Abstract
Pharmacological inhibition of MDM2/4, which activates the critical tumor suppressor p53, has been gaining increasing interest as a strategy for the treatment of acute myeloid leukemia (AML). While clinical trials of MDM2 inhibitors have shown promise, responses have been confined to largely molecularly undefined patients, indicating that new biomarkers and optimized treatment strategies are needed. We previously reported that the microRNA miR-10a is strongly overexpressed in some AML, and demonstrate here that it modulates several key members of the p53/Rb network, including p53 regulator MDM4, Rb regulator RB1CC1, p21 regulator TFAP2C, and p53 itself. The expression of both miR-10a and its downstream targets were strongly predictive of MDM2 inhibitor sensitivity in cell lines, primary AML specimens, and correlated to response in patients treated with both MDM2 inhibitors and cytarabine. Furthermore, miR-10a inhibition induced synergy between MDM2 inhibitor Nutlin-3a and cytarabine in both in vitro and in vivo AML models. Mechanistically this synergism primarily occurs via the p53-mediated activation of cytotoxic apoptosis at the expense of cytoprotective autophagy. Together these findings demonstrate that miR-10a may be useful as both a biomarker to identify patients most likely to respond to cytarabine+MDM2 inhibition and also a druggable target to increase their efficacy.
Subject terms: Acute myeloid leukaemia, Targeted therapies
Introduction
p53 is a critical tumor suppressor protein that has a central role in modulating diverse cellular processes including proliferation, cell cycle, DNA repair, senescence, apoptosis, and autophagy [1]. Approximately half of all human cancers bear inactivating mutations of p53, and p53-deficent mice develop a range of tumors with high penetrance [2]. While common in solid cancers, TP53 mutations are less frequent in hematological malignancies such as AML, in which they are present in only around 5% of patients [3]. In the majority of these cases however it is thought that p53 is repressed by other means, including via the overexpression of its two primary negative regulators, murine double minute 2 homolog (MDM2) and MDM4 [4, 5], or via mutation of nucleophosmin 1 (NPM1) [6] which results in the improper relocalization of the crucial MDM2 inhibitor p14ARF from the nucleoli to the cytoplasm, effectively disabling it [7, 8].
Pharmacological inhibitors of MDM2/4 have therefore recently been developed in an effort to reactivate the p53/Rb tumor suppressor network in cancers in which p53 is functional but repressed [9]. While most clinical trials in AML have been Phase I/II studies focussed on safety and tolerability, some preliminary response data has been reported [9]. Overall efficacy has been promising but variable, with second generation MDM2 inhibitor idasanutlin (RG7388) + cytarabine in relapsed/refractory AML for example reporting response rates of around 60% [10]. Toxicity is common however, with most patients experiencing adverse gastrointestinal and infection-related events, as well as frequent high grade neutropenia and thrombocytopenia [10, 11]. Interestingly, responses have been observed in highly diverse patients, including those in varied risk groups as well as subgroups of relapsed/refractory patients. These findings, while showing promise, highlight the need to develop new predictive biomarkers to identify those patients most likely to benefit as well as non-responders that can be spared toxic side-effects. In addition, the identification of targeted compounds with the potential to synergize with MDM2 inhibition to enable lower effective doses would be beneficial.
We previously reported that the microRNA miR-10a is strongly overexpressed in AML compared to normal bone marrow (up to 80-fold), and also displays subgroup-specific expression [12]. We also provided some evidence that miR-10a may be involved in p53/Rb tumor suppressor/stress response pathway regulation via its repression of a number of the p53/Rb networks’ key genes, including p53 regulator MDM4, Rb regulator RB-Inducible Coiled-Coil 1 (RBCC1), and p21 regulator Transcription Factor AP2-Gamma (TFAP2C) [12, 13]. These findings have suggested that miR-10a may act as an oncomiR via its repression of the p53/Rb network in those ~40–60% of AML patients that strongly overexpress the miRNA. In the current study we hypothesized that miR-10a therefore may have a role in the response to MDM2 inhibitors in AML, and could represent both a valuable therapeutic target for modulating drug responses via de-repression of its key p53/Rb-related targets, as well as a potential biomarker of response.
Materials and methods
Cell lines and transfections
Cell lines were obtained from ATCC and cultured according to their recommendations (www.atcc.org). Cell lines were authenticated by STR profiling and confirmed to be mycoplasma-free immediately prior to use. An hsa-miR-10a mimic, inhibitor, and negative control based on Qiagen miScript sequences (product numbers MSY0000253, MIN0000253, and 1027271 respectively) were cloned into CD713B lentiviral expression plasmids (System Biosciences, CA, USA) and transduced into cultured cells. Successfully transduced cells were selected using 2µG/mL puromycin and underwent FACS to isolate GFP + (positively transduced) clones. miR-10a expression was determined by TaqMan QPCR in each clone, which identified an inhibitor-transduced clone with 94% knockdown of miR-10a expression, and a mimic-transduced clone with 624-fold increase in miR-10a expression, compared to scrambled inhibitor/mimic-transduced cells (data not shown). Cells regularly underwent flow cytometry to confirm purity of transduced cells. A mammalian expression construct containing RB1CC1 (AddGene plasmid #24300) and CDKN1A siRNA (Qiagen siRNA SI03031105) were transiently transfected into recipient cells 24 h prior to drug treatment.
In vitro assays
Drug sensitivity assays were performed using the CellTitre 96 Aqueous One Solution Cell Proliferation Assay Assay (Promega). Growth inhibition following drug treatment was used to generate Combination Index (CI) values calculated by Calcusyn (Biosoft), as previously described [14]. CI values of <1, 1, >1 indicated synergism, additive effect, and antagonism, respectively. The Click-iT Plus EdU Cell Cycle Assay (ThermoFisher Scientific) was performed as per manufacturer’s instructions. Autophagy activation was measured by flow cytometry in cells labeled with the autofluorescent compound monodansylcadaverine (MDC) which binds to autophagic vacuoles, as previously described [15], as well as Western blotting for expression of the autophagy marker LC3-II [16].
mRNA and protein quantitation
miRNA and mRNA were extracted using the mirVana miRNA Isolation Kit (Ambion) and RNEasy Kit (Qiagen), respectively. cDNA was prepared using the SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (mRNA; Life Technologies) or TaqMan MicroRNA Reverse Transcription Kit (miRNA; Applied Biosystems) and measured using the SYBR Select Mastermix (mRNA; Applied Biosystems) or TaqMan Universal PCR Master Mix (miRNA; hsa-miR-10a assay ID 1093, rnu6B assay ID 387; Applied Biosystems). mRNA QPCR primer sequences are shown in Supplementary Table 1. Antibodies used for Western blotting were purchased from Cell Signaling: p21 (12D1 Clone, Cat# 2947), p53 (7F5 Clone, Cat# 2527), and LC3 (D11 clone, Cat# 3868).
Biomarker analyses
Primary AML specimens from the “Sydney” and “Dresden” cohorts (see Supplementary Table 2 for clinicopathological characteristics) were collected at diagnosis with informed consent and approval from the relevant centre’s Human Research Ethics Committees (SVH HREC approval 13/012). NPM1-mutant patients were excluded to minimize bias as they uniquely display both unusually high miR-10a expression [12, 13, 17] while also being strongly associated with a favorable outcome following chemotherapy. mRNA expression data from publicly-available microarray datasets GSE110087 (“Houston”) and GSE12417 (“Munich”) from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) was also used to correlate miR-10a downstream targets to MDM2 inhibitor sensitivity and patient response. Cohorts were chosen for analysis if they included sufficient clinical information and follow-up time, and met a minimum sample size to ensure adequate power based on our previous similar studies [18–21]. A minimum p value approach was used to select a cut-point for each biomarker with p values corrected for multiple testing [22]. Data were analyzed and reported according to ReMARK recommendations for studies on tumor markers [23].
Animal studies
Animal experiments were approved by the Animal Ethics Committee of the Garvan Institute of Medical Research (Sydney, Australia; AEC approval 16_27). To establish AML orthotopic xenografts, 5 × 106 OCI-AML3 cells were injected into the tail veins of randomly-selected 8–12-week-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. 12 days after cell delivery, mice were treated with cytarabine (50 mg/kg I.P.), Nutlin-3a (100 mg/kg oral gavage), a combination of both compounds at the dosages above, or a vehicle control 4 days/week for up to 3 weeks. Ethical endpoint was >20% weight loss or hind limb disability. Investigators were not blinded to treatment group allocation. Eight animals were included per treatment group following a power analysis with variables including effect size estimated based on our previous similar studies [24].
Statistical analysis
Statistical analyses were performed using Prism 9 (Graphpad) and PASW Statistics 18 (SPSS). All results were derived from at least three independent experiments, and all data was confirmed to meet the assumptions of the statistical test used. Quantitative data are presented as mean ± standard deviation (SD) unless otherwise denoted, and all groups compared were confirmed to have similar variances. Student’s T tests were used for the comparison of means of quantitative data between groups. Two-way ANOVA was used to compare dose response curves. Univariate Cox regression analyses and Kaplan–Meier plots were used for survival analyses in which endpoint was overall survival. Multivariate Cox regression analysis was used to test prognostic variables for independence, including miR-10a expression, FAB subtype, FLT3-ITD status, DMNT3a mutation status, and age (<60 vs ≥60). Affymetrix microarray gene expression data was normalized by the MAS5 algorithm as implemented by the ‘affy’ package version 1.56.0 under Bioconductor version 3.6 and R version 3.4.3. p values < 0.05 were considered statistically significant and all tests were two-sided.
Results
miR-10a regulates several members of the p53/Rb network
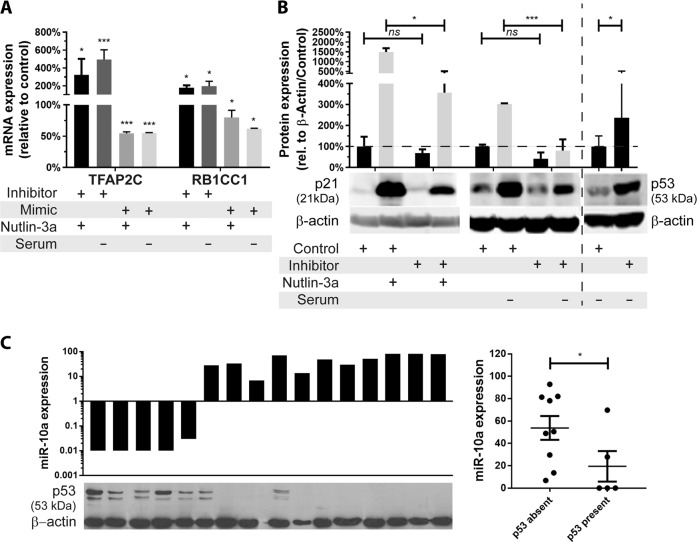
We previously demonstrated that miR-10a directly regulates the p53 regulator MDM4 [13], and also binds to the 3′UTR of two other key p53/Rb network genes: the p21 regulator TFAP2C, and the Rb regulator RB1CC1 [12]. Since these effects may modulate response to MDM2 inhibition, we first wished to confirm whether, like MDM4, miR-10a binding to TFAP2C and RB1CC1 had functional effects, and whether manipulating the intracellular availability of miR-10a would modulate of the expression of these genes and hence activity of the p53/Rb network. In cells in which the p53 network was activated by MDM2 inhibitor Nutlin-3a or stress induction by serum starvation (Supplementary Fig. 1a), inhibition of miR-10a in miR-10a-high OCI-AML3 cells (Supplementary Fig. 1b) resulted in significant upregulation of both TFAP2C and RBCC1, and conversely overexpression in miR-10a-low MV4–11 cells resulted in their significant downregulation (Fig. 1a). As a consequence of miR-10a-mediated modulation of TFAP2C, its transcriptional target p21 was significantly downregulated (4-fold, p < 0.05 for both; Fig. 1b, Supplementary Fig. 1c, d). There exists significant crosstalk between the Rb and p53 pathways, with p53 binding to RBCC1 to upregulate Rb [25], and Rb binding to MDM2 to stabilize and stimulate the apoptotic function of p53 [26]. In accordance, miR-10a inhibition also significantly increased accumulation of p53 itself (Fig. 1b, Supplementary Fig. 1e). A similar trend was found in primary patient specimens, in which a strong negative correlation was observed between endogenous miR-10a levels and p53 levels (Fig. 1c). Together this data confirms that miR-10a modulates several key members of the active p53/Rb stress response network (Supplementary Fig. 1f).
Fig. 1. miR-10a regulates several members of the p53/Rb network.
a mRNA expression of p53 pathway modulators and miR-10a targets TFAP2C and RBCC1 in response to miR-10a inhibition (“+ inhibitor”) in the miR-10a-high (/NPM1c+) OCI-AML3 cell line or miR-10a overexpression (“+ mimic”) in the miR-10a-low (/NPM1wt) MV4–11 cell line in which p53 was stabilized by Nutlin-3a (“+ Nutlin-3a”) or serum deprivation (“– serum”). b The ability of p53 to upregulate p21 was significantly attenuated in OCI-AML3 cells following upregulation of p21 negative regulator TFAP2C in response to miR-10a inhibition. miR-10a inhibition also significantly increased accumulation of p53 itself (2.4-fold). c A negative correlation was observed between endogenous miR-10a levels and p53 protein levels in NPM1wt primary AML patient samples. (*p < 0.05, **p < 0.01; ***p < 0.001 by Student’s T test with error bars representing SD (3–6 replicates per experiment); mRNA and protein expression data displayed relative to B-actin, with the scrambled mimic or inhibitor (control)-transfected cells set at 100% for each comparison.).
Sensitivity to MDM2 inhibitor Nutlin-3a is regulated by several downstream miR-10a gene targets
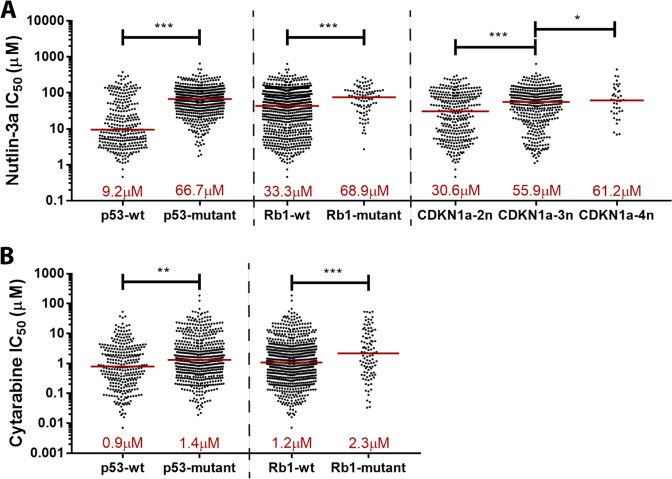
To determine whether these downstream miR-10a target genes were also associated with MDM2 inhibitor sensitivity, we data mined the Genomics of Drug Sensitivity in Cancer (GDSC) database [27], which correlates sensitivity of 265 compounds to genomic features (mutation/loss/gain) across >1000 human cancer cell lines. As expected, loss-of-function mutations in TP53 was the strongest predictor of Nutlin-3a resistance of all genomic features present in the database (p = 1.49 × 10−40; Fig. 2a). RB1 loss-of-function was the 4th strongest predictor (p = 8.12 × 10−7). Mutation in TFAP2C or RB1CC1 was not observed among these cell lines so no correlations could be made for these targets. Cell lines with loss-of-function mutations of TFAP2C target cyclin-dependent kinase inhibitor 1a (CDKN1A) were vanishingly rare in this dataset (with a mutation rate of <0.01%), however the more common amplification of CDKN1A was significantly associated with resistance (p < 0.05 for all). Interestingly, loss-of-function of TP53 and RB1 were also significantly correlated to resistance to AML standard-of-care chemotherapy cytarabine (p = 0.001 and 0.013 respectively; Fig. 2b).
Fig. 2. Sensitivity to MDM2 inhibitor Nutlin-3a is regulated by several downstream miR-10a gene targets.
a, b Correlations between mutation status of miR-10a downstream targets and sensitivity to Nutlin-3a and cytarabine in 888 cancer cell lines (GDSE database). Loss-of-function mutations in p53 and Rb, and chromosomal gain in CKDN1A (D; 2n = diploid, 3n = triploid, 4n = tetraploid) were significantly associated with resistance to Nutlin-3a (a) and cytarabine (b). (*p < 0.05, **p < 0.01; ***p < 0.001 by Student’s T test with error bars representing SD).
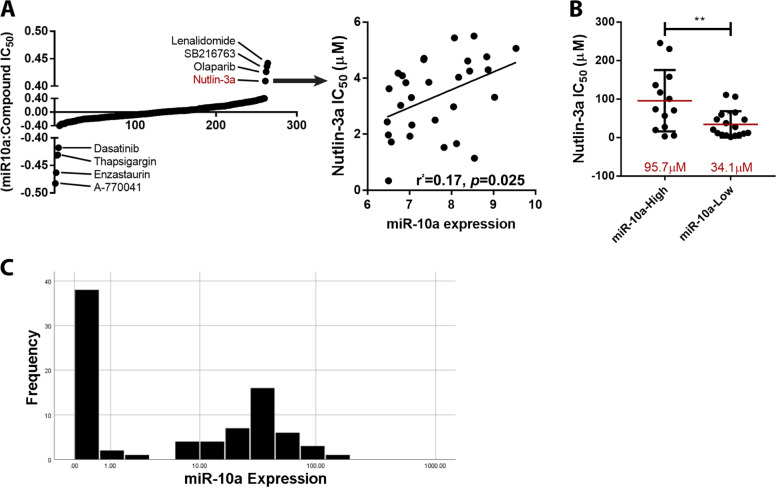
In concordance with these findings, endogenous expression of miR-10a itself in the NCI-60 Human Tumor Cell Line panel [28, 29] was strongly correlated to sensitivity to Nutlin-3a (4th highest correlation of 265 drugs screened in GDSC; Fig. 3a), with a mean IC50 of 34.1 µM for those cell lines that expressed miR-10a below the median level, versus 95.7 µM for those above (p = 0.008; Fig. 3b). Importantly, in consecutively-collected primary AML patient samples (n = 86; Supplementary Table 3), miR-10a displayed a clear bimodal expression, potentially identifying a group of NPM1wt patients (~50%) whose leukemia overexpress miR-10a and are likely to be sensitive to MDM2 inhibition, and a subset (~50%) that express very low miR-10a and may be refractory to this approach (Fig. 3c). Within this cohort, low (<4.0) miR-10a was significantly associated with decreased death (p = 0.024) and wildtype FLT3 (p = 0.007), but there was no significant association with age (p = 0.825), gender (p = 0.781), FAB subtype (p = 0.364), relapse (p = 0.082), or DNMT3A mutation status (p = 0.435; all by Fisher Exact test). Overall this data provides evidence that miR-10a and several of its downstream target genes are key mediators of sensitivity to MDM2 inhibitor Nutlin-3a and AML standard-of-care chemotherapeutic cytarabine.
Fig. 3. miR-10a expression is correlated to MDM2 inhibitor sensitivity in vitro.
a The endogenous expression of miR-10a in the NCI-60 cell line panel was correlated to sensitivity to 265 drugs screened in GDSC and Nutlin-3a was found to have the 4th strongest positive correlation with an r2 value of 0.17 (p < 0.05). b NCI-60 cell lines with endogenous expression below the median level had a mean IC50 of 34.1 µM, versus 95.7 µM for those above. c miR-10a displayed bimodal expression in 86 NPM1wt primary AML specimens assayed at diagnosis. (*p < 0.05, **p < 0.01; ***p < 0.001 by Student’s T test with error bars representing SD).
miR-10a and its downstream targets are biomarkers of MDM2 inhibitor and cytarabine responsiveness in AML patient samples
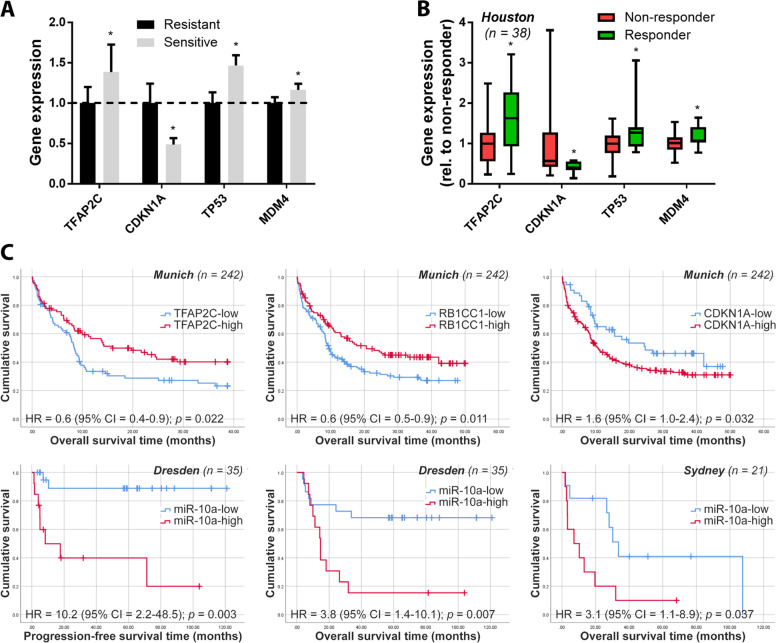
We next determined whether these observations in cell lines could be generalized to primary AML patient samples, potentially identifying miR-10a and/or its downstream gene targets as clinically valuable predictive biomarkers. Sensitivity of 38 primary AML patient samples treated ex vivo with second generation high potency MDM2 inhibitor and Nutlin derivative DS-3032b was correlated to miR-10a target expression (labeled here as the ‘Houston’ cohort; response data, clinical, and molecular features as previously described [30]). As is observed in the clinical setting, sensitivity to MDM2 inhibition varied markedly between these p53-wildtype leukemias in vitro, with primary samples that were sensitive to DS-3032b (defined as exhibiting an IC50 dose in the lower third of the cohort) exhibiting a > 20-fold lower mean IC50 than those that were resistant (upper third; p < 0.001; Supplementary Fig. 2a, b). Consistent with the GDSC data, primary samples that were sensitive to DS-3032b expressed significantly higher levels of TFAP2C, TP53, and MDM4 mRNA (p < 0.05 for all), and significantly lower levels of CDKN1A (52% lower; p < 0.05) compared to non-responsive samples (Fig. 4a). Importantly, similar correlations between expression of these genes and the clinical response of these same patients to DS-3032b during a clinical trial [30] was also observed (p < 0.05 for all targets; Fig. 4b).
Fig. 4. miR-10a and/or its downstream targets are biomarkers of MDM2 inhibitor and cytarabine responsiveness in AML patient samples.
a 38 pre-treatment AML primary patient samples were treated ex vivo with a dose range of MDM2 inhibitor DS-3032b [30]. Samples sensitive to DS-3032b in vitro expressed significantly higher levels of TFAP2C, TP53, and MDM4 mRNA, and significantly lower levels of CDKN1A. b These patients also participated in a Phase I/II clinical trial of DS-3032b and response data was available for the cohort [30]. “Responders” (those that experienced either complete remission (CR), complete remission with incomplete platelet recovery (CRp) or hematological improvement (HI)) displayed analogous expression patterns of miR-10a target expression when compared to non-responders. c TFAP2C, RB1CC1, and CDKN1A were also significantly prognostic in a second independent cohort (‘Munich’; n = 242) treated with intensive cytarabine-based double-induction and consolidation chemotherapy. miR-10a itself was also strongly predictive of outcome in patients treated with cytarabine-based chemotherapy + HSCT. In a subset of patients (n = 35) derived from the SAL AML2003 trial (‘Dresden’), low miR-10a was strongly predictive of both improved progression-free and overall survival. Similar results were observed in an additional independent cohort (‘Sydney’; n = 21), in which low miR-10a was also associated with improved overall survival in patients treated with cytarabine-based induction chemotherapy ± HSCT.
The GDSC data led us to also investigate the predictive value of miR-10a and its targets in patients treated with cytarabine-based chemotherapy in three further independent, retrospective AML patient cohorts, in which miR-10a was measured at diagnosis and correlated to outcome (Fig. 4c). The first cohort (‘Munich’) consisted of 242 cytogenetically-normal AML patients (most of whom were derived from German AMLCG 1999 trial [31]; clinical and molecular features as previously described [32]) given intensive double-induction and consolidation chemotherapy. Patients expressing high levels of TFAP2C or RB1CC1, or low levels of CDKN1A had significantly improved outcome following cytarabine-based chemotherapy compared to other patients (hazard ratios (HR) = 0.6, 0.6, and 1.6, respectively (all p < 0.05). A second cohort (‘Dresden’) comprising NPM1wt patients derived from the Study Alliance Leukemia (SAL) AML2003 trial [33, 34] (n = 35; Supplementary Table 2), which investigated optimal timing of allogeneic hematopoietic stem cell transplantation (HSCT), was used to correlate miR-10a expression itself to outcome. Low miR-10a was strongly predictive of improved relapse-free and overall survival in those patients treated with intensive cytarabine-based induction chemotherapy followed by allogeneic stem cell transplantation in remission (hazard ratio (HR) = 10.2 (p = 0.003) and 3.8 (p = 0.007), respectively). Similar results were shown in a third independent cohort (‘Sydney’; n = 23; Supplementary Table 2) consecutively collected from AML patients at diagnosis, with low miR-10a also associated with improved overall survival in patients treated with cytarabine-based induction chemotherapy ± HSCT (HR = 3.1 (p = 0.037)). Multivariate Cox regression analyses demonstrated that miR-10a expression provided prognostic information independent of FAB subtype, FLT3-ITD status, DMNT3a mutation status, and age (all p < 0.05). Together this data supports the hypothesis that miR-10a and its downstream targets may be valuable predictive biomarkers for both MDM2 inhibitor and cytarabine-based therapies.
miR-10a mediates response to Nutlin-3a and cytarabine in AML
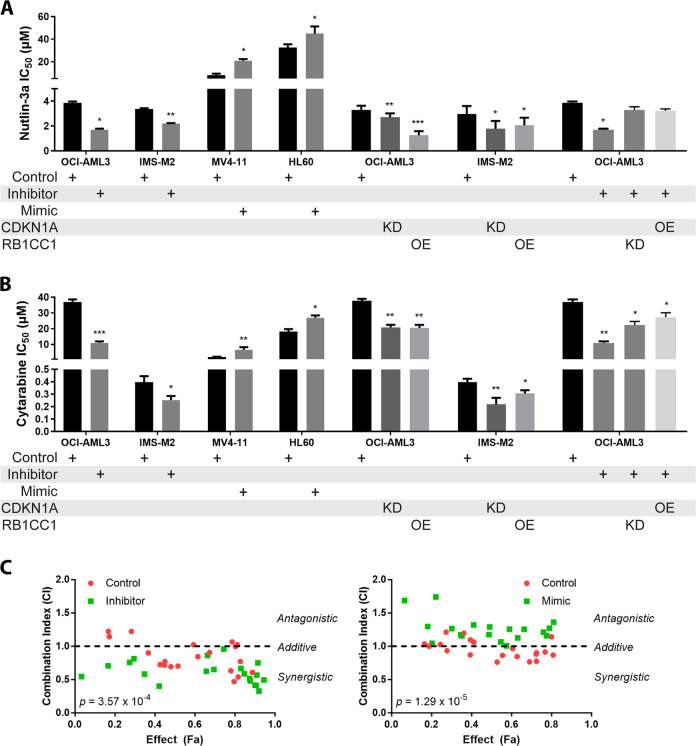
We next hypothesized that modulating miR-10a and/or its targets may in turn modulate sensitivity to MDM2 inhibition and cytarabine (Fig. 5a, b). To investigate this, miR-10a-high AML cell lines OCI-AML3 and IMS-M2 were transduced with a miR-10a inhibitor, resulting in a significant sensitization to both compounds. Conversely, resistance to both compounds could be induced by overexpressing miR-10a in miR-10a-low AML cell line MV4–11, and interestingly also in the miR-10a-low/p53-mutant/Nutlin-3a resistant cell line HL60. The ectopic expression of RBCC1 or knockdown of CDKN1A alone was sufficient to recapitulate these effects in the absence of miR-10a repression. Conversely, the sensitization effects of miR-10a inhibition could be rescued by the knockdown of RB1CC1 or overexpression of CDKN1A. These data further confirm the role of these miR-10a target genes themselves in sensitivity to these compounds.
Fig. 5. miR-10a mediates sensitivity to Nutlin-3a and cytarabine in AML.
miR-10a inhibition in the miR-10a-high AML cell lines OCI-AML3 and IMS-M2 resulted in significant sensitization to Nutlin-3a (a) and cytarabine (b). Conversely, overexpression of miR-10a in miR-10a-low AML cell lines MV4–11 and HL60 led to significant resistance. The exogenous overexpression (OE) of Rb1cc1 or knockdown (KD) of p21 alone were sufficient to recapitulate these effects in OCI-AML3 and IMS-M2 cells, confirming that these miR-10a targets are mediators of Nutlin-3a and cytarabine sensitivity. Conversely, the sensitization effects of miR-10a inhibition could be rescued by the knockdown of Rb1cc1 or overexpression of p21. c OCI-AML3 cells transduced with a miR-10a or scrambled control inhibitor (left) and MV4–11 cells transduced with a miR-10a or scrambled control mimic (right) were treated with a combination of Nutlin-3a and cytarabine at various doses and cell viability determined by MTS assay to quantify fractional inhibition of proliferation (Effect (Fa)). The Combination Index (CI) was calculated to determine antagonistic (CI > 1), additive (CI ≈ 1), or synergistic (CI < 1) effects between the two compounds. Nutlin-3a and cytarabine were strongly additive in miR-10a high OCI-AML3 cells but less so in miR-10a-low MV4–11 cells. Drug interactions became synergistic when miR-10a was simultaneously inhibited (p = 3.57 × 10−4), and antagonistic when miR-10a was overexpressed (p = 1.29 × 10−5). (For statistical testing, each treatment was compared to the control within its own group. *p < 0.05, **p < 0.01; ***p < 0.001 by Student’s T test with error bars representing SD (3–6 replicates per experiment)).
Combination treatment modalities, particularly those that target multiple nodes of the same network, have the potential for increased efficacy, decreased toxicity, and reduced likelihood of drug resistance. We further demonstrated that a combination of Nutlin-3a and cytarabine and was additive in miR-10a-high OCI-AML3 cells (with a mean Combination Index (CI) [14] of 0.85), which became synergistic when miR-10a was also inhibited (mean CI = 0.61; Fig. 5c). This corresponded to a potential mean dose reduction of 4.4-fold for Nutlin-3a and 2.6-fold for cytarabine when the two compounds were combined with miR-10a inhibition, while maintaining equivalent efficacy. Conversely, in miR-10a-low MV4–11 cells the combination was only weakly additive (CI = 0.95), and became antagonistic when miR-10a was overexpressed (CI = 1.30). These data confirm that there may be significant benefit in combining Nutlin-3a, cytarabine, and miR-10a inhibition in miR-10a-overexpressing AML cells, potentially allowing increased efficacy and/or decreased dosage and therefore toxicity while maintaining equivalent anti-leukemic activity.
miR-10a regulates the p53-mediated switch between autophagy and apoptosis
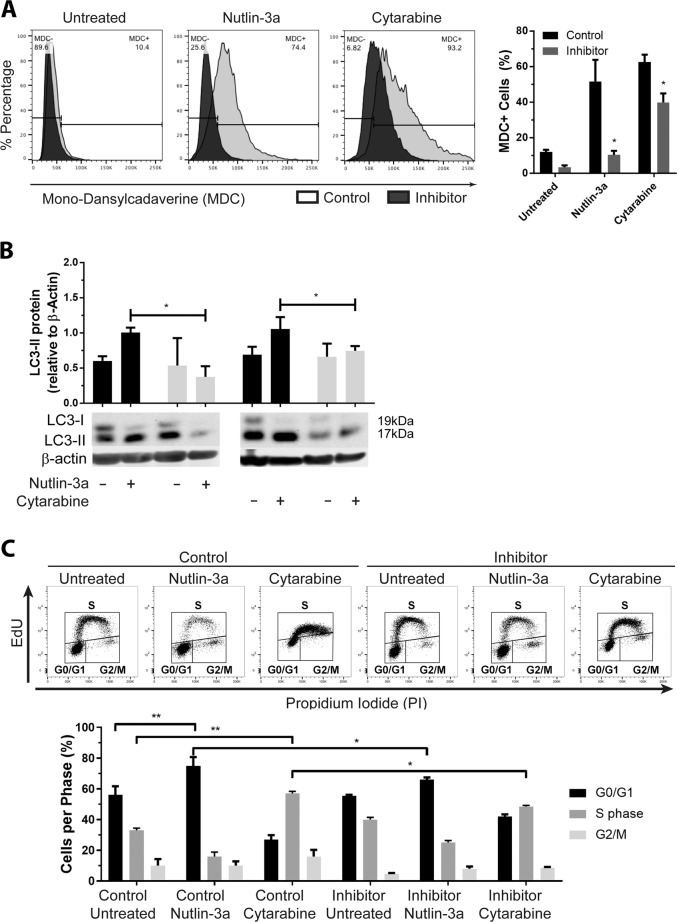
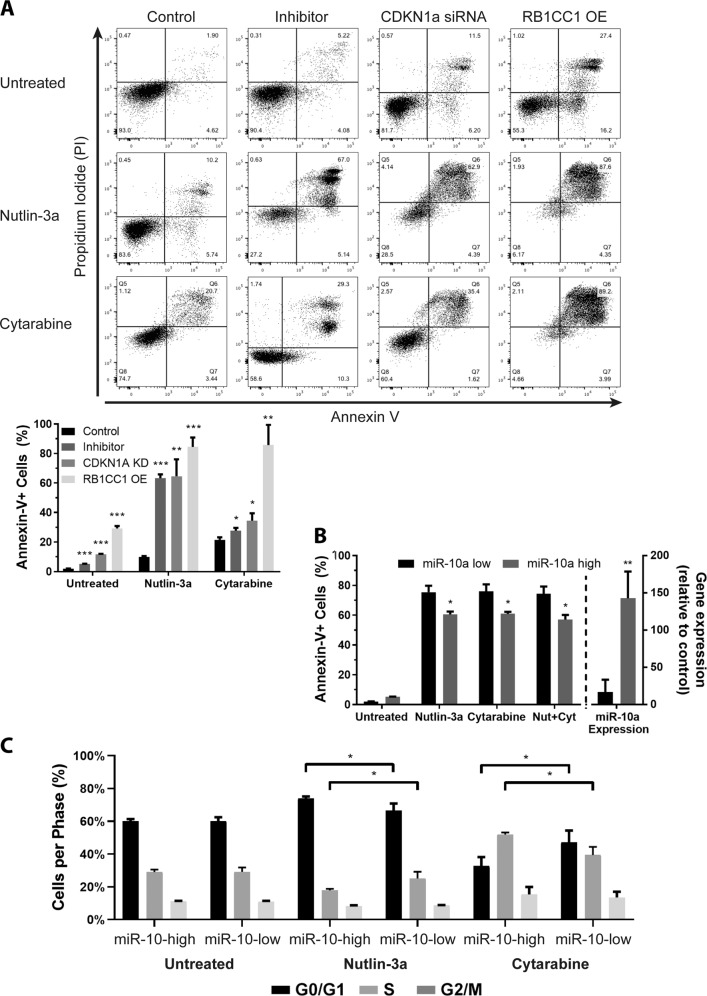
Activation of the p53/Rb-mediated stress response is potentiated by increased expression of miR-10a target RBCC1 via its direct binding to and stabilization of p53, and subsequent upregulation of Rb, p16, and p21 [25]. Excessive accumulation of Rb and p53 in the cytoplasm represses pro-survival autophagy and instead promotes intrinsic apoptosis via their direct interaction with Bcl-2 family proteins [35–37] (Supplementary Fig. 1f). Consistent with miR-10a’s ability to regulate key genes in this network, significantly fewer miR-10a-inhibited OCI-AML3 cells had detectable autophagic vacuoles following treatment with Nutlin-3a or cytarabine as determined by MDC fluorescence (Fig. 6a). Concordantly, these cells also expressed significantly lower levels of autophagy marker LC3-II compared to controls (Fig. 6b, Supplementary Fig. 3a, b). Cytotoxic stress leading to the activation of cytoprotective autophagy is universally associated with cell cycle arrest mediated by downstream miR-10a target p21 [38]. Extended arrest gives the cells sufficient time to repair DNA and recycle organelles before recommencing proliferation, and an inability to do so results in the accumulation of DNA damage and mitotic catastrophe leading to apoptosis [39–41]. OCI-AML3 cells transduced with a scrambled control inhibitor and treated with Nutlin-3a or cytarabine arrested in G0/G1-phase and S-phase, respectively, as has been previously observed [42, 43]. Conversely, those transduced with a miR-10a inhibitor were unable to arrest as normal in response to either compound, with significantly more cells escaping arrest and progressing through each checkpoint (Fig. 6c). The inhibition of autophagy and cell cycle arrest coincided with a significant activation of apoptosis in miR-10a-inhibited, RB1CC1-overexpressing, and p21-knockdown cells treated with Nutlin-3a, and to a more modest extent with cytarabine (Fig. 7a). These effects were also observed in primary AML specimens with low or high endogenous expression of miR-10a treated with a low dose (IC25) of each drug (Fig. 7b, c). Together this data is consistent with miR-10a inhibition potentiating activation of p53/Rb stress response pathway following treatment via RB1CC1 and p21, which favors the activation of apoptosis over autophagy.
Fig. 6. miR-10a regulates the switch between p53-mediated autophagy and apoptosis in AML by repressing autophagy induction.
Cells in which miR-10a was inhibited were significantly less likely to undergo autophagy as measured by MDC fluorescence (a) and LC3-II protein accumulation (b) following treatment with Nutlin-3a or cytarabine. c Scrambled miRNA inhibitor-transfected control cells undergo cell cycle arrest in the G0/G1 and S phases in response to cytarabine and Nutlin-3a treatment, respectively. When miR-10a was inhibited however, cell cycle arrest was attenuated, with significantly increased numbers of cells progressing through G0/G1 and S checkpoints. (*p < 0.05 by Student’s T test with error bars representing SD (3-6 replicates per experiment)).
Fig. 7. miR-10a regulates cell cycle arrest and apoptosis.
a Subsequent to the repression of autophagy and cell cycle arrest observed in miR-10a-inhibited cells in response to Nutlin-3a and cytarabine treatment, cells became apoptotic. These pro-apoptotic effects could be recapitulated in the absence of miR-10a inhibition by knocking down CDKN1a expression, or overexpressing RB1CC1. b, c NPM1wt primary AML specimens with high (n = 4) or low (n = 4) endogenous miR-10 expression (shown on right Y-axis of plot) were treated with a low (IC25) does of Nutlin-3a, cytarabine, or Nutlin-3a+Cytarabine. Those samples with low miR-10a expression exhibited significantly higher levels of apoptosis (as measured by Annexin-V positivity (b)) and had significantly fewer cells arrested (c) in response to drugs compared to specimens with high miR-10a expression. (*p < 0.05, **p < 0.01; ***p < 0.001 by Student’s T test with error bars representing SD (3–6 replicates per experiment)).
A combination of Nutlin-3a, cytarabine, and miR-10a inhibition is an effective therapeutic strategy for miR-10a-high AML in vivo
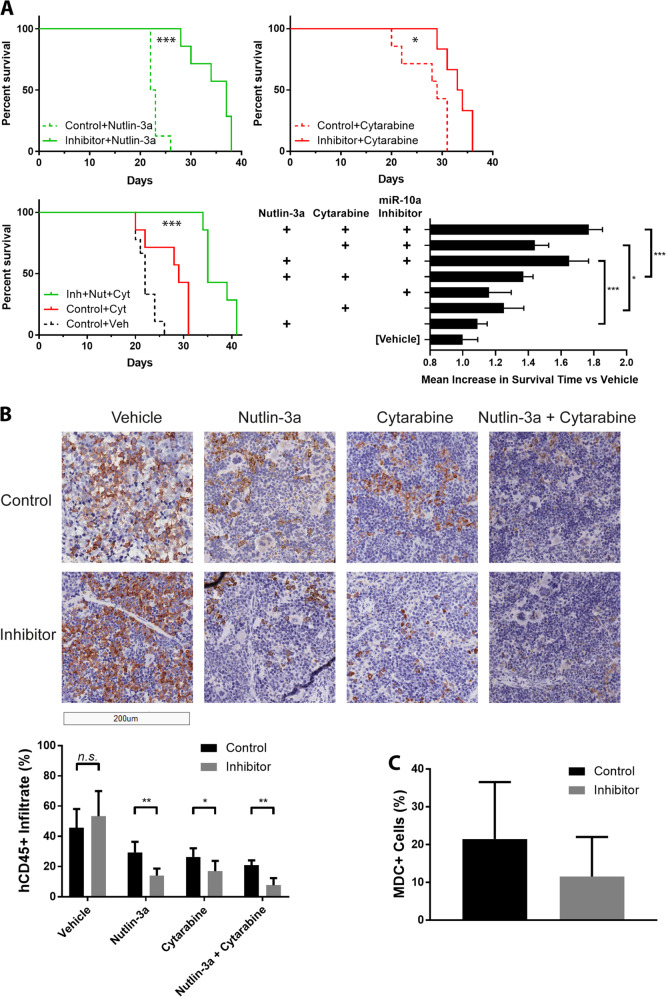
Orthotopic xenografts were established via tail vein injection of 5 × 106 OCI-AML3 cells transduced with a miR-10a inhibitor or scrambled control, resulting in rapid engraftment of aggressive acute leukemia with a mean survival of just 22 days. Twelve days after cell delivery, mice were treated with cytarabine, Nutlin-3a, both compounds, or a vehicle control 4 days/week for up to 3 weeks (n = 6–9 per group). Inhibition of miR-10a markedly improved the survival of animals treated with Nutlin-3a compared to the control inhibitor group (mean survival 34.6 days vs 22.9 days; p < 0.001; Fig. 8a). A similar effect was observed in animals treated with cytarabine (mean survival 33.2 days for miR-10a inhibitor vs 27.4 days for control inhibitor; p = 0.02). Simultaneous treatment with Nutlin-3a, cytarabine, and the miR-10a inhibitor resulted in the longest overall survival (37.1 days), extending survival by 35% in this aggressive in vivo model compared to the “standard-of-care” cytarabine + control inhibitor group (p = 0.001; Supplementary Table 4). Improved survival was consistent with significantly lower human CD45 positive (hCD45+) leukemic cell infiltrate in the spleen of miR-10a inhibited vs control animals collected after 7 days of treatment (all except vehicle p < 0.05; Fig. 8b). In concordance with the in vitro findings, Nutlin-3a/cytarabine-treated leukemic cells in which miR-10a was inhibited displayed approximately 50% lower MDC-positivity indicating repressed autophagy (p = 0.21; Fig. 8c). Together this data confirms that inhibition of miR-10a synergizes with Nutlin-3a + cytarabine and is an effective combination in AML in vivo.
Fig. 8. A combination of Nutlin-3a, cytarabine, and miR-10a inhibition is an effective therapeutic strategy for AML in vivo.
a Inhibition of miR-10a markedly improved the survival of mice bearing OCI-AML3 xenografts compared to the control inhibitor group when treated with both Nutlin-3a (upper left; mean survival 34.6 days vs 22.9 days; p < 0.001) and cytarabine (upper right; mean survival 33.2 days vs 27.4 days; p = 0.02), as visualized in Kaplan–Meier survival plots. A combination of cytarabine, Nutlin-3a, and miR-10a inhibition further increased survival (mean 37.1 days) compared to vehicle-only (mean 22.3 days) and cytarabine + control inhibitor “standard-of-care” controls (lower left; mean 27.4 days; p < 0.001; also see Supplementary Table 4 for pairwise Cox regression comparison p values). Lower right shows mean survival for each treatment group expressed as a fold-change versus vehicle control. Addition of the miR-10a inhibitor significantly increased survival in mice treated with Nutlin-3a (p < 0.001), cytarabine (p = 0.02), and Nutlin-3a + cytarabine (p < 0.001). b This corresponded with significantly reduced hCD45+ leukemic cell infiltrate in the spleen following 7 days of treatment. c Consistent with in vitro findings, while variable, miR-10a inhibited AML cells exhibited a trend towards lower MDC-positivity, indicating a repressed autophagy response (p = 0.21). Images were collected using a Leica Aperio LV1 Real-time Digital Pathology System (20× scan). (*p < 0.05, **p < 0.01; ***p < 0.001 by Student’s T test with error bars representing SD (six 400–600-cell fields quantified per bar)).
Discussion
We show here for the first time that miR-10a and several of its direct and downstream targets are strong biomarkers of sensitivity to MDM2 inhibition and cytarabine in cell lines, primary patient-derived AML cells treated ex vivo, and to clinical response in patients. Initial efforts to identify biomarkers of MDM2 inhibitor response have focussed on MDM2 and p53 themselves [44, 45], however it has since become clear that alone these are not strongly predictive, with one recent study demonstrating that TP53 mutation status had a positive predictive value (PPV) in primary AML specimens of just 62% [30]. This observation is likely due to the finding that p53 can be repressed by a myriad of mechanisms other than mutation [46]. Three more recent studies sought to identify gene expression signatures in cancer cell lines and primary AML samples [30, 47, 48], resulting in multigene signatures ranging from 4 to 1532 genes, and PPVs up to 82% when internally validated on patient-derived tumor PDX models and ex vivo-treated primary cells. Of note, miR-10a downstream targets CDKN1a, MDM4, and TP53 were consistently among the top genes that made up the predictive signatures. Independent validation is required to confirm the predictive power of multi-gene signatures to exclude model overfitting, however, which is a significant risk [21]. An attempt to independently validate the 4-gene signature in a subsequent study [48] for example demonstrated a PPV no better than using TP53 mutation status itself [30], and due to improper training of the 13-gene signature [47] it could not be independently validated at all [49]. Implementing gene expression-based signatures in a clinical setting also requires specialist diagnostic equipment and expertise and is more expensive than measuring the expression of a single biomarker. miRNAs have proven to be effective biomarkers since they modulate many targets, often including several in the same pathway as we have shown here, and can therefore act as a surrogate marker for the activation of multiple nodes of important gene networks [18, 24]. Here validation using independent cell line datasets miR-10a demonstrated a PPV of 86% in identifying lines with Nutlin-3a IC50 below the median, at a specificity of 94%. Data from upcoming clinical trials will allow further independent validation in additional patient cohorts. Also observed here was an association between miR-10a and response to cytarabine-based therapy, with which MDM2 inhibition is synergistic [50]. Previous studies that have investigated miR-10a in this context have reported variable findings [51–54]; for example, while Havelange et al. observed a correlation between miR-10a expression and complete remission following idarubicin/cytarabine-based treatment in de novo AML patients, little effect on cell proliferation or apoptosis was observed when miR-10a was inhibited in a cytarabine-treated p53-wildtype AML cell line [52]. In other models however, miR-10a inhibition is associated with sensitivity to other genotoxic chemotherapeutics including gemcitabine [55], temozolomide [56], and cisplatin [57]. Our data suggests a combination of miR-10a inhibition, MDM2 inhibition, and cytarabine is potently synergistic, however additional work may be required to determine the exact conditions under which miR-10a can modulate cytarabine sensitivity specifically.
MDM2 inhibitors such as Nutlin-3a trigger the rapid intracellular accumulation of p53 resulting in the activation of one (or sometimes both) of the tumor suppressors’ contrasting canonical signaling networks – either cytoprotective, promoting cell cycle arrest, autophagy, and DNA repair, or cytotoxic, promoting cell death via the activation of apoptosis [41, 58]. Cell fate depends heavily on the status of key nodes within these signaling networks [59], of which four – p21, Rb1, MDM4, and p53 itself – we show here are both downstream targets of miR-10a as well as important determinants of cellular sensitivity to Nutlin-3a and cytarabine. miR-10a regulates cell cycle inhibitor p21 by directly binding to and downregulating it’s repressor TFAP2C [12], with miR-10a inhibition ameliorating the ability of p53 to upregulate p21 in response to genotoxic stress (Fig. 1b, c), coinciding with an inability to initiate cell cycle arrest and the subsequent activation of apoptosis (Fig. 7). These observations are consistent with p21’s function as a key determinant of the switch between p53’s cytoprotective and cytotoxic roles. While the normal cellular response to sublethal DNA damage is to trigger extended G1 arrest to allow cells an opportunity for DNA repair, p21 ablation prevents this block and results instead in repeated rounds of asynchronized DNA replication leading to gross nuclear abnormalities, mutation, and polyploidy, followed by the subsequent activation of apoptosis by both transcriptional and post-transcriptional means [39–41, 60].
We also demonstrated that miR-10a directly targets and downregulates RB1CC1 [12], which plays an essential role in the induction of autophagy in mammalian cells [61]. The activation of autophagy normally results in resistance to weak-to-moderate cytotoxic stressors, including sublethal concentrations of chemotherapeutic agents [62]. As with other stress response pathways, autophagy is universally associated with cell cycle arrest, requiring the induction of p21-mediated cellular senescence for successful execution [38]. Here we showed that loss of p21 coincided with the inhibition of autophagy activation, as has been observed previously [63]. Both RB1CC1 and fellow miR-10a target MDM4 play dual roles in response to cell stress. When cell repair pathways are overwhelmed by cytotoxic or genotoxic injury, RB1CC1 switches from its canonical role as an inducer of cytoprotective autophagy to one of classical tumor suppressor and apoptosis inducer [25, 64, 65]. Similarly, MDM4 normally cooperates with MDM2 to mediate p53 degradation [66], but in response to DNA damage it can instead stabilize the secondary structure of TP53 mRNA to promote its transcription [67, 68], as well as compete with MDM2 for p53 binding to stimulate its stabilization and relocalization to the cytoplasm [69] where it can further contribute to apoptosis. For these reasons AML cells that express high endogenous MDM4 levels under genotoxic stress can be highly sensitive to MDM2 inhibition [69]. Our data suggests that these mechanisms are central to the efficacy of the miR-10a inhibitor/MDM2 inhibitor/cytarabine combination and result in a cellular bias away from autophagy and towards apoptosis in both our in vitro and in vivo models.
In AML MDM2 inhibition has shown useful activity as both a single agent and with cytarabine, though thus far in undefined clinical subgroups [70]. The data here suggest a biomarker-driven approach using miR-10a may allow optimal targeting of the most responsive patients in future. While the use of miRNA inhibitors as therapeutic molecules is still in its infancy, there have been a handful of successful non-human primate in vivo studies and clinical trials that have shown promise, combining efficacy with an acceptable toxicity profile [71–73]. Further investigations into the use of the described combined therapy, either to increase efficacy or decrease the dose of either (or both) compound while maintaining equivalent efficacy are therefore warranted. This may represent an efficacious companion biomarker-driven treatment strategy for a significant subgroup of AML patients whose only moderately efficacious first-line therapy has remain largely unchanged for several decades.
Supplementary information
Acknowledgements
The authors would like to thank Brunangelo Falini and colleagues (University of Perugia, Italy) for providing the IMS-M2 cells, and Catalina Palma, Joel Tan, S Park, and Gerhard Ehninger for technical contributions, advice, and clinicopathological data curation. This work was supported by the Arrow Bone Marrow Transplant Foundation (TJM, DDF), St Vincent’s Clinic Foundation (TJM, DDF), the St Vincent’s Hospital Sydney Haematology Department Research Funds (TJM, DDF), and the DFG grant “Collaborative Research Center 655”, Dresden, Project B7 (FS).
Compliance with ethical standards
Conflict of interest
TJM and DDM are named inventors on a patent application arising from the work described.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally:Timothy J. Molloy, David D. Ma
Contributor Information
Timothy J. Molloy, Email: t.molloy@amr.org.au
David D. Ma, Email: d.ma@amr.org.au
Supplementary information
The online version of this article (10.1038/s41375-020-01095-z) contains supplementary material, which is available to authorized users.
References
- 1.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toledo F, Krummel KA, Lee CJ, Liu CW, Rodewald LW, Tang M, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006;9:273–85. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Seipel K, Marques MT, Bozzini MA, Meinken C, Mueller BU, Pabst T. Inactivation of the p53-KLF4-CEBPA Axis in Acute Myeloid Leukemia. Clin Cancer Res. 2016;22:746–56. doi: 10.1158/1078-0432.CCR-15-1054. [DOI] [PubMed] [Google Scholar]
- 6.Colombo E, Martinelli P, Zamponi R, Shing DC, Bonetti P, Luzi L, et al. Delocalization and destabilization of the Arf tumor suppressor by the leukemia-associated NPM mutant. Cancer Res. 2006;66:3044–50. doi: 10.1158/0008-5472.CAN-05-2378. [DOI] [PubMed] [Google Scholar]
- 7.Bolli N, De Marco MF, Martelli MP, Bigerna B, Pucciarini A, Rossi R, et al. A dose-dependent tug of war involving the NPM1 leukaemic mutant, nucleophosmin, and ARF. Leukemia. 2009;23:501–9. doi: 10.1038/leu.2008.326. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 9.Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical overview of MDM2/X-targeted therapies. Front Oncol. 2016;6:7. doi: 10.3389/fonc.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee K, Martinelli G, Vey N, Dickinson MJ, Seiter K, Assouline S, et al. Phase 1/1b study of RG7388, a potent MDM2 antagonist, in acute myelogenous leukemia (AML) patients (Pts) Blood. 2014;124:116. [Google Scholar]
- 11.Ray-Coquard I, Blay JY, Italiano A, Le Cesne A, Penel N, Zhi J, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13:1133–40. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 12.Bryant A, Palma CA, Jayaswal V, Yang YW, Lutherborrow M, Ma DD. miR-10a is aberrantly overexpressed in Nucleophosmin1 mutated acute myeloid leukaemia and its suppression induces cell death. Mol Cancer. 2012;11:8. doi: 10.1186/1476-4598-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ovcharenko D, Stolzel F, Poitz D, Fierro F, Schaich M, Neubauer A, et al. miR-10a overexpression is associated with NPM1 mutations and MDM4 downregulation in intermediate-risk acute myeloid leukemia. Exp Hematol. 2011;39:1030–42. doi: 10.1016/j.exphem.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Foucquier J, Guedj M. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3:e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 16.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–50. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gee HE, Buffa FM, Harris AL, Toohey JM, Carroll SL, Cooper CL, et al. MicroRNA-related DNA repair/cell-cycle genes independently associated with relapse after radiation therapy for early breast cancer. Int J Radiat Oncol Biol Phys. 2015;93:1104–14. doi: 10.1016/j.ijrobp.2015.08.046. [DOI] [PubMed] [Google Scholar]
- 19.Molloy TJ, Bosma AJ, Baumbusch LO, Synnestvedt M, Borgen E, Russnes HG, et al. The prognostic significance of tumour cell detection in the peripheral blood versus the bone marrow in 733 early-stage breast cancer patients. Breast Cancer Res. 2011;13:R61. doi: 10.1186/bcr2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molloy TJ, Devriese LA, Helgason HH, Bosma AJ, Hauptmann M, Voest EE, et al. A multimarker QPCR-based platform for the detection of circulating tumour cells in patients with early-stage breast cancer. Br J Cancer. 2011;104:1913–9. doi: 10.1038/bjc.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molloy TJ, Roepman P, Naume B, van’t Veer LJ. A prognostic gene expression profile that predicts circulating tumor cell presence in breast cancer patients. PLoS ONE. 2012;7:e32426. doi: 10.1371/journal.pone.0032426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman DG, Lausen B, Sauerbrei W, Schumacher M. Dangers of using “optimal” cutpoints in the evaluation of prognostic factors. J Natl Cancer Inst. 1994;86:829–35. doi: 10.1093/jnci/86.11.829. [DOI] [PubMed] [Google Scholar]
- 23.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–91. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pajic M, Froio D, Daly S, Doculara L, Millar E, Graham PH, et al. miR-139-5p modulates radiotherapy resistance in breast cancer by repressing multiple gene networks of DNA repair and ROS defense. Cancer Res. 2018;78:501–15. doi: 10.1158/0008-5472.CAN-16-3105. [DOI] [PubMed] [Google Scholar]
- 25.Chano T, Ikebuchi K, Ochi Y, Tameno H, Tomita Y, Jin Y, et al. RB1CC1 activates RB1 pathway and inhibits proliferation and cologenic survival in human cancer. PLoS ONE. 2010;5:e11404. doi: 10.1371/journal.pone.0011404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol Cell. 1999;3:181–93. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 27.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sokilde R, Kaczkowski B, Podolska A, Cirera S, Gorodkin J, Moller S, et al. Global microRNA analysis of the NCI-60 cancer cell panel. Mol Cancer Ther. 2011;10:375–84. doi: 10.1158/1535-7163.MCT-10-0605. [DOI] [PubMed] [Google Scholar]
- 29.Weinstein JN, Pommier Y. Transcriptomic analysis of the NCI-60 cancer cell lines. C R Biol. 2003;326:909–20. doi: 10.1016/j.crvi.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Ishizawa J, Nakamaru K, Seki T, Tazaki K, Kojima K, Chachad D, et al. Predictive gene signatures determine tumor sensitivity to MDM2 inhibition. Cancer Res. 2018;78:2721–31. doi: 10.1158/0008-5472.CAN-17-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchner T, Berdel WE, Schoch C, Haferlach T, Serve HL, Kienast J, et al. Double induction containing either two courses or one course of high-dose cytarabine plus mitoxantrone and postremission therapy by either autologous stem-cell transplantation or by prolonged maintenance for acute myeloid leukemia. J Clin Oncol. 2006;24:2480–9. doi: 10.1200/JCO.2005.04.5013. [DOI] [PubMed] [Google Scholar]
- 32.Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rollig C, Bornhauser M, Kramer M, Thiede C, Ho AD, Kramer A, et al. Allogeneic stem-cell transplantation in patients with NPM1-mutated acute myeloid leukemia: results from a prospective donor versus no-donor analysis of patients after upfront HLA typing within the SAL-AML 2003 trial. J Clin Oncol. 2015;33:403–10. doi: 10.1200/JCO.2013.54.4973. [DOI] [PubMed] [Google Scholar]
- 34.Schetelig J, Schaich M, Schafer-Eckart K, Hanel M, Aulitzky WE, Einsele H, et al. Hematopoietic cell transplantation in patients with intermediate and high-risk AML: results from the randomized Study Alliance Leukemia (SAL) AML 2003 trial. Leukemia. 2015;29:1060–8. doi: 10.1038/leu.2014.335. [DOI] [PubMed] [Google Scholar]
- 35.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–4. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 36.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilgendorf KI, Leshchiner ES, Nedelcu S, Maynard MA, Calo E, Ianari A, et al. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev. 2013;27:1003–15. doi: 10.1101/gad.211326.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goehe RW, Di X, Sharma K, Bristol ML, Henderson SC, Valerie K, et al. The autophagy-senescence connection in chemotherapy: must tumor cells (self) eat before they sleep? J Pharmacol Exp Ther. 2012;343:763–78. doi: 10.1124/jpet.112.197590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dotto GP. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta. 2000;1471:M43–56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 40.Sohn D, Essmann F, Schulze-Osthoff K, Janicke RU. p21 blocks irradiation-induced apoptosis downstream of mitochondria by inhibition of cyclin-dependent kinase-mediated caspase-9 activation. Cancer Res. 2006;66:11254–62. doi: 10.1158/0008-5472.CAN-06-1569. [DOI] [PubMed] [Google Scholar]
- 41.Waldman T, Lengauer C, Kinzler KW, Vogelstein B. Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature. 1996;381:713–6. doi: 10.1038/381713a0. [DOI] [PubMed] [Google Scholar]
- 42.Miyachi M, Kakazu N, Yagyu S, Katsumi Y, Tsubai-Shimizu S, Kikuchi K, et al. Restoration of p53 pathway by nutlin-3 induces cell cycle arrest and apoptosis in human rhabdomyosarcoma cells. Clin Cancer Res. 2009;15:4077–84. doi: 10.1158/1078-0432.CCR-08-2955. [DOI] [PubMed] [Google Scholar]
- 43.Sampath D, Cortes J, Estrov Z, Du M, Shi Z, Andreeff M, et al. Pharmacodynamics of cytarabine alone and in combination with 7-hydroxystaurosporine (UCN-01) in AML blasts in vitro and during a clinical trial. Blood. 2006;107:2517–24. doi: 10.1182/blood-2005-08-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reis B, Jukofsky L, Chen G, Martinelli G, Zhong H, So WV, et al. Acute myeloid leukemia patients’ clinical response to idasanutlin (RG7388) is associated with pre-treatment MDM2 protein expression in leukemic blasts. Haematologica. 2016;101:e185–188. doi: 10.3324/haematol.2015.139717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kojima K, Konopleva M, Samudio IJ, Shikami M, Cabreira-Hansen M, McQueen T, et al. MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood. 2005;106:3150–9. doi: 10.1182/blood-2005-02-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469:325–46. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 47.Jeay S, Gaulis S, Ferretti S, Bitter H, Ito M, Valat T, et al. A distinct p53 target gene set predicts for response to the selective p53-HDM2 inhibitor NVP-CGM097. Elife. 2015;4:e06498. doi: 10.7554/eLife.06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong H, Chen G, Jukofsky L, Geho D, Han SW, Birzele F, et al. MDM2 antagonist clinical response association with a gene expression signature in acute myeloid leukaemia. Br J Haematol. 2015;171:432–5. doi: 10.1111/bjh.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonkin D. Expression signature based on TP53 target genes doesn’t predict response to TP53-MDM2 inhibitor in wild type TP53 tumors. Elife. 2015;4:e10279. doi: 10.7554/eLife.10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yee K, Martinelli G, Assouline S, Kasner M, Vey N, Kelly KR, et al. Phase 1b study of the MDM2 antagonist RG7112 in combination with 2 doses/schedules of cytarabine. Blood. 2013;122:498. [Google Scholar]
- 51.Bi L, Sun L, Jin Z, Zhang S, Shen Z. MicroRNA-10a/b are regulators of myeloid differentiation and acute myeloid leukemia. Oncol Lett. 2018;15:5611–9.. doi: 10.3892/ol.2018.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Havelange V, Ranganathan P, Geyer S, Nicolet D, Huang X, Yu X, et al. Implications of the miR-10 family in chemotherapy response of NPM1-mutated AML. Blood. 2014;123:2412–5. doi: 10.1182/blood-2013-10-532374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang TJ, Guo H, Zhou JD, Li XX, Zhang W, Ma JC, et al. Bone marrow miR-10a overexpression is associated with genetic events but not affects clinical outcome in acute myeloid leukemia. Pathol Res Pract. 2018;214:169–73. doi: 10.1016/j.prp.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 54.Zhi Y, Xie X, Wang R, Wang B, Gu W, Ling Y, et al. Serum level of miR-10-5p as a prognostic biomarker for acute myeloid leukemia. Int J Hematol. 2015;102:296–303. doi: 10.1007/s12185-015-1829-6. [DOI] [PubMed] [Google Scholar]
- 55.Xiong G, Huang H, Feng M, Yang G, Zheng S, You L, et al. MiR-10a-5p targets TFAP2C to promote gemcitabine resistance in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2018;37:76. doi: 10.1186/s13046-018-0739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shang C, Tang W, Pan C, Hu X, Hong Y. Long non-coding RNA TUSC7 inhibits temozolomide resistance by targeting miR-10a in glioblastoma. Cancer Chemother Pharmacol. 2018;81:671–8. doi: 10.1007/s00280-018-3522-y. [DOI] [PubMed] [Google Scholar]
- 57.Sun W, Ma Y, Chen P, Wang D. MicroRNA-10a silencing reverses cisplatin resistance in the A549/cisplatin human lung cancer cell line via the transforming growth factor-beta/Smad2/STAT3/STAT5 pathway. Mol Med Rep. 2015;11:3854–9. doi: 10.3892/mmr.2015.3181. [DOI] [PubMed] [Google Scholar]
- 58.Javelaud D, Besancon F. Inactivation of p21WAF1 sensitizes cells to apoptosis via an increase of both p14ARF and p53 levels and an alteration of the Bax/Bcl-2 ratio. J Biol Chem. 2002;277:37949–54. doi: 10.1074/jbc.M204497200. [DOI] [PubMed] [Google Scholar]
- 59.Sionov RV, Haupt Y. The cellular response to p53: the decision between life and death. Oncogene. 1999;18:6145–57. doi: 10.1038/sj.onc.1203130. [DOI] [PubMed] [Google Scholar]
- 60.Chou A, Froio D, Nagrial AM, Parkin A, Murphy KJ, Chin VT, et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut. 2018;67:2142–55. doi: 10.1136/gutjnl-2017-315144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar A, Singh UK, Chaudhary A. Targeting autophagy to overcome drug resistance in cancer therapy. Future Med Chem. 2015;7:1535–42. doi: 10.4155/fmc.15.88. [DOI] [PubMed] [Google Scholar]
- 63.Mohapatra P, Preet R, Das D, Satapathy SR, Choudhuri T, Wyatt MD, et al. Quinacrine-mediated autophagy and apoptosis in colon cancer cells is through a p53- and p21-dependent mechanism. Oncol Res. 2012;20:81–91. doi: 10.3727/096504012x13473664562628. [DOI] [PubMed] [Google Scholar]
- 64.Morselli E, Shen S, Ruckenstuhl C, Bauer MA, Marino G, Galluzzi L, et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle. 2011;10:2763–9. doi: 10.4161/cc.10.16.16868. [DOI] [PubMed] [Google Scholar]
- 65.Chano T, Ikegawa S, Kontani K, Okabe H, Baldini N, Saeki Y. Identification of RB1CC1, a novel human gene that can induce RB1 in various human cells. Oncogene. 2002;21:1295–8. doi: 10.1038/sj.onc.1205178. [DOI] [PubMed] [Google Scholar]
- 66.Hernandez-Monge J, Rousset-Roman AB, Medina-Medina I, Olivares-Illana V. Dual function of MDM2 and MDMX toward the tumor suppressors p53 and RB. Genes Cancer. 2016;7:278–87. doi: 10.18632/genesandcancer.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malbert-Colas L, Ponnuswamy A, Olivares-Illana V, Tournillon AS, Naski N, Fahraeus R. HDMX folds the nascent p53 mRNA following activation by the ATM kinase. Mol Cell. 2014;54:500–11. doi: 10.1016/j.molcel.2014.02.035. [DOI] [PubMed] [Google Scholar]
- 68.Medina-Medina I, Garcia-Beltran P, de la Mora-de la Mora I, Oria-Hernandez J, Millot G, Fahraeus R, et al. Allosteric interactions by p53 mRNA govern HDM2 E3 ubiquitin ligase specificity under different conditions. Mol Cell Biol. 2016;36:2195–205. doi: 10.1128/MCB.00113-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan BX, Khoo KH, Lim TM, Lane DP. High Mdm4 levels suppress p53 activity and enhance its half-life in acute myeloid leukaemia. Oncotarget. 2014;5:933–43. doi: 10.18632/oncotarget.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andreeff M, Kelly KR, Yee K, Assouline S, Strair R, Popplewell L, et al. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin Cancer Res. 2016;22:868–76. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christopher AF, Kaur RP, Kaur G, Kaur A, Gupta V, Bansal P. MicroRNA therapeutics: discovering novel targets and developing specific therapy. Perspect Clin Res. 2016;7:68–74. doi: 10.4103/2229-3485.179431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–7. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.