Abstract
The Hippo signaling effector, TEAD1 plays an essential role in cardiovascular development. However, a role for TEAD1 in postmitotic cardiomyocytes (CMs) remains incompletely understood. Herein we reported that TEAD1 is required for postmitotic CM survival. We found that adult mice with ubiquitous or CM-specific loss of Tead1 present with a rapid lethality due to an acute-onset dilated cardiomyopathy. Surprisingly, deletion of Tead1 activated the necroptotic pathway and induced massive cardiomyocyte necroptosis, but not apoptosis. In contrast to apoptosis, necroptosis is a pro-inflammatory form of cell death and consistent with this, dramatically higher levels of markers of activated macrophages and pro-inflammatory cytokines were observed in the hearts of Tead1 knockout mice. Blocking necroptosis by administration of necrostatin-1 rescued Tead1 deletion-induced heart failure. Mechanistically, genome-wide transcriptome and ChIP-seq analysis revealed that in adult hearts, Tead1 directly activates a large set of nuclear DNA-encoded mitochondrial genes required for assembly of the electron transfer complex and the production of ATP. Loss of Tead1 expression in adult CMs increased mitochondrial reactive oxygen species, disrupted the structure of mitochondria, reduced complex I-IV driven oxygen consumption and ATP levels, resulting in the activation of necroptosis. This study identifies an unexpected paradigm in which TEAD1 is essential for postmitotic CM survival by maintaining the expression of nuclear DNA-encoded mitochondrial genes required for ATP synthesis.
Subject terms: Gene regulation, Experimental models of disease
Introduction
Heart disease is a leading cause of death worldwide. Among all cardiomyopathies, dilated cardiomyopathy (DCM), characterized by left ventricular dilatation with deteriorated myocardial contraction, is one of the most common heart diseases [1]. Cell death plays a critical role in the pathogenesis of DCM due to the extremely low self-renewal of postmitotic cardiomyocytes (CMs) [2].
In additional to apoptosis that is a well-established form of programmed cell death in DCM, recent studies have suggested that necroptosis, is another major contributor to the loss of CMs in DCM [2]. In contrast to the well-maintained cell membrane integrity observed in apoptosis, necroptosis is characterized by cell swelling, cell and organelle membrane disruption and cell lysis [3]. Plasma membrane leakage from necrotic cells causes the release of intracellular damage-associated molecular patterns into extracellular milieu which evoke pro-inflammatory responses [4]. Necroptosis is initiated through the inhibition of caspase-8 (CASP8), activation of receptor-interacting serine/threonine-protein kinase (RIPK) 1 and 3 [5]. Subsequently, RIPK1 and RIPK3 form a functional complex with MLKL and PGAM5 [5]. Despite significant progress in defining the core components of necroptosis pathway and some evidence implicating CM necroptosis in DCM [2, 5, 6], the upstream effectors governing necroptosis in adult CMs remain elusive.
As the heart is a high energy-demanding organ, insufficient energy induced by mitochondrial dysfunction has been implicated in DCM and other heart diseases [7, 8]. Functional defects in the mitochondrial electron transport chain (ETC) result in deficient ATP synthesis, over-production of damaging reactive oxygen species (ROS), CM necroptosis, and DCM [9–11]. The ETC comprises complexes I to V, which are encoded by both nuclear and mitochondrial DNA (nDNA or mtDNA) [7]. Mutations in either form of DNA can cause ETC functional defects, leading to a variety of cardiomyopathies [12]. Although the nDNA genes encoding each of subunits of ETC complexes has been identified, transcription factors responsible for the regulation of expression of these genes largely remain to be identified.
The Hippo pathway is evolutionarily conserved and is crucial for heart development and regeneration [13, 14]. YAP1 is one of the core components of Hippo pathway in mammals and has been shown to play a critical role in cardiogenesis [15]. YAP1 exerts its function through binding to a variety of transcription factors due to lack of a DNA binding domain. As the primary binding partners with YAP1, the TEAD family proteins consist of four members (TEAD1-4), all of which bind to a consensus DNA sequence 5’-CATTCC-3’ [16, 17]. We have recently shown that global knockout (KO) of Tead1 in mice leads to early embryonic lethality between E9.5 and E10.5 [18, 19]. Although the importance of TEAD1 in CMs has been established [20–22], the specific role of Tead1 in adult tissue homeostasis especially in the adult heart remains incompletely understood.
Utilizing the Tead1 conditional allele mice we generated [18, 19, 23], we investigated the function of Tead1 in tissue homeostasis by deletion of Tead1 globally in adult mice. We found that global loss of Tead1 in adult mice resulted in a rapid lethality due to acute-onset DCM. Inducible CM-specific loss of Tead1 in adult mice results in a phenotype strikingly similar to that observed in the Tead1 adult global null mutants. Mechanistically, we found deletion of Tead1 in adult CMs specifically induced CM necroptosis, but not apoptosis. Furthermore, we demonstrated that TEAD1 directly regulates a cohort of mitochondrial genes including components of ETC, which have been implicated as direct regulators of cell death.
Methods and materials
Tead1 flox (F) mice were generated as we previously described in detail [18, 19, 23]. The inducible global Cre transgenic mouse strain CAGGCre-ERTM+ were purchased from the Jackson Laboratory. To generate mice of inducible global KO of Tead1 (igKO), we first generated CAGGCre-ERTM+; Tead1F/W mouse, by crossing CAGGCre-ERTM+ male mice with Tead1F/F female mice. Then CAGGCre-ERTM+; Tead1F/W male mice were used to breed with Tead1F/F female mice to obtain CAGGCre-ERTM−; Tead1F/F control and CAGGCre-ERTM+; Tead1F/F experimental mice. Age and gender-matched CAGGCre-ERTM+; Tead1W/W mice serve as an additional control to rule out a potential toxicity mediated by the Cre transgene. All 3 groups of mice were intraperitoneally injected with tamoxifen (TAM) (50 mg/kg/day) for 5 consecutive days at age of 12 weeks old.
To generate Tead1 inducible cardiac-specific KO (icKO) mice, Tead1F/F female mice were bred with cardiac-specific gene Myh6 promoter driven MerCreMer transgenic mice [24]. The resultant Myh6-MerCreMer+; Tead1F/W male mice were then mated with TeadF/F female mice, which gave rise to littermate mice Myh6-MerCreMer-; Tead1F/F (control mice) and Myh6-MerCreMer+; Tead1F/F (icKO; experimental mice). Similarly, age and gender-matched Myh6-MerCreMer+; Tead1W/W mice were used as an additional control. All 3 groups of mice were received TAM injection in the same way as for igKO mice except with lower dose of TAM (25 mg/kg/day). Both male and female mice were used in this study and were maintained in C57BL/6J strain background. The primers for genotyping are described in Online Table I. The RNA-seq data generated in this study have been deposited in the Sequences Read Archive at the NCBI under SRA accession numbers PRJNA575531. The TEAD1 adult mouse heart ChIP-seq data has been deposited in NCBI GEO database under the accession number GSE124008. The use of experimental mice was approved by the Institutional Animal Care and Use Committee and Biosafety committee at Augusta University. A detailed, expanded method section is included in the online Supplemental Material.
Results
Global deletion of Tead1 in adult mice results in rapid lethality due to acute-onset DCM
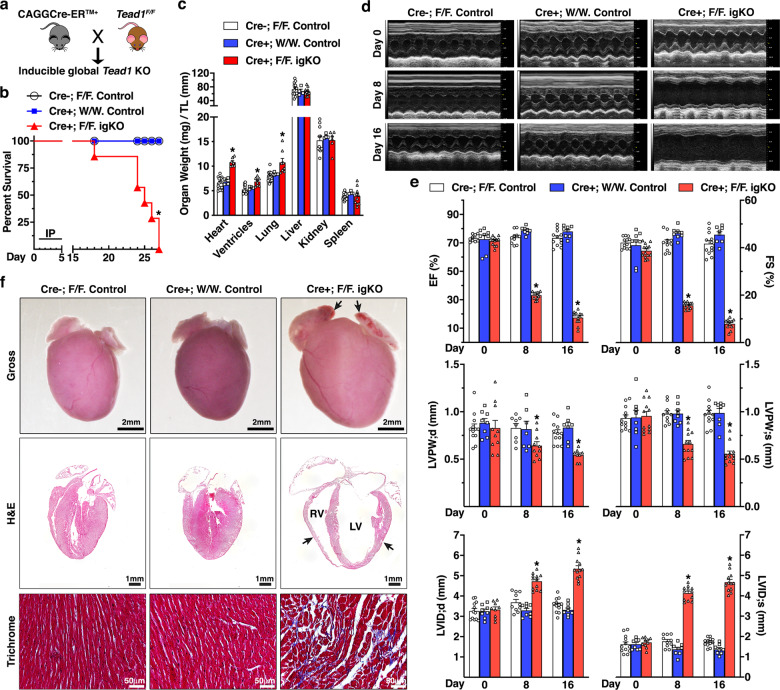
To explore the potential role of TEAD1 in tissue homeostasis in adult mice, we generated inducible Tead1 global KO mice (Online Fig. Ia and Fig. 1a) by crossing the TAM-inducible global Cre mouse (CAGGCre-ERTM+) with Tead1F/F mice. Following TAM administration in adult mice, deletion of Tead1 was validated in diverse tissues by detecting the deleted Tead1 allele (Online Fig. Ib and c) in igKO mice. Western blotting and immunofluorescence (IF) staining further confirmed efficient loss of Tead1 protein in igKO mouse hearts (Online Fig. Id and e). 18 days following the first TAM injection, igKO mice started to display mortality and none of them survived beyond 27 days (Fig. 1b) and had significantly increased ratios of heart, ventricle, and lung, but normal ratios of liver, kidney and spleen, relative to the tibial length, suggesting a cardio-pulmonary phenotype in the Tead1 igKO mice (Fig. 1c). We next performed echocardiography in the igKO and two control groups of mice prior to (day 0, baseline) or day 8 and 16 post-TAM injection (Online Fig. I a). Echocardiography data revealed that all of three groups of mice had indistinguishable cardiac function at day 0. However, Tead1 igKO mice exhibited left ventricle (LV) wall thinning, ventricular chamber dilation and severely progressive impaired cardiac contractility 8 and 16 days following TAM injection as indicated by diminished LV ejection fraction and fractional shortening (Fig. 1d and e). Gross examination of hearts revealed that the Tead1 igKO hearts were dramatically enlarged with thrombi in both atria (Fig. 1f, top panel). Histological analysis further revealed the severe dilation of both right ventricle (RV) and LV with ventricular wall thinning (Fig. 1f, middle panel). Furthermore, trichrome staining showed collagen deposition with disruptive myocardial architecture (Fig. 1f, bottom panel). Taken together, these data demonstrated that global deletion of Tead1 in adult mice results in rapid lethality, acute-onset DCM, and heart failure.
Fig. 1. Global deletion of Tead1 in adult mice leads to rapid lethality due to acute-onset DCM.
a The breeding scheme to generate adult inducible global Tead1 KO mice (igKO) by crossing CAGGCre-ERTM+ mice with Tead1 flox (F) mice. b Survival curve of adult control (CAGGCre-ERTM-; Tead1F/F, N = 17 and CAGGCre-ERTM+; Tead1W/W, N = 18) and Tead1 igKO (CAGGCre-ERTM+; Tead1F/F, N = 12) mice following 5 daily intraperitoneal injections (IP) of tamoxifen. *p < 0.01. Kaplan–Meier analysis. c Control and Tead1 igKO mice were harvested at day 18 following the first tamoxifen injection. The organ weights then were normalized by the tibial length (TL). *p < 0.05 vs. control groups. d Representative echocardiographic M-mode images are shown from the control and Tead1 igKO mice prior (day 0) or at day 8 or 16 following the first tamoxifen injection. e Echocardiographic quantifications of control and Tead1 igKO mice. Shown are left ventricle (LV) ejection fraction (EF) and fractional shortening (FS) (upper panel), LV posterior wall thickness at end-diastole (LVPW;d) or at end-systole (LVPW;s) (middle panel), LV diastolic and systolic internal diameters (LVID;d and LVID;s, bottom panel). *p < 0.05 vs. control groups. f At 18 days following the first tamoxifen injection, hearts from control and Tead1 igKO mice were harvested for gross morphology (upper panel), sectioned for H&E staining (middle panel) and trichrome staining (bottom panel). Note thrombus accumulation in atria (arrows in upper panel), dramatic dilation of both LV and right ventricle (RV) chambers and thinning ventricular wall (arrows in middle panel) in Tead1 igKO mice.
Inducible CM-specific deletion of Tead1 in adult mice recapitulates the phenotype of Tead1 igKO mice
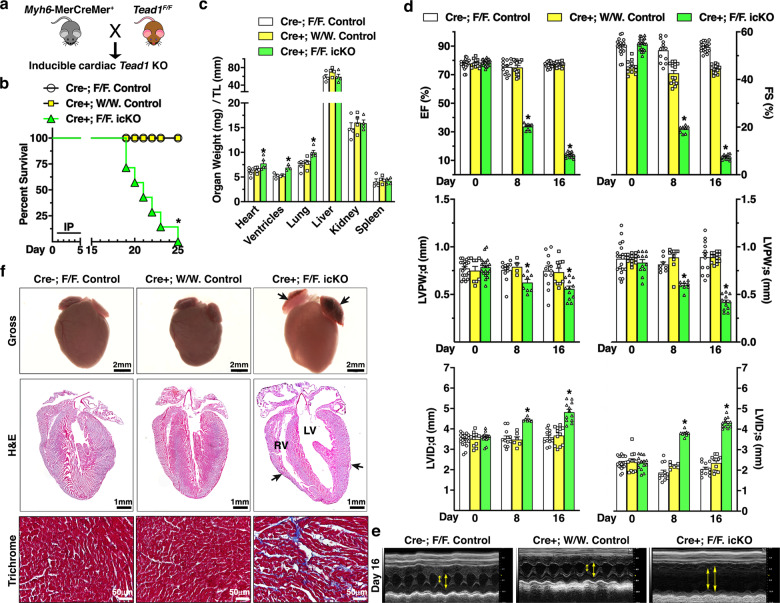
To determine whether the rapid lethality of igKO mice was primarily driven by the deletion of Tead1 in CMs, we generated Tead1 icKO mice by crossing TAM-inducible transgenic mice carrying cardiac-specific gene Myh6 promoter driven MerCreMer with Tead1F/F mice (Online Fig. II a and Fig. 2a). Efficient deletion of Tead1 in adult icKO mouse heart was validated by detecting the recombination of Tead1 allele and by IF staining (Online Fig. II b and c). Strikingly, icKO Tead1 mice showed essentially the same rapidly progressive, lethal DCM phenotype as igKO Tead1 mice (Fig. 2b–f). Together, these data suggest that TEAD1 plays an essential role in maintaining cardiac homeostasis and that ablation of Tead1 in CMs in adult mice leads to a rapid lethality, likely resulting from DCM.
Fig. 2. Mice deficient of Tead1 in postmitotic CMs exhibit rapid lethality due to acute-onset DCM, phenocopying that of Tead1 igKO mice.
a The breeding scheme to generate adult inducible cardiac-specific Tead1 KO mice (icKO) by crossing Myh6-MerCreMer+ mice with Tead1 flox mice. b Survival curve of adult control (Myh6-MerCreMer-; Tead1F/F, N = 15 and Myh6-MerCreMer+; Tead1W/W, N = 18) and Tead1 icKO (Myh6-MerCreMer+; Tead1F/F, N = 14) mice after 5 daily IP injections with tamoxifen. *p < 0.01. Kaplan–Meier analysis. c Control and Tead1 icKO mice were harvested at 18 days following the first tamoxifen injection. The organ weights then were normalized by the tibial length (TL). *p < 0.05 vs. control groups. d Echocardiographic quantifications of cardiac function of control and Tead1 icKO mice by assessing LV EF and FS (upper panel), LVPW;d and LVPW;s (middle panel), LVID;d and LVID;s (bottom panel). *p < 0.05 vs. control groups. e Representative echocardiographic M-mode images are shown from the mice 16 days following the first tamoxifen injection. Double-head arrows mark LV systolic and diastolic internal diameters, respectively. f 18 days following the first tamoxifen injection, hearts from control and Tead1 icKO mice were harvested for gross morphology (upper panel), sectioned for H&E staining (middle panel) and trichrome staining (bottom panel). Similar to Tead1 igKO mouse hearts, deletion of Tead1 in postmitotic CMs leads to thrombus accumulation in atria (arrows in upper panel), dramatic dilation of both LV and RV chambers and thinning ventricular wall (arrows in middle panel).
Deletion of Tead1 in postmitotic CMs of adult mice induces necroptosis and inflammation
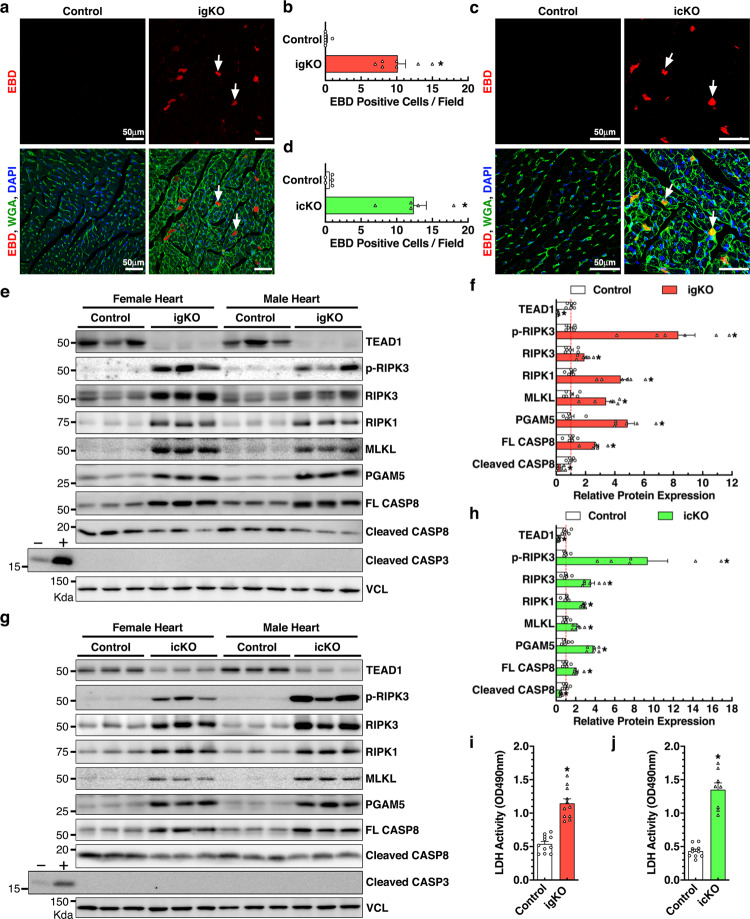
We next explored the cellular mechanism underlying the acute-onset DCM phenotype observed in Tead1-deficient mice. Since CM death plays a critical role in DCM, we first performed TUNEL assays to identify apoptotic CMs. Our data showed almost undetectable CM apoptosis in control, Tead1 igKO and icKO hearts (Online Fig. III a and b). We then sought to evaluate necroptosis, another prevalent CM cell death [6]. Evans blue dye (EBD) assay revealed barely observable signal in control mouse hearts, whereas massive EBD-positive CMs in both Tead1 igKO and icKO hearts (Fig. 3a–d).
Fig. 3. Deletion of Tead1 in postmitotic CMs activates necroptosis.
a–d 17 days after the first tamoxifen injection, Tead1 igKO (a) or icKO (c) mice together with their respective control mice were intraperitoneally injected with one dose of Evans blue dye (EBD; 100 mg/kg) overnight prior to sacrifice. Hearts were then cryosectioned for staining with wheat germ agglutinin (WGA, green) to identify cell membranes and EBD-positive cells (red, arrows). All sections were counter-stained with DAPI to visualize cell nuclei (blue). Average EBD-positive cells per 3 representative sections per mouse (5–7 mice each group) are plotted in panel “b” and “d” for Tead1 igKO and icKO hearts, respectively. *p < 0.01. CMs positive for EBD, indicating loss of cell membrane integrity and viability, are significantly increased in Tead1 igKO and icKO hearts. e–h 18 days following the first tamoxifen injection, hearts from Tead1 igKO (e) or icKO (g) mice together with their respective control mice were harvested for Western blotting as indicated. Vinculin (VCL) serves as the loading control. The band intensity was quantified and plotted as shown in “f” and “h” for Tead1 igKO and icKO groups, respectively. Protein lysates extracted from mouse embryonic fibroblasts treated with (+) or without (–) staurosporine serve as the control for apoptosis as indicated by cleaved caspase 3. 18 days after the first tamoxifen injection, serum was harvested from both control and Tead1 igKO (i) or icKO (j) mice and subjected to lactate dehydrogenase (LDH) activity assay.
Necroptosis is characterized by the inactivation of CASP8 and the activation of the RIPK1/3 necroptotic pathway [4]. Western blot revealed a significant increase in full-length CASP8 and a concomitant decrease in the cleaved (active) CASP8 in both igKO and icKO hearts. Meanwhile, we observed drastically elevated expression of necroptotic pathway components such as RIPK1, RIPK3, MLKL and PGAM5, and activation of phosphorylated RIPK3 [25] (Fig. 3e–h). Consistent with TUNEL staining, Western blotting failed to detect discernible changes in the cleavage of CASP3, a master effector of apoptosis, in Tead1-deficient hearts (Fig. 3e and g). Further lactate dehydrogenase (LDH) assays demonstrated a significant elevation of LDH release in serum from both Tead1 igKO (Fig. 3i) and icKO mice (Fig. 3j). We next isolated CMs from adult Myh6-MerCreMer+; Tead1F/F; mTmG+/− mouse hearts and treated them with 4-hydroxytamoxifen (4-OHT) to delete Tead1 in vitro [26] (Online Fig. IV a and b). Consistent with in vivo findings (Fig. 3), CMs treated with 4-OHT displayed increased cellular damage as indicated by Propidium Iodide (PI) staining (Online Fig. IV c and d) and LDH activity (Online Fig. IV e). Together, these in vivo and in vitro data suggest that Tead1 deficiency induces CM necroptosis.
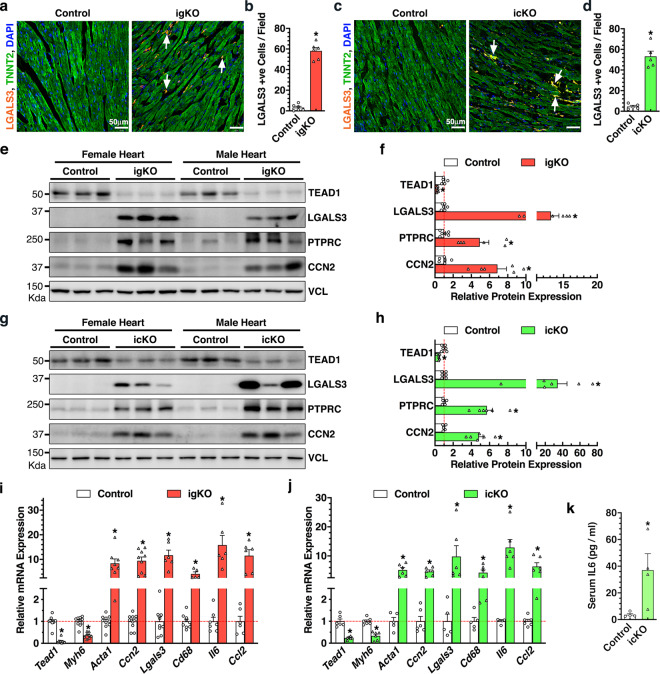
Necrotic cell death evokes inflammatory responses due to the release of intracellular molecules into the surrounding environment [4, 27]. IF staining identified a markedly increase in cells positive for LGALS3 (also known as Mac-2), a marker for the activated macrophages, in both Tead1 igKO (Fig. 4a and b) and icKO hearts (Fig. 4c and d). Western blotting further revealed dramatic elevation of LGALS3 and PTPRC (also known as CD45, a marker for leukocytes, particularly T cells) in Tead1 KO hearts accompanying with upregulated expression of CCN2 (also known as CTGF), a marker of failing fibrotic hearts (Fig. 4e–h). Consistently, qRT-PCR data confirmed that both Tead1 igKO and icKO hearts underwent inflammation as evidenced by increased expression of Lgals3, Cd68, and inflammatory cytokines Il6 and Ccl2 in Tead1 KO hearts, in parallel with downregulated Myh6 and upregulated expression of Acta1 and Ccn2 that indicate heart failure and remodeling (Fig. 4i and j). ELISA assays further demonstrated a significant increased level of IL6 in the serum of icKO mice (Fig. 4k). Taken together, these data suggest that ablation of Tead1 in adult mouse heart induces an inflammatory response, likely triggered by CM necroptosis.
Fig. 4. Deletion of Tead1 in postmitotic CMs in adult mice induces inflammation.
a–d 18 days after the first tamoxifen injection, hearts from Tead1 igKO (a) or icKO (c) mice together with their respective control mice were harvested and sectioned for IF staining of macrophage marker LGALS3 (red, arrows) and CM marker TNNT2 (green). Cell nuclei were counter-stained with DAPI (blue). Average LGALS3-positive cells per field from 3 representative sections per mouse (5-6 mice each group) are plotted in panel “b” and “d” for Tead1 igKO and icKO hearts, respectively. *p < 0.01. e–h 18 days after the first tamoxifen injection, hearts from Tead1 igKO (e) or icKO (g) mice together with their respective control mice were harvested for Western blotting as indicated. Vinculin (VCL) serves as the loading control. The band intensity was quantified and plotted as shown in “f” and “h” for Tead1 igKO and icKO groups, respectively. Same to “e” or “g” except total RNA was extracted from the Tead1 igKO (i) and icKO (j) hearts for qRT-PCR to assess gene expression at the mRNA level. Gene expression in control hearts was set to 1 (red dashed line). *p < 0.05. N = 6–10. k ELISA assays were performed to measure IL6 concentration in the serum of icKO Tead1 or control mice at day 18 post the first tamoxifen injection. N = 4 each group. *p < 0.05.
Inhibition of necroptosis mitigates Tead1 KO-induced DCM
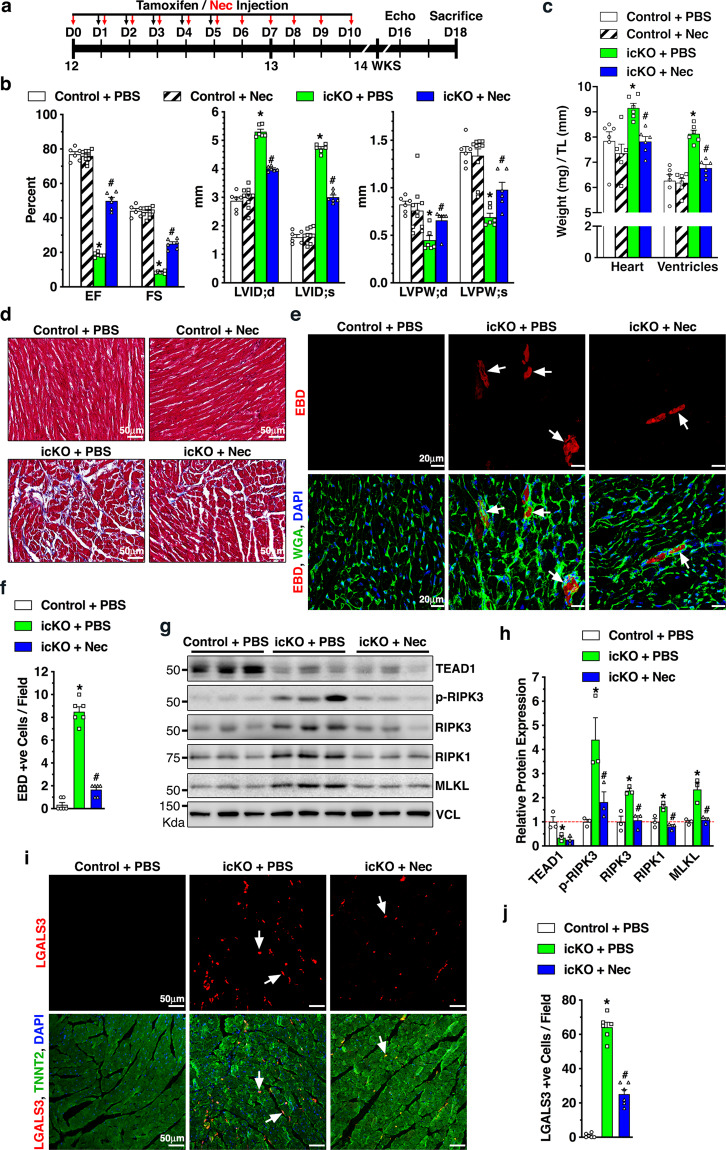
To test whether CM necroptosis is responsible for the DCM phenotype induced by Tead1 deficiency, Myh6-MerCreMer+;Tead1W/W (control) and icKO mice were treated with necrostatin-1 (Nec), a potent agent which prevents necroptotic cell death [28] (Fig. 5a). Mice treated with vehicle (PBS) served as additional controls. Nec treatment did not affect cardiac function in control mice, while significantly attenuated LV chamber dilatation and wall thinning and improved cardiac contractility in icKO mice (Fig. 5b). Consistently, Nec treatment significantly decreased the heart and ventricle weight to tibia length ratio in icKO mice (Fig. 5c). Trichrome staining showed much less collagen deposition in the Nec-treated icKO hearts (Fig. 5d). Furthermore, EBD staining and Western blotting demonstrated that Nec treatment significantly decreased the numbers of EBD-positive CMs (Fig. 5e and f) and attenuated Tead1 ablation-induced necroptotic pathway activation in Tead1 icKO heart (Fig. 5g and h). Additionally, Nec treatment significantly attenuated macrophage infiltration into the heart as evidenced by the fewer LGALS3-positive cells in icKO mice (Fig. 5i and j). Taken together, these results suggest that Tead1 deficiency causes DCM at least in part by activating CM necroptosis.
Fig. 5. Inhibition of necroptosis by Necrostatin-1 (Nec) treatment mitigates Tead1 KO-induced DCM.
a Schematic illustration of Nec treatment (11 times, day 0–10, indicated by red arrows) in Tead1 icKO mice that were generated by 5 daily tamoxifen injections of 12 weeks old mice (Myh6-MerCreMer+; Tead1F/F, day 1–5, indicated by dark arrows). The Tead1 icKO mice treated with the vehicle PBS served as control (icKO + PBS). Myh6-MerCreMer+; Tead1W/W mice treated with PBS (Control + PBS) or Nec (Control + Nec) served as additional controls. b Echocardiographic measurement of cardiac function of 4 groups of mice at day 16 following the first tamoxifen injection. c At day 18 following the first tamoxifen injection, mice were sacrificed and the ratio between heart or ventricle weight and tibial length was calculated. d Similarly, hearts were harvested for trichrome staining to assess extracellular matrix deposition. e 18 h prior to harvest, EBD was injected to mice to assess necroptosis. Heart sections were stained with WGA (green) and necrotic cells were visualized by EBD (red, arrows). Cell nuclei were stained with DAPI (blue). f Quantification of EBD-positive cells per field as shown in “e”. N = 6 mice each group. g Protein lysates were exacted from control or Nec-treated Tead1 icKO mice for Western blotting to assess necroptotic activity in hearts. h Band intensity of Western blots shown in “g” was quantified and plotted as relative expression to control mice treated with PBS (set to 1, red dashed line). i IF staining of LGALS3 (red, arrows) and TNNT2 (green) was performed in heart sections as indicated. DAPI was used to stain nuclei. j Quantification of LGALS3-positive cells per field as shown in “i”. *p < 0.01 vs. “Control + PBS” group; #p < 0.05 vs. “icKO + PBS” group. N = 6.
TEAD1 transcriptionally activates nDNA-encoded mitochondrial genes
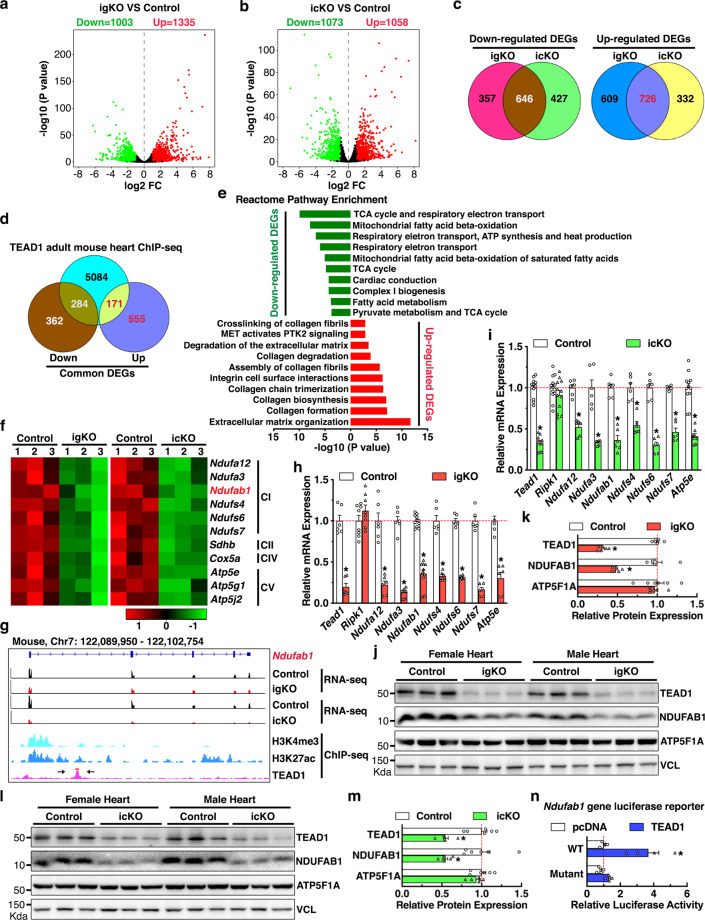
To gain insight into the molecular mechanism underlying TEAD1-mediated function in postmitotic CMs, we sought to identify the TEAD1-dependent genes in CMs by performing RNA-seq using ventricular tissues from both Tead1 igKO and icKO hearts. The analysis revealed 1,003 and 1,335 significantly down- and upregulated genes in Tead1 igKO (Fig. 6a and Online Table II), as well as 1073 and 1058 significantly down- and upregulated genes in icKO hearts (Fig. 6b and Online Table III), respectively, as compared to control hearts. The majority of significantly differentially expressed genes (DEGs, 646 downregulated and 726 upregulated genes) overlapped between Tead1 igKO and icKO hearts (Fig. 6c). To further define the direct targets of TEAD1 in CMs, we analyzed ChIP-seq data of TEAD1 in adult mouse hearts [29, 30]. The results revealed that TEAD1 binds to the promoter region of 5,539 genes in the adult mouse heart (Online Table IV), and 171 and 284 of them were identified to be significantly up- and downregulated in both Tead1 igKO and icKO hearts, respectively (Fig. 6d). Pathway analysis demonstrated that the 171 upregulated genes were mostly associated with pathways related to collagen turnover and extracellular matrix remodeling (Fig. 6e). As shown in Online Fig. V, TEAD1 directly binds to the promoter/enhancer region (defined by H3K4me3 and H3K27ac ChIP-seq data in adult mouse heart from ENCODE) of the 10 upregulated genes involved in the pathway of “extracellular matrix organization”, suggesting upregulation of these matrix genes are direct consequences of Tead1 deficiency, instead of secondary effects of heart failure. In contrast, this analysis unexpectedly revealed that nine out of the top ten pathways associated with the 284 downregulated genes are related to mitochondrial function, especially with complex (C) I biogenesis (Fig. 6e). A substantial portion of nDNA-encoded mitochondrial ETC components were significantly downregulated in both Tead1 igKO and icKO hearts (Fig. 6f) and were directly targeted by TEAD1 in active transcriptional regions (Fig. 6g and Online Fig. VI a–d). Furthermore, six of them were also found to be direct targets of TEAD1 in human HepG2 cells as revealed by ChIP-seq data from ENCODE (Online Fig. VI e). These results suggest TEAD1-mediated direct regulation of nDNA-encoded mitochondrial ETC component genes is a conserved regulatory mechanism between different cell types in mouse and human.
Fig. 6. Identification of the bona fide TEAD1 target genes in postmitotic CMs in mice.
Volcano plots demonstrate differentially expressed genes (DEGs) between control and Tead1 igKO (a) or icKO (b) hearts. Genes changed with fold change (FC) ≥ 2 and FDR < 0.05 are considered significant. c Venn diagrams illustrate the overlap of downregulated (left panel, 646 genes) and upregulated DEGs (right panel, 726 genes) between Tead1 igKO and icKO hearts. d Venn diagram depicts the bona fide TEAD1 target genes (455 in total) in adult mouse CMs as revealed by the overlap between TEAD1 binding genes as identified by ChIP-seq and genes commonly downregulated (284 genes) and upregulated (171 genes) in both Tead1 igKO and icKO hearts. e Reactome pathway enrichment analysis of the TEAD1 bona fide target genes in the adult mouse heart. f Heat maps from RNA-seq data show 11 nDNA-encoded mitochondrial ETC genes that are significantly downregulated in both Tead1 igKO (left panel) and icKO hearts (right panel). g Integrative Genomics Viewer (IGV) tracks of Ndufab1 gene locus to show a representative RNA-seq of Tead1 adult heart KO (igKO and icKO, upper panels) and ChIP-seq of TEAD1, H3K4me3 or H3K27ac modifications (bottom panel) in adult mouse hearts. A pair of arrows points to the TEAD1 binding region in Ndufab1 gene locus with which the DNA fragment was cloned to a luciferase reporter vector. qRT-PCR analysis of expression of TEAD1 target mitochondrial ETC genes in Tead1 igKO (h) and icKO hearts (i) at mRNA level. Gene expression in control hearts was set to 1 (red dashed line). N = 6–12. *p < 0.05. j–m The downregulation of NDUFAB1 at protein level is confirmed in both Tead1 igKO (j) and icKO (l) hearts by Western blot. The band intensity of NDUFAB1 in Western blots of Tead1 igKO and icKO heart samples is quantified and plotted as shown in “k” and “m”, respectively. Protein expression in control hearts is set to 1 (red dashed line). N = 6. *p < 0.05. n Ndufab1 luciferase reporter genes containing WT or mutation of the TEAD1 DNA binding element as shown in panel “g” were co-transfected with TEAD1 expression plasmid for dual luciferase reporter assays. Co-transfection with pcDNA empty plasmid served as control and the reporter activity from this control group was set to 1 (red dashed line). N = 4–6. *p < 0.05.
Among the TEAD1-dependent mitochondrial complex I genes, NDUFAB1 has been previously shown to be essential for human cell viability in vitro and for mouse CM viability in vivo [10, 31]. Ndufbab1 was thus selected for further investigation. qRT-PCR and Western blotting validated the decreased expression of Ndufab1 gene together with other TEAD1-dependent ETC components in both Tead1 igKO and icKO hearts (Fig. 6h–m). In contrast, Ripk1 gene mRNA was not significantly different between Tead1 KO and control hearts, suggesting that the activation of necroptotic pathway in Tead1- deficient CMs is likely at post transcriptional level (Fig. 6h–i). A canonical TEAD1 binding motif were further identified within the TEAD1 peak region in mouse Ndufab1 gene (Fig. 6g) and a 739-bp DNA fragment surrounding the binding site was amplified and cloned into a luciferase reporter vector. Dual luciferase reporter assays revealed that TEAD1 significantly transactivates the WT Ndufab1 gene reporter while mutation of the binding element abolishes the effect (Fig. 6n), suggesting the TEAD1 binding to Ndufab1 gene is required for its transcriptional activation. Taken together, these results suggest an unexpected role of TEAD1 in directly activating nDNA-encoded mitochondrial genes via physically binding to these genes’ promoter/enhancer regions.
To examine the effects of over-expression of Tead1 on mitochondrial gene expression in CMs, we transduced TEAD1 adenovirus into cultured neonatal rat CMs for 3 days. Western blotting analysis showed that over-expression of TEAD1 does not affect endogenous mitochondrial gene expression in neonatal rat CMs (Online Fig. VII), suggesting TEAD1 is not sufficient for the subset of mitochondrial ETC gene expression in cultured CMs.
Loss of Tead1 in CMs leads to mitochondrial dysfunction
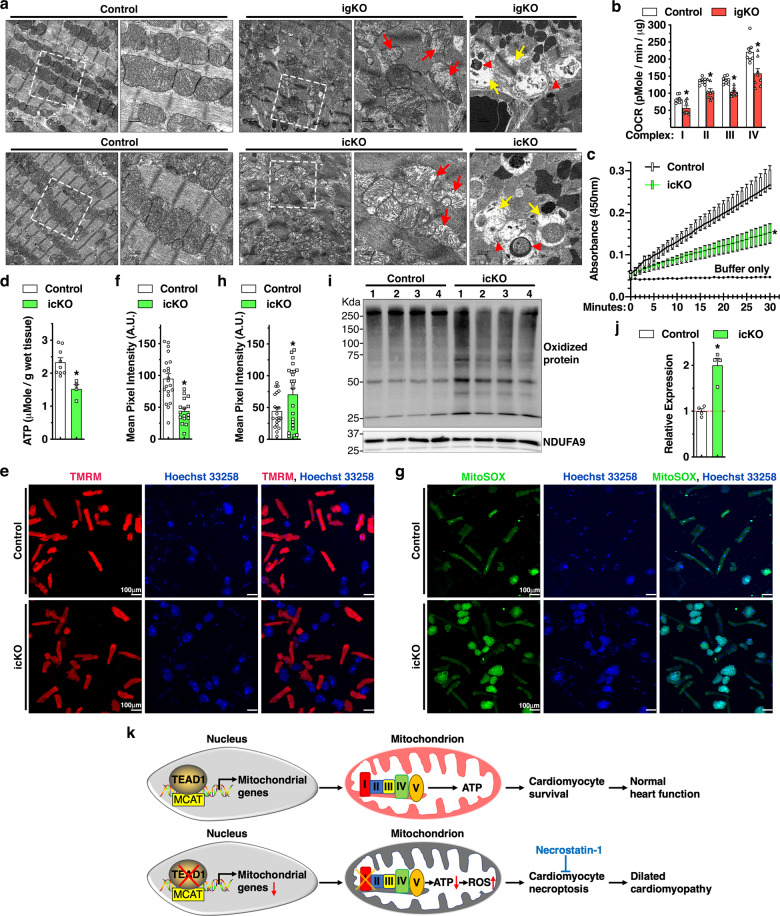
To rule out possible secondary effects of heart failure on mitochondrial morphology and function, we harvested the Tead1 igKO and icKO hearts 6 days following the first tamoxifen injection while the animals were displaying negligible cardiac dysfunction. Electron microscopic examination of the Tead1 KO hearts revealed that mitochondria display a markedly abnormal structure, including mitochondrial cristae degradation and disrupted mitochondrial membrane integrity, apparent loss and degeneration of myofibrils. Furthermore, both Tead1 igKO and icKO hearts display reminiscent of mitochondria and autophagic vacuoles engulfing mitochondria (Fig. 7a). Mitochondrial bioenergetics analysis of isolated mitochondria demonstrated that Tead1 ablation significantly lowers rates of oxygen consumption of mitochondrial ETC complex I-IV (Fig. 7b). Complex I activity assays specifically confirmed that complex I function is significantly impaired in the CMs isolated from Tead1 icKO hearts (Fig. 7c). Importantly, impairment of mitochondrial ETC complex I-IV activity led to decreased ATP production in Tead1 icKO hearts (Fig. 7d). Subsequent TMRM staining demonstrated CMs isolated from Tead1 icKO hearts exhibit decreased mitochondrial membrane potential as evidenced by the significantly weaker TMRM staining compared to controls (Fig. 7e, f). Staining by MitoSOX mitochondrial superoxide indicator further showed that Tead1 deletion significantly promotes mitochondria-produced superoxide content in the live CMs (Fig. 7g, h). As a result, oxidized proteins were significantly increased in Tead1 icKO hearts as revealed by OxyBlot assays (Fig. 7i, j). In neonatal rat CMs, assessment of cell death by PI staining revealed that H2O2 treatment-induced cell death is enhanced after knocking down Tead1 whereas this effect is abolished in the presence of the mitochondria-targeted antioxidant agent MitoTEMPOL, suggesting that the excessive ROS production in Tead1 deficient CMs is, at least in part, responsible for cell death (Online Fig. VIII). Taken together, these data suggest that deletion of Tead1 in CMs inhibits ETC activity, especially complex I function, reduces ATP production and mitochondrial membrane potential, and increases oxidative stress, thereby promoting CM death.
Fig. 7. Loss of Tead1 in CMs leads to mitochondrial dysfunction.
a Representative transmission electron microscopy images demonstrate mitochondrial morphology in control, Tead1 igKO (upper panel) and icKO (bottom panel) hearts. Red and yellow arrows indicate mitochondria that lost cristae and focal areas with myofibrillar lysis, respectively. Red arrow heads mark reminiscent of mitochondrial membrane or damaged mitochondria induced mitophagic vesicles. The boxed area is magnified on the right. b Mitochondrial bioenergetics assays were performed to measure oxygen consumption rate (OCR) in mitochondria isolated from control and Tead1 igKO hearts to determine ETC complex I-IV respiratory activity. N = 4. *p < 0.05. c Protein lysates were isolated from control and Tead1 icKO hearts. Equal amounts of protein were then used to determine complex I activity following the oxidation of NADH to NAD + and the simultaneous reduction of a dye which leads to increased absorbance at OD = 450 nm, over 30 min. Reaction with buffer only serves as the negative control. N = 4–6. *p < 0.05. d ATP bioluminescent assay was performed to measure the level of intracellular ATP in heart tissues isolated from control and Tead1 icKO mice. N = 4–6. *p < 0.05. e-f CMs isolated at day 6 post the first tamoxifen injection from adult control Myh6-MerCreMer+; Tead1W/W or Myh6-MerCreMer+; Tead1F/F icKO mice. Subsequently cells were subjected to incubate with membrane-permeable, voltage-sensitive fluorescent probe TMRM (e, red) to quantify changes in mitochondrial membrane potential in live cells (f). *p < 0.05. g, h Similar to “e”, except that CMs treated with MitoSOX (green) to measure the production of superoxide by mitochondria. Cell nuclei were counter-stained with Hoechst 33258 (blue). The green fluorescence intensity of individual cells was quantified and plotted in “h”. *p < 0.05. i Using heart protein lysates from control or Tead1 icKO mice, Western blot was performed to determine the oxidized proteins that are marked by DNPH derivatization and probed by an anti-DNP antibody. j Quantification of the band intensity shown in “i” and normalized to respective loading control NDUFA9 (set to 1, red dashed line). N = 4. *p < 0.05. k Schematic diagram summarizing the major findings of this study. Transcription factor TEAD1 directly activates expression of nDNA-encoded mitochondrial genes that are essential for mitochondrial ETC activity, ATP production and cell survival in postmitotic CMs (upper panel). Conversely, Tead1 deletion represses mitochondrial genes especially complex I of ETC, leading to reduced ATP production and elevated oxidative stress. As a result, CMs deficient of Tead1 undergo necroptosis, which ultimately leads to DCM that can be partially rescued by the necroptosis blocker necrostatin-1 treatment (bottom panel).
To examine the expression level of Tead1 in response to cardiac injury, we re-analyzed the whole transcriptome RNA-seq dataset (GSE128034) that was generated from mouse left ventricular remote, border and infarction zone at day 7 post left coronary artery ligation which induces myocardial infarction [32]. This unbiased analysis revealed that, compared to remote zone, Tead1 expression is significantly reduced in the infarction zone, in concomitant with down- or up-regulation of Tead1 target mitochondrial genes Ndufab1 and Ndufa3 or matrix gene Col1a1, respectively (Online Fig. IX). These data suggest that downregulation of Tead1 may play a role in cardiac pathogenesis in response to cardiac injury.
In summary, our study demonstrates an unexpected role of the nuclear transcription factor TEAD1 in regulating expression of nDNA-encoded mitochondrial genes that are essential for mitochondrial ETC activity, ATP synthesis and cell survival in postmitotic CMs (Fig. 7k, upper panel). Conversely, Tead1 deletion suppresses mitochondrial genes especially complex I of ETC, leading to reduced ATP production while elevated oxidative stress. As a result, CMs deficient of Tead1 undergo necroptosis and ultimately lead to DCM which can be partially rescued by the treatment of RIPK1 blocker Nec (Fig. 7k, bottom panel).
Discussion
As a prime nuclear mediator of the Hippo pathway, we and others have previously shown that TEAD1 and its co-factor YAP1 are critical for CM proliferation during development in mice [19, 20, 33, 34]. Single CM-specific Yap1 or Yap1/Wwtr1 (a paralogue of Yap1) double KO mice develop cardiac defects and prematurely die primarily due to impaired proliferation and enhanced apoptosis of CMs [35, 36]. In contrast to these prior studies, we found that deletion of Tead1 in adult postmitotic CMs led to rapid development of lethal DCM due to necroptosis but not apoptosis. The underlying mechanism by which Tead1 deficiency kills postmitotic CMs specifically by necroptosis remains to be determined. Our data suggest that it is likely the depletion of intracellular ATP concentration induced by Tead1 ablation in CMs switches on the necroptosis pathway, as previously described in human T cells [37]. In the future it would be interesting to test whether knockout of Ripk3 or Mlkl can mitigate Tead1-deficiency induced DCM phenotype by protecting CMs specifically against necroptosis. Furthermore, although Tead1 is highly and widely expressed in adult mice [38], the igKO Tead1 mice specifically exhibited acute-onset DCM phenotype, most likely due to the high energy demanding of heart for the constant beating. We also noticed that icKO Tead1 mice died earlier than igKO Tead1 mice after tamoxifen injection (Figs. 1 and 2). We speculate that this observation largely attributed to the combined effects of the acute-onset of DCM phenotype driven by Tead1-deficiency and the tamoxifen-induced cardiac toxicity that occurs specifically in Mhy6-MerCreMer mice as previously reported [39].
Recently, Liu et al reported that inducible adult CM-specific Tead1 ablation led to lethal acute-onset DCM, the same phenotype we observed in the current study. However, they proposed that this occurred through a distinct mechanism by which TEAD1 directly targets SERCA2a (encoded by Atp2a2 gene) that is involved in CM excitation-contraction coupling [21]. Atp2a2 mRNA is indeed downregulated in both Tead1 igKO and icKO hearts in our RNA-seq data (Online Table II and III), but our ChIP-seq data did not reveal TEAD1 occupancy within Atp2a2 gene locus in adult mouse heart (Online Table IV). It is likely that TEAD1 binds to Atp2a2 gene locus beyond of 3-kb surrounding transcription starting site that we used for ChIP-seq analysis (Fig. 6). Therefore, it is possible that the combination of downregulation of nuclear DNA-encoded mitochondrial genes that we identified in this study, together with Atp2a2 gene, contributes to the acute-onset DCM phenotype of the Tead1 KO hearts. Notably, there is a discrepancy regarding TEAD1 protein expression in human DCM hearts. Liu et al. reported that TEAD1 tends to be decreased (~20%) in human DCM hearts [21]. Hou et al., however, reported that TEAD1 expression is significantly increased in human DCM hearts [40]. Since both studies used small numbers of human heart specimen, a larger cohort of human DCM heart samples is required to clarify TEAD1 expression in human heart diseases.
It has been reported that YAP1-TEAD1 signaling regulate mitochondrial biogenesis in endothelial cells through regulating Ppargc1a gene [41] and in Drosophila through activating opa1-like (Opa1) and mitochondria assembly regulatory factor (Marf) [42]. Furthermore, TEAD4 has been shown to localize in mitochondria, bind to mtDNA to regulate expression of mtDNA-coded ETC components, thereby regulating oxidative energy metabolism and blastocyst maturation [43]. Together with our current study, these findings collectively suggest that Hippo-YAP-TEAD pathway is important for cell proliferation, maturation or viability by promoting mitochondrial function through directly regulating nDNA- or mtDNA-encoded mitochondrial gene expression. It appears that regulation of mtDNA transcription is unique to TEAD4 due to its ability of localization in the mitochondria [44] while TEAD1 is exclusively localized in CM nuclei (Online Figure I e and Online Figure II c). We speculate that TEAD4 likely contains a unique mitochondrial targeting signal while TEAD1 does not.
Our omics approach identified 6 out of 38 nDNA-coded complex I [45], together with 5 other ETC complex genes as bona fide TEAD1 targets (Fig. 6). Importantly, CM-specific deletion of Ndufab1 in mice caused defective ATP production and elevated ROS levels, leading to progressive DCM and eventual heart failure and lethality [10], which resembles the phenotype of Tead1 icKO mice to some extent although the phenotype observed in CM-specific Ndufab1 KO mice is relatively mild. Our results showed that Ndufab1 expression was significantly reduced in Tead1 deficient hearts and is a direct target of TEAD1 (Fig. 6). We speculate that downregulation of Ndufab1, together with other complex I subunits and additional TEAD1 target genes of ETC complexes (Fig. 6f), contribute to the severe DCM phenotype of Tead1 mutant mice. Future studies are needed to test whether restoration of Ndufab1 gene expression in Tead1 deleted CMs can, at least in part, rescue the mitochondrial dysfunction and DCM phenotype. Interestingly, although Tead1 acts as an activator for multiple nDNA-encoded mitochondrial ETC gene (Fig. 6) or as a repressor for some matrix gene expression in adult mouse CMs (Online Figure V), we found over-expression of Tead1 is not sufficient to induce the expression of mitochondrial ETC genes in cultured neonatal rat CMs (Online Figure VII). Notably, a previous study has demonstrated that TEAD1 over-expression in the mouse heart leads to an age-dependent cardiac dysfunction and heart failure [46]. Together with our current loss-of-function study, we propose that an exquisite range of Tead1 expression in heart is critical for cardiac homeostasis.
Inhibition of mitochondrial complex I and III results in robust formation of mitochondrial ROS and loss of mitochondrial membrane potential [47, 48]. Emerging evidence suggest that over-production of ROS induced by mitochondrial ETC defect leads to mitochondrial permeability transition, which results in matrix swelling, outer membrane rupture and ultimately CM death [49]. Preventing ROS formation has been shown to attenuate myocardial ischemia/reperfusion injury in mice [50]. In this study, we found ablation of Tead1 in CMs significantly increased superoxide content (Fig. 7g-h) due to mitochondrial dysfunction. The excessive ROS produced by the dysfunctional mitochondria could be a cause of necroptosis by activating RIPK1/3-MLKL-PGAM5 necroptotic pathway. It will be interesting to determine whether blocking ROS in vivo can mitigate the DCM phenotype induced by deletion of Tead1 in adult CMs, similar to MitoTEMPOL treatment in vitro (Online Figure VIII) and blocking necroptosis by Nec treatment in vivo (Fig. 5) as we described in this study.
In summary, our study identifies TEAD1 as an essential regulator of mitochondrial gene expression that coordinates mitochondrial activity and survival of postmitotic CMs.
Supplementary information
Acknowledgements
We thank Drs. B. Paul Herring and David Fulton for a critical reading of the manuscript. The authors appreciate Drs. John Johnson, Bobby Thomas at Augusta University, Xiongwen Chen at Temple University and Yang Kevin Xiang at University of California, Davis for their inspiring discussion.
Funding
The work is supported by grants from the National Heart, Lung, and Blood Institute, NIH (R01HL149995 to JZ; R01HL132182 to WC). JZ is a recipient of Established Investigator Award (17EIA33460468) and Transformational Project Award (19TPA34910181) from American Heart Association. KD is supported by a postdoctoral fellowship (19POST34450071) from American Heart Association. IO is supported by a K99 award (K99HL153896) from the National Heart, Lung, and Blood Institute, NIH.
Author contributions
JZ and WZ conceived and supervised the project. JinL and TW designed and performed experiments. KD analyzed RNA-seq and ChIP-seq data. JZ and KD wrote the manuscript. XH, HZ, JS, ZF, GH, WM, JieL, WeW, LW, BA, JX, IO, ZZ, WW, QD, WP, MX, WC and HS performed experiments and/or edited the manuscript.
Data availability
The RNA-seq data generated in this study have been deposited in the Sequences Read Archive at the NCBI under SRA accession numbers PRJNA575531. The TEAD1 adult mouse heart ChIP-seq data has been deposited in NCBI GEO database under the accession number GSE124008. Reagents described in the study will be available upon request.
Compliance with ethical standards
Ethics declarations
Not applicable. No human subjects were involved in this study.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Oberst
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jinhua Liu, Tong Wen
Contributor Information
Wei Zhang, Email: zhangweiliuxin@163.com.
Jiliang Zhou, Email: jizhou@augusta.edu.
Supplementary information
The online version of this article (10.1038/s41418-020-00732-5) contains supplementary material, which is available to authorized users.
References
- 1.McKenna WJ, Maron BJ, Thiene G. Classification, epidemiology, and global burden of cardiomyopathies. Circ Res. 2017;121:722–30. doi: 10.1161/CIRCRESAHA.117.309711. [DOI] [PubMed] [Google Scholar]
- 2.Zhu H, Sun A. Programmed necrosis in heart disease: molecular mechanisms and clinical implications. J Mol Cell Cardiol. 2018;116:125–34. doi: 10.1016/j.yjmcc.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 3.Orogo AM, Gustafsson AB. Cell death in the myocardium: my heart won’t go on. IUBMB Life. 2013;65:651–6. doi: 10.1002/iub.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinlich R, Oberst A, Beere HM, Green DR. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol. 2017;18:127–36. doi: 10.1038/nrm.2016.149. [DOI] [PubMed] [Google Scholar]
- 5.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–14. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 6.Del ReDP, Amgalan D, Linkermann A, Liu Q, Kitsis RN. Fundamental mechanisms of regulated cell death and implications for heart disease. Physiological Rev. 2019;99:1765–817. doi: 10.1152/physrev.00022.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyers DE, Basha HI, Koenig MK. Mitochondrial cardiomyopathy: pathophysiology, diagnosis, and management. Tex Heart Inst J. 2013;40:385–94. [PMC free article] [PubMed] [Google Scholar]
- 8.Goldenthal MJ. Mitochondrial involvement in myocyte death and heart failure. Heart Fail Rev. 2016;21:137–55. doi: 10.1007/s10741-016-9531-1. [DOI] [PubMed] [Google Scholar]
- 9.Adameova A, Goncalvesova E, Szobi A, Dhalla NS. Necroptotic cell death in failing heart: relevance and proposed mechanisms. Heart Fail Rev. 2016;21:213–21. doi: 10.1007/s10741-016-9537-8. [DOI] [PubMed] [Google Scholar]
- 10.Hou T, Zhang R, Jian C, Ding W, Wang Y, Ling S, et al. NDUFAB1 confers cardio-protection by enhancing mitochondrial bioenergetics through coordination of respiratory complex and supercomplex assembly. Cell Res. 2019;29:754–66. doi: 10.1038/s41422-019-0208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu P, Hu S, Jin Q, Li D, Tian F, Toan S, et al. Ripk3 promotes ER stress-induced necroptosis in cardiac IR injury: a mechanism involving calcium overload/XO/ROS/mPTP pathway. Redox Biol. 2018;16:157–68. doi: 10.1016/j.redox.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Hattab AW, Scaglia F. Mitochondrial cardiomyopathies. Front Cardiovasc Med. 2016;3:25. doi: 10.3389/fcvm.2016.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan D. The hippo signaling pathway in development and cancer. Dev cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Liu S, Heallen T, Martin JF. The Hippo pathway in the heart: pivotal roles in development, disease, and regeneration. Nat Rev Cardiol. 2018;15:672–84. doi: 10.1038/s41569-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 15.Wackerhage H, Del ReDP, Judson RN, Sudol M, Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci Signal. 2014;7:re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida T. MCAT elements and the TEF-1 family of transcription factors in muscle development and disease. Arterioscler Thromb Vasc Biol. 2008;28:8–17. doi: 10.1161/ATVBAHA.107.155788. [DOI] [PubMed] [Google Scholar]
- 17.Lin KC, Park HW, Guan KL. Regulation of the Hippo pathway transcription factor TEAD. Trends Biochem Sci. 2017;42:862–72. doi: 10.1016/j.tibs.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen T, Hu S, Yin Q, Yu L, Hu G, Liu J, Zhang W, et al. Characterization of mice carrying a conditional TEAD1 allele. Genesis. 2017;55:e23085. doi: 10.1002/dvg.23085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wen T, Liu J, He X, Dong K, Hu G, Yu L, et al. Transcription factor TEAD1 is essential for vascular development by promoting vascular smooth muscle differentiation. Cell Death Differ. 2019;26:2790–806. doi: 10.1038/s41418-019-0335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu R, Jagannathan R, Li F, Lee J, Balasubramanyam N, Kim BS, et al. Tead1 is required for perinatal cardiomyocyte proliferation. PLoS One. 2019;14:e0212017. doi: 10.1371/journal.pone.0212017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu R, Lee J, Kim BS, Wang Q, Buxton SK, Balasubramanyam N, et al. Tead1 is required for maintaining adult cardiomyocyte function, and its loss results in lethal dilated cardiomyopathy. JCI Insight. 2017;2:e93343. doi: 10.1172/jci.insight.93343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Jagannathan R, Sun L, Li F, Yang P, Lee J, et al. Tead1 is essential for mitochondrial function in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2020;319:H89–99. doi: 10.1152/ajpheart.00732.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osman I, He X, Liu J, Dong K, Wen T, Zhang F, et al. TEAD1 (TEA Domain Transcription Factor 1) promotes smooth muscle cell proliferation through upregulating SLC1A5 (Solute Carrier Family 1 Member 5)-mediated glutamine uptake. Circ Res. 2019;124:1309–22. doi: 10.1161/CIRCRESAHA.118.314187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–5. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 25.Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–23. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asp ML, Martindale JJ, Metzger JM. Direct, differential effects of tamoxifen, 4-hydroxytamoxifen, and raloxifene on cardiac myocyte contractility and calcium handling. PLoS One. 2013;8:e78768. doi: 10.1371/journal.pone.0078768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–89. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CC, Davidson SM, Lim SY, Simpkin JC, Hothersall JS, Yellon DM. Necrostatin: a potentially novel cardioprotective agent? Cardiovasc Drugs Ther. 2007;21:227–33. doi: 10.1007/s10557-007-6035-1. [DOI] [PubMed] [Google Scholar]
- 29.Zhou P, Gu F, Zhang L, Akerberg BN, Ma Q, Li K, et al. Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. eLife. 2017;6:e22039. doi: 10.7554/eLife.22039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akerberg BN, Gu F, VanDusen NJ, Zhang X, Dong R, Li K, et al. A reference map of murine cardiac transcription factor chromatin occupancy identifies dynamic and conserved enhancers. Nat Commun. 2019;10:4907. doi: 10.1038/s41467-019-12812-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–6. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 32.van Duijvenboden K, de Bakker DEM, Man JCK, Janssen R, Gunthel M, Hill MC, et al. Conserved NPPB+ border zone switches from MEF2- to AP-1-driven gene program. Circulation. 2019;140:864–79. doi: 10.1161/CIRCULATIONAHA.118.038944. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Hu G, Liu F, Wang X, Wu M, Schwarz JJ, et al. Deletion of yes-associated protein (YAP) specifically in cardiac and vascular smooth muscle cells reveals a crucial role for YAP in mouse cardiovascular development. Circ Res. 2014;114:957–65. doi: 10.1161/CIRCRESAHA.114.303411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J. An emerging role for Hippo-YAP signaling in cardiovascular development. J Biomed Res. 2014;28:251–4. doi: 10.7555/JBR.28.20140020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA. 2013;110:13839–44. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Re DP, Yang Y, Nakano N, Cho J, Zhai P, Yamamoto T, et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J Biol Chem. 2013;288:3977–88. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185:1481–6. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, et al. A comprehensive mouse transcriptomic BodyMap across 17 tissues by RNA-seq. Sci Rep. 2017;7:4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koitabashi N, Bedja D, Zaiman AL, Pinto YM, Zhang M, Gabrielson KL, et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ Res. 2009;105:12–15. doi: 10.1161/CIRCRESAHA.109.198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou N, Wen Y, Yuan X, Xu H, Wang X, Li F, et al. Activation of Yap1/Taz signaling in ischemic heart disease and dilated cardiomyopathy. Exp Mol Pathol. 2017;103:267–75. doi: 10.1016/j.yexmp.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mammoto A, Muyleart M, Kadlec A, Gutterman D, Mammoto T. YAP1-TEAD1 signaling controls angiogenesis and mitochondrial biogenesis through PGC1alpha. Microvasc Res. 2018;119:73–83. doi: 10.1016/j.mvr.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagaraj R, Gururaja-Rao S, Jones KT, Slattery M, Negre N, Braas D, et al. Control of mitochondrial structure and function by the Yorkie/YAP oncogenic pathway. Genes Dev. 2012;26:2027–37. doi: 10.1101/gad.183061.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar RP, Ray S, Home P, Saha B, Bhattacharya B, Wilkins HM, et al. Regulation of energy metabolism during early mammalian development: TEAD4 controls mitochondrial transcription. Development. 2018;145:dev162644. doi: 10.1242/dev.162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaneko KJ, DePamphilis ML. TEAD4 establishes the energy homeostasis essential for blastocoel formation. Development. 2013;140:3680–90. doi: 10.1242/dev.093799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–7. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 46.Tsika RW, Ma L, Kehat I, Schramm C, Simmer G, Morgan B, et al. TEAD-1 overexpression in the mouse heart promotes an age-dependent heart dysfunction. J Biol Chem. 2010;285:13721–35. doi: 10.1074/jbc.M109.063057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 48.Chen YR, Zweier JL. Cardiac mitochondria and reactive oxygen species generation. Circ Res. 2014;114:524–37. doi: 10.1161/CIRCRESAHA.114.300559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou B, Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. J Clin Investig. 2018;128:3716–26. doi: 10.1172/JCI120849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dhalla NS, Elmoselhi AB, Hata T, Makino N. Status of myocardial antioxidants in ischemia-reperfusion injury. Cardiovascular Res. 2000;47:446–56. doi: 10.1016/s0008-6363(00)00078-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study have been deposited in the Sequences Read Archive at the NCBI under SRA accession numbers PRJNA575531. The TEAD1 adult mouse heart ChIP-seq data has been deposited in NCBI GEO database under the accession number GSE124008. Reagents described in the study will be available upon request.