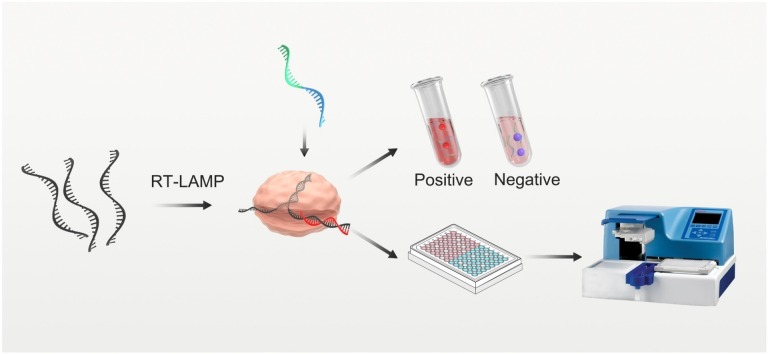
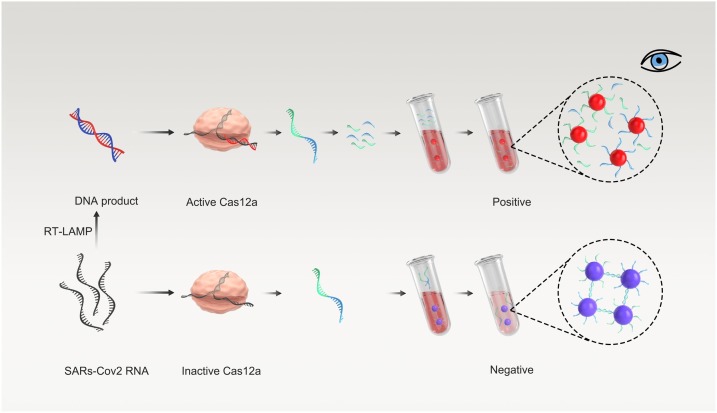
Graphical abstract
Abbreviations: COVID-19, Corona Virus Disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; RT-qPCR, reverse transcription-real time quantitative PCR; POCT, point of care testing; LAMP, loop-mediated isothermal amplification; RPA, recombinase polymerase amplification; HCRs, hybridization chain reactions; NMPA, the Chinese National Medical Products Administration; FDA, American Food and Drug Administration; CRISPR, clustered regularly interspaced short palindromic repeats; AuNP, gold nanoparticle; TCEP, Tris(2-carboxyethyl) phosphine; DMEM, Dulbecco’s modified Eagle’s medium; TEM, transmission electron microscopy
Keywords: Coronavirus disease, Loop-mediated isothermal amplification, CRISPR/Cas, Gold nanoparticle, High-throughput on-site detection
Abstract
The outbreak of corona virus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to a global pandemic. The high infectivity of SARS-CoV-2 highlights the need for sensitive, rapid and on-site diagnostic assays of SARS-CoV-2 with high-throughput testing capability for large-scale population screening. The current detection methods in clinical application need to operate in centralized labs. Though some on-site detection methods have been developed, few tests could be performed for high-throughput analysis. We here developed a gold nanoparticle-based visual assay that combines with CRISPR/Cas12a-assisted RT-LAMP, which is called Cas12a-assisted RT-LAMP/AuNP (CLAP) assay for rapid and sensitive detection of SARS-CoV-2. In optimal condition, we could detect down to 4 copies/μL of SARS-CoV-2 RNA in 40 min. by naked eye. The sequence-specific recognition character of CRISPR/Cas12a enables CLAP a superior specificity. More importantly, the CLAP is easy for operation that can be extended to high-throughput test by using a common microplate reader. The CLAP assay holds a great potential to be applied in airports, railway stations, or low-resource settings for screening of suspected people. To the best of our knowledge, this is the first AuNP-based colorimetric assay coupled with Cas12 and RT-LAMP for on-site diagnosis of COVID-19. We expect CLAP assay will improve the current COVID-19 screening efforts, and make contribution for control and mitigation of the pandemic.
1. Introduction
The outbreak of coronavirus disease 2019 (COVID-19) caused a significant public health concern. Developing a fast and reliable diagnostic method is critical in tracking of pathogen (SARS-CoV-2, severe acute respiratory syndrome coronavirus) transmission, and then interrupt the spread [1]. Large-scale population screening is an effective way to control the pandemic [2]. Until now, reverse transcription-real time quantitative PCR (RT-qPCR) is the golden standard for the diagnosis of SARS-CoV-2 [3]. However, RT-qPCR requires thermal cycling with complicated equipment and trained personnel, and is always performed in the central lab, which is challenging to be applied for point of care testing (POCT). Thus, there is an urgent need for achieving detection of nucleic acid in a rapid, sensitive way based on decentralized diagnostic technologies for diagnosis of SARS-CoV-2.
To meet this demand, isothermal amplification methods, such as loop-mediated isothermal amplification (LAMP) [4,5], recombinase polymerase amplification (RPA) [6], and hybridization chain reactions (HCRs) [7] have been applied to detect the SARS-CoV-2 RNA. Reverse transcription LAMP (RT-LAMP) is especially practical that COVID-19 nucleic acid test kits based on RT-LAMP have been approved by the Chinese National Medical Products Administration (NMPA) and American Food and Drug Administration (FDA). Especially, clustered regularly interspaced short palindromic repeats (CRISPR) and associated protein (CRISPR/Cas) system have been combined with RT-LAMP for molecular diagnosis of COVID-19, and was reported to improve the sensitivity and specificity [[8], [9], [10], [11]]. Cas13a-based SHERLOCK system was first developed by Zhang’s group to detect the Zika virus. Cas12a-based DETECTR system and HOLMES system have also been reported and used to detect viral nucleic acid [[12], [13], [14]]. The nonspecific trans-cleavage activity of Cas12 or Cas13 nuclease can be activated after the recognition of specific target nucleic acids, and thus to cleave the reporter probe [13,15]. Most of these methods used fluorescent probes, which need a special instrument to monitor fluorescent signal [16,17].
Gold nanoparticle (AuNP) has been used for visual assay due to its strong molar absorption coefficient [[18], [19], [20], [21], [22]]. AuNP-based colorimetric method utilizes minimum lab instrument and is promising to be applied in POCT. Cas12a was first coupled with RPA for COVID-19 diagnosis, using AuNP probe for reading out [23]. Then, thermostable Cas12b was employed to combine with RT-LAMP to detect COVID-19 in one pot [8]. The Cas12-based methods relied on lateral flow assays [9,24], centrifuge-based method [25], or magnetic pull-down operation [23], which were difficult for high-throughput tests.
Cas13a was also used for amplification-free detection of COVID-19. Though the fluorescent signal could be collected by a smartphone, this method needs to keep recording the signal for 30 min, which is not suitable for high-throughput screening [26]. A number of other CRISPR/Cas-based detection methods for COVID-19 based on microfluidic chip [27] and fluorescence [28] have also been developed. However, each of them still has its shortcomings including complicated manipulation and time-consuming procedures, or requirement for elaborate instruments. Thus, it is still an unmet need for developing a rapid, sensitive, and cost-effective method, which is capable of high-throughput tests for large-scale population screening.
Here, we have developed an AuNP-based visual assay that combines with Cas12a-assisted RT-LAMP amplification, which is called Cas12a-assisted RT-LAMP/AuNP (CLAP) assay for rapid and sensitive detection of SARS-CoV-2 RNA. The CLAP assay is convenient for operation, which could be extended to high-throughput testing. In our strategy, two AuNP probes which were modified with different DNA (DNA1 and DNA2) could be crosslinked by a linker-ssDNA, resulting in color change (Fig. 1). In the presence of the virus RNA, the specific sequence of viral N gene is reverse-transcribed and amplified by RT-LAMP. The resulting amplicon is sequence-specific recognized by predesigned Cas12a/gRNA. Then the trans-cleavage activity of the Cas12a enzyme could be activated, resulting in the cleavage of linker-ssDNA. Therefore, the AuNP probes could not be cross-linked and the color of AuNPs is still red. While in the absence of virus RNA, the linker-ssDNA, which is designed to complementarily hybridize with AuNP-DNA, will crosslink the AuNP-DNA1/2 mixture and the color will change from red to purple, that can be observed by naked eye or measured by UV–vis spectrum. As far as we know, this is the first AuNP-based colorimetric assay coupled with Cas12 and RT-LAMP for on-site diagnosis of COVID-19.
Fig. 1.
Schematic of the CLAP assay for detecting SARS-CoV-2 by the naked-eye readout.
2. Materials and methods
2.1. Materials
AuNP was obtained from Suzhou Tanfeng Graphene Technology Co., Ltd (Jiangsu, China). Nucleic acid sequences (Table S1) were ordered from Sangon Biotech (Shanghai, China). WarmStart® LAMP Kit (DNA & RNA), Bst 3.0 DNA Polymerase, EnGen® Lba Cas12a (Cpf1) and NEBuffer™ 2.1 were obtained from New England Biolabs (Beijing, China). Tris(2-carboxyethyl) phosphine (TCEP) was obtained from Aladdin (Shanghai, China). SYBR Green Ⅰ was purchased from Solarbio (Beijing, China). BM2000 + DNA Marker was obtained from Biomed (Beijing, China). DEPC water (DNase/RNase free) was obtained from Beyotime (Shanghai, China). TE Buffer and GeneJET RNA Purification Kit were obtained from Thermo Fisher Scientific (Shanghai, China). COVID-19 (Corona Virus Disease 2019) RNA reference material (high concentration) was obtained from the National Institute of Metrology of China.
2.2. Bioconjugation of DNA on AuNPs
DNA functionalized AuNPs were prepared by salt aging method: first, the sulfhydryl DNA1 and DNA2 were reduced by mixing with TCEP at the ratio of 1:100 for 30 min at room temperature, respectively. The AuNPs were incubated with pretreated sulfhydryl DNA at the ratio of 1:200 overnight at room temperature, 2 M of sodium phosphate buffer (2 M of NaCl, 200 mM of Na2HPO4 and NaH2PO4, pH 7.4) was added to the mixture (a total of four salting steps and each time interval is not less than 1 h) to reach a final concentration of 0.2 M. The mixture was incubated overnight at room temperature. Then, the mixture was washed 3 times with 0.01 M Tris-HCl buffer (pH 7.4) through centrifugation (12,000 rpm, 20 min) to remove excess DNA, and the precipitation was dispersed in 0.01 M sodium phosphate buffer (pH 7.4), stored at 4℃.
The AuNP/DNA ratio was calculated using the following method: The UV–vis spectrum of the naked AuNPs and functionalized AuNPs were measured using a Microvolume UV–vis Spectrophotometer (Thermo Fisher, Nanodrop one, China). The extinction coefficients of DNA strands at 260 nm and the AuNPs at 520 nm were used to calculate the ratio. The calculated loading corresponded to ≈75 v-DNAs per AuNP.
2.3. Crosslink of AuNPs by linker-ssDNA
The length of linker-ssDNA was optimized for better AuNP-based visual detection. The assay was performed with 5 nM AuNP-DNA1, 5 nM AuNP-DNA2 and 250 nM linker-ssDNA of different lengths, linker-ssDNA1, linker-ssDNA2, linker-ssDNA3 in a 50 μL reaction system. 5 μL of saturated NaCl solution was added to the reaction to accelerate the combination of AuNP-DNA1/2 and linker-ssDNA. After incubation at room temperature for 5 min, the absorbance of the mixture was measured from 190 nm to 850 nm using a spectrophotometer. Transmission Electron Microscope (Hitachi-HT7800, Japan) was used to confirm whether AuNP was crosslinked by linker-ssDNA.
2.4. Activation of Cas12a/gRNA by targeting DNA
In order to evaluate how much DNA can activate the Cas12a/gRNA, a section of dsDNA and ssDNA of the N gene of SARS-CoV-2 was selected for Cas12a/gRNA fluorescence and visual detection. The reaction was divided into two groups: (1) fluorescence group, in which reporter DNA was added to the reaction system to monitor the reaction dynamics by fluorescence signal; (2) visual detection group, in which linker-ssDNA2 was added to the reaction system to observe the color change by naked eyes. The remaining reaction conditions of the two groups were consistent. 400 nM Cas12a was pre-assembled with 500 nM gRNA in 1× NEBuffer 2.1 for 10 min. at 37 °C. The assays were performed with different concentrations of dsDNA and ssDNA, 500 nM reporter DNA/ linker-ssDNA2, 1× NeBuffer 2.1, 50 nM Cas12a and 62.5 nM gRNA in a 25 μL volume. The reaction condition was set at 37℃, and fluorescence was measured every 1.5 min in the fluorescence group, the experiment was repeated three times. The vision detection group was finally mixed with the pre-mixed AuNP-DNA1/2 solution and then colorimetric detection was performed as described above.
2.5. Real-time RT-LAMP
Reverse transcription amplification reactions were based on the WarmStart® LAMP kit (DNA & RNA) to convert viral RNA into DNA, which has an optimal temperature of 65℃. RNA sequences derived from influenza virus (Flu-HA gene) were used as the negative control. RT-LAMP was prepared as suggested by New England Biolabs with WarmStart LAMP 1× Master Mix, 1× primer mixture (0.2 μM for F3 and B3, 1.6 μM for forward inner and backward inner primers, 0.8 μM for loop forward and loop backward primers), 0.5 × SYBR Green Ⅰ and RNA in a 25 μL volume. RT-LAMP reactions were performed at 65℃ on a Real-time PCR machine (Bio-Rad, CFX96™ Real-Time System, America) with fluorescent measurements every 1.5 min. 2.5 μL product was used for fluorescence and visual detection of Cas12a/gRNA, and the reaction was performed as described above. The RT-LAMP was optimized by adding Bst3.0 DNA polymerase.
2.6. Gel electrophoresi
The products of RT-LAMP were analyzed by gel electrophoresis on a 2% agarose gel in 1x TBE buffer for 30 min. Images were taken using an Imaging System (Bio-Rad, GelDoc Go, America).
2.7. High-throughput CLAP assay
The method can be applied directly in 96-well plate. For high-throughput detection screening, the visual detection reactions were performed in 96-well plate. The RT-LAMP was performed in line 1–3 in 96-well plate at 65 ℃ for 10 min., then 2.5 μL product was used for activation of Cas12a/gRNA in line 4–6 at 37 ℃ for 15 min.. The pre-mixed AuNP-DNA1/2 solution was added and then colorimetric detection was performed at room temperature for 5−10 min.. The color changes could be observed, and the absorbance of the mixture could be scanned at 520 nm and 560 nm using a microplate reader (Bio-Tek, Epoch 2, America).
2.8. Cell culture and RNA extraction
The 293 T cells (a human renal epithelial cell line, from the American Type Culture Collection, No CRL-11268) were transfected with pEF plasmid with N gene of SARS-CoV-2 at a ratio of 2 μg/3 × 105 cells. Infected cells were incubated for 48 h in Dulbecco's modified Eagle's medium (DMEM) supplemented 10 % fetal bovine serum, 0.1 mg streptomycin and 100 units penicillin per mL at 37℃ in a CO2 incubator. Then the RNA was extracted using GeneJET RNA Purification Kit and the concentration was measured using a NanoDrop.
3. Results and discussion
3.1. Visual assay platform based on AuNP probes
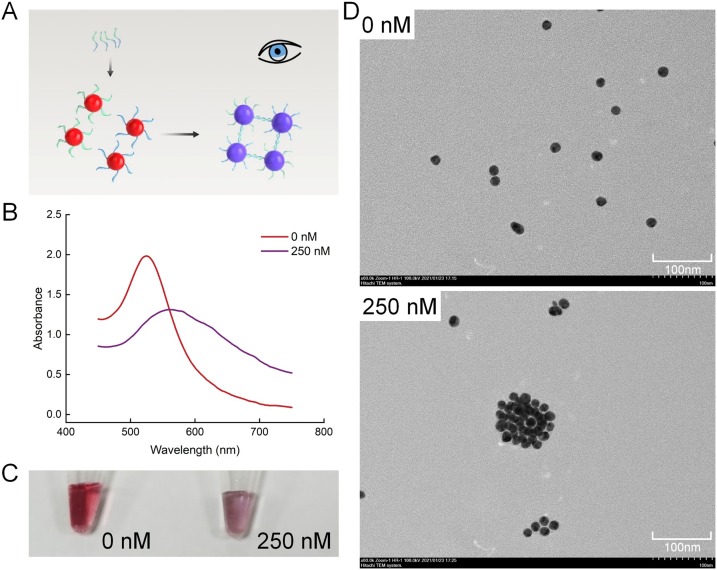
To demonstrate the utility of this approach, we first prepared thiolated DNAs modified AuNPs according to a reported procedure. The calculated surface coverage of the AuNP-DNA1/2 indicated an average loading of 75 DNAs per AuNP. We next evaluated if the length of linker-ssDNA would affect the crosslinking of AuNP-DNA1/2 (Fig. 2A). We tested three different lengths of linker-ssDNA. In all groups, the color turns from red to purple in 5 min. (Fig. S1B). At the same time, the maximum absorption peaks were red-shifted from 520 nm to 560 nm (Fig. S1A). We selected medium-length linker-ssDNA to crosslink AuNP-DNA1/2 mixture at a ratio of 250:5:5 for the following experiments. Under this condition, the UV–vis spectrum showed the characteristic redshift (Fig. 2B) and a concomitant color change (Fig. 2C). The phenomenon was further confirmed by transmission electron microscopy (TEM) (Fig. 2D), which showed that the AuNPs were monomeric in the absence of the linker-ssDNA, and agglomerated significantly in the presence of the linker-ssDNA. In general, we demonstrated that the linker-ssDNA can crosslink AuNPs together in 5 min. and the length of linker-ssDNA has little effect on crosslinking.
Fig. 2.
The optimal length of linker-ssDNA for visual detection. (A) Schematic illustration of visual color changes. (B) The UV–vis spectrum of the AuNP-DNA1/2 mixture before and after the crosslinking. (C) Visual color changes of the samples in (B). (D) TEM images of the samples in (B) (scale bars =100 nm).
3.2. Trans-cleavage activity of CRISPR/Cas12a
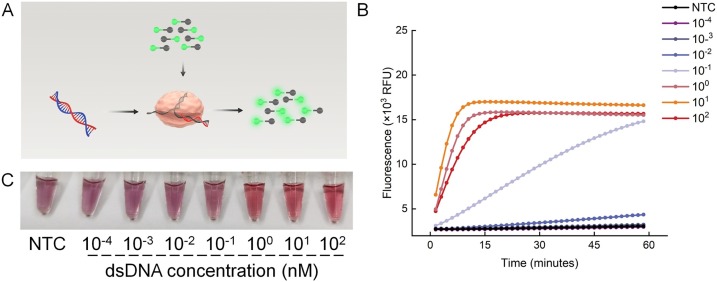
Since the trans-cleavage mechanism of CRISPR/Cas12a has been discovered, it has been widely used to detect DNA or RNA. In our assay, Cas12a/gRNA was designed to recognize the specific sequence of LAMP product. Then the trans-cleavage activity of Cas12a/gRNA is activated and the nearby fluorescent probes can be cleaved nonspecifically (Fig. 3A). To evaluate and optimize the trans-cleavage activity of Cas12a, we used 10-fold sequential dilutions dsDNA and ssDNA containing viral targets with specific sequences to activate the Cas12a/gRNA. We used fluorescent probes or linker-ssDNA induced color change of AuNPs to characterize the activation of Cas12a/gRNA. Obvious fluorescent signals could be observed in 15 min. when the concentration of the dsDNA target was more than 1 nM, indicating 1 nM of dsDNA could effectively activate the Cas12a/gRNA (Fig. 3B). Similarly, when linker-ssDNA was used and mixed with AuNP-DNA1/2, the mixture remained a stable red color for groups with concentrations of more than 1 nM (Fig. 3C), indicating the linker-ssDNA were totally cleaved, which was consistent with the fluorescence analysis. In addition, ssDNA containing viral targets was also used to activate the Cas12a/gRNA (Fig. S2). Fluorescence values of the reaction were measured at 10 min, 20 min and 30 min. Extremely high fluorescence (P < 0.001) was generated when the concentration of ssDNA was as low as 1 nM at 30 min, while 10 nM at 10 min (P < 0.001). These results indicated that about 1 nM of ssDNA or dsDNA could activate the trans-cleavage activity of Cas12a, and Cas12a can cleave all linker-ssDNA in 15 min. in the presence of 1 nM of dsDNA.
Fig. 3.
The trans-cleavage activity of the Cas12a/gRNA system. (A) Schematic diagram of the Cas12a/gRNA system. The activity of the Cas12a/gRNA system using fluorescence detection (B) and visual detection (C) for different concentrations of dsDNA containing viral targets. NTC, no-template control.
3.3. Cas12a assisted RT-LAMP
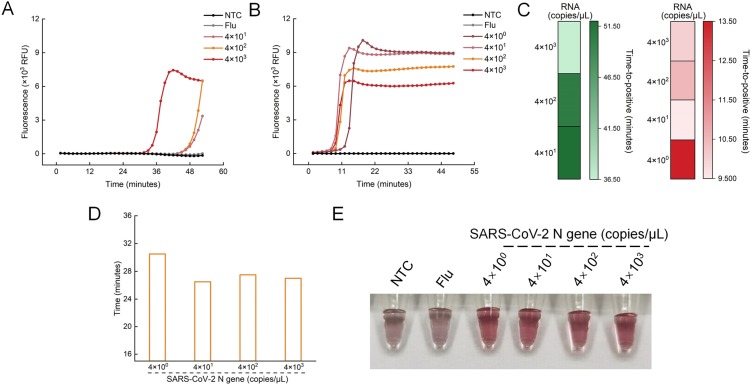
RT-LAMP primer targeting the N gene was used to amplify the specific sequence RNA of SARS-CoV-2. Serially diluted COVID-19 RNA reference materials were used as the template. Amplification curves were observed in all positive groups in about 50 min. (Fig. 4A). We next tested if the Cas12a/gRNA could be activated by the products of RT-LAMP by using the fluorescent probes. Extremely high fluorescence was generated after a 15-min reaction when the viral loads were as low as 40 copies/μL (Fig. S4A). The results of visual detection were consistent (Fig. S4B). The whole experiment took about 80 min., which was too long to be applied in POCT.
Fig. 4.
Optimization of RT-LAMP amplification. Real-time RT-LAMP amplification curves with different concentrations of SARS-CoV-2 N gene before optimization (A) and after optimization (B). (C) Time to positive (TTP) for RT-LAMP amplification before (left) and after (right) optimization (lower value and lighter colors indicate faster amplification). TTP is defined as the time when the fluorescent intensity passes 3000. (D) The time needed for the CLAP assay with different concentrations of SARS-CoV-2. (E) Visual detection of SARS-CoV-2 by CLAP after optimization. NTC, no-template control, Flu, Flu-HA gene.
To accelerate the reaction, Bst 3.0 DNA polymerase, which can improve the LAMP amplification performance, was added to the reaction system. The optimal concentration of Bst 3.0 was about 160 U/mL, which significantly improved the amplification efficiency (Fig. S3). Under the optimal reaction conditions, the RT-LAMP could be completed within 15 min., even at 4 copies/μL (Fig. 4B), which was confirmed by gel electrophoresis results (Fig. S5). As shown in Fig. 4C, Bst 3.0 DNA polymerase greatly shortens the reaction time. The fluorescence monitoring experiment showed that cleavage by Cas12a/gRNA could be completed within 15 min. (Fig. S4C). By combining with AuNP-based visual assay, the CLAP assay could complete the detection of virus RNA in 30−40 min. (Fig. 4D, E).
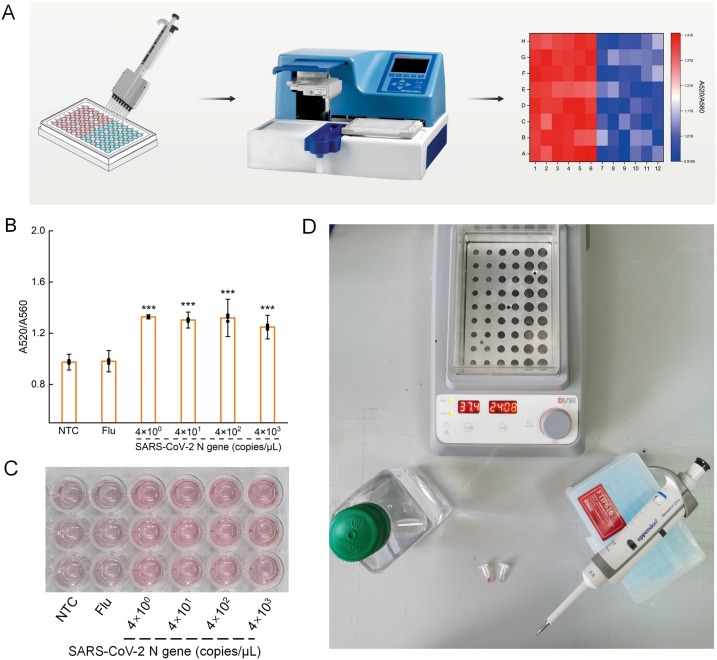
3.4. High-throughput CLAP assay
The CLAP assay is fast and easy for operation, and can be developed in a high-throughput form (Fig. 5A). To demonstrate the feasibility of CLAP for high-throughput assay, we carried out the whole reaction in a 96-well plate, and measured the photometric results using a microplate reader. The A520/A560 of the positive group was obviously higher than that of blank control group and the negative group (P < 0.001) (Fig. 5B), which is consistent with visual detection (Fig. 5C). This indicated that the method had the potential to be used extensively for on-site detection of SARS-CoV-2 with high-throughput capability. Fig. 5D showed the minimum equipment needed for CLAP assay, including tubes, metal bath (37 °C and 65 °C), pipettes, nuclease-free water, and AuNP-DNA1/2 mixture.
Fig. 5.
(A) Schematic of the high-throughput testing. (B) The measured ratio value of A520/A560 using a microplate reader. Data points represent replicates from three independent experiments, and the error bars indicate the mean ± S.E.M., ***P < 0.001 (C) Photograph of visual detection of SARS-CoV-2 with CLAP, which was performed in 96-well plate. (D) The minimum equipment needed to run the CLAP assay. NTC, no-template control, Flu, Flu-HA gene.
3.5. Detection of the viral gene in transfected cells
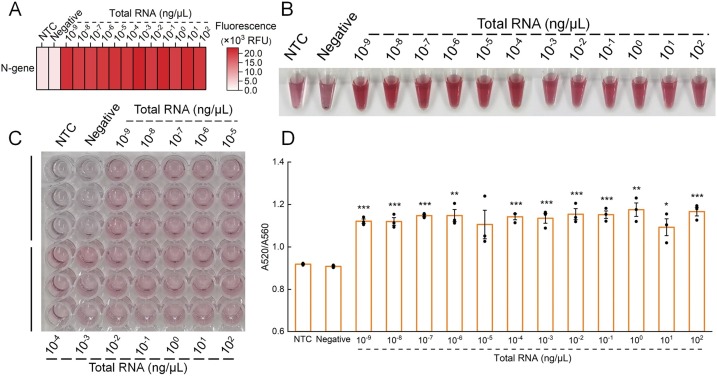
To test whether the CLAP assay could accurately detect the presence of SARS-CoV-2 in real settings, we used cells transfected by SARS-CoV-2 to simulate the infected cells in clinical samples. The total RNA from the transfected cells were extracted and serially diluted. Fig. S6 showed that all positive groups could be amplified by RT-LAMP. Our assay could detect SARS-CoV-2 in all positive samples, but not in the negative sample and bank (Fig. 6A, B), indicating the specificity. By using the CLAP assay, 10−9 ng/μL of total RNA could be detected by naked eye, demonstrating the excellent sensitivity of this method. We also performed the CLAP assay in a 96-well plate to further confirm the high-throughput capability for screening in large quantities, and the results were consistent with those in tubes (Fig. 6C, D).
Fig. 6.
CLAP for detection of SARS-CoV-2 in extracted total RNA from transfected cells. (A) Heat map analysis of SARS-CoV-2 N gene detection results based on Cas12a assisted RT-LAMP with fluorescent signal. (B) Visual detection of the samples using the CLAP assay. CLAP assay in 96-well plate for visual detection (C) of SARS-CoV-2 in diluted extracted total RNA, and the corresponding A520/A560 (D). Each sample was performed with three replicates. The error bars indicate the mean ± S.E.M., *P < 0.05, **P < 0.01, ***P < 0.001. NTC, no-template control.
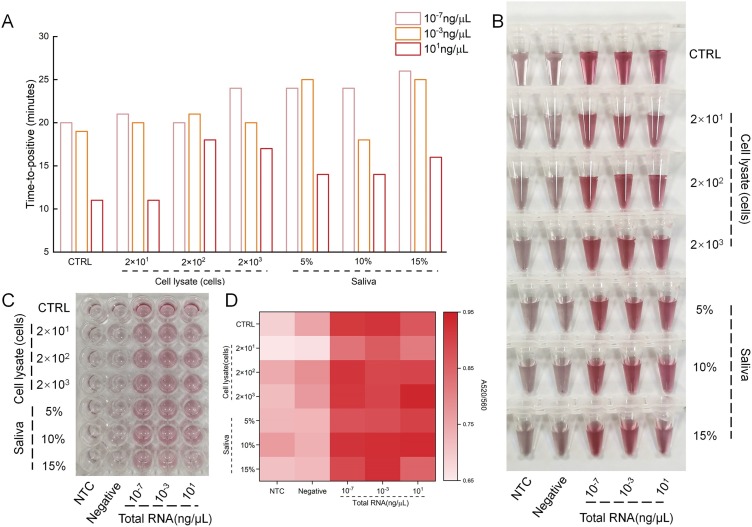
To further explore the possibility of our method in clinical diagnosis, the robustness of our method in complex biosamples (cell lysate and saliva) was tested. The biosamples were spiked in different concentrations of total RNA. As shown in Fig. 7 A, though the group with more biological samples took more time, all the samples could complete the amplification within half an hour. In the subsequent visual detection, the result did not show any differences (Fig. 7B, S7). We also performed the CLAP assay in a 96-well plate, and the results were consistent with those of visual detection (Fig. 7C, D). These results indicated that CLAP could be applied in real samples, and had great potential for high-throughput testing with superior sensitivity and specificity.
Fig. 7.
(A) Time to positive for RT-LAMP amplification in samples spiked with different concentrations of cell lysate and saliva matrixes. (B) Visual detection results of (A). (C) The photograph of visual detection using a 96-well plate. (D) The heatmap showing the A520/A560 read by a microplate reader. The values of A520/A560 are between 0.65-0.95. NTC, no-template control.
4. Conclusions
In summary, we have developed an AuNP-based visual assay that combined with Cas12a-assisted RT-LAMP (CLAP). Under the optimal condition, 4 copies/μL of SARS-CoV-2 RNA could be detected in less than 40 min by naked eye. The innate character of CRISPR/Cas12a enables the CLAP an excellent sensitivity and specificity. The operation of CLAP is highly simplified, which is suitable for POCT application. More importantly, the CLAP could be performed in 96 well plate as a high-throughput test, which has potential to be extended to automatic platform. Given many studies reported that LAMP was compatible with a simple operation of cell lysis for viral nucleic acid extraction, the CLAP assay was very promising for on-site diagnosis of COVID-19 in airports and railway stations in cities, or low-resource settings like rural areas for rapid large-scale population screening. We expect the CLAP assay could be used in both developed countries and rural areas, that will have a significant impact for the control and mitigation of the pandemic.
CRediT authorship contribution statement
Jiasi Wang and Di Wang: contributed to the conceptual design of the research. Yaqin Zhang, Minyan Chen and Jiaqi Chen: performed the most of the experiments. Yaqin Zhang and Jiasi Wang: analyzed the data and wrote the manuscript. Chengrong Liu and Qiming Liang: constructed the plasmids of SARS-CoV-2. Xinyi Luo and Yingying Xue: contributed on cells culture and transfection. Li Zhou, Yu Tao, Mingqiang Li and Jianhua Zhou: helped on data analysis.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
This study is supported by Innovation Capacity Building Project of Jilin Provincial Development and Reform Commission (2021C035-6), the Fundamental Research Funds for the Central Universities (Project No.: 20lgzd28), and the Guangdong Province Science and Technology Innovation Special Fund (International Scientific Cooperation, 2018A050506035).
Biographies
Yaqin Zhang is a master degree candidate in School of Life Sciences, Jilin University, Changchun, China. She obtained her B.S. degree in 2019 from Jilin University. Her research mainly focuses on rapid detection of virus with biosensor.
Minyan Chen is an undergraduate of school of Plant Protection, Jilin Agricultural University, Changchun, China. Her research interests focus on Fungus chemical.
Chengrong Liu is a research associate in Shanghai Jiao Tong University School of Medicine. She received her B.S and M.S degree in Biology from Jilin University. Her research interests focus on the mechanism of human innate immunity on virus infection.
Jiaqi Chen is a fungal science and engineering internal student at Jilin Agricultural University. Her research interests focus on the research and development of fungal chemistry.
Xinyi Luo is a master degree candidate of School of Biomedical Engineering, Sun Yat-sen University, Shenzhen, China. Her research interests focus on development of novel microfluidic chip for diagnosis.
Yingying Xue is a master degree candidate of School of Biomedical Engineering, Sun Yat-sen University, Shenzhen, China. Her research interests focus on design and development of digital isothermal method for precise diagnosis.
Qiming Liang is a professor in Shanghai Jiao Tong University School of Medicine. He received his Ph.D. degree in Biology from Florida State University. His research interests focus on mechanism of human innate immunity on virus infection.
Li Zhou is a professor in Institute of Environmental Research at Greater Bay Area, Guangzhou University. She received her Ph.D. degree in Chemistry from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences. Her research interests focus on development of nanosensor for environmental application.
Yu Tao is a research professor in The Third Affiliated Hospital, Sun Yat-sen University. She received her Ph.D. degree in Chemistry from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences. Her research interests focus on development of nanotechnology in biomedicine.
Mingqiang Li a research professor in The Third Affiliated Hospital, Sun Yat-sen University. He received her Ph.D. degree in Chemistry from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences. His research interests focus on development of novel nanocarrier for drug delivery.
Di Wang graduated from The University of HongKong in 2013, and worked at Jilin University at the same year. Pro. Wang focus his research on the function screening and mechanism investigation of natural compounds. Till now, Pro. Wang has undertaken 16 projects including National Natural Science Foundation of China, Key Scientific and Technological Project in Jilin Province and Industry-University-Research Cooperation Projects. Accumulative scientific research funds reach to RMB 10.0 million. Pro. Wang has published 106 SCI indexed papers as the first responsibility author in Molecular Cancer, Clinical and Translational Medicine, Carbohydrate Polymers, Cell Death & Disease, Expert Opinion on Drug Delivery, Journal of Neuroinflammation, et al, and obtained 10 authorized national patents.
Jianhua Zhou is a full professor in School of Biomedical Engineering, Sun Yat-sen University, Shenzhen, China. He received his Ph.D. degree in Chemistry from The Hong Kong University of Science and Technology. His research interests focus on design and development of novel microfluidic biosensor for biomedical diagnosis.
Jiasi Wang is an associate professor in School of Biomedical Engineering, Sun Yat-sen University, Shenzhen, China. He received his Ph.D. degree in Chemistry from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences. His research interests include nanotechnology in biosensor, digital diagnosis, point of care testing.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2021.130411.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19) a review. JAMA-J. Am. Med. Assoc. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 2.Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., et al. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 3.Luo Z., Ang M.J.Y., Chan S.Y., Yi Z., Goh Y.Y., Yan S., et al. Combating the coronavirus pandemic: early detection, medical treatment, and a concerted effort by the global community. Research. 2020;2020 doi: 10.34133/2020/6925296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L., Wu S., Hao X., Dong X., Mao L., Pelechano V., et al. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020;66:975–977. doi: 10.1093/clinchem/hvaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue G., Li S., Zhang W., Du B., Cui J., Yan C., et al. Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2) Anal. Chem. 2020;92:9699–9705. doi: 10.1021/acs.analchem.0c01032. [DOI] [PubMed] [Google Scholar]
- 7.Wu T.-H., Chang C.-C., Yang C.-H., Lin W.-Y., Ee T.J., Lin C.-W. Hybridization chain reactions targeting the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21093216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joung J., Ladha A., Saito M., Kim N.-G., Woolley A.E., Segel M., et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383 doi: 10.1056/NEJMc2026172. 1492-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38 doi: 10.1038/s41587-020-0513-4. 870-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang B., Xu J., Liu Y., Peng H., Feng W., Cao Y., et al. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology. Anal. Chem. 2020;92:16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- 11.Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., et al. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360 doi: 10.1126/science.aar6245. 436-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S.-Y., Cheng Q.-X., Wang J.-M., Li X.-Y., Zhang Z.-L., Gao S., et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4 doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356 doi: 10.1126/science.aam9321. 438-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W., Yu L., Wen D., Wei D., Sun Y., Zhao H., et al. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. Ebiomedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z., Tian D., Liu Y., Lin Z., Lyon C.J., Lai W., et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J., Wu L., Ren J., Qu X. Visualizing human telomerase activity with primer-modified Au nanoparticles. Small. 2012;8:259–264. doi: 10.1002/smll.201101938. [DOI] [PubMed] [Google Scholar]
- 19.Elghanian R., Storhoff J.J., Mucic R.C., Letsinger R.L., Mirkin C.A. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science (New York, NY) 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 20.Moon C.W., Park J., Hong S.-P., Sohn W., Andoshe D.M., Shokouhimehr M., et al. Decoration of metal oxide surface with {111} form Au nanoparticles using PEGylation. RSC Adv. 2018;8:18442–18450. doi: 10.1039/c8ra03523g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabiee N., Bagherzadeh M., Ghasemi A., Zare H., Ahmadi S., Fatahi Y., et al. Point-of-use rapid detection of SARS-CoV-2: nanotechnology-enabled solutions for the COVID-19 pandemic. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21145126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmadi S., Rabiee N., Fatahi Y., Hooshmand S.E., Bagherzadeh M., Rabiee M., et al. Green chemistry and coronavirus. Sustain. Chem. Pharm. 2021;21:100415. doi: 10.1016/j.scp.2021.100415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y., Hu M., Liu A.-A., Lin Y., Liu L., Yu B., et al. Detection of SARS-CoV-2 by CRISPR/Cas12a-enhanced colorimetry. ACS Sens. 2021;6:1086–1093. doi: 10.1021/acssensors.0c02365. [DOI] [PubMed] [Google Scholar]
- 24.Zhu X., Wang X., Li S., Luo W., Zhang X., Wang C., et al. Rapid, ultrasensitive, and highly specific diagnosis of COVID-19 by CRISPR-based detection. ACS Sens. 2021;6:881–888. doi: 10.1021/acssensors.0c01984. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W.S., Pan J., Li F., Zhu M., Xu M., Zhu H., et al. Reverse transcription recombinase polymerase amplification coupled with CRISPR-Cas12a for facile and highly sensitive colorimetric SARS-CoV-2 detection. Anal. Chem. 2021;93:4126–4133. doi: 10.1021/acs.analchem.1c00013. [DOI] [PubMed] [Google Scholar]
- 26.Fozouni P., Son S., Derby M.Dd.L., Knott G.J., Gray C.N., D’Ambrosio M.V., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184 doi: 10.1016/j.cell.2020.12.001. 323-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J.S., Hsieh K., Chen L., Kaushik A., Trick A.Y., Wang T.-H. Digital CRISPR/Cas-assisted assay for rapid and sensitive detection of SARS-CoV-2. Adv. Sci. 2021;8 doi: 10.1002/advs.202003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou T., Zeng W., Yang M., Chen W., Ren L., Ai J., et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.