Abstract
Literature survey shows that convolutional neural network (CNN)-based pretrained models have been largely used for CoronaVirus Disease 2019 (COVID-19) classification using chest X-ray (CXR) and computed tomography (CT) datasets. However, most of the methods have used a smaller number of data samples for both CT and CXR datasets for training, validation, and testing. As a result, the model might have shown good performance during testing, but this type of model will not be more effective on unseen COVID-19 data samples. Generalization is an important term to be considered while designing a classifier that can perform well on completely unseen datasets. Here, this work proposes a large-scale learning with stacked ensemble meta-classifier and deep learning-based feature fusion approach for COVID-19 classification. The features from the penultimate layer (global average pooling) of EfficientNet-based pretrained models were extracted and the dimensionality of the extracted features reduced using kernel principal component analysis (PCA). Next, a feature fusion approach was employed to merge the features of various extracted features. Finally, a stacked ensemble meta-classifier-based approach was used for classification. It is a two-stage approach. In the first stage, random forest and support vector machine (SVM) were applied for prediction, then aggregated and fed into the second stage. The second stage includes logistic regression classifier that classifies the data sample of CT and CXR into either COVID-19 or Non-COVID-19. The proposed model was tested using large CT and CXR datasets, which are publicly available. The performance of the proposed model was compared with various existing CNN-based pretrained models. The proposed model outperformed the existing methods and can be used as a tool for point-of-care diagnosis by healthcare professionals.
Keywords: COVID-19, CT scan, Chest X-rays, Computer-aided diagnosis, Transfer learning, Deep learning, Meta-classifier, Stacked classifier, Feature fusion
Introduction
The World Health Organisation (WHO) reported that the cases of COVID-19 were first detected in Wuhan City, China [1]. This outbreak caused significant loss of lives in a few days and has become an international concern. With exponential spread across the countries reporting increased confirmed cases followed by deaths characterised the disease as pandemic within a few weeks. In the current situation, COVID-19 risks getting the world off track to achieve Sustainable Development Goals (SDGs), says the World Health statistics 2020 [2]. Affecting millions across the world with more than a million deaths increased the fatality rate of the disease making it epidemic. A recently updated report by the WHO states that there are about 1,599,922 deaths all over the world, out of which 143,355 are from India [3]. Currently, the diagnosis of COVID-19 is based on two types such as molecular and antigen tests to detect the viral genetic material and its protein, respectively. Reverse transcription polymerase chain reaction (RT-PCR), a molecular detection test involves various point-of-care (POC) kits and devices [4]. Shortage in RT-PCR kits during the epidemic situation delays tests on suspected cases and risks the disease control and prevention measures. The professionally laboratory trained individuals can only use the RT-PCR kits and instruments in a laboratory setting. Developing countries with malnourished people not only intensify the risk of the spread among the citizens but also cannot afford such resources in mass. Contrarily, radiology-based diagnosis methods are cost-effective and less time consuming which also identifies the clinical changes in the lungs to be used for early diagnosis [4]. As the mortality rate of COVID-19 is high on late treatments, early diagnosis helps to control the spread of the infection. Onsets of COVID-19 caused by SARS-Cov 2 are similar to pneumonia. The WHO reported that SARS-associated coronavirus was first discovered in 2003 during an outbreak affecting five countries which emerged in China. Later in December 2019, the SARS-CoV-2 virus began to transmit the infection to humans from Rhinolophus bat. The most common symptoms of COVID-19 are fever and cough. Respiratory infections like respiratory viruses and influenza also have similar symptoms associating the difficulty in control of this pandemic [5].
Methods like CXR and CT scan are medical imaging techniques, which are widely used to find the infection spread with the patient’s lungs. Unlike RT-PCR, medical imaging techniques are non-invasive and cost-effective techniques that are available worldwide. CT scans are capable of projecting the organs in a 3-D representation including the thin tissues. It has been widely used for detecting the abnormalities in the lungs such as tumors, excess fluids, and pneumonia. COVID-19 being a similar infection can be well diagnosed using CT scans. Chest X-rays detect and represent the dense tissues in the organs in 2-D view making it easily affordable for hospitals and laboratories all over the world. Proven experiments concluded that CT scans show significant differences between these respiratory diseases including COVID-19 [6]. Also, during the COVID-19 pandemic in Italy, adoption of CXR alongside RT-PCR mitigated the oscillating sensitivity of the RT-PCR test results [7]. Alongside, CXRs were also helpful for extensive pneumonia and critically ill COVID patients on diagnosis and faster treatment [8]. CXRs helps the radiologists to confirm the presence of COVID-19 infection with the pathological information. An epidemic situation not only demands early diagnosis but also faster and most accurate test results with high sensitivity to support treatment. Clinical trials of COVID-19 diagnosis and other similar respiratory infections suggest CT scans and CXRs can be used for early classification of patients and severity checking on the infection spread. CXRs are less costly than CT scans, but most countries have sufficient amounts of CT devices. Both CT scans and CXRs are equally helpful in diagnosing COVID-19 infection and so employing both imaging techniques can help to screen the COVID-19 patients. The CT and CXR images are required and considered to be an efficient method for providing point-of-care diagnosis by healthcare professionals.
Though CXRs and CT scans help in early diagnosis, decision-making by radiologists takes time particularly with the pandemic situations. Developing countries with high population encounters out of proportion between test samples and radiologists during an outbreak. Automating the diagnosis helps the radiologist for faster and accurate decision-making. Computer-aided diagnosis methods are used in several biomedical applications such as diabetic eye disease diagnosis, cancer metastases detection, tuberculosis detection and classification [9]. Advances in machine learning algorithms contributed in classifying CXRs in biomedical applications such as pneumonia, tuberculosis, etc [9]. Deep learning, as ChexNet [10], has been found to perform better and faster than radiologist-level performance. Deep learning techniques for classifying and detecting COVID-19 have been implemented not just with CXRs but also CT scans and blood samples [11]. The main issues with the machine learning method were that these methods are dependent on feature extraction and feature engineering. These are considered as daunting tasks and the performance of the classifier depends on optimal features. However, deep learning has the capability to learn optimal features itself and avoid additional feature extraction and feature engineering [12]. Deep learning-based medical image classification approaches have shown expert-level accuracy and in some tasks such as CT and CXR, deep learning-based CAD approaches could surpass radiologists [12]. CT scan-based and CXR-based identification and detection of COVID-19 have been implemented using pretrained networks such as InceptionV3, VGGNet, InceptionResNetV2, ResNet, etc., and achieved benchmark accuracies as high as 99% [13]. Other novel methods are hybrid learning networks, such as CheXNet pneumonia, have achieved 99.99% accuracy rate in classifying CXRs [14].
Applications of deep learning-based networks for classification or detection usually require large samples of patient COVID-19 and Non-COVID-19 CT or CXR data. To overcome disadvantages of limited data, techniques such as transfer learning and data augmentations are employed. Most existing deep learning techniques employed to classify COVID-19 CXRs or CT scans use the pretrained architectures to transfer and learn high level features from images. Though these methods have achieved higher accuracy rates in testing, the test data samples used to quantify the performance is however limited. Such evaluation might help with test data under control but not in real time. The main challenge is the generalization that is the model’s ability to perform equally with unobserved data that asserts how well a deep learning algorithm will perform in the real time. The network error can be reduced by training and also testing the deep learning algorithm with some large number of samples. Considering these factors, the following are the major contributions of the proposed work:
This work proposes a large-scale learning with stacked ensemble meta-classifier and deep learning-based feature fusion approach for COVID-19 classification.
The performance evaluation of various CNN-based EfficientNet pretrained models were tested for COVID-19 classification using CXR and CT datasets.
The detailed investigation and analysis for all the models were shown and the performance evaluation of all the models reported on large-scale publicly available benchmark datasets of CT and CXR for COVID-19 classification.
The performance of the proposed method was compared with various existing CNN-based pretrained models.
t-distributed stochastic neighbour embedding (t-SNE) visualization approach was employed to ensure that the learned features were meaningful for COVID-19 classifications.
The performance of the CNN-based models were shown for COVID-19 classification using both CT and CXR datasets with the aim to create a robust and generalized system.
The rest of this paper is organized as follows. Section 2 includes the literature survey on COVID-19 classification using CXR and CT images. The proposed architectures and its details are included in Sect. 3. Description of CT and CXR COVID-19 datasets are included in Sect. 4. The metrics used to evaluate the models are discussed in detail in Sect. 5. Results, discussion, and visualization are included in Sect. 6. Finally, conclusions are drawn in Sect. 7.
Literature survey
Medical imaging techniques integrated with artificial intelligence (AI) can be widely used to fight against COVID-19 in the entire pipeline such as image acquisition, screening, and follow-up. Machine learning-based methods has been in practice for healthcare in various types of medical imaging techniques such as CT scans, X rays, Mammograms, and Pathological images. Some of the recent advancements include, a fuzzy classification and segmentation system for analysing mammography images to diagnose breast cancer [15], Deep learning-based malaria detection from blood smear microscopic images with unsupervised learning [16] and so on. With a good quality dataset trained on an efficient deep learning model can assist diagnosis in a productive manner. A survey on the open source datasets availability in COVID-19 reports the importance of the medical images like CT scans and CXRs dataset for diagnosing the infection in a separate section [17]. Shi et al. reviews the datasets used in the AI research for COVID-19 and also summarizes the recent deep learning methods implemented on discriminating the COVID-19 from other diseases using radiological images [18].
The opportunity of digital technologies such as AI with deep learning, blockchain, and big data analytics to fight against COVID-19 is explained by Ting et al. [19]. This enhanced detection and analysis of the disease likely to provide cost-effective and accurate diagnosis helping the developing countries. In a review that depicts the use of recent deep learning and AI algorithms for COVID-19 with the advancements of medical imaging elaborates challenges and open problems [20]. This review suggests improving the quality and quantity of the datasets to implement the deep learning algorithm on CXRs and thoracic CT scans. Furthermore, the importance of classifying the radiological images at the early stage of the disease is well illustrated. Deep transfer learning and self-supervised methods are suggested to mitigate the expensive manual labeling of large samples of data. Application of deep transfer learning techniques for both CT scans and CXRs were studied for several lung infections along with COVID-19 [21]. Automating the diagnosis using AI and deep learning methods were highly recommended by these authors in the epidemic circumstances. To implement an effective deep learning based algorithm for automatic diagnosis, it is important to take care of the biases and quality of the methodology. Most of the existing techniques to diagnose COVID-19 from CT scans and X-rays have major limitation of dataset used for training or mistakes in methodologies introducing biases [22]. It is always suggested to provide a replicable methodology for clinical use and also for external validation.
CT-based methods
Augmenting AI with radiologist’s performance has shown significant improvement in distinguishing COVID-19 CTs from other pneumonia CTs [23]. As differentiating the CT scans of COVID-19 from pneumonia is challenging unlike other lung diseases, assisting radiologists with a deep learning model has improved the performance on an average of 5%. Though the model helped the screening, there is an involvement of bias due to limited samples in the dataset. Also, the performance decline on external validation is noted due to the lack of generalization. Performances of ten different CNNs were compared on classifying COVID-19 CT slices from Non-COVID-19 slices [24]. All the networks used to diagnose the infections were pretrained on the massive ImageNet dataset and the samples used for training and testing are of only 1020 image patches from CT slices. The bias might have been introduced in the models due to small sample-size training and the decrease in performance can lead to overfitting while testing other unobserved diverse data. To avoid overfitting, a novel multi-view representation learning was proposed by Kang et al. [25]. A well-structured latent representation of the extracted features from CT images is learnt to classify COVID-19 infections with better stability, generalization, and robustness.
Extended residual learning with the prior-attention mechanism achieves the state of the art performance in lobe segmentation for automatic pneumonia detection and pneumonia type classification including COVID-19 infection [26]. The main drawback of this model is the vulnerability of misclassifying normal CTs as pneumonia and COVID-19 at early stages. Most common reasons of misclassification in medical images are due to the lack of balance in data samples between positive and negative cases. An online attention mechanism combined with dual sample learning was introduced to overcome the imbalance in COVID-19 and community acquired pneumonia (CAP) [27]. This method performed cross-validation to induce generalization in the model. Some of the insufficient visual evidence from the attention maps resulted in false negatives of COVID-19 scans was noticeable from this method. Another way to deal with the overfitting problem with segmentation is to use a large set of data samples, but this requires the cost of annotating large samples which is time-consuming and not affordable. A weekly supervised method was proposed to classify as well as detect COVID-19 lesions without the need of annotating training samples [28]. With the patient-level annotation the segmentation task is made unsupervised using the activation regions of the supervised deep CNN on classifying COVID-19 CT scans. This method does not include the CAP class and so the level of performance has been under study for similar pneumonia diseases. A similar method for minimizing the manual annotation was proposed by Hu et al. [29], which is a multi-scale learning framework for detecting and classifying the COVID-19 from CAP CT scans. The main limitation of this work is that the model is less separable between classes CAP and COVID-19. Covid CT-Net was introduced along with the ground truth manual annotation of more than 2,000 CT images [30]. The proposed network is an attention CNN and the annotated data was introduced as an open source. The handling of limited data was to combine CT data with other external features. Harmon et al. [31] proposed an AI-based model trained with CT images and non-image information such as other laboratory testing, clinical symptoms, and exposure history. This method was highly successful in finding the probability of a patient being COVID-19 positive but biased towards the COVID-19 in training data and tested against very small numbers of samples of training and testing sets.
Chest X-ray-based methods
Several AI-based methods were proposed to detect and classify the COVID-19 cases from chest X-rays. OptCoNet is a CNN designed to identify COVID-19 CXRs from normal and pneumonia CXRs [32]. A swarm intelligent (SI) optimization algorithm implemented during the training of the CNN helped the network achieve good accuracy. This method has a major difficulty in choosing the right parameters for the optimizer involved. Transfer learning methods can help tackle the training with limited data. Jain et al. [33] examined the performance of three different pretrained CNN networks to initiate the future research on hyperparameter tuning and developing similar detection algorithms. Similarly, five popular CNNs [34] implemented with the transfer learning approach experienced limited CXRs for the COVID-19 class. A multi-task learning method called COVID-MTNet was proposed to detect COVID-19 from CXRs, segmented CXRs and CT scan slices, and localize the infection regions in the medical images using transfer learning approach [35]. Another deep transfer learning approach to detect COVID-19 CXRs is the Deep-COVID [36], which was implemented with four other pretrained networks to overcome the small sample size of the COVID-19 class. To deal with the drawback of limited samples, random patches from the lung-segmented CXRs were extracted [37]. Those extracted patches were trained on a pretrained CNN to obtain the classes. This method also visualizes the working of the CNN through saliency mapping technique. In many deep transfer learning techniques presented, the improvement in performances was suggested commonly based on adding more data and hyperparameter tuning. Kumar et al. [38] proposed a method for improving deep transfer learned features with machine learning classifiers. This method also addressed the limitation of imbalance in data samples between the classes through resampling technique. Another approach for improving a transfer learning approach is proposed by Turkoglu et al. [39], where features from all the layers of the pre-trained network were extracted and combined. From these combined features, only important features were selected based on a feature selection method with which an SVM classifier was used to classify the COVID-19 CXRs. In medical imaging applications, there is a high chance of irregularities between the classes in the dataset. To cope up with such limitations, Abbas et al. [40] proposed DeTrac CNN, which implements class decomposition algorithm on features learned from pretrained architectures. Apart from the implementation of these fine-grained classification techniques, combined cost-sensitive learning technique has outperformed other methods in classification accuracy [40]. Most existing techniques have limitations with respect to limited samples in positive class and also in complete datasets. To deal with disparities between data samples in classes, synthetic augmentation technique using a CycleGAN was proposed [41]. By generating COVID-19 X-rays through GAN, the quality of the generated data is always uncertain.
The aforementioned literature survey shows that various CNN-based pretrained models were employed for COVID-19 classification using CT and CXR datasets. However, most existing methods have the following issues:
Performances of various CNN-based pretrained models were shown on very small datasets of COVID-19 CT and CXR. Due to overfitting, these models might achieve good performances during training, but they might perform poorly with datasets obtained from a variety of different patients.
There are no detailed investigations and analysis of COVID-19 classification.
Most studies on COVID-19 classification using either CT or CXR datasets, but not both.
Here, this study proposes a novel approach based on the EfficientNet pretrained model. The proposed approach is presented and discussed in the following sections.
Proposed architecture for COVID-19 classification
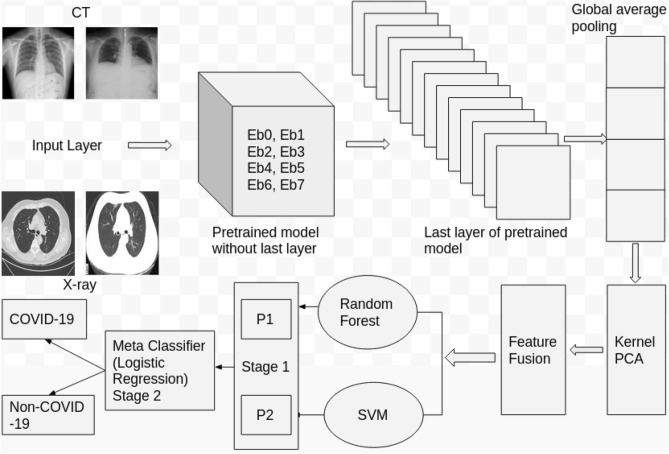
The proposed architecture for COVID-19 classification using CT and CXR datasets is shown in Fig. 1. The architecture uses an EfficientNet [42]-based transfer learning approach. Reusing the weights of trained models in other applications for performing similar tasks is known as transfer learning. This type of learning approach for COVID-19 classification can reduce the training time, faster convergence rate, and achieve optimal performances in detecting patients’ data samples of CT or CXR as either COVID-19 or Non-COVID-19. The EfficientNet model was trained on an ImageNet database, which has more than one million images. This database has 1000 classes. This model learnt rich features which represent images from different classes. To achieve better performances on the ImageNet database, researchers used different CNN architectures that have the same property with different scaling schemes. Scaling means arbitrarily increasing the CNN model depth or width or usage of larger input image resolution for training and evaluation. In this work, these models were reused as a transfer learning approach with the aim to transfer similar performances for the COVID-19 classification. The proposed architecture is composed of 3 steps that are discussed as follows.
Fig. 1.
Proposed architecture for COVID-19 classification
Preprocessing In this step, all images of CT and CXR are resized to pixels and normalization is employed to convert the data into 0–1 range. Here, the value of n depends on the architecture and the value of n for EfficientNetB0 (Eb0), EfficientNetB1 (Eb1), EfficientNetB2 (Eb2), EfficientNetB3 (Eb3), EfficientNetB4 (Eb4), EfficientNetB5 (Eb5), EfficientNetB6 (Eb6), and EfficientNetB7 (Eb7) are 224, 240, 260, 300, 380, 456, 528, and 600, respectively.
Feature extraction Literature survey shows that CNN based pretrained models demonstrated outstanding performances in medical image classification [19]. The detailed investigation and analysis of various CNN-based pretrained models are necessary to identify an optimal model. The performances of the pretrained models varies across tasks using different datasets. Applying various pretrained models and identifying the best model have been considered as an active area of research [19]. In this work we employ various models of EfficientNet architecture for COVID-19 classification using CT and CXR datasets. All models in EfficientNet composed of an input layer, convolution layer, pooling layer, and fully connected layer. An input layer takes an input in the form of tensor and the following convolution layer employs convolution operation to extract features. The features of the convolution layer are called a feature map. To reduce the dimension of the feature map, pooling is used. Fully connected layer serves as an output layer that contains two neurons. One neuron is for COVID-19 and another one is for Non-COVID-19. In a fully connected layer, each neuron has connections to every other neuron of the previous layer.
Classification Finally, the penultimate layer (global average pooling) features of Eb0, Eb1, Eb2, Eb3, E4, Eb5, Eb6, and Eb7 models are extracted. Eb0, Eb1, Eb2, Eb3, E4, Eb5, Eb6, and Eb7 architectures have the same property with different scaling schemes. Thus, each model have the capability to extract its own unique features. Thus, feature fusion approach is employed for the features of Eb0, Eb1, Eb2, Eb3, E4, Eb5, Eb6, and Eb7 architectures. The dimension of extracted features of Eb0, Eb1, Eb2, Eb3, E4, Eb5, Eb6, and Eb7 are 1,280, 1,280, 1,408, 1,536, 1,792, 2,048, 2,304, and 2,560, respectively. We applied the kernel PCA, and dimensionality of extracted features of the model were reduced to 160. The kernel PCA parameters are n_components =160, and kernel = rbf and other parameter values are default values provided by scikit-learn. Finally, the features of Eb0, Eb1, Eb2, Eb3, Eb4, Eb5, E6, and Eb7 models are merged, i.e. , here 160 is the dimension of extracted features of Eb0, Eb1, Eb2, Eb3, Eb4, Eb5, E6, and Eb7 models and 8 denotes the number of EfficientNet models. These were again passed into t-SNE for visualization and shown in Fig. 2a for COVID-19 classification using CT dataset and Fig. 2b for COVID-19 classification using CXR dataset. This figure shows that data samples for both CT and CXR datasets are highly non-linearly separable and require some complex learning classifier. We used stacking, which is a two-stage approach or ensemble approach, where the predictions of a set of models (base classifiers) are aggregated and fed into the second-stage predictor (meta-classifier). This type of classifier has the capability to show better performance than any single model in the ensemble [43]. The base classifiers are the random forest and SVM, while the meta classifier is the logistic regression. For the random forest, we set n_components = 70 and criterion = gini, for SVM, the kernel was set to rbf, and . The logistic regression uses the sigmoid activation function, which outputs 0 or 1, where 0 indicates COVID-19 positive and 1 indicates COVID-19 negative. Other parameter values of SVM, random forest, and logistic regression are default values provided by scikit-learn.
Fig. 2.
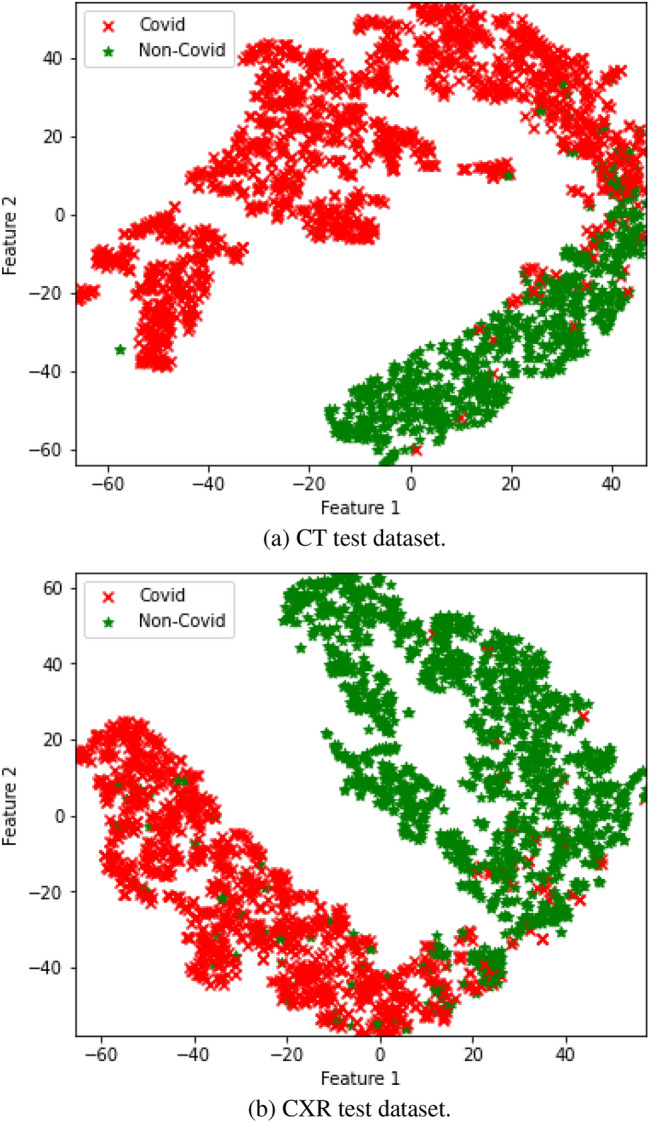
t-SNE feature representation
Data
There are many CT and CXR-based image datasets available publicly but most of the datasets are small. This may be due to security and privacy reasons. These types of datasets can hinder the research in deep learning as well the creation of a more robust and generalized system for COVID-19 classification using CT and CXR images. By considering these issues, recently, Mendeley [44] has made the COVID-19 CT and CXR datasets available publicly for research purposes. This type of datasets helps to develop more robust and generalized COVID-19 classification. This dataset has more numbers of COVID-19 and Non-COVID-19 images for both CT and CXR compared to existing datasets. Various data augmentation methodologies were used by the authors to generate many samples for both CXR and CT. The CT dataset contains 8055 images and the CXR dataset contains 9544 images. The data were randomly divided into training and testing using the scikit-learn1 train-test split methodology. The detailed statistics of both CT and CXR datasets are given in Table 1. Some samples of CT and CXRs datasets are shown in Fig. 3. The first row of figure is of CT data and second row is of CXR. In each row, the first two images are COVID-19 positive and next two are COVID-19 negative.
Table 1.
COVID-19 CT and CXR data information
| Dataset | Type | COVID | Non-COVID | Total |
|---|---|---|---|---|
| CT | Train | 3795 | 1843 | 5638 |
| Test | 1632 | 785 | 2417 | |
| CXR | Train | 2832 | 3848 | 6680 |
| Test | 1212 | 1652 | 2864 |
Fig. 3.
Randomly chosen CT and CXR samples [44]
Performance metrics
The following metrics were used to assess the performances of the proposed model for COVID-19 classification using CT and CXR datasets.
| 1 |
| 2 |
| 3 |
| 4 |
where TP, FP, TN, and FN are true positive, false positive, true negative and false negative, respectively. In this work, TP, FP, TN, and FN are defined as follows:
TP = number of COVID-19 samples predicted accurately as COVID-19.
FN = number of COVID-19 samples incorrectly predicted as non-COVID-19.
FP = number of non-COVID-19 samples incorrectly predicted as COVID-19.
TN = number non-COVID-19 samples predicted accurately as non-COVID-19.
The TP, FP, TN, and FN were obtained from a confusion matrix. It is a table that displays and compares actual values with the predicted values. To assess the performance of the proposed model at each class level, precision, recall, and F1 score were used along with accuracy. The precision, recall, and F1 score are reported for both weighted and macro. Macro is considered as a good metric for precision, recall, and F1 score in the case of imbalanced datasets. Macro metric computes the precision, recall, and F1 score for each class and returns the average without considering the proportion for each class in the CT or CXR dataset. The weighted metric computes the precision, recall, and F1 score for each class and returns the average by considering the proportion for each class in the CT or CXR dataset.
Experiments and results
All the models of this work were implemented using TensorFlow2 with Keras.3 The experiments of all the models were run using Google Colab4 with K80 GPU and 25 GB RAM.
The proposed work for COVID-19 classification using CT and CXRs images was based on the EfficientNet pretrained model. The EfficientNet architecture consists of a compound scaling factor that uniformly scales all dimensions of depth, width, and resolution. It provides a family of models such as Eb0, Eb1, Eb2, Eb3, Eb4, Eb5, Eb6, and Eb7. These were proposed based on a good combination of efficiency and accuracy on a variety of scales of image inputs. The required input image data resolution for Eb0, Eb1, Eb2, Eb3, Eb4, Eb5, Eb6, and Eb7 are , and , respectively. Along with EfficientNet, Xception, VGG16, VGG19, ResNet50, ResNet101, ResNet152, ResNet50V2, ResNet101V2, ResNet152V2, InceptionV3, InceptionResNetV2, MobileNet, MobileNetV2, DenseNet121, DenseNet169, DenseNet201, NASNetMobile, and NASNetLarge pretrained CNN-based architectures were used in this study for COVID-19 classification using CT and CXRs images. For Xception, VGG16, VGG19, ResNet50, ResNet101, ResNet152, ResNet50V2, ResNet101V2, ResNet152V2, InceptionV3, InceptionResNetV2, MobileNet, MobileNetV2, DenseNet121, DenseNet169, DenseNet201, NASNetMobile, and NASNetLarge input image size are , and , respectively. During training, optimizer, learning rate, batch size, and epochs parameter value were set as adam, 0.001, 32, and 20 respectively. The training data samples were shuffled for each epoch during training and the models were initialized with ImageNet pretrained model weights. The trainable parameters for Eb0, Eb1, Eb2, Eb3, Eb4, Eb5, Eb6, and Eb7 are 4,008,829, 6,514,465, 7,702,403, 10,697,769, 17,550,409, 28,342,833, 40,738,009, and 63,789,521, respectively. The trainable parameter details for Xception, VGG16, VGG19, ResNet50, ResNet101, ResNet152, ResNet50V2, ResNet101V2, ResNet152V2, InceptionV3, InceptionResNetV2, MobileNet, MobileNetV2, DenseNet121, DenseNet169, DenseNet201, NASNetMobile, and NASNetLarge are 20,809,001, 14,715,201, 20,024,897, 23,536,641, 42,554,881, 58,221,569, 23,521,409, 42,530,945, 58,189,953, 21,770,401, 54,277,729, 3,208,001, 2,225,153, 6,954,881, 12,486,145, 18,094,849, 4,234,035, and 84,724,183, respectively.
The EfficientNet-based CNN model train accuracy and loss curve for CT and CXR are shown in Figs. 4a, b and 5a, b, respectively. The figures show a steady increase and steady decrease in train accuracy and loss, respectively, on both CT and CXR datasets. Most model trainings converged within 20 epochs. The train and loss curves for other CNN-based pretrained models were made publicly available.5 Most of the CNN-based pretrained models have shown more than 90% train accuracy except ResNet152V2, ResNet101V2, VGG16, and VGG19. However, the performances of these models were not the same during testing. These pretrained models showed poorer performance compared to the EfficientNet and the proposed method based on the EfficientNet during testing. This may be due to overfitting. The existing CNN-based pretrained models were ineffective to detect the variants of CT and CXR samples and unseen CT and CXR samples. To improve the performance of a Deep Convolution Network, model dimensions are scaled. To implement the improvisation of overall performance, it is important to balance the depth, width, and resolution of the architecture through scaling. For most CNN-based pretrained models, such as Xception, VGG, ResNet, Inception, and DenseNet, scaling is to arbitrarily increase CNN depth or width, or to use larger input image resolution for training, validation, and testing. While the manual tuning improves performance in these architectures, the choice of scaling is empirically found and also the increase in performance is trivial. To avoid the aforementioned limitations during scaling up CNNs, EfficientNet model was introduced [42]. This architecture is designed with compound scaling to achieve better model efficiency and performance compared to other standard scaling methods. In addition, EfficientNet models provide higher accuracy for both transfer learning and training from scratch methods with benchmark datasets.
Fig. 4.
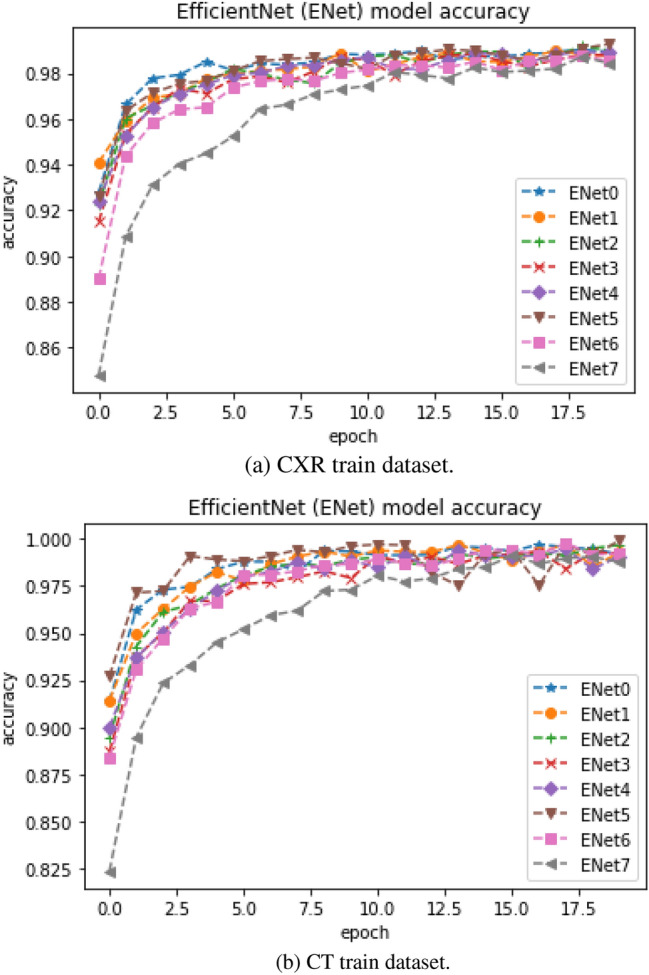
EfficientNet model train accuracy
Fig. 5.
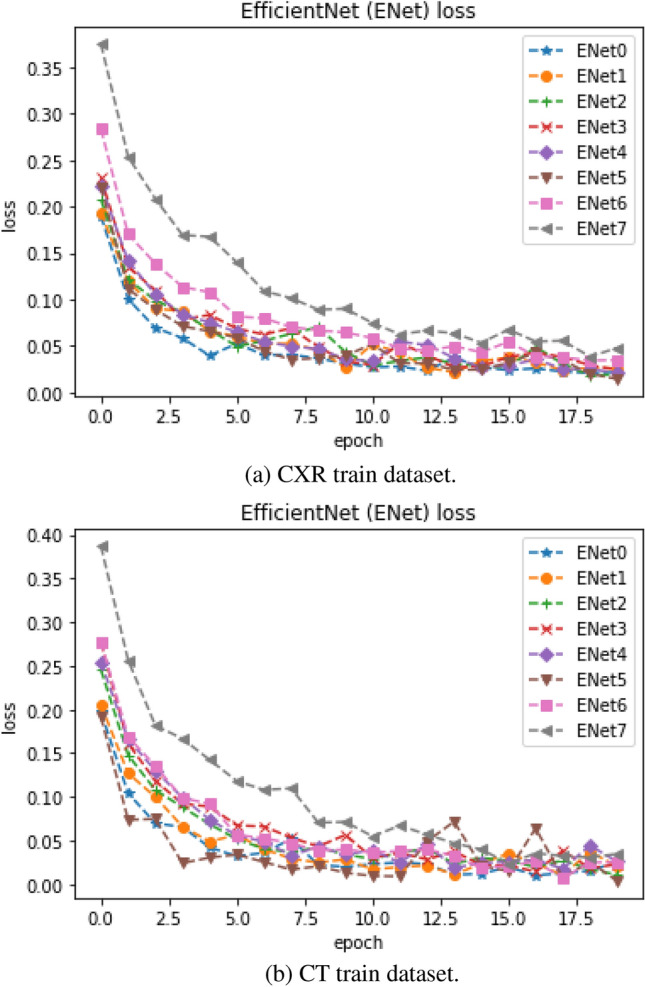
EfficientNet model train loss
We compared the performance of the proposed method with other existing methods for COVID-19 classification using CT and CXR datasets. The results obtained from the proposed method and other CNN-based pretrained models for COVID-19 classification on both CT and CXR datasets are reported in Tables 2 and 4, respectively. Detailed results obtained from the proposed method are shown in Table 3 for the CT dataset and Table 5 for the CXR dataset. Based on the results shown in tables, the proposed method outperformed the other CNN-based pretrained models by having higher values in accuracy, precision, recall, and F1 score. Most importantly, the proposed method has shown better performance than the existing methods in macro precision, macro recall, and macro F1 score. As a result, the proposed model has the capability to handle imbalanced datasets during training, validation, and testing. Particularly, the proposed method has showed weighted and macro F1 score of 0.99 for both CT and CXR test datasets.
Table 2.
Results for COVID-19 classification using CT images
| Model | Accuracy | Type | Precision | Recall | F1-score |
|---|---|---|---|---|---|
| Xception [45] | 0.98 | Macro | 0.97 | 0.98 | 0.97 |
| Weighted | 0.98 | 0.98 | 0.98 | ||
| VGG16 [21] | 0.68 | Macro | 0.34 | 0.50 | 0.40 |
| Weighted | 0.46 | 0.68 | 0.54 | ||
| VGG19 [40] | 0.83 | Macro | 0.81 | 0.85 | 0.82 |
| Weighted | 0.86 | 0.83 | 0.83 | ||
| ResNet50 [46] | 0.86 | Macro | 0.84 | 0.87 | 0.85 |
| Weighted | 0.88 | 0.86 | 0.86 | ||
| ResNet101 [47] | 0.86 | Macro | 0.84 | 0.88 | 0.85 |
| Weighted | 0.88 | 0.86 | 0.86 | ||
| InceptionV3 [33] | 0.95 | Macro | 0.95 | 0.94 | 0.94 |
| Weighted | 0.95 | 0.95 | 0.95 | ||
| InceptionResNetV2 [47] | 0.96 | Macro | 0.96 | 0.95 | 0.96 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| MobileNetV2 [47] | 0.71 | Macro | 0.75 | 0.78 | 0.71 |
| Weighted | 0.83 | 0.71 | 0.72 | ||
| DenseNet121 [36] | 0.89 | Macro | 0.91 | 0.84 | 0.87 |
| Weighted | 0.90 | 0.89 | 0.89 | ||
| DenseNet201 [47] | 0.89 | Macro | 0.91 | 0.84 | 0.87 |
| Weighted | 0.90 | 0.89 | 0.89 | ||
| NASNetMobile [47] | 0.68 | Macro | 0.34 | 0.50 | 0.40 |
| Weighted | 0.46 | 0.68 | 0.54 | ||
| NASNetLarge [47] | 0.68 | Macro | 0.34 | 0.50 | 0.40 |
| Weighted | 0.46 | 0.68 | 0.54 | ||
| ResNet152 | 0.76 | Macro | 0.85 | 0.64 | 0.64 |
| Weighted | 0.81 | 0.76 | 0.71 | ||
| ResNet50V2 | 0.90 | Macro | 0.88 | 0.92 | 0.89 |
| Weighted | 0.92 | 0.90 | 0.90 | ||
| ResNet101V2 | 0.80 | Macro | 0.88 | 0.69 | 0.71 |
| Weighted | 0.84 | 0.80 | 0.77 | ||
| ResNet152V2 | 0.80 | Macro | 0.88 | 0.70 | 0.72 |
| Weighted | 0.84 | 0.80 | 0.78 | ||
| MobileNet | 0.96 | Macro | 0.95 | 0.96 | 0.96 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| DenseNet169 | 0.94 | Macro | 0.95 | 0.91 | 0.92 |
| Weighted | 0.94 | 0.94 | 0.93 | ||
| EfficientNetB0 | 0.98 | Macro | 0.97 | 0.98 | 0.97 |
| Weighted | 0.98 | 0.98 | 0.98 | ||
| EfficientNetB1 | 0.96 | Macro | 0.97 | 0.94 | 0.95 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| EfficientNetB2 | 0.98 | Macro | 0.98 | 0.97 | 0.97 |
| Weighted | 0.98 | 0.98 | 0.98 | ||
| EfficientNetB3 | 0.97 | Macro | 0.95 | 0.97 | 0.96 |
| Weighted | 0.97 | 0.97 | 0.97 | ||
| EfficientNetB4 | 0.96 | Macro | 0.95 | 0.97 | 0.96 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| EfficientNetB5 | 0.98 | Macro | 0.98 | 0.97 | 0.97 |
| Weighted | 0.98 | 0.98 | 0.98 | ||
| EfficientNetB6 | 0.97 | Macro | 0.97 | 0.96 | 0.96 |
| Weighted | 0.97 | 0.97 | 0.97 | ||
| EfficientNetB7 | 0.97 | Macro | 0.96 | 0.96 | 0.96 |
| Weighted | 0.97 | 0.97 | 0.97 | ||
| Proposed | 0.99 | Macro | 0.99 | 0.99 | 0.99 |
| Weighted | 0.99 | 0.99 | 0.99 |
Table 4.
Results for COVID-19 classification using CXR images
| Model | Accuracy | Type | Precision | Recall | F1 score |
|---|---|---|---|---|---|
| Xception [45] | 0.79 | Macro | 0.83 | 0.82 | 0.79 |
| Weighted | 0.86 | 0.79 | 0.79 | ||
| VGG16 [21] | 0.90 | Macro | 0.91 | 0.89 | 0.90 |
| Weighted | 0.91 | 0.90 | 0.90 | ||
| VGG19 [40] | 0.58 | Macro | 0.29 | 0.50 | 0.37 |
| Weighted | 0.33 | 0.58 | 0.42 | ||
| ResNet50 [46] | 0.88 | Macro | 0.90 | 0.87 | 0.88 |
| Weighted | 0.89 | 0.88 | 0.88 | ||
| ResNet101 [47] | 0.80 | Macro | 0.79 | 0.80 | 0.79 |
| Weighted | 0.81 | 0.80 | 0.80 | ||
| InceptionV3 [33] | 0.82 | Macro | 0.84 | 0.84 | 0.82 |
| Weighted | 0.85 | 0.82 | 0.82 | ||
| InceptionResNetV2 [47] | 0.95 | Macro | 0.95 | 0.95 | 0.95 |
| Weighted | 0.95 | 0.95 | 0.95 | ||
| MobileNetV2 [47] | 0.58 | Macro | 0.29 | 0.50 | 0.37 |
| Weighted | 0.33 | 0.58 | 0.42 | ||
| DenseNet121 [36] | 0.91 | Macro | 0.91 | 0.92 | 0.91 |
| Weighted | 0.92 | 0.91 | 0.91 | ||
| DenseNet201 [47] | 0.92 | Macro | 0.93 | 0.92 | 0.92 |
| Weighted | 0.92 | 0.92 | 0.92 | ||
| NASNetMobile [47] | 0.58 | Macro | 0.29 | 0.50 | 0.37 |
| Weighted | 0.33 | 0.58 | 0.42 | ||
| NASNetLarge [47] | 0.42 | Macro | 0.21 | 0.50 | 0.30 |
| Weighted | 0.18 | 0.42 | 0.25 | ||
| ResNet152 | 0.73 | Macro | 0.79 | 0.76 | 0.72 |
| Weighted | 0.81 | 0.73 | 0.72 | ||
| ResNet50V2 | 0.89 | Macro | 0.90 | 0.89 | 0.89 |
| Weighted | 0.90 | 0.89 | 0.89 | ||
| ResNet101V2 | 0.87 | Macro | 0.87 | 0.88 | 0.87 |
| Weighted | 0.88 | 0.87 | 0.87 | ||
| ResNet152V2 | 0.87 | Macro | 0.87 | 0.88 | 0.87 |
| Weighted | 0.88 | 0.87 | 0.87 | ||
| MobileNet | 0.96 | Macro | 0.96 | 0.96 | 0.96 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| DenseNet169 | 0.79 | Macro | 0.80 | 0.78 | 0.78 |
| Weighted | 0.79 | 0.79 | 0.79 | ||
| EfficientNetB0 | 0.95 | Macro | 0.95 | 0.95 | 0.95 |
| Weighted | 0.95 | 0.95 | 0.95 | ||
| EfficientNetB1 | 0.96 | Macro | 0.96 | 0.96 | 0.96 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| EfficientNetB2 | 0.96 | Macro | 0.96 | 0.95 | 0.95 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| EfficientNetB3 | 0.96 | Macro | 0.96 | 0.95 | 0.95 |
| Weighted | 0.96 | 0.96 | 0.96 | ||
| EfficientNetB4 | 0.94 | Macro | 0.94 | 0.93 | 0.94 |
| Weighted | 0.94 | 0.94 | 0.94 | ||
| EfficientNetB5 | 0.97 | Macro | 0.96 | 0.96 | 0.96 |
| Weighted | 0.97 | 0.97 | 0.97 | ||
| EfficientNetB6 | 0.95 | Macro | 0.95 | 0.95 | 0.95 |
| Weighted | 0.95 | 0.95 | 0.95 | ||
| EfficientNetB7 | 0.95 | Macro | 0.95 | 0.95 | 0.95 |
| Weighted | 0.95 | 0.95 | 0.95 | ||
| Proposed | 0.99 | Macro | 0.99 | 0.99 | 0.99 |
| Weighted | 0.99 | 0.99 | 0.99 |
Table 3.
Best performed model results for COVID-19 classification using CT Images
| Class | Precision | Recall | F1 score |
|---|---|---|---|
| COVID-19 | 0.99 | 0.99 | 0.99 |
| Non-COVID-19 | 1.00 | 1.00 | 1.00 |
Table 5.
Best performed model detailed results for COVID-19 classification using CXR images
| Class | Precision | Recall | F1 score |
|---|---|---|---|
| COVID-19 | 0.99 | 0.99 | 0.99 |
| Non-COVID-19 | 1.00 | 1.00 | 1.00 |
Tables 2 and 4 show that the proposed method based on large-scale learning stacked meta-classifier with EfficientNet CNN-based pretrained model classification results are higher for COVID-19 detection using CT and CXR datasets than the EfficientNet CNN-based pretrained models. More importantly, the proposed method has achieved better performances on both datasets compared to the EfficientNet CNN-based pretrained models. As a result, the proposed model is more generalizable compared to the EfficientNet CNN-based model. Moreover, combining SVM with CNN for classification has the capability to show better performance than the softmax function at the last layer of CNN [48]. This is because SVM can produce good decision surfaces when applied to well-behaved feature vectors. Mostly, CNN has the capability to extract optimal invariant feature vectors by passing the raw data into many hidden layers. By considering this all, this work employs the large-scale learning stacked meta-classifier approach for COVID-19 classification using CT and CXR datasets, resulting in better performances than the existing various CNN-based pretrained models, and most importantly outperformed various EfficientNet CNN-based models.
The confusion matrix for the proposed method for COVID-19 classification is shown in Fig. 6b for the CT dataset and Fig. 6a for the CXR dataset. The confusion matrices for other EfficientNet CNN-based pretrained models for COVID-19 classification using the CXR and CT datasets were made publicly available.6 In CXR dataset, 7 samples of COVID-19 were misclassified as Non-COVID-19 and 8 samples of Non-COVID-19 were misclassified as COVID-19 by proposed method. For CT dataset, 6 samples of COVID-19 were misclassified as Non-COVID-19 and 7 samples of Non-COVID-19 were misclassified as COVID-19 by the proposed method. Overall, the proposed method achieved 0.9948 accuracy and 0.0052 misclassification rate for the CXR data, and 0.9946 accuracy and 0.0054 misclassification rate for the CT data.
Fig. 6.
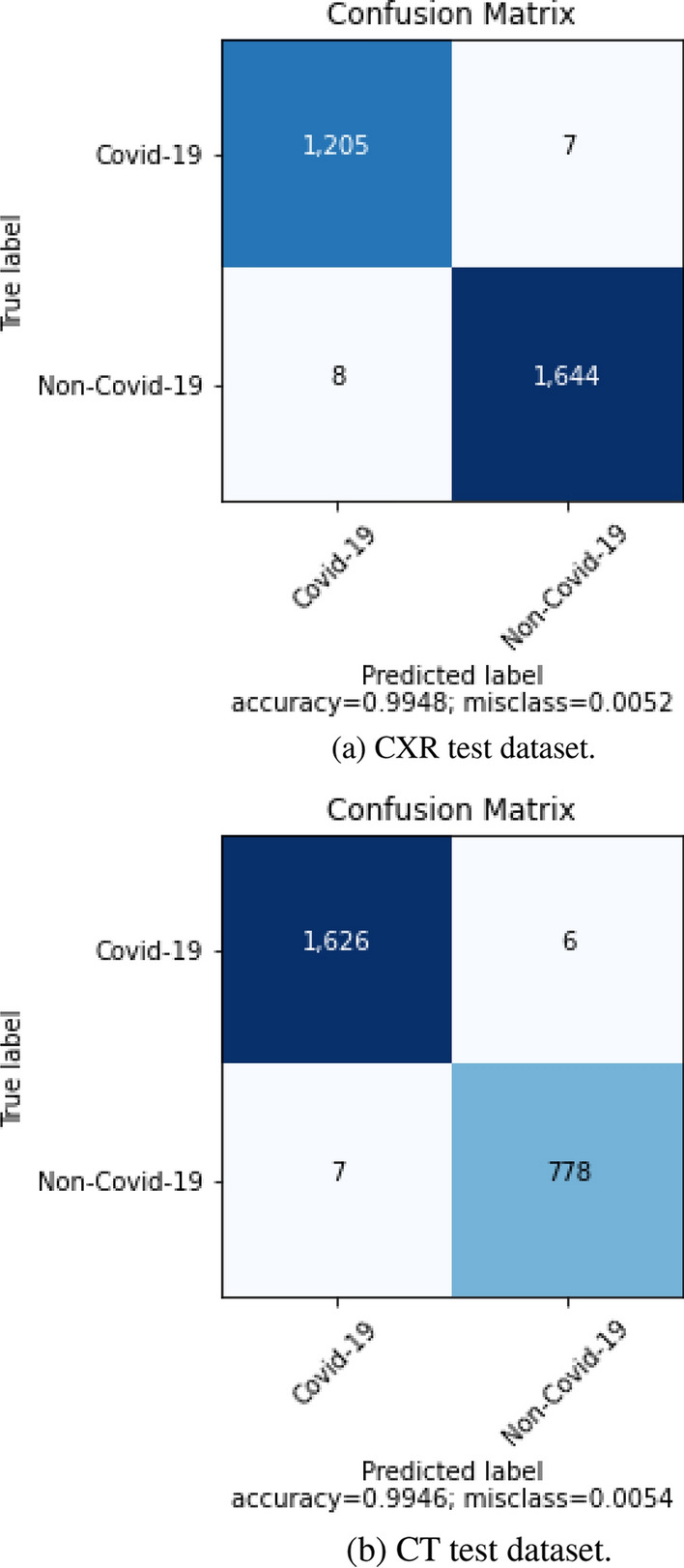
Confusion matrix obtained from the proposed approach for COVID-19 classification
Comparisons with related works
With the aim to develop more generalized and robust system for COVID-19 classification using CT and CXR images, the proposed approach is compared with the various related studies based on pretrained CNN-based models. In the existing studies, the experimental analysis was carried out with a smaller number of COVID-19 samples of CT and CXR. In most of these studies, the datasets were highly imbalanced, and had smaller number of samples for COVID-19 compared to Non-COVID-19. Most of the studies used data augmentation approach to increase the number of data samples of COVID-19. However, in recent study it has been reported that the data augmentation approach was not efficient to handle imbalanced datasets, also data augmentation did not improve the performance for COVID-19 classification [47]. Though most of these datasets are available publicly, in this work, the experiments were run again on large COVID-19 datasets [44] of CT and CXR, and the results were reported in Tables 2 and 4 for CT and X-ray datasets, respectively.
The existing pretrained models for COVID-19 classification are Xception [45], Densenet121 [36], VGG19 [40], InceptionV3 [33], VGG16 [21], Resnet50 [46], ResNet101 [47], InceptionResNetV2 [47], MobileNetV2 [47], DenseNet201 [47], NASNetMobile [47], and NASNetLarge [47]. The performance of the proposed model in terms of accuracy, macro and weighted precision, macro and weighted recall, and macro and weighted F1-score, is superior to all other models.
Generalization is an important aspect in machine learning, which indicates the model’s ability to adapt to new or variants of existing data and it is unseen during creating a model [49]. These samples are from the same distribution as the one used to create the model. The databases of training and testing for both COVID-19 CT and X-ray are completely unseen during creating a model and these were from the same distribution as the one used to create the model. It can be seen from Figs. 4a, b, and 5a, b, EfficientNet models achieved more than 95% accuracy and less than 0.05 loss, and their performances during the testing attained similar performances. Most models achieved more than 95% accuracy. However, this may not be the same with the other CNN-based pretrained models.
This indicates the proposed EfficientNet-based models are more generalizable towards classifying new or variants of existing COVID-19 patient data samples of CT and X-ray.
Visualization
Recently, interpretability is considered as an important concept in deep learning[50 ]. Thus, this work adopts the t-SNE feature visualization that is shown in Fig. 2a for the test dataset of CT, and Fig. 2b for the test dataset of CXR. The penultimate layers of the best performed models such as Ebo, Eb1, Eb2, Eb3, Eb4, Eb5, Eb6, and Eb7 features were extracted for the testing dataset and passed to t-SNE. The t-SNE implicitly used the PCA to reduce the higher dimension of features into two dimensions. The two-dimensional data are the first and second principal components. The parameter value for t-SNE n_components = 2, perplexity = 30.0, early_exaggeration = 12.0, learning_rate = 200.0, n_iter = 1000, n_iter_without_progressint = 300, min_grad_norm = 1e−7, metric=euclidean, init=random, and other parameter values are default values provided by scikit-learn. This practice allowed us to verify the COVID-19 and Non-COVID-19 samples in separate clusters. Though COVID-19 had a distinct pattern compared to Non-COVID-19, still both showed an overlapping region between them. This indicates that further study is required to analyze the misclassified samples and a further development needed to minimize misclassified samples.
Conclusions
Though many deep learning-based CNN pretrained models were proposed for COVID-19 classification using CT and CXR, most of these methods were trained and tested on much smaller numbers of samples of both CT and CXR datasets. This work has proposed a deep learning-based meta-classifier approach for COVID-19 classification. The proposed approach has been investigated on large-scale publicly available CT and CXR datasets. The proposed approach uses EfficientNet-based pretrained models for feature extraction and feature dimensionality reduction. The features can be combined and passed into stacked meta-classifiers. The stacked meta-classifier employs the random forest and SVM for prediction and passes them into the second stage for classification. The second stage uses the logistic regression to classify unlabelled samples into COVID-19 or Non-COVID-19. The proposed model has been shown to outperform other CNN pretrained models.
As an issue for future research, instead of simply combining all the features together, various recent advanced methods of feature fusion methods can be explored to further enhance the performance of the proposed model. In addition, in the stacked classifier, there is no analytical rationale for selecting the random forest and SVM to be used in the first stage and the logistic regression in the second stage. Thus, more investigation of stacked classifiers towards achieving better performance on COVID-19 classification can be considered. This work has not shown experiments for choosing the best possible setting of the deep learning network parameters and network architecture. Instead, the performances were shown for various CNN-based pretrained models for COVID-19 and Non-COVID-19 classification using large-scale CT and CXR datasets. Since the network parameters and architectures have direct impact on the performances, more study is suggested to identify the optimal network parameters and network structures of the pretrained CNN based architectures.
The results of the proposed method show that the best performed model has misclassification samples of COVID-19 and Non-COVID-19 for both CT and X-ray dataset. The misclassification rate of the proposed model on both CT and X-ray dataset is less and the same thing cannot be expected when dealing with real-time data COVID-19 patient samples in future. Thus the proposed approach is very limited in handline imbalanced datasets of COVID-19. This can be avoided using any of the data imbalance approaches and this is mentioned as future work.
Acknowledgements
The authors are thankful to all the reviewers and editors for their valuable suggestions to enhance the depth and quality of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vinayakumar Ravi, Email: vravi@pmu.edu.sa.
Harini Narasimhan, Email: harinin@iitk.ac.in.
Chinmay Chakraborty, Email: cchakrabarty@bitmesra.ac.in.
Tuan D. Pham, Email: tpham@pmu.edu.sa
References
- 1.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mil. Med. Res. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . World Health Statistics 2020: Monitoring Health for the SDGs Sustainable Development Goals. Geneva: World Health Organization; 2020. [Google Scholar]
- 3.World Health Organization.: Coronavirus disease 2019 (COVID-19): situation report. 198 (2020)
- 4.Asrani, P., Eapen, M.S., Chia, C., Haug, G., Weber, H.C., Hassan, M.I., Sohal, S.S.: Diagnostic approaches in COVID-19: clinical updates. Expert Rev. Respir. Med. 1–16 (2020) [DOI] [PubMed]
- 5.Zhao, D., Yao, F., Wang, L., Zheng, L., Gao, Y., Ye, J., Gao, R.: A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin. Infect. Dis. (2020) [DOI] [PMC free article] [PubMed]
- 6.Song, F., Shi, N., Shan, F., et al.: Emerging coronavirus 2019-nCoV pneumonia. Radiology 200–274 (2020). 10.1148/radiol.2020200274
- 7.Schiaffino S, Tritella S, Cozzi A, Carriero S, Blandi L, Ferraris L, Sardanelli F. Diagnostic performance of chest X-ray for COVID-19 pneumonia during the SARS-CoV-2 pandemic in Lombardy, Italy. J. Thorac. Imaging. 2020;35(4):W105–W106. doi: 10.1097/RTI.0000000000000533. [DOI] [PubMed] [Google Scholar]
- 8.Wu, G., Li, X.: Mobile X-rays are highly valuable for critically ill COVID patients. Eur. Radiol. 1 (2020) [DOI] [PMC free article] [PubMed]
- 9.Esteva, A., Chou, K., Yeung, S., Naik, N., Madani, A., Mottaghi, A., Socher, R. Deep learning-enabled medical computer vision. NPJ Digit. Med. 4(1), 1–9 [DOI] [PMC free article] [PubMed]
- 10.Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan, T., Ng, A. Y.: Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. (2017). arXiv preprint arXiv:1711.05225
- 11.Lalmuanawma, S., Hussain, J., Chhakchhuak, L.: Applications of machine learning and artificial intelligence for Covid-19 (SARS-CoV-2) pandemic: a review. Chaos Solitons Fractal 110059 (2020) [DOI] [PMC free article] [PubMed]
- 12.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, Sánchez CI. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Kassani, S.H., Kassasni, P.H., Wesolowski, M.J., Schneider, K.A., Deters, R.: Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning-based approach. (2020). arXiv preprint arXiv:2004.10641 [DOI] [PMC free article] [PubMed]
- 14.Al-Waisy, A.S., Al-Fahdawi, S., Mohammed, M.A., Abdulkareem, K.H., Mostafa, S.A., Maashi, M.S., Garcia-Zapirain, B.: COVID-CheXNet: hybrid deep learning framework for identifying COVID-19 virus in chest X-rays images. Soft. Comput. 1–16 (2020) [DOI] [PMC free article] [PubMed] [Retracted]
- 15.Chowdhary CL, Mittal M, Pattanaik PA, Marszalek Z. An efficient segmentation and classification system in medical images using intuitionist possibilistic fuzzy C-mean clustering and fuzzy SVM algorithm. Sensors. 2020;20(14):3903. doi: 10.3390/s20143903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattanaik PA, Mittal M, Khan MZ. Unsupervised deep learning cad scheme for the detection of malaria in blood smear microscopic images. IEEE Access. 2020;8:94936–94946. doi: 10.1109/ACCESS.2020.2996022. [DOI] [Google Scholar]
- 17.Mohamadou Y, Halidou A, Kapen PT. A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl. Intell. 2020;50(11):3913–3925. doi: 10.1007/s10489-020-01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi, F., Wang, J., Shi, J., Wu, Z., Wang, Q., Tang, Z., Shen, D.: Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. (2020) [DOI] [PubMed]
- 19.Ting DSW, Carin L, Dzau V, Wong TY. Digital technology and COVID-19. Nat. Med. 2020;26(4):459–461. doi: 10.1038/s41591-020-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shuja, J., Alanazi, E., Alasmary, W., Alashaikh, A.: Covid-19 open source data sets: a comprehensive survey. Appl. Intell. 1–30 (2020) [DOI] [PMC free article] [PubMed]
- 21.Perumal, V., Narayanan, V., Rajasekar, S.J.S.: Detection of COVID-19 using CXR and CT images using transfer learning and Haralick features. Appl. Intell. 1–18 (2020) [DOI] [PMC free article] [PubMed]
- 22.Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, Schönlieb CB. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
- 23.Bai, H.X., Wang, R., Xiong, Z., Hsieh, B., Chang, K., Halsey, K., Liao, W.H.: AI augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other etiology on chest CT. Radiology 201491 (2020) [DOI] [PMC free article] [PubMed]
- 24.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. 2020;12:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang H, Xia L, Yan F, Wan Z, Shi F, Yuan H, Shen D. Diagnosis of coronavirus disease 2019 (COVID-19) with structured latent multi-view representation learning. IEEE Trans. Med. Imaging. 2020;39:2606–2614. doi: 10.1109/TMI.2020.2992546. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Bao Y, Wen Y, Lu H, Luo H, Xiang Y, Qian D. Prior-attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans. Med. Imaging. 2020;39:2572–2583. doi: 10.1109/TMI.2020.2994908. [DOI] [PubMed] [Google Scholar]
- 27.Ouyang X, Huo J, Xia L, Shan F, Liu J, Mo Z, Shen D. Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans. Med. Imaging. 2020;39:2595–2605. doi: 10.1109/TMI.2020.2995508. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Zheng C. A Weakly-supervised Framework for COVID-19 Classification and Lesion Localization from Chest CT. IEEE Trans. Med. Imaging. 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 29.Hu S, Gao Y, Niu Z, Jiang Y, Li L, Xiao X, Yang G. Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access. 2020;8:118869–118883. doi: 10.1109/ACCESS.2020.3005510. [DOI] [Google Scholar]
- 30.Yazdani, S., Minaee, S., Kafieh, R., Saeedizadeh, N., Sonka, M.: COVID CT-Net: predicting COVID-19 from chest CT images using attentional convolutional network. (2020). arXiv preprint arXiv:2009.05096
- 31.Harmon SA, Sanford TH, Xu S, Turkbey EB, Roth H, Xu Z, Turkbey B. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goel, T., Murugan, R., Mirjalili, S., Chakrabartty, D.K.: OptCoNet: an optimized convolutional neural network for an automatic diagnosis of COVID-19. Appl. Intell. 1–16 (2020) [DOI] [PMC free article] [PubMed]
- 33.Jain, R., Gupta, M., Taneja, S., Hemanth, D.J.: Deep learning based detection and analysis of COVID-19 on chest X-ray images. Appl. Intell. 1–11 (2020) [DOI] [PMC free article] [PubMed]
- 34.Apostolopoulos, I.D., Mpesiana, T.A.: Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 1 (2020) [DOI] [PMC free article] [PubMed]
- 35.Alom, M.Z., Rahman, M.M., Nasrin, M.S., Taha, T.M., Asari, V.K.: COVID\_MTNet: COVID-19 detection with multi-task deep learning approaches. (2020). arXiv preprint arXiv:2004.03747
- 36.Minaee, S., Kafieh, R., Sonka, M., Yazdani, S., Soufi, G.J.: Deep-COVID: predicting COVID-19 from chest X-ray images using deep transfer learning. (2020). arXiv preprint arXiv:2004.09363 [DOI] [PMC free article] [PubMed]
- 37.Oh Y, Park S, Ye JC. Deep learning COVID-19 features on CXR using limited training data samples. IEEE Trans. Med. Imaging. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, R., Arora, R., Bansal, V., Sahayasheela, V.J., Buckchash, H., Imran, J., Raman, B.: Accurate prediction of COVID-19 using chest X-ray images through deep feature learning model with SMOTE and machine learning classifiers. (2020). medRxiv
- 39.Turkoglu, M.: COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl. Intell. 1–14 (2020) [DOI] [PMC free article] [PubMed]
- 40.Abbas, A., Abdelsamea, M.M., Gaber, M.M.: Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. (2020). arXiv preprint arXiv:2003.13815 [DOI] [PMC free article] [PubMed]
- 41.Li, T., Han, Z., Wei, B., Zheng, Y., Hong, Y., Cong, J.: Robust screening of COVID-19 from chest X-ray via discriminative cost-sensitive learning. (2020). arXiv preprint arXiv:2004.12592
- 42.Tan, M., Le, Q.V.: EfficientNet: rethinking model scaling for convolutional neural networks. (2019). arXiv preprint arXiv:1905.11946
- 43.Wolpert DH. Stacked generalization. Neural Netw. 1992;5(2):241–259. doi: 10.1016/S0893-6080(05)80023-1. [DOI] [Google Scholar]
- 44.El-Shafai, W., Abd El-Samie, F.: Extensive COVID-19 X-ray and CT chest images dataset. Mendeley Data V3 (2020)
- 45.Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput. Methods Progr. Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loey, M., Manogaran, G., Khalifa, N.E.M.: A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput. Appl. 1–13 (2020) [DOI] [PMC free article] [PubMed]
- 47.Pham TD. A comprehensive study on classification of COVID-19 on computed tomography with pretrained convolutional neural networks. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang, F.J., LeCun, Y.: Large-scale learning with SVM and convolutional netw for generic object recognition. In: 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, vol. 10 (2006)
- 49.Bousquet O, Elisseeff A. Stability and generalization. J. Mach. Learn. Res. 2002;2:499–526. [Google Scholar]
- 50.Zebin, T., Rezvy, S.: COVID-19 detection and disease progression visualization: deep learning on chest X-rays for classification and coarse localization. Appl. Intell. 1–12 (2020) [DOI] [PMC free article] [PubMed]