G4 motifs are significantly overrepresented in promoters (Huppert and Balasubramanian 2007), 5′ and 3′ untranslated regions (UTRs) (Huppert et al. 2008), replication origins (Besnard et al. 2012), recombination hotspots (Mani et al. 2009), telomeres (Smith et al. 2011; Moye et al. 2015), and transposable elements (TEs) (Lexa et al. 2014). More recently, substantial evidence supporting genome-wide functions of G4 structures in vivo has been rapidly accumulating (for review, see Varshney et al. 2020). G4 structures participate in regulation of transcription (Baral et al. 2012; Hänsel-Hertsch et al. 2016; Varshney et al. 2020), in the life cycle of active L1 TEs (Sahakyan et al. 2017b), in telomere end protection and maintenance (Smith et al. 2011; Moye et al. 2015), and in local chromatin remodeling (Hänsel-Hertsch et al. 2016). These numerous groups of G4 loci with proven genome-wide functions in vivo are expected to evolve under purifying selection for motif retention and conservation; however, this has so far been shown only for G4s in UTRs (Lee et al. 2020).
The role of G4s in several other biological processes—replication initiation (Valton et al. 2014), mRNA metabolism (Beaudoin and Perreault 2013; Dolinnaya et al. 2016), alternative splicing (Gomez 2004), regulation of noncoding RNAs (Simone et al. 2015), and translation (Wilkie et al. 2003; Babendure et al. 2006; Kumari et al. 2007; Huppert et al. 2008; Bochman et al. 2012; Bugaut and Balasubramanian 2012)—has been shown in vivo for individual loci but still needs to be confirmed genome-wide. To illustrate this point, G4 motifs were shown to be required for starting replication at two chicken replication origins (Valton et al. 2014). Additionally, the insertion of a G4 motif doubled the activity of a replication origin locus in the human genome (Prorok et al. 2019). G4 motifs have been annotated at 91.4% of human replication origins (Besnard et al. 2012); however, whether they are required to start replication genome-wide is yet to be shown.
Some other functions of G4 structures—for example, their participation in recombination (Mani et al. 2009; Boán and Gómez-Márquez 2010), in affecting higher-order chromatin organization at boundaries of topologically associated domains (TADs) (Hou et al. 2019), in regulation of non-L1 TE life cycle (Lexa et al. 2014), in distal interactions between promoters and enhancers (Hou et al. 2019; Williams et al. 2020), and in protecting CpG islands from methylation (Halder et al. 2010; Mao et al. 2018; Jara-Espejo and Peres Line 2020)—remain hypothetical. If G4s in these genomic regions (e.g., in TADs) evolved under purifying selection for motif retention, then such an evolutionary signature would be suggestive of their functionality in these regions.
G4 structures could act as a double-edged sword for the genome. Whereas some G4 structures perform important functions, many G4 structures have the potential to hinder nucleic acid polymerization during replication, transcription, and translation (for review, see Varshney et al. 2020) and to lead to increases in germline (Du et al. 2014; Guiblet et al. 2021) and somatic (Georgakopoulos-Soares et al. 2018) mutations as well as to genomic instability (Cheung et al. 2002; Zhao et al. 2010). Thus, G4 structures that are not functional might be detrimental to the genome and hence might be selected against (Valton and Prioleau 2016). This might explain why G4s are depleted at exons (Huppert and Balasubramanian 2005). Conversely, at functional G4 loci, the high mutability should be constantly combatted by purifying selection, acting to preserve G4 motifs to allow G4 structure formation.
Previous studies have proposed that some G4 loci evolve under natural selection (Valton and Prioleau 2016); however, this has not been analyzed on a genome-wide basis. It has been reported that single-nucleotide polymorphisms (SNPs) disrupting G4 motifs found in promoters were less common than expected by chance (Baral et al. 2012). G4 loci at promoters have also been shown to be conserved across mammals (Verma et al. 2008). Additionally, an enrichment in human SNPs with low minor allele frequencies (MAFs) was found at non-B DNA loci, which include G4 loci, supporting their evolution under purifying selection (Du et al. 2014). Fewer polymorphic SNPs and fixed differences were found at structure-disruptive than other motif positions for G4 loci located in both genic and intergenic regions (Nakken et al. 2009). Finally, it was recently shown that G4 loci located in 5′ and 3′ UTRs and forming on mRNAs were selectively constrained (Lee et al. 2020). However, it remains unknown whether G4 loci located in genic components other than promoters and UTRs and in other functional regions of the genome (e.g., replication origins) evolve under selection.
In this study, we aimed to analyze selection acting at G4 loci genome-wide while taking their thermostability (henceforth termed “stability”) into account. Stability of G4 structures is usually higher than that of B-DNA, but it can vary (for review, see Varshney et al. 2020) depending on the number and lengths of stems and loops, as well as the nucleotide composition of the loops (Kim et al. 2016). Stability is commonly measured by biophysical methods such as circular dichroism (the differential absorption of circular polarized light) or melting temperature (Vorlíčková et al. 2012). Although accurate, these methods are challenging to scale up to the whole genome. Recently, the relative stability of G4 structures has been estimated by G4-seq, namely, by comparing short-read sequencing quality before and after G4 stabilization with potassium (Chambers et al. 2015). Subsequently, a machine-learning model trained on these relative stability estimates and validated on 392 experimentally studied G4 structures was developed as the software tool Quadron, which is used to annotate G4 loci and predict their stability across the human genome (Sahakyan et al. 2017a). Considering stability in studies of G4s is critical because (1) more stable G4s are known to have higher mutability (Guiblet et al. 2021), and (2) we expect G4 stability to affect their function.
Capitalizing on these recent developments, we investigated stability and selection among G4 loci harbored by different genic components (e.g., promoters, UTRs, and protein-coding exons) and nongenic functional regions (e.g., replication origins and enhancers) in the human genome. On the one hand, we hypothesized that purifying selection preserves functional G4 loci by maintaining an optimal stability for the function they perform. On the other hand, as also suggested by others (Piazza et al. 2015; Valton and Prioleau 2016), we hypothesized that stable G4s represent a particular problem for replication, transcription, and translation, and if some such G4s are not functional, then they are not well tolerated and might be underrepresented in the genome. To test these hypotheses, we exploited Quadron, a publicly available software for predicting G4 stability genome-wide (Sahakyan et al. 2017a), and detected the footprints of purifying selection acting on G4 loci using the Hudson–Kreitman–Aguadé (HKA) test (Hudson et al. 1987) and a test based on distortion of the site frequency spectrum.
Results
Uneven density of G4 loci across the genome
We annotated a total of 670,076 G4 loci in the human genome (version hg19) using Quadron (Sahakyan et al. 2017a), a software application that uses a machine-learning approach capitalizing on the decrease in sequencing quality at chemically stabilized G4 loci (Chambers et al. 2015). Approximately half of G4 loci have the potential to form G4 structures on the reference strand; the other half—on its reverse complement. Because the transcribed and nontranscribed DNA strands were considered separately for some genic components, we computed G4 density and coverage per base and not per base pair. This resulted in the genome-wide G4 density of 0.116 loci per kilobase (taking the total length of autosomes on reference and reverse complement strands equal to 2,880,813,286 × 2 bases as a denominator). The mean length of G4 loci across the genome was 36.4 bp, and thus, the genome-wide coverage (the number of bases annotated as G4 loci divided by the total number of bases in reference and reverse complement strands for autosomes) was 4.24 × 10−3 per base. We next studied the distribution of G4 loci (i.e., their density and coverage) among genic components and nongenic functional regions (Supplemental Table S1).
We considered, among genic components, 1-kb regions upstream from the transcription start sites (such regions contain promoters and will be henceforth termed “upstream regions”), CpG islands, 5′ UTRs, protein-coding exons, introns, 3′ UTRs, and 1-kb regions downstream from the transcription termination sites (see Methods). The DNA strand at which a G4 structure can form is important for G4 loci located in transcribed genic components. Therefore, the transcribed (i.e., used as a template for transcription) and nontranscribed strands of 5′ UTRs, protein-coding exons, introns, and 3′ UTRs were considered separately. G4 structures forming on the transcribed DNA strand may suppress transcription, whereas G4 structures on the nontranscribed DNA strand may either facilitate or suppress transcription (for review, see Varshney et al. 2020). G4 loci on the nontranscribed DNA strand of protein-coding exons have the potential to form structures in mRNA and interfere with translation (Shabalina et al. 2006; Kumari et al. 2007; Huppert et al. 2008; Endoh and Sugimoto 2013; Endoh et al. 2013; Rhodes and Lipps 2015); G4 structures forming on either DNA strand may interfere with replication (for review, see Varshney et al. 2020).
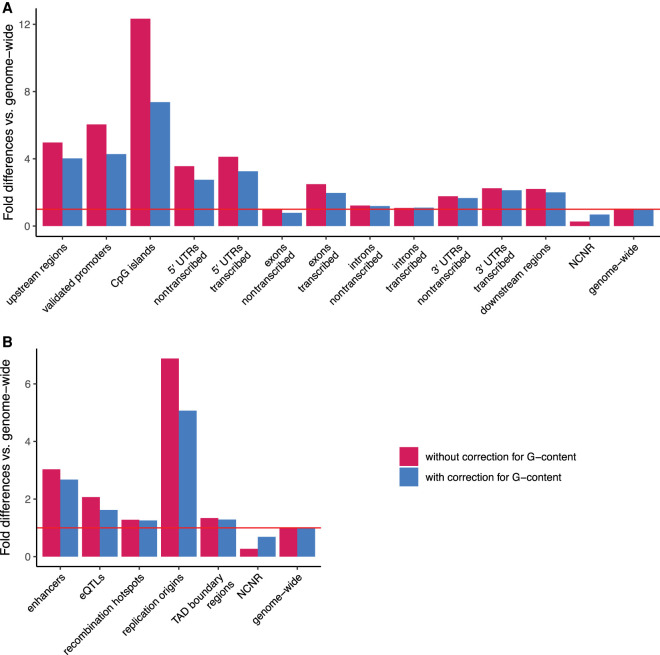
We considered, among the nongenic functional regions, replication origins (Besnard et al. 2012), recombination hotspots (Halldorsson et al. 2019), enhancers (Andersson et al. 2014; Lizio et al. 2015), expression quantitative trait loci (eQTLs) (The GTEx Consortium 2020), and TAD boundary regions (Dixon et al. 2015; Hong and Kim 2017). For comparison, we used density and coverage of G4 loci genome-wide or analyzed these measurements in the noncoding nonrepetitive (NCNR) subgenome (Fig. 2), which comprises the genome left after removing all the genic components and functional regions described above, as well as some other components and regions (e.g., TEs and noncoding RNA; see Methods). The NCNR subgenome, which covers a total of 227 Mb, was used as a control because it is (presumably) neutrally evolving. In it, we annotated 14,437 G4 loci.
Figure 2.
Fold-differences in mean density of G4 loci located at different genic components (A) and nongenic functional regions (B) compared with the genome-wide average and the noncoding nonrepetitive (NCNR) subgenome. Red horizontal line indicates no difference compared with the genome-wide average.
We found that, among the genic components considered (Fig. 2A; Supplemental Table S2A), upstream regions, CpG islands, 5′ UTRs (both transcribed and nontranscribed strands), transcribed strands of exons and of 3′ UTRs, and downstream regions had G4 density at least twofold higher than the genome-wide average (adjusted P < 3.96 × 10−15 in each case, Fisher's exact test; all P-values in the manuscript were corrected for multiple testing using the Bonferroni method). The fold-differences in G4 density for CpG islands, upstream regions, and transcribed strands of 5′ UTRs versus the genome-wide average were particularly high: 12.3, 4.98, and 4.11, respectively. We verified the results for upstream regions, which included putative promoters, by observing similar trends (Fig. 2A; Supplemental Table S2A) in a more limited set of experimentally validated promoters (FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014; Lizio et al. 2015). In contrast, nontranscribed and transcribed strands of introns, nontranscribed strands of exons, and nontranscribed strands of 3′ UTRs had G4 density closer to the genome-wide average (albeit also significantly higher; P < 3.96 × 10−15 for all, Fisher's exact test) (Fig. 2A; Supplemental Table S2A). G4 density in each group of genic components considered was higher than that in the NCNR subgenome (P < 3.96 × 10−15 in each case, Fisher's exact test) (Supplemental Table S2A). For almost all genic components for which we considered G4 density separately for the transcribed and nontranscribed strand (Supplemental Table S3), it was significantly higher in the former than in the latter (P < 8.8 × 10−16), with the only exception of introns. This suggests that G4 structures are disfavored at the level of RNA. Similar trends were observed for G4 coverage (Supplemental Fig. S1; Supplemental Tables S2B, S3).
Some of the observed differences in G4 density and coverage might be explained by the distinct G-content of genic components, for example, by high G-content in upstream regions, CpG islands, and 5′ UTRs. After correcting for G-content (see Methods) (Fig. 2A; Supplemental Tables S1, S2), we still detected significant enrichment in G4 density and coverage for almost all groups of genic components considered (vs. the genome-wide average or the NCNR subgenome; P < 3.96 × 10−15 in each case, Fisher's exact test). However, in some cases this enrichment became less pronounced (Fig. 2A; Supplemental Table S2). For instance, after correcting for G-content, G4 density was 7.37-, 4.03-, and 3.26-fold higher in CpG islands, upstream regions, and transcribed strands of 5′ UTRs than the genome-wide average, respectively (P < 3.96 × 10−15 in each case, Fisher's exact test) (Supplemental Table S2A). Similar trends were observed for G4 coverage (Supplemental Fig. S1; Supplemental Table S2B). After correcting for G-content, the nontranscribed strands of exons had G4 density and coverage 21% and 19% lower than the genome-wide average, respectively (P < 3.96 × 10−15 in each case, Fisher's exact test) (Supplemental Table S2).
Each group of nongenic functional regions studied had significantly higher G4 density than the genome-wide average and the NCNR subgenome (P < 3.96 × 10−15 in each case, Fisher's exact test) (Supplemental Table S2A). Replication origins and enhancers had particularly high G4 density (Fig. 2B): 6.88- and 3.03-fold higher than the genome-wide average, respectively. G4 coverage showed similar trends (Supplemental Fig. S1; Supplemental Table S2B). Correcting for G-content did not affect the results qualitatively but led to smaller differences in G4 density and coverage between nongenic functional regions and genome-wide averages, as well as between nongenic functional regions and the NCNR subgenome (Fig. 2B; Supplemental Table S2).
G4 stability differs among genic regions and nongenic functional regions
The software we used to annotate G4 loci across the genome (Sahakyan et al. 2017a) assigns a predicted stability score to each G4 locus, reflecting the probability and strength of G4 structure formation. G4 stability is affected by the number of G-tetrads, loop length and topology, and sequence composition of the G4 motif and flanking regions (Varshney et al. 2020). Quadron incorporates information about the predicted G4 structure. Quadron is based on a machine learning approach that integrates (1) G4 motif sequence consensus matching with Quadparser, which takes loop size into account (Huppert and Balasubramanian 2005), and (2) G4-seq, which measures the decrease in sequencing quality at chemically stabilized G4 loci (Chambers et al. 2015). The readout of G4-seq can be thought of as a proxy for DNA polymerase stalling caused by G4 structures and thus is indicative of structure formation. In the original publication, Quadron results were validated with 392 in vitro, experimentally verified G4 structures, which included parallel, antiparallel, and mixed G4 structures (Sahakyan et al. 2017a).
The distribution of G4 stability scores in the human genome is bimodal (Supplemental Fig. S2). Based on this distribution and using experimental validations, Sahakyan and colleagues (Sahakyan et al. 2017a) used a threshold stability score of 19 to split G4 loci into two groups. G4 loci with stability scores above 19 can form stable structures, and G4 loci with stability scores equal to or below 19 do not form stable structures. Henceforth, we will refer to the former group as “stable G4 loci” and to the latter group as “unstable G4 loci.” Using this threshold, we designated 342,778 G4 loci as “stable” and 327,298 G4 loci as “unstable.” The median stability score in the human genome was 19.5.
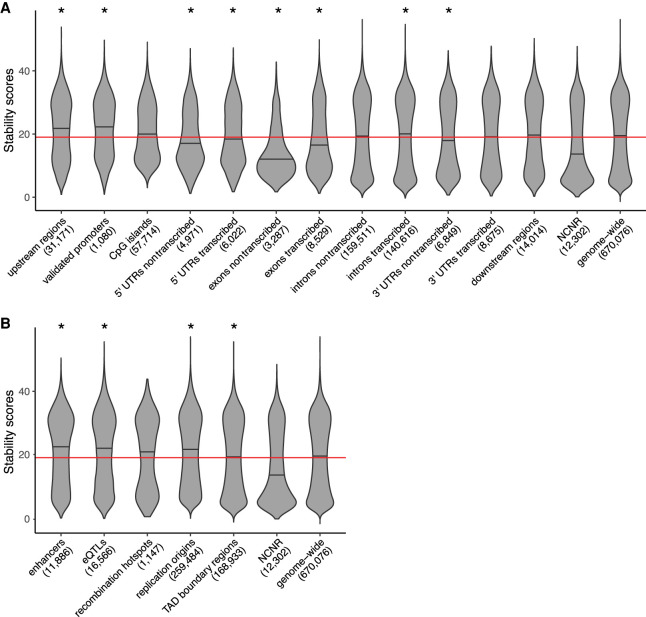
We investigated the distributions of stability scores (Fig. 3) among G4 loci located within the NCNR subgenome, the genic components described in the previous section, enhancers, replication origins, TAD boundary regions, and recombination hotspots, as well as among G4 loci intersecting with eQTLs, which are much shorter than G4 loci (The GTEx Consortium 2013). In the NCNR subgenome, the majority of G4 loci were unlikely to form stable structures; the median G4 stability score was 13.6, lower than the threshold value of 19 and the genome-wide median of 19.5 (Fig. 3A).
Figure 3.
Distribution of stability scores (violin plots) at G4 loci located at different genic components (A) and nongenic functional regions (B) compared with the genome-wide distribution and with the distribution in the NCNR subgenome. Stability scores were obtained with the Quadron software (Sahakyan et al. 2017a). Median values are marked on the violin plots. The number of G4 loci contained completely within components or regions (Supplemental Table S1) is shown in the parentheses. Because eQTLs are always smaller than G4 loci, we plotted the scores of G4 loci only partially intersecting with eQTLs. Red horizontal line indicates stability score of 19 used to differentiate between stable (more than 19) and unstable (19 or fewer) G4 loci. Stars indicate a significant difference between median stability scores in a group of components or regions and that in the rest of the genome.
We next compared the stability score distributions of G4 loci within genic components to other G4 loci genome-wide and to G4 loci within the NCNR subgenome, using the stability threshold of 19 (Fig. 3A; Supplemental Table S4). There were more stable than unstable G4 loci at upstream regions, validated promoters, and CpG islands. Median G4 stability scores at these regions were 21.9, 22.3, and 20.0, which were 1.61-, 1.64-, and 1.47-fold higher than that in the NCNR subgenome, respectively (P < 0.0017 in each case, two-tailed permutation test). The median G4 stability scores in upstream regions and validated promoters were also significantly higher than respective median values in the rest of the genome (P < 0.0017 for both, two-tailed permutation test); the difference in median stability scores between G4s at CpG islands and the rest of the genome was not significant (P = 1.00, two-tailed permutation test). The median G4 stability score in downstream regions (19.8) was 1.45-fold higher than that in the NCNR subgenome (P < 0.0017, two-tailed permutation test) but was not significantly different from that in the rest of the genome (P = 1.00, two-tailed permutation test).
Among genic components for which we could separate transcribed and nontranscribed strands, exons had the most unstable G4 loci and introns had the most stable G4 loci, whereas the stability scores of G4s in 5′ and 3′ UTRs were in between (Fig. 3A; Supplemental Table S4). At exons, most G4 loci were unstable; median stability score was 12.0 on the nontranscribed strands and 16.5 on the transcribed strands, which was 1.13-fold lower and only 1.21-fold higher, respectively, than that in the NCNR subgenome (P < 0.0017 for both, two-tailed permutation test); both values were below stability threshold of 19 and significantly lower than the respective median stability scores in the rest of the genome (P < 0.0017 for both, two-tailed permutation test). In contrast, at introns, there were more stable than unstable G4 loci. The median G4 stability score at the transcribed strands of introns (20.1) was significantly higher than that in the rest of the genome and in the NCNR subgenome (P < 0.0017 for both, two-tailed permutation test), and the median G4 stability score at the nontranscribed strands of introns (19.4) was significantly higher than that in the NCNR (P < 0.0017, two-tailed permutation test) but slightly, albeit significantly, lower than that in the rest of the genome (P < 0.0017, two-tailed permutation test). 5′ UTRs possessed mostly unstable G4 loci; their median stability scores were equal to 17.1 and 18.4 on the nontranscribed and transcribed strands, respectively, which was 1.26- and 1.35-fold higher than that in the NCNR subgenome (P < 0.0017 for both, two-tailed permutation test). Both median values were also significantly lower than the corresponding median values in the rest of the genome (P < 0.0017 for both, two-tailed permutation test). Finally, in 3′ UTRs, the median G4 stability scores were equal to 17.8 and 19.1 on the nontranscribed and transcribed strands, respectively—which was 1.31- and 1.41-fold higher than that in the NCNR subgenome (P < 0.0017 for both, two-tailed permutation test). The median value on the nontranscribed strand was also significantly lower than the median in the rest of the genome (P < 0.0017, two-tailed permutation test). Notably, for each genic group in which we could differentiate between strands, the median G4 stability was significantly lower on the nontranscribed than on the transcribed strand (Supplemental Table S5).
In each group of nongenic functional regions we studied, there were more stable than unstable G4 loci; namely, the median stability scores were higher than the stability threshold of 19 (Fig. 3B; Supplemental Table S4). At enhancers, eQTLs, and replication origins, the median G4 stability scores were particularly high: 22.4, 22.0, and 21.6, which were 1.65-, 1.62-, and 1.59-fold higher than that in the NCNR subgenome, respectively (P < 0.0017 for all; two-sided permutation test) and significantly higher than the corresponding median values in the rest of the genome (P < 0.0017 for all; two-sided permutation test). The median G4 stability score at the TAD boundary regions (19.2) was lower than those at enhancers, eQTLs, and replication origins but was still significantly higher than that in the NCNR subgenome (P < 0.0017; two-tailed permutation test), albeit significantly lower than the median in the rest of the genome (P < 0.0017, two-tailed permutation test). The median G4 stability score at recombination hotspots (20.8) was significantly higher (1.53-fold) than in the NCNR subgenome (P < 0.0017, two-tailed permutation test) but not significantly different from that in the rest of the genome (P = 1.00, two-tailed permutation test).
Selection footprints at G4 loci
An overrepresentation of G4 loci at some genic components and nongenic functional regions suggests that such loci are subject to selective constraints. In particular, we hypothesize that purifying selection toward motif retention and sequence conservation operates at functional G4 loci (e.g., the ones located in promoters). Biologically, balancing selection is not expected to operate at G4 loci, and we hypothesize that positive selection is uncommon at G4s, because it is rare in the human genome in general (Hernandez et al. 2011; Granka et al. 2012). To evaluate these hypotheses, we performed the HKA test (Hudson et al. 1987). We used this particular selection test because it is applicable regardless of the genomic context and thus it allowed us to evaluate patterns of selection at different genomic locations. This test usually contrasts the counts of polymorphic and fixed variants between two groups of regions—one hypothesized to evolve under selection and the other used as a neutrally evolving control. With the HKA test, purifying selection or balancing selection is expected to lead to an enrichment of polymorphisms relative to substitutions compared with a neutral background (Charlesworth and Charlesworth 2003), yielding an odds ratio above one. In contrast, positive selection is expected to lead to a deficit of polymorphisms relative to substitutions compared with a neutral background, yielding an odds ratio below one.
Applying the HKA test to G4 loci, we contrasted the counts of polymorphic and fixed variants at G4 loci with those expected based on the remaining sequences (devoid of G4 loci) of genic or nongenic functional regions that harbor them. We expect that many of the functional genomic regions we study (e.g., promoters) already evolve under purifying selection. If the observed counts of polymorphic variants at G4 loci are not significantly different from expectations based on non-G4 sequences in the same functional regions (significance is evaluated with Fisher's exact test), we have evidence that G4 loci and non-G4 sequences in such regions are evolving under the same selective constraints. In contrast, a significantly greater number of polymorphic variants at G4 loci is indicative of stronger purifying selection acting on G4 loci than on non-G4 sequences. In such cases, the odds ratio of Fisher's exact test (the probability of a site being polymorphic if it is located within a G4 locus) is expected to be significantly greater than one, because mutations are more likely to be polymorphic than fixed at G4 loci evolving under stronger selection than the remaining sequences.
To apply the HKA test to G4 loci, we used SNPs from the Simons Genome Diversity Project (Mallick et al. 2016). This data set was generated from 279 human genomes sequenced at a relatively high depth (∼30×). Single-nucleotide fixed variants were obtained from whole-genome alignments (Blanchette et al. 2004; Harris 2007) of human and orangutan genomes (International Human Genome Sequencing Consortium 2001; Locke et al. 2011). Odds ratios and their 95% confidence intervals were computed for all G4 loci located in a particular group of genic components or nongenic functional regions together (Fig. 4A), as well as separately for stable and unstable G4 loci (Fig. 4B). G4 loci intersecting with eQTLs and experimentally validated promoters were not included in this part of the study because on average they were longer than the components/regions they intersected with and thus did not have an appropriate background to which they could be compared. Recombination hotspots were also excluded from this analysis owing to the lack of mutations in G4s located in these regions.
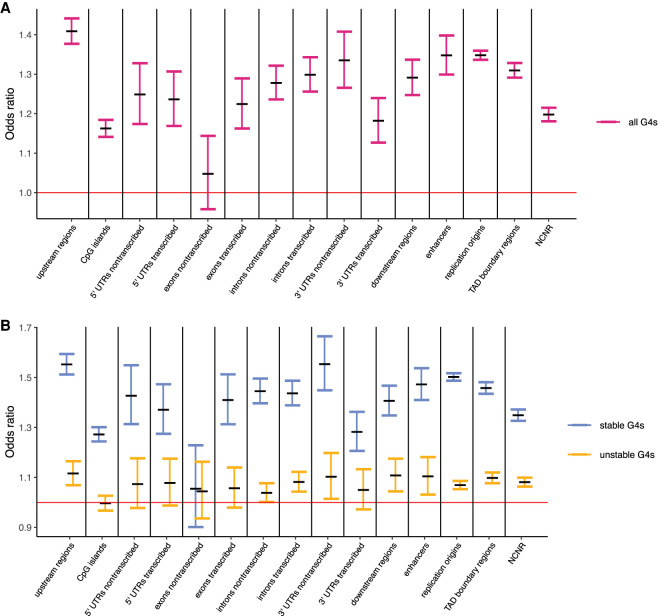
Figure 4.
Odds ratios (and their 95% confidence intervals) of the Fisher's exact test used to evaluate the significance of the Hudson–Kreitman–Aquadé test used to evaluate selection acting on G4 loci located at genic components and nongenic functional regions. Stable and unstable G4 loci are considered together (A) and separately (B). Red line represents an expectation under a null hypothesis of similar selective pressure acting on G4 loci and on the remaining sequences at the components/regions they are located within. If confidence intervals do not overlap the red line, the test is significant. Sample sizes are shown in Supplemental Table S1.
Our results suggest that G4 loci evolved under different selective constraints, depending on which genic components they were located within (Fig. 4A; Supplemental Table S6). Odds ratios were significantly greater than one for G4 loci in upstream (odds ratio = 1.41, P < 3.5 × 10−15, Fisher's exact test) and downstream regions of genes (odds ratio = 1.29, P < 3.5 × 10−15). Odds ratios were significantly greater than one on both the nontranscribed and transcribed strands of 5′ UTRs (odds ratio = 1.25 and 1.24, P = 6.38 × 10−11 and P = 3.76 × 10−12, respectively), as well as on both the nontranscribed and transcribed strands of 3′ UTRs (odds ratio = 1.34 and 1.18, P < 3.5 × 10−15 and P = 2.15 × 10−10, respectively). Odds ratios were also significantly greater than one on both the nontranscribed (odds ratio = 1.28, P < 3.5 × 10−15) and transcribed (odds ratio = 1.30, P < 3.5 × 10−15) strands of introns and on the transcribed strand of exons (odds ratio = 1.22, P = 7.37 × 10−13). In contrast, the odds ratio was not significantly different from one on the nontranscribed strand of exons (odds ratio = 0.94, P = 1.00).
We found that G4 loci located in all three groups of nongenic functional regions analyzed evolved under stronger purifying selection for motif retention than the remaining sequences in these regions (Fig. 4A; Supplemental Table S6). Odds ratios were high in all three cases: for G4 loci located at enhancers (odds ratio = 1.35, P < 3.5 × 10−15), at replication origins (odds ratio = 1.35, P < 3.5 × 10−15), and at TAD boundary regions (odds ratio = 1.31, P < 3.5 × 10−15). This finding suggests that such G4 loci are functional. Although we originally analyzed FANTOM enhancers (Andersson et al. 2014; FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014; Lizio et al. 2015), analyzing ENCODE enhancers (The ENCODE Project Consortium et al. 2020) led to a similar HKA odds ratio (odds ratio = 1.23, P < 3.5 × 10−15) (Supplemental Fig. S3). G4 loci located in the NCNR subgenome also had odds ratio greater than one (odds ratio = 1.20, P < 3.5 × 10−15), suggesting that some of them may, in fact, also evolve under purifying selection and might be functional. NCNR regions with low levels of recombination might be affected by background selection, potentially deflating the HKA odds ratio. However, the HKA odds ratios were not significantly different between NCNR regions with high versus low recombination (Supplemental Fig. S3), and thus, we retained NCNR regions with low levels of recombination in the analysis.
We then used the HKA test to evaluate selection acting on stable and (separately) unstable G4 loci in each group of genic components and nongenic functional regions considered (Fig. 4B; for P-values, see Supplemental Table S6). In almost all cases (except for the nontranscribed strands of exons), the odds ratios were significantly higher for the stable than unstable G4 loci (Fig. 4B; Supplemental Table S6). For stable G4 loci, the odds ratios were almost always (again except for the nontranscribed strands of exons) significantly greater than one (Fig. 4B; Supplemental Table S6). In contrast, for unstable G4 loci, the odds ratios were not significantly different from one in the majority of (nine out of 16) the groups of elements and regions we considered. Thus, stable G4 loci evolved under stronger purifying selection than unstable ones. This difference was not observed between stable and unstable G4s on the nontranscribed strand of exons because neither of them evolved under selective constraints different from these for non-G4 exonic sequences (Fig. 4B).
In principle, a significant HKA test result with odds ratio greater than one is consistent with either purifying selection or balancing selection (Hudson et al. 1987). To distinguish between these two possibilities, we used the Kolmogorov–Smirnov test to compare the site frequency spectrum between G4 loci and the remaining sequences for groups of genic components and nongenic functional regions for which we obtained significant HKA test results. In all cases, we observed a higher prevalence of polymorphic variants with a low MAF for G4 loci than for remaining sequences, strongly suggesting that G4 loci evolved under purifying selection (Supplemental Fig. S4; Supplemental Table S7).
Note that we never detected evidence of positive selection operating at G4 loci (Fig. 4), as the odds ratios were never significantly lower than one. Thus, although some individual G4 loci might be evolving under positive directional selection, we expect such instances to be rare and undetectable in our analysis of groups of G4 loci.
We found that the rate of polymorphism was higher at G4s than outside of G4s in some of the functional regions examined (enhancers and upstream regions) (Supplemental Table S8A). Thus, selective constraints might be more relaxed at G4 versus non-G4 sites in some functional regions, leading to the observed trends. Alternatively, but not exclusively, the increase in polymorphic rate at G4s might be owing to their elevated mutation rate (Guiblet et al. 2021).
If stable G4s are functional, then they are expected to be more conserved (i.e., to have lower human-orangutan divergence) than unstable G4s or non-G4 sites. However, elevated mutation rates at G4s, and at stable G4s in particular (Guiblet et al. 2021), might also affect divergence. To analyze this effect on divergence, we initially focused on the putatively neutral NCNR, for which selection effects should be minimal. We observed (Supplemental Table S8B) that in the NCNR, stable G4s had higher divergence than unstable G4s, and all G4s had higher divergence than non-G4s (odds ratio = 1.64, P < 2.2 × 10−16, Fisher's exact test), likely reflecting effects of mutation. We next contrasted our results for NCNR with those for upstream regions and enhancers (Supplemental Table S8B), in which G4s are likely affected both by elevated mutation rate and by purifying selection owing to functional constraints. In these regions, we again observed higher divergence at stable versus unstable G4s, and at G4s (in general) versus non-G4s. The odds ratios for G4s versus non-G4s in upstream regions and enhancers (1.19 and 1.38, respectively) were lower than that in the NCNR (1.64), suggesting that G4s in upstream regions and enhancers are affected by purifying selection. However, such selection was not strong enough to compensate for the elevated mutation rate.
The elevation of mutation rate at G4s also raises a question of whether the enrichment of polymorphisms at G4s in the HKA test reflects an increase in mutation rate at recently stabilized G4s rather than purifying selection. However, a closer look suggests that the increase in mutation rate alone cannot explain the results of the HKA test, because the majority of G4 loci were conserved in their stability between human and orangutan. Indeed, among 133,144 stable human G4s with corresponding G4 annotations in orangutan, as many as 90% were also stable in orangutan. Thus, the enrichment of polymorphisms relative to substitutions at G4 loci is more likely to reflect natural selection than mutation processes.
Discussion
In this work, we present a comprehensive analysis of G4 loci in the human genome. To the best of our knowledge, our study is the first to analyze G4 enrichment in a single framework: to study the distribution of G4 stability, and to investigate selection acting on G4 loci, as related to functional annotations in the genome. We showed that coverage, density, predicted stability, and selective pressure of G4s depend upon the genic components and nongenic functional regions in which they are located. Our results suggest that natural selection maintains a high density of G4 loci and high stability of G4 structures in some functional regions of the genome, as well as a low density and low stability in others. The situation for each particular group of regions likely depends on the balance between the selective pressure to maintain functional G4s and the cost of harboring such structures.
Our findings strongly support a functional role for several groups of G4s in the genome. We corroborated earlier studies showing an overrepresentation of G4s at most genic components (Huppert and Balasubramanian 2007; Huppert et al. 2008). For the first time, we also showed their strong overrepresentation at several nongenic functional regions (e.g., enhancers), suggesting that G4s play important roles in these regions (Fig. 2). We also found that, as a rule, genic components and nongenic functional regions with strong G4 overrepresentation harbor loci capable of forming stable G4 structures (Fig. 3). Stable G4 loci evolved under stronger purifying selection than non-G4 sequences in most groups of genic components and nongenic functional regions considered (Fig. 4B). In a summary, our results suggest that functional G4s possess the following signatures: They (1) are overrepresented, (2) are usually stable, and (3) evolve under stronger purifying selection than the remaining sequences of the components or regions to which they belong.
We expect functional G4s to have stability optimal for the function they perform. Note that here we are not arguing that they evolve toward maximum stability, as we only analyzed G4s in a binary fashion: stable versus unstable. However, our results suggest that, in most cases, a certain level of stability, namely, sufficient for G4s to be assigned as “stable,” should be reached for G4s to evolve under purifying selection and thus likely to enable their function. Another way of thinking about this is that unstable G4s might form structures only occasionally and are thus functionally inert. Whereas G4 structure formation is sensitive to environment and to temporal signals (Hänsel-Hertsch et al. 2017), our results suggest that this regulation occurs at the level of forming versus not forming stable G4s, which evolve under purifying selection.
Our observation that G4s on the nontranscribed strand of exons and in the NCNR subgenome are underrepresented agrees with the notion that G4 loci can be costly for the genome; that is, they might interfere with and disrupt basic nucleic acid processes, such as replication, transcription, and translation (Rhodes and Lipps 2015), and trigger genomic instability (Piazza et al. 2015) and high mutation rates (Guiblet et al. 2021). Therefore, unless they perform a function and thus evolve under purifying selection, G4 loci capable of forming stable G4 structures are usually removed from the genome. In genic components and functional regions where they do not possess a function, G4s usually are not overrepresented, are unstable, and are not subject to stronger selective pressure than non-G4 parts of the components/regions. These observations allow us to speculate about the functional roles of G4s located in different parts of the genome, which we discuss in light of experimental studies showing or suggesting particular functions of some G4s.
G4 loci in genic components
Upstream regions and promoters
At putative (upstream regions) and experimentally validated promoters, G4 loci were not only strongly overrepresented but also significantly more stable compared with the G4 loci in the rest of the genome and in the NCNR subgenome. In fact, median G4 stability scores at experimentally validated promoters were higher than at any other genic components analyzed. Moreover, we also detected a strong signature of purifying selection acting on G4 loci located at putative promoters, with the highest HKA odds ratio among genic components studied. Selection was particularly strong for G4 loci capable of forming stable structures at putative promoters. These findings corroborate earlier studies showing an enrichment of G4 loci at promoters (Huppert and Balasubramanian 2007) and are in agreement with the growing experimental support of their role in regulating gene expression, likely via binding transcription factors (for review, see Varshney et al. 2020). Indeed, it was recently shown that, in human cells, G4 antibodies colocalize with transcriptionally active chromatin, G4 structures form preferentially at promoters of highly expressed genes, and enhanced G4 formation is associated with increased transcriptional activity (Hänsel-Hertsch et al. 2016a). In another study, G4 structures were mapped to chromatin in upstream regulatory regions of actively transcribed human genes (Kouzine et al. 2017). Additionally, polymorphisms at G4 loci located within promoters were found to affect variation in transcription levels of genes in human populations (Baral et al. 2012).
CpG islands
The G4 loci located at CpG islands, which are frequently found within promoters, had higher coverage (and density) compared with the corresponding mean and median values for the genome, respectively, and evolved under purifying selection. This result is consistent with a proposed functional role of G4 structures at CpG islands: it was hypothesized that such structures contribute to maintaining the nonmethylated status of the CpG islands (Mao et al. 2018).
Downstream regions
We discovered that G4 loci were overrepresented and more stable, and evolved under purifying selection, when located within 1 kb downstream from the transcription termination sites. These results point toward functionality of G4 structures at downstream regions and support a suggestion that such structures aid in demarcating transcription termination (Gromak et al. 2006; Huppert et al. 2008). An enrichment of G4 loci immediately downstream (within 100 bp) from 3′ UTRs was shown in another study (Huppert et al. 2008).
General observations for strand-specific genic components
G4 loci within genic components for which we could differentiate between transcribed and nontranscribed strands showed two common trends. First, except for introns, their G-corrected coverage (and density) was higher on the transcribed than on the nontranscribed strand (Fig. 2A), an observation in line with previous studies (Huppert et al. 2008; Varshney et al. 2020). Second, they were more stable on the transcribed than on the nontranscribed strand (Fig. 3A). Thus, stable G4s are particularly depleted on the nontranscribed DNA strand. Previous studies suggested that G4s located on the nontranscribed DNA strand aid in maintaining the DNA in an open state and thus in transcription reinitiation, whereas those located on the transcribed DNA strand inhibit transcription (for review, see Varshney et al. 2020). Thus, the latter and not the former G4s should be depleted, whereas we observed the opposite. We note that G4s on the nontranscribed DNA strand may also form within the corresponding RNA. In this case, our results are consistent with G4 depletion owing to interference with other processes, for example, translation. Moreover, our study suggests that such structures are avoided in pre-mRNA in general, even for introns that are not part of mRNA.
5′ and 3′ UTRs
Our findings that G4 loci are strongly overrepresented at 5′ UTRs and overrepresented (though not as strongly) at 3′ UTRs are in agreement with another study (Huppert et al. 2008). We made novel observations related to stability of G4s located in 5′ and 3′ UTRs. We found that the median stability values of such G4s were similar to the rest of the genome on the transcribed strand but were lower on the nontranscribed strand, consistent with the general trend we observed across genic components. We detected footprints of purifying selection acting on stable G4s located on both the nontranscribed and transcribed strands of both 5′ and 3′ UTRs. Signatures of selection on G4s located on the nontranscribed strand of 5′ and 3′ UTRs were also found in another recent study (Lee et al. 2020).
The function of G4s capable of forming at UTRs is still being investigated. G4s are frequently located near the beginning of 5′ UTRs in mRNAs and may play a role in transcription initiation (Huppert et al. 2008; Morris et al. 2010). It was previously found that G4 structures form at 5′ UTRs of highly expressed genes and thus might facilitate their transcription (Hänsel-Hertsch et al. 2016). Our selection results also support that the nontranscribed strand of UTRs harbors stable functional G4s, which have the potential to form at the level of RNA. Consistent with this, it was recently shown that G4 loci at UTRs are enriched for RNA-binding protein interactions, arguing for their functionality in RNA (Lee et al. 2020). Nevertheless, we found G4 loci to be less stable on the nontranscribed than on the transcribed strand of 5′ UTRs. This is likely because G4 loci on the nontranscribed strand of 5′ UTRs may repress binding of the translation initiation complex to 5′-cap of mRNAs or disrupt ribosomal scanning of the mRNA toward the start codon (Kumari et al. 2007; Huppert et al. 2008).
Exons
For G4s located in exons, after correcting for G-content we found an overrepresentation on the transcribed strand and a depletion on the nontranscribed strand, consistent with another study (Huppert and Balasubramanian 2005). Our study showed that the median stability of G4s at exons was always lower than that for the rest of the genome, and for G4s on the nontranscribed strand of exons, it was even lower than that for the NCNR subgenome. This observation agrees with another study that also suggested low stability of G4s in exons (Arachchilage et al. 2019). Thus, predominantly stable G4s may not be tolerated in exons, particularly on the nontranscribed strand. Moreover, we did not detect purifying selection acting on either stable or unstable G4s on the nontranscribed strand of exons, suggesting that G4 structures on the nontranscribed strand are not maintained by natural selection. Thus, both stable and unstable G4s are unlikely to be functional, and stable G4s may be strongly avoided, in this group of genic components. Because such G4s can form at the level of RNA, they may hinder mRNA translation elongation through ribosomal frameshifting and stalling (Shabalina et al. 2006; Endoh and Sugimoto 2013; Endoh et al. 2013) and were proposed to be selected against by the context-dependent codon bias (Arachchilage et al. 2019). In fact, G4 stability was found to be proportional to their interference with translation (Endoh et al. 2013). For the transcribed strand of exons, we only detected selection acting on stable G4s. Such G4s may participate in regulation of transcription at the level of DNA, a hypothesis that should be evaluated in future studies.
Introns
We found that coverage and density of G4s located in introns were similar to those in the rest of the genome but higher than those in the NCNR subgenome. We found signatures of purifying selection acting on stable G4 loci located on both the transcribed and nontranscribed strands of introns (and also on unstable G4s on their transcribed strands), arguing for their functionality. Intronic G4s on the nontranscribed strand may participate in transcription regulation, particularly if they are located in the vicinity of the transcription start sites (Eddy and Maizels 2008), or in RNA processing, including alternative splicing (Eddy and Maizels 2008; Marcel et al. 2011; Fisette et al. 2012). Similar to G4s in exons, G4s located on the transcribed strand of introns may be involved in regulating transcription, and further studies should evaluate this possibility experimentally.
G4 loci in nongenic functional regions
Enhancers
Our study indicated overrepresentation, high stability, and evolution under purifying selection for G4 loci at enhancers. In fact, the median G4 stability score at enhancers was higher than that for any other group of genic components or nongenic functional regions we studied. These observations strongly suggest that G4 loci play an important role in maintaining the function of enhancers. These results are particularly novel, as most previous studies focused on the analysis of G4 loci in genic components. One notable exception is a recent study showing the presence of G4s at long G-rich enhancer-associated regions and at the promoters of genes they regulate, suggesting that G4s facilitate enhancer–promoter interactions (Williams et al. 2020).
eQTLs
We found that eQTLs were enriched in G4s and that G4s overlapping with eQTLs were usually highly stable. This result is perhaps expected, as eQTLs frequently include sites of promoters, UTRs, and enhancers (The GTEx Consortium 2020), where we also found an overrepresentation of G4s. Another study recently detected an enrichment of eQTLs at G4s located at UTRs (Lee et al. 2020). Taken together, these observations provide strong evidence for an important role of G4s in regulating gene expression genome-wide.
Replication origins
We found strong overrepresentation, high stability, and purifying selection for G4 loci at replication origins. In fact, replication origins had the highest G4 density among all groups of nongenic functional regions we analyzed. Thus, our results are consistent with the functional role of G4 loci at replication origins genome-wide, confirming previous single-locus studies (e.g., Valton et al. 2014), and support the proposed importance of G4 loci in replication origin firing (Prorok et al. 2019).
TAD boundary regions
G4 loci at TAD boundaries displayed an enrichment in density and coverage, in agreement with another study (Hou et al. 2019). Their median stability score was close to that for the rest of the genome, albeit higher than the median score for the NCNR subgenome. We uncovered footprints of purifying selection operating at both stable and unstable G4 loci at TAD boundary regions. Therefore, our results suggest that G4s in these regions are functional. G4s were shown to have strong insulation ability and were suggested to play a role in delineating TADs (Hou et al. 2019). The fact that they have relatively low stability scores is at odds with the pattern we observed for most other functionally important G4s, which were predominantly stable. One possible explanation for this observation is the current lack of knowledge concerning the precise sizes of TAD boundary regions. An alternative explanation is that some unstable G4s are also functional in the genome.
Recombination hotspots
G4 loci were modestly overrepresented at recombination hotspots. The majority of G4 loci at recombination hotspots were stable; their median stability was not significantly different from that for the rest of the genome but was higher than in the NCNR subgenome. Unfortunately, the currently available data did not allow us to study selection acting on G4s located at recombination hotspots. Thus, our study could not provide evidence of functionality of G4s located at them, and this question should be investigated in the future. G4 structures were shown to be sites of preferential recombination via quadruplex formation in vitro (Boán and Gómez-Márquez 2010) and were suggested to promote meiotic homologous recombination in vivo (for review, see Bochman et al. 2012).
Future directions, study limitations, and conclusions
Our study is the first to report depletion and, on average, low stability of G4 loci in the NCNR subgenome. We propose that stable G4 loci in neutrally evolving regions may be harmful to the genome owing to their association with genomic instability (Piazza et al. 2015) and high mutation rates (Guiblet et al. 2021). We found that some G4 loci in the NCNR subgenome evolved under purifying selection and thus might be functional. This novel observation suggests that G4 loci outside of the genic components and nongenic functional genomic regions studied, as well as outside of the other regions excluded from the NCNR subgenome (e.g., repetitive elements, telomeres), might possess novel, yet unknown, functions—an exciting possibility that should be investigated in future studies.
The results of our study can only be interpreted as genome-wide trends and cannot be extrapolated to individual loci. Recent studies have established that only a subset of annotated G4 loci fold into non-B DNA structures at a given time in vivo (Hänsel-Hertsch et al. 2016; Kouzine et al. 2017). The formation of a G4 structure is not solely determined by its stability but is modulated by supercoiling (over- or underwinding of a DNA strand) and cellular conditions such as ionic concentrations. Because of this transient nature, it is difficult to study the effect of G4 structures at individual loci. Recently developed methods allowing one to investigate genome-wide G4 formation in real time (e.g., permanganate sequencing [Kouzine et al. 2019] and kethoxal-assisted single-stranded sequencing [Wu et al. 2020]) are expected to enable locus-specific analysis of G4 formation, selection, and function in the near future. Such analyses will be critical for uncovering the roles individual G4 loci play in genome function and evolution.
Individual G4 loci may overlap with more than one genomic annotation. As a result, determining what specific G4 functions may be under selection becomes challenging. In our study, only a relatively small proportion of bases in G4 loci intersected with more than one functional annotation (Supplemental Table S9). We aimed to present an exhaustive analysis of G4 overrepresentation, stability, and selection and thus accepted that overlaps in assigning G4s to particular groups of components or regions represent a biological reality. Future studies might take an opposite approach and analyze only groups of G4 loci with nonoverlapping annotations.
In conclusion, our observations on enrichment or depletion, stability, and purifying selection of G4 loci at multiple groups of genic components and nongenic functional genomic regions support a wide variety of their hypothesized and reported functions and disruptive properties (Rhodes and Lipps 2015; Varshney et al. 2020). Moreover, our results suggest that predicted G4 stability scores provide additional insights into functionality of G4 loci, as most functional G4 loci in the genome appear to be stable. Maintaining beneficial, functional G4s, especially those associated with gene expression, must be balanced with the costs of detrimental G4s, which increase mutagenesis and genome instability. Because G4 loci are involved in a wide range of genomic functions and evolve under purifying selection even in otherwise neutrally evolving regions, we propose to classify such loci as functional elements of the genome.
Methods
Data sets
G4 loci and their predicted stability scores were annotated using Quadron (Sahakyan et al. 2017a). We did not consider G4 loci annotated on the two sex chromosomes and on the mitochondrial DNA. The hg19 version of the human genome was used because it has a larger number of annotations of genomic features than the more recent hg38 version.
Multiple data sets were downloaded from the UCSC Genome Browser (Haeussler et al. 2019) for the human reference genome (version hg19). We retrieved the NCBI RefSeq annotations of genic components for protein-coding genes (Pruitt et al. 2014), upstream and downstream regions of genes (each 1 kb long from the transcription start site and the transcription termination site, respectively), 5′ and 3′ UTRs, protein-coding exons, and introns. All exons and introns from NCBI RefSeq, including alternative transcripts, were taken into account. All transcript coordinates were merged in “exonic” (or “intronic”) versus “nonexonic” (or “nonintronic”). We also obtained the annotations of CpG islands (Gardiner-Garden and Frommer 1987), repeats (based on RepeatMasker) (Smit 2004), and eQTLs occurring in multiple tissues (based on GTEx project analyses) (The GTEx Consortium 2019).
The annotations of origins of replication were retrieved from Besnard et al. (2012). The annotations of experimentally defined enhancers and promoters were obtained from the FANTOM5 project (Andersson et al. 2014; FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014; Lizio et al. 2015). In this data set, promoters are annotated at a greater resolution than in the 1-kb upstream regions of genes. However, annotated promoters are on average shorter than G4 motifs (∼20 bp vs. ∼36 bp). As a result, G4 loci only partially intersected with FANTOM5 promoters. The CTCF binding site annotations were retrieved from The ENCODE Project (The ENCODE Project Consortium 2012). The annotations of TAD boundaries were retrieved from Dixon et al. (2015). A TAD boundary and TAD boundary region were defined as the center point between two consecutive TADs and ±150 kb around it, respectively, following the method previously described (Hong and Kim 2017). The sex-averaged recombination rates were retrieved from Halldorsson et al. (2019), and following the same study, recombination hotspots were defined as regions where recombination rates were 10 times higher than the genome-wide average.
The NCNR subgenome was defined as all locations of the genome that did not overlap with the studied genic components or nongenic functional regions, as well as with CTCF binding sites, repetitive elements, noncoding RNAs (The RNA Central Consortium et al. 2017), gaps (including telomeres, centromeres, and heterochromatin) in the reference genome (downloaded from the UCSC Genome Browser) (Haeussler et al. 2019), and 5-kb upstream and downstream regions of genes. Finally, we deleted a 6-kb region (Chr 16: 31,970,000–33,760,000) that contains the immunoglobulin heavy (IGH) locus to remove G4 loci involved in class-switch recombination (Sen and Gilbert 1988).
Density and coverage of G4 loci across the genome
We computed the total number of bases annotated as genic components or nongenic functional regions on the reference strand and then also on its reverse complement of the human genome using BEDTools (Quinlan and Hall 2010). When two or more annotations of the same type (e.g., for exons) overlapped, we selected uniformly at random only one such annotation to avoid counting the same interval multiple times (Supplemental Fig. S5). We ran this process 10 times, and the resulting standard deviations of coverage values were extremely low (Supplemental Table S10); thus, this process did not affect our estimates substantially. 5′ and 3′ UTRs, protein-coding exons, and introns were classified as “nontranscribed” if their coding sequence (i.e., the sequence present in RNA) matched the one of the reference strand and were classified as “transcribed” if it matched the one of the reverse complement strand.
G4 coverage was computed by dividing the number of bases annotated as G4 loci by the total number of bases (including both reference and reverse complement strands) in each group of genic components or nongenic functional regions. G4 density was computed by dividing the number of G4 loci by the total number of bases in each group of genic components or nongenic functional regions. If a G4 locus was not completely contained within a genic component or nongenic functional region, then the fraction of its length that was contained was used in the calculation. We also computed G4 coverage and density corrected by the guanine content, by dividing each of these measurements by the proportion of guanines in each group of genic components or nongenic functional regions. Fisher's exact tests were performed to evaluate the significance of differences in G4 density or coverage (corrected or uncorrected by guanine content) between different groups of genic regions or nongenic functional regions and genome-wide averages or measurements for the NCNR subgenome. Resulting P-values were adjusted for multiple testing using the Bonferroni correction for 18 tests.
Analysis of stability scores in genic components and nongenic functional regions
Stability scores of G4 loci were generated using Quadron (Sahakyan et al. 2017a). The stability scores of G4s overlapping with each group of the studied genic components and nongenic functional regions were pooled with those in the NCNR subgenome. We removed labels indicating whether a G4 locus belonged to a particular genic component or the NCNR subgenome. Next, we randomly assigned these labels to G4s, keeping the number of G4s in a group of genic components (or nongenic functional regions) and in the NCNR subgenome intact, and computed the absolute difference in medians between these two newly labeled sets of G4s. This procedure was performed 10,000 times. To obtain the empirical P-values, we computed the percentile of the original, observed absolute value of the difference in medians to the distribution of absolute value of differences in medians as obtained from 10,000 permuted distributions. The resulting P-values were adjusted for multiple testing using the Bonferroni correction for 17 tests. A similar analysis was performed for each group of genic components or nongenic functional regions and the remaining genome-wide data.
HKA test
To perform the HKA test, we used fixed single-nucleotide variants between human and orangutan, as well as SNPs from the Simons Genome Diversity Project (Mallick et al. 2016). A total of 69,329,877 fixed single-nucleotide variants between human and orangutan genomes was retrieved from the vertebrate MULTIZ 100-way alignment (Blanchette et al. 2004; Miller et al. 2007) obtained from the UCSC Genome Browser (Haeussler et al. 2019). A total of 44,833,480 SNPs from the Simons Genome Diversity Project (Mallick et al. 2016) was acquired from the Seven Bridges Cancer Genomic Cloud (https://cgc.sbgenomics.com/). We discarded singletons and doubletons as they might represent false positives (removing only singletons or all SNPs with MAF <5% did not change results qualitatively). Fixed and polymorphic mutations were (separately) intersected with genic components, nongenic functional regions, and the NCNR subgenome. For each group of genic components or nongenic functional regions, mutations not intersecting with G4 loci were considered as background variation for that group of components or regions. We compared the observed counts of fixed and polymorphic variants at G4 loci to those expected based on the remaining (non-G4) sequences of the components/regions they were harbored by. We used Fisher's exact test to evaluate the significance of an odds ratio of nucleotide variants to be polymorphic if found at G4 loci, as well as the confidence interval of these odds ratios. The resulting P-values were adjusted for multiple testing using the Bonferroni correction for 16 tests.
Site frequency spectrum
The distributions of MAFs of polymorphic mutations from the Simons Genome Diversity Project (Mallick et al. 2016) were compared between G4 loci and the remaining sequences of genic components or nongenic functional regions they are located within. Minor alleles were assumed to be the ones with a lower allele frequency, resulting in a folded site frequency spectrum. We discarded singletons and doubletons as they might represent false positives. Comparisons between the MAF distribution in G4s and the remaining sequences were performed with the two-sample Kolmogorov–Smirnov test. The resulting P-values were adjusted for multiple testing using the Bonferroni correction for 14 tests.
Software availability
All computational tools used in this study are available at GitHub (https://github.com/makovalab-psuG4_Selection) and as Supplemental Codes 1–3.
Supplementary Material
Acknowledgments
We thank Marzia Cremona and Matthias Weissensteiner for helpful discussions and Xiao Sun and Yue Hou for providing the TAD boundary data set. This study was supported by National Institutes of Health (NIH) R01GM136684 and R01CA23715. It was also supported by the Clinical and Translational Sciences Institute, the Institute of Computational and Data Sciences, the Huck Institutes of the Life Sciences, and the Eberly College of Science of the Pennsylvania State University. M.D. was supported by NIH R35GM128590, National Science Foundation (NSF) DEB-1949268, and NSF BCS-2001063. Finally, this research was supported by the CBIOS Predoctoral Training Program awarded to Penn State by the National Institutes of Health (W.M.G. was a trainee). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.269589.120.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–461. 10.1038/nature12787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arachchilage GM, Arachchilage MH, Venkataraman A, Piontkivska H, Basu S. 2019. Stable G-quadruplex enabling sequences are selected against by the context-dependent codon bias. Gene 696: 149–161. 10.1016/j.gene.2019.02.006 [DOI] [PubMed] [Google Scholar]
- Babendure JR, Babendure JL, Ding J-H, Tsien RY. 2006. Control of mammalian translation by mRNA structure near caps. RNA 12: 851–861. 10.1261/rna.2309906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral A, Kumar P, Halder R, Mani P, Yadav VK, Singh A, Das SK, Chowdhury S. 2012. Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals. Nucleic Acids Res 40: 3800–3811. 10.1093/nar/gkr1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin J-D, Perreault J-P. 2013. Exploring mRNA 3′-UTR G-quadruplexes: evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res 41: 5898–5911. 10.1093/nar/gkt265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin J-M, Lemaitre J-M. 2012. Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19: 837–844. 10.1038/nsmb.2339 [DOI] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S. 2013. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5: 182–186. 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AFA, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715. 10.1101/gr.1933104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boán F, Gómez-Márquez J. 2010. In vitro recombination mediated by G-quadruplexes. Chembiochem 11: 331–334. 10.1002/cbic.200900612 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA. 2012. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 13: 770–780. 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaut A, Balasubramanian S. 2012. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res 40: 4727–4741. 10.1093/nar/gks068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. 2015. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 33: 877–881. 10.1038/nbt.3295 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. 2003. Evolution: a very short introduction. Oxford University Press, Oxford. [Google Scholar]
- Cheung I, Schertzer M, Rose A, Lansdorp PM. 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet 31: 405–409. 10.1038/ng928 [DOI] [PubMed] [Google Scholar]
- Chiaromonte F, Weber RJ, Roskin KM, Diekhans M, Kent WJ, Haussler D. 2003. The share of human genomic DNA under selection estimated from human–mouse genomic alignments. Cold Spring Harb Symp Quant Biol 68: 245–254. 10.1101/sqb.2003.68.245 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewicz-Bourget JE, Lee AY, Ye Z, Kim A, Rajagopal N, Xie W, et al. 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinnaya NG, Ogloblina AM, Yakubovskaya MG. 2016. Structure, properties, and biological relevance of the DNA and RNA G-quadruplexes: overview 50 years after their discovery. Biochemistry (Mosc) 81: 1602–1649. 10.1134/S0006297916130034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Gertz EM, Wojtowicz D, Zhabinskaya D, Levens D, Benham CJ, Schäffer AA, Przytycka TM. 2014. Potential non-B DNA regions in the human genome are associated with higher rates of nucleotide mutation and expression variation. Nucleic Acids Res 42: 12367–12379. 10.1093/nar/gku921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy J, Maizels N. 2008. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res 36: 1321–1333. 10.1093/nar/gkm1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583: 699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh T, Sugimoto N. 2013. Unusual −1 ribosomal frameshift caused by stable RNA G-quadruplex in open reading frame. Anal Chem 85: 11435–11439. 10.1021/ac402497x [DOI] [PubMed] [Google Scholar]
- Endoh T, Kawasaki Y, Sugimoto N. 2013. Stability of RNA quadruplex in open reading frame determines proteolysis of human estrogen receptor α. Nucleic Acids Res 41: 6222–6231. 10.1093/nar/gkt286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT). 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette J-F, Montagna DR, Mihailescu M-R, Wolfe MS. 2012. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J Neurochem 121: 763–773. 10.1111/j.1471-4159.2012.07680.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. 1987. CpG islands in vertebrate genomes. J Mol Biol 196: 261–282. 10.1016/0022-2836(87)90689-9 [DOI] [PubMed] [Google Scholar]
- Georgakopoulos-Soares I, Morganella S, Jain N, Hemberg M, Nik-Zainal S. 2018. Noncanonical secondary structures arising from non-B DNA motifs are determinants of mutagenesis. Genome Res 28: 1264–1271. 10.1101/gr.231688.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez D. 2004. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res 32: 371–379. 10.1093/nar/gkh181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granka JM, Henn BM, Gignoux CR, Kidd JM, Bustamante CD, Feldman MW. 2012. Limited evidence for classic selective sweeps in African populations. Genetics 192: 1049–1064. 10.1534/genetics.112.144071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ. 2006. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol 26: 3986–3996. 10.1128/MCB.26.10.3986-3996.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) Project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2020. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369: 1318–1330. 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiblet WM, Cremona MA, Cechova M, Harris RS, Kejnovská I, Kejnovsky E, Eckert K, Chiaromonte F, Makova KD. 2018. Long-read sequencing technology indicates genome-wide effects of non-B DNA on polymerization speed and error rate. Genome Res 28: 1767–1778. 10.1101/gr.241257.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiblet WM, Cremona MA, Harris RS, Chen D, Eckert KA, Chiaromonte F, Huang Y-F, Makova KD. 2021. Non-B DNA: a major contributor to small- and large-scale variation in nucleotide substitution frequencies across the genome. Nucleic Acids Res 49: 1497–1516. 10.1093/nar/gkaa1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, et al. 2019. The UCSC genome browser database: 2019 update. Nucleic Acids Res 47: D853–D858. 10.1093/nar/gky1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder R, Halder K, Sharma P, Garg G, Sengupta S, Chowdhury S. 2010. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol Biosyst 6: 2439–2447. 10.1039/c0mb00009d [DOI] [PubMed] [Google Scholar]
- Halldorsson BV, Palsson G, Stefansson OA, Jonsson H, Hardarson MT, Eggertsson HP, Gunnarsson B, Oddsson A, Halldorsson GH, Zink F, et al. 2019. Characterizing mutagenic effects of recombination through a sequence-level genetic map. Science 363: eaau1043. 10.1126/science.aau1043 [DOI] [PubMed] [Google Scholar]
- Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, et al. 2016. G-quadruplex structures mark human regulatory chromatin. Nat Genet 48: 1267–1272. 10.1038/ng.3662 [DOI] [PubMed] [Google Scholar]
- Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. 2017. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol 18: 279–284. 10.1038/nrm.2017.3 [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. “Improved pairwise alignment of genomic DNA.” PhD thesis, The Pennsylvania State University, University Park, PA. [Google Scholar]
- Hernandez RD, Kelley JL, Elyashiv E, Melton SC, Auton A, McVean G, 1000 Genomes Project, Sella G, Przeworski M. 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924. 10.1126/science.1198878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Kim D. 2017. Computational characterization of chromatin domain boundary-associated genomic elements. Nucleic Acids Res 45: 10403–10414. 10.1093/nar/gkx738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Li F, Zhang R, Li S, Liu H, Qin ZS, Sun X. 2019. Integrative characterization of G-quadruplexes in the three-dimensional chromatin structure. Epigenetics 14: 894–911. 10.1080/15592294.2019.1621140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116: 153–159. 10.1093/genetics/116.1.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. 2005. Prevalence of quadruplexes in the human genome. Nucleic Acids Res 33: 2908–2916. 10.1093/nar/gki609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Balasubramanian S. 2007. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res 35: 406–413. 10.1093/nar/gkl1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Bugaut A, Kumari S, Balasubramanian S. 2008. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res 36: 6260–6268. 10.1093/nar/gkn511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Jara-Espejo M, Peres Line SR. 2020. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J 287: 483–495. 10.1111/febs.15065 [DOI] [PubMed] [Google Scholar]
- Kim M, Kreig A, Lee C-Y, Rube HT, Calvert J, Song JS, Myong S. 2016. Quantitative analysis and prediction of G-quadruplex forming sequences in double-stranded DNA. Nucleic Acids Res 44: 4807–4817. 10.1093/nar/gkw272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Baranello L, Yamane A, Nelson S, Resch W, Kieffer-Kwon K-R, Benham CJ, Casellas R, Przytycka TM, et al. 2017. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst 4: 344–356.e7. 10.1016/j.cels.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzine F, Wojtowicz D, Yamane A, Casellas R, Przytycka TM, Levens DL. 2019. In vivo chemical probing for G-quadruplex formation. Methods Mol Biol 2035: 369–382. 10.1007/978-1-4939-9666-7_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Bugaut A, Huppert JL, Balasubramanian S. 2007. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol 3: 218–221. 10.1038/nchembio864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Kim DS. 2009. Dramatic effect of single-base mutation on the conformational dynamics of human telomeric G-quadruplex. Nucleic Acids Res 37: 3625–3634. 10.1093/nar/gkp216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DSM, Ghanem LR, Barash Y. 2020. Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat Commun 11: 527. 10.1038/s41467-020-14404-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexa M, Steflova P, Martinek T, Vorlickova M, Vyskot B, Kejnovsky E. 2014. Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics 15: 1032. 10.1186/1471-2164-15-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. 2015. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16: 22. 10.1186/s13059-014-0560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang S-P, Wang Z, Chinwalla AT, Minx P, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533. 10.1038/nature09687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. 2015. G4-associated human diseases. EMBO Rep 16: 910–922. 10.15252/embr.201540607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538: 201–206. 10.1038/nature18964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani P, Yadav VK, Das SK, Chowdhury S. 2009. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One 4: e4399. 10.1371/journal.pone.0004399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao S-Q, Ghanbarian AT, Spiegel J, Cuesta SM, Beraldi D, Di Antonio M, Marsico G, Hänsel-Hertsch R, Tannahill D, Balasubramanian S. 2018. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol 25: 951–957. 10.1038/s41594-018-0131-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Tran PLT, Sagne C, Martel-Planche G, Vaslin L, Teulade-Fichou M-P, Hall J, Mergny J-L, Hainaut P, Van Dyck E. 2011. G-quadruplex structures in TP53 intron 3: role in alternative splicing and in production of p53 mRNA isoforms. Carcinogenesis 32: 271–278. 10.1093/carcin/bgq253 [DOI] [PubMed] [Google Scholar]
- Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, Burhans R, King DC, Baertsch R, Blankenberg D, et al. 2007. Twenty-eight-way vertebrate alignment and conservation track in the UCSC genome browser. Genome Res 17: 1797–1808. 10.1101/gr.6761107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. 2010. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc 132: 17831–17839. 10.1021/ja106287x [DOI] [PubMed] [Google Scholar]
- Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, Lovrecz GO, Beck JL, Bryan TM. 2015. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun 6: 7643. 10.1038/ncomms8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakken S, Rognes T, Hovig E. 2009. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res 37: 5749–5756. 10.1093/nar/gkp590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A, Adrian M, Samazan F, Heddi B, Hamon F, Serero A, Lopes J, Teulade-Fichou M-P, Phan AT, Nicolas A. 2015. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J 34: 1718–1734. 10.15252/embj.201490702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnavaia TJ, Marshall CL, Mettler CM, Fisk CL, Todd Miles H, Becker ED. 1978. Alkali metal ion specificity in the solution ordering of a nucleotide, 5′-guanosine monophosphate. J Am Chem Soc 100: 3625–3627. 10.1021/ja00479a070 [DOI] [Google Scholar]
- Prorok P, Artufel M, Aze A, Coulombe P, Peiffer I, Lacroix L, Guédin A, Mergny J-L, Damaschke J, Schepers A, et al. 2019. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat Commun 10: 3274. 10.1038/s41467-019-11104-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, Farrell CM, Hart J, Landrum MJ, McGarvey KM, et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 42: D756–D763. 10.1093/nar/gkt1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Lipps HJ. 2015. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 43: 8627–8637. 10.1093/nar/gkv862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The RNA Central Consortium, Petrov AI, Kay SJE, Kalvari I, Howe KL, Gray KA, Bruford EA, Kersey PJ, Cochrane G, Finn RD, et al. 2017. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res 45: D128–D134. 10.1093/nar/gkw1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan AB, Chambers VS, Marsico G, Santner T, Di Antonio M, Balasubramanian S. 2017a. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci Rep 7: 14535. 10.1038/s41598-017-14017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan AB, Murat P, Mayer C, Balasubramanian S. 2017b. G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat Struct Mol Biol 24: 243–247. 10.1038/nsmb.3367 [DOI] [PubMed] [Google Scholar]
- Sen D, Gilbert W. 1988. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334: 364–366. 10.1038/334364a0 [DOI] [PubMed] [Google Scholar]
- Sen D, Gilbert W. 1990. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 344: 410–414. 10.1038/344410a0 [DOI] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov NA. 2006. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res 34: 2428–2437. 10.1093/nar/gkl287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone R, Fratta P, Neidle S, Parkinson GN, Isaacs AM. 2015. G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett 589: 1653–1668. 10.1016/j.febslet.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Smit AFA. 2004. RepeatMasker Open-3.0. http://www.repeatmasker.org [accessed October 23, 2018].
- Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou M-P, Abramowitz L, et al. 2011. Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 18: 478–485. 10.1038/nsmb.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Brazier JA, Sugimoto N. 2017. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc Natl Acad Sci 114: 9605–9610. 10.1073/pnas.1704258114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton A-L, Prioleau M-N. 2016. G-Quadruplexes in DNA replication: a problem or a necessity? Trends Genet 32: 697–706. 10.1016/j.tig.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Valton A-L, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintomé C, Riou J-F, Prioleau M-N. 2014. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J 33: 732–746. 10.1002/embj.201387506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. 2020. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol 21: 459–474. 10.1038/s41580-020-0236-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. 2008. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J Med Chem 51: 5641–5649. 10.1021/jm800448a [DOI] [PubMed] [Google Scholar]
- Vorlíčková M, Kejnovská I, Sagi J, Renčiuk D, Bednářová K, Motlová J, Kypr J. 2012. Circular dichroism and guanine quadruplexes. Methods 57: 64–75. 10.1016/j.ymeth.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH. 1953. Genetical implications of the structure of deoxyribonucleic acid. Nature 171: 964–967. 10.1038/171964b0 [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci 28: 182–188. 10.1016/s0968-0004(03)00051-3 [DOI] [PubMed] [Google Scholar]
- Williams JD, Houserova D, Johnson BR, Dyniewski B, Berroyer A, French H, Barchie AA, Bilbrey DD, Demeis JD, Ghee KR, et al. 2020. Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop “G4 kissing” interaction. Nucleic Acids Res 48: 5907–5925. 10.1093/nar/gkaa357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Lyu R, You Q, He C. 2020. Kethoxal-assisted single-stranded DNA sequencing captures global transcription dynamics and enhancer activity in situ. Nat Methods 17: 515–523. 10.1038/s41592-020-0797-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Bacolla A, Wang G, Vasquez KM. 2010. Non-B DNA structure-induced genetic instability and evolution. Cell Mol Life Sci 67: 43–62. 10.1007/s00018-009-0131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.