Abstract
Necroptosis is a lytic, inflammatory cell death pathway that is dysregulated in many human pathologies. The pathway is executed by a core machinery comprising the RIPK1 and RIPK3 kinases, which assemble into necrosomes in the cytoplasm, and the terminal effector pseudokinase, MLKL. RIPK3-mediated phosphorylation of MLKL induces oligomerization and translocation to the plasma membrane where MLKL accumulates as hotspots and perturbs the lipid bilayer to cause death. The precise choreography of events in the pathway, where they occur within cells, and pathway differences between species, are of immense interest. However, they have been poorly characterized due to a dearth of validated antibodies for microscopy studies. Here, we describe a toolbox of antibodies for immunofluorescent detection of the core necroptosis effectors, RIPK1, RIPK3, and MLKL, and their phosphorylated forms, in human and mouse cells. By comparing reactivity with endogenous proteins in wild-type cells and knockout controls in basal and necroptosis-inducing conditions, we characterise the specificity of frequently-used commercial and recently-developed antibodies for detection of necroptosis signaling events. Importantly, our findings demonstrate that not all frequently-used antibodies are suitable for monitoring necroptosis by immunofluorescence microscopy, and methanol- is preferable to paraformaldehyde-fixation for robust detection of specific RIPK1, RIPK3, and MLKL signals.
Subject terms: Cell biology, Kinases
Introduction
Cell death by necroptosis is thought to have originated as an ancestral host defense mechanism, which is reflected in the breadth of pathogen-encoded proteins that inhibit the pathway [1–6]. In addition to reported innate immunity functions [7–9], the dysregulation of necroptosis has been implicated in a range of pathologies, including ischemic-reperfusion injuries [10–12], and inflammatory diseases [13–16], including inflammatory bowel disease [17]. Accordingly, there is widespread interest in therapeutically-targeting the pathway to counter human disease. Owing to the recent identification of the terminal effectors of the pathway, RIPK3 (in 2009) [1, 18, 19] and MLKL (in 2012) [20, 21], however, the extent of indications attributable to necroptotic cell death is poorly understood. Precisely defining pathologies impacted by necroptosis has posed a challenge owing to the dearth of antibodies validated to specifically detect members of the pathway and their activated (phosphorylated) forms in fixed cells and tissues. As a result, the contribution of necroptosis to many pathologies remains a subject of ongoing debate [22–28].
Necroptotic cell death signaling is initiated by ligation of death receptors, such as the TNF receptor 1, or pathogen detectors. In cellular contexts where the activities of the Inhibitors of Apoptosis proteins E3 Ubiquitin ligase family and the proteolytic apoptotic effector, Caspase-8, are depleted or compromised, necroptosis ensues. The precise choreography of necroptotic signaling is still emerging, although recent studies have defined key events and checkpoints in the pathway [29–33]. Following pathway induction, RIPK1 autophosphorylation prompts hetero-oligomerization with RIPK3 to form a cytoplasmic platform known as the necrosome [34–36]. Upon RIPK3 activation by autophosphorylation within necrosomes [37, 38], RIPK3 is primed to recruit MLKL from the cytoplasm and phosphorylate the activation loop within the MLKL pseudokinase domain to activate MLKL’s killing function [14, 20, 30, 31, 39–43]. MLKL phosphorylation is thought to provoke a conformational change in the pseudokinase domain that leads to oligomerization and unmasking of the killer N-terminal four-helix bundle (4HB) domain [41, 44–46]. The human MLKL 4HB domain likely engages chaperones to facilitate translocation to the plasma membrane via an actin-, Golgi- and microtubule-dependent mechanism, where MLKL accumulates in hotpots [29, 32]. When a threshold is surpassed, the 4HB domains of MLKL permeabilize the membrane to induce cell death.
While biochemical studies have defined these steps and checkpoints, visualizing the spatiotemporal dynamics of endogenous proteins during necroptosis using microscopy-based approaches has proven challenging in the absence of antibodies that have been validated for target specificity. Similarly, the lack of validated reagents poses a challenge to immunohistochemical staining of patient tissue sections, and therefore attribution of a role for necroptosis in pathologies, because knockout tissue controls are not available. Here, we have established procedures for staining endogenous RIPK1, RIPK3 and MLKL, and their phosphorylated forms, in fixed mouse and human cells. While several frequently-used antibodies were found to be suitable for selectively staining these proteins, as validated by comparisons with cells deficient for each protein, many antibodies exhibited non-specific staining and are therefore unsuitable for immunofluorescence and immunohistochemistry. Our studies also highlight the importance of validating antibody compatibility with fixation methods. In most cases, paraformaldehyde fixation ablated epitopes, whereas methanol fixation enabled specific detection of these proteins. Due to the sequence divergence between mouse and human RIPK3 and MLKL [44, 46–49], it was not possible to specifically-detect proteins across species using a single reagent, which necessitated the development of a new antibody that specifically detected mouse MLKL. Collectively, our studies present a toolbox of selective antibodies that will enable critical analysis of the chronology, checkpoints and kinetics of necroptotic signaling in mouse and human cells and tissues.
Results
Immunofluorescent detection of human MLKL
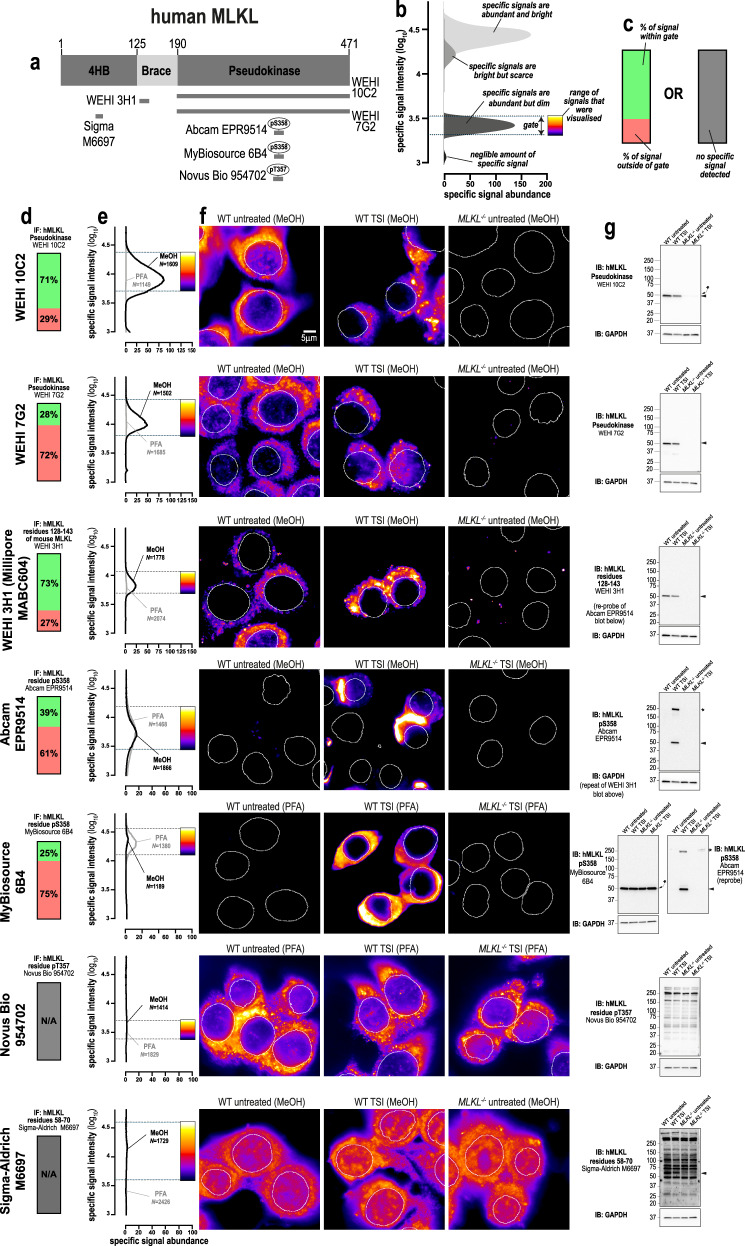
We recently described a monoclonal antibody, clone 10C2 (source: WEHI Antibody Facility, in-house), that recognises an epitope centered on residues 413–471 of human MLKL [32] (Fig. 1a). The initial screen that identified this clone suggested it yielded specific immunosignals when cells were fixed with methanol, but not with paraformaldehyde. Organic solvents such as methanol preserve cells by precipitating proteins, whereas aldehyde-based agents such as paraformaldehyde fix cells by crosslinking lysines and other primary amines. Because human MLKL is lysine-rich (93rd percentile in the cytoplasmic and membrane-associated proteome), paraformaldehyde likely causes widespread crosslinking of MLKL, which in turn masks epitopes and reduces immunoreactivity. As other necroptotic proteins are lysine-rich, we considered whether the choice of fixative was a critical variable for robust immunodetection of MLKL, RIPK3, and RIPK1 in human and mouse cells. Accordingly, we compared the performance of 26 antibodies for immunofluorescent staining of human HT29 cells and mouse dermal fibroblasts (MDFs)—two cellular models that are well-characterized to undergo necroptosis when treated with TNF, Smac-mimetic and IDN-6556 (herein referred to as TSI) [5, 29, 31, 32, 47] (Fig. S1). To quantitatively gauge the performance of each antibody, their immunosignals were characterized in four ways: (1) A ratio of the immunofluorescent signals between a positive and negative control. This produces a signal-to-noise curve that estimates “specific signal abundance” (Fig. 1b). For testing phospho-specific antibodies, wild-type cells undergoing necroptosis were compared to untreated wild-type cells (Fig. S1). For testing all other antibodies, untreated wild-type cells were used as a positive control and relevant knockout cells as a negative control. (2) Micrographs were gated so that the minimum/maximum immunosignals corresponded to the 5th/95th percentiles of the signal-to-noise curve (Fig. 1b). (3) The percentage of total signal that falls within this gate was determined to show how much signal from the positive control was considered specific (Fig. 1c). (4) Antibodies were immunoblotted against positive and negative controls to independently assess their specificity.
Fig. 1. Methanol fixation is optimal for the immunofluorescent detection of human MLKL.
a Human MLKL domain architecture showing the immunogens used to raise the tested anti-MLKL antibodies. b Demonstration of how signal-to-noise ratios were used to quantify the abundance and brightness of specific immunofluorescent signals generated by different antibodies. The 5th and 95th percentile of each signal-to-noise curve defines the gate where specific immunosignals were observed. As indicated by the pseudocolour look-up-table, only immunosignals within this gate were visualised. c Chart exemplifying how the amount of signal within the gate, relative to the total amount of detectable signal, provides another gauge of antibody specificity for immunofluorescence. d Quantitation of the percentage of gated signals for the tested MLKL antibodies. e Quantitation of specific signal abundance produced by the tested MLKL antibodies on methanol-fixed (MeOH) or paraformaldehyde-fixed (PFA) HT29 cells. The number of cells imaged (N) to generate each signal-to-noise curve is shown. f Micrographs of immunofluorescent signals for the tested MLKL antibodies on HT29 cells. As indicated by each pseudocolour look-up-table, only immunosignals within the respective gate in panel e were visualised. Data are representative of n = 3 (clones 10C2, 7G2) and n = 2 (Abcam clone EPR9514, Novus Biological MAB9187/clone 954702, WEHI clone 3H1, MyBiosource clone 6B4 and Sigma-Aldrich M6697) independent experiments. Nuclei were detected by Hoechst 33342 staining and are demarked by white outlines in micrographs. T, TNF; S, Smac mimetic Compound A; I, pan-Caspase inhibitor IDN-6556. g Immunoblot using the tested MLKL antibodies against wild-type and MLKL−/− HT29 cell lysates. Closed arrowheads indicate the main specific band. An asterisks indicate non-specific bands that could otherwise confound data interpretation. Immunoblots were re-probed for GAPDH as loading control.
Using this approach, we confirmed that clone 10C2 specifically detects endogenous human MLKL in methanol-fixed, but not paraformaldehyde-fixed cells (Fig. 1d–f). The 10C2 clone produces a favorable immunostaining profile, where the specific signals are abundant (Fig. 1e) and these specific signals represent the majority of all detectable signals (Fig. 1d). As expected, the immunostaining profile of the 10C2 clone was concentration-dependent; with lower concentrations of 10C2 increasing the percentage of specific signal, albeit at lower signal intensity (Fig. S2A, B). By comparison, higher concentrations of 10C2 yielded more intense specific signals, but with the trade-off being that a greater proportion of total signal falls outside the specific gate (Fig. S2A–F).
We recently developed another MLKL-specific antibody, clone 7G2 (source: in-house), which recognises an epitope centered on the C-lobe of the human MLKL pseudokinase domain [32]. Despite binding a distinct site in human MLKL to 10C2, the 7G2 clone shows similar specificity for human MLKL in immunoblot analyses and only yields specific immunofluorescent signals on methanol-fixed cells (Fig. 1d–g). Notably, clone 7G2 is inferior to clone 10C2 for immunofluorescence studies, because specific signals were less abundant (Fig. 1e) and were a minor fraction of all detectable signals (Fig. 1d).
Similarly, clone 3H1 (source: in-house, available from Millipore as MABC604), which was raised against a sequence within the N-terminal brace helix of MLKL [41], specifically detects endogenous human MLKL in methanol-fixed, but not paraformaldehyde-fixed cells (Fig. 1d–f). However, clone 3H1 is suboptimal for immunofluorescence studies, because its specific signals are less abundant than those produced by clone 10C2 (Fig. 1e).
We observed that clone EPR9514 (source: Abcam), an antibody raised against phospho-S358 of human MLKL [14], which is a hallmark of MLKL activation during necroptosis [20], produced specific and comparable immunofluorescent signals in both methanol- and paraformaldehyde-fixed cells undergoing necroptosis (Fig. 1e, f). As MLKL can be disulfide-crosslinked during necroptosis [50, 51] (Fig. S3A, B), we tested whether fixation in the presence of N-ethylmaleimide (NEM) to prevent further disulfide bonding, or post-fixative treatment with Tris(2-carboxyethyl)phosphine (TCEP) to reduce disulfide bonds, altered the immunofluorescent detection of phospho-MLKL by clone EPR9514. However, neither preventing nor reducing disulfide bonds in fixed cells precluded the immunodetection of MLKL by clone EPR9514 (Fig. S3C, D). Indeed, disulfide-crosslinking may be a by-product of MLKL activation given that recombinant human MLKL oligomerizes in a non-disulfide bond dependent manner (Fig. S3E).
Another antibody raised against phospho-S358 of human MLKL, clone 6B4 (source: MyBiosource), produced immunofluorescent signals in paraformaldehyde-fixed, but not methanol-fixed cells undergoing necroptosis (Fig. 1e, f). However, the inability of clone 6B4 to detect MLKL in immunoblot analyses (Fig. 1g) and its diffuse cytosolic staining (dissimilar to the punctate cytosolic staining produced by the selective clones 10C2, 7G2, 3H1 and EPR9514 in necroptotic cells), raises doubt about the specificity of clone 6B4 for immunofluorescent studies.
Two other anti-human MLKL antibodies were found to be non-specific for immunofluorescent studies (Novus Biological clone 954702 raised against phospho-T357, and the Sigma-Aldrich M6697 polyclonal antibody raised against residues 58–70; Fig. 1a, e), as they both produced equivalent signal intensity and diffuse staining in wild-type and MLKL−/− cells.
Lastly, immunoblotting confirmed that clones 10C2, 7G2, 3H1 (all in-house) and EPR9514 (Abcam) were highly-specific with bands corresponding to human MLKL’s molecular weight of 54 kDa observed in wild-type but not MLKL−/− HT29 cell lysates. In contrast, clone 954702 (Novus Biological MAB9187), clone 6B4 (MyBiosource) and the Sigma-Aldrich M6697 polyclonal antibody were largely non-specific, with prominent bands observed in both wild-type and MLKL−/− HT29 cell lysates (Fig. 1g).
In summary, four of the seven antibodies that were tested selectively recognised human MLKL in methanol-fixed cells (clones 10C2, 7G2, 3H1 and EPR9514) and, accordingly, we recommend methanol fixation for detecting endogenous human MLKL by immunofluorescence. Under these conditions, cytoplasmic forms of MLKL could be detected using the 10C2, 7G2, or 3H1 clones, and phospho-MLKL accumulating at the cell periphery with the EPR9514 clone (Fig. 1f). These data underscore the importance of careful consideration of fixation method when using MLKL as a histological marker of necroptosis.
Immunofluorescent detection of human RIPK3
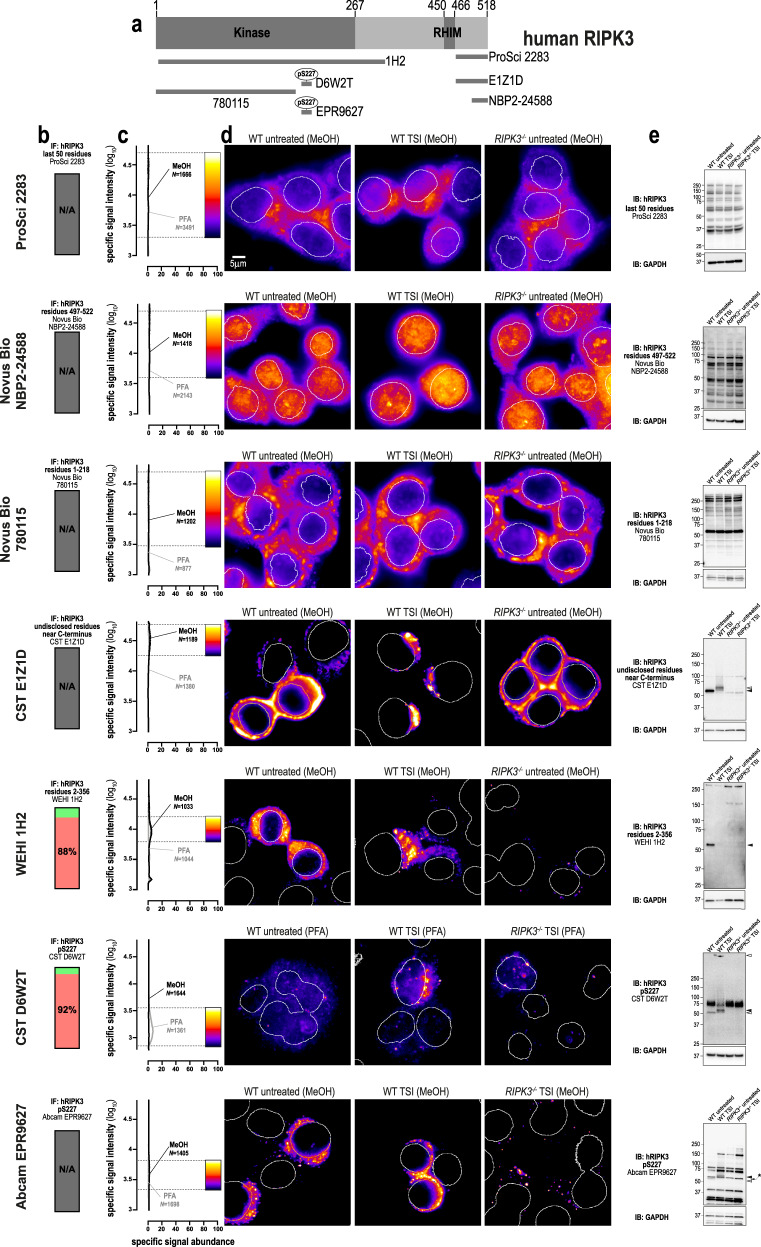
We next assessed seven anti-human RIPK3 antibodies (Fig. 2a). Two antibodies raised against the C-terminus (source: ProSci 2283, Novus Biological NBP2-24588) and one antibody raised against the N-terminus of RIPK3 (source: Novus Biological MAB7604, clone 780115) yielded non-specific signals via both immunofluorescence and immunoblotting (Fig. 2b–e). Another antibody raised against the C-terminus of human RIPK3, clone E1Z1D (source: Cell Signaling Technology), yielded only non-specific signals via immunofluorescence (Fig. 2b–d), despite exhibiting a high degree of specificity toward RIPK3 in immunoblot analyses (Fig. 2e). Our recently described antibody, clone 1H2 (source: in-house) [5], detected RIPK3 via immunofluorescence in methanol-fixed, but not paraformaldehyde-fixed cells. However, most of the immunofluorescent signals produced by 1H2 were non-specific (Fig. 2b), and the remaining signals are specific but of very low abundance (Fig. 2c). Our data also showed that clone D6W2T (source: Cell Signaling Technology; target human RIPK3 phospho-S227) produced specific immunofluorescent signals in paraformaldehyde-fixed cells undergoing necroptosis, but not in methanol-fixed cells (Fig. 2d). For both 1H2 and D6W2T, the low abundance of these specific immunosignals suggests that clone D6W2T should only be used for immunofluorescence under carefully controlled conditions, such as where RIPK3−/− control cells are available for direct comparison. Another caveat is that immunoblotting showed that clone D6W2T not only detected RIPK3 in cells undergoing necroptosis, but also in untreated cells when RIPK3 was presumably not phosphorylated at S227 (Fig. 2e). A similar caveat accompanies another anti-pS227 antibody, clone EPR9627 (source: Abcam), which recognised RIPK3 under both basal and necroptotic conditions via immunoblot analyses (Fig. 2e), and produced non-specific signals via immunofluorescence (Fig. 2b–d). Therefore, while two of the seven antibodies that were tested specifically recognised human RIPK3 in fixed cells, the low abundance of their specific signals complicates use of these antibodies for studying endogenous human RIPK3 via immunofluorescence. At present, more specific antibodies are needed to enable the immunofluorescent detection of human RIPK3.
Fig. 2. Better antibodies are needed for imaging human RIPK3.
a Human RIPK3 domain architecture showing the immunogens or epitopes for the tested anti-RIPK3 antibodies. b Quantitation of the percentage of gated signals for the tested RIPK3 antibodies. c Quantitation of specific signal abundance produced by the tested RIPK3 antibodies on methanol-fixed (MeOH) or paraformaldehyde-fixed (PFA) HT29 cells. The number of cells imaged (N) to generate each signal-to-noise curve is shown. d Micrographs of immunofluorescent signals for the tested RIPK3 antibodies on HT29 cells. As indicated by each pseudocolour look-up-table, only immunosignals within the respective gate in panel c were visualised. Data are representative of n = 3 (Cell Signaling Technology clone D6W2T and in-house clone 1H2) and n = 2 (ProSci 2283, Novus Biological NBP2-24588, CST clone E1Z1D, Abcam clone EPR9627 and MAB7604/clone 780115) independent experiments. Nuclei were detected by Hoechst 33342 staining and are demarked by white outlines in micrographs. e Immunoblot using the tested RIPK3 antibodies against wild-type and RIPK3−/− HT29 cell lysates. Closed arrowheads indicate the main specific band. Open arrowheads indicate other specific bands of interest. An asterisk indicates a non-specific band that could otherwise confound data interpretation. Immunoblots were re-probed for GAPDH as loading control.
Immunofluorescent detection of human RIPK1
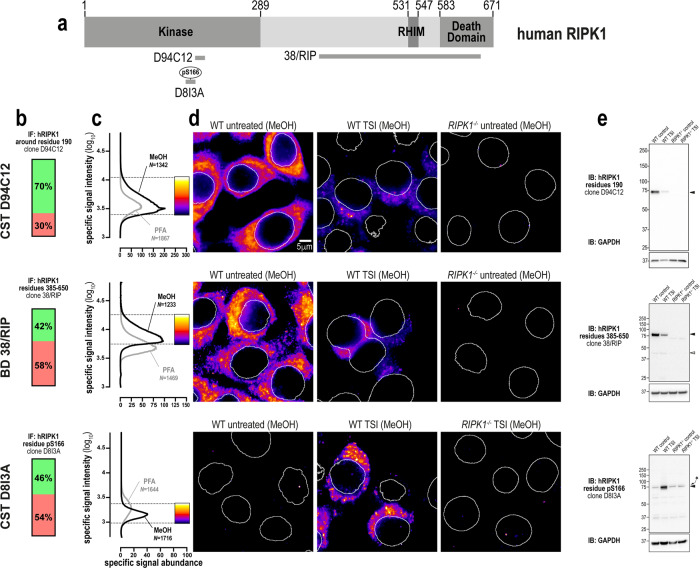
We next tested three monoclonal anti-RIPK1 antibodies: one against the N-terminal domain (source: Cell Signaling Technology clone D94C12), one against the C-terminal region (source: BD Transduction Laboratories clone 38/RIP) and one raised against phospho-S166 (source: Cell Signaling Technology clone D8I3A; Fig. 3a). All three clones yielded specific immunofluorescent signals in methanol- and paraformaldehyde-fixed cells, however, these signals were more abundant in methanol-fixed samples (Fig. 3b–d). All three clones were also specific via immunoblot (Fig. 3e). Importantly, the immunosignals from clones D94C12 and 38/RIP were markedly lower in cells undergoing necroptosis than in unstimulated wild-type cells (Fig. 3d, e). This observation is likely due to RIPK1 undergoing widespread post-translational modification and/or proteasomal degradation during TNF-induced cell death [52, 53]. Thus, despite clones D94C12 and 38/RIP exhibiting a favorable immunostaining profile in unstimulated cells, their decreased reactivity in cells undergoing necroptosis indicates that they should be used judiciously for immunofluorescent labeling of samples for accurate analysis of necroptosis. Conversely, while clone D8I3A preferentially stains RIPK1 in cells undergoing necroptosis, the specific signals that were detected also had low intensity and low abundance (Fig. 3c). In summary, three highly-specific antibodies exist for human RIPK1. However, because their specific signals have very low intensity and abundance in necroptotic cells, their use for the immunofluorescent detection of RIPK1 during necroptosis should be carefully controlled, including side-by-side examination of wild-type and RIPK1−/− control cells.
Fig. 3. Three specific antibodies for imaging endogenous human RIPK1.
a Human RIPK1 domain architecture showing the immunogens or epitopes for the tested anti-RIPK1 antibodies. b Quantitation of the percentage of gated signals for the tested RIPK1 antibodies. c Quantitation of specific signal abundance produced by the tested RIPK1 antibodies on methanol-fixed (MeOH) or paraformaldehyde-fixed (PFA) HT29 cells. The number of cells imaged (N) to generate each signal-to-noise curve is shown. d Micrographs of immunofluorescent signals for the tested RIPK1 antibodies on HT29 cells. As indicated by each pseudocolour look-up-table, only immunosignals within the respective gate in (c) were visualised. Data are representative of n = 2 (Cell Signaling Technology clones D94C12 and D8I3A, BD Transduction laboratories clone 38/RIP) independent experiments. Nuclei were detected by Hoechst 33342 staining and are demarked by white outlines in micrographs. e Immunoblot using the tested RIPK1 antibodies against wild-type and RIPK1−/− HT29 cell lysates. Closed arrowheads indicate the main specific band. Open arrowheads indicate other specific bands of interest. Asterisks indicate non-specific bands that could otherwise confound data interpretation. Immunoblots were re-probed for GAPDH as loading control.
Immunofluorescent detection of mouse MLKL
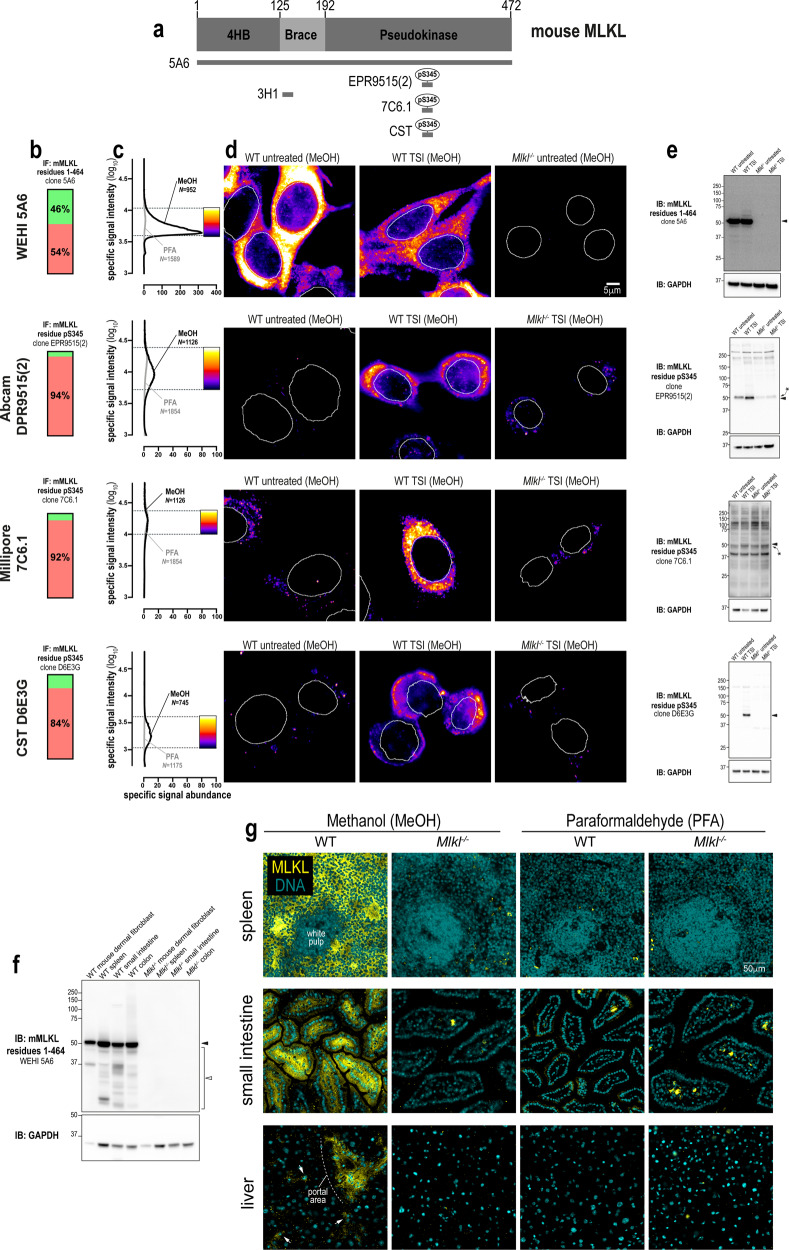
To our knowledge, there are currently no validated monoclonal antibodies for the immunofluorescent detection for mouse MLKL in unstimulated cells. Accordingly, we raised a monoclonal antibody, clone 5A6 (source: in-house), against full-length mouse MLKL (Fig. 4a). The epitope for clone 5A6 resides in the C-terminal domain of mouse MLKL (Fig. S4). As shown in Fig. 4a–d, clone 5A6 produced specific and abundant signals for endogenous mouse MLKL in methanol-fixed, but not paraformaldehyde-fixed cells. Immunoblotting confirmed that clone 5A6 was highly-specific for the detection of endogenous mouse MLKL in cultured cell homogenates (Fig. 4e) and in whole tissue lysates (Fig. 4f). Notably, compared to human MLKL which translocates en masse into cytoplasmic clusters during necroptosis, the relocation of mouse MLKL into cytoplasmic clusters during necroptosis was more subtle. This observation may relate to the notion that human MLKL is recruited to the necrosome in a more stable manner than mouse MLKL [31].
Fig. 4. A new monoclonal antibody to image endogenous mouse MLKL.
a Mouse MLKL domain architecture showing the immunogens or epitopes for the tested anti-MLKL antibodies. b Quantitation of the percentage of gated signals for the tested MLKL antibodies. c Quantitation of specific signal abundance produced by the tested MLKL antibodies on methanol-fixed (MeOH) or paraformaldehyde-fixed (PFA) MDFs. The number of cells imaged (N) to generate each signal-to-noise curve is shown. d Micrographs of immunofluorescent signals for the tested MLKL antibodies on MDFs. As indicated by each pseudocolour look-up-table, only immunosignals within the respective gate in panel c were visualised. Data are representative of n = 3 (in-house clone 5A6) and n = 2 (Abcam clone EPR9515(2), Millipore MABC1158/clone 7C6.1 and Cell Signaling Technology clone D6E3G) independent experiments. Nuclei were detected by Hoechst 33342 staining and are demarked by white outlines in micrographs. Immunoblot using the tested MLKL antibodies against lysates from wild-type versus Mlkl−/− MDFs (e) and from lysates of wild-type versus Mlkl−/− mouse spleen, small intestine and colon (f). Closed arrowheads indicate the main specific band. Asterisks indicate non-specific bands that could otherwise confound data interpretation. Open arrowheads indicate other specific bands of interest. Immunoblots were re-probed for GAPDH as loading control. g Micrographs of clone 5A6 immunosignals on methanol- or paraformaldehyde-fixed sections of the wild-type versus Mlkl−/− mouse spleen, small intestine and liver. Data are representative of n = 2 mice per genotype. In the spleen, MLKL expression was noted in the red pulp, but not the white pulp. In the small intestine, MLKL expression was observed both in epithelial cells and subepithelial cell types in villi. In comparison, MLKL expression was more restricted in the liver; being primarily observed in perivascular regions such as the portal area and in parenchymal cells that resemble Kupffer cells (exemplified by arrows).
Phosphorylation of mouse MLKL at S345 is essential for necroptosis in murine cells [41], and therefore several antibodies have been raised against this phosphosite. We tested three commonly-used antibodies (Fig. 4a–e), namely clones: EPR9515(2) (source: Abcam), 7C6.1 [43] (source: Millipore MABC1158) and D6E3G (source: Cell Signaling Technology). All three clones yielded specific immunosignals in methanol-fixed cells (Fig. 4c), whereas only clone 7C6.1 was compatible with paraformaldehyde fixation. However, the performance of clone D6E3G was superior to the other anti-phospho-MLKL antibodies, because specific signals were more abundant (Fig. 4c) and represented a higher percentage of the total detectable signal (Fig. 4b). This trend was also evident via immunoblot, with clone 7C6.1 being largely non-specific, clone EPR9515(2) being moderately specific and clone D6E3G exhibiting a high degree of specificity toward activated MLKL in necroptotic cells (Fig. 4e).
In summary, clones 5A6 and D6E3G allow the immunofluorescent detection of mouse MLKL under resting and necroptotic conditions, respectively. As with human MLKL, methanol is the fixative of choice for robust immunofluorescent detection of endogenous mouse MLKL. This is an important consideration for the field, as use of a non-crosslinking fixative such as methanol, rather than a crosslinking fixative such as paraformaldehyde, was critical for the immunofluorescent detection of endogenous MLKL by clone 5A6 in several mouse tissues including spleen, small intestine and liver (Fig. 4g).
Immunofluorescent detection of mouse RIPK3
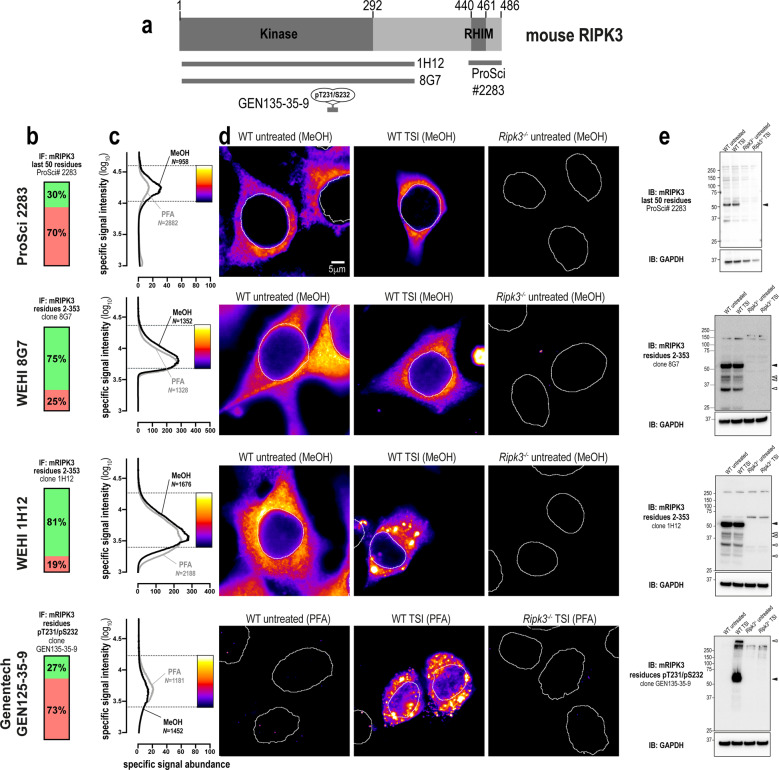
There are numerous well-validated antibodies raised against mouse RIPK3, including a polyclonal antibody against the C-terminus (source: ProSci 2283), and monoclonal antibodies recognising phospho-T231/phospho-S232 (source: Genentech clone GEN135-35-9 [54]) and the extended kinase domain (residues 2–353) of mouse RIPK3 (clone 8G7 [5], in-house; Fig. 5a). These antibodies yielded specific signals via both immunofluorescence and immunoblot analyses (Fig. 5b–e). Interestingly, while phospho-RIPK3 concentrates into cytoplasmic clusters during necroptosis, this relocation event was not detected by other, anti-total RIPK3 antibodies (Fig. 5d). This finding suggests that the epitopes for RIPK3 detection are obscured via protein-protein interactions during the chronology of necroptotic signaling events, or alternatively that other post-translational or conformational changes in RIPK3 may obscure epitope availability. To further probe these steps in the pathway, we generated another mouse RIPK3 antibody, clone 1H12 (source: in-house; Fig. 5b, c), which recognises the translocation of mouse RIPK3 into cytoplasmic clusters during necroptosis (Fig. 5d). Notably, monoclonal antibodies raised against the kinase domain of RIPK3, 8G7, and 1H12, detected several species of lower molecular weight than full length RIPK3 by immunoblot. These signals are likely to be spliced isoforms of mouse RIPK3 harboring the kinase domain antigen (residues 2–353), which are therefore not detected by antibodies directed toward the C-terminus of the full-length mouse RIPK3 isoform, such as ProSci 2283. In summary, there are several excellent options for the immunofluorescent detection of endogenous mouse RIPK3. Notably, unlike MLKL and RIPK1, the choice of fixative has no major bearing on the ability to detect mouse RIPK3 by immunofluorescence (Fig. 5c).
Fig. 5. Unlike human RIPK3, mouse RIPK3 is highly amenable to detection by immunofluorescence and immunoblotting.
a Mouse RIPK3 domain architecture showing the immunogens or epitopes for the tested anti-RIPK3 antibodies. b Quantitation of the percentage of gated signals for the tested RIPK3 antibodies. c Quantitation of specific signal abundance produced by the tested RIPK3 antibodies on methanol-fixed (MeOH) or paraformaldehyde-fixed (PFA) MDFs. The number of cells imaged (N) to generate each signal-to-noise curve is shown. d Micrographs of immunofluorescent signals for the tested RIPK3 antibodies on MDFs. As indicated by each pseudocolour look-up-table, only immunosignals within the respective gate in (c) were visualised. Data are representative of n = 3 (in-house clone 8G7 and Genentech clone GEN135-35-9) and n = 2 (ProSci 2283, in-house clone 1H12) independent experiments. Nuclei were detected by Hoechst 33342 staining and are demarked by white outlines in micrographs. e Immunoblot using the tested RIPK3 antibodies against lysates from wild-type and Ripk3−/− MDFs. Closed arrowheads indicate the main specific band. Open arrowheads indicate other specific bands of interest. Immunoblots were re-probed for GAPDH as loading control.
Immunofluorescent detection of mouse RIPK1
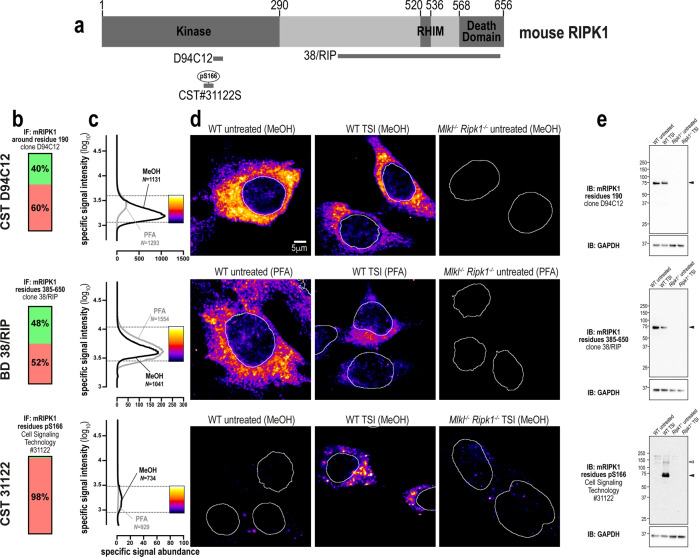
RIPK1 exhibits greater sequence identity between mouse and human orthologs than either RIPK3 or MLKL [44]. In keeping with this, both clone D94C12 (source: Cell Signaling Technology) and clone 38/RIP (source: BD Transduction Laboratories), which were raised against human RIPK1, also specifically detected mouse RIPK1 via immunofluorescence and immunoblotting (Fig. 6a–e). A polyclonal antibody raised against phospho-S166 of mouse RIPK1 (31122; source: Cell Signaling Technology) was also found to specifically detect RIPK1 via immunoblot (Fig. 6e) and in methanol-fixed, but not paraformaldehyde-fixed, cells undergoing necroptosis (Fig. 6b–d).
Fig. 6. Three specific antibodies for imaging endogenous mouse RIPK1.
a Mouse RIPK1 domain architecture showing the immunogens or epitopes for the tested anti-RIPK1 antibodies. b Quantitation of the percentage of gated signals for the tested RIPK1 antibodies. c Quantitation of specific signal abundance produced by the tested RIPK1 antibodies on methanol-fixed (MeOH) or paraformaldehyde-fixed (PFA) MDFs. The number of cells imaged (N) to generate each signal-to-noise curve is shown. d Micrographs of immunofluorescent signals for the tested RIPK1 antibodies on MDFs. As indicated by each pseudocolour look-up-table, only immunosignals within the respective gate in panel c were visualised. Data are representative of n = 2 (Cell Signaling Technology clones D94C12 and 31122, and BD Transduction Laboratories clone 38/RIP) independent experiments. Nuclei were detected by Hoechst 33342 staining and are demarked by white outlines in micrographs. e Immunoblot using the tested RIPK1 antibodies against lysates from wild-type and Ripk1−/− MDFs. Closed arrowheads indicate the main specific band. Open arrowheads indicate other specific bands of interest. Immunoblots were re-probed for GAPDH as loading control.
In summary, three highly-specific antibodies exist for mouse RIPK1. However, as observed in human cells, necroptosis in mouse cells causes a marked reduction in RIPK1 levels (Fig. 6d, e). Thus, the specific immunosignals from the antibodies tested are of low intensity and abundance in necroptotic cells. Therefore, careful gating of signals is recommended for the study of mouse RIPK1 in necroptotic cells.
Antibody cocktails for imaging endogenous necroptotic signaling
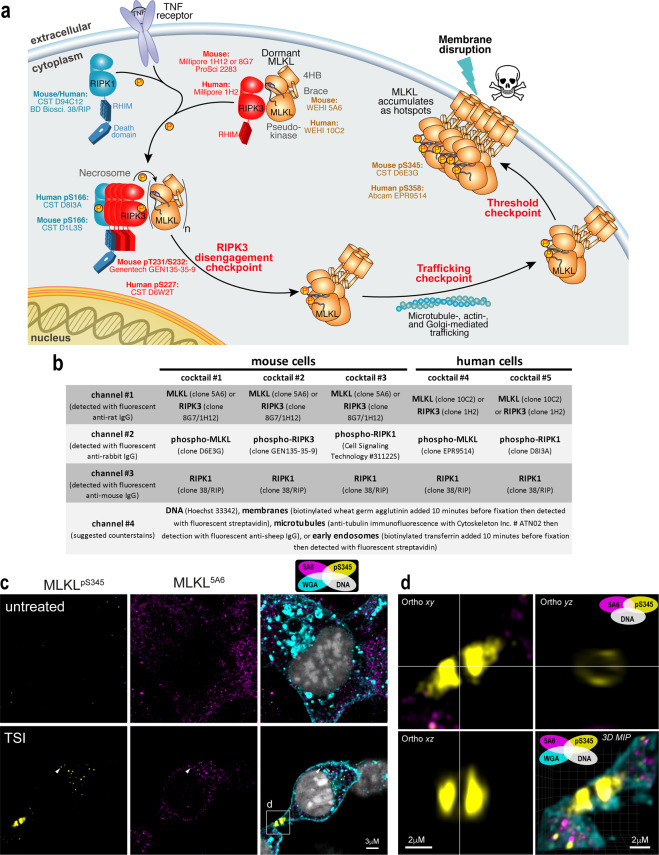
Having validated and compared the performance of numerous reagents, we now propose different antibody combinations that can be used to study endogenous necroptotic signaling (Fig. 7a, b). Because these recommendations are made to facilitate the investigation of endogenous necroptotic signaling in fixed cells, we also provide an ImageJ macro to generate signal-to-noise curves for the gating of specific immunosignals (Supplementary File 1). To exemplify the advantages of visualising endogenous necroptotic events in situ, we co-stained MDFs for non-phosphorylated (clone 5A6; source: in-house) and phosphorylated MLKL (clone D6E3G; source: Cell Signaling Technology). Similar to what was described for human MLKL [32], mouse MLKL was observed to concentrate into cytoplasmic clusters prior to necroptotic cell death, albeit less pronounced than that observed for human MLKL in necroptotic HT29 cells (arrowhead; Fig. 7c). Also similar to what was described for human MLKL [32], phosphorylated MLKL was observed to form focal structures at the plasma membrane, rather than docking uniformly with the cell periphery (boxed region in Fig. 7c, d). The MLKL-specific signals of clone 5A6 and the pS345-specific signals of clone D6E3G (source: CST) overlap at the cytoplasmic clusters, but not at the plasma membrane (Fig. 7c, d), suggesting a signaling chronology analogous to the activation of human MLKL, which can be dissected via immunofluorescence using different antibodies [32]. Remarkably, although smaller than the hotspots of phosphorylated human MLKL that form in necroptotic HT29 cells, junctional accumulations of phosphorylated mouse MLKL were also apparent in necroptotic MDFs (Fig. 7d and Supplementary Video 1). These data suggest that both mouse and human MLKL assemble into macromolecular structures at the plasma membrane. Moreover, as this accumulation occurs preferentially at sites of intercellular contact, it may be an intrinsic and penultimate feature of necroptotic signaling.
Fig. 7. Optimised antibody cocktails for visualising endogenous necroptotic signaling in fixed human and mouse cells.
a Cartoon summary of the currently-understood chronology of TNF-induced necroptosis. Recommendations of validated antibodies for immunostaining various steps in the necroptotic pathway are provided. b Summary of validated antibody cocktails and counterstains that can be multiplexed to examine endogenous necroptotic signaling in fixed human and mouse cells. Successful detection of specific signals relies on fixation in methanol, rather than crosslinking fixatives such as paraformaldehyde. c Two-dimensional Airyscan micrographs of Wheat Germ Agglutinin (WGA)-stained membranes, Hoechst-stained DNA and anti-MLKL immunosignals from clone 5A6 and clone D6E3G on methanol-fixed wild-type MDFs that had been left untreated or TSI-treated for 60 min. Arrowhead exemplifies the small clusters of MLKL that form during necroptosis. The box indicates the junctional accumulation of phospho-MLKL. d Three-dimensional orthogonal projections and maximum intensity projection (MIP) of the boxed region from (c) showing a ring-like structure adopted by phospho-MLKL at the WGA-stained plasma membrane. Accompanied by Supplementary Video 1.
Discussion
Here, we profiled the immunofluorescent staining of MLKL, RIPK3, and RIPK1 in fixed human and mouse cells with a panel of 26 antibodies. While 18 of 26 antibodies were capable of detecting their respective necroptotic protein target in fixed wild-type cells and not their knockout counterparts, only 14 of 26 antibodies exhibited favorable signal-to-noise for robust immunostaining (as summarized in Fig. 7a).
The choice of fixative was a critical variable for the immunofluorescent detection of necroptotic proteins. With the exception of phospho-RIPK3, immunoreactivity was increased in methanol-fixed cell monolayers and tissue sections. These observations have ramifications for the experimental and clinical immunohistological detection of cell death, which is almost exclusively performed on aldehyde-fixed samples. Thus, we recommend the use of methanol fixation for high resolution immunofluorescence of necroptosis; all but one of the antibodies (source: Cell Signaling Technology, clone D6W2T) in the suggested toolbox in Fig. 7a are compatible with methanol fixation.
Our data underscore the importance of using controlled experimental conditions to maximise the identification of specific immunosignals. We used wild-type cells that express high levels of MLKL, RIPK3 and RIPK1, which have a well-characterised response to necroptotic stimuli, and knockout (for non-phospho-targets) or unstimulated wild-type cells (for phospho-sites) as negative controls to define the signal-to-noise profile of each antibody (Fig. 1b). In addition, it is advisable to titrate the antibody-in-question to determine the optimal trade-off between immunofluorescent signal specificity, intensity and abundance (Fig. S2). Furthermore, it is also helpful if different antibodies directed toward the same target yield concordant immunofluorescent signals, and if accompanying immunoblot data confirm their specificity. This combined approach was important for deducing that clone D6E3G (source: Cell Signaling Technology) is the best option for detecting phospho-MLKL in mouse cells (Fig. 4) and that clone EPR9514 (source: Abcam) is the best option for detecting phospho-MLKL in human cells (Fig. 1).
It is concerning that the specificity of some antibodies could not be verified. For instance, while numerous studies use the ProSci 2283 antibody to detect human RIPK3 [55–57], we found that it only recognised mouse RIPK3. Clone 6B4 (source: MyBiosource), M6697 (source: Sigma-Aldrich) and clone 7C6.1 (source: Millipore) have been widely used to respectively detect human MLKL and mouse MLKL [43, 58–64], however these reagents exhibited a low degree of specificity in our hands. Without exception in this study, antibodies that exhibited a low degree of specificity via immunoblot also performed poorly in immunofluorescence analyses. Accordingly, we propose that antibodies should be screened for their specificity for the protein target by immunoblot before proceeding with immunofluorescence studies. Nonetheless, as we did not exhaustively test the performance of these antibodies for immunofluorescence using a wide variety of fixatives, blocking agents, and detergents, we cannot categorically state that they are non-selective. Moreover, as ProSci 2283 (reported to detect mouse and human RIPK3) and Sigma-Aldrich M6697 (reported to detect human MLKL) are polyclonal antibodies, batch-to-batch variation may explain differences in their specificity between studies. We note that the majority of monoclonal antibodies that yielded the most abundant and specific signals were raised against recombinant full-length protein or component domains, rather than unfolded partial domains or peptides. Phosphosite-specific antibodies differ in this regard; these were raised using linear phosphopeptides and their specificity likely reflects the unstructured and solvent-exposed nature of the sequences in which they reside. In general, phosphoprotein antibodies appeared to produce less specific and/or less intense immunofluorescent signals than non-phosphoprotein antibodies. However, the vast differences in abundance between the phospho- and total protein levels can account for these observations, and do not necessarily reflect deficiencies in phospho-specific antibody specificity or affinities for their targets.
Broadly, the specific immunosignals described in this study provide consensus about three key compartmentalisation events that underlie necroptotic signaling [65]. First, the core necroptotic proteins – MLKL, RIPK3 and RIPK1 –predominantly reside in the cytosol under basal conditions as small puncta. These puncta may represent a preassembled structure, such as a membrane-less organelle. Second, RIPK1, RIPK3, and MLKL then coalesce into large cytoplasmic clusters that preferentially locate to the perinuclear space, co-stain for the presence of phospho-RIPK1, -RIPK3 and -MLKL (Figs. 1–7 and ref. [32]). These clusters form in TSI-treated HT29 cells subsequent to the phosphorylation of MLKL [32]. Third, as necroptotic signaling continues, phospho-MLKL is trafficked from clusters to the plasma membrane [29] where it accumulates into supramolecular assemblies called “hotspots” [32]. Our data indicate that phospho-MLKL disengages from necrosomal RIPK3-containing clusters, because neither RIPK1 nor RIPK3 were observed to accumulate at the plasma membrane during necroptosis in mouse or human cells (Figs. 2–3 and 5–6). This disengagement step, and formation of supramolecular assemblies MLKL at the plasma membrane, are likely to be checkpoints in both mouse and human necroptotic signaling.
Methods
Materials
Primary antibodies and their working concentrations (or dilution when stock concentrations were unknown) used for immunoblotting/immunofluorescence were: rat anti-human MLKL (clone 10C2; produced in-house [32]; 1 mg.L−1/5 mg.L−1, to be available from Millipore as MABC1635), rat anti-human MLKL (clone 7G2; produced in-house [32]; 1 mg.L−1/5 mg.L−1, to be available from Millipore as MABC1636), rat anti-mouse/human/rat/horse MLKL (clone 3H1; produced in-house [41] and available as Millipore MABC604; 1 mg.L−1/5 mg.L−1), rabbit anti-phospho-S358 human MLKL (Abcam ab187091; clone EPR9514; lot GR212667-42; 0.3 mg.L−1/3 mg.L−1), mouse anti-phospho-S358 human MLKL (MyBioSource MBS9419709; clone 6B4; lot 6711; 1 mg.L−1/5 mg.L−1), mouse anti-phospho-T357 human MLKL (Novus Biological MAB9187; clone 954702; lot CHKM0219071; 0.5 mg.L−1/2.5 mg.L−1), rabbit anti-human MLKL (Sigma-Aldrich M6697; lot O26M4758; 1:1000/1:200), rabbit anti-RIPK3 (ProSci 2283; lot 8337-1903; 1 mg.L−1 / 5 mg.L−1), rabbit anti-human RIPK3 (Novus Biological NBP2-24588; 1 mg.L−1/5 mg.L−1), mouse anti-human RIPK3 (Novus Biological; clone 780115; lot CGTV0119091; 0.5 mg.L−1/2.5 mg.L−1), rat anti-human RIPK3 (clone 1H2; produced in-house [5], to be available as Millipore MABC1640; 1 mg.L−1/5 mg.L−1), rabbit anti-phospho-S227 human RIPK3 (Cell Signaling Technology; clone D6W2T; 1:2000/1:200), rabbit anti-human RIPK3 (Cell Signaling Technology; clone E1Z1D, 1:2000/1:200), rabbit anti-phospho-S227 human RIPK3 (Abcam; clone EPR9627; lot GR313767-14; 0.5 mg.L−1/5 mg.L−1), rabbit anti-mouse or human RIPK1 (Cell Signaling Technology; clone D94C12; 1:2000/1:200), mouse anti-mouse or human RIPK1 (BD Biosciences; clone 38/RIP; lot 9038922; 0.25 mg.L−1/2.5 mg.L−1), rabbit anti-phospho-S166 human RIPK1 (Cell Signaling Technology; clone D8I3A; 1:2000/1:200), rat anti-mouse MLKL (clone 5A6; produced in-house; 1 mg.L−1/5 mg.L−1, to be available from Millipore as MABC1634), rabbit anti-phospho-S345 mouse MLKL (Abcam; clone EPR9515(2); lot GR246882-35; 0.75 mg.L−1/7.5 mg.L−1), mouse anti-phospho-S345 mouse MLKL (Millipore MABC1158; clone 7C6.1; lot 3471964; 1:2000/1:200), rabbit anti-phospho-S345 mouse MLKL (Cell Signaling Technology; clone D6E3G; 1:2000/1:200), rat anti-mouse RIPK3 (clone 8G7; produced in-house [5], to be available from Millipore as MABC1595; 1 mg.L−1/5 mg.L−1), rat anti-mouse RIPK3 (clone 1H12; produced in-house; 1 mg.L−1/5 mg.L−1), rabbit anti-phospho-T231/S232 mouse RIPK3 (Genentech; clone GEN135-35-9 [54]; lot PUR73907; 2.5 mg.L−1/10 mg.L−1), rabbit anti-phospho-S166 mouse RIPK1 (Cell Signaling Technology 31122; 1:2000/1:200), mouse anti-GAPDH (Millipore MAB374; 1:2000/N/A). Secondary antibodies for immunoblotting were: horseradish peroxidase (HRP)-conjugated goat anti-rat IgG (Southern Biotech 3010-05), HRP-conjugated goat anti-mouse IgG (Southern Biotech 1010-05), and HRP-conjugated goat anti-rabbit IgG (Southern Biotech 4010-05). All secondary antibodies for immunoblotting were used at a dilution of 1:10000. Secondary immunofluorescence detection reagents were: AlexaFluor647-conjugated donkey anti-rabbit IgG (ThermoFisher Scientific A31573), AlexaFluor568-conjugated donkey anti-rabbit IgG (ThermoFisher Scientific A10042), AlexaFluor568-conjugated donkey anti-mouse IgG (ThermoFisher Scientific A10037), AlexaFluor-594 donkey anti-rat IgG (ThermoFisher Scientific A-21209), AlexaFluor488-conjugated donkey anti-rat IgG (ThermoFisher Scientific A21208). All secondary antibodies for immunofluorescence were used at a dilution of 1:1000. Bond-Breaker TCEP solution (ThermoFisher Scientific 77720). N-Ethylmaleimide (Sigma-Aldrich E3876).
Mice
All animal procedures were approved by and conducted in accordance with the Animal Ethics Committee of the Walter and Eliza Hall Institute, Australia. Mice lacking functional alleles for MLKL (Mlkl−/−) have been described previously [41]. All mice were bred on a C57BL/6 J genetic background.
5A6 and 1H12 antibody production
Antibodies were generated at the Walter and Eliza Hall Institute Monoclonal Antibody Facility by immunizing Wistar rats with recombinant full-length mouse MLKL [41] (for clone 5A6) or recombinant mouse RIPK3 residues 2–353 (for clone 1H12 and the previously-reported clone 8G7 [5]) that was expressed and purified from Sf21 insect cells using the baculovirus expression system, before splenocytes were fused with SP2/O mouse myeloid cells and arising hybridoma lines cloned. The specificity of clone 5A6 for mouse MLKL was validated via immunoblot analyses as exemplified in Figs. 4e and S4. The specificity of clone 1H12 for mouse RIPK3 was validated via immunoblot analyses as exemplified in Fig. 5e.
Cell lines
HT29 cells were provided by Mark Hampton (University of Otago). RIPK1−/−, RIPK3−/−, and MLKL−/− HT29 cells have been previously reported [31, 32, 66]. MDF lines were generated in-house from the tails of wild-type C57BL/6 J, Mlkl−/−41 and Ripk3−/− [67] mice and immortalized by SV40 large T antigen as reported previously [39, 41]. The sex and precise age of these animals were not recorded, although our MDFs are routinely derived from tails from 8-week-old mice. Mlkl−/−Ripk1−/− MDFs were derived from the belly/back dermis of Mlkl−/−Ripk1−/− E19.5 mice [16] and immortalized as described above. MDF lines were generated in accordance with protocols approved by the Walter and Eliza Hall Institute of Medical Research Animal Ethics Committee. Cell line identities were not further validated, although their morphologies and responses to necroptotic stimuli were consistent with their stated origins. Cell lines were routinely monitored to confirm they were mycoplasma-free.
Cell culture
HT29 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Life Technologies) containing 8% v/v fetal calf serum (FCS), 2 mM L-Glutamine/-GlutaMAX (ThermoFisher Scientific 35050061), 50 U ml−1 penicillin and 50 U ml−1 streptomycin (G/P/S) under humidified 10% CO2 at 37 °C.
Cell treatment
Cells were seeded into Ibi-treated 8-well μ-slides (Ibidi 80826) in media containing 8% v/v FCS and G/P/S at 3.0 × 104 cells per well for HT29 and 0.25 ×104 cells per well for MDFs. Cells were left to adhere overnight then treated in media containing 1% v/v FCS and G/P/S and supplemented with the following stimuli: 100 ng/mL recombinant human TNF-α-Fc (produced in-house as in ref. [68]), 500 nM Smac mimetic/Compound A (provided by Tetralogic Pharmaceuticals; as in ref. [69]), 5 μM IDN-6556 (provided by Tetralogic Pharmaceuticals). Unless stipulated, HT29 cells were stimulated for 7.5 h with TSI and MDF cells were stimulated for 90 min with TSI.
LDH release
Colorimetric LDH release assay kit (Promega G1780) was performed according to manufacturer’s instructions.
Whole cell/tissue lysis, SDS-PAGE & immunoblot
Cells were lysed in ice-cold RIPA buffer (10 mM Tris-HCl pH 8.0, 1 mM EGTA, 2 mM MgCl2, 0.5% v/v Triton X100, 0.1% w/v Na deoxycholate, 0.5% w/v SDS and 90 mM NaCl) supplemented with 1x Protease & Phosphatase Inhibitor Cocktail (Cell Signaling Technology 5872 S) and 100 U/mL Benzonase (Sigma-Aldrich E1014). Tissues were lysed in ice-cold RIPA buffer to a concentration of 50 mg/ml (w/v) with a stainless steel bead using a Qiagen TissueLyser II (1 min at 30 Hz). Whole cell/tissue lysates were boiled for 10 min in 1× SDS Laemmli sample buffer (126 mM Tris-HCl, pH 8, 20% v/v glycerol, 4% w/v SDS, 0.02% w/v bromophenol blue, 5% v/v 2-mercaptoethanol), and resolved on 1.5 mm NuPAGE 4–12% Bis-Tris gels (ThermoFisher Scientific NP0335BOX) using MES Running buffer (ThermoFisher Scientific NP000202) or Bio-Rad Criterion TGX 4–15% gels (Bio-Rad 5678085) using 1x TGS buffer (Bio-Rad 1610772). After transfer onto nitrocellulose, membranes were blocked in 5% w/v skim milk powder in TBS-T, probed with primary antibodies (see Materials above) then the appropriate HRP-conjugated secondary antibody (see Materials above) and signals revealed by enhanced chemiluminescence (Merck P90720) on a ChemiDoc Touch Imaging System (Bio-Rad). Between each probe, membranes were incubated in stripping buffer (200 mM glycine pH 2.9, 1% w/v SDS, 0.5 mM TCEP) for 30 min at room temperature then re-blocked.
Subcellular fractionation, BN-PAGE & immunoblot
HT29 cells or MDFs were seeded into 6-well plates (1.0 × 106 cells per well) in media containing 8% v/v FCS and G/P/S and equilibrated overnight under humidified 10% CO2 at 37 °C conditions. Cells were then treated in media containing 1% FCS and G/P/S supplemented with the agonists (as indicated above). Cells were fractionated into cytoplasmic and membrane fractions [39]. Cells were permeabilized in MELB buffer (20 mM HEPES pH 7.5, 100 mM KCl, 2.5 mM MgCl2 and 100 mM sucrose, 0.025% v/v digitonin, 2 μM N-ethyl maleimide, phosphatase and protease inhibitors). Crude membrane and cytoplasmic fractions were separated by centrifugation (5 min 11,000 g), and fractions prepared in buffers to a final concentration of 1% w/v digitonin. The samples were resolved on a 4–16% Bis-Tris Native PAGE gel (ThermoFisher), transferred to polyvinylidene difluoride (Merck IPVH00010). After transfer, membranes were destained (in 50% (v/v) methanol, 25% (v/v) acetic acid), denatured (in 6 M Guanidine hydrochloride, 10 mM Tris pH6.8, 5 mM β-mercaptoethanol), blocked in 5% skim milk (Diploma), and probed in the same manner as above.
Protein production and purification
Full-length human and mouse MLKL and the C-terminal, pseudokinase domain of mouse and human MLKL were expressed in Sf21 insect cells using the Bac-to-Bac system (ThermoFisher Scientific) and purified using established procedures [31, 41, 45]. Briefly, proteins were expressed with an N-terminal, TEV protease-cleavable GST (full length human MLKL) or His6 (full length mouse MLKL and pseudokinase domains) tags and captured from lysates using glutathione resin (UBP Bio) or HisTag Ni-NTA resin (Roche) respectively. Proteins were cleaved on-resin from the GST tag or off-resin for His6 tags using His-tagged TEV protease, before protease was removed by Ni-NTA chromatography (HisTag resin, Roche). Protein was concentrated by centrifugal ultrafiltration and applied to a Superdex-200 (GE Healthcare) size exclusion chromatography column; protein was eluted in 20 mM HEPES pH 7.5, 200 mM NaCl, 5% v/v glycerol. Protein containing fractions were spin concentrated to 5–10 mg/mL, aliquoted, snap frozen in liquid N2 and stored at –80 °C until required.
Immunofluorescence on fixed cells
Cells in 8-well μ-Slides (Ibidi 80826) were ice-chilled for 3 min, then washed in ice-cold Dulbecco’s PBS (dPBS; ThermoFisher Scientific 14190144), then fixed for 30 min in either ice-cold methanol or ice-cold 4% w/v PFA. Cells were washed twice in ice-cold dPBS, then blocked in ice-cold Tris-balanced salt solution with 0.05% v/v Triton-X100 (TBS-T) supplemented with 10% v/v donkey serum (Sigma-Aldrich D9663) for >1 h. Cells were incubated in primary antibodies (see Materials above) overnight at 4 °C in TBS-T with 10% v/v donkey serum. Cells were washed twice in TBS-T then incubated in the appropriate secondary antibodies supplemented with 0.1μg/mL Hoechst 33342 (ThermoFisher Scientific H3570) for 3 h at room temperature with gentle rocking. Cells were washed four times in ice-cold TBS-T then stored at 4 °C until imaged. Where indicated, to demarcate the plasma membrane, 2μL of biotinylated wheat germ agglutinin (Sigma-Aldrich L5142) was added to each well 10 min before fixation. The fixed wheat germ agglutinin was then detected via the addition of 1:1000 dilution of DyLight650-conjugated streptavidin (ThermoFisher Scientific 84547) during the secondary antibody incubation step.
Immunofluorescence on fixed tissues
Tissues were dissected post-mortem from adult male wild-type or Mlkl−/− mice of 9–30 weeks of age, submerged in ice-cold Milestone freezing medium, then frozen using PrestoCHILL device (Milstone; settings of 2 min at −40 °C). 8 μm sections were cut on a cryostat and air-dried onto Superfrost slides (ThermoFisher Scientific) for 30 min at room temperature. Sections were fixed for 30 min in either ice-cold 4% (w/v) paraformaldehyde or ice-cold methanol, then rinsed twice in ice-cold Dulbecco’s PBS and blocked in Tris-balanced salt solution with 0.05% v/v Triton-X100 (TBS-T) supplemented with 10% v/v donkey serum (Sigma-Aldrich D9663) overnight at 4 °C. Sections were incubated overnight at 4 °C in 5 mg.L−1 of clone 5A6 in TBS-T with 10% v/v donkey serum. Sections were washed three times in TBS-T then incubated overnight at 4 °C in a 1:1000 dilution of AlexaFluor594-conjugated donkey anti-rat IgG (ThermoFisher Scientific) supplemented with 0.1 μg/mL Hoechst 33342 (ThermoFisher Scientific H3570) in TBS-T with 10% v/v donkey serum. Sections were washed four times in ice-cold TBS-T, mounted in DAKO fluorescent mounting media (DAKO S3023) and kept in the dark at room temperature until being imaged.
Two-dimensional epifluorescence microscopy of immunostained cells
Samples in TBS-T were imaged on an Inverted Axio Observer.Z1 microscope (Zeiss) with the following specifications: C-Apochromat 40x/1.20 W autocorr UV VIS IR lens, HXP 120 V excitation source, AlexaFluor647 and DyLight650 imaged with a λExcitation = 625–655 nm; λbeamsplitter = 660 nm; λEmission = 665–715 nm filter, AlexaFluor568 imaged with a λExcitation = 532–544 nm; λbeamsplitter = 560 nm; λEmission = 573–613 nm, AlexaFluor488 imaged with a λExcitation = 450–490 nm; λbeamsplitter = 495 nm; λEmission = 500–550 nm, Hoechst 33342 imaged with a λExcitation = 359–371 nm; λbeamsplitter = 395 nm; λEmission = 397-∞ nm, a sCMOS PCO.edge 4.2 camera, ZEN blue 2.5 pro capture software and ImageJ 1.53c post-acquisition processing software [70]. Typically, for each independent experiment, 5–10 randomly selected fields were captured per treatment group, whereby only the Hoechst 33342 signal was visualised prior to multi-channel acquisition. To ensure consistent signal intensities across independent experiments, the same excitation, emission and camera settings were used throughout this study.
Two-dimensional epifluorescence microscopy of immunostained tissues
Immunostained tissue sections were imaged on a Vectra Polaris Imaging System (Akoya Biosciences). Acquisition Software: Vectra Polaris v.1.0. Resolution:0.5μm/pixel (20x objective). LED light source. Default filters for DAPI MSI (1 ms exposure) and Texas Red (590 ms exposure) were used. Post-acquisition image processing was performed using QuPATH software v.0.2.3 [71].
Airyscan microscopy
Fixed immunostained cells in 8-well μ-Slides (Ibidi 80827) were subjected to super-resolution 3-dimensional Airyscan microscopy on an Inverted LSM 880 platform (Zeiss) equipped with the following specifications: a 63x/1.4 N.A. PlanApo DIC M27 oil immersion objective (Zeiss), 405-, 488-, 568-, and 640-nm laser lines, and radially-stacked Airyscan GaASP detectors set to SR-mode, 405-, 488-, 561-, 633-nm laser lines and ZEN black 2.3 SP1 FP3 v.14.0 capture software. Image stacks were acquired with a z-step size of 159 nm. Super-resolution deconvolution was performed using the automated ‘3D AiryScan Processing’ function of ZEN blue software.
Hotspot quantitation
Micrographs (captured as in two-dimensional epifluorescence microscopy) were opened in ImageJ v.1.53c [70]. A rolling ball filter of 7 was applied and phospho-S358 MLKL immunosignals thresholded (≥7000 units) and objects segmented using the “Analyze>Analyze Particles” tool. Segmented objects with size 0.5–100 μm2 and feret diameter>2 (i.e., elliptical objects) were considered hotspots. The number of segmented objects per 100 cells was taken as an index of hotspot occurrence. The mean size of the segmented objects was taken as an index of hotspot size. The fluorescence intensity of each segmented object divided by its size was taken as an index of hotspot intensity.
Supplementary information
Acknowledgements
We thank the Walter and Eliza Hall Institute Monoclonal Antibody Facility for their assistance generating clones 3H1, 10C2, 7G2, 1H2, 5A6, 8G7 and 1H12; and Cathrine Hall for assistance generating Mlkl−/− Ripk1−/− MDF lines. We acknowledge the WEHI histology facility for hosting the Vectra Polaris Imaging System and Emma Pan for acquiring the accompanying data. We thank: the laboratory of Tracy Putoczki for use of their Qiagen TissueLyser II device; Kim Newton and Vishva Dixit for sharing Ripk3−/− mice; and Michelle Kelliher for the Ripk1−/− mice used to generate the Ripk1−/−Mlkl−/− strain.
Funding
We are grateful to the Australian National Health and Medical Research Council for fellowship (JMH, 1142669; EDH, 1159488; JMM, 1172929), grant (1124735, 1124737, 1105023 and 2002965), and infrastructure (IRIISS 9000653) support, with additional support from the CASS Foundation (ALS) and the Victorian Government Operational Infrastructure Support scheme. We acknowledge the support for AVJ from an Australian Research Training Program Scholarship.
Author contributions
Investigation: ALS, KP, CF, JMH, LW, JR, AVJ, CRH, XG, SNY Supervision and Methodology: ALS, JMH, KLR, EDH and JMM; Conceptualization: ALS, EDH, and JMM; Writing – Original Draft: ALS and JMM; Writing – Review & Editing: all authors.
Compliance with ethical standards
Conflict of interest
ALS, CF, KP, JMH, SNY, and JMM contribute to a project developing necroptosis inhibitors in collaboration with Anaxis Pharma. The other authors declare no competing interests.
Ethics
All animal procedures were approved by and conducted in accordance with the Animal Ethics Committee of the Walter and Eliza Hall Institute, Australia.
Materials requests
All materials will be provided under Materials Transfer Agreement upon request to JMM (jamesm@wehi.edu.au).
Footnotes
Edited by G. Melino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
André L. Samson, Email: samson.a@wehi.edu.au
James M. Murphy, Email: jamesm@wehi.edu.au
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-021-00742-x.
References
- 1.Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–23. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo H, Omoto S, Harris PA, Finger JN, Bertin J, Gough PJ, et al. Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe. 2015;17:243–51. doi: 10.1016/j.chom.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearson JS, Giogha C, Muhlen S, Nachbur U, Pham CL, Zhang Y, et al. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat Microbiol. 2017;2:16258. doi: 10.1038/nmicrobiol.2016.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson JS, Murphy JM. Down the rabbit hole: is necroptosis truly an innate response to infection? Cell Microbiol. 2017;19:e12750. doi: 10.1111/cmi.12750. [DOI] [PubMed] [Google Scholar]
- 5.Petrie EJ, Sandow JJ, Lehmann WIL, Liang LY, Coursier D, Young SN, et al. Viral MLKL homologs subvert necroptotic cell death by sequestering cellular RIPK3. Cell Rep. 2019;28:3309–3319 e3305. doi: 10.1016/j.celrep.2019.08.055. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher-Etherington A, Nobre L, Nightingale K, Antrobus R, Nichols J, Davison AJ, et al. Human cytomegalovirus protein pUL36: a dual cell death pathway inhibitor. Proc Natl Acad Sci USA. 2020;117:18771–9. doi: 10.1073/pnas.2001887117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitur K, Wachtel S, Brown A, Wickersham M, Paulino F, Penaloza HF, et al. Necroptosis promotes staphylococcus aureus clearance by inhibiting excessive inflammatory signaling. Cell Rep. 2016;16:2219–30. doi: 10.1016/j.celrep.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shubina M, Tummers B, Boyd DF, Zhang T, Yin C, Gautam A, et al. Necroptosis restricts influenza A virus as a stand-alone cell death mechanism. J Exp Med. 2020;217:e20191259. doi: 10.1084/jem.20191259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson J, Loh Z, Ullah MA, Lynch JP, Werder RB, Collinson N, et al. Respiratory syncytial virus infection promotes necroptosis and HMGB1 release by airway epithelial cells. Am J Respir Crit Care Med. 2020;201:1358–71. doi: 10.1164/rccm.201906-1149OC. [DOI] [PubMed] [Google Scholar]
- 10.Muller T, Dewitz C, Schmitz J, Schroder AS, Brasen JH, Stockwell BR, et al. Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell Mol Life Sci. 2017;74:3631–45. doi: 10.1007/s00018-017-2547-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pefanis A, Ierino FL, Murphy JM, Cowan PJ. Regulated necrosis in kidney ischemia-reperfusion injury. Kidney Int. 2019;96:291–301. doi: 10.1016/j.kint.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Luedde M, Lutz M, Carter N, Sosna J, Jacoby C, Vucur M, et al. RIP3, a kinase promoting necroptotic cell death, mediates adverse remodelling after myocardial infarction. Cardiovasc Res. 2014;103:206–16. doi: 10.1093/cvr/cvu146. [DOI] [PubMed] [Google Scholar]
- 13.Hildebrand JM, Kauppi M, Majewski IJ, Liu Z, Cox AJ, Miyake S, et al. A missense mutation in the MLKL brace region promotes lethal neonatal inflammation and hematopoietic dysfunction. Nat Commun. 2020;11:3150. doi: 10.1038/s41467-020-16819-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Sun L, Su L, Rizo J, Liu L, Wang LF, et al. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol Cell. 2014;54:133–46. doi: 10.1016/j.molcel.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Rickard JA, Anderton H, Etemadi N, Nachbur U, Darding M, Peltzer N, et al. TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. Elife. 2014;3:e03464. doi: 10.7554/eLife.03464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rickard JA, O’Donnell JA, Evans JM, Lalaoui N, Poh AR, Rogers TW, et al. RIPK1 regulates RIPK3-MLKL driven systemic inflammation and emergency hematopoiesis. Cell. 2014;157:1175–88. doi: 10.1016/j.cell.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 17.Pierdomenico M, Negroni A, Stronati L, Vitali R, Prete E, Bertin J, et al. Necroptosis is active in children with inflammatory bowel disease and contributes to heighten intestinal inflammation. Am J Gastroenterol. 2014;109:279–87. doi: 10.1038/ajg.2013.403. [DOI] [PubMed] [Google Scholar]
- 18.He S, Wang L, Miao L, Wang T, Du F, Zhao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–11. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–6. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–27. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA. 2012;109:5322–7. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Perera ND, Chiam MDF, Cuic B, Wanniarachchillage N, Tomas D, et al. Necroptosis is dispensable for motor neuron degeneration in a mouse model of ALS. Cell Death Differ. 2020;27:1728–39. doi: 10.1038/s41418-019-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dermentzaki G, Politi KA, Lu L, Mishra V, Perez-Torres EJ, Sosunov AA, et al. Deletion of Ripk3 prevents motor neuron death in vitro but not in vivo. eNeuro. 2019;6:ENEURO.0308-18.2018. doi: 10.1523/ENEURO.0308-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez S, Varfolomeev E, Brendza R, Stark K, Tea J, Imperio J, et al. Genetic inactivation of RIP1 kinase does not ameliorate disease in a mouse model of ALS. Cell Death Differ. 2020. 10.1038/s41418-020-00625-7. [DOI] [PMC free article] [PubMed]
- 25.Dara L, Johnson H, Suda J, Win S, Gaarde W, Han D, et al. Receptor interacting protein kinase 1 mediates murine acetaminophen toxicity independent of the necrosome and not through necroptosis. Hepatology. 2015;62:1847–57. doi: 10.1002/hep.27939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Diaz S, Preaudet A, Samson AL, Nguyen PM, Fung KY, Garnham AL, et al. Necroptosis is dispensable for the development of inflammation-associated or sporadic colon cancer in mice. Cell Death Differ. 2020. 10.1038/s41418-020-00673-z. [DOI] [PMC free article] [PubMed]
- 27.Webster JD, Kwon YC, Park S, Zhang H, Corr N, Ljumanovic N, et al. RIP1 kinase activity is critical for skin inflammation but not for viral propagation. J Leukoc Biol. 2020;107:941–52. doi: 10.1002/JLB.3MA1219-398R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton K, Dugger DL, Maltzman A, Greve JM, Hedehus M, Martin-McNulty B, et al. RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 2016;23:1565–76. doi: 10.1038/cdd.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrie EJ, Birkinshaw RW, Koide A, Denbaum E, Hildebrand JM, Garnish SE, et al. Identification of MLKL membrane translocation as a checkpoint in necroptotic cell death using Monobodies. Proc Natl Acad Sci USA. 2020;117:8468–75. doi: 10.1073/pnas.1919960117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrie EJ, Czabotar PE, Murphy JM. The structural basis of necroptotic cell death signaling. Trends Biochem Sci. 2019;44:53–63. doi: 10.1016/j.tibs.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Petrie EJ, Sandow JJ, Jacobsen AV, Smith BJ, Griffin MDW, Lucet IS, et al. Conformational switching of the pseudokinase domain promotes human MLKL tetramerization and cell death by necroptosis. Nat Commun. 2018;9:2422. doi: 10.1038/s41467-018-04714-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samson AL, Zhang Y, Geoghegan ND, Gavin XJ, Davies KA, Mlodzianoski MJ, et al. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat Commun. 2020;11:3151. doi: 10.1038/s41467-020-16887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng Y, Sandow JJ, Czabotar PE, Murphy JM. The regulation of necroptosis by post-translational modifications. Cell Death Differ. 2021. 10.1038/s41418-020-00722-7. [DOI] [PMC free article] [PubMed]
- 34.Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–50. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mompean M, Li W, Li J, Laage S, Siemer AB, Bozkurt G, et al. The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell. 2018;173:1244–53. e1210. doi: 10.1016/j.cell.2018.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X, Yin J, Starovasnik MA, Fairbrother WJ, Dixit VM. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J Biol Chem. 2002;277:9505–11. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- 37.Cook WD, Moujalled DM, Ralph TJ, Lock P, Young SN, Murphy JM, et al. RIPK1- and RIPK3-induced cell death mode is determined by target availability. Cell Death Differ. 2014;21:1600–12. doi: 10.1038/cdd.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orozco S, Yatim N, Werner MR, Tran H, Gunja SY, Tait SW, et al. RIPK1 both positively and negatively regulates RIPK3 oligomerization and necroptosis. Cell Death Differ. 2014;21:1511–21. doi: 10.1038/cdd.2014.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hildebrand JM, Tanzer MC, Lucet IS, Young SN, Spall SK, Sharma P, et al. Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc Natl Acad Sci USA. 2014;111:15072–7. doi: 10.1073/pnas.1408987111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy JM. The killer pseudokinase mixed lineage kinase domain-like protein (MLKL) Cold Spring Harb Perspect Biol. 2020;12:a036376. doi: 10.1101/cshperspect.a036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–53. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 42.Tanzer MC, Tripaydonis A, Webb AI, Young SN, Varghese LN, Hall C, et al. Necroptosis signalling is tuned by phosphorylation of MLKL residues outside the pseudokinase domain activation loop. Biochemical J. 2015;471:255–65. doi: 10.1042/BJ20150678. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez DA, Weinlich R, Brown S, Guy C, Fitzgerald P, Dillon CP, et al. Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 2016;23:76–88. doi: 10.1038/cdd.2015.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies KA, Fitzgibbon C, Young SN, Garnish SE, Yeung W, Coursier D, et al. Distinct pseudokinase domain conformations underlie divergent activation mechanisms among vertebrate MLKL orthologues. Nat Commun. 2020;11:3060. doi: 10.1038/s41467-020-16823-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy JM, Lucet IS, Hildebrand JM, Tanzer MC, Young SN, Sharma P, et al. Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochemical J. 2014;457:369–77. doi: 10.1042/BJ20131270. [DOI] [PubMed] [Google Scholar]
- 46.Tanzer MC, Matti I, Hildebrand JM, Young SN, Wardak A, Tripaydonis A, et al. Evolutionary divergence of the necroptosis effector MLKL. Cell Death Differ. 2016;23:1185–97. doi: 10.1038/cdd.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies KA, Tanzer MC, Griffin MDW, Mok YF, Young SN, Qin R, et al. The brace helices of MLKL mediate interdomain communication and oligomerisation to regulate cell death by necroptosis. Cell Death Differ. 2018;25:1567–80. doi: 10.1038/s41418-018-0061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dondelinger Y, Hulpiau P, Saeys Y, Bertrand MJM, Vandenabeele P. An evolutionary perspective on the necroptotic pathway. Trends Cell Biol. 2016;26:721–32. doi: 10.1016/j.tcb.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Newton K, Manning G. Necroptosis and Inflammation. Annu Rev Biochem. 2016;85:743–63. doi: 10.1146/annurev-biochem-060815-014830. [DOI] [PubMed] [Google Scholar]
- 50.Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J, et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol. 2014;16:55–65. doi: 10.1038/ncb2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, Li W, Ren J, Huang D, He WT, Song Y, et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 2014;24:105–21. doi: 10.1038/cr.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alturki NA, McComb S, Ariana A, Rijal D, Korneluk RG, Sun SC, et al. Triad3a induces the degradation of early necrosome to limit RipK1-dependent cytokine production and necroptosis. Cell Death Dis. 2018;9:592. doi: 10.1038/s41419-018-0672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson DS, Gabrielyan A, Feltham R. RIPK1 ubiquitination: evidence, correlations and the undefined. Semin Cell Dev Biol. 2021;109:76–85. doi: 10.1016/j.semcdb.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 54.Newton K, Wickliffe KE, Maltzman A, Dugger DL, Strasser A, Pham VC, et al. RIPK1 inhibits ZBP1-driven necroptosis during development. Nature. 2016;540:129–33. doi: 10.1038/nature20559. [DOI] [PubMed] [Google Scholar]
- 55.Xu X, Kalac M, Markson M, Chan M, Brody JD, Bhagat G, et al. Reversal of CYLD phosphorylation as a novel therapeutic approach for adult T-cell leukemia/lymphoma (ATLL) Cell Death Dis. 2020;11:94. doi: 10.1038/s41419-020-2294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C, Li J, Yu L, Zhang Z, Xu F, Jiang L, et al. Regulation of RIP3 by the transcription factor Sp1 and the epigenetic regulator UHRF1 modulates cancer cell necroptosis. Cell Death Dis. 2017;8:e3084. doi: 10.1038/cddis.2017.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Ni HM, Dorko K, Kumer SC, Schmitt TM, Nawabi A, et al. Increased hepatic receptor interacting protein kinase 3 expression due to impaired proteasomal functions contributes to alcohol-induced steatosis and liver injury. Oncotarget. 2016;7:17681–98. doi: 10.18632/oncotarget.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, Che L, He C, Huang J, Guo N, Shi J, et al. Drp1 and RB interaction to mediate mitochondria-dependent necroptosis induced by cadmium in hepatocytes. Cell Death Dis. 2019;10:523. doi: 10.1038/s41419-019-1730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ali M, Mocarski ES. Proteasome inhibition blocks necroptosis by attenuating death complex aggregation. Cell Death Dis. 2018;9:346. doi: 10.1038/s41419-018-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong YN, Guy C, Olauson H, Becker JU, Yang M, Fitzgerald P, et al. ESCRT-III acts downstream of MLKL to regulate necroptotic. Cell Death Its Conséq Cell. 2017;169:286–300 e216. doi: 10.1016/j.cell.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon S, Bogdanov K, Kovalenko A, Wallach D. Necroptosis is preceded by nuclear translocation of the signaling proteins that induce it. Cell Death Differ. 2016;23:253–60. doi: 10.1038/cdd.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–43. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 63.Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. 2014;124:3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang T, Yin C, Boyd DF, Quarato G, Ingram JP, Shubina M, et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell. 2020;180:15–29. e1113. doi: 10.1016/j.cell.2020.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Samson AL, Garnish SE, Hildebrand JM, Murphy JM. Location, location, location: A compartmentalized view of TNF-induced necroptotic signaling. Sci Signal. 2021;14:eabc6178. doi: 10.1126/scisignal.abc6178. [DOI] [PubMed] [Google Scholar]
- 66.Tanzer MC, Khan N, Rickard JA, Etemadi N, Lalaoui N, Spall SK, et al. Combination of IAP antagonist and IFNgamma activates novel caspase-10- and RIPK1-dependent cell death pathways. Cell Death Differ. 2017;24:481–91. doi: 10.1038/cdd.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Mol Cell Biol. 2004;24:1464–9. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bossen C, Ingold K, Tardivel A, Bodmer JL, Gaide O, Hertig S, et al. Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J Biol Chem. 2006;281:13964–71. doi: 10.1074/jbc.M601553200. [DOI] [PubMed] [Google Scholar]
- 69.Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–93. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 70.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.