Abstract
An increasing number of outbreaks due to resistant non-albicans Candida species have been reported worldwide. Between 2014 and 2016, Candida isolates causing invasive candidiasis were recovered in a Mexican hospital. Isolates were identified to species level and antifungal susceptibility was determined. In the time period studied, 74 invasive candidiasis cases were identified, with 38% (28/74) caused by Candida parapsilosis, out of which 54% (15/28) were fluconazole resistant. The ERG11 gene was sequenced for 12 recoverable fluconazole-resistant C. parapsilosis isolates and SNPs identified. The 12 isolates had one common silent point mutation in ERG11 (T591C) and seven isolates had an additional (A395T) mutation, which corresponded to Y132F. Four of the isolates carrying this mutation were closely related within the same cluster by microsatellite typing. This is the first report of an invasive candidiasis outbreak in Mexico due to azole-resistant C. parapsilosis associated with the Y132F substitution.
Keywords: Candida parapsilosis, ERG11, Y132F, outbreak, Mexico
Introduction
In the past decade, an increasing number of publications have reported outbreaks and cases of infection due to azole-, polyene-, and/or echinocandin-resistant non-albicans Candida species.1–3 Probably the best known is the global emergence of Candida auris. However, invasive infections due to another antifungal resistant Candida species are also reported to be increasing globally. Azole-resistant Candida parapsilosis clonal outbreaks have occurred in Brazil,4 India,5 Turkey,6 Italy,7 South Africa,8 South Korea,9 Kuwait,10 China,11 and the United States.12 In addition, a report from Iran identified not only azole-resistant C. parapsilosis but also a case of multidrug-resistant C. parapsilosis.13
In Latin America, Candida parapsilosis is the most frequent species of the non-albicans Candida group and the second most common cause of invasive candidiasis (IC) after Candida albicans.14C. parapsilosis has maintained a low rate of antifungal resistance, especially for azoles, where the resistance rate is less than 7%.15–18 Mutations in, and/or overexpression of, ERG11 and overexpression of MDR1 and CDR1 have been reported previously as important molecular mechanisms associated with azole resistance C. parapsilosis.19–21
In Mexico, there have been no reports on outbreaks of azole-resistant Candida parapsilosis nor associated azole resistance mechanisms. The aim of this study was to investigate ERG11 single nucleotide polymorphisms (SNPs) in isolates of C. parapsilosis obtained during an outbreak of IC due to azole-resistant C. parapsilosis in a Mexican hospital.
Methods
Study design
This is an observational and retrospective study. We recovered clinical isolates causing IC in the Hospital General Dr. Manuel Gea Gonzalez in Mexico City, a 220-bed general hospital with a 10-bed ICU unit, between January 2014 and December 2016. Clinical data were retrieved retrospectively using a standardized format. Clinical data obtained were age, gender, date of admission, date of discharge, previous diseases, year and month of isolation, medical service during the diagnosis of IC, previous surgeries, APACHE II score, time using a central venous catheter (CVC), use of antifungal prior to IC, antifungal treatment, and clinical outcome.
This study was approved by the Hospital General Dr. Manuel Gea Gonzalez Institutional Review Board (IRB): number 36-16-2020.
Clinical isolate identification and antifungal susceptibility testing
Candida isolates recovered from storage were initially identified by the local microbiology laboratory using Vitek2 (Biomerieux, France). This system was also used to determine fluconazole and voriconazole antifungal drug susceptibility. EUCAST clinical breakpoints22 were used to define fluconazole resistance: >4 µg/ml for C. albicans, C. parapsilosis, and C. krusei, and for species without breakpoints the same cutoff was used. For C. glabrata isolates, the cut-off for fluconazole resistance was >32 µg/ml. For resistant isolates of C. parapsilosis, fluconazole and voriconazole susceptibility was confirmed using EUCAST E.DEF 7.3.1 broth microdilution antifungal susceptibility testing for yeasts.22
PCR amplification of ITS2 region and ERG11 gene
Genomic DNA of each fluconazole-resistant C. parapsilosis isolate was extracted as previously reported.23 To amplify the ITS2 region, primers specific for this region were designed. ITS region DNA sequences (SSU-ITS-5.8-ITS2-LSU) for 15 different Candida species (C. albicans, C. sake, C. parapsilosis, C. metapsilosis, C. orthopsilosis, C. glabrata, C. tropicalis, C. utilis, C. dubliniensis, C. krusei, C. guilliermondii, C. rugose, C. famata, C. lusitaniae, C. ciferri) were obtained from Genbank (http://www.ncbi.nlm.nih.gov/genbank/), ISHAM ITS database (http://its.mycologylab.org/), and the UNITE repository (www.unite.ut.ee/analysis.php). DNA sequences were aligned using ClustalW2 Omega (www.ebi.ac.uk/Tools/msa/clustalo/) and regions suitable as primers were identified. The quality of the primer pair was analyzed using the IDT tool (www.idtdna.com/calc/analyzer). The ITS2 primer pair used was ITS2F (5′-3′) TCGAATCTTTGAACGCACATTG and ITS2R (5′-3′) GCTTTTGCCGCTTCACTC, amplifying a predicted PCR product between 233 and 409 bp depending on the species.
An ERG11 primer pair was also designed based on the ERG11 gene sequence of C. parapsilosis ATCC 22 019 (Genbank accession number GQ302972.1). ERG11 primer pair sequences were ERG11F (5′-3′) TGGGTACAACTACTTTATGAC and ERG11R (5′-3′) CCATTGAAGTATAATCAAC, amplifying a predicted PCR product of 1518 bp.
To amplify the ITS2 region and the ERG11 gene, PCR reactions were set up using KAPA HiFi hotstart ReadyMix (Roche Diagnostics Ltd, Welwyn garden, UK) following the manufacturer's instructions. The final primer concentration in the reaction was 0.4 μM, with 50 ng genomic DNA template. PCR cycles were one cycle of initial denaturation 95°C for 3 min, 30 cycles of denaturation 98°C for 20 s, annealing temperature 52°C for 15 s, extension 72°C for 30 s, and one cycle of final extension 72°C for 1 min in a Primo base Techne Thermal Cycler (Cole-Parmer, Staffordshire, UK). PCR products were analyzed by 1% agarose gel. PCR products generated for the ITS2 region and ERG11 were purified using the QIAquick® PCR Purification Kit (Qiagen, Manchester UK) prior to DNA sequencing.
DNA sequencing and analysis
DNA sequencing was performed by DNA Sequencing & Services (MRC I PPU, School of Life Sciences, University of Dundee, Scotland, www.dnaseq.co.uk) using Applied Biosystems Big-Dye version 3.1 chemistry on an Applied Biosystems model 3730 automated capillary DNA sequencer. PCR products were sequenced in both directions using the same primer pairs used in PCR reactions.
Species and SNP identification
The ITS2 PCR product DNA sequences were analyzed using SeqMan Pro (DNASTAR, Madison, WI USA), then the sequences were compared to three ITS/nucleotide databases (GENBANK, ISHAM ITS database http://its.mycologylab.org/and UNITE repository https://unite.ut.ee/repository.php) to identify the species.
The ERG11 DNA sequences were aligned against the C. parapsilosis ATCC 22 019 GQ302972.1 reference sequence. Sequence alignment, evaluation of the quality of the sequences, and identification of SNPs were done using SNAPgene version 4.3.10 (GSL Biotech LLC, Chicago, USA).
To localize the putative amino acid changes in the C. parapsilosis Erg11 (14-alpha sterol demethylase) protein sequences, the C7EXA5 sequence (https://www.uniprot.org) was used as a template, which is derived from the EMBL nucleotide sequence of C. parapsilosis ATCC 22 019 GQ302972.1. ExPASY bioinformatics resource tool portal (https://web.expasy.org/docs/expasy_ref.html) and Clustal Omega portal (https://www.ebi.ac.uk/Tools/msa/clustalo/) were used for aligning and translation analysis.
Microsatellite genotyping
Four previously reported C. parapsilosis specific microsatellite loci named CP1, CP4, CP6, and B5 were used to analyze the fluconazole-resistant C. parapsilosis isolates.24 PCR reactions, settings, and products were performed as mentioned above. Annealing temperatures varied between 58°C and 60°C depending on the primer pair. After PCR, samples were prepared with the Agilent DNA 1000 kit and analyzed in a 2100 Agilent bioanalyzer instrument using 2011 Expert Software (Agilent Technologies LDA UK Limited, Cheshire, UK). C. parapsilosis ATCC 90 018 was also analyzed for the same loci and it was used as a reference strain. The software “Genepop 4.7.5 on the Web” (https://genepop.curtin.edu.au) was used to analyze the characteristics of the microsatellite loci such as the number of alleles, allele frequency, and number of genotypes. Genetic distance and clustering analysis were performed and a dendrogram was constructed using DendroUPGMA (http://genomes.urv.cat/UPGMA/).
Statistical analysis
Descriptive clinical data are presented as proportions, median, and interquartile range (IQR) as required. Statistical analysis for the proportion of resistant isolates was performed with the χ2 test for trend using IBM SPSS version 25 software (IBM, NY, USA) and a P value ≤ 0.05 was considered statistically significant. Plots were created with GraphPad Prism 8 (Graphpad Software, CA, USA).
Results
Characteristics of the invasive candidiasis cases and identification of an outbreak of fluconazole-resistant C. parapsilosis
In the Hospital General Dr. Manuel Gea Gonzalez, Mexico City, the number of new cases of IC were one to three per year prior to 2014 (2010-2013), all due to C. albicans. Between 2014 and 2016, a total of 74 cases were diagnosed (at least 20 cases per year). In this hospital, surveillance of infections is carried out with endemic channels. As the incident cases of IC from 2014 to 2016 were above the 75th percentile, this was considered an outbreak.
During the study period, most of the invasive infections occurred in ICU/ER services (43%, 32/74), and the median length of stay in hospital was 15 days (IQR 10–30 days). Most isolates were recovered from blood (69/74, 93%), with the remaining five cases identified in kidney abscess (n = 1) and peritoneal/abdominal liquid, including dialysis (n = 4). Concomitant bacteremia was found in 15 cases (22%, 15/68), with the most frequent bacterial isolates being Escherichia coli (35%, 6/17) and Enterococcus sp. (35%, 6/17). Complete medical information was only available for 44 patients. Diabetes (18/44, 41%) was the condition most frequently found, 64% (28/44) had undergone surgery, 34% (15/44) had a previous hospital admission, and 9% (4/44) had been transferred from a different hospital in the 72 h before diagnosis of invasive candidiasis. Antifungal use prior to the IC event was found in 9% (4/44) of the cases. All patients with candidaemia had a follow-up blood culture after initiation of antifungal therapy and no persistent fungemia was documented. The mortality rate among the 44 patients was <10% (Table 1).
Table 1.
General characteristics of the invasive candidiasis cases studied between 2014–2016
| General characteristics | n = 74 (%) | FCZ resistant C. parapsilosis n = 15 (%)+ | FCZ susceptible C. parapsilosis n = 14 (%)++ |
|---|---|---|---|
| Gender (male) | 45 (64) | 8 (53) | 9 (64) |
| Age (Years) | 47 (27-58) | 46 (29-58) | 4 (0-43) |
| Previous diseases/condition (n = 44) | |||
| Diabetes | 18 (41) | 2 (18)+ | 1 (14)++ |
| Newborn | 8 (18) | 1 (7) | 7 (50) |
| Chronic renal failure | 5 (12) | 1 (7) | 0 (0) |
| Cancer | 4 (9) | 1 (7) | 1 (7) |
| Lung disease | 2 (4) | 1 (7) | 0 |
| HIV | 1 (2) | 0 | 0 |
| Transfer from other hospital (n = 44) | 4 (9) | 1 (9)+ | 0++ |
| Previous surgery (n = 44) | 28 (64) | 5 (45)+ | 3 (43)++ |
| Previous admission (n = 44) | 15 (34) | 2 (18)+ | 1 (14)++ |
| Service of identification | |||
| ICU or emergency service* | 32 (43) | 10 (67) | 4 (29) |
| Medical | 28 (38) | 4 (27) | 8 (57) |
| Surgical | 14 (19) | 1 (6) | 2 (14) |
| APACHE II score (median, IQR) | 21 (17-27) | 23 (15-29)+ | 13 (5-21)++ |
| Central venous catheter (days) (median, IQR) | 11 (7-25) | 10 (7-19)+ | 6 (3-11)++ |
| Previous antifungal use (n = 44) | 4 (9) | 1 (9)+ | 0 (0) ++ |
| Length of stay in hospital before diagnosis (days)** (median, IQR) | 15 (10-30) | 29 (21-54) | 14 (8.5-19) |
| Concomitant bacteraemia** | 15 (22) | 1 (7) | 1 (7) |
| Specimen | |||
| Blood | 69 (93.2) | 15 (100) | 13 (93) |
| Peritoneal liquid/secretion | 3 (4.1) | 0 | 1 (7) |
| Dialysis liquid | 1 (1.4) | 0 | 0 |
| Renal abscess | 1 (1.4) | 0 | 0 |
| Antifungal management (n = 38) | |||
| None | 2 (5) | – | – |
| AmBD | 7 (18) | 2 (29)# | 2 (50)## |
| Fluconazole | 6 (16) | 1 (14)# | 1 (25)## |
| Caspofungin | 22 (58) | 4 (57)# | 1 (25)## |
| Combined treatment | 1 (3) | - | - |
| Mortality rate (n = 44) | 4 (9) | 1 (9)+ | 1 (14)++ |
*ICU isolation means identification of Candida species during ICU stay (at least 48 h after ICU stay and 48 h after discharge).
** Information from 68 patients was available.
n = 11, ++n = 7; #n = 7, ##n = 4.
All 74 Candida isolates were identified by Vitek2 at the local clinical laboratory (Mexico City). The majority of isolates causing IC were C. parapsilosis (39%, 29/74), followed by C. albicans (23%, 17/74) and C. glabrata (19%, 14/74; Figure 1A).
Figure 1.
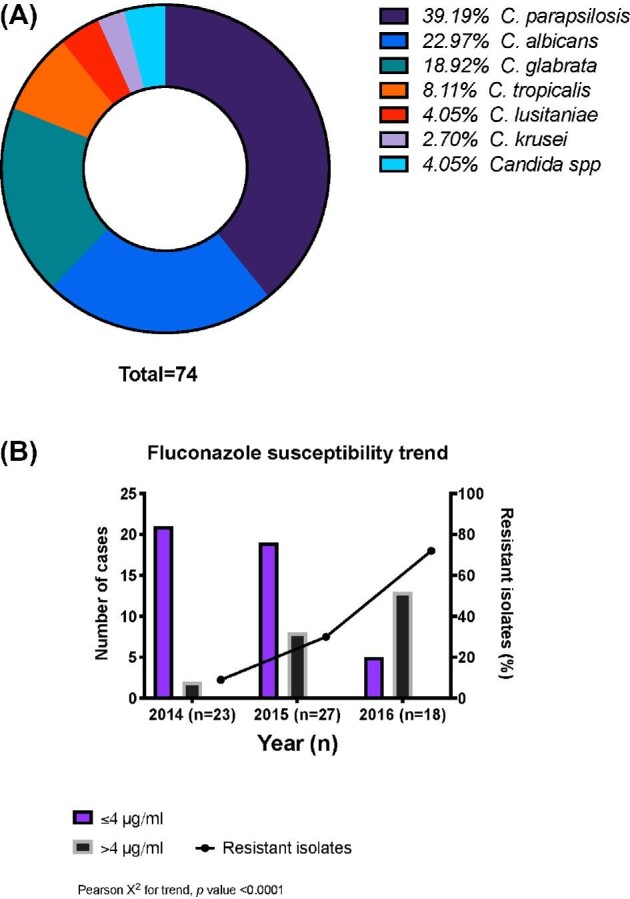
Fluconazole resistance and species distribution of outbreak isolates. A total of 74 isolates were obtained during the IC outbreak and were identified to species level by Vitek2 during the study period (A). Fluconazole resistance measured by Vitek2 for all the Candida isolates obtained during the study period was plotted by year (B).
Antifungal drug susceptibility testing was also performed by Vitek2 for 68 of 74 isolates, with six isolates not tested due to lack of material. Twenty-three of the 68 isolates tested (34%) were considered resistant to fluconazole. In 2014, fluconazole resistance was <10%; however, in 2016, this increased to >60% (Figure 1B). C. parapsilosis accounted for most of the resistant isolates (15/29, 52%), followed by C. tropicalis (2/5, 40%) and C. glabrata (3/13, 23%). The fluconazole MIC range determined by Vitek for C. parapsilosis isolates was 1–32 µg/ml, while none of the C. glabrata isolates had MICs <1 µg/ml and the three resistant C. glabrata isolates had MICs ≥32 µg/ml. Two C. krusei isolates were considered intrinsically resistant to fluconazole.
SNPs identified in C. parapsilosis fluconazole-resistant isolates
Twelve of 15 fluconazole resistant C. parapsilosis isolates were recovered for further analysis, with the other three not recoverable from storage. All were identified as C. parapsilosis sensu stricto by ITS2 sequencing. Fluconazole MICs determined by broth microdilution ranged between 8 and 32 µg/ml and for voriconazole MICs between 0.125 and 2 µg/ml. Voriconazole resistance was 42% (5/12), where resistance was defined as an MIC above 0.25 µg/ml (Table 2).22
Table 2.
Fluconazole and voriconazole susceptibility of Candida parapsilosis blood isolates and associated ERG11 SNPs
| Isolate # (specimen, year) | Service of identification | Fluconazole MIC (µg/ml) | Voriconazole MIC (µg/ml) | ERG11 SNPs+ | Amino Acid change++ | SNP Zygosity |
|---|---|---|---|---|---|---|
| 2 | Medical | 16 | 0.25 | ND | ND | ND |
| (blood, 2014) | ||||||
| 16 | ICU/ER | 8 | 0.125 | T591C | I197I | Homozygous |
| (blood, 2014) | ||||||
| 23 | Medical | 32 | 2 | T591C | I197I | Homozygous |
| (blood, 2014)* | A395T | Y132F | Homozygous | |||
| 34 | ICU/ER | 32 | 0.25 | ND | ND | ND |
| (blood, 2015) | ||||||
| 42 | Medical | 16 | 0.25 | T591C | I197I | Homozygous |
| (blood, 2015) | ||||||
| 44 | Medical | 16 | 0.5 | T591C | I197I | Homozygous |
| (blood, 2015) | A395T | Y132F | Heterozygous | |||
| 47 | Surgical | 32 | 0.5 | T591C | I197I | Homozygous |
| (blood, 2015) | A395T | Y132F | Heterozygous | |||
| 48 | ICU/ER | 8 | 0.125 | T591C | I197I | Homozygous |
| (blood, 2015) | ||||||
| 56 | ICU/ER | 8 | 0.125 | T591C | I197I | Homozygous |
| (blood, 2015) | ||||||
| 53 | Medical | 8 | 0.125 | ND | ND | ND |
| (blood, 2016) | ||||||
| 66 | ICU/ER | 16 | 1 | T591C | I197I | Homozygous |
| (blood, 2016) | ||||||
| 67 | ICU/ER | 32 | 0.25 | T591C | I197I | Homozygous |
| (blood, 2016) | A395T | Y132F | Heterozygous | |||
| 68 | ICU/ER | 16 | 0.25 | T591C | I197I | Homozygous |
| (blood, 2016) | A395T | Y132F | Heterozygous | |||
| 76 | ICU/ER | 16 | 1 | T591C | I197I | Homozygous |
| (blood, 2016) | A395T | Y132F | Heterozygous | |||
| 78 | ICU/ER | 8 | 0.25 | T591C | I197I | Homozygous |
| (blood, 2016) | A395T | Y132F | Heterozygous |
Twelve of 15 fluconazole resistant Candida parapsilosis isolates were recovered and analysed.
*Isolate 23 was the only resistant isolate recovered from a patient transferred from another hospital.
**Isolate 47 was the only one recovered from a patient having received prior antifungal drug therapy.
Change in the nucleotide sequence of ERG11 gene compared with C. parapsilosis ATCC 22 019 accession number GQ302972.1.
++Amino acid changes compared to the 14-alpha sterol demethylase (Erg11) C7EXA5 sequence.
ND = not determined, ICU = intensive care unit, ER = emergency room.
The ERG11 gene of all 12 C. parapsilosis isolates was amplified and sequenced. Compared to the reference ATCC 22 019 ERG11 sequence, one common silent SNP was identified at T591C in all isolates. A second mutation was found in seven of the isolates at A395T, corresponding with Y132F, which is a missense mutation. Six of the isolates carrying this mutation were heterozygous at this position and only one was homozygous for the A395T SNP. The six isolates had similar MICs for fluconazole and voriconazole, with two isolated in July 2015 and the other four between April and August 2016. The presence of the same two mutations in the six isolates means that they are likely to be closely related. The isolate with the homozygous mutation at Y132F was initially recovered from a patient who transferred from another hospital before the number of azole-resistant C. parapsilosis cases started to increase in 2014 (Table 2).
Microsatellite typing showed closely related azole-resistant isolates
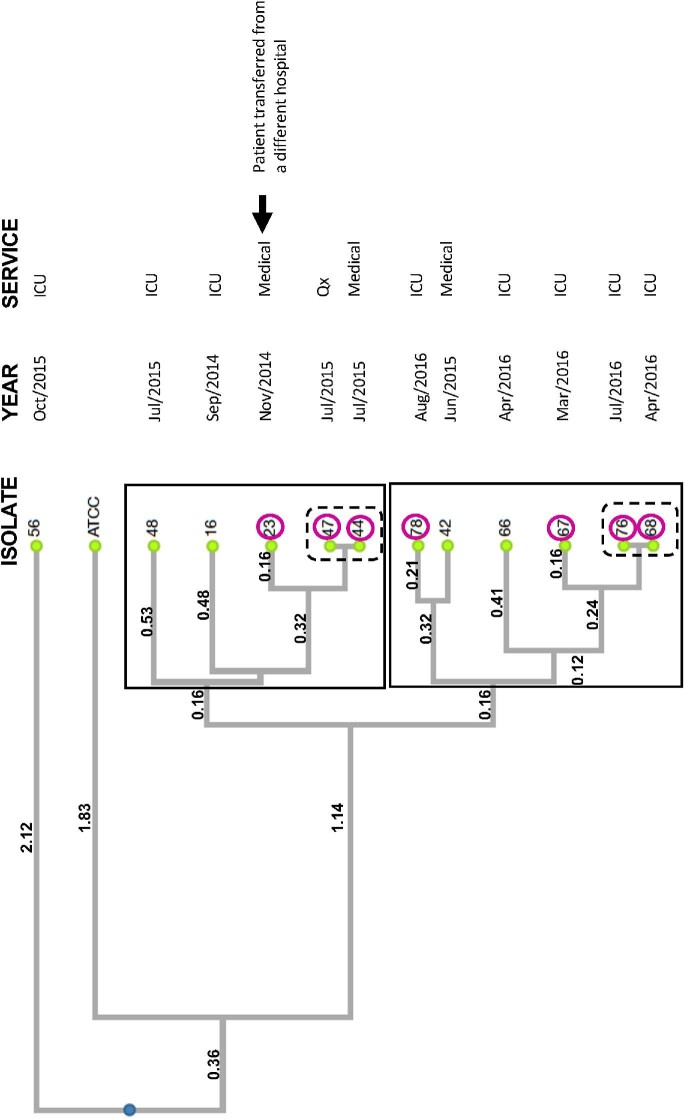
Genotyping analysis identified 10 different but closely related genotypes in the azole-resistant C. parapsilosis isolates (Table 3). Two closely related clusters and one outlier isolate of azole-resistant C. parapsilosis were found (Figure 2). Isolates were considered closely related and included within a cluster if they shared at least five of eight alleles and/or at least three of four loci among them. All included isolates shared these characteristics except for isolate 42, which was included in cluster 2 as it shared the same B5 locus as isolate 78. Cluster 1 contained only isolates recovered between 2014 and 2015, including the isolate recovered from the patient transferred from a different hospital (isolate 23). Two pairs of isolates (isolates 44 and 47, and isolates 68 and 76) were identical by microsatellite typing and localized one pair per cluster. The identical pair in cluster 1, isolates 44, 47 were closely related to the isolate 23 (Figure 2). Meanwhile, five isolates in cluster 2 (isolates 66, 67, 68, 76, and 78) matched three of four loci and had either one or two mismatches in the B5 locus (Table 3). These same five isolates were all recovered in 2016 and were isolated from the ICU/ER; four of these isolates contained the Y132F mutation (Table 3, Figure 2).
Table 3.
Microsatellite typing results of 12 azole-resistant C. parapsilosis blood isolates
| Isolate | Year | CP1* | CP4* | CP6* | B5* |
|---|---|---|---|---|---|
| ATCC 90 018 | Reference strain (1992) | 234/234 | 245/245 | 261/336 | 147/169 |
| 16 | 2014 | 234/234 | 236/360 | 291/366 | 149/171 |
| 23 | 2014 | 234/234 | 230/357 | 291/366 | 147/171 |
| 42 | 2014 | 234/234 | 230/363 | 291/366 | 143/167 |
| 44 | 2015 | 234/234 | 230/366 | 288/375 | 147/171 |
| 47 | 2015 | 234/234 | 230/366 | 288/375 | 147/171 |
| 48 | 2015 | 234/234 | 242/372 | 288/369 | 147/171 |
| 56 | 2015 | 234/234 | 197/197 | 288/369 | 139/165 |
| 66 | 2016 | 234/234 | 233/375 | 297/372 | 147/171 |
| 67 | 2016 | 234/234 | 233/375 | 297/372 | 145/171 |
| 68 | 2016 | 234/234 | 233/375 | 294/372 | 145/169 |
| 76 | 2016 | 234/234 | 233/375 | 294/372 | 145/169 |
| 78 | 2016 | 234/234 | 233/375 | 294/372 | 143/167 |
Identical isolates in bold.
The numbers indicate the fragment size in base pairs of the different alleles obtained with the listed marker.
Figure 2.
Clustering of azole-resistant C. parapsilosis isolates during the studied outbreak. Dendrogram showing clusters among 12 azole-resistant C. parapsilosis blood isolates based on microsatellite multilocus genotyping. Distance matrix was based on Eucledian coefficient. Genetic distances are in Newick format and clustering was performed by the UPGMA method. Cophenetic Correlation Coefficient (CP) = 0.96. Clusters are within boxes, identical isolates are within dashed line boxes, isolates within circles contain the Y132F mutation.
Discussion
Our study reports an outbreak of IC due to fluconazole-resistant C. parapsilosis isolates carrying a Y132F mutation in Erg11. Although C. parapsilosis is expected to be fluconazole-susceptible, since 2015 several publications worldwide have reported outbreaks due to azole-resistant C. parapsilosis and, in Latin America, this resistant species has already been reported in Brazil.4 These studies also reported the same ERG11 SNP (A395T) identified in our study.4–13
The Y132F mutation has been identified as directly contributing to reduced azole susceptibility in both C. auris25 and C. tropicalis.26 The mutation is located in a predicted catalytic site interacting with fluconazole and appears to contribute to modifying the sterol composition in the fungal cell membrane.19 Berkow et al. showed that this mutation was associated with higher levels of 14-methyl fecosterol and lanosterol/obtusifoliol, suggesting that the Y132F change modifies the function of the lanosterol 14-alpha-demethylase enzyme.19 Expressing the Y312F ERG11 variant in S. cerevisiae was sufficient to raise the fluconazole MIC from 8 to 128 µg/ml.5
In South Korea, Choi et al. reported that Y312F C. parapsilosis isolates had a higher propensity for clonal transmission and also persisted in hospitals for several years (as shown by microsatellite typing).9 In South Africa, the mutation was found only in fluconazole-resistant C. parapsilosis and not found in susceptible ones.8 This mutation was also linked with an outbreak in Brazil and was only found in azole-resistant isolates.4
The increase in fluconazole-resistant species could be related to the previous use of this antifungal agent. We only had evidence for prior fluconazole exposure in one patient with a resistant isolate in our study, although the proportion could be higher because complete clinical information was not available for all samples.
The spreading and persistence of this resistant species within the hospital could have several explanations other than previous exposure to fluconazole. The transmission, spreading, and persistence in healthcare settings has been documented recently for C. auris and its persistence relies on remaining as a colonizer of hospital environment/surfaces, allowing patient to patient transmission or fomite/device to patient transmission.27,28 Similar to C. auris, C. parapsilosis has always been known for its capacity to persist and to be transmitted by hand carriage.21 In a recent study, C. parapsilosis persisted for up to 28 days on plastic surfaces, twice the time that C. auris persisted.29 Maintaining hospital active surveillance programs to identify outbreaks like the current one, including antifungal use as part of stewardship programs, and improving infection control measures are key strategies to prevent the spread and increase of azole-resistant Candida species.
In Mexico, fluconazole remains the first choice for empirical treatment in fungal infections according to a recent report from our group.30 Azoles remain an important affordable therapeutic option for IC and the only option to date for oral and ambulatory therapy, especially for C. parapsilosis due to concerns about intrinsic higher echinocandins minimal inhibitory concentrations.31 The increasing rate of azole-resistant Candida species will further limit the already low number of therapeutical options available to treat invasive infections.
To our knowledge, this is the first time that an IC outbreak due to azole-resistant C. parapsilosis associated with the Y132F mutation has been reported in Mexico, thus contributing to mapping antifungal resistance distribution worldwide. The finding of Y132F in these isolates does not mean that there may not be other coexisting mechanisms of antifungal resistance, such as overexpression of CDR1 and/or MDR1, but these were not evaluated in this study.
Acknowledgment
We thank DNA Sequencing & Services (MRC I PPU, School of Life Sciences, University of Dundee, Scotland, www.dnaseq.co.uk) for DNA sequencing. DECL was funded by a Wellcome Trust Strategic Award for Medical Mycology and Fungal Immunology (097377) PhD studentship and MP was funded by the Medical Research Council Centre for Medical Mycology at the University of Aberdeen (MR/N006364/1) for MRes studentship. DMM was funded by NC3Rs (BB/P02050X/1) and BBSRC (BB/P02050X/1).
Contributor Information
Dora E Corzo-Leon, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen, UK; Department of Infectious Diseases and Epidemiology, Hospital General Dr. Manuel Gea González, Mexico City, México.
Mark Peacock, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen, UK; MRC Centre for Medical Mycology, University of Exeter, Exeter, UK.
Patricia Rodriguez-Zulueta, Department of Infectious Diseases and Epidemiology, Hospital General Dr. Manuel Gea González, Mexico City, México.
Grace J Salazar-Tamayo, Department of Infectious Diseases and Epidemiology, Hospital General Dr. Manuel Gea González, Mexico City, México; Department of Infectious Diseases and Epidemiology, Hospital de SOLCA, Quito, Ecuador.
Donna M MacCallum, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Foresterhill, Aberdeen, UK.
Declaration of interest
PRZ has been speaker for Pfizer. All the other authors have no conflict of interest.
References
- 1. Lone SA, Ahmad A. Candida auris - the growing menace to global health. Mycoses. 2019; 62: 620. [DOI] [PubMed] [Google Scholar]
- 2. Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin Set al. . Multidrug-resistant Candida haemulonii and C. auris, tel aviv, Israel. Emerg Infect Dis. 2017; 23: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perlin DS, Rautemaa-Richardson R, Alastruey-Izquierdo A. The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect Dis. 2017; 17: e383–92. [DOI] [PubMed] [Google Scholar]
- 4. Thomaz DY, De Almeida JN, Lima GME, De Oliveira Nunes M, Camargo CH, De Carvalho Grenfell Ret al. . An azole-resistant Candida parapsilosis outbreak: clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front Microbiol. 2018; 9: 2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh A, Singh PK, de Groot T, Kumar A, Mathur P, Tarai Bet al. . Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J Antimicrob Chemother. 2019; 74: 1260–1268. [DOI] [PubMed] [Google Scholar]
- 6. Arastehfar A, Daneshnia F, Hilmioğlu-Polat S, Fang W, Yaşar M, Polat Fet al. . First report of candidemia clonal outbreak caused by emerging fluconazole-resistant Candida parapsilosis isolates harboring Y132F and/or Y132F+K143R in Turkey. Antimicrob Agents Chemother. 2020; 64: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martini C, Torelli R, de Groot T, De Carolis E, Morandotti GA, De Angelis Get al. . Prevalence and clonal distribution of azole-resistant Candida parapsilosis isolates causing bloodstream infections in a large Italian hospital. Front Cell Infect Microbiol. 2020; 10: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Magobo RE, Lockhart SR, Govender NP. Fluconazole‐resistant Candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, Mycoses. 2020; 63:471–477. [DOI] [PubMed] [Google Scholar]
- 9. Choi YJ, Kim Y-J, Yong D, Byun J, Kim TS, Chang YSet al. . Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F Mutation in ERG11 Gene, Emerg Infect Dis. 2018; 24: 1768–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asadzadeh M, Ahmad S, Al-Sweih N, Khan Z. Epidemiology and molecular basis of resistance to fluconazole among clinical Candida parapsilosis isolates in Kuwait. Microb Drug Resist. 2017; 23: 966–972. [DOI] [PubMed] [Google Scholar]
- 11. Zhang L, Yu SY, Chen SCA, Xiao M, Kong F, Wang Het al. . Molecular characterization of Candida parapsilosis by microsatellite typing and emergence of clonal antifungal drug resistant strains in a multicenter surveillance in China. Front Microbiol. 2020; 11: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grossman NT, Pham CD, Cleveland AA, Lockhart SR. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob Agents Chemother. 2015; 59: 1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arastehfar A, Daneshnia F, Najafzadeh MJ, Hagen F, Mahmoudi S, Salehi Met al. . Evaluation of molecular epidemiology, clinical characteristics, antifungal susceptibility profiles, and molecular mechanisms of antifungal resistance of Iranian Candida parapsilosis species complex blood isolates. Front Cell Infect Microbiol. 2020; 10: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nucci M, Queiroz-Telles F, Alvarado-Matute T, Tiraboschi IN, Cortes J, Zurita Jet al. . Epidemiology of candidemia in Latin America: A Laboratory-Based Survey. PLoS One 2013; 8: e59373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lyon GM, Karatela S, Sunay S, Adiri Y. Antifungal susceptibility testing of candida isolates from The Candida surveillance study. J Clin Microbiol. 2010; 48: 1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio Vet al. . Results from the artemis disk global antifungal surveillance study, 1997 to 2007: A 10.5-year analysis of susceptibilities of candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J Clin Microbiol. 2010; 48: 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida Species. Front Microbiol. 2017; 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Castanheira M, Messer SA, Rhomberg PR, Pfaller MA. Antifungal susceptibility patterns of a global collection of fungal isolates: results of the SENTRY antifungal surveillance program (2013). Diagn Microbiol Infect Dis. 2016; 85: 200–204. [DOI] [PubMed] [Google Scholar]
- 19. Berkow EL, Manigaba K, Parker JE, Barker KS, Kelly SL, Rogers PD. Multidrug Transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob Agents Chemother. 2015; 59: 5942–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Souza ACR, Fuchs BB, Pinhati HMS, Siqueira RA, Hagen F, Meis JFet al. . Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella larvae. Antimicrob Agents Chemother. 2015; 59: 6581–6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tóth R, Nosek J, Mora-Montes HM, Gabaldon T, Bliss JM, Nosanchuk JDet al. . Candida parapsilosis : from genes to the bedside. Clin Microbiol Rev. 2019; 32: 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arendrup M, et al. EUCAST antifungal MIC method for yeasts. EUCAST Defeinitive Doc EDEF 731 2017; (December): 1–21. https://eucast.org/astoffungi/methodsinantifungalsusceptibilitytesting/susceptibility_testing_of_yeasts/ and it was consulted on February 2019. [Google Scholar]
- 23. Hoffman CS, Winston F.. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformaion of Escherichia coli. Gene. 1987; 57: 267–272. [DOI] [PubMed] [Google Scholar]
- 24. Sabino R, Sampaio P, Rosado L, Stevens DA, Clemons KV, Pais C. New polymorphic microsatellite markers able to distinguish among Candida parapsilosis sensu stricto isolates. J Clin Microbiol. 2010; 48: 1677–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Healey KR, Kordalewska M, Ortigosa CJ, Singh A, Berrío I, Chowdhary Aet al. . Limited ERG11 mutations identified in isolates of candida auris directly contribute to reduced azole susceptibility. Antimicrob Agents Chemother. 2018; 62: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan J, Zhang J, Chen W, Sun Y, Wan Z, Li Ret al. . The A395T mutation in ERG11 gene confers fluconazole resistance in Candida tropicalis causing candidemia. Mycopathologia. 2015; 179: 213–218. [DOI] [PubMed] [Google Scholar]
- 27. Chowdhary A, Sharma C, Meis JF. Candida auris: A rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 2017; 13: e1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eyre DW, Madder H, Moir I, Moroney R, Butcher L, Morgan Met al. . A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018; 379: 1322–1331. [DOI] [PubMed] [Google Scholar]
- 29. Welsh RM, Bentz ML, Shams A, Houston H, Lyons A, Rose LJet al. . Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017; 55: 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corzo-León DE, Perales-Martínez D, Martin-Onraet A, Rivera-Martínez N, Camacho-Ortiz A, Villanueva-Lozano H. Monetary costs and hospital burden associated with the management of invasive fungal infections in Mexico: a multicenter study. Braz J Infect Dis. 2018; 22: 360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner Let al. . Clinical practice guideline for the management of candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016; 62: e1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]