Abstract
To investigate the relationship between thrombomodulin (THBD) gene single nucleotide polymorphisms (SNPs) and susceptibility to sepsis and the occurrence and prognosis of acute kidney injury (AKI) in sepsis patients.
The genotypes of THBD gene rs1962, rs3176123, and rs1042580 in 178 sepsis patients with AKI, 243 sepsis patients without AKI (No AKI), and 103 healthy controls were analyzed by direct sequencing. Enzyme-linked immunosorbent assay (ELISA) was used to detect the plasma THBD protein levels. Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of plasma THBD levels in sepsis, AKI, and death of sepsis patients.
The C allele carriers of THBD gene rs1962 were more likely to develop AKI and sepsis than the T allele carriers (OR = 1.61, 95% CI: 1.18–2.19, P < .01; OR = 2.16, 95% CI: 1.42–3.29, P < .01). The rs3176123 G allele was associated with an increased risk of AKI in sepsis patients (OR = 1.41, 95% CI: 1.06–1.88, P = .02), the G allele had a significant association with a higher risk of sepsis susceptibility (OR = 1.91, 95% CI: 1.33–2.75, P < .01). Sepsis patients of rs1042580 C allele had a lower risk of AKI than those of T allele (OR = 0.58, 95% CI: 0.37–0.91, P = .02), the C allele was related to a reduced risk of sepsis susceptibility (OR = 0.38, 95% CI: 0.26–0.55, P < .01). The THBD gene rs1962, rs3176123, and rs1042580 TGT haplotype was linked to higher risk of AKI in patients with sepsis (OR = 1.96, 95%CI: 1.14–3.38, P = .02). Sepsis patients with the THBD gene rs1962 TC + CC genotype had a higher risk of death than those with TT genotype (OR = 10.93, 95%CI: 5.05–26.96, P < .01), but there was no significant difference in the risk of death in sepsis patients with different genotypes at rs3176123 and rs1042580 (P > .05).
The THBD gene rs1962, rs3176123, and rs1042580 SNPs are significantly associated with sepsis susceptibility and the risk of AKI. The rs1962 SNP is related to the risk of death in sepsis patients.
Keywords: acute kidney injury, sepsis, single nucleotide polymorphism, thrombomodulin
1. Introduction
Sepsis is defined as a life-threatening organ dysfunction due to a dysregulated host response to infection,[1] and acute kidney injury (AKI) is the most common organ dysfunction in sepsis. Previous studies have shown that sepsis patients with AKI accounted for 26% to 50% of hospitalized patients,[2] with a higher proportion of intensive care patients.[3] The occurrence of AKI in sepsis patients prolonged hospitalization time, increased hospitalization costs, and the risk of death in patients, which was an independent risk factor for death among ICU patients.[4,5] Studies have reported that the high mortality rate of AKI in sepsis patients is not only related to the severity of the disease, but also to the early prevention of AKI.[6] Therefore, it is particularly important to predict AKI in patients with sepsis early and take preventive measures as soon as possible.
Thrombomodulin (THBD) is a glycoprotein mainly expressed on the surface of endothelial cells, which is also expressed in platelets, monocytes, smooth muscle cells, cancer cells, and cardiomyocytes. It has anti-inflammatory and anticoagulant effects. When sepsis occurs, changes in its structure and function lead to changes in coagulation, anticoagulation system imbalance, and anti-inflammatory effects, and the genetic factors also contribute to the abnormal structure and function of THBD.[6,7] The THBD gene is located on human chromosome 20p11.21 with a length of 6.1 kb, containing 1 exon and no intron. There are several types of THBD site mutations:
-
1.
Mutation of coding gene sequence, including G236C, G543A, C1418T, G1456T, G127A, G129C, C1483T, C1502T, and 1689 insertion T[8];
-
2.
Mutation of the distal sequence of regulatory genes[9];
-
3.
Mutation of the proximal sequence of regulatory genes, including G-33A, C-133A, and -10GG/AT.[10]
The correlation between THBD gene polymorphism and the occurrence of sepsis and AKI has not been reported.The THBD gene Tag single nucleotide polymorphisms (SNPs) rs1962, rs3176123, and rs1042580 from the 1000 Genomes Project database (http://www.1000genomes.org/) were selected in our study (Figure S1, Supplemental Digital Content), based on the minor allele frequency >0.05 of these SNPs and relevant researches. It has been reported that the rs1962 SNP was associated with the risk of death of steroid-refractory graft-versus-host disease,[11] SNP at rs3176123 was related to the THBD expression and graft-versus-host disease,[12] the rs1042580 served as a predictor for warfarin bleeding complications under international normalized ratio.[13]
In this study, we analyzed the correlation between the THBD gene rs1962, rs3176123, and rs1042580 SNPs and the susceptibility to sepsis in Chinese Han people and the risk of AKI.
2. Materials and methods
2.1. Samples
A total of 421 patients with sepsis treated in the ICU of the Zibo Central Hospital and Tiantai people's Hospital were enrolled in this study, including 178 AKI patients and 243 No AKI patients. The diagnostic criteria of sepsis referred to International Sepsis Definitions Conference.[14] In accordance with the diagnostic criteria of Kidney Disease: Improving Global Outcomes (KDIGO),[15] AKI was diagnosed accompanied by one of the following indicators:
-
1.
The absolute value of Scr rises ≥ 26.5 μmol/L within 48 hours;
-
2.
Scr increased 1.5 times to baseline within 7 days;
-
3.
Urine output <0.5 mL/kg/h for more than 6 hours.
The sepsis patients with AKI were divided into a death group (56 cases) and a survival group (122 cases) according to whether the AKI patients died 30 days after the onset. Totally 103 healthy subjects who had no physical relationship with sepsis patients were randomly selected as the control group. A self-made questionnaire was used to collect the clinical data of the subjects such as age, body mass index (BMI), gender, acute physiology and chronic health evaluation III score,[16] Simplified Acute Physiology Score II,[17] Sequential Organ Failure Assessment score,[18] source of infection and pathogens. Exclusion criteria:
-
1.
<18 years old or >80 years old;
-
2.
Patients with cerebral infarction;
-
3.
Cancer patients;
-
4.
Patients with immune system deficiency diseases.
This study was approved by the Medical Ethics Committee of Zibo Central Hospital and Tiantai people's Hospital, and all subjects signed an informed consent.
2.2. Genomic DNA extraction and sequencing
Peripheral venous blood (2 mL) was collected from the subjects, and genomic DNA was isolated from the blood using QIAamp DNA blood mini kit (Qiagen, Valencia, CA) according to the supplier's instructions. Primer 3.0 was used to design SNP site amplification primers on basis of the SNP sequence information in dbSNP (https://www.ncbi.nlm.nih.gov/snp/) and then synthesized by Shanghai Tianhao Biological Company (Shanghai, China). Primers for rs1962 were as follows: forward primer: 5’-AGCTTTGAAAATCAGAGATGGTGC-3’; reverse primer: 5’-TATGAGATGCATGGAGGGCTG-3’. Primers for rs3176123 were: forward primer: 5’-AACTGTCTCCTGGGAGGTGT-3’; reverse primer: 5’-ACAGTGTTGAAAATGTTCAGAAGG-3’. Primers for rs1042580 were: forward primer: 5’-AGCAACAAAATTCAGGCTCACT-3’; reverse primer: 5’-GGCCATTTGCTTTTTCACCAGA-3’. The extracted genomic DNA was used as a template for PCR amplification. The reaction mixture contained 12.5 μL of Premix Taq, 10 mM of each forward and reverse primer, 20 ng of genomic DNA, and the volume was made up to 25 μL with sterilized purified water. The PCR conditions were 94 °C for 5 minutes, followed by 30 cycles of 94 °C for 25 seconds, 58 °C for 25 seconds, 72 °C for 25 seconds and then 72 °C for 10 minutes. The PCR products were sequenced by ABI 3730xl sequencer (Applied Biosystems, Foster City, CA), and the SNP genotypes of the subjects were determined according to the sequencing results.
2.3. Enzyme-linked immunosorbent assay (ELISA)
The venous blood of the subjects was collected, stored at room temperature for 2 hours, centrifuged at 1000 r/min for 20 minutes, and the plasma was separated. Human THBD ELISA Kit (Abcam, Cambridge, UK) was utilized to detect plasma THBD protein levels, and the standard curve method was applied to quantify THBD levels. All the operations were performed in strict accordance with the instructions.
2.4. Statistical analysis
Continuous variables were expressed as mean ± standard deviation, and t test and 1-way analysis of variance were used for statistical analysis. Categorical variables were represented by frequency, and the χ2 test was used to analyze differences between groups. Hardy Weinberg equilibrium was tested by χ2 test. Logistic regression was performed to assess the correlation between the SNP of the THBD gene at rs1962, rs3176123, and rs1042580 and the susceptibility to sepsis and the risk of AKI, the odds ratio (OR) and 95% confidence interval (CI) were calculated, and the age, BMI and gender were adjusted. Haploview4.1 software was utilized to analyze the linkage disequilibrium of rs1962, rs3176123, and rs1042580 of THBD gene and construct haplotypes. The value of plasma THBD levels in the diagnosis of sepsis and AKI and the death of sepsis patients was evaluated by the receiver operating characteristic (ROC) curve with a cut-off value = sensitivity + specificity − 1. All tests were 2-tailed, and P < .05 was considered as statistically significant.
3. Results
3.1. Comparison of clinical data
The clinical data of 178 AKI patients, 243 No AKI patients, and 103 healthy controls were shown in Table 1. There were no statistical differences in age, BMI, and gender in the 3 groups (P > .05). The acute physiology and chronic health evaluation III score, Simplified Acute Physiology Score II score, and Sequential Organ Failure Assessment of AKI patients were significantly higher than those of No AKI patients, and the difference was statistically significant (P < .05). No significant difference was observed in the source of infection by the type of pathogens between AKI and No AKI patients (P > .05).
Table 1.
Comparison of clinical data between AKI patients, No AKI patients, and healthy controls.
| Characteristics | AKI (n = 178) | No AKI (n = 243) | Control (n = 103) |
| Age | 62.31 ± 7.87 | 62.62 ± 7.08 | 62.58 ± 8.82 |
| BMI (kg/m2) | 26.18 ± 1.74 | 26.24 ± 2.90 | 26.30 ± 1.87 |
| Gender | |||
| Male | 101 (56.74%) | 140 (57.61%) | 58 (56.31%) |
| Female | 77 (43.26%) | 103 (42.39%) | 45 (43.69%) |
| APACHE III score | 86.49 ± 8.43∗ | 67.36 ± 10.12 | |
| SAPS II score | 46.69 ± 10.60∗ | 39.51 ± 9.88 | |
| SOFA | 6.65 ± 1.46∗ | 4.81 ± 1.98 | |
| Source of infection, n (%) | |||
| Respiratory tract infection | 130 (73.03%) | 182 (74.90%) | |
| Primary bloodstream infection | 64 (35.96%) | 85 (34.98%) | |
| Wound infection | 29 (16.29%) | 41 (16.87%) | |
| Abdominal infection | 66 (37.08%) | 89 (36.63%) | |
| Urinary tract infection | 71 (39.89%) | 98 (40.33%) | |
| Catheter-associated infection | 32 (17.98%) | 38 (15.64%) | |
| Others | 26 (14.61%) | 30 (12.35%) | |
| Pathogens (positive blood cultures) (n [%]) | |||
| Gram-negative | 68 (38.20%) | 99 (40.74%) | |
| Gram-positive | 35 (19.66%) | 32 (13.17%) | |
| Mixed Gram-negative and -positive | 55 (30.90%) | 84 (34.57%) | |
| Fungus | 20 (11.24%) | 27 (11.11%) |
AKI = acute kidney injury, BMI = body mass index, APACHE = acute physiology and chronic health evaluation, SAPS = simplified acute physiology score, SOFA = sequential organ failure assessment.
P < .05, compared with No AKI.
3.2. Correlation between THBD gene polymorphism and sepsis susceptibility and AKI risk
The genotypes of the THBD gene rs1962, rs3176123, and rs1042580 in the control group in this study were in line with Hardy Weinberg equilibrium (P = .11, .13, .54) (Table 2).
Table 2.
Correlation of TM gene, SNP genotypes, and allele frequency with sepsis susceptibility and AKI risk.
| AKI (n = 178) | No AKI (n = 243) | Control (n = 103) | OR (95%CI)†,∗ | P | OR (95%CI)‡,∗ | P∗ | |
| rs1962 | |||||||
| TT | 101 (56.74%) | 154 (63.37%) | 78 (75.73%) | 1.00 | 1.00 | ||
| TC | 42 (23.60%) | 70 (28.81%) | 21 (20.39%) | 0.92 (0.58–1.45) | .79 | 1.63 (0.96–2.77) | .09 |
| CC | 35 (19.66%) | 19 (7.82%) | 4 (3.88%) | 2.81 (1.52–5.18) | <.01 | 4.13 (1.45–11.76) | <.01 |
| TC + CC | 77 (43.26%) | 89 (36.63%) | 25 (24.27%) | 1.32 (0.89–1.96) | .20 | 2.03 (1.24–3.32) | <.01 |
| TT + TC | 143 (80.34%) | 224 (92.18%) | 99 (96.12%) | 1.00 | 1.00 | ||
| CC | 35 (19.66%) | 19 (7.82%) | 4 (3.88%) | 2.89 (1.59–5.24) | <.01 | 3.64 (1.29–10.30) | .02 |
| T | 244 (68.54%) | 378 (77.78%) | 177 (85.92%) | 1.00 | 1.00 | ||
| C | 112 (31.46%) | 108 (22.22%) | 29 (14.08%) | 1.61 (1.18–2.19) | <.01 | 2.16 (1.42–3.29) | <.01 |
| rs3176123 | |||||||
| TT | 85 (47.75%) | 133 (54.73%) | 67 (65.05%) | 1.00 | 1.00 | ||
| TG | 51 (28.65%) | 73 (30.04%) | 29 (28.16%) | 1.09 (0.70–1.71) | .78 | 1.31 (0.81–2.14) | .33 |
| GG | 42 (23.60%) | 37 (15.23%) | 7 (6.80%) | 1.78 (1.06–2.99) | .04 | 3.47 (1.53–7.88) | <.01 |
| TG + GG | 93 (52.25%) | 110 (45.27%) | 36 (34.95%) | 1.32 (0.90–1.95) | .19 | 1.73 (1.11–2.71) | .02 |
| TT + TG | 136 (76.40%) | 206 (84.77%) | 96 (93.20%) | 1.00 | 1.00 | ||
| GG | 42 (23.60%) | 37 (15.23%) | 7 (6.80%) | 1.72 (1.05–2.81) | .04 | 3.17 (1.42–7.09) | <.01 |
| T | 221 (62.08%) | 339 (69.75%) | 163 (79.13%) | 1.00 | 1.00 | ||
| G | 135 (37.92%) | 147 (30.25%) | 43 (20.87%) | 1.41 (1.06–1.88) | .02 | 1.91 (1.33–2.75) | <.01 |
| rs1042580 | |||||||
| TT | 151 (84.83%) | 187 (76.95%) | 58 (56.31%) | 1.00 | 1.00 | ||
| TC | 24 (13.48%) | 45 (18.52%) | 37 (35.92%) | 0.66 (0.39–1.13) | .17 | 0.32 (0.20–0.52) | <.01 |
| CC | 3 (1.69%) | 11 (4.53%) | 8 (7.77%) | 0.34 (0.09–1.23) | .15 | 0.30 (0.12–0.75) | .02 |
| TC + CC | 27 (15.17%) | 56 (23.05%) | 45 (43.69%) | 0.60 (0.36–0.99) | .06 | 0.32 (0.20–0.50) | <.01 |
| TT + TC | 175 (98.31%) | 232 (95.47%) | 95 (92.23%) | 1.00 | 1.00 | ||
| CC | 3 (1.69%) | 11 (4.53%) | 8 (7.77%) | 0.36 (0.10–1.32) | .18 | 0.41 (0.17–1.00) | .08 |
| T | 326 (91.57%) | 419 (86.21%) | 153 (74.27%) | 1.00 | 1.00 | ||
| C | 30 (8.43%) | 67 (13.79%) | 53 (25.73%) | 0.58 (0.37–0.91) | .02 | 0.38 (0.26–0.55) | <.01 |
AKI = acute kidney injury, OR = odds ratio, CI = confidence interval.
The risk of AKI.
The risk of sepsis susceptibility.
Adjusted age, BMI, gender.
There was no correlation between the rs1962 TC genotype and susceptibility to sepsis and the risk of developing AKI in patients with sepsis (P = .79, .09). CC genotype and recessive model (CC vs TT + TC) were associated with increased risk of AKI in sepsis patients (OR = 2.81, 95% CI: 1.52–5.18, P < .01; OR = 2.89, 95% CI: 1.59–5.24, P < .01). CC genotype, dominant model (TC + CC vs TT), and recessive model (CC vs TT + TC) were related to a higher risk of sepsis susceptibility (OR = 4.13, 95% CI: 1.45–11.76, P < .01; OR = 2.03, 95% CI: 1.24–3.32, P < .01; OR = 3.64, 95% CI: 1.29–10.30, P = .02). Carriers of C allele have a higher risk of AKI and susceptibility to sepsis than those of T allele (OR = 1.61, 95% CI: 1.18–2.19, P < .01; OR = 2.16, 95% CI: 1.42–3.29, P < .01).
No significant correlation was found between the TG genotype and dominant model (TG + GG vs TT) at rs3176123 and the risk of AKI in patients with sepsis (P = .78, 0.19), while the GG genotype, recessive model (GG vs TT + TG) and G allele were related to an increased risk of developing AKI in sepsis patients (OR = 1.78, 95% CI: 1.06–2.99, P = .04; OR = 1.72, 95% CI: 1.05–2.81, P = .04; OR = 1.41, 95% CI: 1.06–1.88, P = .02). There was no significant correlation between the TG genotype at rs3176123 and the risk of sepsis susceptibility (P = .33). GG genotype, dominant model (TG + GG vs TT), recessive model (GG vs TT + TG), and G allele had a significant relationship with elevated risk of sepsis susceptibility (OR = 3.47, 95% CI: 1.53–7.88, P < .01; OR = 1.73, 95% CI: 1.11–2.71, P = .02; OR = 3.17, 95% CI: 1.42–7.09, P < .01; OR = 1.91, 95% CI: 1.33–2.75, P < .01).
The TC genotype, CC genotype, dominant model (TC + CC vs TT), and recessive model (CC vs TT + TC) of rs1042580 were not markedly related to the risk of having AKI in patients with sepsis (P = .17, 0.15, 0.06, 0.18). The C allele carriers were less likely to develop AKI than T allele carriers among sepsis patients (OR = 0.58, 95% CI: 0.37–0.91, P = .02), but the TC genotype, CC genotype, dominant model (TC + CC vs TT), and C allele were associated with reduced risk of sepsis susceptibility (OR = 0.32, 95% CI: 0.20–0.52, P < .01; OR = 0.30, 95% CI: 0.12–0.75, P = .02; OR = 0.32, 95% CI: 0.20–0.50, P < .01; OR = 0.38, 95% CI: 0.26–0.558, P < .01). Recessive model (CC vs TT +TC) was not significantly correlated with sepsis susceptibility (P = .08).
3.3. The stratified analysis of the correlation between the risk of AKI in sepsis patients and THBD gene polymorphism
A stratified analysis of the age, BMI, and gender of sepsis patients was conducted, and the results showed that there was no significant change in the risk of AKI in TC genotype carriers at rs1962 in population aged< 60 years (P = .22), while the carriers with CC genotype had a higher risk of having AKI compared with those with TT genotype (OR = 3.21, 95% CI: 1.42–7.26, P < .01). However, in patients with sepsis aged ≥60 years, there was no significant difference in the risk of AKI in carriers with different genotypes at rs1962 (P > .05). In sepsis patients with BMI < 26 kg/m2 and BMI ≥ 26 kg/m2, an elevated risk of developing AKI was found in carriers of CC genotype at rs1962 (OR = 2.47, 95% CI: 1.09–5.57, P = .04; OR = 3.44, 95% CI: 1.33–8.87, P = .02). No significant difference was observed in the risk of AKI among sepsis patients with different genotypes at rs1962 in males, but there was a significantly higher risk of AKI in sepsis patients with CC genotype at rs1962 in females (OR = 2.17, 95% CI: 1.49–2.68, P < .01) (Table 3).
Table 3.
The stratified analysis of the correlation between TM gene rs1962 genotype frequency and AKI risk.
| AKI(n = 178) | No AKI(n = 243) | OR (95%CI)∗ | P | |
| Age | ||||
| <60 | ||||
| TT | 56 (58.66%) | 75 (62.50%) | 1.00 | |
| TC | 16 (16.67%) | 35 (29.17%) | 0.61 (0.31–1.22) | .22 |
| CC | 24 (25.00%) | 10 (8.33%) | 3.21 (1.42–7.26) | <.01 |
| ≥60 | ||||
| TT | 45 (54.88%) | 79 (64.23%) | 1.00 | |
| TC | 26 (31.71%) | 35 (28.46%) | 1.30 (0.70–2.44) | .50 |
| CC | 11 (13.41%) | 9 (7.32%) | 2.15 (0.83–5.57) | .18 |
| BMI (kg/m2) | ||||
| <26 | ||||
| TT | 48 (53.93%) | 79 (60.77%) | 1.00 | |
| TC | 23 (25.84%) | 39 (30.00%) | 0.97 (0.52–1.82) | .93 |
| CC | 18 (20.22%) | 12 (9.23%) | 2.47 (1.09–5.57) | .04 |
| ≥26 | ||||
| TT | 53 (59.55%) | 75 (66.37%) | 1.00 | |
| TC | 19 (21.35%) | 31 (27.43%) | 0.87 (0.44–1.70) | .81 |
| CC | 17 (19.10%) | 7 (6.19%) | 3.44 (1.33–8.87) | .02 |
| Gender | ||||
| Male | ||||
| TT | 59 (58.42%) | 83 (59.29%) | 1.00 | |
| TC | 28 (27.72%) | 43 (30.71%) | 0.92 (0.51–1.64) | .88 |
| CC | 14 (13.86%) | 14 (10.00%) | 1.41 (0.62–3.17) | .54 |
| Female | ||||
| TT | 42 (54.55%) | 71 (68.93%) | 1.00 | |
| TC | 14 (18.18%) | 27 (26.21%) | 0.88 (0.41–1.86) | .88 |
| CC | 21 (27.27%) | 5 (4.85%) | 2.17 (1.49–2.68) | <.01 |
AKI = acute kidney injury, BMI = body mass index, OR = odds ratio, CI = confidence interval.
Adjusted age, BMI, gender.
We found that the risk of AKI was significantly increased only in female sepsis patients with the GG genotype at rs3176123 (OR = 2.81, 95% CI: 1.30–6.11, P = .01), and there was no significant change in the AKI risk of carriers with different genotypes at rs3176123 in the population stratified by other clinical data (P > .05) (Table 4).
Table 4.
The stratified analysis of the correlation between TM gene rs3176123 genotype frequency and AKI risk.
| AKI (n = 178) | No AKI (n = 243) | OR (95%CI)∗ | P | |
| Age | ||||
| <60 | ||||
| TT | 46 (47.92%) | 66 (55.00%) | 1.00 | |
| TG | 25 (26.04%) | 36 (30.00%) | 0.99 (0.53–1.88) | .99 |
| GG | 25 (26.04%) | 18 (15.00%) | 1.99 (0.98–4.07) | .08 |
| ≥60 | ||||
| TT | 39 (47.56%) | 67 (54.47%) | 1.00 | |
| TG | 26 (31.71%) | 37 (30.08%) | 1.21 (0.64–2.29) | .68 |
| GG | 17 (20.73%) | 19 (15.45%) | 1.54 (0.72–3.30) | .36 |
| BMI (kg/m2) | ||||
| <26 | ||||
| TT | 44 (49.44%) | 74 (56.92%) | 1.00 | |
| TG | 22 (24.72%) | 36 (27.69%) | 1.03 (0.54–1.97) | .93 |
| GG | 23 (25.84%) | 20 (15.38%) | 1.93 (0.96–3.92) | .10 |
| ≥26 | ||||
| TT | 41 (46.07%) | 59 (52.21%) | 1.00 | |
| TG | 29 (32.58%) | 37 (32.74%) | 1.13 (0.60–2.12) | .83 |
| GG | 19 (21.35%) | 17 (15.04%) | 1.61 (0.75–3.46) | .31 |
| Gender | ||||
| Male | ||||
| TT | 53 (52.48%) | 79 (56.43%) | 1.00 | |
| TG | 31 (30.69%) | 39 (27.86%) | 1.19 (0.67–2.13) | .68 |
| GG | 17 (16.83%) | 22 (15.71%) | 1.15 (0.56–2.37) | .84 |
| Female | ||||
| TT | 32 (41.56%) | 54 (52.43%) | 1.00 | |
| TG | 20 (25.97%) | 34 (33.01%) | 0.99 (0.49–2.01) | .98 |
| GG | 25 (32.47%) | 15 (14.56%) | 2.81 (1.30–6.11) | .01 |
AKI = acute kidney injury, BMI = body mass index, OR = odds ratio, CI = confidence interval.
Adjusted age, BMI, gender.
No significant correlation was observed between sepsis patients with different genotypes at rs1042580 and the risk of AKI (P > .05) (Table 5).
Table 5.
The stratified analysis of the correlation between TM gene rs1042580 genotype frequency and AKI risk.
| AKI (n = 178) | No AKI (n = 243) | OR (95%CI)∗ | P | |
| Age | ||||
| <60 | ||||
| TT | 84 (87.50%) | 88 (73.33%) | 1.00 | |
| TC | 12 (12.50%) | 25 (20.83%) | 0.50 (0.24–1.07) | .10 |
| CC | 0 (0%) | 7 (5.83%) | \ | \ |
| ≥60 | ||||
| TT | 67 (81.71%) | 99 (80.49%) | 1.00 | |
| TC | 12 (14.63%) | 20 (16.26%) | 0.89 (0.41–1.93) | .92 |
| CC | 3 (3.66%) | 4 (3.25%) | 1.11 (0.24–5.11) | .90 |
| BMI (kg/m2) | ||||
| <26 | ||||
| TT | 76 (85.39%) | 105 (80.77%) | 1.00 | |
| TC | 11 (12.36%) | 21 (16.15%) | 0.72 (0.33–1.59) | .54 |
| CC | 2 (2.25%) | 4 (3.08%) | 0.69 (0.12–3.87) | .67 |
| ≥26 | ||||
| TT | 75 (84.27%) | 82 (72.57%) | 1.00 | |
| TC | 13 (14.61%) | 24 (21.24%) | 0.59 (0.28–1.25) | .23 |
| CC | 1 (1.12%) | 7 (6.19%) | 0.16 (0.02–1.30) | .11 |
| Gender | ||||
| Male | ||||
| TT | 86 (85.15%) | 108 (77.14%) | 1.00 | |
| TC | 13 (12.87%) | 27 (19.29%) | 0.61 (0.29–1.24) | .23 |
| CC | 2 (1.98%) | 5 (3.57%) | 0.50 (0.10–2.65) | .66 |
| Female | ||||
| TT | 65 (84.42%) | 79 (76.70%) | 1.00 | |
| TC | 11 (14.29%) | 18 (17.48%) | 0.74 (0.33–1.68) | .61 |
| CC | 1 (1.30%) | 6 (5.83%) | 0.20 (0.02–1.73) | .22 |
AKI = acute kidney injury, BMI = body mass index, OR = odds ratio, CI = confidence interval.
Adjusted age, BMI, gender.
3.4. Haplotype analysis
Haploview 4.1 (http://www.broad.mit.edu/mpg/haploview) was used to analyze the correlation between the haplotypes of THBD gene rs1962, rs3176123, and rs1042580 and the risk of AKI (Table 6). The results exhibited that the TGT haplotype was associated with an increased risk of developing AKI in sepsis patients (OR = 1.96, 95% CI: 1.14–3.38, P = .02), while TTC and CTT haplotypes were not significantly correlated with the risk of AKI (P > .05) (Table 6).
Table 6.
Correlation between the haplotypes of TM gene rs1962, rs3176123, rs1042580, and the risk of AKI.
| Haplotype | AKI (n = 178) | No AKI (n = 243) | OR (95%CI)∗ | P |
| TTT | 28.65% | 31.69% | 1.00 | |
| TTC | 19.66% | 30.86% | 0.71 (0.41–1.20) | .25 |
| CTT | 22.47% | 20.86% | 1.18 (0.69–2.04) | .64 |
| TGT | 29.21% | 16.46% | 1.96 (1.14–3.38) | .02 |
AKI = acute kidney injury, OR = odds ratio, CI = confidence interval.
Adjusted age, BMI, gender.
3.5. Correlation between plasma THBD level and sepsis AKI
ELISA analysis of plasma THBD levels in sepsis patients and healthy controls showed that plasma THBD level in sepsis patients was significantly higher than that in control group (P < .01, Fig. 1A). The area under the curve (AUC) of the ROC of plasma THBD level to diagnose sepsis was 0.79 (95% CI: 0.74–0.846, P < .01), and the cut-off value was 5.65 ng/mL (Fig. 1B). The plasma THBD level in patients with sepsis was higher than that in patients with No AKI, with significant difference (P < .01, Fig. 1C). The AUC of plasma THBD level for diagnosis of AKI in sepsis was 0.61 (95% CI: 0.55–0.66, P < .01), and the cut-off value was 6.65 ng/mL (Fig. 1D).
Figure 1.
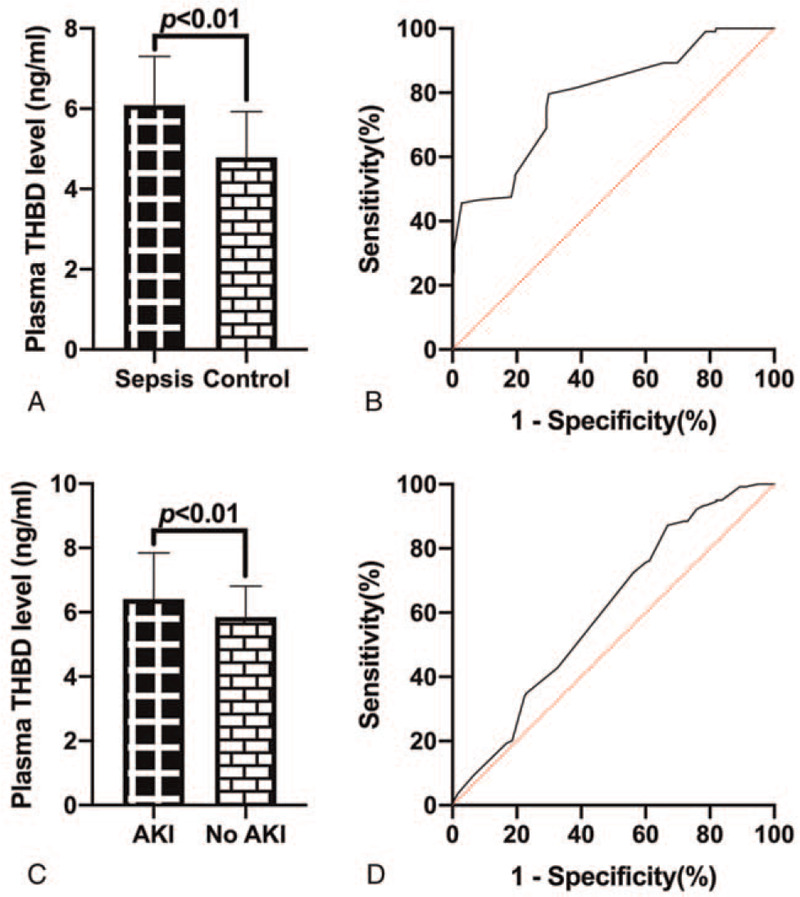
ELISA analysis of plasma THBD levels. (A) The plasma THBD level of sepsis patients were higher than that of the control group. (B) ROC of plasma THBD levels for diagnosis of sepsis. (C) Plasma THBD levels of sepsis patients with AKI were significantly higher than those of No AKI patients. (D) ROC of plasma THBD levels for diagnosis of sepsis patients who develop AKI.
3.6. Correlation between THBD gene polymorphism and plasma THBD expression level
Further analysis results showed that the THBD gene rs1962, rs3176123, and rs1042580 SNPs were significantly associated with plasma THBD levels, rs1962 T > C, rs3176123 T > G had a significant relation with elevated plasma THBD levels (all P < .01, Fig. 2A–E), and rs1042580 T > C was related to lower plasma THBD levels (all P < .01, Figs. 2C, F).
Figure 2.
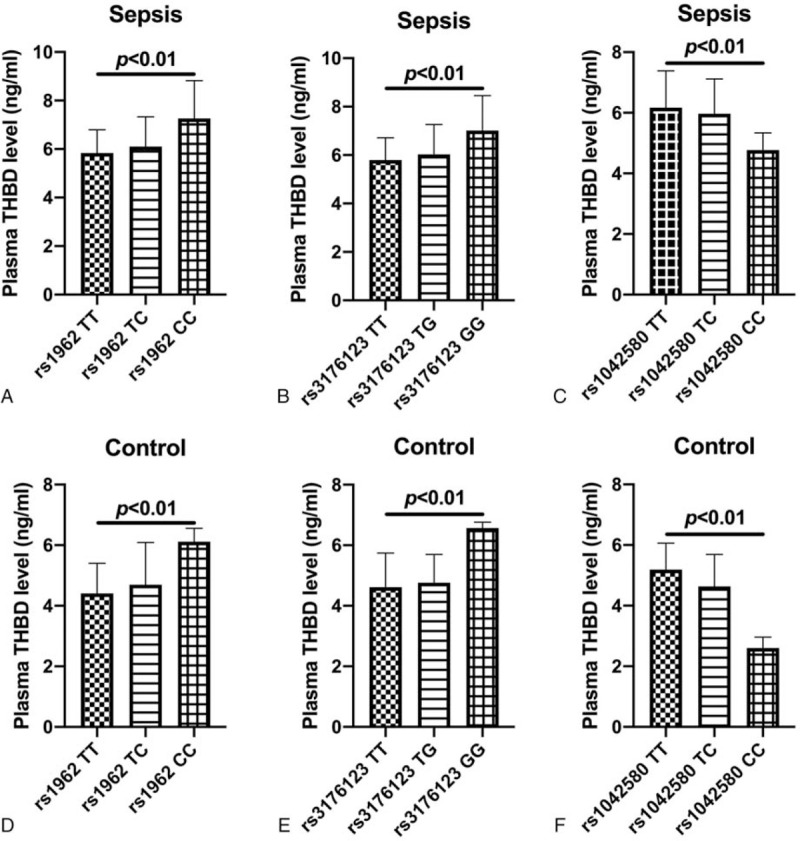
Comparison of plasma THBD levels in subjects with different genotypes at THBD gene rs1962, rs3176123, and rs1042580. (A) Comparison of plasma THBD levels in sepsis patients with different genotypes of THBD gene rs1962; (B) Comparison of plasma THBD levels in sepsis patients with different genotypes of THBD gene rs3176123; (C) Comparison of plasma THBD levels in sepsis patients with different genotypes of THBD gene rs1042580; (D) Comparison of plasma THBD levels in controls with different genotypes of THBD gene rs1962; (E) Comparison of plasma THBD levels in subjects with different genotypes of THBD gene rs3176123; (F) Comparison of plasma THBD levels in controls with different genotypes of THBD gene rs1042580.
3.7. Correlation between plasma THBD levels and death in patients with sepsis
A total of 56 deaths occurred and 122 patients survived after follow-up in sepsis patients. We found that the plasma THBD levels of patients who died of sepsis were markedly higher compared with those who survived (P < .01, Fig. 3A), and the AUC of ROC of plasma THBD levels to diagnose death in sepsis patients was 0.79 (95% CI: 0.72–0.86, P < .01), and the cut-off value was 7.60 ng/mL (Fig. 3B).
Figure 3.
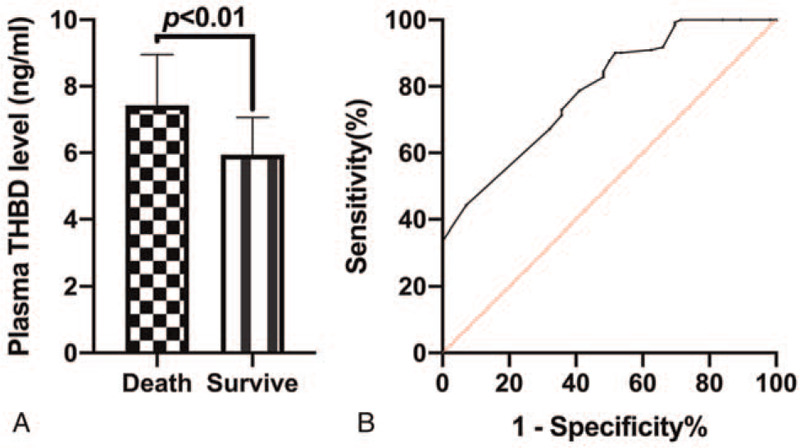
Correlation between plasma THBD levels and death of sepsis patients. (A) Comparison of plasma THBD levels between patients who died of sepsis and those who survived sepsis; (B) ROC curve of plasma THBD levels to diagnose death in sepsis patients.
3.8. Correlation between THBD gene polymorphism and prognosis of sepsis patients
The results showed that the TC + CC genotype of THBD gene rs1962 had a higher risk of death than the TT genotype in sepsis patients (OR = 10.93, 95% CI: 5.05–26.96, P < .01). However, there was no significant difference in the risk of death in sepsis patients with different genotypes at rs3176123 and rs1042580 (P > .05) (Table 7).
Table 7.
Comparison of death risk of sepsis patients with different genotypes of TM gene rs1962, rs3176123, and rs1042580.
| Death (n = 56) | Survive (n = 122) | OR (95%CI)∗ | P | |
| rs1962 | ||||
| TT | 6 (10.71%) | 95 (77.87%) | 1.00 | |
| TC + CC | 50 (89.29%) | 27 (22.13%) | 10.93 (5.05–26.96) | <.01 |
| rs3176123 | ||||
| TT | 21 (37.50%) | 64 (52.46%) | 1.00 | |
| TG + GG | 35 (62.50%) | 58 (47.54%) | 1.84 (0.96–3.51) | .09 |
| rs1042580 | ||||
| TT | 49 (87.50%) | 102 (83.61%) | 1.00 | |
| TC + CC | 7 (12.50%) | 20 (15.85%) | 0.73 (0.29–1.84) | .66 |
OR = odds ratio, CI = confidence interval.
Adjusted age, BMI, gender.
4. Discussion
Our study found that the THBD gene rs1962 C allele, rs3176123 G allele, and rs1042580 T allele were associated with sepsis susceptibility and increased risk of sepsis AKI, suggesting that these alleles were high risk factors for sepsis and AKI. Besides, the TGT haplotype was related to an elevated risk of developing AKI in sepsis patients. Although the rs3176123 and rs1042580 SNPs were not significantly correlated with death risk in the sepsis patients, there was significant relation between the rs1962 SNP and the risk of death in patients with sepsis. The above results indicated that the rs1962, rs3176123, and rs1042580 SNPs of the THBD gene were associated with sepsis and the risk of sepsis AKI.
Sepsis is a life-threatening organ dysfunction caused by a host's dysfunctional response to infection.[1] The pathogenesis of sepsis is very complicated, and various factors such as abnormal coagulation, endothelial cell damage and genetics are involved in the mechanism. Whereas, the specific pathogenesis remains unclear at home and abroad.[19–21] The waterfall release of numerous inflammatory mediators during body infection and sepsis mediated the adhesion of leukocytes and endothelial cells, which comprehensively activated the inflammation and coagulation, eventually leading to tissue damage and multiple organ dysfunction syndrome.[22] Vascular endothelial cells can release inflammatory factors, and they are also the targets of inflammatory factors.[23] Studies have revealed that patients with sepsis can develop endothelial dysfunction at an early stage, participate in organ dysfunction, and affect the prognosis of patients.[24,25]
THBD is secreted by vascular endothelial cells, which is involved in the activation of protein C system.[26,27] A large amount of THBD is released into the blood when vascular endothelial cells are damaged, thus the increased plasma THBD level can better reflect the damage of vascular endothelial cells. In recent years, several studies have shown that the plasma THBD level is regarded as a sensitive and specific marker of vascular endothelial cell damage, verifying that THBD is an independent predictor of sepsis AKI.[28] In this study, ROC analysis results showed that plasma THBD level was a potential diagnostic marker for sepsis, with high AUC value of 0.79. Meanwhile, plasma THBD level was a marker of sepsis AKI, and the AUC value was 0.61, which was consistent with the study by De Ceunynck K et al.[28]
In this study, we found that the risk of AKI and sepsis susceptibility in carriers of the THBD gene rs1962 C allele, rs3176123 G allele, and rs1042580 T allele were significantly increased since rs1962, rs3176123 and rs1042580 are located in the 3’ untranslated region of the THBD gene. The correlation between these SNPs and the disease has been confirmed, while the specific reason remains unclear. Therefore, the effect of these SNPs on THBD expression level, structure and function needs further study. The authors speculated that these SNPs may be located in the key regions of THBD gene expression regulation. The allelic transformation may affect the THBD expression regulation and amplify the impact on the disease. It has been demonstrated that the THBD gene rs3176123 polymorphism was significantly associated with a higher risk of deep vein thrombosis and soluble THBD levels in male patients,[29] which confirmed the author's speculation. Whereas, some studies have revealed that these SNPs had no significant correlation with the disease risk. Auro et al[30] have shown that the THBD rs1042580, rs1962 and rs3176123 polymorphism did not significantly contribute to the risk of cardiovascular events.
Based on these different research results, the authors speculated that the disease may result from the complex internal and external factors. It is known that the occurrence of sepsis is closely related to the internal genetic factors and the external environmental factors. In this study, the stratified analysis of the clinical data of the subjects was conducted, and the results also confirmed the authors’ speculation that the clinical data of the subjects plays a crucial role in sepsis AKI. Our results suggest that polymorphism analysis of key genes in specific populations is an important means to predict the risk of AKI in patients with sepsis in clinical work.
There are some shortcomings in this study. Firstly, we found that the sepsis patients with TC + CC genotype of THBD gene rs1962 had a higher risk of death than those with TT genotype, but there was no significant difference in the risk of death in sepsis patients with different genotypes at rs3176123 and rs1042580, which may need further validation in a large sample. Secondly, sepsis is caused by both endogenous and exogenous exposures (nongenetic factors), and there are few studies in this study that address exogenous exposures in sepsis, and further research on the combined effects of endogenous and exogenous factors may be needed to be of clinical value. Further, the specific reasons for the correlation between the rs1962, rs3176123 and rs1042580 SNPs and THBD expression level needs in vitro analysis. Fourth, the lack of evidence for functional studies of THBD gene SNP loci needs to be compensated by further studies. Fifth, although we performed some statistical tests in this study, we have not performed multiple testing, which is one of the limitations of this study.
In summary, we confirmed that the SNPs of THBD gene rs1962, rs3176123 and rs1042580 were significantly associated with susceptibility to sepsis and the risk of AKI. Moreover, the rs1962 SNP was linked to the risk of death in sepsis patients.
Author contributions
Conceptualization: Qin Li.
Data curation: Qin Li.
Formal analysis: Wenjuan Yang.
Investigation: Wenjuan Yang, Xifeng Sun.
Methodology: Liuqian Bao.
Resources: Keming Zhao.
Software: Keming Zhao.
Supervision: Liuqian Bao.
Visualization: Keming Zhao.
Writing – original draft: Qin Li, Xifeng Sun.
Writing – review & editing: Liuqian Bao.
Supplementary Material
Footnotes
Abbreviations: AKI = acute kidney injury, ROC = receiver operating characteristic, SNPs = single nucleotide polymorphisms, THBD = thrombomodulin, UTR = untranslated region.
How to cite this article: Li Q, Yang W, Zhao K, Sun X, Bao L. Thrombomodulin gene polymorphism and the occurrence and prognostic value of sepsis acute kidney injury. Medicine. 2021;100:26(e26293).
This work was supported by grants from Key R&D Projects in Zibo City (Policy Guidance) (2019ZC010140).
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
Supplemental digital content is available for this article.
References
- [1].Singer M, Deutschman C, Seymour C, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016;315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alobaidi R, Basu R, Goldstein S, et al. Sepsis-associated acute kidney injury. Semin Nephrol 2015;35:02–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Remuzzi G, Horton RJL. Acute renal failure: an unacceptable death sentence globally. Lancet 2013;382:2041–2. [DOI] [PubMed] [Google Scholar]
- [4].Metnitz P, Krenn C, Steltzer H, et al. Effect of acute renal failure requiring renal replacement therapy on outcome in critically ill patients. Crit Care Med 2002;30:2051–8. [DOI] [PubMed] [Google Scholar]
- [5].Rhodes A, Evans L, Alhazzani W, et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med 2017;43:304–77. [DOI] [PubMed] [Google Scholar]
- [6].Bellomo R, Cass A, Cole L, et al. Intensity of continuous renal-replacement therapy in critically ill patients. N Engl J Med 2009;361:1627–38. [DOI] [PubMed] [Google Scholar]
- [7].Ogawa Y, Yamakawa K, Ogura H, et al. Recombinant human soluble thrombomodulin improves mortality and respiratory dysfunction in patients with severe sepsis. J Trauma Acute Care Surg 2012;72:1150–7. [DOI] [PubMed] [Google Scholar]
- [8].Kunz G, Ireland H, Stubbs P, et al. Identification and characterization of a thrombomodulin gene mutation coding for an elongated protein with reduced expression in a kindred with myocardial infarction. Blood 2000;95:569–76. [PubMed] [Google Scholar]
- [9].Le Flem L, Mennen L, Aubry M, et al. Thrombomodulin promoter mutations, venous thrombosis, and varicose veins. Arterioscler Thromb Vasc Biol 2001;21:445–51. [DOI] [PubMed] [Google Scholar]
- [10].Ireland H, Kunz G, Kyriakoulis K, et al. Thrombomodulin gene mutations associated with myocardial infarction. Circulation 1997;96:15–8. [DOI] [PubMed] [Google Scholar]
- [11].Rachakonda S, Penack O, Dietrich S, et al. Single-nucleotide polymorphisms within the thrombomodulin gene (THBD) predict mortality in patients with graft-versus-host disease. J Clin Oncol 2014;32:3421–7. [DOI] [PubMed] [Google Scholar]
- [12].Nomoto H, Takami A, Espinoza J, et al. A donor thrombomodulin gene variation predicts graft-versus-host disease development and mortality after bone marrow transplantation. Int J Hematol 2015;102:460–70. [DOI] [PubMed] [Google Scholar]
- [13].An S, Lee K, Chang B, et al. Association of gene polymorphisms with the risk of warfarin bleeding complications at therapeutic INR in patients with mechanical cardiac valves. J Clin Pharm Ther 2014;39:314–8. [DOI] [PubMed] [Google Scholar]
- [14].Lorente L, Martín M, Plasencia F, et al. The 372 T/C genetic polymorphism of TIMP-1 is associated with serum levels of TIMP-1 and survival in patients with severe sepsis. Crit Care 2013;17:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 2012;120:179–84. [DOI] [PubMed] [Google Scholar]
- [16].Wang C, Shang M, Feng L, et al. Correlation between APACHE III score and sleep quality in ICU patients. J Int Med Res 2019;47:3670–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Granholm A, Møller M, Krag M, et al. Predictive performance of the Simplified Acute Physiology Score (SAPS) II and the Initial Sequential Organ Failure Assessment (SOFA) Score in acutely ill intensive care patients: post-Hoc analyses of the SUP-ICU Inception Cohort Study. PLoS One 2016;11:e0168948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jones A, Trzeciak S, JJCcm Kline. The Sequential Organ Failure Assessment score for predicting outcome in patients with severe sepsis and evidence of hypoperfusion at the time of emergency department presentation. Crit Care Med 2009;37:1649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang M, Cai S, JJIjoms Su. The pathogenesis of sepsis and potential therapeutic targets. Int J Mol Sci 2019;20:5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chang JJTj. Sepsis and septic shock: endothelial molecular pathogenesis associated with vascular microthrombotic disease. Thromb J 2019;17:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hendrickson C, Matthay M. Endothelial biomarkers in human sepsis: pathogenesis and prognosis for ARDS. Pulm Circ 2018;8: 2045894018769876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marshall JJCcm. Inflammation, coagulopathy, and the pathogenesis of multiple organ dysfunction syndrome. Crit Care Med 2001;29:S99–106. [DOI] [PubMed] [Google Scholar]
- [23].Wei F, Liu S, Luo L, et al. Anti-inflammatory mechanism of ulinastatin: Inhibiting the hyperpermeability of vascular endothelial cells induced by TNF-( via the RhoA/ROCK signal pathway. Int Immunopharmacol 2017;46:220–7. [DOI] [PubMed] [Google Scholar]
- [24].Coletta C, Módis K, Oláh G, et al. Endothelial dysfunction is a potential contributor to multiple organ failure and mortality in aged mice subjected to septic shock: preclinical studies in a murine model of cecal ligation and puncture. Crit Care 2014;18:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee W, Slutsky AS. Sepsis and endothelial permeability. N Engl J Med 2010;363:689–91. [DOI] [PubMed] [Google Scholar]
- [26].Wolter J, Schild L, Bock F, et al. Thrombomodulin-dependent protein C activation is required for mitochondrial function and myelination in the central nervous system. J Thromb Haemost 2016;14:2212–26. [DOI] [PubMed] [Google Scholar]
- [27].Ikezoe TJJoic. Thrombomodulin/activated protein C system in septic disseminated intravascular coagulation. J Intensive Care 2015;3:01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].De Ceunynck K, Peters C, Jain A, et al. PAR1 agonists stimulate APC-like endothelial cytoprotection and confer resistance to thromboinflammatory injury. Proc Natl Acad Sci USA 2018;115:E982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sugiyama S, Hirota H, Kimura R, et al. Haplotype of thrombomodulin gene associated with plasma thrombomodulin level and deep vein thrombosis in the Japanese population. Thromb Res 2007;119:35–43. [DOI] [PubMed] [Google Scholar]
- [30].Auro K, Komulainen K, Alanne M, et al. Thrombomodulin gene polymorphisms and haplotypes and the risk of cardiovascular events: a prospective follow-up study. Arterioscler Thromb Vasc Biol 2006;26:942–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


