Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a beta coronavirus that belongs to the Coronaviridae family. SARS-CoV-2 is an enveloped spherical-shaped virus. The ribonucleic acid (RNA) is oriented in a 5’-3’direction which makes it a positive sense RNA virus, and the RNA can be read directly as a messenger RNA. The nonstructural protein 14 (nsp14) has proofreading activity which allows the rate of mutations to stay low. A change in the genetic sequence is called a mutation. Genomes that differ from each other in genetic sequence are called variants. Variants are the result of mutations but differ from each other by one or more mutations. When a phenotypic difference is demonstrated among the variants, they are called strains. Viruses constantly change in two different ways, antigenic drift and antigenic shift. SARS-CoV-2 genome is also prone to various mutations that led to antigenic drift resulting in escape from immune recognition. The Center of Disease Control and Prevention (CDC) updates the variant strains in the different classes. The classes are variant of interest, variant of concern and variant of high consequence. The current variants included in the variant of interest by the USA are: B.1.526, B.1.525, and P.2; and those included in the variant of concern by the USA are B.1.1.7, P.1, B.1.351, B.1.427, and B.1.429. The double and triple mutant variants first reported in India have resulted in a massive increase in the number of cases. Emerging variants not only result in increased transmissibility, morbidity and mortality, but also have the ability to evade detection by existing or currently available diagnostic tests, which can potentially delay the diagnosis and treatment, exhibit decreased susceptibility to treatment including antivirals, monoclonal antibodies and convalescent plasma, possess the ability to cause reinfection in previously infected and recovered individuals, and vaccine breakthrough cases in fully vaccinated individuals. Hence, continuation of precautionary measures, genomic surveillance and vaccination plays an important role in the prevention of spread, early identification of variants, prevention of mutations and viral replication, respectively.
Keywords: COVID-19, SARS-CoV-2, Variants, Vaccines, Mutations, Double mutant variant, Triple mutant variant, Vaccine breakthrough cases
Introduction
Natural origin and course
Viruses innately have the ability to mutate constantly and lead to variants. Some variants emerge and disappear while some persist. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a beta coronavirus that belongs to the Coronaviridae family. The family is composed of single-stranded positive ribonucleic acid (RNA) viruses [1]. Coronaviruses have four genera, and the alpha and beta genera have viruses known to cause human disease. They are zoonotic viruses that can be transmitted from animal to human; and the first time that occurs, it is referred to as a spillover event. The SARS-CoV-2 has been found to be closely related to coronaviruses found in the bat population and to the SARS-CoV [2]. Two coronaviruses found in bat populations, RaTG13 and RmYN02 were found to have 96.2% and 93.3% sequence homology, respectively with SARS-CoV-2 [3, 4]. Coronaviruses in Malayan pangolins have also been found to have sequence homology to SARS-CoV-2 [5]. The zoonotic source for SARS-CoV-2 is yet to be established. The bat and pangolin coronaviruses lack the polybasic cleavage site and mutations in the spike (S) protein, which SARS-CoV-2 possesses making the theory of human to human transmission at undetectable rates post spillover, a possibility for the virus to have acquired these genomic features prior to starting off the pandemic [6].
December 31, 2019 was the day the World Health Organisation (WHO) China Country Office was made aware of cases of pneumonia of an unknown etiology occurring in the city of Wuhan in the Hubei Province of China [7]. The virus once isolated from the airway epithelial cells of the infected patients was temporarily assigned the name 2019-nCoV [8]. Once it was determined that the virus is related to SARS-CoV, it was designated the name SARS-CoV-2 by the Coronavirus Research Group (CSG) of the International Committee for the classification of viruses on February 11, 2020 [9].
Genetics and pathogenesis
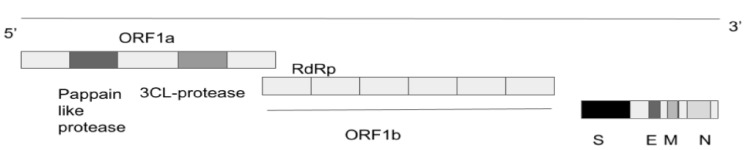
SARS-CoV-2 is an enveloped spherical-shaped virus [1]. It has four structural proteins and 16 nonstructural proteins. The structural proteins are the nucleocapsid (N) protein, the membrane (M), the S protein and the envelope (E) protein. The RNA is oriented in a 5’-3’direction which makes it a positive sense RNA virus, and the RNA can be read directly as a messenger RNA (mRNA, Fig. 1). The RNA replicase is encoded at the 5’ terminal end. The nonstructural protein 14 (nsp14) has proofreading activity which allows the rate of mutations to stay low. The S protein causes the attachment of the virus to the host cell at the angiotensin-converting enzyme 2 (ACE2) receptor, which is present on the membrane of the host cell. The ACE2 receptors are found in abundance on alveolar cells. The attachment causes fusion of the viral lipid membrane with the cell membrane of the host thus internalising the virus. The host machinery translates the viral RNA and leads to the production of the replicase and structural proteins of the virus. The replicase is cleaved into nonstructural proteins of which RNA-dependent RNA polymerase (RdRp) is one of them. Viral replication and amplification is carried out and assembly of the virions is carried out in the host cell endoplasmic reticulum and Golgi apparatus. During the process of replication, errors can occur in the genome leading to mutations which give rise to variants. The virions are finally released out of the cell by exocytosis [10, 11].
Figure 1.
SARS-CoV-2 genome. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; N: nucleocapsid; M: membrane; S: spike; E: envelope; RdRp: RNA-dependent RNA polymerase; ORF: open reading frame.
Coronavirus Disease 2019 (COVID-19) Mutations and Variants
A change in the genetic sequence is called a mutation (Table 1 [12]). Genomes that differ from each other in genetic sequence are called variants. Variants can differ from each other by one or more mutations. When a phenotypic difference is demonstrated among the variants, they are called strains [13]. In 2008 a global science initiative and primary source, Global Influenza Surveillance and Response System (GISAID) was established to provide open access to genomic data of influenza viruses. In January 2020, the SARS-CoV-2 genome sequences were made available on GISAID [14]. Nextstrain curates and analyses the genetics of SARS-CoV-2, and is a collaborative effort of researchers between Seattle, USA and Basel, Switzerland [12].
Table 1. SARS-CoV-2 Mutations [12].
| Mutation | Mutation location | Role of the mutation | Variant |
|---|---|---|---|
| S:N501 | RDB | May increase ACE2 binding. Tests in Pfizer-BioNTech and Moderna vaccinated individuals suggest reduction in neutralisation. | UK (20I/501Y.V1) |
| South Africa (20H/501Y.V2) | |||
| Brazil (20J/501Y.V3) | |||
| S:E484 | RDB | May increase ACE2 binding. | South Africa (20H/501Y.V2) |
| Brazil (20J/501Y.V3) | |||
| Brazil (20B/S.484K) | |||
| S:H69- | Spike N-terminal domain | May alter recognition by antibodies. | UK (20I/501Y.V1)/B.1.1.7 |
| S:Q677 | Near to both outside the furin building pocket; important for S1/S2 cleavage | Hypothetically thought to influence the S1/S2 cleavage. | 20G (20C-US clade) |
| S:Y453F | RDB | May increase ACE2 binding. | Cluster 5 “mink” variant seen in minks in the Netherlands |
| Confer resistance to the antibody in the Regeneron cocktail. | |||
| S:S477 | RDB | Slight increase in ACE2 binding. | |
| Confer resistance to antibody and some convalescent sera and a modest increase in infectivity. | |||
| S:L18F | Spike N-terminal domain | Reduction in binding for monoclonal antibody. | South Africa (20H/501Y.V2) |
| Brazil (20J/501Y.V3) | |||
| S:Y144- | Spike N-terminal domain | Associated with antibody escape. | UK (20I/501Y.V1), 20A/S:484K |
| S:H655 | Brazil (20J/501Y.V3) | ||
| S:P681 | Near the furin cleavage site | It may reduce antibody recognition. | UK (20I/501Y.V1) |
| S:K417 | RDB | It may escape antibody binding and decrease binding to ACE2 receptor. | South Africa (20H/501Y.V2) |
| Brazil (20J/501Y.V3) | |||
| ORF1a:S3675 | It is a three amino acids deletion in ORF1a at positions 3675-5677. | 20C/S:484K and 20A/S:484K |
RDB: receptor binding domain; ACE2: angiotensin-converting enzyme 2; ORF: open reading frame.
The interagency of the USA government has divided the SARS-CoV-2 variants into three classes. Based on the scientific evidence, the variant status can sometimes be escalated or deescalated and hence, the Center of Disease Control and Prevention (CDC) will update the variant strains in the different classes (Table 2) [15]. According to the report of April 21, 2021, the classes are: variant of interest, variant of concern, and variant of high consequence.
Table 2. Overview of SARS-CoV-2 Variants [15].
| Signs and symptoms | Original variant | Variant of interest | Variant of concern | Variant of high consequence |
|---|---|---|---|---|
| Symptoms | ||||
| Cough | 28% | 35% | ||
| Fatigue/weakness | 29% | 32% | ||
| Headache | 30% | 32% | ||
| Muscle aches | 21% | 25% | ||
| Sore throat | 19% | 22% | ||
| Fever | 20% | 22% | ||
| Loss of taste | 19% | 16% | ||
| Loss of smell | 19% | 15% | ||
| Disease transmissibility | Yes | No | Yes | Yes |
| Disease severity | Less | Less | More | More |
| Diagnostic testing failures | No | No | No | Yes |
| Treatment | Potential reduction in neutralization by monoclonal antibody treatments | B.1.1.7 variants have minimal, while P.1, B.1.351 variants have moderate and B.1.427, B.1.429 have significant impact on EUA therapeutics and monoclonal antibody treatments, respectively. | ||
| Vaccine effectiveness | Pfizer, Moderna, and Janssen are effective | Potential reduction in neutralization by post-vaccination sera | Minimal impact on neutralization by post vaccination sera for B.1.1.7 variant, while other variants have moderate reduction in neutralization by post-vaccination sera. | Significant impact on neutralization by post vaccination sera |
| Notification to WHO and CDC | No | No | Yes | Yes |
WHO: World Health Organisation; CDC: Center of Disease Control and Prevention; EUA: Emergency Use Authorization.
Variant of interest
This class includes variants with specific markers that are associated with changes to receptor binding, decreased antibody neutralization developed against previous infection or vaccination, decreased efficacy of treatments, potential diagnostic impact or predicted increase in transmissibility of disease severity. This class also requires increased sequence surveillance, increased laboratory characterization and epidemiological analysis to check the disease transmissibility and severity, the risk of reinfection and protection against vaccination. The current variants included in the variant of interest by the USA are B.1.526, B.1.525, and P.2. All these variants share a common mutation D614G; and evidence shows that the variants with this mutation spread disease faster than that of the variants without it. In late January, early February 2020 the then circulating strain was replaced by the D614G mutation carrying strain. The codon 614 replaces aspartic acid with glycine in the amino acid at this S protein site [16].
Variant of concern
This class includes variants which demonstrated high disease transmissibility, more disease severity including hospitalizations and deaths, remarkable decrease in antibody neutralization, decreased effectiveness of treatments, and diagnostic detection failure. This class also requires increased efforts to control spread by developing testing kits, increased research to ascertain the vaccine and treatments efficacy against the variant. The current variants included in the variant of concern by the USA are B.1.1.7, P.1, B.1.351, B.1.427, and B.1.429. Similar to variant of interest class, the variants in this class also share a common mutation D614G, which spreads rapidly than variants without the mutation.
Variant of high consequence
This class includes variants which have evidence that the preventive and medical measures have a significantly decreased effectiveness compared to previously circulating variants. There are no variants in this class [17].
The COVID-19 Variants
The following are the different variants identified in different countries. Of these, the three important variants which have rapidly become dominant are B.1.1.7, B.1.351, and P.1.
UK variant (B.1.1.7 or 20I/501Y.V1)
B.1.1.7, also known as VOC202012/01 was detected in September, 2020 in the UK. It has 23 mutations compared to the original strain found in Wuhan, China. Eight of these mutations were found to be in the S protein. Its notable mutations are N501Y, 69/70 deletion and P681H. The N501Y mutation appears to allow for the S protein to bind more tightly to the ACE2 receptor [18].
It is 40-80% more transmissible [19]. According to the report by Davies et al, nearly 5,000 out of 17,452 COVID-19 deaths during the months September to February were due to this variant. They also estimated that the mortality was approximately 55% higher when compared to other variants [20]. Similarly, the scientific reports in January 2021 indicated that there was an increased rate of death with this variant [21]. As of April 1, 2021, there were 12,505 reported cases across 51 jurisdictions [22]. It has been identified in 82 countries [23]. There are studies going on to determine the effectiveness of antivirals and anti-inflammatory medication in the treatment of UK strain [24].
South African variant (B.1.351or 20H/501Y.V2)
The B.1.351 variant, also known as 501Y.V2 was first identified as early as October 2020 in Nelson Mandela Bay in South Africa. By December 2020 it was also detected in Zambia. By April 1 2021, there were 323 reported cases in 31 jurisdictions in the USA. There are 23 mutations with 17 amino acid changes but the notable mutations in this variant are K417N, E484K, and N501Y on the S protein. It is suggested to have increased transmissibility and most commonly seen in young people without underlying diseases [22].
The mutation E484K in this variant mediates the antibody escape is the main reason for the reduced sensitivity to vaccines [25]. The mRNA vaccines (Pfizer and Moderna) were authorized in the USA before the identification of this strain in the country [26]. According to the latest studies, these two vaccines elicited lower neutralizing antibodies than that of the previous strains. Novavax, Janssen, and Astra-Zeneca conducted trials in South Africa that have dominant B.1.351 mutated strains. These studies demonstrated the lower vaccine efficacy compared to that of the other variants where this strain was not dominant [27].
Brazilian variant (P.1 or 20J/501Y.V3)
P.1 variant also known as B.1.1.28.1 was first detected in North Brazil in the city of Manaus in the Amazonas state in December 2020. By January 2021, it was identified just outside Tokyo at Haneda airport among four travelers from Brazil during routine screening of passengers in samples analysed at National Institute of Infectious Diseases (NIID) in Japan. As of April 1, 2021, there were 224 reported cases in 22 jurisdictions in the USA [22]. It has 35 mutations with 17 amino acid changes. Its notable mutations are K417T, E484K, and N501Y [22]. In a study by Naveca et al, it was found that this variant is 2.2 times higher transmissible that led to a few cases of reinfection who recovered from COVID-19, and almost has a similar rate infection in the younger (18 - 59 years old) and older (> 60 years old) patients [28, 29]. B.1.351 and P.1 consist of similar receptor binding mutations and hence, the vaccine efficacy against P.1 strain is assumed to be similar to B.1.351. As the studies demonstrated reduced vaccine efficacy against B.1.351, it is likely that the efficacy against P. 1 strain is reduced [30]. Sinovac Biotech has initiated the clinical trials which demonstrated that the CoronoVac vaccine is 50% effective in preventing infection with the P.1 variant in Brazil [31].
Brazilian variant (B.1.1.28)
The Brazilian variant B.1.1.28 was initially found in the state of Rio de Janeiro in Brazil, and is the P2 lineage. Unlike P.1 variant, B.1.1.28 variant has only one notable E484K mutation in the S protein [32]. The therapeutic and vaccine efficacy has not been reported yet. Similar to B.1.351 and P.1 variants, the vaccine might be ineffective against this variant [33].
US Midwest variant (20C-US or COH.20G/501Y)
It was detected in Ohio followed by other Midwest states in December 2020 and January 2021. This variant has mutations on the S protein (Q677H), M protein (A85S) and on the N protein (D377Y). Another variant with the mutation S N501Y, a marker of the B.1.1.7, with no other associated mutations with that strain has been identified [34]. Currently there is not any evidence of increased transmissibility or virulence for this variant.
US San Francisco Bay Area variant (B.1.427 and B.1.429)
These variants were first identified in California in February, 2021. The notable mutations in B.1.427 are L452R and D614G; while mutations in B.1.429 are S131, W152C, L452R and D614G. Both variants have 20% increased risk of transmissibility and reduction in therapeutic efficacy [15]. It was identified last year across the USA and Europe. It is a variant with a mutation in the S protein. In January 2021 it rose rapidly as the cause of cases across several counties in California [35].
US Southern California variant (CAL.20C)
It was first seen in July 2020 in Southern California and detected again amongst population samples of the same region in October 2020. Its notable mutations are ORF1a: I4205V, ORF1b: D1183Y, S: S13I; W152C and L452R. The binding of the S protein could be made easier by the latter three mutations [36].
B.1.526 (20C/S:484K) and B.1.525 (20A/S:484K)
These variants were first identified in New York, USA. The notable mutations are E484K and S477N. While E484K decreases antibody response, S477N increases the attachment process [34].
Double mutant variant (B.1.617)
This variant is first detected in India. As two mutations are seen in the same virus, this variant is called a “double mutant” variant. There was a significant increase in COVID-19 cases in India. The first case in the USA was identified in San Francisco on April 5, 2021. The notable mutations are E484Q and L452R. These variants are at increased risk of transmission and also resistant to vaccination. According to Indian Council of Medical Research Virology Lab, Bharat Biotech’s COVAXIN vaccine has been found to effectively neutralize the infection, and is 78% effective against the double mutant variant [37-39].
Triple mutant variant (B.1.618)
In addition to E484Q and L425R in double mutant variants, the new triple variant discovered on April 20 2021, is characterized by the deletion of two amino acids, H146del and Y145del in the S protein. As of April 21 2021, a total of 1,189 samples were tested positive in Maharashtra, Delhi, West Bengal and Chhattisgarh, India. Similar to other variants, triple mutant variants have higher transmissibility. Data show that two of the three mutations in this variant are resistant to antibodies and also possess the ability to escape the body’s natural acquired immunity to COVID-19, and as such, do not know much about the vaccine effectiveness [40, 41].
20A.EU1/ S:A222V
The 20A.EU1 variant has non-terminal domain (NTD) mutations which do not play a direct role in receptor binding or membrane fusion. This variant was initially identified on 20 June, 2020 in Spain but rapidly spread across Europe and many countries [42].
20A.EU2
The 20A.EU2 variant was found in France in June 2020 and has become the second dominant variant in Europe. The notable mutations are S477N, E484K, and N501Y, which demonstrated slight increase in ACE2 binding, resistance to multiple antibodies and convalescent sera. They confer modest increase in infectivity as measured by soluble mACE2 [43].
20A/S:439K
The 20A/S:439K variant was initially found in Ireland. This variant has S:N439K mutation with the deletions of amino acids at positions 69 and 70 of S proteins that results in an increase in ACE2 binding, resistance to antibodies and convalescent plasma [12].
20A/S:98F
The 20A/S:98F variant has S:98F mutation which was found predominantly in Belgium and Netherlands [12].
20C/S:80Y
The 20C/S:80Y variant had 18 nucleotide mutations, possibly related to apolipoprotein B editing complex (APOBEC)-like editing within the host which are found in at least 10 countries in Europe [12].
20B/S:626S
The 20B/S:626S variant has S:626S mutation. This variant is found in 15 countries of Europe that is predominantly seen in Norway, Denmark, and the UK [12].
20B/S:1122L
The 20B/S:1122L variant has S:V1122L mutation and is found predominantly in Sweden, Norway, and Denmark [12].
N440K
According to the latest report, another new variant N440K with the mutation in the S protein has emerged, which resulted in the sudden increase in cases in Andhra Pradesh, India. The Center for Cellular and Molecular Biology found that this variant has enhanced binding to ACE2 receptors, 10 to 1,000 folds more transmissible and resistant to class 3 monoclonal antibodies C135 and REGN10987. There are several documented cases of reinfection with the presence of anti-SARS-CoV-2 antibodies indicating the possibility of loss of neutralizing activity of antibodies elicited by vaccines [44, 45].
COVID-19 Vaccines Versus the Variants
At the time of this paper, there were 91 COVID-19 vaccines in the clinical development stage and 184 COVID-19 vaccines in pre-clinical developmental stage [46]. Several platforms have been utilized in the development of these vaccines which include protein subunit, viral vector (replicating and non-replicating), deoxyribonucleic acid (DNA), inactivated virus, RNA, virus like particle and live attenuated. As of now, the Food and Drug Administration (FDA) has approved three vaccinations for emergency use in the USA, and data suggest that the mRNA vaccines reasonably provide protection against the B.1.1.7 COVID-19 variant. The mRNA vaccines, Pfizer and Moderna were authorized in the USA before the identification of the South African variant (B.1.351 or 20H/501Y.V2) strain in the country. According to the latest studies, these two vaccines elicited lower neutralizing antibodies than that of the previous strains. Novavax, Janssen, and Astra-Zeneca conducted trials in South Africa that have dominant B.1.351 mutated strains. These studies demonstrated the lower vaccine efficacy compared to that of the other variants where this strain was not dominant [47]. Vaccine efficacy for Brazilian variant (B.1.1.28) has not yet been reported.
B.1.351 and P.1 consist of similar receptor binding mutations and hence, the vaccine efficacy against P.1 strain is assumed to be similar to B.1.351. As the studies demonstrated reduced vaccine efficacy against B.1.351, it is likely that the efficacy against P. 1 strain is reduced [47]. Sinovac Biotech has initiated the clinical trials which demonstrated that the CoronoVac vaccine is 50% effective in preventing infection with the P.1 variant in Brazil [47]. B.1.526 and B.1.525 have a reduction in vaccine efficacy [15]. Further studies are required to study the efficacy of the current approved vaccines against the double mutant variant [40, 47]. According to Indian Council of Medical Research Virology Lab, Bharat Biotech’s COVAXIN vaccine has been found to effectively neutralize the infection, and is 78% effective against the double mutant variant [39-41].
COVID-19 Vaccine Challenges
Though COVID-19 vaccines are an effective critical tool, none of them are a 100% effective in the prevention of COVID-19 illness. A small percentage of the fully immunized population will still get sick to varying degrees from COVID-19 disease. They are known as vaccine breakthrough cases. A person who has SARS-CoV-2 RNA or antigen detected on a respiratory specimen collected on or after 14 days after completing the primary series of a US FDA-authorized COVID-19 vaccine fits the definition of a vaccine breakthrough case. More than 75 million people have been fully vaccinated as of April 13, 2021 since December 14, 2020. During the same duration, there have been 5,814 vaccine breakthrough cases reported across the USA. Forty-five percent of the total cases were female, 29% were asymptomatic, 45% were over 60 years of age, 7% of the total cases were hospitalized, and 1% died [48]. There have been several hurdles in general to vaccine administration. Pregnant and breastfeeding women, immunocompromised, diverse race and ethnicities have been underrepresented populations in vaccines studies. Vaccine refusal has been an issue physicians have had to deal with even in pre-pandemic times [49]. There has been growing COVID-19 vaccine hesitancy [50]. Equitable vaccine distribution, storage requirements and cost are factors that will have a significant impact in areas across the world.
Conclusions
The emerging variants not only result in increased transmissibility, morbidity and mortality but also have the ability to evade detection by diagnostic tests, exhibit decreased susceptibility to treatment including antivirals, monoclonal antibodies and convalescent plasma, possess the ability to cause reinfection in previously recovered and vaccinated individuals. Children and immunocompromised individuals are at increased risk of developing multisystem inflammatory syndrome or long-haul COVID [51, 52]. The longer the virus propagates the chance of mutations are higher. Vaccine breakthrough cases are often undercounted and fully vaccinated populations should still practice preventative measures. At the time of this paper 40.7% of the total population in the USA was fully vaccinated. The USA ranks eighth in terms of percentage of population vaccinated. Israel ranks first with 57% of the total population vaccinated followed by Bahrain and Aruba [53, 54]. Continuation of precautionary measures including masking, hand hygiene, isolation and quarantine along with other public health mitigation strategies, genomic surveillance and vaccination plays an important role in prevention of spread; early identification of variants, prevention of mutations and viral replication, respectively, and still form the cornerstone of the action plan to control the COVID-19 pandemic [55].
Acknowledgments
None to declare.
Financial Disclosure
The authors declare that they do not have a financial relationship with any commercial entity that has an interest in the subject of this manuscript. No grants were obtained for this study.
Conflict of Interest
The authors declare that they do not have a conflict of interest.
Author Contributions
DV, RV, GM, SVM and PA performed literature search. DV and RV drafted the manuscript. DV, RV, GM, SVM and PA did critical editing. DV and RV contributed to the illustrations. DV and RV carefully supervised this manuscript preparation and writing.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
Abbreviations
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- RNA
ribonucleic acid
- WHO
World Health Organisation
- CSG
Coronavirus Research Group
- N
nucleocapsid
- M
membrane
- S
spike
- E
envelope
- nsp14
nonstructural protein 14
- ACE2
angiotensin-converting enzyme 2
- RdRp
RNA-dependent RNA polymerase
- GISAID
Global Influenza Surveillance and Response System
- NIID
National Institute of Infectious Diseases
- CDC
Center of Disease Control and Prevention
- NTD
non-terminal domain
- mRNA
messenger RNA
- DNA
deoxyribonucleic acid
References
- 1.Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11(1):5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Emma B. Hodcroft. 2021. “CoVariants: SARS-CoV-2 Mutations and Variants of Interest.” https://covariants.org/
- 3.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 5.Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG. et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583(7815):282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 6.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pneumonia of unknown cause - China. World Health Organization. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/. Published January 30, 2020. Accessed February 9, 2021.
- 8. WHO. Novel coronavirus (2019-nCoV) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed Jan 20, 2020.
- 9.Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA. et al. Severe acute respiratory syndrome-related coronavirus: The species and its viruses - a statement of the Coronavirus Study Group. bioRxiv. doi: 10.1101/2020.02.07.937862. https://www.biorxiv.org/content/10.1101/2020.02.07.937862v1. Published January 1, 2020. Accessed February 9, 2021. [DOI]
- 10.Atri D, Siddiqi HK, Lang JP, Nauffal V, Morrow DA, Bohula EA. COVID-19 for the Cardiologist: Basic Virology, Epidemiology, Cardiac Manifestations, and Potential Therapeutic Strategies. JACC Basic Transl Sci. 2020;5(5):518–536. doi: 10.1016/j.jacbts.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. StatPearls. Treasure Island (FL): StatPearls Publishing; Features, evaluation, and treatment of coronavirus. October 4, 2020. [PubMed] [Google Scholar]
- 12. Emma B. Hodcroft. 2021. “CoVariants: SARS-CoV-2 Mutations and Variants of Interest.” https://covariants.org/
- 13.Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2 - what do they mean? JAMA. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 14. SARS-CoV-2 Variants. World Health Organization. https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/. Published January 22, 2021. Accessed May 3, 2021.
- 15. SARS-CoV-2 Variants of Concern. Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html. Accessed May 3, 2021.
- 16. Europe PMC. https://europepmc.org/article/PMC/PMC7839510. Accessed April 22, 2021.
- 17. Virginia Department of Health. Go to Virginia Department of Health. https://www.vdh.virginia.gov/coronavirus/variants/. Accessed June 10, 2021.
- 18. How the flu virus can change: "Drift" and "Shift". Centers for Disease Control and Prevention. https://www.cdc.gov/flu/about/viruses/change.htm. Published October 15, 2019. Accessed April 23, 2021.
- 19.Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A. et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. Centers for Disease Control and Prevention. MMWR Morb Mortal Wkly Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies NG, Jarvis CI. CMMID COVID-19 Working Group. Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(7858):270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. NERVTAG - GOV.UK. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/961037/NERVTAG_note_on_B.1.1.7_severity_for_SAGE_77__1_.pdf. Accessed April 24, 2021.
- 22. US COVID-19 Cases Caused by Variants. Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/transmission/variant-cases.html. Accessed April 24, 2021.
- 23. Aine.otoole. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Virological. https://virological.org/t/tracking-the-international-spread-of-sars-cov-2-lineages-b-1-1-7-and-b-1-351-501y-v2/592. Published February 4, 2021. Accessed April 24, 2021.
- 24. kevin_stank. Gilead CEO expects remdesivir to effectively treat Covid strains found in UK, South Africa. CNBC. https://www.cnbc.com/2021/01/11/remdesivir-likely-works-against-covid-strains-found-in-uk-south-africa-gilead-ceo.html. Published January 11, 2021. Accessed April 24, 2021.
- 25.Planas D, Bruel T, Grzelak L, Guivel-Benhassine F, Staropoli I, Porrot F, Planchais C. et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 26.Vasireddy D, Atluri P, Malayala SV, Vanaparthy R, Mohan G. Review of COVID-19 Vaccines Approved in the United States of America for Emergency Use. J Clin Med Res. 2021;13(4):204–213. doi: 10.14740/jocmr4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rita Rubin MA. Figuring Out Whether COVID-19 Vaccines Protect Against Variants. JAMA. https://jamanetwork.com/journals/jama/fullarticle/2777785. Published April 6, 2021. Accessed April 24, 2021. [Google Scholar]
- 28. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new Variant of Concern P.1. Home. https://www.researchsquare.com/article/rs-275494/v1. Published February 25, 2021. Accessed April 24, 2021.
- 29. Robert Glatter MD. P.1 Variant, dominant strain in Brazil, reported in New York. Forbes. https://www.forbes.com/sites/robertglatter/2021/03/21/p1-variant-dominant-strain-in-brazil-reported-in-new-york/?sh=19c9a91e1883. Published March 23, 2021. Accessed April 24, 2021.
- 30. Existing vaccines may protect against the Brazilian coronavirus variant. University of Oxford. https://www.ox.ac.uk/news/2021-03-18-existing-vaccines-may-protect-against-brazilian-coronavirus-variant. Accessed April 24, 2021.
- 31. COVID-19 vaccine effective against new variant in Brazil. Articles - COVID19 vaccine effective against new variant in Brazil - Emerging Pathogens Institute - University of Florida. https://epi.ufl.edu/articles/covid19-vaccine-effective-against-new-variant-in-brazil.html. Accessed April 24, 2021.
- 32. Michieli AG. #Genomic characterization of a novel #SARS-CoV-2 #lineage from #Rio de Janeiro, #Brazil (J Virol., abstract). ETIDIoH. https://etidioh.wordpress.com/2021/03/02/genomic-characterization-of-a-novel-sars-cov-2-lineage-from-rio-de-janeiro-brazil-j-virol-abstract/. Published March 2, 2021. Accessed April 24, 2021.
- 33. Update on SARS-CoV-2 Variants. GVN. https://gvn.org/update-on-sars-cov-2-variants-020521/. Published February 5, 2021. Accessed April 24, 2021.
- 34. Coronavirus mutations and variants: what does it mean? SRHD. https://srhd.org/news/2021/coronavirus-mutations-and-variants-what-does-it-mean. Published April 1, 2021. Accessed April 24, 2021.
- 35. Callaway E. Multitude of coronavirus variants found in the US - but the threat is unclear. Nature News. https://www.nature.com/articles/d41586-021-00564-4. Published March 5, 2021. Accessed May 3, 2021.
- 36.Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Coronavirus: ‘Double mutant’ Covid variant found in India. BBC News. https://www.bbc.com/news/world-asia-india-56507988. Published March 25, 2021. Accessed June 10, 2021.
- 38. Choi J. First case of India's 'double mutant' COVID-19 variant found in San Francisco Bay Area. TheHill. https://thehill.com/homenews/state-watch/546491-first-case-of-indias-double-mutant-covid-19-variant-found-in-san. Published April 5, 2021. Accessed April 24, 2021.
- 39. Service TN. COVAXIN works against double mutant; reduces hospitalisation, shows Phase 3 interim data. Tribuneindia News Service. https://www.tribuneindia.com/news/nation/covaxin-works-against-double-mutant-shows-78-100-efficacy-against-severe-covid-phase-3-interim-data-242191. Accessed April 24, 2021.
- 40. Desk W. 'Triple mutant' coronavirus variant discovered in India? What it means. The Week. https://www.theweek.in/news/india/2021/04/21/triple-mutant-coronavirus-variant-discovered-india-what-it-means.html. Published April 21, 2021. Accessed April 24, 2021.
- 41. CNBCTV18.com share, CNBCTV18.com. COVID-19: Triple mutation strain of virus raises concern, says report. cnbctv18.com. https://www.cnbctv18.com/healthcare/covid-19-triple-mutation-strain-of-virus-raises-concern-says-report-9008191.htm. Accessed April 24, 2021.
- 42. medRxiv. https://www.medrxiv.org/content/medrxiv/early/2020/09/03/2020.09.01.20186304.full.pdf. Accessed April 24, 2021. https://doi.org/10.1101/2020.09.01.20186304.
- 43.Liu Z, VanBlargan LA, Bloyet LM. et al. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Preprint. bioRxiv. 2020:2020.11.06.372037. doi: 10.1101/2020.11.06.372037. Published Nov 8, 2020. [DOI] [Google Scholar]
- 44.Rani PR, Imran M, Lakshmi JV. et al. Symptomatic reinfection of SARS-CoV-2 with spike protein variant N440K associated with immune escape [published online ahead of print, 2021 Apr 5] J Med Virol. 2021 doi: 10.1002/jmv.26997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv. bioRxiv COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv. https://connect.biorxiv.org/relate/content/181.?page=1496. Accessed May 3, 2021.
- 46. Draft landscape and tracker of COVID-19 candidate vaccines. World Health Organization. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. Accessed May 3, 2021.
- 47. Rita Rubin MA. Figuring Out Whether COVID-19 Vaccines Protect Against Variants. JAMA. https://jamanetwork.com/journals/jama/fullarticle/2777785. Published April 6, 2021. Accessed May 3, 2021.
- 48. COVID-19 Breakthrough Case Investigations and Reporting. Centers for Disease Control and Prevention. https://www.cdc.gov/vaccines/covid-19/health-departments/breakthrough-cases.html. Published April 30, 2021. Accessed May 3, 2021.
- 49.Vasireddy D, Yusi D, Berrak SG, Lichtenberger J. Factors affecting Refusal rates of the Birth dose of Hepatitis B vaccine: A single Center study. Journal of Pediatric Infection. 2014;8(4):159–164. doi: 10.5152/ced.2014.1920. [DOI] [Google Scholar]
- 50.Guidry JPD, Laestadius LI, Vraga EK, Miller CA, Perrin PB, Burton CW, Ryan M. et al. Willingness to get the COVID-19 vaccine with and without emergency use authorization. Am J Infect Control. 2021;49(2):137–142. doi: 10.1016/j.ajic.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Science Brief: Emerging SARS-CoV-2 Variants. Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html. Accessed May 3, 2021.
- 52. Contributor IDSA, 3 KLMDF, 3 JAF. COVID "Mega-variant" and eight criteria for a template to assess all variants. Science Speaks: Global ID News. https://sciencespeaksblog.org/2021/02/02/covid-mega-variant-and-eight-criteria-for-a-template-to-assess-all-variants/. Published February 21, 2021. Accessed May 3, 2021.
- 53. CDC COVID Data Tracker. Centers for Disease Control and Prevention. https://covid.cdc.gov/covid-data-tracker/. Accessed June 1, 2021.
- 54. Ritchie Rand data: H. Coronavirus (COVID-19) Vaccinations - Statistics and Research. Our World in Data. https://ourworldindata.org/covid-vaccinations. Accessed June 1, 2021.
- 55. T. T. Shimabukuro and Others, Others EHand, F. P. Polack and Others. New SARS-CoV-2 Variants - Clinical, Public Health, and Vaccine Implications: NEJM. New England Journal of Medicine. https://www.nejm.org/doi/full/10.1056/NEJMc2100362. Published April 21, 2021. Accessed May 3, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the article.