Abstract
Introduction
Cough is a common symptom of COVID-19 and other respiratory illnesses. However, objectively measuring its frequency and evolution is hindered by the lack of reliable and scalable monitoring systems. This can be overcome by newly developed artificial intelligence models that exploit the portability of smartphones. In the context of the ongoing COVID-19 pandemic, cough detection for respiratory disease syndromic surveillance represents a simple means for early outbreak detection and disease surveillance. In this protocol, we evaluate the ability of population-based digital cough surveillance to predict the incidence of respiratory diseases at population level in Navarra, Spain, while assessing individual determinants of uptake of these platforms.
Methods and analysis
Participants in the Cendea de Cizur, Zizur Mayor or attending the local University of Navarra (Pamplona) will be invited to monitor their night-time cough using the smartphone app Hyfe Cough Tracker. Detected coughs will be aggregated in time and space. Incidence of COVID-19 and other diagnosed respiratory diseases within the participants cohort, and the study area and population will be collected from local health facilities and used to carry out an autoregressive moving average analysis on those independent time series. In a mixed-methods design, we will explore barriers and facilitators of continuous digital cough monitoring by evaluating participation patterns and sociodemographic characteristics. Participants will fill an acceptability questionnaire and a subgroup will participate in focus group discussions.
Ethics and dissemination
Ethics approval was obtained from the ethics committee of the Centre Hospitalier de l’Université de Montréal, Canada and the Medical Research Ethics Committee of Navarre, Spain. Preliminary findings will be shared with civil and health authorities and reported to individual participants. Results will be submitted for publication in peer-reviewed scientific journals and international conferences.
Trial registration number
Keywords: COVID-19, respiratory infections, epidemiology
Strengths and limitations of this study.
This is the first study to evaluate the utility of artificial intelligence models and smartphone applications for cough detection at population level.
The studied approach has the potential to be rapidly scalable even within underdeveloped public health systems.
Qualitative methods will improve the understanding of barriers and facilitators affecting the uptake of cough detection on smartphone applications and participation in acoustic surveillance programmes.
Recorded coughs will be annotated with clinical diagnoses (when available) and will contribute to the training of cough-based disease-specific screening and diagnostic tools.
Success of this study is contingent on large-scale enrolment and high participant retention since a small sample size might not be sufficient to appropriately correlate the study population’s cough trends and their relationship with the incidence of respiratory diseases at population level.
Introduction
Real-time tracking of the COVID-19 pandemic and detection of the emergence of novel variants or other respiratory pathogens represent challenges for public health authorities globally. Nonetheless, understanding local epidemiology is essential to disease control efforts. This is particularly true now that the world has moved into a COVID-19 endemic phase, with periodical outbreaks in multiple locations superimposed over ongoing community transmission. The ability to appropriately monitor disease incidence, however, is frequently limited by a lack of testing capacity, delays in health-seeking behaviours, as well as complexities related to timely aggregation of actionable surveillance data by public health authorities.
Although asymptomatic cases of COVID-19 and other respiratory communicable diseases are well described, cough remains a common symptom of COVID-19.1 2 A meta-analysis including data from over 24 000 adults with confirmed COVID-19 revealed cough as the second most prevalent symptom, being reported by 57% (CI: 54% to 60%) of all patients.3 Additionally, cough is a key event in the transmission of COVID-19 and other respiratory pathogens.4
Given the intrinsic subjectivity of existing cough monitoring tools, mostly based on self-reported questionnaires, there is a great interest in developing an automated system to objectively register coughs. Attempts to achieve this date back to the 1950s, but until recently, important technological constraints limited significant advances.5
Even now, detecting and classifying sounds as coughs without the input of a human observer still represent a remarkable challenge. However, automatically detecting cough is now possible thanks to the development of acoustics and artificial intelligence (AI) models which can be incorporated into wearable devices. This has allowed the development of several alternatives, whose widespread deployment beyond a research context remains limited due to problems related to portability, the need to continuously record sound (which may compromise patients’ privacy), inconsistent sensitivity and specificity, high false positivity rates and little financial incentives.5
Some of these limitations might be addressed by incorporating similar systems on smartphone applications, an approach that has just recently begun to be widely explored.6 7 In brief, putative cough sounds (manifested as short and explosive noises) are recognised and captured by smartphones. Machine learning models trained on hundreds of thousands of annotated sounds are then used to distinguish cough from other sounds.
We propose that COVID-19 surveillance and early outbreak recognition can be enhanced by digitally monitoring cough and detecting changes in its incidence at population level. We hypothesise such changes precede individual symptoms recognition, healthcare seeking behaviours, diagnosis and data aggregation within conventional disease surveillance systems. Indeed, digital cough monitoring data can be aggregated in real time providing constantly updated information on the emergence and activity of communicable respiratory diseases within populations. This approach would be of particular value in low-income and middle-income countries where (a) capturing passive data through health services is insufficient given unequal access to healthcare, and (b) active case detection and contact tracing capacity is limited.8 9
To our knowledge, digital cough monitoring for early detection of respiratory disease outbreaks has never been performed. Provided the participation of a critical mass of active users is achieved, monitoring of aggregated cough data could provide a simple and inexpensive surrogate indicator for overall respiratory infection incidence, similar to, but more accurate than wastewater monitoring.10 This information could in turn guide public health interventions. If proven successful in the specific case of COVID-19, this study would establish a template for early and disease-agnostic detection of emerging pathogens, therefore contributing to health system’s epidemic preparedness.
Primary research question
Can digital cough surveillance predict the incidence of respiratory diseases at population level in Navarra, Spain?
Methods
Study design
This is a prospective observational study which will take place between November 2020 and October 2021 (figure 1). Participants will be recruited in (a) the Cendea de Cizur, a municipality composed by a cluster of villages south of the city of Pamplona, (b) the neighbouring town of Zizur Mayor, in the Chartered Community of Navarra (Spain), as well as (c) in the different campuses of the University of Navarra (Pamplona), which collides with the municipalities.
Figure 1.
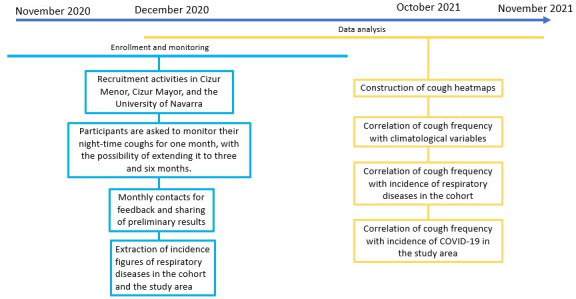
Study design, timeline and monitoring plan for the study’s primary objective.
All these communities are located within a 5 km range from each other, and there is a considerable geographical and social overlap between them, as the University of Navarra is the second most important employer in the area.
The 4000 people living in the Cendea de Cizur are served by a public health centre which receives 45 000 outpatient visits per year. Of these, approximately 12% are associated with respiratory diseases. Furthermore, the Clínica Universidad de Navarra is the main private healthcare provider in the region and offers medical care to a significant proportion of the population, as well as university students and workers. Both centres offer COVID-19 PCR testing. Together, those two healthcare facilities cover most of the population’s healthcare needs and have digitalised medical record systems that facilitate the retrieval of medical data concerning the diagnosis of respiratory diseases among participants.
Recruitment strategies will include direct solicitation, community meetings, videos and advertisements in social media and university communication platforms. Those willing to participate will attend individual sessions with a study coordinator for counselling and training using the Hyfe Cough Tracker (Hyfe) application, and for obtention of informed consent. Information consent forms for adults and minors are available as online supplemental materials 1 and 2, respectively. Participants will consent to be contacted regularly via email and telephone by the study team and will also fill an enrolment questionnaire (online supplemental material 3) where demographic and respiratory disease medical data will be collected.
bmjopen-2021-051278supp001.pdf (239.1KB, pdf)
bmjopen-2021-051278supp002.pdf (248KB, pdf)
bmjopen-2021-051278supp003.pdf (95.9KB, pdf)
Primary objective
To assess the value of digital cough monitoring and acoustic surveillance in predicting COVID-19 and other respiratory disease incidence at population level.
Secondary objective
To assess barriers and facilitators to participation in an acoustic surveillance programme using a smartphone-based digital cough monitoring application.
Eligibility criteria
To be included in the study, participants must (1) be 13 years old or older, (2) own a smartphone, (3) be willing to install and use the Hyfe digital cough monitoring application, (4) accept and comply with Hyfe privacy policy and terms of use, and (5) grant access to their medical records. To optimise population representativity, participants also need to (6) visit the University of Navarra on a regular basis, either as a student, worker or patient of the university clinic, or (7) have a general interest in the study, and (8) currently reside in Navarra. These criteria aim at ensuring a large sample size and the availability of needed medical records while concentrating recruitment in a geographically focused area. Accounting for differences between smartphone models’ capabilities, on recruitment, participants will also have to complete a microphone calibration process within the application. This is to make sure their smartphones can accurately detect cough-like sounds. Participants who fail to complete this process will be excluded from the study.
Participant retention
To ensure participants’ retention and promote continuous cough recording, we will use videos posted on social media, as well as emailed to participants, text messages and regular push notifications sent via the Hyfe application on participants’ smartphones. Participants will also receive monthly emails with information regarding the current stage of the study and high-level preliminary results.
Digital cough monitoring
The cough detection application used in this study is Hyfe Cough Tracker (Hyfe) (https://www.hyfeapp.com/). Hyfe runs in the background of smartphones’ operating systems, listens for and records short snippets (<0.5 s) of explosive, putative cough sounds and then classifies them as cough or non-cough, using a convolutional neural network model. Cough sounds are automatically matched with time and Global Positioning System coordinates which can be jittered to ensure participants’ privacy. Hyfe is a research tool that has collected over 5 million putative cough sounds, of which over 1 000 000 have been classified as coughs or not by a human listener in order to train the AI model.
The analytical performance of Hyfe for cough detection (referred to whether the application is sensitive and specific for the classification of recorded sounds as coughs or non-coughs) was confirmed using this study’s preliminary results from a subgroup of participants recruited in the Cendea de Cizur between November and December 2020.
Approximately 500 participants were contacted from an existing mailing list used in a previous epidemiological study, of these, 57 were enrolled. While participants were initially requested to keep Hyfe running continuously, they were later instructed to restrict their use to at least 6 hours during night-time, for 30 days, following complaints of increased battery consumption.
During this period, nearly 700 000 putative cough sounds were registered, of which 119 876 were classified by human observers, revealing a sensitivity of 96.34% and a specificity of 96.54%, when using a cough-positivity threshold of 0.85 (figure 2).
Figure 2.
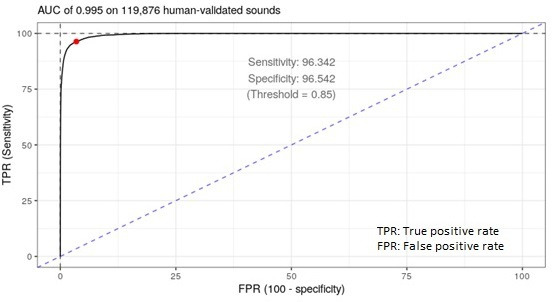
Receiver operating characteristic analysis showing an area under the curve (AUC) of 0.995 for the classification of cough in participants recruited between November and December 2020.
These values, however, must be interpreted carefully, as specificity measured at the explosive sound level does not necessarily provide a full picture of diagnostic accuracy, particularly in loud environments, where the probability of an explosive sound being a cough is lower, and the false positivity rate might be considerably higher. Similarly, sensitivity does not account for non-explosive sounds that are not captured by the system, and of which a non-zero number might be coughs.
On enrolment, a study coordinator will assist participants in installing the Hyfe application. Once turned on, it will continuously monitor participants’ cough. Participants can turn the application on and off at will but will be instructed to keep it active for at least 6 hours or while they sleep, every day for a minimum period of 30 days. In an adaptive design aimed at improving retention, participants will have the possibility of extending their participation to a 3-month and 6-month period.
Local epidemiology of respiratory disease
Digital medical data systems at the Zizur’s health centre and the Clínica Universidad de Navarra will be reviewed monthly to establish the incidence of the following respiratory or cough-associated diseases in the cohort of participants: COVID-19, influenza, respiratory syncytial virus, pneumonia, asthma, bronchitis, pharyngitis, chronic cough, chronic obstructive pulmonary disease, gastro-oesophageal reflux disease, other non-specific respiratory tract infections. Specifically, for COVID-19, daily incidence figures for the study area (Cendea de Cizur, Zizur Mayor and Pamplona) and regional population will also be obtained from local health authorities. Data will be aggregated and used to build local epidemic curves of respiratory diseases during the study period.
Barriers and facilitators to participation
A participation cascade informing on the number of solicited, enrolled and actively recording participants will be tracked in real time throughout the study. The total number of user-hours using Hyfe will also be monitored in real time.
All participants will be asked to fill a satisfaction survey in which they will be asked about the usability of the app, the problems that they ran into and their likelihood of using the app in the future. We will follow up with a group of 25 participants who are willing to attend a focus group discussion (FGD) to better understand the barriers and facilitators affecting participation in the cough surveillance system. The 25 participants will include some of whom deleted the application early on, who stopped using it after a month and who had low satisfaction scores.
Patient and public involvement
This study pilots an experimental syndromic surveillance approach at population level. The municipal authorities of Cendea de Cizur were informed about the nature of the study and provided feedback on the best approach to recruit local participants. Similarly, representatives of the University of Navarra’s medical services collaborated on the design of the study protocol including the review of recruitment strategies as well as data collection, aggregation and reporting plans. No anticipated participants were involved in the study design. Study findings are to be reported in both English and Spanish to participants and key stakeholders at individual, municipality and university levels during the surveillance phase.
Data analysis and sample size calculations
Syndromic surveillance as a surrogate marker of respiratory disease activity
Given the uncertainty around the incidence of the various respiratory diseases including COVID-19, throughout the study period, it is impossible to establish a target sample size sufficient to either confirm or confute with statistical significance that cough monitoring at population level can predict incidence of respiratory diseases. We will endeavour to recruit the highest number of participants possible in order to achieve the best representation of the population.
First, cough data will be aggregated in time and space to create cough incidence curves and geospatial heat maps reflecting the frequency and density of cough among the studied population (figures 3 and 4). Second, epidemic curves reflecting the incidence of targeted respiratory illnesses within the cohort of participants and the number of COVID-19 cases diagnosed in the study area and population will be generated with data collected from the Clínica Universidad de Navarra and Zizur’s health centre, as well as aggregated epidemiological records obtained from regional health authorities, respectively. Coughs per person-hour and clinical diagnoses data will be superposed in time. Finally, we will carry out an autoregressive moving average (ARIMA) analysis to compare the incidence of confirmed and forecasted respiratory diseases (including COVID-19) with the frequency of cough among study participants, measured as coughs per person-hour. ARIMA analysis is a type of time series analysis that uses past data to forecast the likely future behaviour of a variable. In brief, the variable of interest is regressed to its own lagged values, and autoregression and partial autocorrelation functions are used to model the stochastic nature of a time series.11 Comparison and prediction analyses will be performed using epidemic curves from both the cohort of participants and in the entire study area and population.
Figure 3.
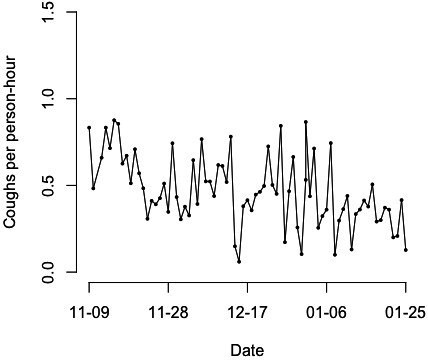
Coughs per person-hour registered in participants recruited between November 2020 and January 2021.
Figure 4.
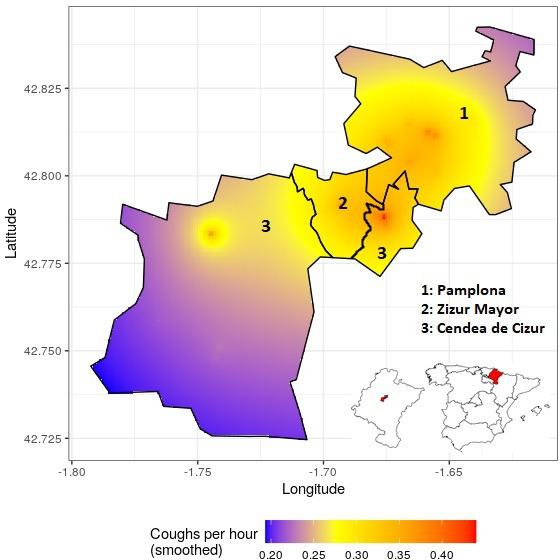
Heat map of registered cough episodes in the municipalities of Zizur Mayor, Cendea de Cizur, and Pamplona between November 2020 and March 2021 (Cendea de Cizur is an incontiguous municipality).
Perceptions and willingness to participate in syndromic surveillance
Mixed methods will be used to assess individual barriers and facilitators to participation in an acoustic surveillance programme. Among all eligible individuals in the Cendea de Cizur, Zizur Mayor and in the University of Navarra who are invited to participate in the study, we will assess how many do install the cough detection application and keep using it through the course of the study. Participants corresponding to specific user profiles based on duration and regularity of cough recording will be identified. We will then descriptively analyse the sociodemographic characteristics and baseline health conditions of participants belonging to the different user profiles.
A subsample of participants will be contacted once a month, by telephone or text messaging, to obtain feedback from their experience using the app.
To assess population uptake of Hyfe, as well as barriers and facilitators to participation, a convenience subsample of 25 participants will be recruited for FGDs. Following similar designs, FGDs will consist of two parts.12 The first one will be aimed at understanding a participant’s awareness of their cough frequency and temporality patterns, as well as the perceived importance of measuring these elements. Questions asked in this part will include: (1) How much attention do you pay to your coughs on a daily basis?; (2) How important is it for you to know the number of times you cough per day?; (3) Will keeping track of your coughs help you improve your health? The second one will focus on the participant’s experience using Hyfe, as well as possible recommendations for developers to improve it. This part will include the following general questions: (1) What do you like about the app?, (2) What do you think of this app compared with other health apps?, (3) What doesn’t work well?, (4) What keeps you committed (or not) to using the app?, (5) What do you think the purpose of the app is?, (6) What advice do you have for the developers?
Risks and privacy
Recording sounds implies specific ethical and privacy concerns. These are, however, addressable at different levels. First, sound snippets recorded are too short (<0.5 s) to capture conversations or background sound. Participants can withdraw from the study at any time, and the application can be turned off or removed from smartphones freely. Our consent process explicitly describes exactly what is and what is not recorded. The use of pre-generated unique identifiers to install and use the smartphone application ensures that only investigators can link cough data to personal identifiers. Contact information collected from participants will be kept in physical forms stored under lock and password-protected files at the University of Navarra. Only the principal investigator, study coordinator and research assistant will have access to this information. Data transferred to other researchers will only include unique identifiers for individual participants, but no other personal information. All acoustic data and metadata collected by the application are stored in encrypted servers physically located in the USA. Data storage and access protocols are compliant with General Data Protection Regulation, and only non-identifiable data information is stored by Hyfe.
Ethics and dissemination
This study protocol was approved by the ethics committee of the Centre Hospitalier de l’Université de Montréal, Canada (reference numbers 2021-9247 20.253 and 2021-9270 20.226) and the Medical Research Ethics Committee of Navarra, Spain (reference number PI107/2020). Any modifications to the approved protocol would be resubmitted to both committees.
Preliminary summary results of this study will be regularly shared with participants via email and through focal meetings. Final results will also be disseminated in open-access scientific journals and international conferences. A two-page summary of results will be prepared in Spanish and posted on the municipal and university website, and shared with local civil and health authorities, as well as with individual participants.
Discussion
The current COVID-19 pandemic highlights the incapacity of existing surveillance networks to rapidly curb the impact of emerging respiratory pathogens in the current globalised world.13 This is particularly true in low-income and middle-income countries, where diagnostic and contact tracing efforts are limited by their crippling costs, and where epidemiological data aggregation infrequently translates into actionable information because of delays inherent to disaggregated and poorly digitalised health information systems. Continuous, individual-based, and passive monitoring of cough among entire populations could represent inexpensive large-scale surveillance networks contributing to the early detection of outbreaks and enabling prioritised and focal delivery of limited resources.
AI, and more specifically machine learning models, are increasingly used in disease surveillance and can maximise the impact of limited available resources.14 Integration of such models with accurate digital epidemiological data was shown to provide reliable, near real-time estimations of influenza disease incidence.15 The widespread distribution of smartphones coupled with the development of AI-enabled sound classification models now represent another new potential breakthrough in public health and epidemic preparedness. Beyond being able to detect cough, most smartphones have integrated geolocation functions. This allows the integration of acoustic records with geospatial and temporal data and increases the range of applications of acoustic surveillance systems for disease control.16
Apart from participating in a collective disease tracking and eradication effort, participants monitoring their cough also benefit from objective feedback on their own symptomatology. During a pandemic, such feedback could trigger appropriate healthcare seeking behaviours and self-quarantine further helping to limit disease transmission at community level.
Implementing such an innovative approach to disease surveillance is contingent on overcoming substantial challenges. Evidence suggests that users are typically willing to install mobile apps with a clearly perceived health benefit. But this is often limited by issues such as increased battery drainage, or disruption of daily activities by alerts and notifications.17 Specifically, for acoustic surveillance apps, protecting user’s privacy must also be a priority, in order to create the social trust needed to achieve a high uptake of these systems. To guarantee this, users must be able to clearly understand the nature of these applications, as well as to control their functioning at will. This means that the quality of the data recorded will greatly depend on individual behaviours.
Despite proven technical capacity and tremendous potential in complementing surveillance systems, whether listed challenges can be overcome and whether digital cough monitoring can provide actionable public health information remain unknown. This study will address those questions. It will also generate operational expertise and qualitative knowledge on the facilitators to be exploited, and the barriers to be addressed in order to maximise uptake and impact prior to implementation on a wider scale.
Limitations and potential challenges
Enrolment and retention are expected to represent the major limitations of this project. Acoustic syndromic surveillance tools might be perceived as threatening for the privacy of potential participants. Furthermore, the quantity of cough data recorded will heavily rely on the regular use of the system by participants. The diversity of respiratory and non-respiratory conditions that can cause cough, as well as the consumption of medications to treat them, can make it difficult to link changes in cough frequency with epidemiological data. Finally, asymptomatic or mild infections of SARS-CoV-2 and other respiratory pathogens mean that a proportion of infected patients will not contribute to the cough-based surveillance system and are unlikely to seek medical attention, reducing available cough and epidemiological data necessary for the analysis. Those limitations and challenges reflect well the impediments to larger scale deployment of cough-based syndromic surveillance for any respiratory disease hence making this innovative study an ideal stress test for such approach.
Supplementary Material
Acknowledgments
The authors want to thank the Municipality of the Cendea de Cizur, the University of Navarra and all participants for their trust and desire to participate in citizen science and altruistic population-level syndromic surveillance.
Footnotes
Twitter: @SimonGLapierre
SGL and CC contributed equally.
Contributors: Conceptualisation—JCG-F, JBrew, DHD, NU, LYT, SGL and CC. Data curation—JCG-F, JBrew, NU and CC. Formal analysis—JCG-F, JBrew, NU, SGL and CC. Funding acquisition—SGL. Investigation—JCG-F, JC, AFM, VO, IB, JBartolome and CC. Methodology—JCG-F, JBrew, NU, SGL and CC. Supervision—JCG-F, SGL and CC. Writing of the original draft—JCG-F, CC and SGL. All authors have read, reviewed, discussed, and approved the final manuscript and their respective representation in the authorship.
Funding: This study was funded by the Patrick J McGovern Foundation (grant name: 'Early diagnosis of COVID-19 by utilising Artificial Intelligence and Acoustic Monitoring'). CC received salary support from Unitaid (grant name: BOHEMIA). ISGlobal acknowledges support from the Spanish Ministry of Science and Innovation through the 'Centro de Excelencia Severo Ochoa 2019–2023' Programme (grant number: CEX2018-000806-S), and support from the Generalitat de Catalunya through the CERCA programme.
Disclaimer: The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Hyfe had no role in the decision to submit this protocol for publication.
Competing interests: JB is the CEO of Hyfe.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not required.
References
- 1.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim G-u, Kim M-J, Ra SH. Clinical characteristics of asymptomatic and symptomatic patients with mild COVID-19. Clinical Microbiology and Infection 2020;26:948.e1–948.e3. 10.1016/j.cmi.2020.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant MC, Geoghegan L, Arbyn M, et al. The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): a systematic review and meta-analysis of 148 studies from 9 countries. PLoS One 2020;15:e0234765-e. 10.1371/journal.pone.0234765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal A, Bhardwaj R. Probability of COVID-19 infection by cough of a normal person and a super-spreader. Phys Fluids 2021;33:031704. 10.1063/5.0041596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall JI, Lozano M, Estrada-Petrocelli L, et al. The present and future of cough counting tools. J Thorac Dis 2020;12:5207–23. 10.21037/jtd-2020-icc-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasheed J, Jamil A, Hameed AA, et al. A survey on artificial intelligence approaches in supporting frontline workers and decision makers for the COVID-19 pandemic. Chaos Solitons Fractals 2020;141:110337. 10.1016/j.chaos.2020.110337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kvapilova L, Boza V, Dubec P, et al. Continuous sound collection using Smartphones and machine learning to measure cough. Digit Biomark 2019;3:166–75. 10.1159/000504666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeling RW, Wedderburn CJ, Garcia PJ, et al. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis 2020;20:e245–9. 10.1016/S1473-3099(20)30517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury R, Luhar S, Khan N, et al. Long-term strategies to control COVID-19 in low and middle-income countries: an options overview of community-based, non-pharmacological interventions. Eur J Epidemiol 2020;35:743–8. 10.1007/s10654-020-00660-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farkas K, Hillary LS, Malham SK, et al. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Curr Opin Environ Sci Health 2020;17:14–20. 10.1016/j.coesh.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.SL H, Xie M. The use of ARIMA models for reliability forecasting and analysis. Computers & Industrial Engineering 1998;35:213–6. [Google Scholar]
- 12.Nouvet E, Knoblauch AM, Passe I, et al. Perceptions of drones, digital adherence monitoring technologies and educational videos for tuberculosis control in remote Madagascar: a mixed-method study protocol. BMJ Open 2019;9:e028073. 10.1136/bmjopen-2018-028073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization . Who coronavirus (COVID-19) Dashboard, 2021. Available: https://covid19.who.int/ [Accessed 06 Mar 2021].
- 14.Naseem M, Akhund R, Arshad H, et al. Exploring the potential of artificial intelligence and machine learning to combat COVID-19 and existing opportunities for LMIC: a scoping review. J Prim Care Community Health 2020;11:215013272096363. 10.1177/2150132720963634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santillana M, Nguyen AT, Louie T, et al. Cloud-Based electronic health records for real-time, region-specific influenza surveillance. Sci Rep 2016;6:25732. 10.1038/srep25732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukundarajan H, Hol FJH, Castillo EA, et al. Using mobile phones as acoustic sensors for high-throughput mosquito surveillance. Elife 2017;6:e27854. 10.7554/eLife.27854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liss DT, Serrano E, Wakeman J, et al. "The Doctor Needs to Know": Acceptability of Smartphone Location Tracking for Care Coordination. JMIR Mhealth Uhealth 2018;6:e112. 10.2196/mhealth.9726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2021-051278supp001.pdf (239.1KB, pdf)
bmjopen-2021-051278supp002.pdf (248KB, pdf)
bmjopen-2021-051278supp003.pdf (95.9KB, pdf)


