Abstract
Background
CHK1 is considered an oncogene with overexpression in numerous cancers. However, CHK1 signalling regulation in hepatocellular carcinoma (HCC) remains unclear.
Methods
CHEK1 mRNA, protein, pri-miR-195 and miR-195 expression in HCC tissue was determined by qPCR, WB and IF staining assay. Survival analyses in HCC with high- and low-CHEK1 mRNA expression was performed using TCGA database. Relative luciferase activity was investigated in HCC cells transfected with p-CHEK1 3’UTR. Apoptosis was detected by TUNEL assay. NK and CD8+ T cells were analysed by flow cytometry.
Results
CHK1 is increased in human HCC tumours compared with non-cancerous liver. High CHK1 predicts worse prognosis. IFN-γ suppresses CHK1 via IRF-1 in HCC cells. The molecular mechanism of IRF-1 suppressing CHK1 is post-transcriptional by promoting miR-195 binding to CHEK1 mRNA 3’UTR, which exerts a translational blockade. Upregulated IRF-1 inhibits CHK1, which induces apoptosis of HCC cells. Likewise, CHK1 inhibition augments cellular apoptosis in HCC tumours. This effect may be a result of increased tumour NK cell infiltration. However, IRF-1 expression or CHK1 inhibition also upregulates PD-L1 expression via increased STAT3 phosphorylation.
Conclusions
IRF-1 induces miR-195 to suppress CHK1 protein expression. Both increased IRF-1 and decreased CHK1 upregulate cellular apoptosis and PD-L1 expression in HCC.
Subject terms: Hepatocellular carcinoma, Oncogenes
Background
Hepatocellular carcinoma (HCC) is the second leading cause of cancer death worldwide.1 Only 30% of HCC patients are detected at early stage in the USA with potential curative therapies including surgical resection, liver transplantation or ablative therapies.2–4 Approximate 60% of patients present in advanced stage are eligible for local-regional treatment or systemic therapies.5 With the development of molecular targeted therapeutics and immunotherapies in HCC, the prognosis of these patients has improved.6 However, resistance to therapy is common, and further understanding of the molecular mechanisms involved in HCC tumorigenesis and treatment resistance is needed.
Checkpoint kinase 1 (CHK1) was initially identified as a serine/threonine kinase and functions as a key component in the DNA damage response (DDR) pathway and cell cycle checkpoints.7,8 When ATM (ataxia telangiectasia mutated) and ATR (ATM and rad3-related) kinases are activated by DNA damage and replication stress, activated ATR phosphorylates and induces CHK1 activation. CHK1 transduces checkpoint signal to hold cell cycle arrest. This delay allows time for DNA to repair damage, or induce apoptosis if damage is severe. Overexpression of CHK1 contributes to resistance of anti-tumour chemotherapy or radiotherapy in numerous tumours including breast, colon, gastric and liver cancer.8,9
Targeting of CHK1 is potentially beneficial for patients with malignant tumours. However, DNA damage or repair defects caused by defective CHK1 may upregulate PD-L1 expression in tumour cells, which help tumour cells to escape anti-tumour immunity via increased exhaustion of effector T cells.10–15 Additionally, CHK1 activation allows phosphorylation of key regulators related to cellular proliferation and apoptosis.7,16,17 Therefore, it is important to study the regulation of CHK1 signalling in HCC.
Interferon regulatory factor 1 (IRF-1) is a master transcription factor in the interferon pathway and plays a key role in oncogenesis and anti-tumour immunity.18 IRF-1 promotes apoptosis of cancer cells by regulating apoptotic signalling pathways.19–22 Our previous study showed that IFN-γ regulated autophagy via IRF-1 in HCC cells.23 However, the correlation between IRF-1 and CHK1 in HCC cells remains unknown.
MicroRNAs (miRs) are small non-coding RNA molecules of 20–30 nucleotides that negatively regulate gene expression by binding to 3’ untranslated region (3’UTR) of target mRNAs.24 This modulation is involved in diverse physiological and pathological processes, including cellular apoptosis, proliferation, as well as carcinogenesis and tumour progression.25 MiR-15/16/195/424 /497 family is grouped together for their common seed sequence. They are identified as tumour suppressor miRs that modulate apoptotic pathways in chronic lymphocytic leukaemia, gastric, colon, liver and prostate cancers.26–29 Meanwhile, CHK1 expression regulated by miR-195 and other family members has been shown in diverse cancers.9,30–32
In this study, we show that CHK1 is overexpressed in human HCC tumours and serves as a biomarker for predicting prognosis. IFN-γ suppresses CHK1 expression via IRF-1. The molecular mechanism of IRF-1 regulating CHK1 is dependent on miR-195 binding to CHK1 3’-UTR to elicit translational blockade of CHK1 protein expression. Furthermore, increased IRF-1 inhibited CHK1 expression and induced cellular apoptosis in HCC cells. CHK1 inhibitor has anti-tumour effects on murine HCC characterised by augmented apoptosis induced by infiltrating NK cells. However, IRF-1 expression or CHK1 inhibition also upregulates PD-L1 expression via phosphorylated STAT3 in HCC, which may promote escape from anti-tumour immunity. These data identify important signalling pathways in HCC resistance, and provide new insight into the molecular mechanism of IRF-1 regulating CHK1 in the tumour setting.
Methods
Patient samples
Thirty paired HCC and adjacent liver tissues were acquired from patients that underwent hepatectomy at the Liver Cancer Center of University of Pittsburgh School of Medicine (Pittsburgh, PA, USA). All individuals provided written informed consent and human tissues were obtained in accordance with the University of Pittsburgh Institutional Review Board (IRB) approved protocol (No. MOD08010372/PRO08010372).
Cell line and reagents
Hepatic cancer cell lines Hepa1-6, Huh-7, Hep3B and HepG2 were purchased from ATCC (Manassas, VA, USA). All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza, Alpharetta, GA, USA), containing 10% heat-inactivated foetal bovine serum (FBS) (Clontech, Mountain View, CA, USA), 100 U/ml penicillin, 100 µg/ml streptomycin, 15 mmol/l HEPES and 200 mmol/l L-glutamine. Cells were incubated at 37 °C in a humidified incubator containing 5% CO2. Recombinant mouse and human IFN-γ was purchased from R&D (Minneapolis, MN, USA). The CHK1 inhibitor prexasertib (HY-18174) and STAT3 phosphorylation (Y705) inhibitor stattic (HY-13818) were purchased from MedChemExpress (Monmouth Junction, NJ, USA).
Mice and animal experiments
Female 5–6 weeks-old wild-type (WT) C57BL/6 (B6) mice (H-2b) and IRF-1 deficient mice (IRF-1−/−, IRF-1 knockout [KO], B6 background, H-2b) were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice were housed in a specific pathogen-free environment under a temperature and light-controlled room with free access to food and water. Animal experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (Protocol No. 18012053).
Three IRF-1 KO mice and three WT B6 mice were used in the experiments 2 and 3. Anaesthesia was carried out with isoflurane (Piramal Critical Care, PA, USA). Under anaesthesia, mice were sacrificed by cervical dislocation. Livers were harvested for analysis.
A total of 26 WT B6 mice were enrolled in experiment 5. Ten mice were first used and 16 mice were used in the repeated experiment. B6 mice were injected with 1 × 107 Hepa1-6 cells subcutaneously in the left flank containing 100 µl growth factor depleted Matrigel (CORNING, MA, USA). Tumour size was calculated every 3 days by calliper. Tumour size = L × W × ((L + W)/2). At 7 days after Hepa1-6 cell injection, mice were randomised by tumour size and body weight and received vehicle or CHK1 inhibitor prexasertib. Under anaesthesia, vehicle or prexasertib (10 mg/kg) was administered by subcutaneous injection in a volume of 200 µl twice daily for 2 days followed by 5 days rest and repeated weekly for 3 weeks. Body weight was recorded every 3 days during the treatment. After 3 weeks, mice were sacrificed under anaesthesia by cervical dislocation. Tumours were collected for single-cell suspension for flow cytometry or frozen with OCT, and 5-µM sections were cut.
Adenovirus infection
An E1- and E3-deficient adenoviral vector carrying the mouse or human AdIRF-1, AdLacZ or Adψ5 cDNA was constructed as previously described.22,33 Hepa1-6 and Huh-7 cells were infected with adenoviral concentration of 50 MOI for 48 h. Forty-eight hours after infection, cells were harvested and then total RNA and total protein and nuclear protein were extracted to determine the protein expression, respectively.
Real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reversely transcribed to single-stranded cDNA with RNA to cDNA EcoDry™ Premix Kit (Takara, Kusatsu, Shiga Prefecture, Japan). Real-time polymerase chain reaction (PCR) was performed using SYBR Premix Kit (Takara) on the ABI Stepone PCRSystem (Applied Biosystems, Foster City, CA, USA). Relative expression of each gene was normalised to β-actin mRNA or GAPDH mRNA for mouse and human studies, respectively. Primer sequences used are provided in Supplemental Table 1.
The expressions of both pri-miRNA and mature miRNA were determined by TaqMan miRNA assays according to the manufacture’s protocol. The cDNA templates for mature miRNA were prepared using the TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems, Foster City, CA, USA). The cDNA templates for pri-miRNA were prepared by RNA to cDNA EcoDry™ Premix Kit. The diluted reverse transcription reaction products were mixed with TaqMan Universal PCR Master Mix II, no UNG (Applied Biosystems) and primers for real-time PCR. U6 snRNA was used as normalisation. The miR-15a-5p, miR-16-5p, miR-195-5p, pri-miR-195, miR-424-5p, miR-497-5p and U6 snRNA primers were purchased from Applied Biosystems.
Western blot
Total cell lysates and nuclear proteins were extracted as previously described.34 A total of 30-ug nuclear protein or 60-ug cell lysate was electrophoresed on 10% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. The membranes were incubated with a 1:1000 dilution of primary antibodies overnight. Antibodies used for western blot were antibodies against IRF-1 (#8478), CHK1 (#2360), phospho-STAT3 (Tyr705) (#9145), STAT3 (#9139), PARP (#9532), human PD-L1 (#13684) and LaminA/C (#4777) (Cell signaling technology, MA, USA); anti β-actin (ab8227) and mouse PD-L1 (ab213480) antibodies (abcam, MA, USA); IRDye 800CW anti-mouse and 680RD anti-rabbit secondary antibodies (LI-COR, Lincoln, USA). After wash with Tris-buffered saline and Tween-20 (TBST) for three times, membranes were incubated with a 1:5000 dilution of secondary antibody for 30 min and scanned by Li-Cor odyssey. The β-actin (actinb) or LaminA/C protein was used as standardisation of cell lysate or nuclear protein, respectively.
Immunofluorescent staining and flow cytometry
Immunofluorescent staining was performed according to our previous study.35 Tissues were fixed with 2% paraformaldehyde in phosphate-buffered saline (PBS) for 2 h, and then dehydrated by 30% sucrose in PBS overnight. HCC cell lines were cultured on coverslips, fixed with 4% paraformaldehyde in PBS for 15 min. The tissues or cells were permeabilised with 0.1% Triton X-100 and 10% FBS in PBS for 30 min at room temperature, and incubated with primary CHK1 antibody (sc-8408, Santa Cruz Biotech, CA, USA) or NK1.1 antibody (Clone: PK136, Biolegend) for 1 h, which was diluted in a 1:100 ratio. Next, Alexa Fluor 546 anti-mouse secondary antibody was applied with F-actin antibody (Abcam) counterstain. After washing with PBS, the slides were stained with 4',6-diamidine-2'-phenylindole dihydrochloride (DAPI) and mounted, and then observed with an Olympus Fluoview FV1000 III microscope (Olympus, Tokyo, Japan). Mean grey value (arbitrary units, AU) was used to quantify immunofluorescent staining for CHK1 by Image J (1.52 a; java 1.8.0_112).
Single-cell suspension was prepared from harvested tumour tissues or cells and staining to standard protocol for flow cytometry with anti-human CD274 (B7-H1, PD-L1) antibody (clone: MIH3, Biolegend), anti-mouse CD4 (Clone: GK1.5, BD HorizonTM), CD8a (Clone: 53-6.7, BD HorizonTM), NK1.1 (Clone: PK136, BD PharmingenTM) and IFN-γ (Clone: XMG1.2 RUO, BD HorizonTM) antibodies. For detecting IFN-γ, single cells were cultured for 3 h with PMA (100 ng/ml), ionomycin (1000 ng/ml; both from Sigma–Aldrich), and GolgiPlug (1 μl/ml; BD Biosciences). Intracellular IFN-γ staining was executed after permeabilisation using intracellular staining kits from BD Biosciences and eBioscience. Flow cytometry was used to analyse cellular apoptosis by APC Annexin V Apoptosis Detection Kit with PI (640932, BioLegend). The data were acquired on BD™ LSR II and analysed with FlowJo software (version 10.6.1).
In situ cell death detection
The apoptotic cells were detected by TUNEL assay kit (11684795910, R&D) according to the standard protocol. The percentage of TUNEL-positive cells was quantified by the mean rate of TUNEL-positive cells from five high-powered fields.
Transfection
The human IRF-1 siRNA (sc-35706) and control siRNA (sc-37007) were purchased from Santa Cruz Biotech. The mirVanaTM miRNA mimic of has-miR-195-5p were acquired from Ambion (Austin, TX, USA). The cells were seeded in a 6-well plate and the following day transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Serum-free medium was replaced with growth medium after 8 h. Forty-eight hours after transfection, the protein level assay was performed.
Luciferase assay
The plasmid psiCHECK2-CHEK1 3’UTR was a gift from Judy Lieberman (Addgene plasmid # 29478).36 The β-gal reporter control plasmid was used to normalise the transfection efficiency. Huh-7 and Hepa1-6 were cultured in 6-well plate and co-transfected with β-gal and either psiCHECK2 empty vector (Promega, Madison, WI, USA) or CHEK1 3’UTR luciferase reporter constructs using lipofectamine 2000 (Invitrogen). Serum-free medium was replaced with growth medium after 6 h. Twenty-four hours after transfection, cells were induced by IFN-γ or infected with either Adψ5 or AdIRF-1 for 24 h. Forty-eight hours after transfection, the cells were lysed. Relative luciferase and β-galactosidase activities were measured with the reporter lysis buffer and luciferase substrate (Promega). The relative luciferase unit (RLU) was measured using the Dual-Luciferase Report Assay (BioTek, Winooski, VT, USA).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8 software (GraphPad Software Inc, San Diego, CA). To test for statistical significance, the t-test was used to compare between different groups. Results are collective data from two to four repeat experiments. For survival curves, log-rank test was used to compare between individual groups. For all analyses, p < 0.05 was considered significant. Data are described by mean values ± standard deviation (SD), unless otherwise specified.
Results
CHK1 in human HCC served as a biomarker for HCC prognosis
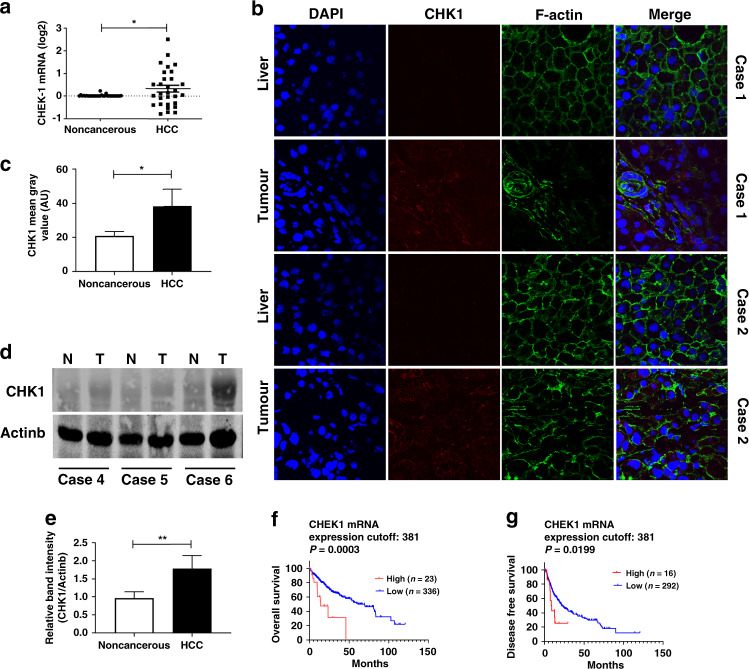
To investigate the expression of CHK1 in patients diagnosed with HCC, we analysed CHEK1 mRNA and protein expression in resected HCC tumours and adjacent liver tissue from patients. Our results showed that CHEK1 mRNA was increased in 13/30 (43%) of the HCC tumours and is decreased in 9/30 (30%) cases compared to non-cancerous background liver (Fig. 1a). In addition, CHK1 protein level determined by immunofluorescent (red) staining was upregulated in the tumour tissue compared to background liver in the HCC patients with high CHEK1 mRNA (Fig. 1b, c). Likewise, CHK1 protein expression measured by western blot was increased in the tumour compared to non-cancerous liver in these patients (Fig. 1d, e).
Fig. 1. CHK1 is overexpressed in some HCC tumours.
a CHEK1 mRNA expression is determined by qPCR in tumour and adjacent background liver from HCC patients (n = 30). Each data point represents one patient. b Representative immunofluorescent images (×400 magnification) are shown for CHK1 protein level (red staining) in tumour and non-cancerous liver from two HCC patients. c Quantification of CHK1 by mean grey value (arbitrary units, AU) is shown for tumour and non-cancerous liver (n = 3). Data represent mean ± SD, *p < 0.05. d Representative western blot images are shown for CHK1 protein expression in tumour (T) and non-cancerous liver (N) from three HCC patients. e Quantification of CHK1 in tumour and background liver from HCC patients (n = 6). Data represent mean ± SD, **p < 0.01. The clinical significance of CHEK1 mRNA expression in HCC overall survival (f) and disease-free survival (g) are, respectively, evaluated through Kaplan–Meier analysis.
Since our data reflected a relatively small sample size, we analysed The Cancer Genome Atlas (TCGA) database through UALCAN.37 The CHEK1 gene expressions in HCC tumours were higher than those of normal (371 tumour vs 50 normal, p = 1.62E-12) (Supplementary Fig. 1A). Furthermore, to explore whether CHK1 was a biomarker for predicting clinical outcome of HCC patients, we analysed TCGA database through UALCAN. HCC patients with high CHEK1 expression predicted significantly worse overall survival compared to those with low and medium CHEK1 level (90 patients vs 275 patients, p < 0.0001) (Supplementary Fig. 1B). We also analysed data from TCGA database through cBioPortal.38 Although only ~6% of HCC patients in cBioPortal showed high CHEK1 mRNA expression, this subset had a particularly poor prognosis (Fig. 1f, g).
IFN-γ repressed CHK1 via IRF-1 in HCC cells
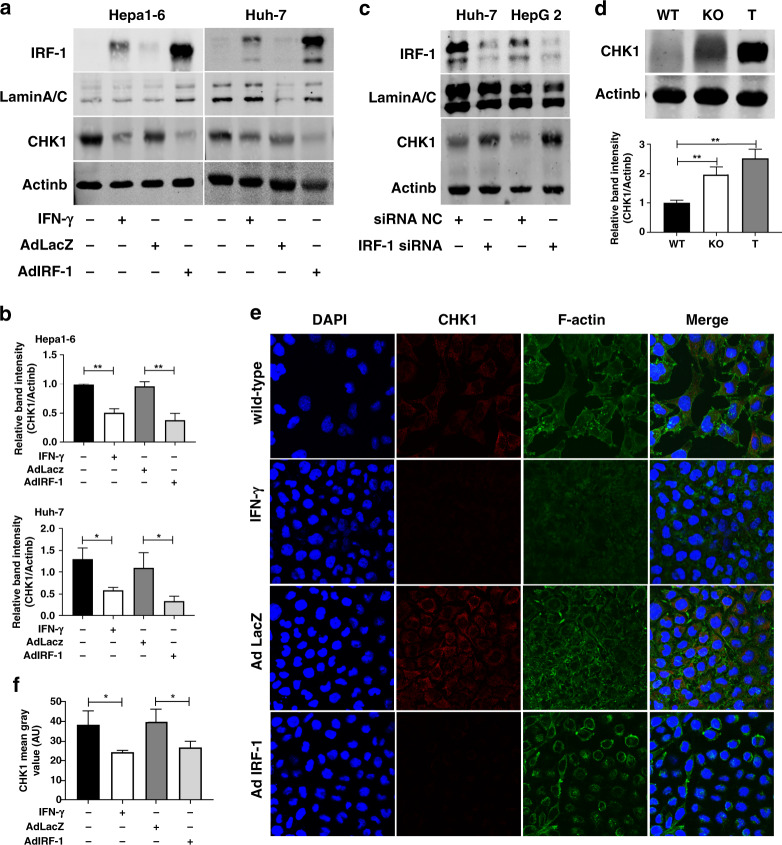
Previously we had shown that IRF-1 mRNA expression was downregulated in the human HCC tumour compared to background liver,39 and that IFN-γ induced autophagy and growth inhibition in HCC mediated by IRF-1.23 Since CHK1 had been shown to have an important role in HCC signalling, we investigated the interaction of IRF-1 and CHK1 in HCC cells. IFN-γ upregulated IRF-1 expression and decreased basal CHK1 total protein expression in murine Hepa1-6 and human Huh-7 HCC cells (Fig. 2a, b). Likewise, exogenous expression of IRF-1 by AdIRF-1 (but not AdLacZ) transduction also downregulated CHK1 expression (Fig. 2a, b). Immunofluorescent staining for CHK1 (red) in HCC cells induced by IFN-γ or infected by AdIRF-1 confirmed that IRF-1 suppressed basal CHK1 protein expression (Fig. 2e, f). In contrast, silencing of endogenous IRF-1 expression with IRF-1 siRNA in human Huh-7 and HepG2 HCC cells increased basal CHK1 protein expression (Fig. 2c).
Fig. 2. IFN-γ represses CHK1 through IRF-1 in HCC cells.
a Nuclear IRF-1 protein and whole cell lysate for CHK1 protein expression are measured by western blot in Hepa1-6 cells and Huh-7 cells induced by mouse IFN-γ (50 u/ml)/human IFN-γ (100 u/ml) for 24 h, or infected by mouse/human AdIRF-1 (50 MOI) for 48 h, respectively. b Quantification of CHK1 protein expression in Hepa1-6 cells (upper) and Huh-7 cells (lower) are from three independent experiments. Data represent mean ± SD. c IRF-1 protein (40 µg) and CHK1 protein expression are detected by western blot in Huh-7 cells and HepG2 cells transfected with human IRF-1 siRNA or negative control (NC) for 48 h. d Basal CHK1 protein expression (upper) is determined by western blot in liver from IRF-1−/− (KO) or wild-type (WT) B6 mouse, and in Hepa1-6 tumour from WT B6 mouse (T). Quantification of CHK1 protein expression (lower) is from three mice, respectively. e Immunofluorescent images (×400 magnification) of CHK1 (red staining) are shown for Hepa1-6 cells induced by mouse IFN-γ (50 u/ml) for 24 h or infected by mouse AdIRF-1 (50 MOI) for 48 h. f Quantification of CHK1 (AU) from three independent experiments are shown for Hepa1-6 cells induced by IFN-γ or infected by either AdLacZ or AdIRF-1. Data represent mean ± SD. Western blot and immunofluorescent results shown are representative image from three independent experiments.
To decide whether IRF-1 depressed CHK1 in vivo, we injected Hepa1-6 cells subcutaneously into B6 wild-type (WT) mice. CHK1 protein expression is low in normal (WT) liver tissue, increased in IRF-1 KO liver tissue, and markedly increased in subcutaneously grown Hepa1-6 tumours in vivo (T) (Fig. 2d). These findings were consistent with the notion that basal IRF-1 expression constitutively repressed hepatic CHK1 expression.
IFN-γ suppressed CHK1 via IRF-1 by inducing miR-195 in HCC cells
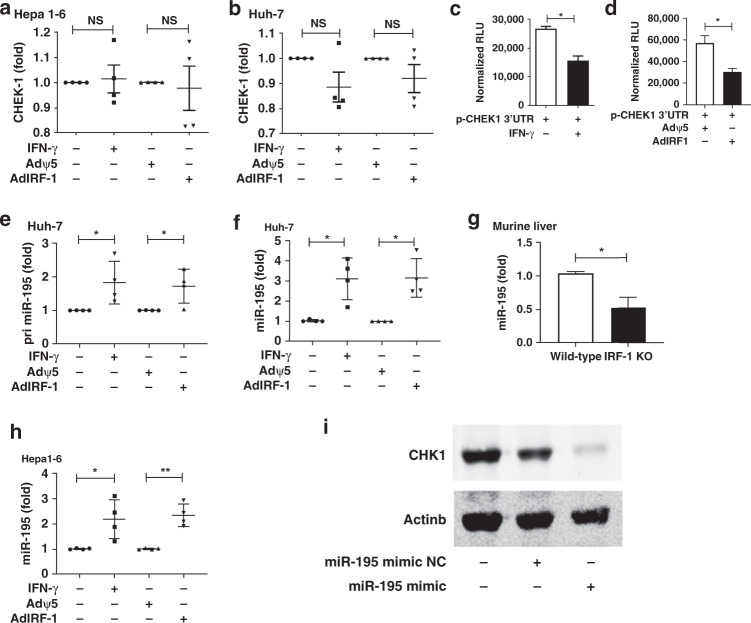
To investigate how IRF-1 suppressed CHK1, we first asked whether IRF-1 regulates CHK1 via a transcriptional mechanism. We analysed CHEK1 mRNA expression by qPCR in HCC cells stimulated by IFN-γ or infected with AdIRF-1 and saw no change in levels of CHEK1 mRNA expression in murine Hepa1-6 (Fig. 3a) or human Huh-7 (Fig. 3b) cells. Since CHK1 protein was diminished by IFN-γ or IRF-1, we investigated for possible post-transcriptional regulation of CHK1 expression by IRF-1. We used a luciferase reporter plasmid containing the CHEK1 3’UTR. After transfection of murine Hepa1-6 cells with p-CHEK1 3’UTR, IFN-γ stimulation or AdIRF-1 (but not Adψ5) transduction significantly decreased CHEK1 luciferase reporter activity (Fig. 3c, d). These data are consistent with post-transcriptional inhibition of CHEK1 mRNA.
Fig. 3. IFN-γ and IRF-1 suppresses CHK1 via miR-195.
CHEK1 mRNA expression is determined by qPCR in Hepa1-6 cells (a) and Huh-7 cells (b) induced by mouse IFN-γ (50 u/ml)/human IFN-γ (100 u/ml) for 24 h, or mouse/human AdIRF-1 (50 MOI) for 48 h, respectively. CHEK1 3’UTR luciferase reporter activities (normalised RLU) are shown in Hepa1-6 cells induced by mouse IFN-γ for 24 h (c) or infected by mouse AdIRF-1 for 24 h (d). Primary miR-195 level (e) and mature miR-195 level (f) are detected by qPCR in Huh-7 cells induced by human IFN-γ (100u/ml) for 24 h or infected by human AdIRF-1(50 MOI) for 48 h. g Basal miR-195 level is measured by qPCR in liver from IRF-1 KO or wild-type B6 mice (n = 3). h MiR-195 level is determined by qPCR in Hepa1-6 cells induced by murine IFN-γ (50u/ml) for 24 h or infected by mouse AdIRF-1 (50 MOI) for 48 h. i CHK1 protein level is measured by western blot in Huh-7 cells transfected with miR-195 mimic or negative control (NC). Each data point (a, b, e, f and h) represents an individual experiment (n = 4). Data represent mean ± SD, *p < 0.05, **p < 0.01. Luciferase assay shown are representative of two individual experiments with similar results. Western blot results shown are representative image of three experiments.
Common post-transcriptional regulation included miRNA binding to the 3’UTR of mRNA. The miR-15/16/195/424/497 family members had been shown to modulate apoptotic pathways and decrease CHK1 protein levels in tumour cells. Therefore, we investigated whether IRF-1 upregulates miR-15 miRNA family members. Both IFN-γ and AdIRF-1, but not Adψ5, upregulated primary (pri-miR-195, Fig. 3e) and mature miR-195 (Fig. 3f) levels in Huh-7 cells. However, miR-15a, miR-16, miR-424 and miR-497 expression were not significantly altered by IFN-γ or AdIRF-1 (Supplementary Fig. 2a–d). Meanwhile, basal miR-195 levels were significantly decreased in IRF-1 KO livers compared to WT (Fig. 3g). These results show that endogenous IRF-1 serves to induce basal hepatic miR-195 since the IRF-1 KO livers had significantly lower basal miR-195 levels. Moreover, IFN-γ stimulation or AdIRF-1 (not Adψ5) transduction induced miR-195 levels in murine Hepa1-6 cells (Fig. 3h).
To confirm that IFN-γ was inducing miR-195 in a transcriptional manner, we blocked transcription with Actinomycin D, and this abrogated the IFN-γ mediated induction of pri-miR-195 and miR-195 in Huh-7 cells (Supplemental Fig. 2e). Furthermore, we analysed two kilobases in the 5’-upstream flanking region of the human miR-195 gene using PROMO bioinformatics software. We identified two putative IRF-1 response elements (IRE1 and IRE2) conserved in both human and murine sequence in the miR-195 promoter at −765 and −220 nucleotides in the 5’-flanking region (Supplemental Fig. 2F).
Since IFN-γ/IRF-1 pathway induced miR-195, and since IFN-γ/IRF-1 inhibited CHEK1 in a post-transcriptional manner, we looked for specific miR-195 binding sites in the CHEK1 mRNA 3’-UTR.
Using TargetScan, miRDB and microrna.org database, we identified a conserved miR-195 binding site at nucleotides 31 to 45 in the CHEK1 mRNA 3’-UTR (Supplemental Fig. 2G). This binding site was contained in our p-CHEK1 3’UTR construct via alignment using the Nucleotide BLAST. To show a functional role for miR-195 in regulating endogenous CHK1 protein expression, miR-195 mimic was added to human Huh-7 HCC cells and markedly decreased constitutive CHK1 protein levels (Fig. 3i). As negative control (NC), miR-195 mimic NC had no significant effect on basal CHK1 protein levels (Fig. 3i). Taken together, these results show that IFN-γ/IRF-1 promotes miR-195 binding to CHEK1 mRNA 3’-UTR to decrease CHK1 protein expression.
Increased IRF-1 and decreased CHK1 induces apoptosis in HCC cells
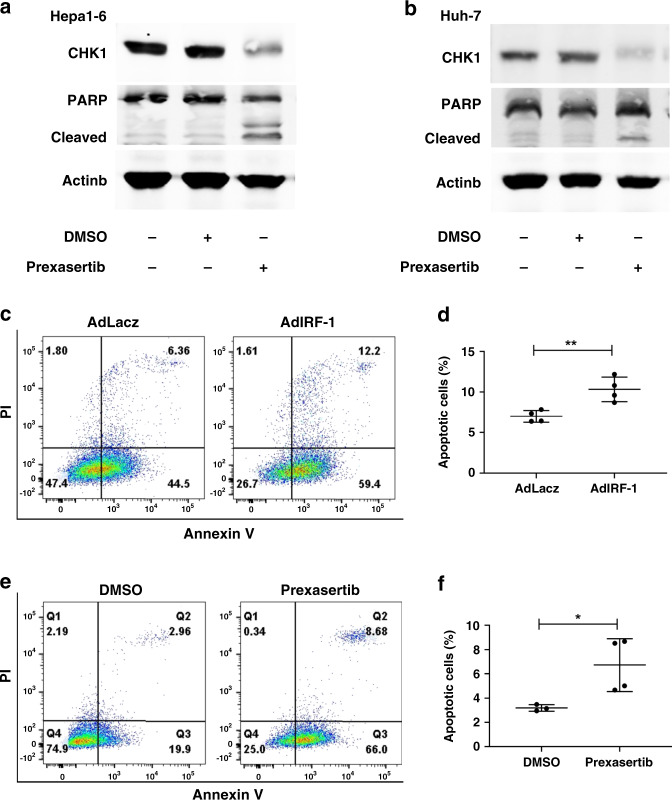
Since CHK1 had been shown to suppress cellular apoptosis,17 we investigated whether CHK1 inhibition with the small molecule inhibitor induced apoptosis in HCC cells. Prexasertib is a known selective, ATP-competitive, small molecular inhibitor of CHK1. It can decrease CHK1 protein expression.40,41 Prexasertib inhibited CHK1 protein levels in Hepa1-6 (Fig. 4a) and Huh-7 cells (Fig. 4b) promoted cleaved PARP expression, a marker of apoptosis. Likewise, AdIRF-1 (but not AdLacZ) also increased cleaved PARP (data not shown). Furthermore, flow cytometry assay showed significantly increased apoptosis in Hepa1-6 cells infected with AdIRF-1 (but not AdLacZ) or treated by prexasertib, respectively (Fig. 4c–f).
Fig. 4. Increased IRF-1 and decreased CHK1 induce apoptosis of HCC cells.
Full length and cleaved PARP protein expressions are determined by western blot in Hepa1-6 cells (a) and Huh-7 cells (b) induced by prexasertib with dose of 1 µM and 5 nM for 24 h, respectively. c Representative images of FACS analysis of late apoptotic (Annexin V+PI+) Hepa1-6 cell rate infected by AdLacz or AdIRF-1 (50 MOI) for 48 h are shown. e Representative image of FACS analysis of late apoptotic Hepa1-6 cell rate treated by DMSO or prexasertib with dose of 1 µM for 24 h are shown. d, f The statistical summary of apoptotic Hepa1-6 cell rate is shown with t-test (n = 4). Data represent mean ± SD, *p < 0.05, **p < 0.01. Western blot image shown are representative of three experiments. FACS assay shown are representative of four independent experiments.
CHK1 inhibitor prexasertib augmented apoptosis of HCC and was associated with NK cell tumour infiltration
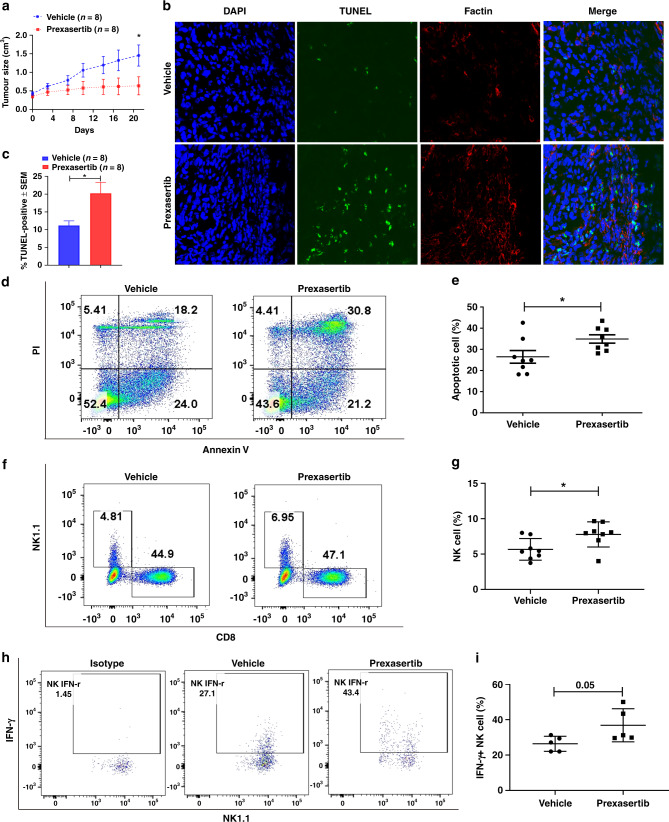
In order to investigate whether CHK1 inhibitor had anti-tumour effects in HCC by inducing an immune response, we injected murine Hepa1-6 cells subcutaneously into B6 mice with/without CHK1 inhibitor prexasertib. HCC tumour growth inhibition was actually observed in 6/8 mice with prexasertib therapy (Supplementary Fig. 3A). The HCC tumour growth curve showed that prexasertib had anti-tumour effects compared to vehicle control (Fig. 5a). TUNEL assay showed increased apoptosis (green staining) in HCC tumours treated by prexasertib (Fig. 5b, c). Likewise, flow cytometry showed significantly augmented apoptosis in HCC tumours after prexasertib treatment (Fig. 5d, e). Furthermore, infiltrating NK cells (but not CD8+T cells) were increased in tumours after prexasertib therapy (Fig. 5f, g, Supplementary Fig. 3B). Immunofluorescent staining for NK1.1 also showed that prexasertib treatment promoted NK cell tumour infiltration (Supplemental Fig. 3D). No significant changes in murine body weight were found during the treatment period (Supplementary Fig. 3C).
Fig. 5. Prexasertib augments cellular apoptosis of murine HCC through promoting tumour infiltrative NK cells.
a Tumour growth curves (Mean ± SEM) of murine HCC model treated by vehicle (n = 8) or prexasertib (10mg/kg, bid, 2 of 7 days, n = 8) are shown. b Representative images of TUNEL assay for cellular apoptosis (green staining) in tumours are shown. c Quantification of the percentage of TUNEL-positive stained cells in tumour treated by vehicle (n = 8) or prexasertib (n = 8). d Representative images of FACS analysis of late apoptotic (Annexin V+PI+) cell rate in single-cell suspensions prepared from tumours treated by vehicle or prexasertib are shown. f Representative image of FACS analysis of NK1.1+ or CD8+ T cell rate in tumours treated by vehicle or prexasertib are shown. The statistical summary of apoptotic tumour cell rate (e) and NK cell rate (g) in tumours treated by vehicle (n = 8) or prexasertib (n = 8) are shown with t-test. h Representative image of FACS analysis of INF-γ+ NK cell rate in tumours treated by vehicle or prexasertib are shown. The statistical summary of IFN-γ+ NK cell rate (i) in tumours treated by vehicle (n = 5) or prexasertib (n = 5) are shown with t-test. Data represent mean ± SD, *p < 0.05.
To decide whether increased NK cells have anti-tumour function in murine HCC treated by prexasertib, we checked the IFN-γ expression in these infiltrating NK cells. The flow cytometry showed modestly increased IFN-γ levels in NK cells from prexasertib-treated HCC (Fig. 5h, i). Therefore, the effect of CHK1 inhibition augmenting cellular apoptosis in murine HCC may in part be caused by increased tumour NK cell infiltration.
Increased IRF-1 and decreased CHK1 induced PD-L1 expression of HCC cells through increased phosphorylated STAT3
We were puzzled why only 6 of 8 HCC tumours were inhibited with prexasertib treatment, which suggested additional signalling pathways were active. IRF-1 has many effects including activation of CD274 (PD-L1 gene) transcription.39,42 IRF-1 phosphorylates STAT3, and phosphorylated STAT3 upregulates IRF-1 via a positive feedback loop.14,42 STAT3 promotes PD-L1 expression in some cancer cells.43,44 The upregulated PD-L1 in cancer cells attenuates anti-tumour immunity by inhibiting effector T cell proliferation, survival, cytokine production, and other effector functions. PD-L1 can also protect tumour cells from the cytotoxic effects of type I and type II interferons and result in reduced NK cell-mediated responses and enhance tumour aggressiveness.45,46
Whether repressed CHK1 upregulates PD-L1 expression remains controversial.12,14,15 One study showed that activated ATR-CHK1 phosphorylated STAT1/3 to upregulate IRF-1 and PD-L1 expression,14 while another observed that repressed CHK1 induced PD-L1 via STING (stimulator of interferon gene) mediated IRF-3.12,15 Therefore, we explored whether upregulated PD-L1 in HCC cells induced by prexasertib attenuates anti-tumour immunity of NK cells.
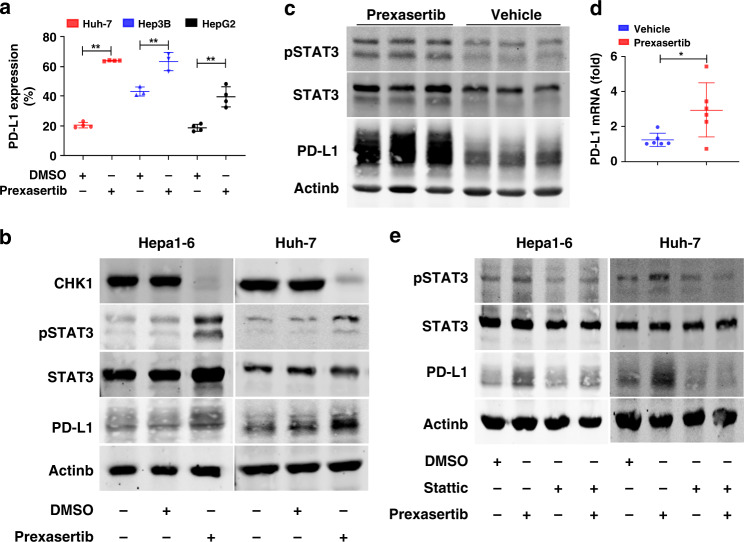
We first analysed STAT3 and PD-L1 and CHEK1 mRNA expression in HCC from the TCGA database through cBioPortal.38 STAT3 and PD-L1 mRNA expression were significantly positively correlated (Spearman: 0.288, p = 2.70 × 10−8). In contrast, STAT3 significantly negatively correlated with CHEK1 mRNA expression (Spearman: −0.24, p = 4.168 × 10−6). We treated a panel of human HCC cell lines with prexasertib for 24 h and analysed PD-L1 expression by flow cytometry. Prexasertib significantly enhanced PD-L1 expression in all cell lines (Fig. 6a, Supplementary Fig. 4A). Furthermore, either adIRF-1 or prexasertib enhanced phosphorylated STAT3 and PD-L1 protein expression in HCC cells (Supplementary Fig. 4B, Fig. 6b). As for in vivo murine HCC tumours, phosphorylated STAT3 and PD-L1 protein expression, as well as PD-L1 mRNA level increased in the prexasertib-treated animals (Fig. 6c, d). Likewise, apparently enhanced phosphorylated STAT3 and PD-L1 level were found in the tumours nonresponsive to prexasertib (Supplementary Fig. 4C).
Fig. 6. Increased IRF-1 and decreased CHK1 induce PD-L1 expression of HCC cells through increased phosphorylated STAT3.
a PD-L1 expressions are measured by flow cytometry in Huh-7, Hep3B and HepaG2 treated by prexasertib with dose of 10 nM for 24 h, respectively. Each data point represents one experiment (n = 3–4). Data represent mean ± SD, **p < 0.01. b CHK1, pSTAT3, STAT3 and PD-L1 protein expression are measured by western blot in Hepa1-6 and Huh-7 cells treated by prexasertib with dose of 1 µM or 10 nM for 24 h, respectively. c pSTAT3, STAT3 and PD-L1 protein expression are detected by western blot in tumours treated by prexasertib (n = 3) or vehicle (n = 3). d PD-L1 mRNA expression is determined by qPCR in tumours treated by prexasertib (n = 6) or vehicle (n = 6). Each data point represents one mouse. Data represent mean ± SD, *p < 0.05. e pSTAT3, STAT3 and PD-L1 are detected by western blot in Hepa1-6 and Huh-7 HCC cells treated with prexasertib and/or Stattic (10 µM) for 24 h. Representative western blot images shown are from three individual experiments.
To confirm CHK1 inhibition upregulated PD-L1 via STAT3 phosphorylation, we treated HCC cells with stattic which inhibits STAT3 phosphorylation (Y705). We found that PD-L1 protein expression was depressed in Hepa1-6 and Huh-7 cells treated with stattic (Fig. 6e). Furthermore, inhibition of STAT3 phosphorylation attenuated the prexasertib upregulated PD-L1 expression in the HCC cells (Fig. 6e). Taken together, CHK1 inhibition upregulated PD-L1 expression in HCC cells in part through STAT3 phosphorylation.
A summary figure in HCC tumour cells under an inflammatory tumour microenvironment shows that IRF-1 downregulates CHK1 by inducing miR-195, which exerts a translational blockade on CHK1.
IFN-γ mediated IRF-1/miR-195 expression decreases CHK1, which leads to enhanced apoptosis of HCC cells. IRF-1 is pleiotropic and also phosphorylates STAT3 to induce PD-L1, which is also observed by CHK1 direct inhibition (Supplementary Fig. 5).
Discussion
In this study, major findings are: (1) CHK1 expression is increased in human HCC tumours and high-CHK1 expression predicts poor prognosis. (2) IRF-1 or IFN-γ suppresses CHK1 expression in HCC cell lines, and basal IRF-1 expression constitutively represses hepatic CHK1 expression in vivo. (3) IFN-γ/IRF-1 mediates CHK1 inhibition in a post-transcriptional manner by inducing miR-195 binding to CHEK1 mRNA 3’-UTR, which produces a translational blockade of CHK1 protein. (4) Both increased IRF-1 and decreased CHK1 can augment apoptosis in murine or human HCC. (5) CHK1 inhibitor prexasertib has anti-tumour effects in murine HCC by increasing tumour apoptosis, mediated in part by increased infiltrating NK cells. (6) Increased IRF-1 or CHK1 blockade can promote PD-L1 expression in HCC cells, regulated in part by phosphorylated STAT3 signalling.
In this study, we show that CHK1 is induced in HCC tumour tissue and that upregulated CHK1 predicts poor clinical outcome of HCC patients. These findings confirm results from a previously published report.16 Since CHK1 is an oncogene, preclinical and clinical trials using ATR-CHK1 pathway inhibitors alone or with chemotherapy have been performed in solid malignant tumours.40,41,47,48 Furthermore, experimental studies have been reported using ATR-CHK1 pathway inhibitors to synergise with immune checkpoint inhibitors in tumours.10–12,15 However, the molecular pathways accounting for CHK1 regulation in HCC are not well-delineated. Our findings shed important new information about the role of the IFN-γ/IRF-1 signalling cascade in regulating CHK1 mediated HCC carcinogenesis.
IRF-1 is a primary transcription factor in the IFN-γ pathway and functions as a tumour suppressor gene promoting apoptosis of cancer cells by modulating key regulators in the apoptotic pathway. Additionally, CHK1 plays an important role in maintaining DNA stability. Overexpressed CHK1 in numerous malignant tumours will assist tumour cell escape from apoptosis caused by chemotherapy or radiotherapy. Our findings show that IRF-1 and CHK1 cooperate to modulate apoptotic pathways in HCC. Except for promoting apoptosis, it was previously reported that IRF-1 suppressed carcinogenesis by reverting c-myc induced cell proliferation and c-myc upregulated CHK1 transcriptional activity.49–51 Thus, it is reasonable that IRF-1 signalling suppresses CHK1 expression and promotes apoptosis of HCC cells.
MiR-195 is one member of miR-15 family that is a conserved small non-coding RNA modulating apoptotic signalling pathway. Our results show that miR-195, but not other members of the miR-15 family, is induced by IFN-γ/IRF-1 signalling and binds to CHEK1 mRNA 3’-UTR producing a post-transcriptional blockade of CHK1 protein in HCC tumour cells. Decreased miR-195 has been observed in a number of cancers, including HCC, colorectal cancer, gastric cancer and lung cancer.52 Analysis of the TCGA database through UALCAN has also shown that miR-195 expression in HCC tumours was lower than normal liver (369 tumour vs 49 normal, p = 1.11E-16).37 The function of miR-195 suppressing CHK1 through binding to CHEK1 mRNA 3’-UTR has been found in colon, gastric and lung cancers.30–32 Our findings extend these observations to HCC and also identify the important role of endogenous IRF-1 in inducing basal miR-195 with two identified IRF-1 response elements (IRE) in the miR-195 promoter region. However, the direct evidence that IRF-1 promotes miR-195 by binding to the specific IRE will be required through future studies.
CHK1 inhibition with prexasertib decreases murine HCC tumour growth in vivo mediated by increasing cleaved PARP and enhanced tumour apoptosis. Increased NK cell tumour infiltration and IFN-γ expression were also observed after CHK1 inhibition with prexasertib. NK cells are pivotal sources of IFN-γ production in the tumour microenvironment. IFN-γ activates IRF-1 signalling, which induces miR-195 and leads to blockade of CHK1 protein expression. However, our findings also show that both IRF-1 expression and CHK1 inhibition induce PD-L1 expression mediated in part by phosphorylated STAT3 activation. The increased PD-L1 expression in HCC can help cancer cells escape from immune surveillance. Hence, IRF-1 expression or CHK1 inhibition can be considered a double-edged sword in tumour biology with increased tumour cell apoptosis vs. attenuation of anti-tumour immunity. These observations lend credence to the rationale for combining molecularly targeted therapeutics CHK1 inhibitors with immune checkpoint blockade to improve therapeutic response rates.
Our study shows that prexasertib exerts anti-tumour effects on murine HCC by increasing infiltrative NK cells, but not CD8+T cells, which differs from previous report where CD8+T cells were required for anti-tumour effects of CHK1 inhibitors in a murine small cell lung cancer model.12,15 NK cells exert innate lymphocyte cytotoxic activity against cancer cells by releasing cytokines (including IFN-γ) and chemokines to activate immune response.46,53 Meanwhile, PD-L1 in cancer cells with decreased MHC I expression can also inhibit NK cell-mediated tumour immunity.46 Immunotherapy of HCC with magnetic PD-1 peptide nanoparticles enhanced the activity of NK cells against HCC cells.54 Others have shown that liver resident NK cells control anti-viral activity of hepatic T Cells via the PD-1/PD-L1 axis.55 Therefore, our results suggest a possible involvement of NK cells in suppressing HCC tumour. However, further studies are required to fully characterise the role of NK cells.
In summary, our data demonstrate CHK1 is increased in human HCC tumours and high-CHK1 levels predicts worse clinical outcome. IFN- γ suppresses CHK1 via IRF-1 in HCC cells. The molecular mechanism of IRF-1 suppressing CHK1 is through promoting miR-195 to bind to CHEK1 mRNA 3’-UTR. Upregulated IRF-1 and inhibited CHK1 induce apoptosis of HCC cells. CHK1 inhibitor augments cellular apoptosis in HCC tumours as well. This effect may be a result of increased infiltrative NK cells. However, IRF-1 and CHK1 inhibitor also upregulate PD-L1 expression via increased phosphorylated STAT3. These findings provide new insights into IRF-1 pathway in modulating CHK1 through small non-coding RNAs, as well as overcoming resistance to molecularly targeted therapeutics in HCC.
Supplementary information
Author contributions
Y.Y. and D.A.G. designed the research. Y.Y., L.Z., Q.D., X.C., K.D. and Y.G. performed research and also analysed the data. Y.Y. and D.A.G. wrote the paper. All authors edited and approved the submission of this work.
Ethics approval and consent to participate
Human tissue samples were obtained in accordance with the University of Pittsburgh Institutional Review Board (IRB) approved protocol (No. MOD08010372/PRO08010372). Animal experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee (Protocol No. 18012053).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Funding information
This work was supported by the NIH Grant (HHSN276201200017C and P30DK120531-01, D.A.G.), and the Guangxi Natural Science Foundation (2017GXNSFAA198014 and 2020GXNSFAA297008, Y.Y.).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yihe Yan, Leting Zheng.
Contributor Information
Yihe Yan, Email: yiheyan@hotmail.com.
David A. Geller, Email: gellerda@upmc.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-021-01337-6.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Sadeghi S, Bejjani A, Finn RS. Systemic therapy for primary liver tumors: cholangiocarcinoma and hepatocellular carcinoma. Surg. Oncol. Clin. N. Am. 2019;28:695–715. doi: 10.1016/j.soc.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Wildi S, Pestalozzi BC, Mccormack L, Clavien PA. Critical evaluation of the different staging systems for hepatocellular carcinoma. Br. J. Surg. 2004;91:400–408. doi: 10.1002/bjs.4554. [DOI] [PubMed] [Google Scholar]
- 4.Clavien PA, Lesurtel M, Bossuyt PM, Gores GJ, Langer B, Perrier A, et al. Recommendations for liver transplantation for hepatocellular carcinoma: an international consensus conference report. Lancet Oncol. 2012;13:e11–e22. doi: 10.1016/S1470-2045(11)70175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montironi C, Montal R, Llovet JM. New drugs effective in the systemic treatment of hepatocellular carcinoma. Clin. Liver Dis. 2019;14:56–61. doi: 10.1002/cld.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marquardt JU, Saborowski A, Czauderna C, Vogel A. The changing landscape of systemic treatment of advanced hepatocellular carcinoma: new targeted agents and immunotherapies. Target Oncol. 2019;14:115–123. doi: 10.1007/s11523-019-00624-w. [DOI] [PubMed] [Google Scholar]
- 7.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin. Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Hunter T. Roles of Chk1 in cell biology and cancer therapy. Int. J. Cancer. 2014;134:1013–1023. doi: 10.1002/ijc.28226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pouliot LM, Chen YC, Bai J, Guha R, Martin SE, Gottesman MM, et al. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012;72:5945–5955. doi: 10.1158/0008-5472.CAN-12-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bever KM, Le DT. DNA repair defects and implications for immunotherapy. J. Clin. Invest. 2018;128:4236–4242. doi: 10.1172/JCI122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shevtsov M, Sato H, Multhoff G, Shibata A. Novel approaches to improve the efficacy of immuno-radiotherapy. Front. Oncol. 2019;9:156. doi: 10.3389/fonc.2019.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9:646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouw KW, Konstantinopoulos PA. From checkpoint to checkpoint: DNA damage ATR/Chk1 checkpoint signalling elicits PD-L1 immune checkpoint activation. Br. J. Cancer. 2018;118:933–935. doi: 10.1038/s41416-018-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sato H, Niimi A, Yasuhara T, Permata TBM, Hagiwara Y, Isono M, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat. Commun. 2017;8:1751. doi: 10.1038/s41467-017-01883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sen T, Della Corte CM, Milutinovic S, Cardnell RJ, Diao L, Ramkumar K, et al. Combination treatment of the oral CHK1 inhibitor, SRA737, and low-dose gemcitabine enhances the effect of programmed death ligand 1 blockade by modulating the immune microenvironment in SCLC. J. Thorac. Oncol. 2019;14:2152–2163. doi: 10.1016/j.jtho.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen G, et al. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J. Clin. Invest. 2012;122:2165–2175. doi: 10.1172/JCI61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 19.Bowie ML, Troch MM, Delrow J, Dietze EC, Bean GR, Ibarra C, et al. Interferon regulatory factor-1 regulates reconstituted extracellular matrix (rECM)-mediated apoptosis in human mammary epithelial cells. Oncogene. 2007;26:2017–2026. doi: 10.1038/sj.onc.1210013. [DOI] [PubMed] [Google Scholar]
- 20.Hong S, Kim HY, Kim J, Ha HT, Kim YM, Bae E, et al. Smad7 protein induces interferon regulatory factor 1-dependent transcriptional activation of caspase 8 to restore tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis. J. Biol. Chem. 2013;288:3560–3570. doi: 10.1074/jbc.M112.400408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stang MT, Armstrong MJ, Watson GA, Sung KY, Liu Y, Ren B, et al. Interferon regulatory factor-1-induced apoptosis mediated by a ligand-independent fas-associated death domain pathway in breast cancer cells. Oncogene. 2007;26:6420–6430. doi: 10.1038/sj.onc.1210470. [DOI] [PubMed] [Google Scholar]
- 22.Kim PK, Armstrong M, Liu Y, Yan P, Bucher B, Zuckerbraun BS, et al. IRF-1 expression induces apoptosis and inhibits tumor growth in mouse mammary cancer cells in vitro and in vivo. Oncogene. 2004;23:1125–1135. doi: 10.1038/sj.onc.1207023. [DOI] [PubMed] [Google Scholar]
- 23.Li P, Du Q, Cao Z, Guo Z, Evankovich J, Yan W, et al. Interferon-gamma induces autophagy with growth inhibition and cell death in human hepatocellular carcinoma (HCC) cells through interferon-regulatory factor-1 (IRF-1) Cancer Lett. 2012;314:213–222. doi: 10.1016/j.canlet.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 27.Cutrona G, Matis S, Colombo M, Massucco C, Baio G, Valdora F, et al. Effects of miRNA-15 and miRNA-16 expression replacement in chronic lymphocytic leukemia: implication for therapy. Leukemia. 2017;31:1894–1904. doi: 10.1038/leu.2016.394. [DOI] [PubMed] [Google Scholar]
- 28.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int. J. Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Wang X, Deng J, Zheng H, Liu W, Chen S, et al. p53-dependent upregulation of miR-16-2 by sanguinarine induces cell cycle arrest and apoptosis in hepatocellular carcinoma. Cancer Lett. 2019;459:50–58. doi: 10.1016/j.canlet.2019.05.042. [DOI] [PubMed] [Google Scholar]
- 30.Kim C, Hong Y, Lee H, Kang H, Lee EK. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett. 2018;412:264–271. doi: 10.1016/j.canlet.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 31.Bargiela-Iparraguirre J, Prado-Marchal L, Fernandez-Fuente M, Gutierrez-Gonzalez A, Moreno-Rubio J, Munoz-Fernandez M, et al. CHK1 expression in Gastric Cancer is modulated by p53 and RB1/E2F1: implications in chemo/radiotherapy response. Sci. Rep. 2016;6:21519. doi: 10.1038/srep21519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Zhang Y, Ma X, Pertsemlidis A. miR-195 potentiates the efficacy of microtubule-targeting agents in non-small cell lung cancer. Cancer Lett. 2018;427:85–93. doi: 10.1016/j.canlet.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokota S, Yoshida O, Dou L, Spadaro AV, Isse K, Ross MA, et al. IRF-1 promotes liver transplant ischemia/reperfusion injury via hepatocyte IL-15/IL-15Ralpha production. J. Immunol. 2015;194:6045–6056. doi: 10.4049/jimmunol.1402505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ueki S, Dhupar R, Cardinal J, Tsung A, Yoshida J, Ozaki KS, et al. Critical role of interferon regulatory factor-1 in murine liver transplant ischemia reperfusion injury. Hepatology. 2010;51:1692–1701. doi: 10.1002/hep.23501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Q, Zhang X, Liu Q, Zhang X, Bartels CE, Geller DA. Nitric oxide production upregulates Wnt/beta-catenin signaling by inhibiting Dickkopf-1. Cancer Res. 2013;73:6526–6537. doi: 10.1158/0008-5472.CAN-13-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan, Y., Zheng, L., Du, Q., Yan, B. & Geller, D. A. Interferon regulatory factor 1 (IRF-1) and IRF-2 regulate PD-L1 expression in hepatocellular carcinoma (HCC) cells. Cancer Immunol. Immunother. 69, 1891–1903 (2020). 10.1007/s00262-020-02586-9. [DOI] [PMC free article] [PubMed]
- 40.Parmar K, Kochupurakkal BS, Lazaro JB, Wang ZC, Palakurthi S, Kirschmeier PT, et al. The CHK1 inhibitor prexasertib exhibits monotherapy activity in high-grade serous ovarian cancer models and sensitizes to PARP inhibition. Clin. Cancer Res. 2019;25:6127–6140. doi: 10.1158/1078-0432.CCR-19-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowery CD, Dowless M, Renschler M, Blosser W, Vanwye AB, Stephens JR, et al. Broad spectrum activity of the checkpoint kinase 1 inhibitor prexasertib as a single agent or chemopotentiator across a range of preclinical pediatric tumor models. Clin. Cancer Res. 2019;25:2278–2289. doi: 10.1158/1078-0432.CCR-18-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding L, Chen X, Xu X, Qian Y, Liang G, Yao F, et al. PARP1 suppresses the transcription of PD-L1 by poly(ADP-Ribosyl)ating STAT3. Cancer Immunol. Res. 2019;7:136–149. doi: 10.1158/2326-6066.CIR-18-0071. [DOI] [PubMed] [Google Scholar]
- 44.Koh J, Jang JY, Keam B, Kim S, Kim MY, Go H, et al. EML4-ALK enhances programmed cell death-ligand 1 expression in pulmonary adenocarcinoma via hypoxia-inducible factor (HIF)-1alpha and STAT3. Oncoimmunology. 2016;5:e1108514. doi: 10.1080/2162402X.2015.1108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault MC, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Invest. 2018;128:4654–4668. doi: 10.1172/JCI99317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karnitz LM, Zou L. Molecular pathways: targeting ATR in cancer therapy. Clin. Cancer Res. 2015;21:4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Angius, G., Tomao, S., Stati, V., Vici, P., Bianco, V. & Tomao, F. Prexasertib, a checkpoint kinase inhibitor: from preclinical data to clinical development. Cancer Chemother. Pharmacol. 85, 9–20 (2020). 10.1007/s00280-019-03950-y. [DOI] [PubMed]
- 49.Tanaka N, Ishihara M, Taniguchi T. Suppression of c-myc or fosB-induced cell transformation by the transcription factor IRF-1. Cancer Lett. 1994;83:191–196. doi: 10.1016/0304-3835(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 50.Kroger A, Dallugge A, Kirchhoff S, Hauser H. IRF-1 reverts the transformed phenotype of oncogenically transformed cells in vitro and in vivo. Oncogene. 2003;22:1045–1056. doi: 10.1038/sj.onc.1206260. [DOI] [PubMed] [Google Scholar]
- 51.Wang WJ, Wu SP, Liu JB, Shi YS, Huang X, Zhang QB, et al. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013;73:1219–1231. doi: 10.1158/0008-5472.CAN-12-1408. [DOI] [PubMed] [Google Scholar]
- 52.Yu W, Liang X, Li X, Zhang Y, Sun Z, Liu Y, et al. MicroRNA-195: a review of its role in cancers. Onco Targets Ther. 2018;11:7109–7123. doi: 10.2147/OTT.S183600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee, M. H., Liu, K. H., Thomas, J. L., Chen, J. R. & Lin, H. Y. Immunotherapy of hepatocellular carcinoma with magnetic PD-1 peptide-imprinted polymer nanocomposite and natural killer cells. Biomolecules. 9, 651 (2019). [DOI] [PMC free article] [PubMed]
- 55.Zhou J, Peng H, Li K, Qu K, Wang B, Wu Y, et al. Liver-resident NK cells control antiviral activity of hepatic T cells via the PD-1-PD-L1 axis. Immunity. 2019;50:403–17 e4. doi: 10.1016/j.immuni.2018.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.