Abstract
The dataset herein indicated the novelty of the article entitled “Dataset on the use of MGIDI in screening drought-tolerant wild wheat accessions at the early growth stage”. Data were gathered during 2018–2019 on a set of wild wheat germplasm under two control and water deficit stress conditions. One hundred and forty-six accessions belonging to Ae. tauschii, Ae. cylindrica, and Ae. crassa were assessed under optimal glasshouse conditions to screen the drought-tolerant samples at the early growth stage. Nine drought tolerance and susceptibility indices along with the multi-trait genotype-ideotype distance index (MGIDI) were used to visualize the dataset. The obtained data can highlight the potential of the MGIDI index in accelerating screening of a large number of plant materials using multiple traits or selection indices in crop breeding programs, especially at the early growth stage.
Keywords: Drought tolerance, Wild wheat, MGIDI index, Multivariate analysis
Specifications Table
| Subject | Data analysis (Agricultural and Biological Science) |
| Specific subject area | Agronomy and Crop Science |
| Type of data | Tables and Figures |
| How data were acquired | Data were obtained by a study conducted under controlled greenhouse conditions on a set of wild progenitors of wheat belonging to three Aegilops species, such as Ae. cylindrica, Ae. crassa, and Ae. tauschii. Data tables and figures were obtained by calculating nine drought tolerance and susceptibility indices using iPASTIC software. The multi-trait genotype-ideotype distance index (MGIDI) was used to rank the accessions based on information of multiple indices. The MGIDI index was calculated in the R software using the ‘metan’ package. |
| Data format | Raw |
| Parameters for data collection | The conditions considered for data collection were controlled greenhouse conditions of the experiment. |
| Description of data collection | A total of 146 accessions of three Aegilops accessions were investigated under two control and water deficit stress treatments (Field capacity [FC] = 95 ± 5% and 30 ± 5%, respectively). A factorial experiment was performed in a randomized complete block design with three replications in a research greenhouse at the Agronomy and Plant Breeding Department, Tehran University, Karaj, Iran, during the 2018–2019 growing seasons. Thirty days after sowing and water deficit treatment, shoot dry biomass was recorded in all samples. Based on both dry biomass under control (Yp) and water deficit treatment (Ys) of tested samples, nine drought tolerance and susceptibility indices, including tolerance index (TOL), mean productivity (MP), geometric mean productivity (GMP), harmonic mean (HM), stress susceptibility index (SSI), stress tolerance index (STI), yield index (YI), yield stability index (YSI), and relative drought index (RSI) were calculated. |
| Data source location | Department of Agronomy and Plant Breeding, Agricultural College, University of Tehran, Karaj |
| Data accessibility | The raw data associated to this article are provided on Mendeley dataset http://dx.doi.org/10.17632/4tw2hrrdfp.1. |
| Related research article | [1] A. Pour-Aboughadareh, M. Omidi, M.R. Naghavi, A. Etminan, A.A. Mehrabi, P. Poczai, H. Bayat, Effect of water deficit stress on seedling biomass and physio-chemical characteristics in different species of wheat possessing the D genome, Agronomy. 9 (2019) 522. https://doi.org/10.3390/agronomy9090522 |
Value of the Data
-
•
The dataset analyzed in this work indicates an overview of the potential of some wild relatives for increasing drought tolerance in wheat breeding programs. Indeed, this dataset provides information for wheat breeders on the breeding capacity of this germplasm to further studies on tolerance mechanisms and discovering new tolerance-related genes or even alleles in alien genomes.
-
•
The presented drought tolerance and susceptibility indices are important mathematical parameters that are widely used by researchers. However, the multi-trait genotype-ideotype distance index (MGIDI) was used to make a unique index for facilitating the selection of superior genotypes.
-
•
This dataset can highlight the applicability of the MGIDI index in accelerating the screening of a large number of plant materials using multiple traits or growth parameters in crop breeding programs, especially at the early growth stage.
1. Data Description
Climate change is expected to result in fluctuations in precipitation patterns, including enhanced severity and accelerated frequency of droughts. Under these circumstances, agricultural production is mainly affected by water limitation. Indeed, water deficit is one of the destructive abiotic stresses that decrease crop growth and yield performance in many areas across the world [2]. Wild relatives of wheat are a valuable genetic resource that harbors many genes related to various abiotic stresses. Aegilops species plays an important role in wheat domestication and improvement breeding programs due to its interesting potential [3], [4], [5], [6]. Hence, this germplasm can be a benchmark for wheat breeding programs.
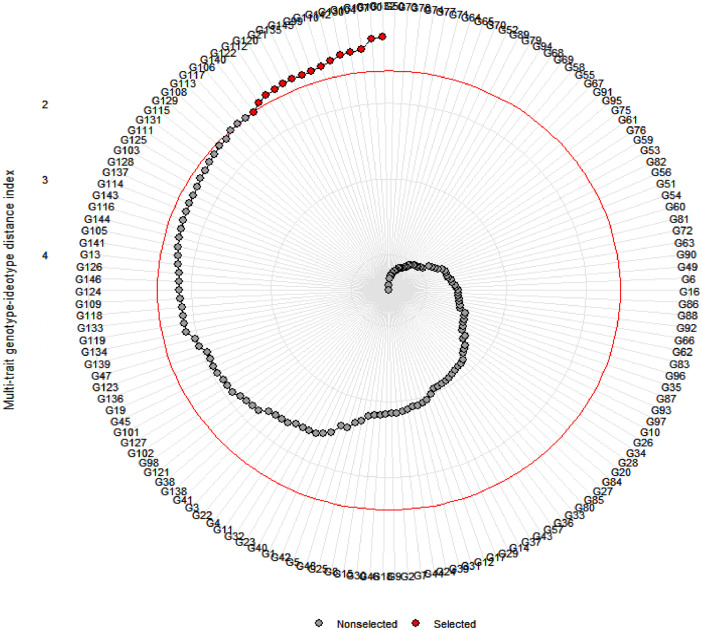
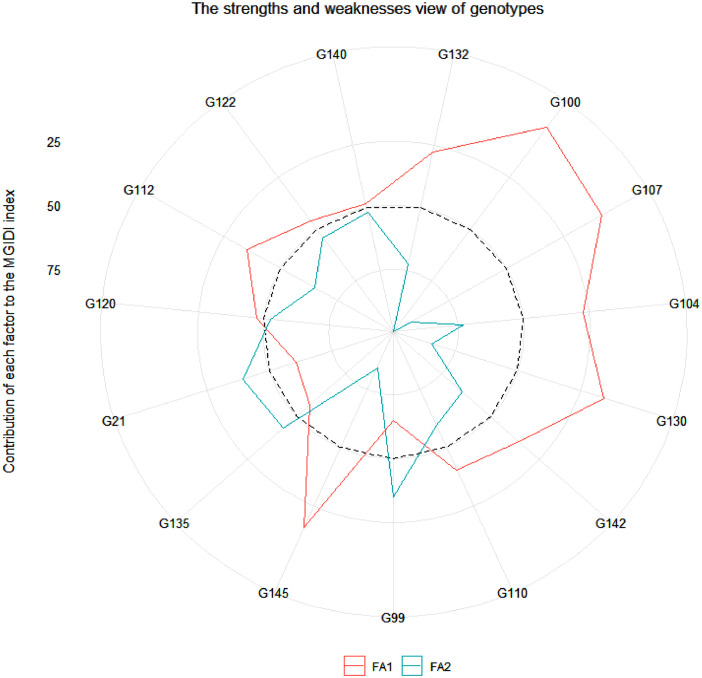
The dataset is presented in two tables and two figures that describe the ability of the MGIDI index to screen for drought-tolerant accessions. Table 2 presents drought tolerance and susceptibility indices in the 146 investigated Aegilops accessions. Based on the indices values in Table 2, accession numbers G150, G153, G157, G160, G164, G175, G180, G183, G184, and G195 form Ae. crassa showed the lowest percent reduction in their shoot dry matter due to water deficit stress compared with control conditions. Table 3 shows the ranking pattern of the investigated accessions based on each calculated index. This table also shows the average sum of ranks and standard deviation of ranks (SD) through Yp, Ys, and other indices. As shown in Table 3, accession numbers G150, G153, G154, G157, G160, G164, G180, G182, G192, and G195 were selected as the top 10 tolerant accessions. Fig. 1 presents the results of screening of the investigated Aegilops accessions based on the MGIDI index. In Fig. 1, the red circle shows the cutpoint according to the selection pressure (SI = 10%). The MGIDI index identified 15 samples as more desirable accessions than others. Except for one accession (G71) that belongs to Ae. tauschii, other selected accessions belong to Ae. crassa. Fig. 2 shows the strengths and weaknesses view of the selected genotypes as shown as the proportion of each factor on the computed MGIDI index. Thus, the MGIDI index can identify the best drought-tolerant genotypes at the early growth stage. The raw data associated to this article are provided on Mendeley dataset.
Table 2.
Calculated drought tolerance and susceptibility indices in 146 Aegilops accessions.
| Code | Yp | Ys | RC | TOL | MP | GMP | HM | SSI | STI | YI | YSI | RSI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 0.73 | 0.32 | 55.63 | 0.41 | 0.53 | 0.48 | 0.45 | 1.01 | 0.55 | 1.09 | 0.44 | 0.99 |
| G2 | 0.73 | 0.28 | 60.99 | 0.44 | 0.51 | 0.45 | 0.41 | 1.11 | 0.48 | 0.96 | 0.39 | 0.87 |
| G3 | 0.75 | 0.34 | 54.29 | 0.41 | 0.54 | 0.50 | 0.47 | 0.99 | 0.59 | 1.16 | 0.46 | 1.02 |
| G4 | 0.73 | 0.33 | 54.98 | 0.40 | 0.53 | 0.49 | 0.46 | 1.00 | 0.56 | 1.12 | 0.45 | 1.00 |
| G5 | 0.69 | 0.30 | 57.02 | 0.39 | 0.49 | 0.45 | 0.42 | 1.04 | 0.48 | 1.01 | 0.43 | 0.95 |
| G6 | 0.85 | 0.30 | 64.48 | 0.55 | 0.58 | 0.51 | 0.45 | 1.17 | 0.60 | 1.03 | 0.36 | 0.79 |
| G7 | 0.76 | 0.30 | 60.63 | 0.46 | 0.53 | 0.47 | 0.43 | 1.10 | 0.53 | 1.01 | 0.39 | 0.87 |
| G8 | 0.72 | 0.29 | 59.30 | 0.42 | 0.50 | 0.46 | 0.41 | 1.08 | 0.48 | 0.99 | 0.41 | 0.90 |
| G9 | 0.69 | 0.27 | 60.38 | 0.42 | 0.48 | 0.43 | 0.39 | 1.10 | 0.44 | 0.93 | 0.40 | 0.88 |
| G10 | 0.79 | 0.27 | 65.91 | 0.52 | 0.53 | 0.46 | 0.40 | 1.20 | 0.50 | 0.91 | 0.34 | 0.76 |
| G11 | 0.74 | 0.33 | 55.05 | 0.41 | 0.54 | 0.50 | 0.46 | 1.00 | 0.58 | 1.13 | 0.45 | 1.00 |
| G12 | 0.72 | 0.27 | 62.14 | 0.45 | 0.50 | 0.44 | 0.40 | 1.13 | 0.46 | 0.93 | 0.38 | 0.84 |
| G13 | 0.74 | 0.38 | 48.71 | 0.36 | 0.56 | 0.53 | 0.50 | 0.89 | 0.65 | 1.28 | 0.51 | 1.14 |
| G14 | 0.71 | 0.26 | 63.38 | 0.45 | 0.49 | 0.43 | 0.38 | 1.15 | 0.43 | 0.88 | 0.37 | 0.81 |
| G15 | 0.74 | 0.29 | 60.27 | 0.44 | 0.51 | 0.46 | 0.42 | 1.10 | 0.50 | 0.99 | 0.40 | 0.88 |
| G16 | 0.74 | 0.23 | 69.37 | 0.51 | 0.48 | 0.41 | 0.35 | 1.26 | 0.39 | 0.77 | 0.31 | 0.68 |
| G17 | 0.70 | 0.26 | 62.30 | 0.43 | 0.48 | 0.43 | 0.38 | 1.13 | 0.42 | 0.89 | 0.38 | 0.84 |
| G18 | 0.72 | 0.28 | 61.00 | 0.44 | 0.50 | 0.45 | 0.40 | 1.11 | 0.47 | 0.95 | 0.39 | 0.87 |
| G19 | 0.68 | 0.34 | 50.15 | 0.34 | 0.51 | 0.48 | 0.45 | 0.91 | 0.53 | 1.15 | 0.50 | 1.11 |
| G20 | 0.70 | 0.24 | 65.34 | 0.46 | 0.47 | 0.41 | 0.36 | 1.19 | 0.40 | 0.83 | 0.35 | 0.77 |
| G21 | 0.70 | 0.39 | 43.71 | 0.31 | 0.55 | 0.53 | 0.50 | 0.80 | 0.64 | 1.34 | 0.56 | 1.25 |
| G22 | 0.70 | 0.32 | 54.20 | 0.38 | 0.51 | 0.48 | 0.44 | 0.99 | 0.53 | 1.09 | 0.46 | 1.02 |
| G23 | 0.71 | 0.32 | 55.32 | 0.39 | 0.51 | 0.47 | 0.44 | 1.01 | 0.52 | 1.07 | 0.45 | 0.99 |
| G24 | 0.67 | 0.27 | 60.30 | 0.40 | 0.47 | 0.42 | 0.38 | 1.10 | 0.41 | 0.90 | 0.40 | 0.88 |
| G25 | 0.73 | 0.30 | 58.98 | 0.43 | 0.51 | 0.47 | 0.42 | 1.07 | 0.51 | 1.01 | 0.41 | 0.91 |
| G26 | 0.66 | 0.23 | 65.14 | 0.43 | 0.44 | 0.39 | 0.34 | 1.18 | 0.35 | 0.78 | 0.35 | 0.77 |
| G27 | 0.75 | 0.26 | 64.75 | 0.48 | 0.50 | 0.44 | 0.39 | 1.18 | 0.46 | 0.89 | 0.35 | 0.78 |
| G28 | 0.70 | 0.24 | 65.62 | 0.46 | 0.47 | 0.41 | 0.36 | 1.19 | 0.39 | 0.82 | 0.34 | 0.76 |
| G29 | 0.66 | 0.25 | 61.93 | 0.41 | 0.46 | 0.41 | 0.37 | 1.13 | 0.39 | 0.85 | 0.38 | 0.85 |
| G30 | 0.72 | 0.28 | 60.34 | 0.43 | 0.50 | 0.45 | 0.41 | 1.10 | 0.47 | 0.96 | 0.40 | 0.88 |
| G31 | 0.73 | 0.28 | 61.92 | 0.45 | 0.50 | 0.45 | 0.40 | 1.13 | 0.47 | 0.94 | 0.38 | 0.85 |
| G32 | 0.67 | 0.31 | 53.44 | 0.36 | 0.49 | 0.46 | 0.42 | 0.97 | 0.48 | 1.05 | 0.47 | 1.03 |
| G33 | 0.65 | 0.25 | 62.33 | 0.41 | 0.45 | 0.40 | 0.36 | 1.13 | 0.37 | 0.83 | 0.38 | 0.84 |
| G34 | 0.73 | 0.24 | 66.62 | 0.48 | 0.48 | 0.42 | 0.36 | 1.21 | 0.41 | 0.82 | 0.33 | 0.74 |
| G35 | 0.85 | 0.32 | 62.85 | 0.53 | 0.58 | 0.52 | 0.46 | 1.14 | 0.62 | 1.07 | 0.37 | 0.83 |
| G36 | 0.64 | 0.25 | 61.15 | 0.39 | 0.45 | 0.40 | 0.36 | 1.11 | 0.37 | 0.84 | 0.39 | 0.86 |
| G37 | 0.68 | 0.25 | 62.90 | 0.43 | 0.47 | 0.42 | 0.37 | 1.14 | 0.40 | 0.86 | 0.37 | 0.82 |
| G38 | 0.69 | 0.33 | 52.39 | 0.36 | 0.51 | 0.48 | 0.45 | 0.95 | 0.53 | 1.11 | 0.48 | 1.06 |
| G39 | 0.75 | 0.29 | 61.42 | 0.46 | 0.52 | 0.47 | 0.42 | 1.12 | 0.50 | 0.98 | 0.39 | 0.86 |
| G40 | 0.71 | 0.32 | 55.16 | 0.39 | 0.51 | 0.47 | 0.44 | 1.00 | 0.52 | 1.07 | 0.45 | 1.00 |
| G41 | 0.72 | 0.33 | 54.22 | 0.39 | 0.53 | 0.49 | 0.45 | 0.99 | 0.56 | 1.12 | 0.46 | 1.02 |
| G42 | 0.68 | 0.31 | 55.34 | 0.38 | 0.49 | 0.46 | 0.42 | 1.01 | 0.48 | 1.03 | 0.45 | 0.99 |
| G43 | 0.72 | 0.26 | 63.64 | 0.46 | 0.49 | 0.43 | 0.38 | 1.16 | 0.43 | 0.88 | 0.36 | 0.81 |
| G44 | 0.70 | 0.27 | 61.23 | 0.43 | 0.49 | 0.44 | 0.39 | 1.11 | 0.44 | 0.92 | 0.39 | 0.86 |
| G45 | 0.68 | 0.34 | 50.29 | 0.34 | 0.51 | 0.48 | 0.45 | 0.91 | 0.53 | 1.15 | 0.50 | 1.10 |
| G46 | 0.70 | 0.28 | 60.23 | 0.42 | 0.49 | 0.44 | 0.40 | 1.10 | 0.45 | 0.94 | 0.40 | 0.88 |
| G47 | 0.68 | 0.35 | 49.49 | 0.34 | 0.51 | 0.49 | 0.46 | 0.90 | 0.55 | 1.17 | 0.51 | 1.12 |
| G48 | 0.68 | 0.29 | 56.64 | 0.38 | 0.49 | 0.45 | 0.41 | 1.03 | 0.46 | 1.00 | 0.43 | 0.96 |
| G49 | 0.82 | 0.27 | 66.99 | 0.55 | 0.55 | 0.47 | 0.41 | 1.22 | 0.52 | 0.92 | 0.33 | 0.73 |
| G50 | 0.59 | 0.14 | 76.24 | 0.45 | 0.36 | 0.29 | 0.22 | 1.39 | 0.19 | 0.47 | 0.24 | 0.53 |
| G51 | 0.67 | 0.19 | 71.28 | 0.48 | 0.43 | 0.36 | 0.30 | 1.30 | 0.30 | 0.65 | 0.29 | 0.64 |
| G52 | 0.67 | 0.16 | 75.63 | 0.51 | 0.42 | 0.33 | 0.26 | 1.38 | 0.26 | 0.56 | 0.24 | 0.54 |
| G53 | 0.63 | 0.18 | 70.98 | 0.45 | 0.41 | 0.34 | 0.29 | 1.29 | 0.27 | 0.62 | 0.29 | 0.64 |
| G54 | 0.78 | 0.23 | 70.23 | 0.55 | 0.50 | 0.42 | 0.36 | 1.28 | 0.42 | 0.78 | 0.30 | 0.66 |
| G55 | 0.61 | 0.17 | 71.92 | 0.44 | 0.39 | 0.32 | 0.27 | 1.31 | 0.24 | 0.58 | 0.28 | 0.62 |
| G56 | 0.61 | 0.19 | 69.06 | 0.42 | 0.40 | 0.34 | 0.29 | 1.26 | 0.27 | 0.64 | 0.31 | 0.69 |
| G57 | 0.55 | 0.30 | 45.72 | 0.25 | 0.42 | 0.40 | 0.39 | 0.83 | 0.38 | 1.01 | 0.54 | 1.21 |
| G58 | 0.55 | 0.18 | 66.42 | 0.36 | 0.37 | 0.32 | 0.28 | 1.21 | 0.23 | 0.62 | 0.34 | 0.75 |
| G59 | 0.54 | 0.21 | 60.71 | 0.33 | 0.37 | 0.34 | 0.30 | 1.10 | 0.26 | 0.72 | 0.39 | 0.87 |
| G60 | 0.50 | 0.29 | 41.77 | 0.21 | 0.39 | 0.38 | 0.37 | 0.76 | 0.34 | 0.98 | 0.58 | 1.29 |
| G61 | 0.57 | 0.19 | 67.02 | 0.38 | 0.38 | 0.33 | 0.28 | 1.22 | 0.25 | 0.63 | 0.33 | 0.73 |
| G62 | 0.64 | 0.21 | 67.29 | 0.43 | 0.42 | 0.37 | 0.31 | 1.22 | 0.31 | 0.71 | 0.33 | 0.73 |
| G63 | 0.61 | 0.20 | 66.94 | 0.41 | 0.40 | 0.35 | 0.30 | 1.22 | 0.28 | 0.68 | 0.33 | 0.73 |
| G64 | 0.59 | 0.16 | 72.74 | 0.43 | 0.37 | 0.31 | 0.25 | 1.32 | 0.22 | 0.54 | 0.27 | 0.61 |
| G65 | 0.65 | 0.16 | 75.69 | 0.49 | 0.40 | 0.32 | 0.25 | 1.38 | 0.24 | 0.54 | 0.24 | 0.54 |
| G66 | 0.65 | 0.21 | 67.85 | 0.44 | 0.43 | 0.37 | 0.31 | 1.23 | 0.31 | 0.70 | 0.32 | 0.71 |
| G67 | 0.67 | 0.18 | 73.76 | 0.49 | 0.42 | 0.34 | 0.28 | 1.34 | 0.27 | 0.59 | 0.26 | 0.58 |
| G68 | 0.59 | 0.17 | 71.09 | 0.42 | 0.38 | 0.32 | 0.26 | 1.29 | 0.23 | 0.58 | 0.29 | 0.64 |
| G69 | 0.54 | 0.19 | 64.86 | 0.35 | 0.36 | 0.32 | 0.28 | 1.18 | 0.23 | 0.64 | 0.35 | 0.78 |
| G70 | 0.51 | 0.19 | 62.89 | 0.32 | 0.35 | 0.31 | 0.28 | 1.14 | 0.23 | 0.64 | 0.37 | 0.82 |
| G71 | 0.55 | 0.17 | 69.64 | 0.38 | 0.36 | 0.30 | 0.26 | 1.27 | 0.21 | 0.57 | 0.30 | 0.67 |
| G72 | 0.63 | 0.20 | 68.62 | 0.43 | 0.41 | 0.35 | 0.30 | 1.25 | 0.29 | 0.67 | 0.31 | 0.70 |
| G73 | 0.70 | 0.14 | 79.57 | 0.55 | 0.42 | 0.31 | 0.24 | 1.45 | 0.23 | 0.48 | 0.20 | 0.45 |
| G74 | 0.61 | 0.15 | 74.88 | 0.45 | 0.38 | 0.30 | 0.24 | 1.36 | 0.21 | 0.52 | 0.25 | 0.56 |
| G75 | 0.55 | 0.19 | 65.70 | 0.36 | 0.37 | 0.32 | 0.28 | 1.19 | 0.24 | 0.64 | 0.34 | 0.76 |
| G76 | 0.59 | 0.18 | 68.77 | 0.40 | 0.38 | 0.33 | 0.28 | 1.25 | 0.25 | 0.62 | 0.31 | 0.69 |
| G77 | 0.54 | 0.17 | 69.32 | 0.38 | 0.35 | 0.30 | 0.25 | 1.26 | 0.21 | 0.56 | 0.31 | 0.68 |
| G78 | 0.54 | 0.17 | 69.27 | 0.37 | 0.35 | 0.30 | 0.25 | 1.26 | 0.21 | 0.56 | 0.31 | 0.68 |
| G79 | 0.55 | 0.18 | 67.52 | 0.37 | 0.36 | 0.31 | 0.27 | 1.23 | 0.23 | 0.60 | 0.32 | 0.72 |
| G80 | 0.58 | 0.27 | 53.57 | 0.31 | 0.42 | 0.39 | 0.36 | 0.97 | 0.36 | 0.90 | 0.46 | 1.03 |
| G81 | 0.57 | 0.21 | 63.33 | 0.36 | 0.39 | 0.35 | 0.31 | 1.15 | 0.28 | 0.71 | 0.37 | 0.81 |
| G82 | 0.63 | 0.19 | 70.29 | 0.44 | 0.41 | 0.34 | 0.29 | 1.28 | 0.27 | 0.63 | 0.30 | 0.66 |
| G83 | 0.56 | 0.23 | 58.09 | 0.32 | 0.39 | 0.36 | 0.33 | 1.06 | 0.30 | 0.79 | 0.42 | 0.93 |
| G84 | 0.61 | 0.24 | 60.56 | 0.37 | 0.43 | 0.38 | 0.35 | 1.10 | 0.34 | 0.82 | 0.39 | 0.88 |
| G85 | 0.61 | 0.25 | 59.54 | 0.37 | 0.43 | 0.39 | 0.35 | 1.08 | 0.35 | 0.84 | 0.40 | 0.90 |
| G86 | 0.64 | 0.20 | 67.87 | 0.43 | 0.42 | 0.36 | 0.31 | 1.23 | 0.30 | 0.69 | 0.32 | 0.71 |
| G87 | 0.56 | 0.24 | 56.89 | 0.32 | 0.40 | 0.37 | 0.34 | 1.03 | 0.31 | 0.82 | 0.43 | 0.96 |
| G88 | 0.56 | 0.23 | 59.79 | 0.34 | 0.39 | 0.36 | 0.32 | 1.09 | 0.30 | 0.77 | 0.40 | 0.89 |
| G89 | 0.54 | 0.18 | 66.85 | 0.36 | 0.36 | 0.31 | 0.27 | 1.22 | 0.22 | 0.61 | 0.33 | 0.74 |
| G90 | 0.53 | 0.25 | 53.23 | 0.28 | 0.39 | 0.36 | 0.34 | 0.97 | 0.30 | 0.83 | 0.47 | 1.04 |
| G91 | 0.54 | 0.20 | 63.36 | 0.34 | 0.37 | 0.32 | 0.29 | 1.15 | 0.24 | 0.66 | 0.37 | 0.81 |
| G92 | 0.62 | 0.21 | 66.83 | 0.42 | 0.42 | 0.36 | 0.31 | 1.22 | 0.30 | 0.70 | 0.33 | 0.74 |
| G93 | 0.59 | 0.23 | 61.02 | 0.36 | 0.41 | 0.37 | 0.33 | 1.11 | 0.32 | 0.78 | 0.39 | 0.87 |
| G94 | 0.55 | 0.18 | 67.76 | 0.37 | 0.36 | 0.31 | 0.27 | 1.23 | 0.23 | 0.60 | 0.32 | 0.72 |
| G95 | 0.63 | 0.17 | 72.48 | 0.45 | 0.40 | 0.33 | 0.27 | 1.32 | 0.25 | 0.58 | 0.28 | 0.61 |
| G96 | 0.64 | 0.22 | 66.41 | 0.43 | 0.43 | 0.37 | 0.32 | 1.21 | 0.32 | 0.73 | 0.34 | 0.75 |
| G97 | 0.55 | 0.25 | 53.83 | 0.30 | 0.40 | 0.37 | 0.35 | 0.98 | 0.32 | 0.86 | 0.46 | 1.03 |
| G98 | 0.68 | 0.33 | 51.75 | 0.35 | 0.51 | 0.48 | 0.45 | 0.94 | 0.53 | 1.12 | 0.48 | 1.07 |
| G99 | 0.71 | 0.40 | 43.86 | 0.31 | 0.55 | 0.53 | 0.51 | 0.80 | 0.66 | 1.35 | 0.56 | 1.25 |
| G100 | 0.65 | 0.45 | 30.15 | 0.20 | 0.55 | 0.54 | 0.53 | 0.55 | 0.69 | 1.54 | 0.70 | 1.55 |
| G101 | 0.60 | 0.38 | 37.33 | 0.22 | 0.49 | 0.47 | 0.46 | 0.68 | 0.53 | 1.27 | 0.63 | 1.39 |
| G102 | 0.79 | 0.40 | 49.62 | 0.39 | 0.59 | 0.56 | 0.53 | 0.90 | 0.73 | 1.35 | 0.50 | 1.12 |
| G103 | 0.62 | 0.43 | 30.63 | 0.19 | 0.52 | 0.51 | 0.51 | 0.56 | 0.61 | 1.45 | 0.69 | 1.54 |
| G104 | 0.66 | 0.42 | 36.23 | 0.24 | 0.54 | 0.52 | 0.51 | 0.66 | 0.64 | 1.42 | 0.64 | 1.42 |
| G105 | 0.76 | 0.40 | 47.63 | 0.36 | 0.58 | 0.55 | 0.52 | 0.87 | 0.70 | 1.35 | 0.52 | 1.16 |
| G106 | 0.67 | 0.38 | 43.32 | 0.29 | 0.53 | 0.51 | 0.49 | 0.79 | 0.60 | 1.29 | 0.57 | 1.26 |
| G107 | 0.65 | 0.44 | 31.89 | 0.21 | 0.54 | 0.53 | 0.52 | 0.58 | 0.66 | 1.49 | 0.68 | 1.51 |
| G108 | 0.69 | 0.38 | 45.53 | 0.32 | 0.54 | 0.51 | 0.49 | 0.83 | 0.61 | 1.28 | 0.54 | 1.21 |
| G109 | 0.66 | 0.36 | 45.92 | 0.30 | 0.51 | 0.49 | 0.46 | 0.84 | 0.55 | 1.21 | 0.54 | 1.20 |
| G110 | 0.63 | 0.51 | 19.59 | 0.12 | 0.57 | 0.57 | 0.56 | 0.36 | 0.75 | 1.72 | 0.80 | 1.79 |
| G111 | 0.65 | 0.38 | 40.93 | 0.26 | 0.51 | 0.50 | 0.48 | 0.74 | 0.57 | 1.29 | 0.59 | 1.31 |
| G112 | 0.66 | 0.40 | 39.27 | 0.26 | 0.53 | 0.51 | 0.50 | 0.71 | 0.61 | 1.35 | 0.61 | 1.35 |
| G113 | 0.74 | 0.39 | 46.94 | 0.35 | 0.56 | 0.54 | 0.51 | 0.85 | 0.67 | 1.32 | 0.53 | 1.18 |
| G114 | 0.61 | 0.45 | 25.78 | 0.16 | 0.53 | 0.52 | 0.52 | 0.47 | 0.64 | 1.53 | 0.74 | 1.65 |
| G115 | 0.66 | 0.38 | 42.88 | 0.28 | 0.52 | 0.50 | 0.48 | 0.78 | 0.58 | 1.28 | 0.57 | 1.27 |
| G116 | 0.71 | 0.37 | 47.90 | 0.34 | 0.54 | 0.52 | 0.49 | 0.87 | 0.62 | 1.26 | 0.52 | 1.16 |
| G117 | 0.64 | 0.40 | 38.16 | 0.25 | 0.52 | 0.50 | 0.49 | 0.69 | 0.59 | 1.35 | 0.62 | 1.37 |
| G118 | 0.68 | 0.35 | 48.09 | 0.33 | 0.52 | 0.49 | 0.46 | 0.87 | 0.56 | 1.20 | 0.52 | 1.15 |
| G119 | 0.68 | 0.35 | 47.71 | 0.32 | 0.52 | 0.49 | 0.46 | 0.87 | 0.56 | 1.20 | 0.52 | 1.16 |
| G120 | 0.67 | 0.39 | 41.84 | 0.28 | 0.53 | 0.51 | 0.50 | 0.76 | 0.61 | 1.33 | 0.58 | 1.29 |
| G121 | 0.63 | 0.34 | 45.44 | 0.28 | 0.48 | 0.46 | 0.44 | 0.83 | 0.50 | 1.16 | 0.55 | 1.21 |
| G122 | 0.67 | 0.39 | 42.09 | 0.28 | 0.53 | 0.51 | 0.49 | 0.77 | 0.61 | 1.31 | 0.58 | 1.29 |
| G123 | 0.60 | 0.39 | 34.23 | 0.20 | 0.49 | 0.48 | 0.47 | 0.62 | 0.54 | 1.33 | 0.66 | 1.46 |
| G124 | 0.62 | 0.39 | 38.00 | 0.24 | 0.50 | 0.49 | 0.48 | 0.69 | 0.56 | 1.30 | 0.62 | 1.38 |
| G125 | 0.62 | 0.42 | 31.22 | 0.19 | 0.52 | 0.51 | 0.50 | 0.57 | 0.61 | 1.43 | 0.69 | 1.53 |
| G126 | 0.65 | 0.36 | 44.80 | 0.29 | 0.51 | 0.49 | 0.47 | 0.81 | 0.55 | 1.22 | 0.55 | 1.23 |
| G127 | 0.62 | 0.35 | 43.82 | 0.27 | 0.49 | 0.47 | 0.45 | 0.80 | 0.51 | 1.19 | 0.56 | 1.25 |
| G128 | 0.63 | 0.39 | 37.32 | 0.23 | 0.51 | 0.50 | 0.48 | 0.68 | 0.57 | 1.33 | 0.63 | 1.39 |
| G129 | 0.69 | 0.38 | 45.19 | 0.31 | 0.53 | 0.51 | 0.49 | 0.82 | 0.60 | 1.27 | 0.55 | 1.22 |
| G130 | 0.64 | 0.46 | 27.24 | 0.17 | 0.55 | 0.54 | 0.53 | 0.50 | 0.68 | 1.57 | 0.73 | 1.62 |
| G131 | 0.63 | 0.41 | 35.14 | 0.22 | 0.52 | 0.50 | 0.49 | 0.64 | 0.59 | 1.38 | 0.65 | 1.44 |
| G132 | 0.66 | 0.43 | 34.69 | 0.23 | 0.55 | 0.54 | 0.52 | 0.63 | 0.67 | 1.47 | 0.65 | 1.45 |
| G133 | 0.61 | 0.40 | 33.94 | 0.21 | 0.50 | 0.49 | 0.48 | 0.62 | 0.57 | 1.36 | 0.66 | 1.47 |
| G134 | 0.61 | 0.41 | 32.67 | 0.20 | 0.51 | 0.50 | 0.49 | 0.59 | 0.58 | 1.38 | 0.67 | 1.50 |
| G135 | 0.69 | 0.39 | 43.06 | 0.30 | 0.54 | 0.52 | 0.50 | 0.78 | 0.63 | 1.34 | 0.57 | 1.26 |
| G136 | 0.76 | 0.37 | 51.51 | 0.39 | 0.57 | 0.53 | 0.50 | 0.94 | 0.65 | 1.25 | 0.48 | 1.08 |
| G137 | 0.65 | 0.38 | 41.55 | 0.27 | 0.51 | 0.49 | 0.48 | 0.76 | 0.57 | 1.28 | 0.58 | 1.30 |
| G138 | 0.60 | 0.36 | 40.00 | 0.24 | 0.48 | 0.46 | 0.45 | 0.73 | 0.50 | 1.22 | 0.60 | 1.33 |
| G139 | 0.66 | 0.35 | 47.35 | 0.31 | 0.50 | 0.48 | 0.46 | 0.86 | 0.54 | 1.18 | 0.53 | 1.17 |
| G140 | 0.67 | 0.38 | 43.24 | 0.29 | 0.53 | 0.51 | 0.49 | 0.79 | 0.60 | 1.29 | 0.57 | 1.26 |
| G141 | 0.62 | 0.39 | 36.51 | 0.23 | 0.51 | 0.49 | 0.48 | 0.66 | 0.57 | 1.33 | 0.63 | 1.41 |
| G142 | 0.66 | 0.41 | 38.18 | 0.25 | 0.53 | 0.52 | 0.50 | 0.69 | 0.63 | 1.38 | 0.62 | 1.37 |
| G143 | 0.66 | 0.37 | 43.36 | 0.28 | 0.51 | 0.49 | 0.47 | 0.79 | 0.57 | 1.26 | 0.57 | 1.26 |
| G144 | 0.64 | 0.38 | 40.50 | 0.26 | 0.51 | 0.49 | 0.48 | 0.74 | 0.56 | 1.28 | 0.59 | 1.32 |
| G145 | 0.64 | 0.42 | 34.01 | 0.22 | 0.53 | 0.52 | 0.51 | 0.62 | 0.63 | 1.43 | 0.66 | 1.47 |
| G146 | 0.63 | 0.37 | 41.23 | 0.26 | 0.50 | 0.49 | 0.47 | 0.75 | 0.55 | 1.26 | 0.59 | 1.31 |
Table 3.
Ranking pattern of the 146 Aegilops accessions based on the shoot biomass-based indices.
| Code | Yp | Ys | TOL | MP | GMP | HM | SSI | STI | YI | YSI | RSI | AR | SD |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 21 | 60 | 93 | 35 | 52 | 58 | 70 | 52 | 60 | 70 | 70 | 58.3 | 19.1 |
| G2 | 21 | 77 | 121 | 62 | 75 | 76 | 87 | 75 | 77 | 87 | 87 | 76.8 | 23.7 |
| G3 | 11 | 51 | 93 | 15 | 31 | 42 | 64 | 31 | 51 | 64 | 64 | 47.0 | 24.2 |
| G4 | 18 | 57 | 90 | 24 | 42 | 52 | 65 | 42 | 57 | 65 | 65 | 52.5 | 20.3 |
| G5 | 45 | 71 | 89 | 75 | 76 | 72 | 73 | 76 | 71 | 73 | 73 | 72.2 | 10.3 |
| G6 | 1 | 67 | 144 | 4 | 26 | 59 | 105 | 26 | 67 | 105 | 105 | 64.5 | 46.8 |
| G7 | 9 | 69 | 130 | 33 | 61 | 66 | 85 | 61 | 69 | 85 | 85 | 68.5 | 30.9 |
| G8 | 28 | 74 | 103 | 69 | 73 | 73 | 76 | 73 | 74 | 76 | 76 | 72.3 | 17.2 |
| G9 | 47 | 82 | 99 | 89 | 85 | 83 | 83 | 85 | 82 | 83 | 83 | 81.9 | 12.6 |
| G10 | 4 | 86 | 141 | 25 | 70 | 79 | 112 | 70 | 86 | 112 | 112 | 81.5 | 39.8 |
| G11 | 13 | 55 | 97 | 19 | 34 | 48 | 66 | 34 | 55 | 66 | 66 | 50.3 | 24.2 |
| G12 | 25 | 82 | 123 | 74 | 81 | 82 | 95 | 81 | 82 | 95 | 95 | 83.2 | 23.4 |
| G13 | 15 | 33 | 64 | 8 | 11 | 18 | 50 | 11 | 33 | 50 | 50 | 31.2 | 19.8 |
| G14 | 31 | 91 | 124 | 84 | 87 | 89 | 103 | 87 | 91 | 103 | 103 | 90.3 | 22.8 |
| G15 | 16 | 73 | 120 | 46 | 69 | 70 | 80 | 69 | 73 | 80 | 80 | 70.5 | 25.1 |
| G16 | 14 | 109 | 140 | 86 | 95 | 102 | 131 | 95 | 109 | 131 | 131 | 103.9 | 34.8 |
| G17 | 41 | 90 | 114 | 91 | 88 | 90 | 96 | 88 | 90 | 96 | 96 | 89.1 | 17.6 |
| G18 | 26 | 79 | 116 | 73 | 79 | 78 | 88 | 79 | 79 | 88 | 88 | 79.4 | 21.1 |
| G19 | 55 | 53 | 57 | 57 | 56 | 55 | 53 | 56 | 53 | 53 | 53 | 54.6 | 1.7 |
| G20 | 35 | 100 | 131 | 92 | 93 | 96 | 109 | 93 | 100 | 109 | 109 | 97.0 | 23.5 |
| G21 | 38 | 20 | 42 | 14 | 12 | 15 | 35 | 12 | 20 | 35 | 35 | 25.3 | 11.7 |
| G22 | 36 | 61 | 80 | 49 | 58 | 62 | 62 | 58 | 61 | 62 | 62 | 59.2 | 10.6 |
| G23 | 34 | 63 | 83 | 52 | 64 | 65 | 68 | 64 | 63 | 68 | 68 | 62.9 | 12.0 |
| G24 | 63 | 88 | 92 | 94 | 90 | 88 | 81 | 90 | 88 | 81 | 81 | 85.1 | 8.6 |
| G25 | 20 | 68 | 110 | 44 | 66 | 68 | 75 | 66 | 68 | 75 | 75 | 66.8 | 21.9 |
| G26 | 73 | 108 | 108 | 99 | 102 | 105 | 108 | 102 | 108 | 108 | 108 | 102.6 | 10.3 |
| G27 | 11 | 89 | 135 | 64 | 82 | 85 | 106 | 82 | 89 | 106 | 106 | 86.8 | 31.2 |
| G28 | 37 | 102 | 131 | 93 | 94 | 97 | 110 | 94 | 102 | 110 | 110 | 98.2 | 23.1 |
| G29 | 68 | 95 | 98 | 96 | 96 | 93 | 94 | 96 | 95 | 94 | 94 | 92.6 | 8.3 |
| G30 | 27 | 77 | 113 | 72 | 77 | 77 | 82 | 77 | 77 | 82 | 82 | 76.6 | 19.7 |
| G31 | 19 | 80 | 126 | 66 | 78 | 80 | 93 | 78 | 80 | 93 | 93 | 80.5 | 25.6 |
| G32 | 65 | 65 | 63 | 78 | 74 | 67 | 59 | 74 | 65 | 59 | 59 | 66.2 | 6.6 |
| G33 | 78 | 98 | 96 | 97 | 98 | 99 | 97 | 98 | 98 | 97 | 97 | 95.7 | 5.9 |
| G34 | 23 | 101 | 135 | 87 | 91 | 95 | 115 | 91 | 101 | 115 | 115 | 97.2 | 28.5 |
| G35 | 2 | 63 | 142 | 2 | 18 | 49 | 98 | 18 | 63 | 98 | 98 | 59.2 | 46.3 |
| G36 | 87 | 96 | 87 | 98 | 99 | 98 | 90 | 99 | 96 | 90 | 90 | 93.6 | 4.8 |
| G37 | 52 | 93 | 109 | 95 | 92 | 91 | 100 | 92 | 93 | 100 | 100 | 92.5 | 14.5 |
| G38 | 45 | 59 | 68 | 52 | 57 | 60 | 57 | 57 | 59 | 57 | 57 | 57.1 | 5.5 |
| G39 | 10 | 76 | 133 | 38 | 67 | 71 | 92 | 67 | 76 | 92 | 92 | 74.0 | 31.6 |
| G40 | 33 | 62 | 83 | 50 | 63 | 64 | 67 | 63 | 62 | 67 | 67 | 61.9 | 12.3 |
| G41 | 24 | 56 | 86 | 34 | 46 | 53 | 63 | 46 | 56 | 63 | 63 | 53.6 | 16.4 |
| G42 | 50 | 66 | 78 | 75 | 72 | 69 | 69 | 72 | 66 | 69 | 69 | 68.6 | 7.2 |
| G43 | 28 | 91 | 129 | 81 | 86 | 87 | 104 | 86 | 91 | 104 | 104 | 90.1 | 24.7 |
| G44 | 39 | 85 | 107 | 84 | 84 | 84 | 91 | 84 | 85 | 91 | 91 | 84.1 | 16.4 |
| G45 | 53 | 53 | 59 | 56 | 55 | 54 | 54 | 55 | 53 | 54 | 54 | 54.5 | 1.8 |
| G46 | 39 | 80 | 102 | 79 | 83 | 81 | 79 | 83 | 80 | 79 | 79 | 78.5 | 14.7 |
| G47 | 50 | 50 | 55 | 44 | 50 | 50 | 51 | 50 | 50 | 51 | 51 | 50.2 | 2.5 |
| G48 | 55 | 72 | 82 | 83 | 80 | 74 | 71 | 80 | 72 | 71 | 71 | 73.7 | 7.8 |
| G49 | 3 | 84 | 145 | 12 | 62 | 75 | 119 | 62 | 84 | 119 | 119 | 80.4 | 44.8 |
| G50 | 124 | 146 | 122 | 140 | 146 | 146 | 145 | 146 | 146 | 145 | 145 | 141.0 | 9.1 |
| G51 | 62 | 121 | 134 | 100 | 111 | 120 | 137 | 111 | 121 | 137 | 137 | 117.4 | 22.2 |
| G52 | 60 | 141 | 139 | 110 | 125 | 138 | 143 | 125 | 141 | 143 | 143 | 128.0 | 24.9 |
| G53 | 93 | 128 | 125 | 115 | 122 | 124 | 135 | 122 | 128 | 135 | 135 | 123.8 | 12.0 |
| G54 | 6 | 106 | 143 | 68 | 89 | 100 | 133 | 89 | 106 | 133 | 133 | 100.5 | 39.1 |
| G55 | 111 | 136 | 116 | 126 | 130 | 136 | 138 | 130 | 136 | 138 | 138 | 130.5 | 9.3 |
| G56 | 108 | 122 | 103 | 119 | 121 | 121 | 128 | 121 | 122 | 128 | 128 | 120.1 | 8.0 |
| G57 | 133 | 69 | 22 | 106 | 97 | 86 | 42 | 97 | 69 | 42 | 42 | 73.2 | 33.9 |
| G58 | 135 | 128 | 71 | 136 | 133 | 131 | 114 | 133 | 128 | 114 | 114 | 121.5 | 18.8 |
| G59 | 140 | 112 | 52 | 133 | 124 | 117 | 86 | 124 | 112 | 86 | 86 | 106.5 | 26.2 |
| G60 | 146 | 75 | 11 | 124 | 104 | 92 | 27 | 104 | 75 | 27 | 27 | 73.8 | 45.2 |
| G61 | 127 | 126 | 79 | 132 | 128 | 126 | 120 | 128 | 126 | 120 | 120 | 121.1 | 14.5 |
| G62 | 88 | 113 | 111 | 105 | 110 | 112 | 121 | 110 | 113 | 121 | 121 | 111.4 | 9.4 |
| G63 | 115 | 118 | 93 | 118 | 118 | 119 | 118 | 118 | 118 | 118 | 118 | 115.5 | 7.5 |
| G64 | 122 | 142 | 105 | 134 | 141 | 143 | 140 | 141 | 142 | 140 | 140 | 135.5 | 11.7 |
| G65 | 79 | 143 | 137 | 117 | 132 | 140 | 144 | 132 | 143 | 144 | 144 | 132.3 | 19.5 |
| G66 | 81 | 115 | 118 | 103 | 109 | 113 | 124 | 109 | 115 | 124 | 124 | 112.3 | 12.4 |
| G67 | 66 | 134 | 138 | 107 | 120 | 129 | 141 | 120 | 134 | 141 | 141 | 124.6 | 22.3 |
| G68 | 121 | 137 | 101 | 130 | 135 | 137 | 136 | 135 | 137 | 136 | 136 | 131.0 | 11.0 |
| G69 | 142 | 125 | 61 | 141 | 134 | 128 | 107 | 134 | 125 | 107 | 107 | 119.2 | 23.3 |
| G70 | 145 | 122 | 49 | 146 | 138 | 130 | 99 | 138 | 122 | 99 | 99 | 117.0 | 28.8 |
| G71 | 132 | 138 | 81 | 143 | 143 | 139 | 132 | 143 | 138 | 132 | 132 | 132.1 | 17.6 |
| G72 | 96 | 119 | 115 | 113 | 117 | 118 | 126 | 117 | 119 | 126 | 126 | 117.5 | 8.4 |
| G73 | 41 | 145 | 146 | 111 | 136 | 145 | 146 | 136 | 145 | 146 | 146 | 131.2 | 31.7 |
| G74 | 115 | 144 | 127 | 131 | 142 | 144 | 142 | 142 | 144 | 142 | 142 | 137.7 | 9.4 |
| G75 | 131 | 124 | 70 | 135 | 130 | 125 | 111 | 130 | 124 | 111 | 111 | 118.4 | 18.2 |
| G76 | 123 | 130 | 90 | 129 | 127 | 127 | 127 | 127 | 130 | 127 | 127 | 124.0 | 11.4 |
| G77 | 138 | 139 | 77 | 144 | 144 | 141 | 130 | 144 | 139 | 130 | 130 | 132.4 | 19.2 |
| G78 | 140 | 140 | 75 | 145 | 145 | 142 | 129 | 145 | 140 | 129 | 129 | 132.6 | 20.2 |
| G79 | 135 | 132 | 74 | 138 | 137 | 134 | 122 | 137 | 132 | 122 | 122 | 125.9 | 18.3 |
| G80 | 125 | 87 | 43 | 108 | 100 | 94 | 60 | 100 | 87 | 60 | 60 | 84.0 | 25.1 |
| G81 | 126 | 113 | 66 | 127 | 119 | 116 | 101 | 119 | 113 | 101 | 101 | 109.3 | 17.1 |
| G82 | 98 | 127 | 119 | 116 | 123 | 123 | 134 | 123 | 127 | 134 | 134 | 123.5 | 10.4 |
| G83 | 130 | 105 | 50 | 123 | 112 | 109 | 74 | 112 | 105 | 74 | 74 | 97.1 | 25.1 |
| G84 | 110 | 102 | 73 | 104 | 103 | 104 | 84 | 103 | 102 | 84 | 84 | 95.7 | 12.1 |
| G85 | 109 | 97 | 72 | 101 | 101 | 101 | 77 | 101 | 97 | 77 | 77 | 91.8 | 13.2 |
| G86 | 91 | 117 | 112 | 109 | 113 | 115 | 125 | 113 | 117 | 125 | 125 | 114.7 | 9.7 |
| G87 | 129 | 102 | 48 | 121 | 108 | 106 | 72 | 108 | 102 | 72 | 72 | 94.5 | 24.9 |
| G88 | 128 | 110 | 54 | 124 | 116 | 111 | 78 | 116 | 110 | 78 | 78 | 100.3 | 24.0 |
| G89 | 139 | 131 | 67 | 142 | 140 | 133 | 117 | 140 | 131 | 117 | 117 | 124.9 | 21.5 |
| G90 | 144 | 98 | 30 | 128 | 114 | 107 | 58 | 114 | 98 | 58 | 58 | 91.5 | 35.5 |
| G91 | 142 | 120 | 56 | 137 | 129 | 122 | 102 | 129 | 120 | 102 | 102 | 114.6 | 23.9 |
| G92 | 102 | 116 | 100 | 112 | 115 | 114 | 116 | 115 | 116 | 116 | 116 | 112.5 | 5.9 |
| G93 | 120 | 107 | 65 | 114 | 107 | 108 | 89 | 107 | 107 | 89 | 89 | 100.2 | 15.6 |
| G94 | 133 | 133 | 76 | 138 | 139 | 135 | 123 | 139 | 133 | 123 | 123 | 126.8 | 18.0 |
| G95 | 100 | 135 | 128 | 122 | 126 | 132 | 139 | 126 | 135 | 139 | 139 | 129.2 | 11.4 |
| G96 | 85 | 111 | 106 | 102 | 105 | 110 | 113 | 105 | 111 | 113 | 113 | 106.7 | 8.2 |
| G97 | 135 | 93 | 39 | 120 | 106 | 103 | 61 | 106 | 93 | 61 | 61 | 88.9 | 29.6 |
| G98 | 49 | 57 | 62 | 60 | 59 | 61 | 56 | 59 | 57 | 56 | 56 | 57.5 | 3.5 |
| G99 | 32 | 16 | 45 | 9 | 9 | 10 | 37 | 9 | 16 | 37 | 37 | 23.4 | 14.2 |
| G100 | 79 | 3 | 6 | 10 | 4 | 3 | 4 | 4 | 3 | 4 | 4 | 11.3 | 22.6 |
| G101 | 117 | 37 | 14 | 80 | 60 | 47 | 17 | 60 | 37 | 17 | 17 | 45.7 | 32.1 |
| G102 | 5 | 18 | 85 | 1 | 2 | 4 | 52 | 2 | 18 | 52 | 52 | 26.5 | 28.9 |
| G103 | 106 | 7 | 4 | 36 | 21 | 13 | 5 | 21 | 7 | 5 | 5 | 20.9 | 29.9 |
| G104 | 73 | 10 | 19 | 20 | 13 | 9 | 14 | 13 | 10 | 14 | 14 | 19.0 | 18.2 |
| G105 | 8 | 16 | 69 | 3 | 3 | 7 | 46 | 3 | 16 | 46 | 46 | 23.9 | 23.4 |
| G106 | 58 | 29 | 37 | 30 | 28 | 28 | 33 | 28 | 29 | 33 | 33 | 33.3 | 8.7 |
| G107 | 82 | 5 | 10 | 16 | 8 | 6 | 7 | 8 | 5 | 7 | 7 | 14.6 | 22.5 |
| G108 | 43 | 33 | 47 | 21 | 22 | 25 | 41 | 22 | 33 | 41 | 41 | 33.5 | 9.7 |
| G109 | 68 | 45 | 41 | 52 | 48 | 46 | 43 | 48 | 45 | 43 | 43 | 47.5 | 7.5 |
| G110 | 94 | 1 | 1 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 9.8 | 27.9 |
| G111 | 83 | 31 | 27 | 47 | 37 | 35 | 24 | 37 | 31 | 24 | 24 | 36.4 | 17.0 |
| G112 | 73 | 15 | 24 | 30 | 23 | 20 | 21 | 23 | 15 | 21 | 21 | 26.0 | 16.1 |
| G113 | 16 | 26 | 60 | 7 | 7 | 11 | 44 | 7 | 26 | 44 | 44 | 26.5 | 18.8 |
| G114 | 111 | 4 | 2 | 27 | 14 | 8 | 2 | 14 | 4 | 2 | 2 | 17.3 | 32.1 |
| G115 | 71 | 35 | 33 | 40 | 33 | 34 | 30 | 33 | 35 | 30 | 30 | 36.7 | 11.7 |
| G116 | 30 | 39 | 58 | 17 | 19 | 26 | 48 | 19 | 39 | 48 | 48 | 35.5 | 14.2 |
| G117 | 86 | 18 | 21 | 37 | 30 | 24 | 19 | 30 | 18 | 19 | 19 | 29.2 | 19.9 |
| G118 | 53 | 47 | 53 | 41 | 44 | 45 | 49 | 44 | 47 | 49 | 49 | 47.4 | 3.7 |
| G119 | 57 | 46 | 50 | 43 | 45 | 44 | 47 | 45 | 46 | 47 | 47 | 47.0 | 3.8 |
| G120 | 58 | 24 | 31 | 23 | 20 | 21 | 28 | 20 | 24 | 28 | 28 | 27.7 | 10.7 |
| G121 | 100 | 51 | 34 | 88 | 71 | 63 | 40 | 71 | 51 | 40 | 40 | 59.0 | 21.6 |
| G122 | 63 | 27 | 31 | 29 | 25 | 23 | 29 | 25 | 27 | 29 | 29 | 30.6 | 11.0 |
| G123 | 119 | 24 | 8 | 75 | 53 | 40 | 11 | 53 | 24 | 11 | 11 | 39.0 | 34.4 |
| G124 | 104 | 28 | 18 | 69 | 47 | 37 | 18 | 47 | 28 | 18 | 18 | 39.3 | 26.9 |
| G125 | 107 | 8 | 5 | 38 | 24 | 17 | 6 | 24 | 8 | 6 | 6 | 22.6 | 29.9 |
| G126 | 77 | 43 | 38 | 59 | 49 | 43 | 38 | 49 | 43 | 38 | 38 | 46.8 | 11.9 |
| G127 | 103 | 48 | 29 | 82 | 65 | 57 | 36 | 65 | 48 | 36 | 36 | 55.0 | 22.6 |
| G128 | 97 | 22 | 17 | 52 | 36 | 31 | 16 | 36 | 22 | 16 | 16 | 32.8 | 24.2 |
| G129 | 48 | 37 | 44 | 25 | 27 | 30 | 39 | 27 | 37 | 39 | 39 | 35.6 | 7.4 |
| G130 | 91 | 2 | 3 | 11 | 5 | 2 | 3 | 5 | 2 | 3 | 3 | 11.8 | 26.4 |
| G131 | 98 | 13 | 13 | 42 | 32 | 22 | 13 | 32 | 13 | 13 | 13 | 27.6 | 25.5 |
| G132 | 67 | 6 | 16 | 12 | 6 | 5 | 12 | 6 | 6 | 12 | 12 | 14.5 | 17.8 |
| G133 | 113 | 14 | 9 | 66 | 38 | 32 | 9 | 38 | 14 | 9 | 9 | 31.9 | 32.5 |
| G134 | 114 | 11 | 7 | 60 | 35 | 27 | 8 | 35 | 11 | 8 | 8 | 29.5 | 32.7 |
| G135 | 44 | 20 | 40 | 17 | 15 | 16 | 31 | 15 | 20 | 31 | 31 | 25.5 | 10.4 |
| G136 | 7 | 42 | 87 | 6 | 10 | 19 | 55 | 10 | 42 | 55 | 55 | 35.3 | 26.7 |
| G137 | 83 | 35 | 28 | 51 | 40 | 36 | 26 | 40 | 35 | 26 | 26 | 38.7 | 16.6 |
| G138 | 117 | 44 | 20 | 90 | 68 | 56 | 22 | 68 | 44 | 22 | 22 | 52.1 | 31.7 |
| G139 | 70 | 49 | 46 | 64 | 54 | 51 | 45 | 54 | 49 | 45 | 45 | 52.0 | 8.2 |
| G140 | 60 | 29 | 36 | 32 | 29 | 29 | 32 | 29 | 29 | 32 | 32 | 33.5 | 9.0 |
| G141 | 105 | 22 | 15 | 62 | 39 | 33 | 15 | 39 | 22 | 15 | 15 | 34.7 | 27.6 |
| G142 | 71 | 11 | 23 | 22 | 16 | 14 | 20 | 16 | 11 | 20 | 20 | 22.2 | 16.7 |
| G143 | 76 | 41 | 35 | 47 | 41 | 39 | 34 | 41 | 41 | 34 | 34 | 42.1 | 12.0 |
| G144 | 90 | 32 | 24 | 57 | 43 | 38 | 23 | 43 | 32 | 23 | 23 | 38.9 | 20.1 |
| G145 | 89 | 9 | 12 | 28 | 17 | 12 | 10 | 17 | 9 | 10 | 10 | 20.3 | 23.5 |
| G146 | 94 | 39 | 26 | 71 | 51 | 41 | 25 | 51 | 39 | 25 | 25 | 44.3 | 21.9 |
Fig. 1.
Genotype ranking in ascending order for the MGIDI index. The selected genotypes based on this index are shown in red. The central red circle represents the cutpoint according to the selection pressure.
Fig. 2.
Strengths and weaknesses view of the selected genotypes is shown as the proportion of each factor on the computed MGIDI index. The smallest the proportion explained by a factor (closer to the external edge), the closer the traits within that factor are to the ideotype. The dashed line indicates the theoretical value if all factors had contributed equally.
2. Experimental Design, Materials and Methods
2.1. Plant materials
The plant materials consisted of 146 accessions form Ae. tauschii (DD-genome) Ae. cylindrica (DDCC-genome), and Ae. crassa (DDMM-genome). Additional information on the studied materials is shown in supplementary Table S1. These accessions were accessed from Ilam University Genbank (IUGB).
2.2. Experimental design
Before sowing, seeds of all accessions were stored to 4 °C for 72 h for break dormancy. Five seeds per accession were sown in plastic pots that were filled with a mixture of sand, soil, and humus (1:1:1). All pots were arranged in a factorial experimental based on a randomized complete block in three replications under an optimal growing photoperiod (16 h light, 8 h dark) and temperature (25 °C day, 20±2 °C night) conditions. Seedlings were exposed to the following water treatments at the three-leaf growth stage: (1) well-watered (full field capacity [FC] = 95 ± 5%) as the control and (2) water-stressed (FC = 30 ± 5%) conditions. Thirty days after sowing and applying stress treatment, shoot samples were harvested and exposed to 70 °C for 72 h to measure shoot dry matter.
2.3. Data collection
The data used in this work were collected by measuring Yp and Ys as the shoot dry matter under control and stress conditions, respectively. Nine drought tolerance and susceptibility indices (Table 1), including tolerance index (TOL), mean productivity (MP), geometric mean productivity (GMP), harmonic mean (HM), stress susceptibility index (SSI), stress tolerance index (STI), yield index (YI), yield stability index (YSI), and relative drought index (RSI), were then calculated using iPASTIC software [7].
Table 1.
Mathematical formulas of nine drought tolerance and susceptibility indices.
| No. | Index | Formula | Pattern of selection | Reference |
|---|---|---|---|---|
| 1 | Tolerance Index | Minimum value | [8] | |
| 2 | Mean Productivity | Maximum value | [8] | |
| 3 | Geometric Mean Productivity | Maximum value | [9] | |
| 4 | Harmonic Mean | Maximum value | [10] | |
| 5 | Stress Susceptibility Index | Minimum value | [11] | |
| 6 | Stress Tolerance Index | Maximum value | [9] | |
| 7 | Yield Index | Maximum value | [12] | |
| 8 | Yield Stability Index | Maximum value | [13] | |
| 9 | Relative Stress Index | Maximum value | [14] |
2.4. Statistical analysis
The multi-trait genotype-ideotype distance index (MGIDI) was used to rank the accessions based on information of multiple indices as proposed by Olivoto and Nardino [15]. In the first step, each trait (rXij) was rescaled using the following equation:
Where and are the original minimum and maximum values for the index j, respectively; and are the new minimum and maximum values for index j after rescaling, respectively; and is the original value for jth index of the ith accession. The values for and were chosen as follows: for the indices in which positive gains are desired, = 0 and = 100 should be used, while for the indices in which negative gains are desired, = 100 and = 0 should be used [15]. In the next step, a factor analysis (FA) was performed to account for the dimensionality reduction of the data and relationships structure. This analysis was performed according to following model:
where F is a g × f matrix with the factorial score; Z is a g × p matrix with the rescaled means; A is a p × f matrix of canonical loading, and R is a p × p correlation matrix between the indices. Furthermore, g, f, and p indicate the number of accessions, factor retained, and calculated indices, respectively. In the third step, a [1 × p] vector was considered as the ideotype matrix. In the last step, Euclidean distance between the scores of accessions and the ideal accessions was computed as the MGIDI index using following equation:
where γij is the score of the ith accession in the jth factor (i = 1, 2,…,t; j = 1,2,…,f ), being t and f the number of accessions and factors, respectively; and γj is the jth score of the ideal accession. The accession with the lowest MGIDI is then closer to the ideal accession and thus indicates desired values for all the calculated indices. The selection differential for all traits was performed considering a selection intensity of approximately 10%. Data manipulation and the index computation were performed in the R software using the ‘metan’ package [15].
Ethics Statement
The paper is not currently being considered for publication elsewhere.
CRediT Author Statement
Alireza Pour-Aboughadareh: Conceptualization, Methodology, Software, Data curtion, Writing original draft, Investigation; Peter Poczai: Visualization, Writing review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.107096.
Contributor Information
Alireza Pour-Aboughadareh, Email: a.poraboghadareh@ut.ac.ir.
Peter Poczai, Email: peter.poczai@helsinki.fi.
Appendix. Supplementary materials
References
- 1.Pour-Aboughadareh A., Omidi M., Naghavi M.R., Etminan A., Mehrabi A.A., Poczai P., Bayat H. Effect of water deficit stress on seedling biomass and physio-chemical characteristics in different species of wheat possessing the D genome. Agronomy. 2019;9:522. doi: 10.3390/agronomy9090522. [DOI] [Google Scholar]
- 2.Liu P.Martre, Ewert F., Porter J.R., Challinor A.J., Muller C., Ruane A.C., Waha K., Thorburn P.J., Aggarwal P.K. Global wheat production with 1.5 and 2.0 C above pre-industrial warming. Glob. Chang. Biol. 2019;25(4):1428–1444. doi: 10.1111/gcb.14542. [DOI] [PubMed] [Google Scholar]
- 3.Suneja Y., Gupta A.K., Bains N.S. Stress adaptive plasticity: Aegilops tauschii and Triticum dicoccoides as potential donors of drought associated morpho-physiological traits in wheat. Front. Plant Sci. 2019;10:211. doi: 10.3389/fpls.2019.00211. 0.3389/fpls.2019.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadi J., Pour-Aboughadareh A., Fabriki Ourang S., Khalili P., Poczai P. Unraveling salinity stress responses in ancestral and neglected wheat species at early growth stage: a baseline for utilization in future wheat improvement programs. Physiol. Mol. Biol. Plants. 2020;26:537–549. doi: 10.1007/s12298-020-00768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pour-Aboughadareh A., Omidi M., Naghavi M.R., Etminan A., Mehrabi A.A., Poczai P. Wild relatives of wheat respond well to water deficit stress: a comparative study of antioxidant enzyme activities and their encoding gene expression. Agriculture. 2020;10:415. doi: 10.3390/agriculture10090415. [DOI] [Google Scholar]
- 6.Zhao X., Bai S., Li L., Han X., Li J., Zhu Y., Fang Y., Zhang D., Li S. Comparative transcriptome analysis of two Aegilops tauschii with contrasting drought tolerance by RNA-Seq. Int. J. Mol. Sci. 2020;21:3595. doi: 10.3390/ijms21103595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pour-Aboughadareh A., Yousefian M., Moradkhani H., Moghaddam Vahed M., Poczai P., Siddique K.H.M. iPASTIC: an online toolkit to estimate plant abiotic stress indices. Appl Plant Sci. 2019;7(7):e11278. doi: 10.1002/aps3.11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosielle A.A., Hambling J. Theoretical aspects of selection for yield in stress and non-stress environments. Crop. Sci. 1981;21:943–946. doi: 10.2135/cropsci1981.0011183X002100060033x. [DOI] [Google Scholar]
- 9.Fernandez G.C.J. (1992) Effective selection criteria for assessing stress tolerance. In: Kuo, C.G. (Ed.), Proceedings of the International Symposium on Adaptation of Vegetables and Other Food Crops in Temperature and Water Stress, Publication. Tainan, Taiwan.
- 10.Bidinger F.R., Mahalakshmi V., Rao G.D. Assessment of drought resistance in pearl millet (Pennisetum americanum (L.) Leeke). II. Estimation of genotype response to stress. Aust. J. Agric. Res. 1987;38:49–59. doi: 10.1071/AR9870049. [DOI] [Google Scholar]
- 11.Fischer R.A., Maurer R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978;29:897–912. doi: 10.1071/AR9780897. [DOI] [Google Scholar]
- 12.Gavuzzi P., Rizza F., Palumbo M., Campaline R.G., Ricciardi G.L., Borghi B. Evaluation of field and laboratory predictors of drought and heat tolerance in winter cereals. Can. J. Plant. Sci. 1997;77:523–531. doi: 10.4141/P96-130. [DOI] [Google Scholar]
- 13.Bouslama M., Schapaugh W.T. Stress tolerance in soybean. Part 1: evaluation of three screening techniques for heat and drought tolerance. Crop. Sci. 1984;24:933–937. doi: 10.2135/cropsci1984.0011183X002400050026x. [DOI] [Google Scholar]
- 14.Fischer R.A., Wood T. Drought resistance in spring wheat cultivars ІІІ. Yield association with morphological traits. Aust. J. Agric. Res. 1979;30:1001–1020. doi: 10.1071/AR9791001. [DOI] [Google Scholar]
- 15.T. Olivoto, M. Nardino, MGIDI: toward an effective multivariate selection in biological experiments. Bioinformatics (2020), doi: 10.1093/bioinformatics/btaa981. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.