Abstract
Recently, we conducted genome-wide association studies of type 2 diabetes (T2D) in a Japanese population, which identified 20 novel T2D loci that were not associated with T2D in Europeans. Moreover, nine novel missense risk variants, such as those of PAX4, were not rare in the Japanese population, but rare in Europeans. We report in silico structural analysis of ethnic-specific variants of PAX4, which suggests the pathogenic effect of these variants.
Subject terms: Genome-wide association studies, Sequence annotation
Large-scale meta-analyses of genome-wide association studies (GWAS) of type 2 diabetes (T2D) have been performed recently1–4. In particular, Mahajan et al.3 reported T2D GWAS meta-analysis data on ~900,000 individuals of European ancestry, increasing the association signals in over 240 loci associated with T2D. Recently, we conducted an extensive T2D GWAS meta-analysis in a Japanese population and identified 28 new loci4, highlighting the value of genetic research in ethnically diverse populations. As an example of clinical differences between ethnic populations, East Asians are more likely to develop T2D at a lower body mass index than Europeans5, who have been reported to show a higher insulin response and lower insulin sensitivity.
We demonstrated that the biological pathway of the maturity-onset diabetes of the young (MODY), monogenic diabetes characterized by reduced β-cell function, showed the most significant association with T2D in both Japanese and European populations4, by performing pathway analysis of GWAS summary data in the two populations1,4 using Pascal software. Here we focused on protein-coding genes involved in the MODY pathway mapping nearest to lead variants at T2D loci, as previously reported6, and investigated ethnic differences in the associations of these loci with T2D between Japanese and European populations3,4. As suggested by a previous study4, most of these lead variants at T2D loci in either the Japanese or European population showed at least nominally significant (P < 0.05) associations in the alternative population, even if not genome-wide significant, except for variants that were rare or monomorphic in one population such as NKX6-1 rs201597274, HNF1A rs187150787, and PAX4 rs2233580 in the Japanese population and HNF1A rs56348580 in the European population. Interestingly, there were several loci such as GCK where lead variants in these populations were independent of each other (Supplementary Tables 1 and 2). As an example of a locus specific to East Asians, a previously unreported missense T2D variant of PAX4 NM_006193:c.574C>A:p.(Arg192Ser) reached genome-wide significance4. This variant was located at the same amino acid as another established independent T2D variant NM_006193:c.575G>A:p.(Arg192His) (Table 2 in ref. 4). These two PAX4 variants were not in linkage disequilibrium (JPT: r2 < 0.01). PAX4 encodes a transcription factor that is important for β-cell development, and rare mutations of this gene are suggested to cause a subtype of MODY (OMIM #612225). Arg192 of PAX4 is located in the homeodomain, which is a DNA-binding domain conserved in a large family of transcription factors. Barrera et al.7 reported that PAX4 variants Arg192His and Arg192Ser caused alteration in DNA-binding specificity in experiments using protein-binding microarrays. In addition, it was reported that the PAX4 p.Arg192His mutant demonstrated decreased repression of the transcription of target genes involved in the maintenance of β-cell function compared with wild-type PAX4 in an in vitro study8. We annotated PAX4 variants Arg192His and Arg192Ser with prediction tools via wANNOVAR (http://wannovar.wglab.org/), which suggested that these variants would be deleterious (Supplementary Table 3). Furthermore, we conducted an in silico structural analysis of the pathogenic effect of these PAX4 variants, which revealed a decrease of the PAX4 homeodomain stability and reduced DNA binding by this domain in both variants (Figs. 1, 2 and Supplementary Note).
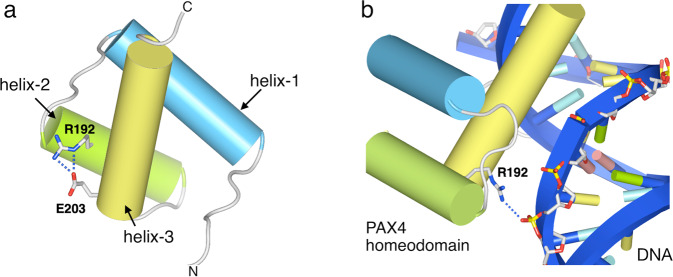
Fig. 1. Structural model of the homeodomain in human PAX4.
a The human PAX4 homeodomain is represented as cylinders, with the Arg192 and Glu203 residues being shown as sticks. The PAX4 homeodomain consists of three helices: helix-1 (light-blue), helix-2 (green), and helix-3 (yellow). Arg192 located on helix-2 is predicted to form salt-bridges with Glu203 located on helix-3. b Binding of the human PAX4 homeodomain to double-stranded DNA. DNA is shown by ribbon representation in blue. Arg192 of PAX4 (sticks) is predicted to bind with the phosphate backbone (sticks) of DNA.
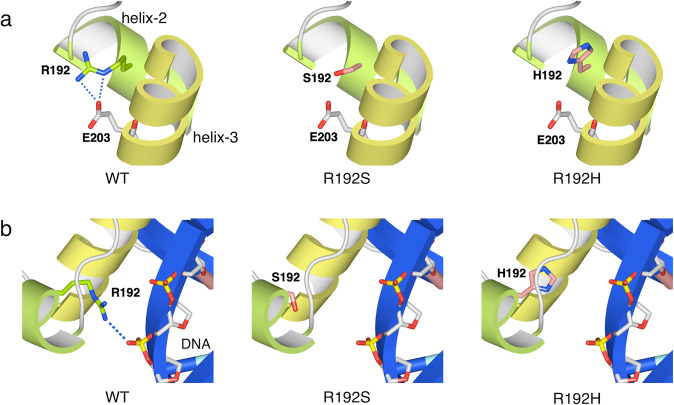
Fig. 2. Structural analysis of two PAX4 variants, p.Arg192Ser, and p.Arg192His.
PAX4 are shown by ribbon representation, with the key residues of PAX4 as sticks. The helix-2 and the helix-3 of PAX4 are shown in green and yellow, respectively. Wild-type Arg192 is green, while the Ser192 and His192 mutants are light red. a In wild-type PAX4, Arg192 forms salt-bridges with Glu203 (dotted blue line). In both mutants (p.Arg192Ser and p.Arg192His), the formation of salt-bridges with Glu203 is disrupted. b DNA is shown by ribbon representation in blue, with the phosphate backbone as sticks. In wild-type PAX4, Arg192 directly binds to the phosphate backbone of DNA (dotted blue line). In both mutants (p.Arg192Ser and p.Arg192His), binding with the phosphate backbone of DNA is disrupted.
We performed a functional study of INS-1 832/13 cells to assess the impact of these PAX4 variants, as described in Supplementary Note. Briefly, INS-1 832/13 cells were transfected with an equal amount of plasmid containing PAX4 wild-type and variants (p.Arg192Ser and p.Arg192His). Then, total RNA obtained from the INS-1 832/13 cells was reverse-transcribed into cDNA using the TaqMan Fast Advanced Cells-to-CT Kit (ThermoFisher SCIENTIFIC). Quantitative PCR was performed using the QuantStudio 7 Flex Real-Time PCR System (ThermoFisher Scientific). In the previous reports, PAX4 was suggested to maintain the integrity of the endoplasmic reticulum (ER) thus protecting β-cells from apoptosis9 as well as promoting the expression of the genes involved in β-cell survival, such as Calr encoding for calreticulin, a Ca2+ chaperone of the ER8. In the present study, we demonstrated that the transcription levels of Calr were decreased not only in INS-1 832/13 cells overexpressing PAX4 p.Arg192His, as previously reported8, but also in those overexpressing p.Arg192Ser, compared to those overexpressing wild-type PAX4 (Supplementary Fig. S1), which also suggested a pathogenic role for the PAX4 variants.
To physiologically classify T2D loci, cluster analyses have been performed using data on metabolic traits1,2,10. Mahajan et al.2 performed a hierarchical cluster analysis of loci associated with T2D in ethnically diverse populations, generating three main clusters associated with BMI/dyslipidemia, insulin secretion, and insulin action. The cluster related to reduced insulin secretion contained a number of important loci involved in β-cell function as shown in their Supplementary Fig. 62, with several lead variants, such as those of PAX4 (p.Arg192His) and HNF1A, that showed distinct minor allele frequency (MAF) spectra and ethnically different associations (Supplementary Tables 10 and 11 in ref. 2). Such differences could lead to distinct proportions of the clusters of T2D loci with different underlying biological pathways2,10, which may result in variable proportions of subgroups of T2D patients classified based on their genetics in ethnically diverse populations. For example, the PAX4 variants are specific to East Asians4, which presumably increase the proportion of the cluster of T2D loci with biological pathways influencing β-cell function in East Asian populations.
In a recent review article11, the pronounced ethnic heterogeneity associated with ethnic-specific risk variants (e.g., PAX4 p.Arg192His and HNF1A p.Glu508Lys12) was suggested to be relatively unusual and not to explain observed ethnic differences in the presentation of T2D. It was also suggested that rare ethnic-specific variants identified through sequencing studies may be more important in this regard11. Although T2D GWAS meta-analysis recently performed in a Japanese population identified no less than 20 novel T2D loci4 that were not associated with T2D in Europeans1,3 (Supplementary Table 3 in ref. 4). Ethnic differences in the associations of a number of new loci with T2D may be affected by several factors, such as differences in allele frequency and diverse patterns of linkage disequilibrium in these populations as suggested recently13. For example, we identified nine novel missense variants such as those of PAX4 (p.Arg192Ser) and GLP1R (p.Arg131Gln)4 that were in linkage disequilibrium with the lead variants of T2D loci and not rare in the Japanese population, but rare or monomorphic in Europeans (Supplementary Table 6 in ref. 4).
Considering the differences in sample size between these studies3,4, more large-scale genetic studies of East Asian populations, such as a recently reported extensive T2D GWAS meta-analysis14, are needed to fully elucidate similarities and differences in the pathogenesis of T2D between them. In conclusion, our new GWAS data4 suggested that large-scale single ethnic genetic studies could be useful for identifying ethnic-specific risk variants including relatively common ones, providing new insights into the genetic heterogeneity of T2D in diverse ethnic groups. The new information thus obtained would facilitate the development of tailored therapy for this disease in various populations.
Supplementary information
Acknowledgements
We acknowledge all authors of our GWAS meta-analysis of T2D in a Japanese population4 for their contribution to advancing genetic research. This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (grant 19K16534 to J.H.). This research was also supported by Advanced Genome Research and Bioinformatics Study to Facilitate Medical Innovation (GRIFIN) in Platform Program for Promotion of Genome Medicine (P3GM) of AMED.
Author contributions
J.H., T.Kato, and N.S. designed the study, conducted the research, and wrote the manuscript. J.H., K.S., F.M., T.Kato, T.T. and N.S. contributed to data acquisition. Y.O., M.H., N.S., T.Y. and T.Kadowaki supervised the study. All authors contributed to and approved the final version of the manuscript.
HGV database
The relevant data from this Data Report are hosted at the Human Genome Variation Database at: 10.6084/m9.figshare.hgv.3018 and 10.6084/m9.figshare.hgv.3021.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nobuhiro Shojima, Email: nshojima-tky@umin.ac.jp.
Toshimasa Yamauchi, Email: tyamau@m.u-tokyo.ac.jp.
Takashi Kadowaki, Email: t-kadowaki@toranomon.kkr.or.jp.
Supplementary information
The online version contains supplementary material available at 10.1038/s41439-021-00156-8.
References
- 1.Scott RA, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan A, et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahajan A, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K, et al. Identification of 28 new susceptibility loci for type 2 diabetes in the Japanese population. Nat. Genet. 2019;51:379–386. doi: 10.1038/s41588-018-0332-4. [DOI] [PubMed] [Google Scholar]
- 5.Ntuk UE, Gill JM, Mackay DF, Sattar N, Pell JP. Ethnic-specific obesity cutoffs for diabetes risk: cross-sectional study of 490,288 UK biobank participants. Diabetes Care. 2014;37:2500–2507. doi: 10.2337/dc13-2966. [DOI] [PubMed] [Google Scholar]
- 6.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrera LA, et al. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science. 2016;351:1450–1454. doi: 10.1126/science.aad2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sujjitjoon J, et al. PAX4 R192H and P321H polymorphisms in type 2 diabetes and their functional defects. J. Hum. Genet. 2016;61:943–949. doi: 10.1038/jhg.2016.80. [DOI] [PubMed] [Google Scholar]
- 9.Mellado-Gil JM, et al. PAX4 preserves endoplasmic reticulum integrity preventing beta cell degeneration in a mouse model of type 1 diabetes mellitus. Diabetologia. 2016;59:755–765. doi: 10.1007/s00125-016-3864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udler MS, et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med. 2018;15:e1002654. doi: 10.1371/journal.pmed.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barroso I, McCarthy MI. The genetic basis of metabolic disease. Cell. 2019;177:146–161. doi: 10.1016/j.cell.2019.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrada K, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spracklen CN, et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature. 2020;582:240–245. doi: 10.1038/s41586-020-2263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The relevant data from this Data Report are hosted at the Human Genome Variation Database at: 10.6084/m9.figshare.hgv.3018 and 10.6084/m9.figshare.hgv.3021.