Abstract
Human parainfluenza virus type 3 (HPIV3), a member of the Paramyxoviridae family, can cause lower respiratory disease in infants and young children. The phosphoprotein (P) of HPIV3 is an essential cofactor of the viral RNA-dependent RNA polymerase large protein (L). P connects nucleocapsid protein (N) with L to initiate genome transcription and replication. Sumoylation influences many important pathways of the target proteins, and many viral proteins are also themselves sumoylated. In this study, we found that the P of HPIV3 could be sumoylated, and mutation of K492 and K532 to arginine (PK492R/K532R) failed to be sumoylated within P, which enhances HPIV3 minigenome activity. Biochemical studies showed that PK492R/K532R had no effect on its interactions with N, formation of homo-tetramers and formation of inclusion bodies. Finally, we found that incorporation of K492R/K532R into a recombinant HPIV3 (rHPIV3-PK492R/K532R) increased viral production in culture cells, suggesting that sumoylation attenuates functions of P and down-regulates viral replication.
Keywords: Human parainfluenza virus type 3 (HPIV3), Phosphoprotein, Sumoylation, Replication, Viral replication
Introduction
Human parainfluenza virus type 3 (HPIV3) belongs to the Paramyxoviridae family and can cause lower respiratory disease in infants and young children (Moscona 2005). However, no effective vaccine and antivirals have been developed or licensed. The genome of HPIV3 encodes nucleoprotein (N), phosphoprotein (P), RNA-dependent RNA polymerase large protein (L), matrix protein (M), and two spike glycoproteins hemagglutinin-neuraminidase protein (HN) and fusion protein (F) (Banerjee et al. 1991). N encapsidates viral genome RNA to form N-RNA templates, and P, as a cofactor of L, recruits the L to N-RNA template for initiation of genome transcription and replication (Durbin et al. 1997; Galinski 1991). In addition, P functions as a chaperone to interact with N to prevent the nonspecific aggregation of N and N binding to cellular RNAs (Chen et al. 2007; Howard and Wertz 1989). Moreover, interaction of N and P provides the minimal requirement for the formation of inclusion bodies which are critical for viral replication (Zhang et al. 2013), and functionally deficient P severely affects the formation of inclusion bodies (Zhang et al. 2018).
Sumoylation is similar to the ubiquitination process (Matunis et al. 1996), but the functional effect of sumoylation and ubiquitination could be completely diverse (Gill 2004). Effect of sumoylation is largely dependent on the function of its target protein (Hay 2005). There are four SUMO isoforms, SUMO1, SUMO2, SUMO3, and SUMO4 with an approximate molecular mass of 12 kDa in human. SUMO1 is quite dissimilative with SUMO2/3 with about 47% sequence identity. A main difference between SUMO1 and SUMO2/3 is that SUMO2/3 can form polymeric SUMO chains and SUMO1 can be as a chain terminator to incorporate into poly-SUMO2/3 chains (Everett et al. 2013). Nonetheless, all SUMO proteins are activated and conjugated by the same enzymes (Flotho and Melchior 2013).
Given that the sumoylation is a highly versatile and dynamic regulation process in the cellular environment and regulates many important pathways like transcription and innate immunity (Everett et al. 2013), many viral proteins are themselves sumoylated. For instance, enterovirus 71 3C protein can be sumoylated along with its degradation, consequently reducing viral replication (Chen et al. 2011); Sumoylation of the HPIV5 P protein is important for viral growth (Sun et al. 2011). Collectively, referring to our previous research basis (Zhang et al. 2018), we further found that phosphoprotein of HPIV3 can be sumoylated and this modification may serve as a defense mechanism of host cells to limit replication of HPIV3.
Materials and Methods
Cells and Viruses
HEK293T were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 15% fetal bovine serum (FBS) at 37 °C in a CO2 incubator. HeLa, LLC-MK2 (monkey kidney cell line), and A549 (human lung adenocarcinoma epithelial cells) were cultured in DMEM medium containing 8% FBS at 37 °C in a CO2 incubator. HPIV3 (NIH 47885) and rHPIV3-PK492R/K532R, were propagated in LLC-MK2 cells by inoculation at a multiplicity of infection (MOI) of 0.1. Recombinant vaccinia virus (vTF7-3) expressing T7 RNA polymerase was amplified in HeLa cells by inoculation at an MOI of 0.1.
Plasmid Constructs
The plasmids pGEM4-N, pGEM4-P, pGEM4-L, HPIV3-MG(-), and pOCUS-HPIV3, which encode untagged N, P, L, HPIV3 minigenome and HPIV3 genome, and HA-tagged P, Myc-tagged P and Myc-tagged N, which were cloned in pCAGGS, pCDNA3.0 and pGADT7, have been described previously (Hoffman and Banerjee 2000; Zhang et al. 2013). cDNAs encoding P mutants were amplified by PCR-based cloning techniques using pCAGGS-HA-P as a template and were cloned in pCAGGS and pGADT7. PCR products of the mutational HPIV3 genome (HPIV3-PK492R/K532R) were cloned into pOCUS-HPIV3 and digested with SalI and Eco91I via ligase-independent cloning. The plasmids pcDNA3.0-Cer-SUMO1, pcDNA3.0-Cer-SUMO1-AA, pcDNA3.0-Cer-SUMO2/3, pcDNA3.0-Ubc9, pcDNA3.0-Flag-SENP1, pcDNA3.0-FLAG-SENP1-mut were kindly provided by Prof. Ke Xu (College of Life Sciences. Wuhan University, China) as previously described (Han et al. 2014). All constructs were verified by DNA sequencing.
Virus Infection and Plaque Assay
Cells were cultured at a density of 70%–80% in 12-well or 24-well plates at 37 °C overnight and HPIV3 was added in wells at an MOI of 1.0 for 2 h and then the medium was replaced with fresh medium with 4% FBS. For the plaque assay, LLC-MK2 cells were cultured in 24-well plates at 37 °C overnight and virus stock was serially diluted tenfold up to 105, and cells were infected with 400 µL of the dilutions for 2 h. Then, the medium was replaced with methylcellulose, and the plates were incubated at 37 °C and 5% CO2 for an additional 3–4 days until visible viral plaque was detected. After being stained with crystal violet, the plaques were counted to calculate the viral titers.
Western Blot Analysis
Cells were harvested and lysed with 80 µL–200 µL lysis buffer (50 mmol/L Tris–HCl [pH 7.4], 1% Triton X-100, 150 mmol/L NaCl, 1 mmol/L EDTA [pH 8.0] and 0.1% sodium dodecyl sulfate [SDS] with a protease inhibitor cocktail) for 30 min on ice. The supernatants were collected via centrifugation at 12,000 ×g at 4 °C for 30 min and then were boiled with SDS-PAGE loading buffer at 100 °C for 10 min and resolved via 10% SDS polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were transferred to nitrocellulose membranes, then were incubated with 5% milk in phosphate buffered saline (PBS) with 0.1% Tween 20 (PBST) for being blocked for 1 h, after that, the membranes were incubated with primary antibodies overnight and then incubated with secondary antibodies for another 1 h and detected on a Fujifilm LAS-4000 imaging system. The following primary antibodies were used: mouse anti-HA (1:10,000, Sigma), rabbit anti-HA (1:10,000, Sigma), mouse anti-Myc (1:5000, Sigma), rabbit anti-Myc (1:2500, Sigma), mouse anti-Flag (1:2500, Sigma), rabbit anti-SUMO1 (1:10,000, ABclonal), rabbit anti-Ubc9 (1:2500, ABclonal), rabbit anti-GFP (1:10,000, Santa Cruz), mouse anti-HN (1:1000, Abcam), mouse anti-GAPDH (1:2500, Santa Cruz). The secondary antibodies used HRP-conjugated goat anti-mouse immunoglobulin (IgG) (1:5000) and goat anti-rabbit IgG (1:5000).
Immunoprecipitation and Co-immunoprecipitation Assay
To precipitate HA tagged P, cells were harvested and lysed with 200 μL lysis buffer supplemented with 10 mmol/L N-ethylmaleimide (Sigma). The supernatants were collected via centrifugation and were extracted 40 µL for being boiled with SDS-PAGE loading buffer at 100 °C for 10 min as input. The rest of supernatants were precleared by incubation with 20 µL protein G-Sepharose 4 Fast Flow medium for 1 h at 4 °C with rotation. The precleared supernatants were incubated with mouse anti-HA tag antibody for 4 h at 4 °C with rotation. Then the samples were mixed with protein G-Sepharose 4 Fast Flow medium and incubated overnight at 4 °C with rotation. The beads were then washed five times with lysis buffer and boiled with SDS-PAGE loading buffer, and the bound P and sumoylation P were analyzed via Western blot.
To explore the interaction between N and P mutants and oligomerization of P mutants, appropriate plasmids were transfected, and Myc tagged N or P was precipitated with anti-Myc agarose. Co-precipitated N or P was detected via Western blot analysis.
In Vivo HPIV3 Minigenome Assay
The in vivo HPIV3 minigenome assay was performed as described previously (Hoffman and Banerjee 2000) with minor modifications. HeLa cells were cultured at a density of 90% in 12-well at 37 °C overnight and incubated with HPIV3 at an MOI of 1 for 1 h. pGADT7-P (62.5 ng) or P mutants were transfected in the presence of pcDNA3.0-N (125 ng), pGEM4-L (100 ng), and a plasmid encoding the HPIV3 minigenome (50 ng) by Lipofectamine 2000 (Invitrogen). The transfection medium was replaced with DMEM containing 4% FBS 5 h later. At 24 h post-transfection, the cells were harvested and lysed in 150 μL lysis buffer. We extracted 20 μL aliquots to assess luciferase activity according to the manufacturer’s instructions. All assays were repeated at least three times for accuracy.
Immunofluorescence Analysis
HeLa cells were cultured on coverslips in 24-well plates overnight. After appropriate plasmids transfected, cells were treated at 24 h later. Cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 for 20 min at room temperature. After being blocked with 3% bovine serum albumin (BSA) for 30 min, cells were incubated with primary antibodies diluted in 1% BSA at 4 °C overnight and secondary antibodies diluted in 1% BSA at room temperature for another 1 h. After staining with 1 mg/mL 4′ 6-diamidino-2-phenylindole (DAPI) in PBS, cells were examined by using a Leica confocal microscope.
Statistical Analysis
Data of HPIV3 minigenome assay were represented as mean ± SD in the way of importing into GraphPad Prism software (Version 6.01). All results are expressed as mean ± SD of at least three independent experiments (n ≥ 3). The P value was calculated using an unpaired Student’s t test. In all tests, P > 0.05 was considered non-statistically significant (n.s.), and P < 0.05 was considered statistically significant, marked as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Results
The P Is Sumoylated by SUMO1/SUMO2/SUMO3
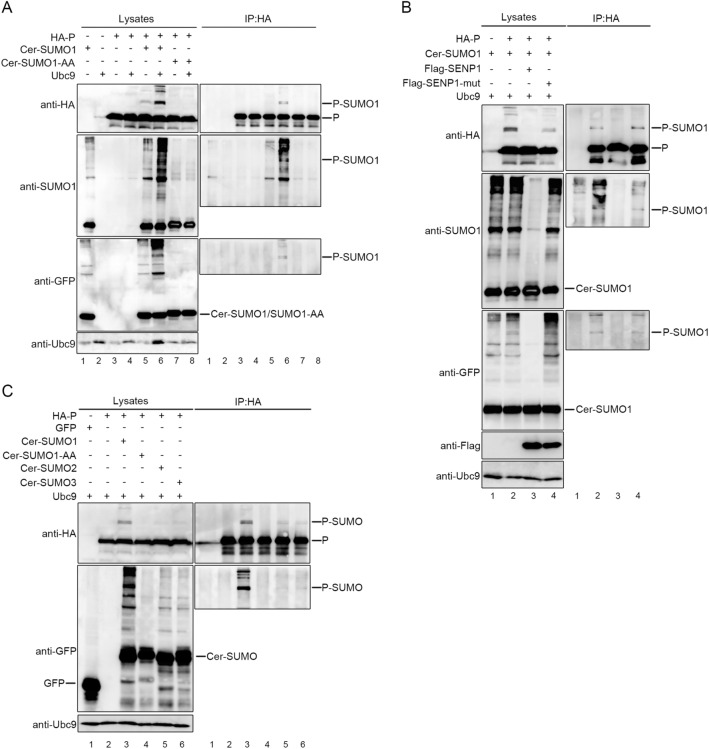
To investigate whether co-factor of L, P, could be sumoylated and test the conjugation of SUMO isoforms SUMO1 and SUMO2/3 to P, exogenous Ubc9 were co-transfected with HA-tagged P together with either Cer-SUMO1 or Cer-SUMO1-AA (conjugation deficient SUMO1 variant). At 36 h post-transfection, HEK-293T cells were lysed and the partial lysates were immunoprecipitated as protocol described. The precipitated proteins and the lysates were further analyzed via Western blot assay with anti-HA polyclonal antibody, anti-SUMO1 antibody and anti-GFP antibody. With adequate contrast, we observed characteristic band of 130 kDa which reflected the modification of P (90 kDa) by Cer-SUMO1 (40 kDa), while modified P could be hardly detected in the absence of exogenous Ubc9, and Ubc9 significantly increased the modification of P with three different antibodies both in lysates and precipitated immune complex (lanes 5 and 6, Fig. 1A). In contrast, this characteristic 130-kDa band was absent from cells expressing Cer-SUMO1-AA in spite of expression of exogenous Ubc9 (lanes 7 and 8, Fig. 1A), suggesting that the P can be sumoylated with SUMO1 but not SUMO1-AA. It is worth noting that two close bands were detected when P was sumoylated, which implies that there might exist two sumoylated sites within P (lane 6, Fig. 1A). Next, we sought to know whether P could be desumoylated by SENP1, which is mainly involved in the removal of SUMO1 from modified proteins, Cer-SUMO1, and Ubc9 were co-expressed with SENP1 or with functionally defective SENP1 (SENP1-mut) in 293T cells. The bands of sumoylated P visibly disappeared in the SENP1-expressing cells but not in the SENP-mut-expressing cells (Fig. 1B). When Cer-SUMO2 or Cer-SUMO3 were co-expressed with P and Ubc9, SUMO2/3-modified P could be also been observed but profiles were much weaker than SUMO1-modified P (Fig. 1C). Taken all these data together, our results show that HPIV3 P could be sumoylated by SUMO1 as well as SUMO2/3, predominantly SUMO1-modified, and sumoylation modification of P could be removed by SENP1.
Fig. 1.
P of HPIV3 can be sumoylated in 293T cells. A 293T cells were co-transfected with exogenous Ubc9 and HA-tagged P together with either Cer-SUMO1 or Cer-SUMO1-AA, as indicated. The cells were lysed at 36 h post-transfection, and P was immunoprecipitated with protein G-Sepharose 4 Fast Flow medium and anti-HA monoclonal antibody, The P were immunoblotted with anti-HA polyclonal antibodies. Cer-SUMO1 was examined in separated blots with an anti-SUMO1 polyclonal antibody and anti-GFP polyclonal antibody. B 293T cells were co-transfected with HPIV3 P and Ubc9 together with Cer-SUMO1, SENP1, or SENP1-mut, as indicated. P was immunoprecipitated as described in (A), and the precipitated proteins and lysates were further analyzed by SDS-PAGE and immunoblotting with anti-HA polyclonal antibodies; Cer-SUMO1 was detected in separated blots with an anti-SUMO1 polyclonal antibody and anti-GFP polyclonal antibody; Anti-Flag monoclonal antibody was used to examine the expression of SENP1 and SENP1-mut. C HPIV3 P and Ubc9 were co-transfected with GFP (as a control), Cer-SUMO1, Cer-SUMO1-AA, Cer-SUMO2, and Cer-SUMO3, as indicated; P was immunoprecipitated, and the proteins were further analyzed by SDS-PAGE and immunoblotting with anti-HA polyclonal antibodies and anti-GFP polyclonal antibodies. Input of the whole-cell lysates was detected by immunoblotting with anti-Ubc9 antibody to show the expression of Ubc9.
Replication of HPIV3 Is Suppressed with the Incremental Sumoylation Level of P
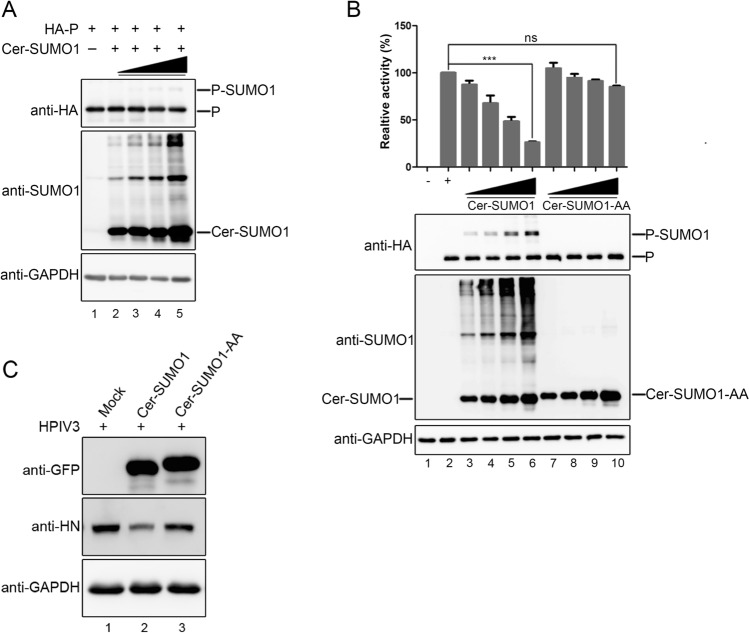
Next, we want to know whether the sumoylation of P regulates the replication of HPIV3. We first estimated the stability of P when P was sumoylated. Hela cells were transfected with HA-tagged P together with the increasing concentration of Cer-SUMO1, and results showed that the sumoylation of P were enhanced along with increasing exogenous SUMO1, but there is no significant change in expression levels of P (Fig. 2A). Then, we further investigated whether the RNA synthesis function was affected via enhancing the sumoylation level of P via HPIV3 minigenome system, and found that the luciferase activity was declined in a dose-depended manner with increasing the expression of Cer-SUMO1, but not expression of SUMO1-AA (Fig. 2B). To further confirm this result, A549 cells were transfected with Cer-SUMO1 or Cer-SUMO1-AA, at 24 h post-transfection, cells were infected with HPIV3 at an MOI of 1 for additional 24 h, and results showed that HN expression of HPIV3 decreased in the presence of SUMO1 expressed but not SUMO1-AA (Fig. 2C), suggesting that the incremental sumoylation level of P could inhibit the replication of HPIV3.
Fig. 2.
The incremental sumoylation level of P inhibits replication of HPIV3. A HeLa cells were transfected with HA-tagged P and increasing amounts of Cer-SUMO1. At 24 h post-transfection, cells were collected and lysed. Samples were further analyzed by SDS-PAGE and immunoblotting with anti-HA monoclonal antibodies and anti-SUMO1 polyclonal antibodies, and with anti-GAPDH monoclonal antibodies to show the loading controls. B vTF7-3-infected HeLa cells were transfected with plasmids encoding N (250 ng), P (62.5 ng), L (100 ng), and the minigenome (50 ng) with increasing amounts of Cer-SUMO1 or Cer-SUMO1-AA (125, 250, 500, and 1000 ng). The relative luciferase expression level of cells transfected with wild-type P was defined as 100% and mock was transfected without P. The expression of P and Cer-SUMO1 or Cer-SUMO1-AA were detected via Western blot analysis with anti-HA and anti-SUMO1 antibodies. GAPDH was used as a loading control. C A549 cells were transfected with plasmids encoding Cer-SUMO1 or Cer-SUMO1-AA. At 24 h post-transfection, the cells were infected with HPIV3 at an MOI of 1. At 48 h post-transfection, the cells were harvested and lysed for Western blot analysis using anti-HN, and anti-GAPDH monoclonal antibodies, and using anti-GFP polyclonal antibodies to detect the expression of Cer-SUMO1 or Cer-SUMO1-AA.***P < 0.001; ns, non-statistically significant.
Two or More Sumoylated Sites Are Existing in the C-Terminus of P
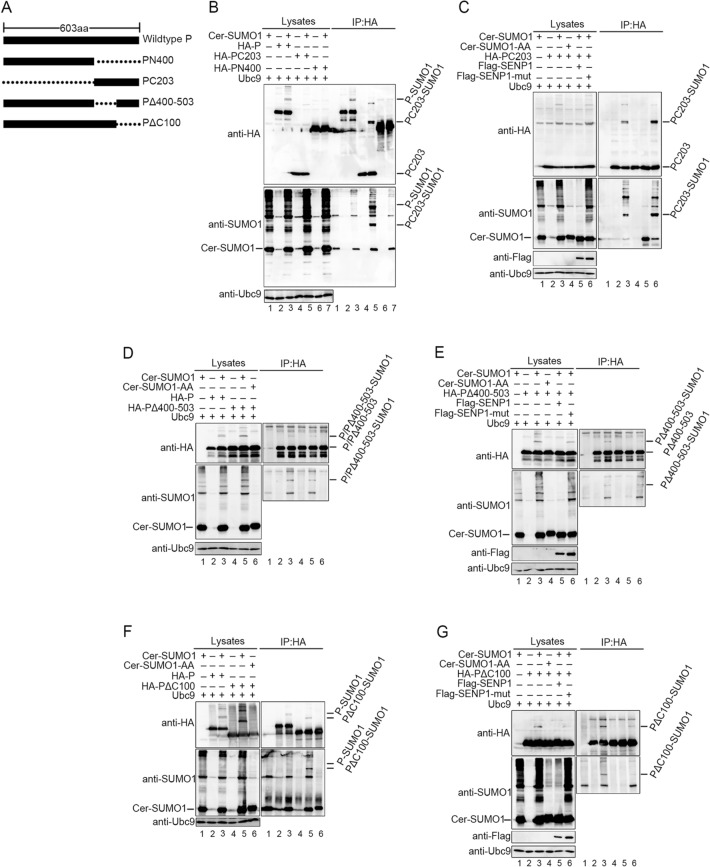
As it had been affirmed that P could be sumoylated, we continued to find out precise sites of P for the conjugation of SUMO. Two HA-tagged P truncated mutants (Fig. 3A), N-terminal 400 amino acids of P (PN400) and C-terminal 203 amino acids of P (PC203) were respectively co-expressed with Cer-SUMO1 and Ubc9 in 293T cells, and specific bands for sumoylated P were detected when wild type P and PC203 expressed, but not PN400 (Fig. 3B). As PC203 were co-expressed with SUMO1 and SENP1 or SENP1-mut, modified P mutants were removed by SENP1 but not SENP1-mut (Fig. 3C), suggesting that PC203 could still be sumoylated and the modified sites locate in C-terminal 203 amino acids within P. Then, we constructed another two truncated mutants, P lacked C-terminal 400–503 amino acids (PΔ400–503) and P lacked C-terminal 100 amino acids (PΔC100) (Fig. 3A). These two HA-tagged mutants were respectively co-expressed with Cer-SUMO1 and Ubc9 in 293T cells, and the result shows that both of the two mutants still could be sumoylated (Fig. 3D, 3F), but the difference is that the sumoylated bands within the mutants turn into one single band, which replies that one of the modified sites might be removed due to truncation of P. To further confirm that these two mutants could be indeed modified by sumoylation, PΔ400–503 or PΔC100 were co-expressed with SUMO1 or SENP1 or SENP1-mut, and we found that sumoylation modification within PΔ400–503 or PΔC100 was also removed by SENP1, but not SENP1-mut (Fig. 3E, 3G), suggesting that at least two sumoylated sites exist in the C-terminus of P, one locates in the region of 400–503 amino acids and another is in the C-terminal 100 amino acids of P.
Fig. 3.
P is sumoylated at C-terminal lysine residues. A Schematic representation of full-length HPIV3 P and its mutants used. B 293T cells were transfected with HA tagged wild type P, N-terminal 400 amino acids of P (PN400) or C-terminal 203 amino acids (PC203) of P and Cer-SUMO1, as well as exogenous Ubc9. At 36 h post-transfection, cells were lysed and P and its mutants was immunoprecipitated with protein G-Sepharose 4 Fast Flow medium and anti-HA monoclonal antibody, The P and mutants were immunoblotted with anti-HA polyclonal antibodies. Cer-SUMO1 was detected in separate blots with an anti-SUMO1 polyclonal antibody, and Ubc9 were detected with anti-Ubc9 antibody. C 293T cells were transfected with HA tagged PC203 and Ubc9 together with Cer-SUMO1, Flag-SENP1/SENP1-mut, as indicated. PC203 was immunoprecipitated as described before, and the precipitated proteins and lysates were further analyzed by SDS-PAGE and immunoblotting with anti-HA polyclonal antibodies. Cer-SUMO1 was detected in separate blots with an anti-SUMO1 polyclonal antibody. Anti-Flag monoclonal antibody was used to detect the expression of SENP1 and SENP1-mut. D, F Two truncated mutants, P lacked C-terminal 400–503 amino acids (PΔ400–503) and P lacked C-terminal 100 amino acids (PΔC100), were constructed with HA tag. To investigate if the two mutants could still be sumoylated, the experimental method was similar with (B). E, G To investigate if the sumoylation of PΔ400–503 and PΔC100 could be removed by SENP1, the experimental method was similar with (C).
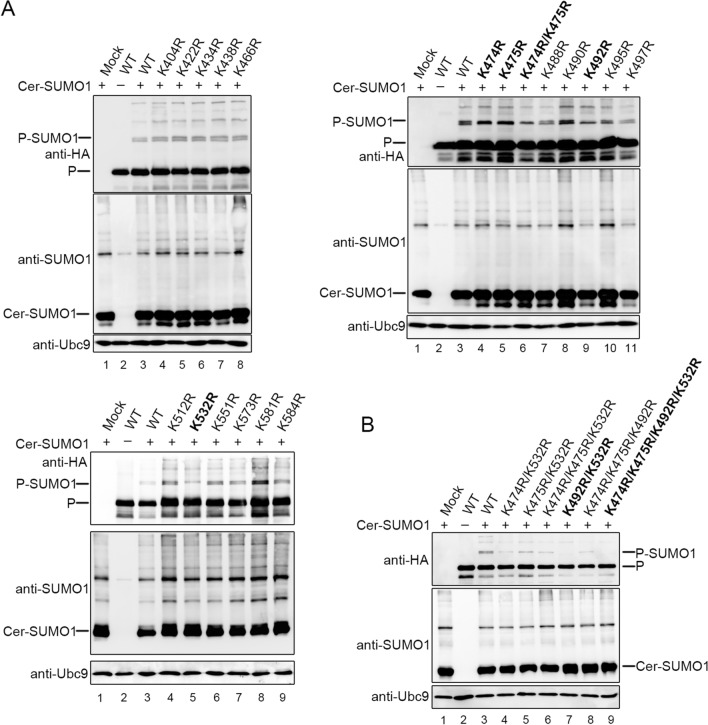
The Lysine Residues at Positions 492 and 532 of the C-Terminus of P Are Sumoylated
To identify the accurate sumoylation sites of P, all 18 lysine residues within C-terminal 203 amino acids of P were individually replaced with arginines, among which 12 of 18 residues are in the region of 400–503 amino acids and 6 of 18 residues are in the region 503–603 amino acids. We also included a double mutation (K474 and K475) to prevent complementary sumoylation caused by adjacent lysine residues. Then, these HA-tagged mutants were co-expressed with Cer-SUMO1 and Ubc9 in 293T cells, in contrast to wild type P, the bands of sumoylated mutants K474R, K475R, K474R/K475R, K492R and K532R have turned into one single band, suggesting that some of the point mutants lost part of abilities of being sumoylated (Fig. 4A). Further, we constructed a series of mutants with double, triple or quadruple point mutations, and these HA-tagged mutants were co-expressed with Cer-SUMO1 and Ubc9 in 293T cells. The result shows that only the double point mutations of lysines at positions 492 and 532 to arginines (PK492R/K532R) completely abolished the sumoylation of P (Fig. 4B). These data suggest that K492 and K532 are SUMO acceptor sites of P and losing one of the residues could affect part of the sumoylation abilities.
Fig. 4.
K492 and K532 residues of P are SUMO acceptor sites. A 18 lysine residues of the PC203 were individually replaced with arginines as well as a double mutation (K474 and K475). HA tagged P and P mutants were co-transfected with Cer-SUMO1 and Ubc9 in 293T cells. At 36 h post-transfection, cells were lysed and the lysates were analyzed to detect sumoylation of P/P mutants by SDS-PAGE and immunoblotting with anti-HA polyclonal antibodies, anti-SUMO1 polyclonal antibodies and anti-Ubc9 polyclonal antibodies. B Numbers of double mutations and triple or tetradic were constructed, as indicated. The same method as (A) was carried out to show the sumoylation of those mutants.
PK492R/K532R Enhances Viral RNA Synthesis
To investigate the difference of function between wild type P and PK492R/K532R, Hela cells were transfected with P and PK492R/K532R in the similar expression level in HPIV3 minigenome system, respectively. The luciferase activities increased when P was replaced with PK492R/K532R (Fig. 5A), and the luciferase activities hardly declined with an increasing amount of exogenous SUMO1 (Fig. 5B), suggesting that sumoylation modification of P limits its function in RNA synthesis.
Fig. 5.
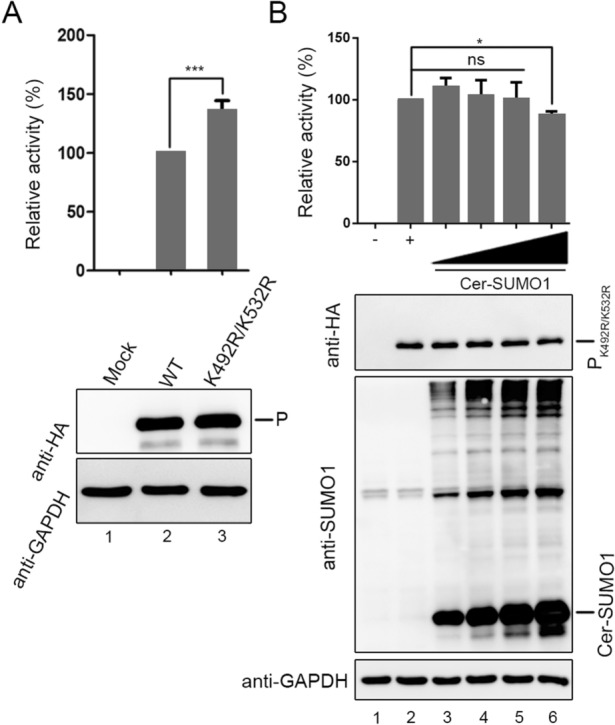
The K492R/K532R mutation within P enhance viral minigenome activities. A vTF7-3-infected HeLa cells were transfected with plasmids encoding N (250 ng), P or PK492R/K532R (62.5 ng), L (100 ng), and the minigenome (50 ng). The relative luciferase expression level of cells transfected with wild-type P was defined as 100% and mock was transfected without P. B vTF7-3-infected HeLa cells were transfected with plasmids encoding N (250 ng), PK492R/K532R (62.5 ng), L (100 ng), and the minigenome (50 ng) with increasing amounts of Cer-SUMO1 (125, 250, 500, and 1000 ng). The relative luciferase expression level of cells transfected with PK492R/K532R was defined as 100% and mock was transfected without PK492R/K532R.***P < 0.001; *P < 0.05; ns, non-statistically significant.
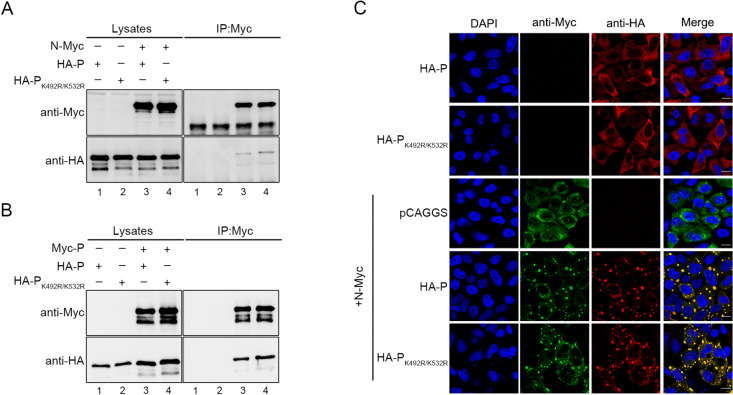
PK492R/K532R Does Not Affect N-P Interaction, Oligomerization of P and Formation of Inclusion Bodies
N-P interaction, as well as oligomerization of P, is essential for HPIV3 RNA synthesis. To study the mechanism of the defect of PK492R/K532R in RNA synthesis, N and P or PK492R/K532R interaction were examined using co-immunoprecipitation in transfected cells. PK492R/K532R can bind to N, similarly to wild type P (Fig. 6A). In addition, PK492R/K532R also can interact with wild type P (Fig. 6B), indicating that there is no defect in N-P interactions or oligomerization formation of P. Furthermore, we found that inclusion bodies which are critical for viral RNA synthesis formed by co-expression of N and PK492R/K532R were the same as co-expression of N and wild type P (Fig. 6C), suggesting that PK492R/K532R does not affect N-P interaction, oligomerization of P and formation of inclusion bodies.
Fig. 6.
The K492R/K532R mutation does not affect N-P interaction or P oligomerization formation as well as the formation of inclusion bodies. A 293T cells were transfected with N-Myc and HA-P/PK492R/K532R, mock was transfected with individual HA-P/PK492R/K532R. At 36 h post-transfection, cells were lysed and N-Myc was immunoprecipitated with anti-Myc agarose. The precipitated proteins and lysates were further analyzed by SDS-PAGE and immunoblotting with anti-HA polyclonal antibodies and anti-Myc polyclonal antibodies. B As similar with (A), 293T cells were transfected with Myc-P and HA-P/PK492R/K532R, mock was transfected with individual HA-P/PK492R/K532R. At 36 h post-transfection, cells were lysed and Myc-P was immunoprecipitated with anti-Myc agarose. The precipitated proteins and lysates were further analyzed by SDS-PAGE and immunoblotting with anti-HA polyclonal antibodies and anti-Myc polyclonal antibodies. C HeLa cells were transfected with plasmids encoding HA-P/PK492R/K532R or N-Myc alone or together. At 24 h posttransfection, cells were fixed and stained with antibodies against HA and Myc. The subcellular locations were visualized via confocal microscopy as described in Materials and Methods. DAPI was used for the nuclear staining. Scale bars, 10 μm.
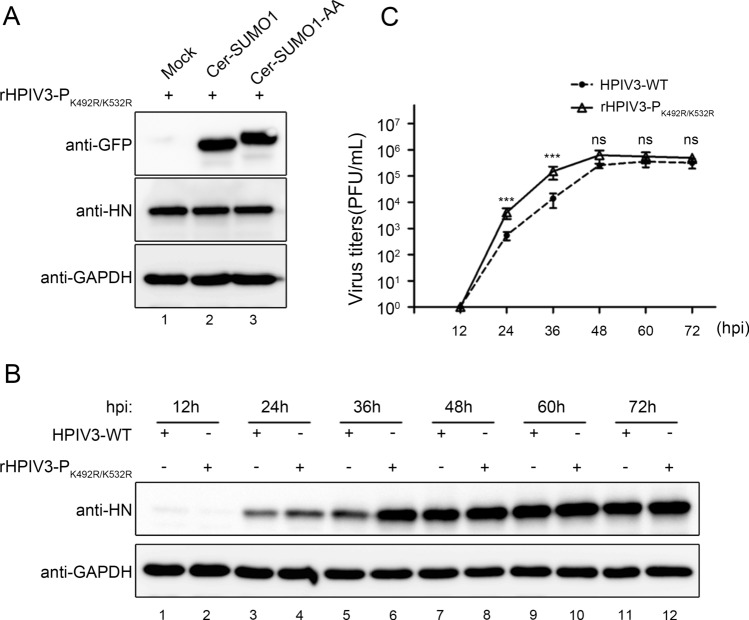
Viral Replication Is Enhanced in the Sumoylation-Defective rHPIV3-PK492R/K532R
We then examined whether the sumoylation-defective P affects viral production by analysing infection of the wild type HPIV3 and recombinant HPIV3-PK492R/K532R. We found that exogenous SUMO1 expression had no effect on the HN level in rHPIV3-PK492R/K532R-infected cells (Fig. 7A). Furthermore, A549 cells were infected with HPIV3 and rHPIV3-PK492R/K532R, and viral HN level and virus titers were examined at different time points, and results showed that HN levels in rHPIV3-PK492R/K532R-infected cells were higher than in HPIV3-infected cells (Fig. 7B) at 36 h post infection (hpi), and replicative kinetic of rHPIV3-PK492R/K532R showed markedly faster with virus titers being up to almost tenfold higher than HPIV3 at 24 and 36 hpi (Fig. 7C), suggesting that the sumoylation of P reduced viral replication and delayed viral growth cycle.
Fig. 7.
Sumoylation of P reduced viral replication and slackened viral growth cycle. A As described in (2C), wild type HPIV3 was replaced with rHPIV3- PK492R/K532R, and the other steps are the same. B, C A549 cells were infected with HPIV3-WT and rHPIV3- PK492R/K532R viruses at an MOI of 0.01. At 12, 24, 36, 48, 60 and 72 hpi, the supernatants were harvested for the plaque assay to determine virus growth curves, and the cells were harvested and lysed to analyze viral HN level via western blotting. HN protein were detected with anti-HN monoclonal antibodies and GAPDH were detected with anti-GAPDH antibodies to show loading controls. Standard errors were calculated from three independent experiments.***P < 0.001; ns, non-statistically significant.
Discussion
Herein, we present evidence demonstrating that P of HPIV3 was sumoylated by SUMO1 as well as SUMO2/3 (Fig. 1). By truncation and multiple point mutations, we found that K492 and K532 are SUMO accepted sites of P (Figs. 3 and 4). The sumolyation-defective mutant of P shows higher replication activity than wild type P (Fig. 5A), but compared with wild type P, there was no significant difference in N-P interaction, oligomerization ability, and the formation ability of inclusion bodies (Fig. 6). In addition, rHPIV3-PK492R/K532R could be rescued and showed more potent replication ability (Fig. 7).
In the past decade, increasing research have focused on the sumoylation modification of various viral proteins, and many viral proteins such as nonstructural protein 5 of DENV (Su et al. 2016), 3D protein of enterovirus 71 (Liu et al. 2016), VP1 of Avibirnavirus (Wu et al. 2019), can utilize SUMO system to enhance viral replication. Sumoylated NP of influenza A had aberrant nucleus and cytoplasm trafficking dynamics, which influenced viral growth (Han et al. 2014). But there are exceptions, for instance, 3C of enterovirus can be sumoylated and this modification results in degradation of 3C associated with a decline in viral replication and cell apoptosis (Chen et al. 2011), which indicates that sumoylation of viral proteins also has antiviral effect.
The effects of paramyxovirus P post-translational modifications such as sumoylation appear to be quite different. P of PIV5 can be sumoylated in the N-terminal 254 lysine, and the recombinant PIV5 with de-sumoylated P is weaker than wild type virus (Sun et al. 2011), which is much different with HPIV3. In our study, although we found P of HPIV3 could be sumoylated at two precise sites and influenced viral replication, the detailed mechanism is still unknown. It is possible that the sumoylation of P might influence its interaction with other cellular factors that regulate viral replication.
Acknowledgements
We thank Prof. Ke Xu (College of Life Sciences, Wuhan University, China) for kindly providing plasmids. This research is supported by grants from National Key R&D Program of China (2017YFA0505801), the National Natural Science Foundation of China (81825015, 81871650 and 31630086), National Science and Technology Major Project (2018ZX10101004), and the Natural Science Foundation of Hubei Province Innovation Group (2017CFA022), and Advanced Customer Cultivation Project of Wuhan National Biosafety Laboratory (2019ACCP-MS06).
Author Contributions
QC and MC designed the experiments and interpreted the results. QC, WJH and XYW performed experiments. QC and MC drated the manuscript. MC finalized the article. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Banerjee AK, Barik S, De BP. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-J. [DOI] [PubMed] [Google Scholar]
- Chen M, Ogino T, Banerjee AK. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J Virol. 2007;81:13478–13485. doi: 10.1128/JVI.01244-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S-C, Chang L-Y, Wang Y-W, Chen Y-C, Weng K-F, Shih S-R, Shih H-M. Sumoylation-promoted enterovirus 71 3C degradation correlates with a reduction in viral replication and cell apoptosis. J Biol Chem. 2011;286:31373–31384. doi: 10.1074/jbc.M111.254896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin AP, Siew JW, Murphy BR, Collins PL. Minimum protein requirements for transcription and RNA replication of a minigenome of human parainfluenza virus type 3 and evaluation of the rule of six. Virology. 1997;234:74–83. doi: 10.1006/viro.1997.8633. [DOI] [PubMed] [Google Scholar]
- Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol. 2013;11:400–411. doi: 10.1038/nrmicro3015. [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Galinski MS. Paramyxoviridae: transcription and replication. Adv Virus Res. 1991;39:129–162. doi: 10.1016/S0065-3527(08)60794-0. [DOI] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Han Q, Chang C, Li L, Klenk C, Cheng J, Chen Y, Xia N, Shu Y, Chen Z, Gabriel G, Sun B, Xu K. Sumoylation of influenza A Virus Nucleoprotein Is Essential For Intracellular Trafficking And Virus Growth. J Virol. 2014;88:9379–9390. doi: 10.1128/JVI.00509-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hoffman MA, Banerjee AK. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology. 2000;269:201–211. doi: 10.1006/viro.2000.0223. [DOI] [PubMed] [Google Scholar]
- Howard M, Wertz G. Vesicular stomatitis virus RNA replication: a role for the NS protein. J Gen Virol. 1989;70:2683–2694. doi: 10.1099/0022-1317-70-10-2683. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng Z, Shu B, Meng J, Zhang Y, Zheng C, Ke X, Gong P, Hu Q, Wang H. SUMO modification stabilizes enterovirus 71 polymerase 3D to facilitate viral replication. J Virol. 2016;90:10472–10485. doi: 10.1128/JVI.01756-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J Clin Investig. 2005;115:1688–1698. doi: 10.1172/JCI25669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C-I, Tseng C-H, Yu C-Y, Lai MMC. SUMO modification stabilizes dengue virus nonstructural protein 5 to support virus replication. J Virol. 2016;90:4308–4319. doi: 10.1128/JVI.00223-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Xu P, He B. Sumoylation of the P protein at K254 plays an important role in growth of parainfluenza virus 5. J Virol. 2011;85:10261–10268. doi: 10.1128/JVI.00389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yang H, Ji G, Zheng T, Zhang Y, Deng T, Zheng X, Zhou J, Hu B (2019) SUMO1 modification facilitates avibirnavirus replication by stabilizing polymerase VP1. J Virol 93 [DOI] [PMC free article] [PubMed]
- Zhang S, Chen L, Zhang G, Yan Q, Yang X, Ding B, Tang Q, Sun S, Hu Z, Chen M. An amino acid of human parainfluenza virus type 3 nucleoprotein is critical for template function and cytoplasmic inclusion body formation. J Virol. 2013;87:12457–12470. doi: 10.1128/JVI.01565-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cheng Q, Luo C, Yin L, Qin Y, Chen M. An alanine residue in human parainfluenza virus type 3 phosphoprotein is critical for restricting excessive N0-P interaction and maintaining N solubility. Virology. 2018;518:64–76. doi: 10.1016/j.virol.2018.02.008. [DOI] [PubMed] [Google Scholar]