Abstract
Microbiota plays a prominent role in periodontal disease, but the canine oral microbiota and how dental chews may affect these populations have been poorly studied. We aimed to determine the differences in oral microbiota of adult dogs consuming dental chews compared with control dogs consuming only a diet. Twelve adult female beagle dogs (mean age = 5.31 ± 1.08 yr) were used in a replicated 4 × 4 Latin square design consisting of 28-d periods. Treatments (n = 12/group) included: diet only (CT); diet + Bones & Chews Dental Treats (BC; Chewy, Inc., Dania Beach, FL); diet + Dr. Lyon’s Grain-Free Dental Treats (DL; Dr. Lyon’s, LLC, Dania Beach, FL); and diet + Greenies Dental Treats (GR; Mars Petcare US, Franklin, TN). Each day, one chew was provided 4 h after mealtime. On day 27, breath samples were analyzed for total volatile sulfur compound concentrations using a Halimeter. On day 0 of each period, teeth were cleaned by a veterinary dentist blinded to treatments. Teeth were scored for plaque, calculus, and gingivitis by the same veterinary dentist on day 28 of each period. After scoring, salivary (SAL), subgingival (SUB), and supragingival (SUP) samples were collected for microbiota analysis using Illumina MiSeq. All data were analyzed using SAS (version 9.4) using the Mixed Models procedure, with P < 0.05 considered significant. All dogs consuming chews had lower calculus coverage and thickness, pocket depth and bleeding, plaque thickness, and halitosis compared with CT. In all sites of collection, CT dogs had a higher relative abundance of one or more potentially pathogenic bacteria (Porphyromonas, Anaerovorax, Desulfomicrobium, Tannerella, and Treponema) and lower relative abundance of one or more genera associated with oral health (Neisseria, Corynebacterium, Capnocytophaga, Actinomyces, Lautropia, Bergeyella, and Moraxella) than those fed chews. DL reduced Porphyromonas in SUP and SUB samples. DL and GR reduced Treponema in SUP samples. DL increased Corynebacterium in all sites of collection. BC increased Corynebacterium in SAL samples. DL and GR increased Neisseria in SAL samples. DL increased Actinomyces in the SUB sample. GR increased Actinomyces in SAL samples. Our results suggest that the dental chews tested in this study may aid in reducing periodontal disease risk in dogs by beneficially shifting the microbiota inhabiting plaque and saliva of a dog’s oral cavity. These shifts occurred over a short period of time and were correlated with improved oral health scores.
Keywords: canine health, next-generation sequencing, oral microbiome, periodontal disease
Introduction
The development of periodontal disease is caused by bacterial plaque accumulation on the periodontium (Dewhirst et al., 2012; Davis et al., 2013; Deng and Swanson, 2015) and subsequent changes in the oral microbiota, which plays a vital role in the pathogenesis of the disease (Hennet and Harvey, 1991; Mandel et al., 1993; Tatakis and Kumar, 2005; Gomes et al., 2015). It is the most common disease in dogs, with 44% to 64% of dogs being affected by periodontitis to some degree (Gad, 1968; Hamp et al., 1984; Hoffmann and Gaengler, 1996; Lund et al., 1999; Klein, 2000; Butković et al., 2001; Kyllar and Witter, 2005; Kortegaard et al., 2008; O′Neill et al., 2014). After scaling and polishing a tooth, a glycoprotein layer (pellicle) begins to form within a few seconds of exposure to saliva, with plaque-forming bacteria adhering to the pellicle and allowing proliferation. Within 24 h, a thin layer of plaque covers the entire tooth, except in areas where it has been removed by natural dietary abrasion. Bacterial proliferation creates a rough surface to which more bacteria may adhere. As the plaque thickens and extends into the subgingival sulcus, oxygen is depleted and anaerobic bacteria proliferate. Within 48 h of plaque deposition, calculus formation may occur. Calcium present in saliva is deposited into plaque, creating calculus, which adds to the rough surface favoring the accumulation of more plaque and irritation of gingival tissues. The persistence of supragingival (SUP) plaque and subgingival (SUB) plaque, and their products, around the teeth, can cause inflammation in adjacent gingival tissues (gingivitis) and induce host immune responses (Fedi et al., 1985; Lindhe, 1989; Genco, 1990; Watson, 1994; Logan, 2006).
Because of its role in periodontitis, it is important to continually remove plaque from teeth. This may be accomplished by mechanical removal (brushing, scaling, and dental chews) and/or by using chemical additives (chlorhexidine gluconate/polyphosphates/immunoglobulin Y) for the prevention and control of periodontal disease (Gorrel and Bierer, 1999; Gorrel et al., 1999; Brown and McGenity, 2005; Hennet et al., 2006b; Clarke et al., 2011; Shofiqur et al., 2011; Albuquerque et al., 2012; Quest, 2013; Harvey et al., 2015; Lacerda and Alessi, 2015; Adepu et al., 2018; Oba et al., 2018; Stella et al., 2018). Dental chews have been proven to be an easy and highly accepted method for the removal of SUP plaque accumulation, with chews having varied shapes and sizes with or without additives being sold commercially (Gorrel and Rawlings, 1996; Rawlings et al., 1998; Gorrel and Bierer, 1999; Gorrel et al., 1999; Brown and McGenity, 2005; Hennet et al., 2006b; Clarke et al., 2011; Quest, 2013; Garanayak et al., 2019; Carroll et al., 2020; Ruparell et al., 2020).
It is through disturbances to the equilibrium of the oral microbiota community that periodontal disease is believed to be initiated. Therefore, understanding changes in the oral microbiota community, and the effects that the consumption of dental chews may have, is of interest. In a previous study, Ruparell et al. (2020) demonstrated that the daily feeding of an oral care chew to dogs increased the proportion of health-associated bacteria vs. bacteria associated with periodontal disease in SUP plaque compared with those fed no chew. Gram-negative bacterial species (Bergeyella, Moraxella, and Porphyromonas) are believed to be predominant in healthy dogs, whereas Gram-positive anaerobic species (Actinomyces, Peptostreptococcus, and Peptostreptococcaceae) predominate in diseased animals (Davis et al., 2013). With the progression from healthy gingiva to mild periodontitis, there is a reduction in the abundance of particular Gram-negative species, namely Bergeyella zoohelcum COT-186, Moraxella sp. COT-017, Pasteurellaceae sp. COT-080, and Neisseria shayeganii COT-090 (Wallis et al., 2015).
Previous studies have demonstrated that the information generated by bacteriological analysis of the oral cavity is highly dependent on the location, or habitat, in the mouth as well as sampling technique (Loomer, 2004; Rober et al., 2008; Segata et al., 2012). Additionally, although a previous study evaluated the effects of a dental chew on the oral microbiota of dogs, they only collected and evaluated SUP plaque samples, tested only one dental chew, and did not evaluate oral health outcomes (Ruparell et al., 2020). The current study aimed to determine the differences in three oral microbiota habitats (salivary [SAL], SUP plaque, and SUB plaque) and health outcomes of dogs consuming three commercially available dental chews. We hypothesized that dental chew consumption would reduce the relative abundance of pathogenic bacterial taxa and increase the relative abundance of bacterial taxa associated with oral health. To our knowledge, this is the first study to evaluate the effects of commercial dental chews on the SAL, SUP, and SUB plaque of dogs.
Materials and Methods
Animals
Twelve adult female beagle dogs (mean age = 5.31 ± 1.08 yr; mean body weight [BW] = 13.12 ± 1.39 kg) were used in a replicated 4 × 4 Latin square design. All procedures were approved by the University of Illinois Institutional Animal Care and Use Committee prior to experimentation (IACUC #18067). Dogs were housed individually in pens (1.0 m wide by 1.8 m long) in a humidity- and temperature-controlled animal facility. Dogs had access to fresh water at all times and were fed once daily to maintain BW. When allotted to dental chew treatments, food intake was adjusted to compensate for the energy provided by the chews. Dogs were weighed once weekly prior to feeding, with a BW of all dogs remaining constant throughout the study. Dogs were fed at 0800 hours each morning and were given 1 h to consume their food. Leftover food was weighed each day to calculate intake. Four hours after eating their diet, dogs receiving a dental chew were given the chews. Prior to the start of the study, a dental evaluation was performed by a veterinary dentist to confirm the presence and integrity of all teeth to be scored to confirm trial eligibility. The experiment consisted of four 28-d periods. On day 0 of each period, dogs had teeth cleaned and polished by a veterinary dentist. The same blinded veterinary dentist scored teeth on day 28 of each period, then ultrasonically cleaned and polished the teeth so that day 28 of each period served as day 0 for the subsequent period. Breath samples were measured for malodor on day 27 of each period.
All dogs were fed a commercial diet (American Journey Salmon & Sweet Potato Recipe, American Journey, LLC, Dania Beach, FL) throughout the study. No additional treats, chew toys or other dental interventions were permitted for the duration of the study. No active anti-plaque or calculus substances were included in chew formulations. Dogs were allotted to one of four treatments in each experimental period:
Diet only (control, CT)
Diet + Bones & Chews Dental Treats, Chewy, Inc., Dania Beach, FL (BC)
Diet + Dr. Lyon’s Grain-Free Dental Treats, Dr. Lyon’s, LLC, Dania Beach, FL (DL)
Diet + Greenies Dental Treats, Mars Petcare US, Franklin, TN (GR)
Diet and dental treat ingredients, analyzed chemical composition, and additional information are provided in our previous publication (Carroll et al., 2020).
Anesthesia methods
All dogs had their food withheld for at least 12 h prior to anesthesia but were allowed water until transportation. The hair over the left or right cephalic vein was clipped, the site was aseptically prepared, and a 20-gauge intravenous catheter was placed in the cephalic vein for the administration of sedative and anesthetic agents and intravenous fluids. Following catheterization, butorphanol (0.3 mg/kg) was administered intravenously and dogs were pre-oxygenated. Anesthesia was then induced with etomidate with or without a co-induction of midazolam (0.3 mg kg−1) or lidocaine (2 mg kg−1). Dogs were orotracheally intubated and transferred to isoflurane delivered in oxygen to maintain anesthesia. Intravenous fluids (Lactated ringer’s solution) were run at 5 mL/kg/h throughout anesthesia and active heating with a forced-air warmer was provided to maintain normothermia. Cardiovascular and respiratory function was monitored continuously using an anesthetic multiparameter monitor. Supplementary anesthetic agents and cardiovascular support were administered as needed based on the decision of the attending veterinary anesthesiologist.
Salivary pH
SAL pH was measured using pH strips (Fisherbrand Plastic pH Strips; pH range: 0 to 14), on the same day and time of dental scoring, and all dogs had their food withheld for at least 12 h prior the salivary pH measurements, using two strips on each side per dog (4 total). The SAL pH reported was the mean of the four strips. SAL samples were collected where it naturally pools (in the cheek pouch and under the tongue) for 30 s.
Halitosis measurement
On day 27, breath samples were analyzed for total volatile sulfur compound concentrations using a Halimeter (Interscan Corp, Simi Valley, CA). Halimeter measurements were conducted 3 h after dental chew administration. Halitosis measurements were obtained for each dog using a clean plastic straw (clean straw was used for each measurement) as an extension of the Halimeter air drawing hose. The highest reading of volatile sulfur compounds over a period of approximately 30 s was displayed by the Halimeter and recorded. The machine was allowed to return to 0 (about 60 to 120 s) before the next measurement was taken. Each dog was measured three times and a mean score was calculated.
Dental scoring
On day 28 of each period, gingivitis, plaque, and calculus scoring were conducted by a board-certified veterinary dentist according to a modified version of previous scoring systems (Mühlemann and Son, 1971; Gorrel et al., 1999). For each measurement, the fourth premolar and first molar teeth on the upper (maxilla) and lower (mandible) jaw were scored. These two teeth were chosen because they are the teeth from which plaque samples for microbiota analysis were collected. Thus, the dental score data would be more compatible with the sample collection site.
To assess gingivitis, after an initial visual evaluation of the gingiva, a periodontal probe (Williams model, Cislak Manufacturing, Inc., Niles, IL) was placed subgingivally on the buccal side of each tooth, and values were assigned via visual assessment of inflammation and bleeding, if present, upon probing. Each tooth was graded by the average of the three scores obtained per tooth. The score for each dog is the mean score for all teeth scored. Plaque levels were evaluated by using Trace Disclosing Solution (Young Dental, Earth City, MO) to cover the teeth, followed by a gentle rinse of water to remove the excess. Plaque was hence revealed and subsequently scored for coverage and thickness according to Gorrel et al. (1999) using the anatomical landmarks described in Hennet et al. (2006a) to divide the teeth into gingival and occlusal portions. The gingival and occlusal values for each tooth were averaged to obtain a tooth total score. The average plaque coverage was multiplied by the average of plaque thickness to obtain a whole mouth mean calculus score for each animal. Calculus scores were based on the visual assessment of coverage and thickness on the mesial, buccal, and distal portions of the tooth. The tooth score is the average of the scores for each of the three tooth surfaces. The average of calculus coverage was multiplied by the average of calculus thickness to obtain a whole mouth mean calculus score for each animal.
Pocket depth was based on height from the bottom of pocket to gingival margin: <2 mm = normal sulcus; >2 and <3 mm = slight; >3 and <5 mm = moderate; and >5 mm = severe. Bleeding on probing was measured based on visual assessment of bleeding after insertion of a probe into the base of the sulcus or pocket (0 = normal-appearing gingiva and no bleeding upon probing; 1 = no color or contour changes, but bleeding upon probing; 2 = bleeding on probing, color change (reddening), and no edema; 3 = bleeding on probing, color change, and mild inflammatory edema; 4 = bleeding on probing, color change, and severe inflammatory edema; and 5 = spontaneous bleeding on probing, color change, and very severe inflammatory edema with or without ulceration). The tooth score is the average of pocket depth and bleeding on probing for each tooth. The average of pocket depth was multiplied by bleeding on probing to obtain a whole mouth mean pocket score for each animal. The sum of gingivitis score, plaque score, calculus score, and pocket score was used to calculate the oral health score (OHS).
Saliva and plaque sample collection
Once scored, plaque (SUP and SUB plaque) and SAL samples were collected for microbiota analysis and the teeth surfaces were cleaned. SAL samples were collected using two swabs (P-151; DNA Genotek, Ottawa, ON, Canada) per dog according to the manufacturer’s guidelines. SAL samples were collected where it naturally pools (in the cheek pouch and under the tongue) for 30 s. Swabs were placed into the manufacturer’s tube and shaken vigorously 10 times to thoroughly mix samples. Samples remained in the collection tubes at room temperature during the collection and then were moved to −20 °C until analysis. Teeth were assessed using a sterile periodontal probe on the gingival margin and sweeping along the base of the crown. SUB and SUP plaque samples were collected from the fourth premolar and first molar mandibular teeth and the fourth premolar and first molar maxillary teeth. Plaque samples were placed into sterile 2.0 mL cryovials (CryoELITE, Wheaton, Millville, NJ, USA) and immediately placed on dry ice until storage at −80 °C, where they were stored until analysis.
Microbiota analysis
Total DNA from SAL and plaque samples were extracted using Mo-Bio PowerSoil Kits (MO BIO Laboratories, Inc., Carlsbad, CA), followed by quantification of extracted DNA using a Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY). The quality of extracted DNA was assessed by electrophoresis using agarose gels (E-Gel EX Gel 1%; Invitrogen, Carlsbad, CA). Bacterial 16S rRNA gene amplicons of 252 bp from the V4 region were generated using a Fluidigm Access Array (Fluidigm Corporation, South San Francisco, CA) with Roche High Fidelity Fast Start Kit (Roche, Indianapolis, IN). The primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) that target the 252-bp fragment of the V4 region were used for amplification (primers synthesized by IDT Corporation, Coralville, IA; Caporaso et al., 2012). The quality of the amplicons was assessed using a Fragment Analyzer (Advanced Analytics, Ames, IA) followed by amplicon size selection using electrophoresis and a Qiagen Gel Purification Kit (Qiagen, Valencia, CA). The appropriate profile and average size of purified amplicons were then confirmed using a Bioanalyzer (Agilent Technologies, Santa Clara, CA). Amplicons were sequenced using the Illumina sequencing platform on a MiSeq using v3 reagents (Illumina Inc., San Diego, CA) at the W. M. Keck Center for Biotechnology at the University of Illinois.
Quantitative Insights Into Microbial Ecology (QIIME 2 2018.8; (Caporaso et al., 2010) was used to process the sequence data. Sequence data with quality value ≥ 20 derived from the sequencing process were demultiplexed. Sequences were clustered into operational taxonomic units (OTU) using UCLUST (Edgar, 2010) through an open-reference OTU picking strategy against the Greengenes 13_8 reference database (DeSantis et al., 2006) with a 97% similarity threshold. Singletons and OTU that had <0.01% of the total observation were discarded. Alpha diversity was estimated using observed OTU. Beta diversity was calculated using weighted and unweighted UniFrac (Lozupone and Knight, 2005) distance measures and presented with principal coordinates analysis (PCoA) plots.
Statistical analysis
All data were analyzed using SAS (version 9.4, SAS Institute, Cary, NC) using the Mixed Models procedure with dog being considered a random effect, and source was considered a fixed effect. Data normality was checked using the univariate procedure and Shapiro–Wilk statistic, with log transformation being used when normal distribution was lacking. If after the logarithmic transformation of the data, the data did not reach normality, the data were analyzed using the npar1way procedure and Wilcoxon statistic. Correlations coefficients were calculated using the Pearson correlation coefficients. Data were reported as means with P < 0.05 considered significant.
Results
Dental scoring and salivary pH
All dental scores and salivary pH are presented in Table 1. Dogs in the CT group had greater calculus coverage (3.14 vs. 1.62 to 2.16, P < 0.0001), pocket depth (1.81 vs. 1.62 to 1.67, P = 0.002) and bleeding (2.23 vs. 1.19 to 1.33, P = 0.01), halitosis (110 vs. 55.4 to 58.6 parts per billion, P < .0001), and scores for calculus (3.21 vs. 1.53 to 2.06; 0 [low] to 12 [maximum], P < 0.0001), plaque (9.60 vs. 6.62 to 7.86; 0 [low] to 12 [maximum], P < 0.0001), pocket (4.06 vs. 2.03 to 2.25, P = 0.003), and oral health (18.1 vs. 11.5 to 13.3, P < 0.0001) than those consuming any of the dental chews (BC, DL, and GR). Calculus thickness tended to be greater for CT dogs (1.01) than those consuming chews. Calculus thickness also tended to be greater in dogs consuming BC (0.93) or GR (0.96) and lower for the ones consuming DL (0.93). Plaque coverage (P = 0.005) was higher in CT dogs (3.74) compared with those consuming GR (3.16). Plaque thickness (P < 0.0001) was greater in CT dogs (2.57) than dogs consuming chews, with those consuming BC (2.30) having greater plaque thickness than those consuming DR (2.00). Salivary pH and gingivitis score were not different among treatments.
Table 1.
Dental scoring and salivary pH from healthy adult dogs consuming dental chews or diet alone
| Item | CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n=11) |
SEM | P-value |
|---|---|---|---|---|---|---|
| Calculus coverage | 3.14a | 2.16b | 1.62b | 1.83b | 0.19 | <0.0001 |
| Calculus thickness | 1.01x | 0.93y | 0.93z | 0.96y | 0.02 | 0.01 |
| Plaque coverage | 3.74a | 3.42ab | 3.35ab | 3.16b | 0.11 | 0.005 |
| Plaque thickness | 2.57a | 2.30b | 2.00c | 2.07bc | 0.08 | <0.0001 |
| Pocket depth | 1.81a | 1.67b | 1.62b | 1.63b | 0.06 | 0.002 |
| Pocket bleeding | 2.23a | 1.31b | 1.33b | 1.19b | 0.29 | 0.01 |
| Salivary pH | 7.78 | 7.75 | 7.69 | 7.81 | 0.08 | 0.71 |
| Halitosis, ppb | 110a | 56.1b | 55.4b | 58.6b | 8.34 | <0.0001 |
| Calculus score1 | 3.21a | 2.06b | 1.53b | 1.77b | 0.22 | <0.0001 |
| Plaque score1 | 9.60a | 7.86b | 6.75b | 6.62b | 0.40 | <0.0001 |
| Pocket score | 4.06a | 2.25b | 2.22b | 2.03b | 0.54 | 0.003 |
| Gingivitis score | 1.20 | 1.11 | 1.09 | 1.11 | 0.03 | 0.07 |
| OHS | 18.1a | 13.3b | 11.6b | 11.5b | 0.98 | <0.0001 |
1Plaque and calculus scores ranged from 0 (low) to 12 (maximum).
a–cMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
x–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Canine oral microbiome composition
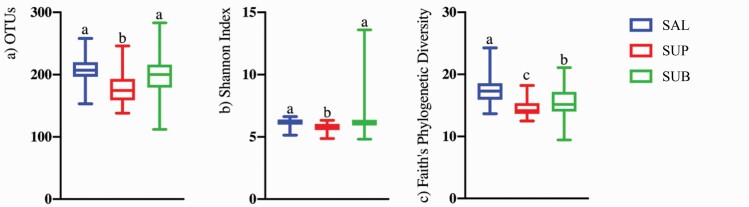
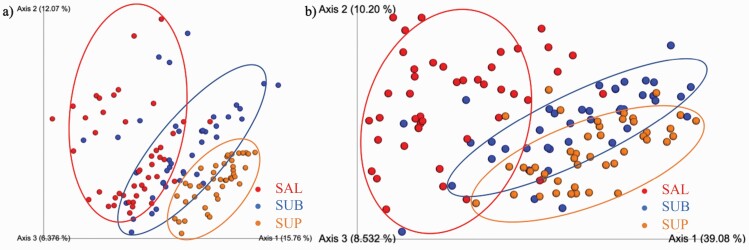
Sequences are available at the NCBI sequence read archive (https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA694668. Illumina sequencing produced a total of 6,872,261 16S rRNA amplicon sequences, with an average of 52,062 sequences per sample after quality filtering. Analyses were conducted with all samples rarified to a level of 26,240 sequences. Alpha- and beta-diversity indices were affected by sample type (habitat). Species richness differed among habitats, with SAL and SUB samples having higher (P < 0.0001) observed OTU (Figure 1a) and Shannon index values (Figure 1b) than SUP samples. Faith’s phylogenetic diversity was higher for SAL than plaque samples, with SUB samples being higher than SUP samples (Figure 1c, P < .0001). Weighted (Figure 2a) and unweighted (Figure 2b) PCoA plots showed how the samples clustered according to oral habitat (P < 0.001). In both plots, the SAL and SUP sample clusters were nearly completely separated, with the SUB samples having an overlap with each.
Figure 1.
Bacterial alpha diversity indices of canine salivary and plaque samples as assessed by the observed OTUs (a), Shannon Index (b), and Faith’s phylogenetic diversity (c). Groups with different superscripts differ (P < 0.01). SAL (n = 44), SUB (n = 43), and SUP (n = 44).
Figure 2.
Principal coordinates analysis (PCoA) plots of weighted (a) and unweighted (b) UniFrac distances of oral microbial communities performed on the 97% OTU abundance matrix using Quantitative Insights Into Microbial Ecology (QIIME). SAL (n = 44), SUB (n = 43), and SUP (n = 44).
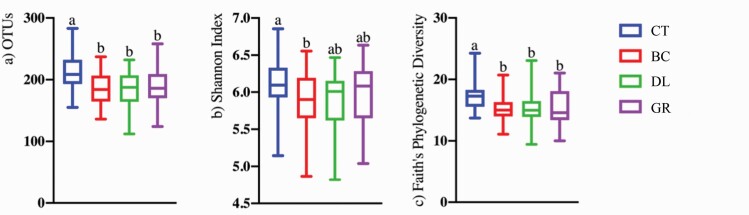
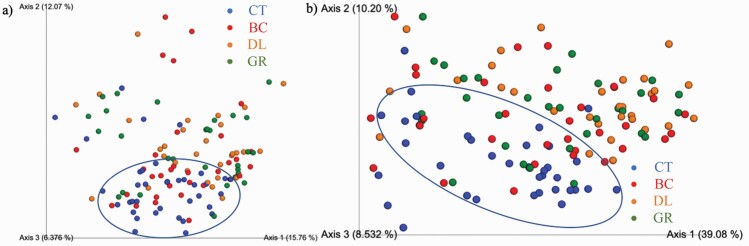
Alpha- and beta-diversity indices were also affected by the treatment group. Species richness differed among treatments, with dogs consuming CT having the highest observed OTU (Figure 3a) and Faith’s phylogenetic diversity (Figure 3c) than those consuming any of the chew treatments (BC, DL, and GR; P < 0.05). Shannon index values were higher for CT dogs than BC dogs, with DL and GR dogs having intermediate values (Figure 3b, P < 0.05). Weighted (Figure 4a) and unweighted (Figure 4b) PCoA plots showed that CT dogs clustered together and separate from those consuming chews (BC, DL, and GR; P < 0.01).
Figure 3.
Bacterial alpha diversity indices of dogs consuming dental chews or diet alone as assessed by the observed OTUs (a), Shannon Index (b), and Faith’s phylogenetic diversity (c). Groups with different superscripts differ (P < 0.05). CT (n = 36), BC (n = 33), DL (n = 30), and GR (n = 32).
Figure 4.
Principal coordinates analysis (PCoA) plots of weighted (a) and unweighted (b) UniFrac distances of oral microbial communities of dogs consuming dental chews or diet alone performed on the 97% OTU abundance matrix using Quantitative Insights Into Microbial Ecology (QIIME). CT (n = 36), BC (n = 33), DL (n = 30), and GR (n = 32).
The oral microbiota measured in this study contained a diverse array of bacteria, including the detection of 12 phyla, with 3 phyla (Bacteroidetes, Proteobacteria, and Firmicutes) accounting for more than 70% of the sequences (Table 2). Oral habitat had a significant influence on the bacterial phyla (Table 2), with SAL samples having higher relative abundances of Firmicutes, Spirochaetes, and Tenericutes than plaque samples, and SUB samples having higher relative abundances of Firmicutes, Spirochaetes, and Tenericutes than SUP samples (P < 0.0001). Relative abundances of Bacteroidetes and SR1 were higher in SAL samples than plaque samples, with SUP samples having a higher relative abundance of Bacteroidetes and SR1 than SUB samples (P < 0.01). Relative abundance of GN02 was higher in SUB and SUP samples than SAL samples (P = 0.003). Relative abundance of Proteobacteria was higher in SUP samples than the other samples, with SUB samples having a higher relative abundance of Proteobacteria than SAL samples (P < 0.001). Relative abundance of Actinobacteria was higher in SUB samples than SUP and SAL samples, with SUP samples had a higher relative abundance of Actinobacteria than SAL samples (P < 0.001). Finally, SAL and SUB samples had a higher relative abundance of Fusobacteria than SUP samples (P < 0.001).
Table 2.
Oral bacterial phyla (relative abundance, %) present in plaque (SUP and SUB plaque) and SAL samples from healthy adult dogs consuming dental chews or diet alone
| Source | Statistics | ||||
|---|---|---|---|---|---|
| Phyla | SAL (n = 44) |
SUB (n = 43) |
SUP (n = 44) |
SEM | P-value |
| Actinobacteria | 3.11z | 8.95x | 7.83y | 0.53 | <0.0001 |
| Bacteroidetes | 32.21a | 21.95c | 24.79b | 1.03 | <0.0001 |
| Chlorobi | 1.11 | 0.98 | 1.33 | 0.18 | 0.11 |
| Firmicutes | 15.86a | 12.59b | 9.12c | 0.85 | <0.0001 |
| Fusobacteria | 5.29a | 6.38a | 2.17b | 0.71 | <0.0001 |
| GN02 | 0.78b | 1.30a | 1.25a | 0.22 | 0.003 |
| Proteobacteria | 28.31c | 42.16b | 48.98a | 1.41 | <0.0001 |
| SR1 | 3.14x | 2.38z | 2.71y | 0.38 | 0.005 |
| Spirochaetes | 5.47x | 2.28y | 1.01z | 0.37 | <0.0001 |
| Synergistetes | 0.22 | 0.45 | 0.44 | 0.06 | 0.09 |
| TM7 | 0.08 | 0.05 | 0.05 | 0.01 | 0.08 |
| Tenericutes | 3.87a | 0.34b | 0.06c | 0.32 | <0.0001 |
a–cMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
x–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Oral habitat had a significant influence on the bacterial genera (Table 3), with relative abundances of Actinomyces, Leucobacter, Capnocytophaga, Bergeyella, and Neisseria being lower in SAL samples than the other oral habitats (P < 0.0001). Relative abundances of Corynebacterium, Desulfomicrobium, and Moraxella were higher in SUB samples, followed by SUP samples, and lowest in SAL samples (P < 0.05). Relative abundances of Euzebya, Proteocatella, Fusibacter, and Leptotrichia were higher in SUB samples than the other oral habitats (P < 0.001). Relative abundances of Bacteroides, Arcobacter, and Enhydrobacter were higher in SUP samples, followed by SAL samples, and lower in SUB samples (P < 0.05). Relative abundance of Paludibacter was higher in SUB samples, followed by SUP samples, and lower in SAL samples (P = 0.0001). Relative abundances of Porphyromonas and Prevotella were higher in SAL samples, followed by SUP samples, and lower in SUB samples (P < 0.0001). Relative abundance of Gemella was higher in SAL samples, followed by SUB samples, and lower in SUP samples (P < 0.0001). Relative abundance of Streptococcus was higher in SUP samples than SAL samples (P = 0.03). Relative abundances of Peptococcus and Filifactor were higher in SAL samples, followed by SUB samples, and lower in SUP samples (P < 0.001). Relative abundances of Anaerovorax, Lautropia, and Lampropedia were higher in SUP samples, followed by SUB samples, and lowest in SAL samples (P < 0.01). Relative abundances of Peptostreptococcus, Parvimonas, Fusobacterium, and Campylobacter were lower in SUP samples than the other oral habitats (P < 0.0001). Relative abundances of Treponema and Acholeplasma were higher in SAL samples, followed by SUP samples, and lowest in SUB samples (P < 0.0001).
Table 3.
Oral bacterial genera (relative abundance, %) that were significantly different between treatments present in plaque (SUP and SUB plaque) and SAL samples from healthy adult dogs consuming dental chews or diet alone
| Genus | Source | Statistics | |||
|---|---|---|---|---|---|
| SAL (n = 44) |
SUB (n = 43) |
SUP (n = 44) |
SEM | P-value | |
| Actinomyces | 2.02b | 2.83a | 3.13a | 0.23 | <0.0001 |
| Corynebacterium | 0.26c | 3.53a | 2.09b | 0.31 | <0.0001 |
| Leucobacter | 0.43b | 1.48a | 1.70a | 0.27 | <0.0001 |
| Euzebya | 0.40b | 0.81a | 0.31b | 0.07 | <0.0001 |
| Bacteroides | 1.04y | 0.95z | 2.24x | 0.18 | <0.0001 |
| Paludibacter | 0.22y | 0.72x | 0.18z | 0.09 | 0.0001 |
| Porphyromonas | 19.11x | 12.30z | 12.86y | 1.22 | <0.0001 |
| Prevotella | 8.25x | 1.84z | 2.24y | 0.42 | <0.0001 |
| Capnocytophaga | 0.34b | 1.31a | 1.49a | 0.14 | <0.0001 |
| Bergeyella | 0.98b | 2.42a | 2.99a | 0.38 | <0.0001 |
| Gemella | 0.78a | 0.11b | 0.02c | 0.07 | <0.0001 |
| Streptococcus | 0.33b | 0.71ab | 1.29a | 0.27 | 0.03 |
| Peptococcus | 2.38a | 1.04b | 0.32c | 0.26 | <0.0001 |
| Filifactor | 0.46x | 0.42y | 0.24z | 0.06 | 0.0002 |
| Peptostreptococcus | 0.45a | 0.28a | 0.10b | 0.09 | 0.0001 |
| Proteocatella | 0.20b | 0.78a | 0.22b | 0.06 | <0.0001 |
| Fusibacter | 1.33b | 2.02a | 1.45b | 0.15 | 0.001 |
| Anaerovorax | 0.14z | 0.30y | 0.26x | 0.05 | 0.004 |
| Helcococcus | 0.42a | 0.04b | 0.05b | 0.06 | <0.0001 |
| Parvimonas | 0.48a | 0.49a | 0.07b | 0.10 | <0.0001 |
| Fusobacterium | 3.55a | 5.10a | 2.04b | 0.44 | <0.0001 |
| Leptotrichia | 0.10b | 1.08a | 0.09b | 0.12 | <0.0001 |
| Lautropia | 0.29c | 0.64b | 1.12a | 0.12 | <0.0001 |
| Lampropedia | 0.29z | 1.14y | 2.41x | 0.16 | <0.0001 |
| Neisseria | 1.73b | 4.32a | 3.47a | 0.39 | <0.0001 |
| Desulfomicrobium | 0.67z | 1.90x | 1.12y | 0.26 | 0.02 |
| Arcobacter | 1.89y | 0.94z | 2.22x | 0.47 | 0.02 |
| Campylobacter | 1.78a | 1.52a | 0.63b | 0.14 | <0.0001 |
| Enhydrobacter | 5.21y | 4.77z | 9.73x | 0.86 | <0.0001 |
| Moraxella | 4.49z | 11.59x | 11.59y | 1.05 | <0.0001 |
| Treponema | 5.11x | 2.19y | 0.94z | 0.35 | <0.0001 |
| Acholeplasma | 2.01a | 0.09b | 0.04c | 0.15 | <0.0001 |
a–cMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
x–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Chew treatments also had a significant influence on the bacterial phyla of SAL (Table 4), SUP (Table 5), and SUB (Table 6) samples. In SAL samples (Table 4), relative abundance of Actinobacteria was higher in dogs consuming DL or GR than those consuming CT (P < 0.01). Relative abundance of Fusobacteria was higher in dogs consuming BC than those consuming CT (P = 0.03). Relative abundance of Spirochaetes was higher in dogs consuming CT than those consuming any of the chews (P < 0.001). In SUP samples (Table 5), relative abundance of Actinobacteria was higher in dogs consuming DL, followed by dogs consuming GR and BC, and lower in dogs consuming CT (P < 0.0001). Relative abundance of Bacteroidetes was higher in dogs consuming CT, BC, or GR than dogs consuming DL (P = 0.001). Relative abundance of Firmicutes was higher in dogs consuming CT, followed by dogs consuming BC, and lower in dogs consuming DL, and dogs consuming GR had an intermediate value between dogs consuming CT or BC (P < 0.0001). In SUB samples (Table 6), relative abundance of Proteobacteria was lower in dogs consuming CT than those consuming all chew treatments (P < 0.01). Relative abundance of Actinobacteria was higher in dogs consuming DL than those consuming all other treatments (P < 0.0001). Relative abundance of Bacteroidetes was higher in dogs consuming CT than those consuming DL (P = 0.03). Relative abundance of Firmicutes was higher in dogs consuming CT than those consuming BC or DL (P < 0.0001).
Table 4.
Oral bacterial phyla (relative abundance, %) that were significantly different between treatments present in SAL samples from healthy adult dogs consuming dental chews or diet alone
| Phyla | SAL | Statistics | ||||
|---|---|---|---|---|---|---|
| CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n = 11) |
SEM | P-value | |
| Actinobacteria | 1.66b | 2.64ab | 4.11a | 4.21a | 0.55 | 0.002 |
| Fusobacteria | 3.98b | 6.57a | 5.20ab | 5.27ab | 1.08 | 0.03 |
| Spirochaetes | 8.58a | 5.19b | 3.64b | 4.19b | 0.74 | <0.0001 |
| Synergistetes | 0.37w | 0.13z | 0.15y | 0.21x | 0.05 | 0.01 |
| TM7 | 0.03b | 0.04ab | 0.13a | 0.10a | 0.02 | <0.0001 |
a–bMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
w–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Table 5.
Oral bacterial phyla (relative abundance, %) that were significantly different between treatments present in SUP plaque samples from healthy adult dogs consuming dental chews or diet alone
| Phyla | SUP | Statistics | ||||
|---|---|---|---|---|---|---|
| CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n = 11) |
SEM | P-value | |
| Actinobacteria | 5.01c | 7.56b | 11.50a | 8.06b | 0.70 | <0.0001 |
| Bacteroidetes | 24.57a | 26.29a | 20.62b | 27.06a | 1.44 | 0.001 |
| Chlorobi | 2.49a | 1.20ab | 0.55c | 0.88b | 0.31 | <0.0001 |
| Firmicutes | 12.30a | 8.99b | 5.11c | 9.45ab | 0.87 | <0.0001 |
| GN02 | 0.56c | 1.32ab | 2.35a | 0.96bc | 0.29 | 0.0001 |
| SR1 | 1.95b | 3.06ab | 3.60a | 2.36ab | 0.55 | 0.02 |
| Spirochaetes | 1.71a | 1.01b | 0.48b | 0.64b | 0.18 | <0.0001 |
| Synergistetes | 0.98a | 0.27b | 0.15b | 0.30b | 0.07 | <0.0001 |
| TM7 | 0.02z | 0.05y | 0.06w | 0.06x | 0.01 | 0.001 |
| Tenericutes | 0.07a | 0.10ab | 0.02bc | 0.02c | 0.02 | 0.001 |
a–cMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
w–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Table 6.
Oral bacterial phyla (relative abundance, %) that were significantly different between treatments present in SUB plaque samples from healthy adult dogs consuming dental chews or diet alone
| Phyla | SUB | Statistics | ||||
|---|---|---|---|---|---|---|
| CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n = 10) |
SEM | P-value | |
| Actinobacteria | 6.18b | 7.76b | 14.6a | 8.18b | 1.11 | <0.0001 |
| Bacteroidetes | 24.10a | 22.61ab | 17.99b | 22.78ab | 1.84 | 0.03 |
| Chlorobi | 2.16a | 0.83b | 0.28b | 0.43b | 0.26 | <0.0001 |
| Firmicutes | 16.44a | 11.93b | 8.26b | 13.05ab | 1.06 | <0.0001 |
| GN02 | 0.55b | 1.29a | 1.80a | 1.75a | 0.39 | 0.001 |
| Proteobacteria | 36.43b | 46.22a | 46.37a | 40.24ab | 2.31 | 0.002 |
| Spirochaetes | 3.90a | 1.77b | 0.97b | 2.11b | 0.47 | <0.0001 |
| Synergistetes | 1.13a | 0.23b | 0.07b | 0.23b | 0.11 | <0.0001 |
a–bMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
Treatments had a significant influence on bacterial genera of SAL (Table 7), SUP (Table 8), and SUB (Table 9) samples. In general, in all oral habitats (SAL, SUB, and SUP), CT had lower relative abundances of Capnocytophaga, Neisseria, and Moraxella compared with one or more of the dental chew treatments (BC, DL, and GR) and higher relative abundance of Treponema compared with one or more dental chew treatments. In plaque samples (SUB and SUP), CT had lower relative abundances of Bergeyella and Pasteurella compared with one or more dental chew treatments and higher relative abundances of Paludibacter, Porphyromonas, Filifactor, Anaerovorax, Desulfomicrobium, Desulfovibrio, Enhydrobacter, and TG5 compared with one or more dental chew treatments. In SAL and SUP samples, CT had a lower relative abundance of Leucobacter compared with one or more dental chew treatments. In SAL samples, CT had lower relative abundances of Actinomyces, Corynebacterium, Euzebya, Proteocatella, p-75-a5, Enhydrobacter, and TG5 compared with one or more dental chew treatments and higher relative abundances of Arcobacter and Wolinella compared with one or more dental chew treatments. In SUP samples, in general, CT had a lower relative abundance of Lautropia compared with one or more dental chew treatments and higher relative abundances of Tannerella, Fusibacter, Propionivibrio, Campylobacter, Enhydrobacter, and Acholeplasma compared with one or more dental chew treatments. In SUB samples, CT had lower relative abundances of Euzebya, Tannerella, and Fusibacter compared with one or more dental chew treatments.
Table 7.
Oral bacterial genera (relative abundance, %) that were significantly different between treatments present in SAL samples from healthy adult dogs consuming dental chews or diet alone
| Genus | SAL | Statistics | ||||
|---|---|---|---|---|---|---|
| CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n = 11) |
SEM | P-value | |
| Actinomyces | 1.21b | 1.90ab | 2.17ab | 2.85a | 0.39 | 0.003 |
| Corynebacterium | 0.06b | 0.15a | 0.59a | 0.26ab | 0.11 | 0.004 |
| Leucobacter | 0.21b | 0.37ab | 0.74a | 0.41ab | 0.13 | 0.01 |
| Euzebya | 0.16b | 0.23b | 0.58a | 0.66a | 0.14 | 0.004 |
| Capnocytophaga | 0.12c | 0.23bc | 0.60a | 0.43b | 0.10 | 0.0001 |
| Proteocatella | 0.03b | 0.12ab | 0.43a | 0.23a | 0.08 | 0.005 |
| p-75-a5 | 0.13b | 0.14ab | 0.26ab | 0.36a | 0.08 | 0.02 |
| Lautropia | 0.28ab | 0.08b | 0.47a | 0.35a | 0.11 | 0.003 |
| Neisseria | 0.66b | 1.88ab | 2.51a | 2.11a | 0.49 | 0.0004 |
| Arcobacter | 2.68a | 2.71b | 1.09b | 0.97b | 0.77 | 0.01 |
| Wolinella | 2.55a | 1.36b | 0.74b | 0.46b | 0.43 | 0.002 |
| Enhydrobacter | 9.35w | 3.70y | 3.21z | 4.05x | 1.58 | 0.047 |
| Moraxella | 2.58b | 4.75ab | 5.39a | 5.63a | 1.17 | 0.02 |
| Treponema | 8.11a | 4.84b | 3.36b | 3.83b | 0.68 | <0.0001 |
| TG5 | 0.37w | 0.13y | 0.15z | 0.21x | 0.05 | 0.01 |
a–cMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
w–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Table 8.
Oral bacterial genera (relative abundance, %) that were significantly different between treatments present in SUP samples from healthy adult dogs consuming dental chews or diet alone
| Genus | SUP | Statistics | ||||
|---|---|---|---|---|---|---|
| CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n = 11) |
SEM | P-value | |
| Corynebacterium | 0.91b | 1.40b | 4.21a | 2.19b | 0.44 | <0.0001 |
| Leucobacter | 1.05b | 2.21a | 2.28ab | 1.41ab | 0.44 | 0.01 |
| Paludibacter | 0.25a | 0.15ab | 0.14b | 0.15ab | 0.04 | 0.01 |
| Porphyromonas | 14.92a | 15.37a | 8.46b | 11.69ab | 2.03 | 0.01 |
| Tannerella | 0.58a | 0.19b | 0.17b | 0.27b | 0.09 | 0.001 |
| Capnocytophaga | 0.65c | 1.51b | 2.38a | 1.59b | 0.21 | <0.0001 |
| Bergeyella | 1.16b | 2.92a | 4.21a | 3.99a | 0.67 | 0.0002 |
| Streptococcus | 1.43b | 0.35b | 0.55b | 2.89a | 0.65 | 0.01 |
| Peptococcus | 0.30ab | 0.65a | 0.15ab | 0.22b | 0.15 | 0.04 |
| Filifactor | 0.33a | 0.41a | 0.08b | 0.12b | 0.06 | 0.0002 |
| Fusibacter | 2.28a | 1.65b | 0.59c | 1.09c | 0.20 | <0.0001 |
| Anaerovorax | 0.36a | 0.31a | 0.12b | 0.22ab | 0.05 | 0.001 |
| Leptotrichia | 0.07ab | 0.07ab | 0.17a | 0.06b | 0.03 | 0.01 |
| Lautropia | 0.61b | 1.04a | 1.78a | 1.14a | 0.24 | 0.001 |
| Neisseria | 1.72b | 3.28a | 3.92a | 5.20a | 0.62 | <0.0001 |
| Propionivibrio | 0.10a | 0.05b | 0.03b | 0.04b | 0.01 | 0.001 |
| Desulfomicrobium | 1.99a | 0.86b | 0.53b | 0.91b | 0.24 | <0.0001 |
| Desulfovibrio | 1.64a | 0.55b | 0.05b | 0.34b | 0.22 | 0.0001 |
| Campylobacter | 0.93a | 0.79a | 0.45ab | 0.31b | 0.11 | 0.0003 |
| Pasteurella | 0.86z | 1.42x | 1.62w | 0.92y | 0.21 | 0.03 |
| Enhydrobacter | 15.00a | 9.69b | 4.86c | 8.09bc | 1.50 | <0.0001 |
| Moraxella | 6.30b | 13.81a | 14.35a | 12.61a | 1.95 | 0.004 |
| Treponema | 1.56a | 0.98ab | 0.46b | 0.58b | 0.18 | 0.0001 |
| TG5 | 0.95a | 0.26b | 0.14b | 0.29b | 0.07 | <0.0001 |
| Acholeplasma | 0.06a | 0.09a | 0.02ab | 0.02b | 0.02 | 0.01 |
a–cMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
w–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Table 9.
Oral bacterial genera (relative abundance, %) that were significantly different between treatments present in SUB samples from healthy adult dogs consuming dental chews or diet alone
| Genus | SUB | Statistics | ||||
|---|---|---|---|---|---|---|
| CT (n = 12) |
BC (n = 11) |
DL (n = 10) |
GR (n = 10) |
SEM | P-value | |
| Actinomyces | 2.32b | 2.60ab | 4.32a | 2.20b | 0.44 | 0.01 |
| Corynebacterium | 2.34b | 2.71b | 6.60a | 2.98b | 0.73 | <0.0001 |
| Euzebya | 0.53b | 0.58ab | 1.23a | 0.99ab | 0.18 | 0.02 |
| Paludibacter | 0.89a | 0.96ab | 0.20b | 0.80ab | 0.26 | 0.03 |
| Porphyromonas | 16.12a | 11.24ab | 9.20b | 11.97ab | 2.10 | 0.03 |
| Tannerella | 0.56w | 0.30x | 0.15z | 0.23y | 0.06 | 0.001 |
| Capnocytophaga | 0.46b | 1.82a | 1.44a | 1.63a | 0.36 | 0.0003 |
| Bergeyella | 0.87b | 2.96a | 2.56a | 3.58a | 0.57 | <0.0001 |
| Filifactor | 0.70a | 0.43ab | 0.16b | 0.30ab | 0.13 | 0.01 |
| Fusibacter | 3.04w | 1.90x | 1.04z | 1.86y | 0.31 | 0.001 |
| Anaerovorax | 0.48a | 0.22ab | 0.14b | 0.33ab | 0.09 | 0.01 |
| Lautropia | 0.38b | 0.85a | 0.85a | 0.53ab | 0.22 | 0.001 |
| Lampropedia | 0.77b | 1.24ab | 1.86a | 0.76b | 0.26 | 0.01 |
| Neisseria | 2.12b | 5.47a | 5.43a | 4.50a | 0.77 | <0.0001 |
| Desulfomicrobium | 3.55a | 1.19b | 0.69b | 2.00ab | 0.57 | 0.0001 |
| Desulfovibrio | 1.47a | 0.23b | 0.05b | 0.07b | 0.22 | 0.001 |
| Pasteurella | 0.72b | 1.05ab | 1.41a | 0.85ab | 0.20 | 0.02 |
| Enhydrobacter | 7.59a | 4.20b | 2.78b | 3.75b | 1.06 | 0.003 |
| Moraxella | 4.16b | 16.16a | 13.87a | 13.14a | 1.46 | <0.0001 |
| Treponema | 3.74a | 1.72b | 0.91b | 2.06b | 0.45 | <0.0001 |
| TG5 | 1.11a | 0.23b | 0.07b | 0.23b | 0.11 | <0.0001 |
a–bMeans with different superscripts within a row differ by Tukey’s test (P < 0.05).
w–zMeans with different superscripts within a row differ by Wilcoxon’s test (P < 0.05).
Several bacterial phyla and genera were significantly (P < 0.05) correlated with OHS. Relative abundances of the phyla Chlorobi and Firmicutes were positively correlated with periodontal disease (increased OHS), salivary pH, plaque, calculus, gingivitis, and pocket scores (Tables 10). Relative abundances of Spirochaetes and Synergistetes were positively correlated with periodontal disease (increased OHS), plaque, calculus, gingivitis, and pocket scores. Relative abundance of GN02 was negatively correlated with periodontal disease (decrease OHS), salivary pH, plaque, calculus, gingivitis, and pocket scores. Relative abundances of Actinobacteria and TM7 were negatively correlated with periodontal disease, plaque, and calculus score. Relative abundance of SR1 was negatively correlated with periodontal disease, plaque, and gingivitis score. Relative abundance of Tenericutes was negatively correlated with periodontal disease, calculus, and pocket score.
Table 10.
Correlation coefficients (r) between OHS, plaque scores, calculus scores, gingivitis scores, pocket scores, pH, and bacterial phyla1
| Phyla | OHS | Plaque score | Calculus score | Gingivitis score | Pocket score | Salivary pH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | r | P-value | r | P-value | r | P-value | |
| Actinobacteria | −0.25 | 0.0043 | −0.27 | 0.0015 | −0.29 | 0.0007 | −0.15 | 0.0967 | −0.15 | 0.0824 | −0.004 | 0.9630 |
| Chlorobi | 0.42 | <0.0001 | 0.46 | <0.0001 | 0.46 | <0.0001 | 0.18 | 0.0410 | 0.28 | 0.0014 | 0.08 | 0.3547 |
| Firmicutes | 0.32 | 0.0002 | 0.23 | 0.0082 | 0.32 | 0.0002 | 0.32 | 0.0002 | 0.30 | 0.0006 | 0.19 | 0.0281 |
| GN02 | −0.46 | <0.0001 | −0.44 | <0.0001 | −0.43 | <0.0001 | −0.41 | <0.0001 | −0.35 | <0.0001 | −0.24 | 0.0050 |
| SR1 | −0.17 | 0.0475 | −0.26 | 0.0027 | −0.15 | 0.0861 | −0.18 | 0.0417 | −0.08 | 0.3757 | −0.23 | 0.0092 |
| Spirochaetes | 0.34 | <0.0001 | 0.35 | <0.0001 | 0.33 | 0.0001 | 0.18 | 0.0435 | 0.26 | 0.0033 | −0.09 | 0.3309 |
| Synergistetes | 0.55 | <0.0001 | 0.52 | <0.0001 | 0.55 | <0.0001 | 0.38 | <0.0001 | 0.46 | <0.0001 | 0.04 | 0.6639 |
| TM7 | −0.24 | 0.0052 | −0.28 | 0.0012 | −0.26 | 0.0031 | −0.09 | 0.3292 | −0.15 | 0.0872 | −0.03 | 0.7247 |
| Tenericutes | −0.19 | 0.0342 | −0.13 | 0.1410 | −0.19 | 0.0333 | −0.15 | 0.0893 | −0.17 | 0.0467 | 0.04 | 0.6391 |
1Bold correlation coefficients (r) with P < 0.05, underlined r with strong correlation (r > 0.5), and italic r with moderate correlation (0.3 < r < 0.5).
Relative abundances of the genera Actinomyces, p-75-a5, and Moraxella were negatively correlated with periodontal disease (OHS), plaque, calculus, and pocket scores (Table 11). Relative abundances of Capnocytophaga, Bergeyella, and Neisseria were negatively correlated with periodontal disease (OHS), plaque, calculus, gingivitis, and pocket scores. Relative abundance of Leucobacter was negatively correlated with plaque and calculus scores. Relative abundance of Euzebya was negatively correlated with periodontal disease (OHS) and calculus scores. Relative abundance of Pasteurella was negatively correlated with periodontal disease (OHS), plaque, calculus, gingivitis, and pocket scores, and salivary pH. Relative abundances of Tannerella, Desulfomicrobium, Desulfovibrio, Arcobacter, and Treponema were positively correlated with periodontal disease (OHS), plaque, calculus, gingivitis, and pocket scores. Relative abundances of Fusibacter, Anaerovorax, and Enhydrobacter were positively correlated with periodontal disease (OHS), plaque, calculus, and pocket scores. Relative abundance of Bacteroides was positively correlated with gingivitis score. Relative abundance of Porphyromonas was positively correlated with plaque score. Relative abundance of Filifactor was positively correlated with periodontal disease (OHS), plaque, and calculus scores. Relative abundance of Peptostreptococcus was positively correlated with periodontal disease (OHS), gingivitis, and pocket scores. Relative abundance of Helcococcus was positively correlated with calculus score. Relative abundance of Propionivibrio was positively correlated with periodontal disease (OHS), plaque, calculus, and gingivitis scores, and salivary pH. Relative abundances of Campylobacter and Wolinella were positively correlated with periodontal disease (OHS) and plaque scores. Finally, relative abundance of Wolinella was negatively correlated with salivary pH.
Table 11.
Correlation coefficients (r) between OHS, plaque scores, calculus scores, gingivitis scores, pocket scores, pH, and bacteria genera1
| Genera | OHS | Plaque score | Calculus score | Gingivitis score | Pocket score | Salivary pH | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | P-value | r | P-value | r | P-value | r | P-value | r | P-value | r | P-value | |
| Actinomyces | −0.23 | 0.0088 | −0.23 | 0.0096 | −0.21 | 0.0149 | −0.08 | 0.3564 | −0.19 | 0.0335 | 0.14 | 0.1228 |
| Leucobacter | −0.16 | 0.0752 | −0.27 | 0.0017 | −0.29 | 0.0009 | −0.14 | 0.1036 | 0.03 | 0.7657 | −0.16 | 0.0735 |
| Euzebya | −0.17 | 0.0482 | −0.15 | 0.0796 | −0.23 | 0.0095 | −0.10 | 0.2361 | −0.14 | 0.1119 | −0.02 | 0.8458 |
| Bacteroides | 0.11 | 0.2039 | 0.04 | 0.6364 | 0.13 | 0.1487 | 0.18 | 0.0446 | 0.13 | 0.1333 | 0.11 | 0.2235 |
| Porphyromonas | 0.13 | 0.1405 | 0.21 | 0.0141 | 0.16 | 0.0694 | −0.02 | 0.8477 | 0.03 | 0.773 | −0.04 | 0.6771 |
| Tannerella | 0.30 | 0.0006 | 0.31 | 0.0004 | 0.29 | 0.0007 | 0.18 | 0.0447 | 0.23 | 0.0097 | −0.03 | 0.7589 |
| Capnocytophaga | −0.31 | 0.0003 | −0.28 | 0.001 | −0.29 | 0.0008 | −0.25 | 0.0046 | −0.28 | 0.0012 | −0.01 | 0.9275 |
| Bergeyella | −0.37 | <0.0001 | −0.43 | <0.0001 | −0.46 | <0.0001 | −0.21 | 0.0150 | −0.20 | 0.0224 | −0.08 | 0.3892 |
| Filifactor | 0.19 | 0.0273 | 0.27 | 0.0022 | 0.21 | 0.0137 | 0.10 | 0.2558 | 0.09 | 0.2906 | 0.00 | 0.9554 |
| Peptostreptococcus | 0.18 | 0.0385 | 0.13 | 0.1447 | 0.17 | 0.0516 | 0.18 | 0.0418 | 0.17 | 0.0489 | 0.11 | 0.206 |
| Fusibacter | 0.35 | <0.0001 | 0.33 | <0.0001 | 0.33 | <0.0001 | 0.15 | 0.0853 | 0.30 | 0.0005 | −0.11 | 0.2075 |
| Anaerovorax | 0.36 | <0.0001 | 0.28 | 0.0013 | 0.26 | 0.0032 | 0.15 | 0.0871 | 0.38 | <0.0001 | −0.10 | 0.2708 |
| Helcococcus | 0.10 | 0.2633 | 0.11 | 0.2104 | 0.17 | 0.0492 | 0.03 | 0.7674 | 0.02 | 0.8587 | 0.05 | 0.5449 |
| p-75-a5 | −0.26 | 0.0028 | −0.26 | 0.0027 | −0.17 | 0.0465 | −0.05 | 0.5721 | −0.23 | 0.0072 | 0.09 | 0.3106 |
| Neisseria | −0.39 | <0.0001 | −0.35 | <0.0001 | −0.37 | <0.0001 | −0.29 | 0.0007 | −0.34 | <0.0001 | 0.01 | 0.8773 |
| Propionivibrio | 0.23 | 0.0075 | 0.29 | 0.0008 | 0.28 | 0.0012 | 0.24 | 0.0063 | 0.12 | 0.1782 | 0.22 | 0.0117 |
| Desulfomicrobium | 0.31 | 0.0003 | 0.34 | <0.0001 | 0.35 | <0.0001 | 0.31 | 0.0004 | 0.21 | 0.0176 | 0.13 | 0.1356 |
| Desulfovibrio | 0.48 | <0.0001 | 0.44 | <0.0001 | 0.46 | <0.0001 | 0.28 | 0.0012 | 0.41 | <0.0001 | −0.03 | 0.7020 |
| Arcobacter | 0.36 | <0.0001 | 0.21 | 0.0154 | 0.28 | 0.0012 | 0.34 | <0.0001 | 0.39 | <0.0001 | −0.06 | 0.5209 |
| Campylobacter | 0.23 | 0.0087 | 0.27 | 0.0015 | 0.17 | 0.0561 | 0.07 | 0.4537 | 0.17 | 0.0524 | −0.01 | 0.9036 |
| Wolinella | 0.19 | 0.0310 | 0.20 | 0.0255 | 0.15 | 0.094 | 0.01 | 0.9413 | 0.16 | 0.0607 | −0.23 | 0.0071 |
| Pasteurella | −0.35 | <0.0001 | −0.23 | 0.0077 | −0.33 | 0.0001 | −0.35 | <0.0001 | −0.33 | <0.0001 | −0.27 | 0.0016 |
| Enhydrobacter | 0.31 | 0.0004 | 0.36 | <0.0001 | 0.38 | <0.0001 | 0.15 | 0.0881 | 0.17 | 0.0457 | −0.12 | 0.1745 |
| Moraxella | −0.33 | 0.0001 | −0.38 | <0.0001 | −0.35 | <0.0001 | −0.10 | 0.2447 | −0.22 | 0.0119 | 0.11 | 0.2017 |
| Treponema | 0.34 | <0.0001 | 0.35 | <0.0001 | 0.33 | 0.0001 | 0.17 | 0.0475 | 0.25 | 0.0037 | −0.08 | 0.3533 |
1Bold correlation coefficients (r) with P < 0.05 and italics r with moderate correlation (0.3 < r < 0.5).
Discussion
The importance of plaque removal is well known and there are several ways to achieve this (Gorrel and Bierer, 1999; Gorrel et al., 1999; Brown and McGenity, 2005; Hennet et al., 2006b; Clarke et al., 2011; Shofiqur et al., 2011; Albuquerque et al., 2012; Quest, 2013; Harvey et al., 2015; Lacerda and Alessi, 2015; Adepu et al., 2018; Oba et al., 2018; Stella et al., 2018). In the past, it was proven that dental chews could contribute to the maintenance of dental hygiene and periodontal health (Gorrel and Rawlings, 1996; Rawlings et al., 1998; Gorrel and Bierer, 1999; Gorrel et al., 1999; Brown and McGenity, 2005; Hennet et al., 2006b; Clarke et al., 2011; Quest, 2013; Garanayak et al., 2019; Carroll et al., 2020; Ruparell et al., 2020), showing that the daily addition of chews to a dry diet was effective in reducing plaque and calculus accumulation on the tooth surfaces and also reducing the severity of gingivitis and oral malodor as compared with feeding the dry diet only (Gorrel et al., 1999). Another study reported that dogs receiving dental chews (dental chew twice daily immediately after food) or 0.2% w/v chlorhexidine (application of chlorhexidine on the buccal surface of the tooth by soaked cotton twice daily after the food was consumed) for 28 d showed lower plaque deposits, and the group receiving both dental chews and chlorhexidine showed that all animals remained free from fresh plaque deposits (Garanayak et al., 2019). In a similar study, daily administration of a dental chew (daily vegetable chew given 4 to 8 h after feeding) for 28 d reduced halitosis, gingivitis, plaque, and calculus accumulation (Clarke et al., 2011). Another study tested the effectiveness of a different dental chew (one per day, fed 5 to 6 h after daily meal) over 4 mo, which resulted in a reduction in plaque deposition (17.3%) and calculus accumulation (45.8%) when compared with dogs fed a dry diet only (Hennet et al., 2006b). Another study tested two dental chews (with or without 0.2% of a natural antimicrobial agent) and showed that dogs fed a single daily dental chew 1 h before feeding the standard diet for 4 wk had less gingivitis, plaque, and calculus compared with dogs in the control group fed the diet only (Brown and McGenity, 2005). When this same group tested the same dental chew or 0.2% w/v chlorhexidine gluconate, it was shown that the daily addition of dental chews to a dry diet resulted in lower gingivitis and calculus compared with a dry diet alone. Plus, the addition of chlorhexidine to the chew resulted in less plaque accumulation (Rawlings et al., 1998). Similarly, in the present study, all chew treatments resulted in lower calculus coverage and thickness, pocket depth and bleeding, plaque thickness, and halitosis compared with CT. Plaque coverage was lower in one chew treatment (GR) compared with CT. As discussed in our previous publication (Carroll et al., 2020), the ingredient composition and/or the shape of the dental chew may affect their hardness and abrasiveness. GR contains wheat gluten and has a semimoist consistency, providing chewing resistance and increased contact with the teeth. Additionally, GR contains fiber sources (oat fiber) that may also support a scrubbing effect during mastication of the dental chew. The hardness of GR (58.45 N) was lower compared with BC (140.5 N) and DL (61.21 N), however. The semimoist consistency and the fiber may be a justification for GR to be more effective in removing the oral plaque compared with other treatments.
The association between bacterial taxa and measures of oral health or periodontal disease is of interest, but few studies have investigated this relationship in dogs (Riggio et al., 2011; Davis et al., 2013; Di Bello et al., 2014; Wallis et al., 2015; Davis, 2016). A previous study collected SUB plaque from 223 dogs (72 with healthy gingiva, 77 with gingivitis, and 74 with mild periodontitis) and reported that Porphyromonas was the most abundant genus in all disease stages, particularly in health, along with Moraxella and Bergeyella. Peptostreptococcus, Actinomyces, and Peptostreptococcaceae were the most abundant genera in mild periodontitis. Additionally, a healthy canine plaque was reported to be dominated by Gram-negative bacterial species, whereas Gram-positive anaerobic species predominated in disease (Davis et al., 2013).
Another study collected SUB plaque from 52 miniature schnauzer dogs, reporting that a group of aerobic Gram-negative species, including B. zoohelcum, Moraxella sp., Pasteurellaceae sp., and N. shayeganii, decreased as a proportion as teeth progressed to mild periodontitis. Furthermore, Capnocytophaga cynodegmi, Corynebacterium, Capnocytophaga sp., Neisseria animaloris, Pasteurellaceae bacterium, N. shayeganii, Lautropia sp., and Desulfovibrio sp. also decreased in proportion as periodontal disease progressed. Bacterial associations that were positively associated with the development of early periodontitis were Peptostreptococcaceae bacterium COT-077, Clostridiales bacterium, Erysipelotrichaceae bacterium, Porphyromonas sp., Peptostreptococcus sp., and Treponema sp. (Wallis et al., 2015).
In another study, dental plaque was collected using sterile swabs from the gingivae of healthy dogs and animals with gingivitis, and SUB plaque was collected from dogs with periodontitis (three samples per group) for routine bacterial culture. It was identified that the prevalent species identified in the normal, gingivitis, and periodontitis groups were uncultured bacteria (12.5% of isolates), Bacteroides heparinolyticus/Pasteurella dagmatis (10.0%), and Actinomyces canis (19.4%), respectively, with the predominant species identified as Pseudomonas sp. (30.9% of clones analyzed), Porphyromonas cangingivalis (16.1%), and Desulfomicrobium orale (12.0%) in the normal, gingivitis, and periodontitis groups, respectively (Riggio et al., 2011). Another study aimed to evaluate the association of red-complex bacteria (Treponema denticola, Tannerella forsythia, and Porphyromonas gingivalis), which have a major role in the etiology of periodontal disease in humans, with periodontal disease in dogs, reporting that dogs with gingivitis or periodontitis were more likely to be infected with T. forsythia and P. gingivalis (Di Bello et al., 2014). In the present study, five of the previous health-associated genera (Actinomyces, Capnocytophaga, Bergeyella, Neisseria, and Moraxella) were also associated with a healthier mouth (lower OHS), and four of the previous disease-associated genera (Tannerella, Peptostreptococcus, Desulfomicrobium, and Treponema) were associated with periodontal disease (higher OHS). However, Desulfovibrio was associated with periodontal disease (higher OHS) instead of a healthier mouth.
Although some have associated microbiota and OHS, few have tested the efficacy of dental chews on oral microbiota. In a previous study that evaluated the effects of a dental chew on SUP plaque microbiota of dogs, researchers reported that the daily consumption of the chew resulted in an increase in the proportion of six health-associated taxa (Klebsiella, Propionibacterium, Catonella, Corynebacterium mustelae, Prevotella, and TM7) but only three disease-associated taxa (Parvimonas, Actinomyces, and Treponema) compared with controls. In contrast, eight disease-associated (Fretibacterium, Helcococcus, Clostridium, Desulfomicrobium orale, Anaerovorax, Bacterodia bacterium, Neisseria canis, and Pelistega) and only one health-associated taxa (Desulfovibrio) were shown to be increased as a proportion in control dogs than those fed an oral care chew (Ruparell et al., 2020).
In the present study, one disease-associated genus (Tannerella) was lower in plaque (SUB and SUP) samples in dogs fed all chew treatments than controls. Moreover, one disease-associated genus (Treponema) was lower in SUB and SAL samples of dogs fed all chew treatments than controls. One disease-associated genus (Porphyromonas) was lower in plaque (SUP and SUB) samples of dogs consuming DL than controls. Lastly, one disease-associated genus (Treponema) was lower in SUP samples of dogs consuming DL or GR. Tannerella is more associated with various forms of periodontal disease, including gingivitis and periodontitis, than with health. Some of the virulence factors that have been identified in Tannerella are their proteolytic enzymes, which can degrade host periodontal tissues, activate host degradative enzymes, modify host cell proteins to expose cryptotopes for bacterial colonization, and cleave components involved in innate and adaptive immunity, thus reducing host immunity and activating components involved in clotting and fibrinolysis (Dzink et al., 1988; Holt and Bramanti, 1991; Listgarten et al., 1993; Grossi et al., 1994, 1995; Tanner et al., 1998; Sharma et al., 2005; Tanner and Izard, 2006; Potempa and Pike, 2009; Sharma, 2010). Treponema is also associated with periodontitis, necrotizing ulcerative gingivitis, and acute pericoronitis. Treponema adheres to epithelial cells and fibroblasts as well to other bacteria, and their bacterial products can cause cell damage and the release of cellular deleterious factors to the periodontal environment, contributing to the progression of periodontal diseases (Dawson and Ellen, 1990; Weinberg and Holt, 1990; Haapasalo et al., 1991, 1996; Baehni et al., 1992; Keulers et al., 1993; Ellen et al., 1994; De Filippo et al., 1995; Riviere et al., 1996; Peters et al., 1999; Sela, 2001). Porphyromonas is predominant isolates from the plaque of dogs with gingivitis and periodontitis (Watson, 1994; Gorrel and Rawlings, 1996; Allaker et al., 1997; Isogai et al., 1999) and is the bacterial species most often associated with periodontal disease in dogs (Fournier et al., 2001). This organism possesses a number of pathogenic properties (fimbriae, proteases, hemagglutinins, and lipopolysaccharide), including the ability to adhere to and colonize the oral surfaces and to invade periodontal tissues (Hamada et al., 1998; Holt et al., 1999; Amano, 2003). Therefore, the lower relative abundance of those bacterial genera may indicate the beneficial use of dental chews to prevent the progression of periodontal diseases.
Four health-associated genera (Bergeyella, Neisseria, Capnocytophaga, and Moraxella) were higher in plaque (SUB and SUP) samples of dogs fed all chew treatments than controls. One health-associated genus (Lautropia) was also higher in SUP samples of dogs fed all chew treatments than in controls. Furthermore, one health-associated genus (Corynebacterium) was higher in all samples sites of dogs consuming DL than dogs in all other treatments and higher in SAL samples of dogs consuming BC. One health-associated genus (Actinomyces) was higher in SUB samples of dogs consuming DL than dogs consuming CT and higher in SAL samples of dogs consuming GR than controls. One health-associated genus (Neisseria) was higher in SAL samples of dogs consuming GR or DL than controls.
Alpha diversity represents species richness and may be measured by Faith’s phylogenetic diversity and the Shannon diversity index. A lower richness is represented by a lower Shannon diversity index (Shannon, 1948). Faith’s phylogenetic diversity is the sum of all branch lengths of the OTU in the sample (Faith, 2006), with more unique OTU having a higher Faith’s phylogenetic diversity. Past studies have shown that OTU and Shannon diversity index were higher in dogs with mild periodontitis compared with healthy dogs (Davis et al., 2013), although a difference is not always observed (Wallis et al., 2015). In humans, periodontal disease is correlated with an increase in microbial community diversity (Abusleme et al., 2013; Pyysalo et al., 2019). In contrast to a previous study that was unable to show significant differences in alpha diversity between the dental chew and control groups (Ruparell et al., 2020), the data from the present study demonstrated lower diversity indices of dogs consuming dental chews than controls. This finding suggests that a 28-d time frame is sufficient to show a benefit from the consumption of dental chews. This reduction in alpha diversity from consumption of dental chew would be deemed as a desirable outcome.
Beta diversity is the overall change in the community between samples, which can be assessed using weighted (quantitative) and unweighted (qualitative; presence or absence) measures (Lozupone et al., 2007). In a previous study, a discrete clustering of healthy and mild periodontitis samples was observed, while gingivitis samples overlapped both healthy and mild periodontitis clusters (Davis et al., 2013). In the present study, it was possible to identify distinct clusters of controls and chew treatments. This again demonstrates that 28 d of chew consumption is capable of changing the beta diversity of oral microbiota populations, significantly modifying the microbiota and making it distinct from that of control animals.
In a previous study using six toy Chihuahua dogs, different diet types (dry, soft, and dental diet), and preventive means of periodontitis (toothbrushing and enzymatic chewing strips) for 8 wk showed that none of the diets and dental hygiene products provided full mouth elimination of periodontal health problems. Dental chewing strips significantly decreased dental plaque, calculus, and gingivitis scores, but only on carnassial teeth (Capík, 2010). In the present study, all dental chews were able to reduce halitosis and calculus, plaque, and pocket scores, although it was not able to reduce gingivitis scores. In our previous study (Carroll et al., 2020), the maxillary incisor 3, canine, premolar 3, premolar 4, molar 1 and the mandibular canine, premolar 3, premolar 4, and molar 1 were scored. Plaque coverage and thickness were lower for dogs consuming DL and GR compared with CT, calculus coverage was lower for all chews compared with CT, and halitosis on day 14 was lower for dogs consuming DL compared with control, and on day 27 was lower for all chew treatments compared with control. Calculus thickness and gingivitis were not affected by treatment, however. Although diets or chews may be somewhat effective, they do not replace regular brushing and cleanings. Therefore, properly timed professional dental prophylaxis plays an important role in the prevention of periodontal diseases (Polkowska and Orzelski, 2003; Fichtel et al., 2005; Capík, 2010).
In conclusion, this study has shown that shifts in microbiota, together with OHS, can be used to demonstrate the efficacy of dental chews, revealing that bacterial communities within plaque and saliva can be influenced toward health within a short period of time. In all collection sites, control dogs had a higher presence of one or more potentially pathogenic bacteria (Porphyromonas, Anaerovorax, Desulfomicrobium, Tannerella, and Treponema) and a lower presence of one or more genera associated with oral health (Neisseria, Corynebacterium, Capnocytophaga, Actinomyces, Lautropia, Bergeyella, and Moraxella) than dogs fed chew treatments. These results suggest that the dental chews tested in this study may aid in reducing the risk of periodontal disease in dogs.
Acknowledgments
The funding for this study was provided by the United States Department of Agriculture Hatch Grant (ILLU-538-937). We want to thank Kelly M. Sieja for assistance in salivary and plaque sample collection.
Glossary
Abbreviations
- BC
diet + Bones & Chews Dental Treats (Chewy Inc. Dania Beach FL)
- BW
body weight
- CT
diet only (control)
- DL
diet + Dr. Lyon’s Grain-Free Dental Treats (Dr. Lyon’s, LLC, Dania Beach FL)
- GR
diet + Greenies Dental Treats (Mars Petcare US Franklin TN)
- OHS
oral health score
- OTU
operational taxonomic units
- PCoA
principal coordinates analysis
- SAL
salivary source
- SUB
subgingival source
- SUP
supragingival source
Conflict of interest statement
All authors have no conflicts of interest.
Literature Cited
- Abusleme, L., Dupuy A. K., Dutzan N., Silva N., Burleson J. A., Strausbaugh L. D., Gamonal J., and Diaz P. I.. . 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7:1016–1025. doi: 10.1038/ismej.2012.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adepu, R., Raghavender K. B. P., Kumar V. G., and Ramesh N.. . 2018. A clinical study on the incidence of periodontal diseases in dogs and their surgical management. Pharm. Innov. J. 7:290–292. [Google Scholar]
- Albuquerque, C., Morinha F., Requicha J., Martins T., Dias I., Guedes-Pinto H., Bastos E., and Viegas C.. . 2012. Canine periodontitis: the dog as an important model for periodontal studies. Vet. J. 191:299–305. doi: 10.1016/j.tvjl.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Allaker, R. P., de Rosayro R., Young K. A., and Hardie J. M.. . 1997. Prevalence of Porphyromonas and Prevotella species in the dental plaque of dogs. Vet. Rec. 140:147–148. doi: 10.1136/vr.140.6.147 [DOI] [PubMed] [Google Scholar]
- Amano, A. 2003. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J. Periodontol. 74:90–96. doi: 10.1902/jop.2003.74.1.90 [DOI] [PubMed] [Google Scholar]
- Baehni, P. C., Song M., McCulloch C. A., and Ellen R. P.. . 1992. Treponema denticola induces actin rearrangement and detachment of human gingival fibroblasts. Infect. Immun. 60:3360–3368. doi: 10.1128/IAI.60.8.3360-3368.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. Y., and McGenity P.. . 2005. Effective periodontal disease control using dental hygiene chews. J. Vet. Dent. 22:16–19. doi: 10.1177/089875640502200102 [DOI] [PubMed] [Google Scholar]
- Butković, V., Šehič M., Stanin D., Šimpraga M., Capak D., and Kos J.. . 2001. Dental diseases in dogs: a retrospective study of radiological data. Acta Vet. Brno. 70:203–208. doi: 10.2754/avb200170020203 [DOI] [Google Scholar]
- Capík, I. 2010. Periodontal health vs. various preventive means in toy dog breeds. Acta Vet. Brno. 79:637–645. doi: 10.2754/avb201079040637 [DOI] [Google Scholar]
- Caporaso, J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., . et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., . et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. doi: 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, M. Q., Oba P. M., Sieja K. M., Alexander C., Lye L., de Godoy M. R. C., He F., Somrak A. J., Keating S. C. J., Sage A. M., . et al. 2020. Effects of novel dental chews on oral health outcomes and halitosis in adult dogs. J. Anim. Sci. 98:1–7. doi: 10.1093/jas/skaa274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, D. E., Kelman M., and Perkins N.. . 2011. Effectiveness of a vegetable dental chew on periodontal disease parameters in toy breed dogs. J. Vet. Dent. 28:230–235. doi: 10.1177/089875641102800403 [DOI] [PubMed] [Google Scholar]
- Davis, E. M. 2016. Gene sequence analyses of the healthy oral microbiome in humans and companion animals. J. Vet. Dent. 33:97–107. doi: 10.1177/0898756416657239 [DOI] [PubMed] [Google Scholar]
- Davis, I. J., Wallis C., Deusch O., Colyer A., Milella L., Loman N., and Harris S.. . 2013. A cross-sectional survey of bacterial species in plaque from client owned dogs with healthy gingiva. Semple, M. G., editor. PLoS One. 8:1–12. doi: 10.1371/journal.pone.0083158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, J. R., and Ellen R. P.. . 1990. Tip-oriented adherence of Treponema denticola to fibronectin. Infect. Immun. 58:3924–3928. doi: 10.1128/IAI.58.12.3924-3928.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo, A. B., Ellen R. P., and McCulloch C. A.. . 1995. Induction of cytoskeletal rearrangements and loss of volume regulation in epithelial cells by Treponema denticola. Arch. Oral Biol. 40:199–207. doi: 10.1016/0003-9969(95)98809-d [DOI] [PubMed] [Google Scholar]
- Deng, P., and Swanson K. S.. . 2015. Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Br. J. Nutr. 113(Suppl):S6–17. doi: 10.1017/S0007114514002943 [DOI] [PubMed] [Google Scholar]
- DeSantis, T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G. L.. . 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst, F. E., Klein E. A., Thompson E. C., Blanton J. M., Chen T., Milella L., Buckley C. M., Davis I. J., Bennett M. L., and Marshall-Jones Z. V.. . 2012. The canine oral microbiome. Ravel, J., editor. PLoS One. 7:e36067. doi: 10.1371/journal.pone.0036067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bello, A., Buonavoglia A., Franchini D., Valastro C., Ventrella G., Greco M. F., and Corrente M.. . 2014. Periodontal disease associated with red complex bacteria in dogs. J. Small Anim. Pract. 55:160–163. doi: 10.1111/jsap.12179 [DOI] [PubMed] [Google Scholar]
- Dzink, J. L., Socransky S. S., and Haffajee A. D.. . 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316–323. doi: 10.1111/j.1600-051x.1988.tb01590.x [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Ellen, R. P., Dawson J. R., and Yang P. F.. . 1994. Treponema denticola as a model for polar adhesion and cytopathogenicity of spirochetes. Trends Microbiol. 2:114–119. doi: 10.1016/0966-842x(94)90597-5 [DOI] [PubMed] [Google Scholar]
- Faith, D. P. 2006. The role of the phylogenetic diversity measure, PD, in bio-informatics: getting the definition right. Evol. Bioinform Online. 2:117693430600200. doi: 10.1177/117693430600200008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedi, P. F., Vernino A. R., and Gray J. L.. . 1985. Etiology of periodontal disease. In: Fedi, P. F., editor. The periodontic syllabus. Philadelphia (PA):Lea & Febiger; p. 13–18. [Google Scholar]
- Fichtel, T., Crha M., and Langerová E.. . 2005. The efficacy of methods used for enamel treatment in the dog: direct impact of damaged surface on the plaque reestablishment – REM study. In: Pavlica, Z., editor. Proceedings of the 14th European Congress of Veterinary Dentistry;September 22 to 25, 2005; Ljubljana (Slovenia): Veterinary University of Ljubljana; p. 81–85. [Google Scholar]
- Fournier, D., Mouton C., Lapierre P., Kato T., Okuda K., and Ménard C.. . 2001. Porphyromonas gulae sp. nov., an anaerobic, gram-negative coccobacillus from the gingival sulcus of various animal hosts. Int. J. Syst. Evol. Microbiol. 51(Pt 3): 1179–1189. doi: 10.1099/00207713-51-3-1179 [DOI] [PubMed] [Google Scholar]
- Gad, T. 1968. Periodontal disease in dogs. J. Periodontal Res. 3: 268–272. doi: 10.1111/j.1600-0765.1968.tb01929.x [DOI] [PubMed] [Google Scholar]
- Garanayak, N., Das M., Patra R. C., Biswal S., and Panda S. K.. . 2019. Effect of age on dental plaque deposition and its control by ultrasonic scaling, dental hygiene chew, and chlorhexidine (0.2%w/v) in dogs. Vet. World 12:1872–1876. doi: 10.14202/vetworld.2019.1872-1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genco, R. J. 1990. Pathogenesis and host responses in periodontal disease. In: Genco, R., Goldma H., and Cohen D., editors. Contemporary periodontics. Vol. 14. St. Louis (MO): The CV Mosby Company; p. 184–193. [Google Scholar]
- Gomes, B. P., Berber V. B., Kokaras A. S., Chen T., and Paster B. J.. . 2015. Microbiomes of endodontic-periodontal lesions before and after chemomechanical preparation. J. Endod. 41:1975–1984. doi: 10.1016/j.joen.2015.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrel, C., and Bierer T. L.. . 1999. Long-term effects of a dental hygiene chew on the periodontal health of dogs. J. Vet. Dent. 16:109–113. doi: 10.1177/089875649901600302 [DOI] [PubMed] [Google Scholar]
- Gorrel, C., and Rawlings J.. . 1996. The role of a “dental hygiene chew” in maintaining periodontal health in dogs. J. Vet. Dent. 1:31–34. [Google Scholar]
- Gorrel, C., Warrick J., and Bierer T. L.. . 1999. Effect of a new dental hygiene chew on periodontal health in dogs. J. Vet. Dent. 16:77–81. doi: 10.1177/089875649901600203 [DOI] [PubMed] [Google Scholar]
- Grossi, S. G., Genco R. J., Machtei E. E., Ho A. W., Koch G., Dunford R., Zambon J. J., and Hausmann E.. . 1995. Assessment of risk for periodontal disease. II. Risk indicators for alveolar bone loss. J. Periodontol. 66:23–29. doi: 10.1902/jop.1995.66.1.23 [DOI] [PubMed] [Google Scholar]
- Grossi, S. G., Zambon J. J., Ho A. W., Koch G., Dunford R. G., Machtei E. E., Norderyd O. M., and Genco R. J.. . 1994. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J. Periodontol. 65:260–267. doi: 10.1902/jop.1994.65.3.260 [DOI] [PubMed] [Google Scholar]
- Haapasalo, M., Hannam P., McBride B. C., and Uitto V. J.. . 1996. Hyaluronan, a possible ligand mediating Treponema denticola binding to periodontal tissue. Oral Microbiol. Immunol. 11:156–160. doi: 10.1111/j.1399-302x.1996.tb00351.x [DOI] [PubMed] [Google Scholar]
- Haapasalo, M., Singh U., McBride B. C., and Uitto V. J.. . 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230–4237. doi: 10.1128/IAI.59.11.4230-4237.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, S., Amano A., Kimura S., Nakagawa I., Kawabata S., and Morisaki I.. . 1998. The importance of fimbriae in the virulence and ecology of some oral bacteria. Oral Microbiol. Immunol. 13:129–138. doi: 10.1111/j.1399-302x.1998.tb00724.x [DOI] [PubMed] [Google Scholar]
- Hamp, S.-E., Olsson S.-E., Farsø-Madsen K., Viklands P., and Fornell J.. . 1984. A macroscopic and radiological investigation of dental diseases of the dog. Vet. Radiol. 25:86–92. doi: 10.1111/j.1740-8261.1984.tb01916.x [DOI] [Google Scholar]
- Harvey, C., Serfilippi L., and Barnvos D.. . 2015. Effect of frequency of brushing teeth on plaque and calculus accumulation, and gingivitis in dogs. J. Vet. Dent. 32:16–21. doi: 10.1177/089875641503200102 [DOI] [PubMed] [Google Scholar]
- Hennet, P. R., and Harvey C. E.. . 1991. Aerobes in periodontal disease in the dog: a review. J. Vet. Dent. 8:9–11. doi: 10.1177/089875649100800103 [DOI] [PubMed] [Google Scholar]
- Hennet, P., Servet E., Salesse H., and Soulard Y.. . 2006a. Evaluation of the Logan & Boyce plaque index for the study of dental plaque accumulation in dogs. Res. Vet. Sci. 80:175–180. doi: 10.1016/j.rvsc.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Hennet, P., Servet E., and Venet C.. . 2006b. Effectiveness of an oral hygiene chew to reduce dental deposits in small breed dogs. J. Vet. Dent. 23:6–12. doi: 10.1177/089875640602300101 [DOI] [PubMed] [Google Scholar]
- Hoffmann, T., and Gaengler P.. . 1996. Epidemiology of periodontal disease in poodles. J. Small Anim. Pract. 37:309–316. doi: 10.1111/j.1748-5827.1996.tb02396.x [DOI] [PubMed] [Google Scholar]
- Holt, S. C., and Bramanti T. E.. . 1991. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit. Rev. Oral Biol. Med. 2:177–281. doi: 10.1177/10454411910020020301 [DOI] [PubMed] [Google Scholar]
- Holt, S. C., Kesavalu L., Walker S., and Genco C. A.. . 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168–238. doi: 10.1111/j.1600-0757.1999.tb00162.x [DOI] [PubMed] [Google Scholar]
- Isogai, H., Kosako Y., Benno Y., and Isogai E.. . 1999. Ecology of genus Porphyromonas in canine periodontal disease. J. Vet. Med. Ser. B. 46:467–473. doi: 10.1046/j.1439-0450.1999.00249.x [DOI] [PubMed] [Google Scholar]
- Keulers, R. A., Maltha J. C., Mikx F. H., and Wolters-Lutgerhorst J. M.. . 1993. Attachment of Treponema denticola strains to monolayers of epithelial cells of different origin. Oral Microbiol. Immunol. 8:84–88. doi: 10.1111/j.1399-302x.1993.tb00550.x [DOI] [PubMed] [Google Scholar]
- Klein, T. 2000. Predisposing factors and gross examination findings in periodontal disease. Clin. Tech. Small Anim. Pract. 15:189–196. doi: 10.1053/svms.2000.22244 [DOI] [PubMed] [Google Scholar]
- Kortegaard, H. E., Eriksen T., and Baelum V.. . 2008. Periodontal disease in research beagle dogs – an epidemiological study. J. Small Anim. Pract. 49:610–616. doi: 10.1111/j.1748-5827.2008.00609.x [DOI] [PubMed] [Google Scholar]
- Kyllar, M., and Witter K.. . 2005. Prevalence of dental disorders in pet dogs. Vet. Med. (Praha). 50:496–505. doi: 10.17221/5654-vetmed [DOI] [Google Scholar]
- de Lacerda, M. S., and Alessi A. C.. . 2015. Effect of ultrasonic scaling of teeth involved with periodontal disease in dogs: a scanning electron microscopic study. Biosci. J. 31:1488–1495. doi: 10.14393/BJ-v31n5a2015-26502 [DOI] [Google Scholar]
- Lindhe, J. 1989. Pathogenesis of plaque-associated periodontal disease. In: Lindhe, J., editor. Textbook of clinical periodontology. 2nd ed. Copenhagen (Denmark):WB Saunders; p. 189–205. [Google Scholar]
- Listgarten, M. A., Lai C. H., and Young V.. . 1993. Microbial composition and pattern of antibiotic resistance in subgingival microbial samples from patients with refractory periodontitis. J. Periodontol. 64:155–161. doi: 10.1902/jop.1993.64.3.155 [DOI] [PubMed] [Google Scholar]
- Logan, E. I. 2006. Dietary influences on periodontal health in dogs and cats. Vet. Clin. North Am. Small Anim. Pract. 36:1385–401, ix. doi: 10.1016/j.cvsm.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Loomer, P. M. 2004. Microbiological diagnostic testing in the treatment of periodontal diseases. Periodontol. 2000 34:49–56. doi: 10.1046/j.0906-6713.2002.003424.x [DOI] [PubMed] [Google Scholar]
- Lozupone, C. A., Hamady M., Kelley S. T., and Knight R.. . 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1585. doi: 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C., and Knight R.. . 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E. M., Armstrong P. J., Kirk C. A., Kolar L. M., and Klausner J. S.. . 1999. Health status and population characteristics of dogs and cats examined at private veterinary practices in the United States. J. Am. Vet. Med. Assoc. 214:1336–1341. [PubMed] [Google Scholar]
- Mandel, E., Machtou P., and Torabinejad M.. . 1993. Clinical diagnosis and treatment of endodontic and periodontal lesions. Quintessence Int. 24:135–139. [PubMed] [Google Scholar]
- Mühlemann, H. R., and Son S.. . 1971. Gingival sulcus bleeding – a leading symptom in initial gingivitis. Helv. Odontol. Acta 15:107–113 [PubMed] [Google Scholar]
- O′Neill, D. G., Church D. B., McGreevy P. D., Thomson P. C., and Brodbelt D. C.. . 2014. Prevalence of disorders recorded in dogs attending primary-care veterinary practices in England. Rosenfeld, C. S., editor. PLoS One. 9:e90501. doi: 10.1371/journal.pone.0090501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba, P. M., Devito F. C., Santos J. P. F., Stipp R. N., Gomes M. de O. S., Carciofi A. C., and Brunetto M. A.. . 2018. Effects of passive immunization by anti-gingipain IgY on the oral health of cats fed kibble diets. J. Vet. Dent. 35:275–280. doi: 10.1177/0898756418814010 [DOI] [Google Scholar]
- Peters, S. R., Valdez M., Riviere G., and Thomas D. D.. . 1999. Adherence to and penetration through endothelial cells by oral treponemes. Oral Microbiol. Immunol. 14:379–383. doi: 10.1034/j.1399-302x.1999.140609.x [DOI] [PubMed] [Google Scholar]
- Polkowska, I., and Orzelski M.. . 2003. The non-surgical treatment of periodontal disease in carnivores and maintenance therapy. In: Gracias, M., editor. Proceedings of the 12th European Congress of Veterinary Dentistry;September 25 to 28, 2003; Pisa (Italy): University of Pisa; p. 75–76. [Google Scholar]
- Potempa, J., and Pike R. N.. . 2009. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 1:70–87. doi: 10.1159/000181144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyysalo, M. J., Mishra P. P., Sundström K., Lehtimäki T., Karhunen P. J., and Pessi T.. . 2019. Increased tooth brushing frequency is associated with reduced gingival pocket bacterial diversity in patients with intracranial aneurysms. PeerJ. 7:e6316. doi: 10.7717/peerj.6316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quest, B. W. 2013. Oral health benefits of a daily dental chew in dogs. J. Vet. Dent. 30:84–87. doi: 10.1177/089875641303000203 [DOI] [PubMed] [Google Scholar]
- Rawlings, J. M., Gorrel C., and Markwell P. J.. . 1998. Effect on canine oral health of adding chlorhexidine to a dental hygiene chew. J. Vet. Dent. 15:129–134. doi: 10.1177/089875649801500303 [DOI] [PubMed] [Google Scholar]
- Riggio, M. P., Lennon A., Taylor D. J., and Bennett D.. . 2011. Molecular identification of bacteria associated with canine periodontal disease. Vet. Microbiol. 150:394–400. doi: 10.1016/j.vetmic.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Riviere, G. R., Thompson A. J., Brannan R. D., McCoy D. E., and Simonson L. G.. . 1996. Detection of pathogen-related oral spirochetes, Treponema denticola, and Treponema socranskii in dental plaque from dogs. J. Vet. Dent. 13:135–138. doi: 10.1177/089875649601300401 [DOI] [PubMed] [Google Scholar]
- Rober, M., Quirynen M., Haffajee A. D., Schepers E., and Teughels W.. . 2008. Intra-oral microbial profiles of beagle dogs assessed by checkerboard DNA–DNA hybridization using human probes. Vet. Microbiol. 127:79–88. doi: 10.1016/j.vetmic.2007.08.007 [DOI] [PubMed] [Google Scholar]
- Ruparell, A., Warren M., Staunton R., Deusch O., Dobenecker B., Wallis C., O′Flynn C., McGenity P., and Holcombe L. J.. . 2020. Effect of feeding a daily oral care chew on the composition of plaque microbiota in dogs. Res. Vet. Sci. 132:133–141. doi: 10.1016/j.rvsc.2020.05.001 [DOI] [PubMed] [Google Scholar]
- Segata, N., Haake S. K., Mannon P., Lemon K. P., Waldron L., Gevers D., Huttenhower C., and Izard J.. . 2012. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13:R42. doi: 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela, M. N. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399–413. doi: 10.1177/10454411010120050301 [DOI] [PubMed] [Google Scholar]
- Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- Sharma, A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000 54:106–116. doi: 10.1111/j.1600-0757.2009.00332.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A., Inagaki S., Honma K., Sfintescu C., Baker P. J., and Evans R. T.. . 2005. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J. Dent. Res. 84:462–467. doi: 10.1177/154405910508400512 [DOI] [PubMed] [Google Scholar]
- Shofiqur, R. A. K. M., Ibrahim E.-S. M., Isoda R., Umeda K., Nguyen V. S., and Kodama Y.. . 2011. Effect of passive immunization by anti-gingipain IgY on periodontal health of dogs. Vet. Sci. Dev. 1:8. doi: 10.4081/vsd.2011.e8 [DOI] [Google Scholar]
- Stella, J. L., Bauer A. E., and Croney C. C.. . 2018. A cross-sectional study to estimate prevalence of periodontal disease in a population of dogs (Canis familiaris) in commercial breeding facilities in Indiana and Illinois. Staffieri, F., editor. PLoS One. 13:e0191395. doi: 10.1371/journal.pone.0191395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, A. C., and Izard J.. . 2006. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000 42: 88–113. doi: 10.1111/j.1600-0757.2006.00184.x [DOI] [PubMed] [Google Scholar]
- Tanner, A., Maiden M. F., Macuch P. J., Murray L. L., and R. L.Kent, Jr. 1998. Microbiota of health, gingivitis, and initial periodontitis. J. Clin. Periodontol. 25:85–98. doi: 10.1111/j.1600-051x.1998.tb02414.x [DOI] [PubMed] [Google Scholar]
- Tatakis, D. N., and Kumar P. S.. . 2005. The etiology and pathogenesis of periodontitis. Dent. Clin. North Am. 49:491–516. doi: 10.1016/j.cden.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Wallis, C., Marshall M., Colyer A., O′Flynn C., Deusch O., and Harris S.. . 2015. A longitudinal assessment of changes in bacterial community composition associated with the development of periodontal disease in dogs. Vet. Microbiol. 181:271–282. doi: 10.1016/j.vetmic.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Watson, A. D. 1994. Diet and periodontal disease in dogs and cats. Aust. Vet. J. 71:313–318. doi: 10.1111/j.1751-0813.1994.tb00905.x [DOI] [PubMed] [Google Scholar]
- Weinberg, A., and Holt S. C.. . 1990. Interaction of Treponema denticola TD-4, GM-1, and MS25 with human gingival fibroblasts. Infect. Immun. 58:1720–1729. doi: 10.1128/IAI.58.6.1720-1729.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]