Zhou, Vachon, et al. identify HSPG Syndecan as a novel core organizer of the cholinergic synapse in C. elegans. Syndecan/SDN-1 acts by bridging extracellular matrix components with intracellular scaffolding proteins to recruit alpha7-like acetylcholine receptors at postsynaptic sites.
Abstract
The extracellular matrix has emerged as an active component of chemical synapses regulating synaptic formation, maintenance, and homeostasis. The heparan sulfate proteoglycan (HSPG) syndecans are known to regulate cellular and axonal migration in the brain. They are also enriched at synapses, but their synaptic functions remain more elusive. Here, we show that SDN-1, the sole orthologue of syndecan in C. elegans, is absolutely required for the synaptic clustering of homomeric α7-like acetylcholine receptors (AChRs) and regulates the synaptic content of heteromeric AChRs. SDN-1 is concentrated at neuromuscular junctions (NMJs) by the neurally secreted synaptic organizer Ce-Punctin/MADD-4, which also activates the transmembrane netrin receptor DCC. Those cooperatively recruit the FARP and CASK orthologues that localize α7-like-AChRs at cholinergic NMJs through physical interactions. Therefore, SDN-1 stands at the core of the cholinergic synapse organization by bridging the extracellular synaptic determinants to the intracellular synaptic scaffold that controls the postsynaptic receptor content.
Introduction
Chemical synapses are specialized cellular junctions that support directional transfer of information between excitable cells. In addition, glial cells were identified as important synaptic players, leading to the notion of the “tripartite synapse” (Araque et al., 1999; Halassa et al., 2007). However, the ECM, which fills the synaptic cleft and the perisynaptic extracellular space, also emerged as an active element that orchestrates synaptogenesis, synaptic maintenance, and synaptic function, thus leading to the concept of the “tetrapartite synapse” (Dityatev and Rusakov, 2011; Dityatev et al., 2006; Ferrer-Ferrer and Dityatev, 2018). The synaptic ECM, also referred to as the synaptomatrix (Dani and Broadie, 2012; Heikkinen et al., 2014), forms a dense and composite environment through which signaling between pre- and postsynaptic partners necessarily and consequently impacts all aspects of synaptic biology (Heikkinen et al., 2014; Kurshan et al., 2014).
Within the synaptomatrix, some glycoproteins such as heparan sulfate (HS) proteoglycans (HSPGs) fulfill the role of bona fide synaptic organizers (Condomitti and de Wit, 2018; Yuzaki, 2018). HSPGs are either secreted or membrane-associated core proteins bearing covalently linked chains of long disaccharide repeats (Sarrazin et al., 2011). HSPGs bind multiple partners and can promote or regulate synaptogenesis (Song and Kim 2013). The founding member of this diverse group is agrin, a large HSPG secreted by motoneurons at the vertebrate neuromuscular junctions (NMJs), which is absolutely required for NMJ differentiation and postsynaptic clustering of acetylcholine receptors (AChRs; Burden et al., 2018; Li et al., 2018; Swenarchuk, 2019). Since then, an increasing number of HSPGs, such as the glypicans 4 and 6, have been demonstrated to function as synaptic regulators (Allen et al., 2012; Condomitti et al., 2018; reviewed in Condomitti and de Wit, 2018).
Syndecans are a class of transmembrane HSPGs also shown to play roles at synapses (Saied-Santiago and Bülow, 2018). In Drosophila melanogaster, syndecan localizes to glutamatergic NMJs. It competes with the glypican Dallylike for binding to the LAR-family receptor protein tyrosine phosphatase dLAR and promotes the growth of presynaptic terminals (Johnson et al., 2006). Although syndecan was initially reported to be acting cell autonomously in the motoneuron terminal, further evidence suggests that it might be provided postsynaptically by muscle cells (Nguyen et al., 2016). In mammals, the syndecan family has four members. Syndecan 2 is expressed in neurons of young and adult rats (Hsueh and Sheng, 1999) and progressively concentrates at asymmetric synapses in most regions of the adult rat brain (Hsueh et al., 1998). It can promote the formation of dendritic spines in cell culture (Ethell and Yamaguchi, 1999; Ethell et al., 2000; Lin et al., 2007), notably by facilitating FGF22 targeting to dendritic filopodia and spines (Hu et al., 2016). Syndecan 3 is also highly expressed in the developing and adult brain but is almost exclusively detected in axonal tracts (Hsueh and Sheng, 1999). Inactivation of Sdc3 does not cause global abnormalities of the brain anatomy (Kaksonen et al., 2002), but the laminar structure of the cerebral cortex is perturbed as a result of impaired radial migration (Hienola et al., 2006). No modification of synaptic density or basal transmission was reported in the hippocampus. However, Sdc3−/− mice have enhanced long-term potentiation in the CA1 area and impaired hippocampus-dependent memory behaviors (Kaksonen et al., 2002), suggesting that syndecan 3 is required for synaptic plasticity.
In Caenorhabditis elegans, the sole syndecan member, SDN-1, is expressed and functions in the nervous system (Minniti et al., 2004; Rhiner et al., 2005). Indirect evidence suggests that SDN-1 carries the HSs that are highly enriched at NMJs (Attreed et al., 2012). We therefore used the C. elegans NMJ as a genetically tractable model to investigate further the synaptic function of SDN-1. In C. elegans, each body-wall muscle cell receives excitatory and inhibitory innervation from cholinergic and GABAergic motoneurons (Fig. 1 A). The cholinergic versus GABAergic identity of postsynaptic domains is specified by the anterograde synaptic organizer Ce-Punctin/MADD-4 (Pinan-Lucarré et al., 2014). This evolutionarily conserved ECM protein belongs to the poorly characterized ADAMTS-like proteins that contain multiple thrombospondin type 1 repeat and immunoglobulin domains as well as structurally unsolved domains in common with the ADAMTS family (Apte, 2009). The functions of its vertebrate orthologues Punctin1/ADAMTSL1 and Punctin2/ADAMTSL3 are ill defined, but Punctin2 is expressed in the brain and was identified as a susceptibility gene for schizophrenia (Dow et al., 2011).
Figure 1.
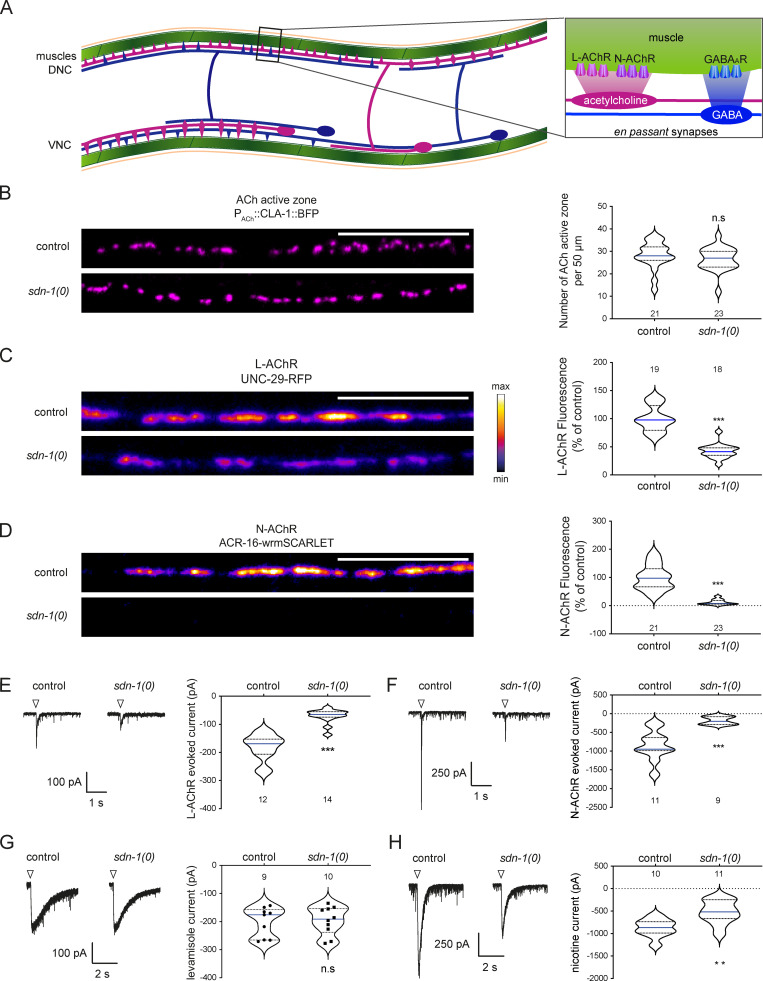
SDN-1 controls AChR synaptic content.(A) Schematics of neuromuscular innervations in C. elegans. Inset: Excitatory (cholinergic) and inhibitory (GABAergic) synapses innervating a muscle cell. DNC, dorsal nerve cord; VNC, ventral nerve cord. (B) Confocal detection and quantification of cholinergic active zones using the CLA-1-BFP marker under control of the Punc-17 promoter. (C and D) Confocal detection and quantification of L-AChR (UNC-29-RFP knock-in; C) and N-AChR (ACR-16-wrmScarlet knock-in; D) fluorescence. (E and F) Representative traces of isolated L-AChR (E) or total AChR (F) synaptic currents evoked by a 10-ms optogenetic stimulation (arrowheads) of cholinergic motoneurons. Isolation of L-AChR currents was achieved by blocking N-AChR with DHβE. (G and H) Representative traces and quantification of peak currents evoked by pressure application of 0.1 mM levamisole (G) or nicotine (H) on muscle cells. Arrowheads mark the 100-ms application onsets. Individual values are shown as dots in E and F. In this figure and all other figures, confocal images are sums of Z-stacks acquired along the dorsal nerve cord by a spinning disk confocal microscope; anterior is to the left. Scale bars, 10 µm (B–D). For the quantification of fluorescence levels, data were normalized to the mean value of the control group. In this figure and all other figures, data distribution in each group is presented as violin plots showing lower and upper quartiles (dotted lines) and median value (blue line). The number of animals analyzed is indicated on the violin plot. See also Fig. S1.
Ce-punctin generates long (L) and short (S) isoforms by the use of alternative promoters. The combination of isoforms present in the synaptic cleft controls the identity of the postsynaptic domains (Pinan-Lucarré et al., 2014). Punctin L is exclusively secreted by cholinergic neurons and triggers the postsynaptic localization of two types of ionotropic AChRs: (1) levamisole-sensitive AChRs (L-AChRs), which are heteromeric and can be activated by the nematode-specific agonist levamisole; and (2) nicotine-sensitive AChRs (N-AChRs), which are homomeric, activated by nicotine, and evolutionarily very close to the α7 AChRs expressed in the mammalian brain. L-AChRs interact with an extracellular scaffold (Gally et al., 2004; Gendrel et al., 2009; Rapti et al., 2011) to form microclusters that are positioned at synapses by Punctin L. The Punctin-dependent clustering of N-AChRs relies on a distinct uncharacterized pathway. The short isoform Punctin S is secreted by both cholinergic and GABAergic motoneurons. It triggers the postsynaptic clustering of type A GABA receptors (GABAARs) at GABAergic NMJs. At cholinergic NMJs, it associates with Punctin L and inhibits the inappropriate recruitment of GABAARs by Punctin L. Punctin S controls the clustering of GABAARs by two convergent molecular pathways (for review, see Zhou and Bessereau 2019). First, Punctin S binds and clusters the synaptic adhesion molecule NLG-1/neuroligin in front of GABAergic boutons, which controls the synaptic localization of GABAARs (Maro et al., 2015; Tu et al., 2015). Second, it binds, recruits, and likely activates the netrin receptor UNC-40/DCC, which controls the synaptic content of GABAARs (Tu et al., 2015). UNC-40/DCC nucleates an intracellular scaffold by physically interacting with FRM-3, a FERM (p4.1, ezrin, Radixin, and Moesin) protein orthologous to FARP1/2 (Zhou et al., 2020). FRM-3 multimerizes and recruits LIN-2, the orthologue of CASK (calcium calmodulin-dependent serine/threonine kinase), which might provide a hub to physically connect the GABAARs, FRM-3/FARP and NLG-1/neuroligin. Except NLG-1, these molecules are also present at cholinergic NMJs but their precise function was not characterized.
Here, we show that SDN-1/syndecan is concentrated at NMJs by Punctin and modulates the content of receptors at excitatory and inhibitory NMJs. Remarkably, SDN-1 is the core component supporting the clustering of homomeric N-AChRs. Through physical interaction with LIN-2/CASK and FRM-3/FARP, SDN-1 stabilizes a DCC-dependent synaptic scaffold at cholinergic NMJs, which in turn recruits N-AChRs. Since all these proteins are evolutionarily conserved, these results provide a framework to analyze the molecular mechanisms controlling the localization of α7 AChRs in mammalian neurons.
Results
Syndecan controls the synaptic localization of α7-like N-AChRs
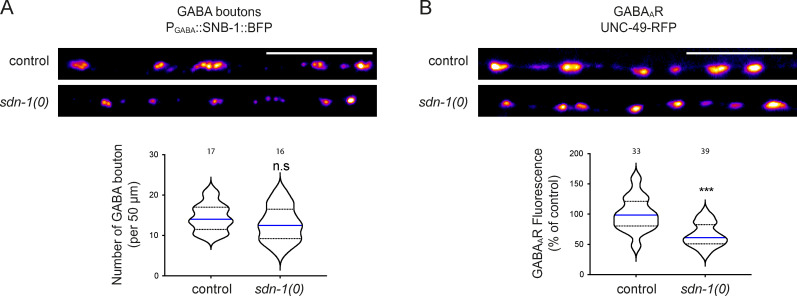
Previous studies suggested that SDN-1 localizes in synapse-rich regions of the C. elegans nervous system (Attreed et al., 2012). To identify putative functions of SDN-1 at synapses, we analyzed NMJs in sdn-1(zh20) null mutants, hereinafter referred to as sdn-1(0). The number of cholinergic or GABAergic NMJs was not changed in sdn-1(0) mutants (Figs. 1 B and S1 A). By contrast, we observed strong alterations of postsynaptic receptor distribution. First, the synaptic content of heteromeric L-AChRs was decreased by ∼60% in sdn-1(0) mutants based on the fluorescence of the UNC-29-RFP subunit expressed from a knock-in allele (Fig. 1 C). The remaining receptors were still clustered at synapses. Second, homomeric N-AChRs were almost undetectable based on ACR-16-wrmScarlet fluorescence (Fig. 1 D). Third, synaptic GABAAR content was also reduced, albeit to a lesser extent, with an ∼35% decrease of UNC-49-RFP fluorescence at synapses (Fig. S1 B).
Figure S1.
SDN-1 has a moderate effect on GABAAR synaptic content.(A) Confocal detection and quantification of the number of GABAergic presynaptic active zones, using the CLA-1-BFP marker under the control of Punc-47 promoter, in control and sdn-1(0) mutant animals. (B) Confocal detection and quantification of GABAARs (UNC-49-RFP knock-in) in control and sdn-1(0) animals. Scale bars, 10 µm.
We subsequently focused our analysis on SDN-1–dependent localization of AChRs and performed electrophysiological analysis of AChRs in muscle cells. We first evoked synaptic release of acetylcholine by optogenetic stimulation of cholinergic motoneurons. The L-AChR–dependent response was diminished by 60% in sdn-1(0) mutants (Fig. 1 E), while the N-AChR–dependent response was reduced by 80% (Fig. 1 F). Such decrease could be due to a redistribution of receptors outside of the synapses or a decrease in functional receptors at the plasma membrane. To test these hypotheses, we pressure-applied selective agonists of L- or N-AChRs. The response to levamisole was similar in sdn-1(0) and wild-type animals (Fig. 1 G), indicating that in the absence of syndecan, L-AChRs are less efficiently clustered at synapses but remain present at the plasma membrane. By contrast, the response to nicotine was decreased by 40% in sdn-1(0) mutants (Fig. 1 H). This decrease was much smaller than the almost complete disappearance of N-AChR–dependent synaptic responses. Therefore, it suggests that syndecan has two functions regarding N-AChRs. First, it is necessary for clustering N-AChRs at synapses. Second, it would stabilize N-AChRs at the plasma membrane.
Syndecan/SDN-1 is a synaptic protein at NMJs
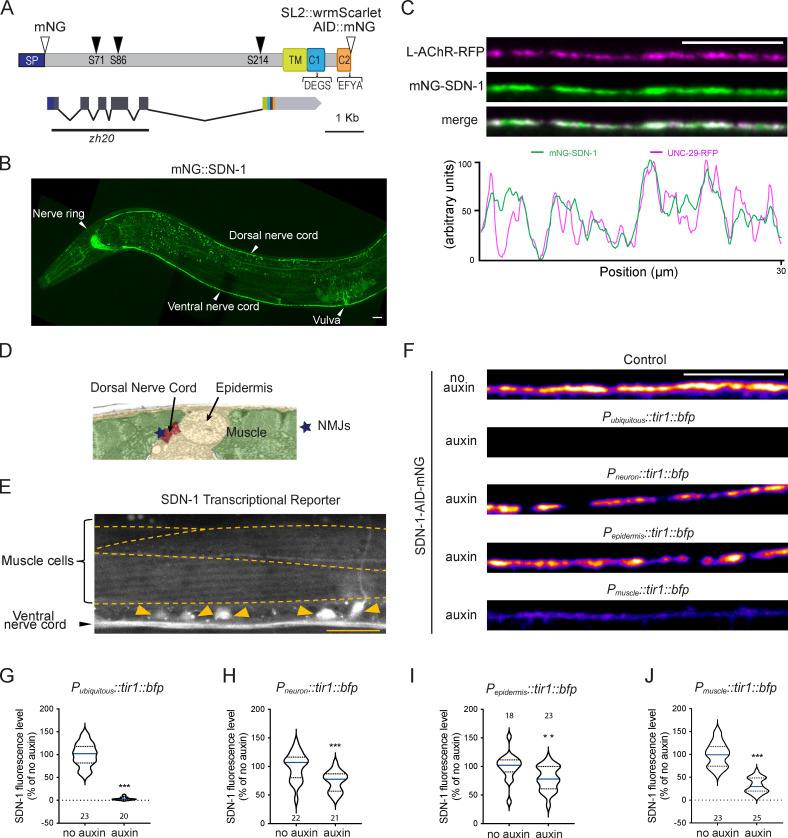
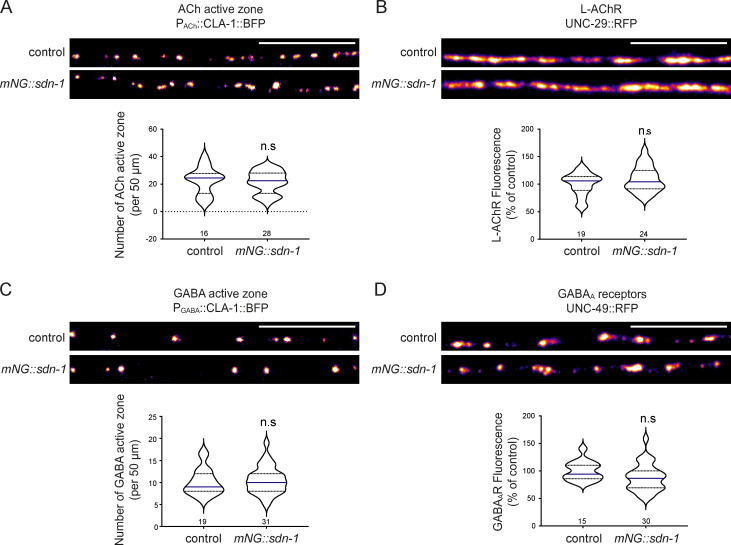
To define the precise localization of SDN-1, we inserted the fluorescent protein mNeonGreen (mNG) at the N terminus of SDN-1 using CRISPR-Cas9 (Fig. 2 A). In adult worms, mNG-SDN-1 was detected at high level in the nerve ring and along the ventral and dorsal nerve cords (Fig. 2 B). The fusion protein mNG-SDN-1 appeared functional based on the absence of abnormal visible phenotypes of knock-in animals and the normal content of AChRs and GABAARs at NMJs as compared with the wild type (Fig. S2).
Figure 2.
SDN-1 is enriched at NMJs and is mainly provided by postsynaptic muscle cells.(A) Predicted structure of the SDN-1 protein (288 aa) and the sdn-1 locus (2,921 bp). SP, signal peptide; TM, transmembrane; C1, domain containing the ezrin-binding motif DEGS; C2, domain containing the PDZ-binding motif EFYA. Three putative GAG anchoring sites (black arrowheads) have been predicted on serine 71, 86, and 214. Open arrowheads indicate the insertion sites of fluorescent tags in knock-in strains: mNG, AID-mNG, and SL2::wormScarlet (transcriptional reporter SL2-wrmScarlet). The black line shows the location of the 1,258-bp deletion in sdn-1(zh20) mutant allele, referred to as sdn-1(0). Boxes represent exons. (B) Confocal detection of mNG-SDN-1 in an adult worm. (C) Confocal detection and fluorescence profiles of mNG-SDN-1 and L-AChRs (UNC-29-RFP) along the dorsal cord. (D) Electronic microscopy image of the dorsal part of a C. elegans coronal section. Modified from SW-Worm Viewer, slice 273 (Altun et al., 2021; http://www.wormatlas.org/SW/SW.php). (E) Confocal detection of the transcriptional reporter SDN-1::SL2::wrmScarlet. Arrowheads indicate cell bodies of motor neurons. Dotted lines delineate borders of body-wall muscle cells. (F) Confocal detection of SDN-1-AID-mNG from dorsal nerve cord in animals grown in the absence (control) or presence of auxin to induce SDN-1 depletion in all tissues (Peft-3::tir-1-bfp), neurons (Prab-3::tir-1-bfp), epidermis (Pdpy-7::tir-1-bfp), or body-wall muscles (Pmyo-3::tir-1-bfp). (G–J) Quantification of SDN-1-AID-mNG fluorescence levels when SDN-1 was depleted in all tissues (G), epidermis (H), neurons (I), or body-wall muscles (J). Scale bars, 10 µm. See also Figs. S2, S3, and S4.
Figure S2.
mNG insertion in the sdn-1 locus has no effect on NMJ organization.(A) Confocal detection and quantification of the number of cholinergic presynaptic active zones, using the CLA-1-BFP marker under the control of Punc-17 promoter, in control and mNG-sdn-1 knock-in animals. (B) Confocal detection and quantification of L-AChRs (UNC-29-RFP) in control and mNG-sdn-1 animals. (C) Confocal detection and quantification of the number of GABAergic active zones, using the CLA-1-BFP marker under Punc-47 promoter, in control and mNG-sdn-1 animals. (D) Confocal detection and quantification of GABAARs (UNC-49-RFP) in control and mNG-sdn-1 animals. Scale bars, 10 µm.
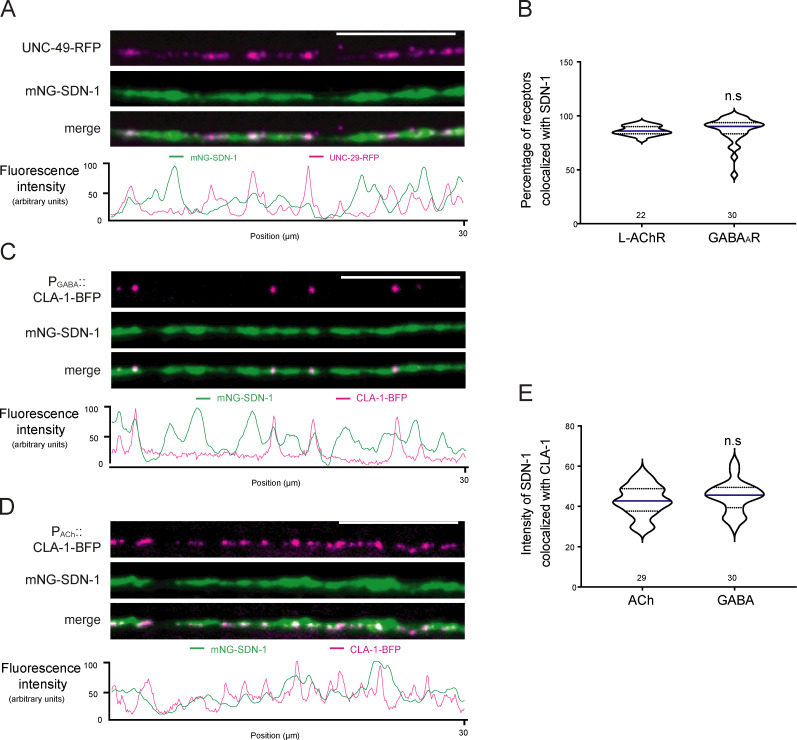
To test if mNG-SDN-1 was enriched at synapses, we used knock-in strains expressing L-AChRs or GABAAR tagged with red fluorescent proteins. At high magnification, mNG-SDN-1 looked highly concentrated at NMJs and both ACh and GABAA receptors almost completely colocalized with syndecan (Fig. 2 C; and Fig. S3, A and B). We also measured the SDN-1 density at each synaptic type using the presynaptic marker CLA-1/Clarinet (Xuan et al., 2017), which accurately delineates active zones (Fig. S3, C and D), and we observed that SDN-1 was equally abundant at excitatory and inhibitory NMJs (Fig. S3 E).
Figure S3.
SDN-1 is equally abundant at excitatory and inhibitory synapses.(A) Confocal detection and fluorescence profiles of mNG-SDN-1 and GABAARs (UNC-49-RFP) along the dorsal cord. (B) Percentage of L-AChRs and GABAARs colocalized with mNG-SDN-1, calculated using Manders’ coefficient on processed images. (C and D) Confocal detection and fluorescence profiles of mNG-SDN-1 and the presynaptic active zone marker CLA-1-BFP under the control of either the GABAergic neuron-specific promoter Punc-47 (C) or the cholinergic neuron-specific promoter Punc-17 (D). (E) Mean intensity of the mNG-SDN-1 signal at cholinergic and GABAergic synapses. Scale bars, 10 µm.
Synaptic syndecan is mainly expressed by postsynaptic muscle cells
NMJs are established between neurons and muscles and are in close contact with epidermal cells (Fig. 2 D). Using traditional transgenic transcriptional reporters, previous studies showed that sdn-1 was expressed in epidermis and neurons, but not in body-wall muscle cells (Minniti et al., 2004; Rhiner et al., 2005). To further investigate the sdn-1 expression pattern, we engineered a chromosomal bi-cistronic reporter by inserting an SL2-wrmScarlet cassette in the sdn-1 locus after the end of its coding sequence (Fig. 2 A). In agreement with previous results, red fluorescence was detected in most tissues, including epidermis and motoneurons, but wrmScarlet was also readily detected in body-wall muscles (Fig. 2 E).
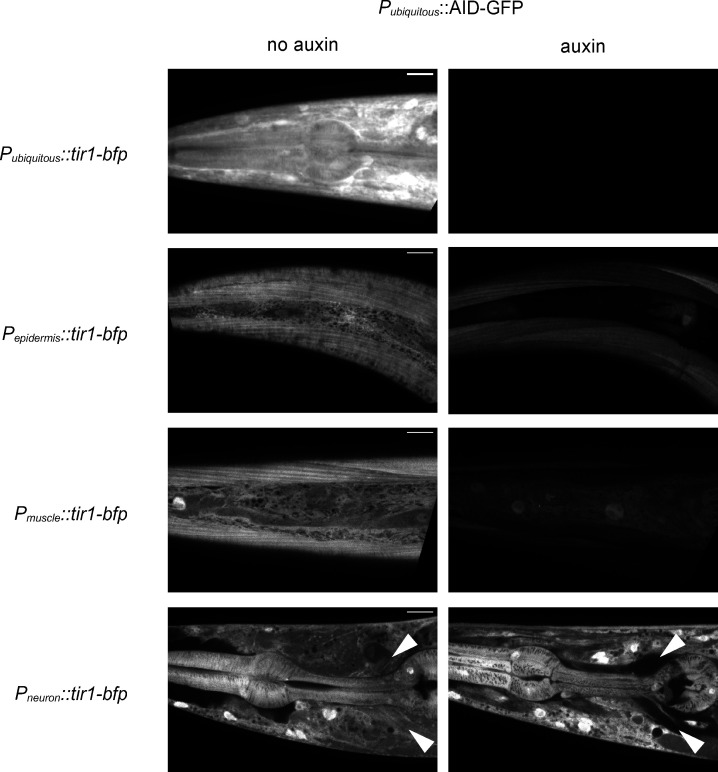
To identify the source of synaptic syndecan, we performed tissue-specific degradation of SDN-1. Using CRISPR-Cas9, we fused an auxin-inducible degron (AID; Zhang et al., 2015) linked to mNG at the SDN-1 C terminus (Fig. 2 A), and we expressed the TIR-1 E3-ligase under various tissue-specific promoters (Fig. S4). SDN-1-AID-mNG properly localized at synapses (Fig. 2 F), similarly to mNG–SDN-1 (Fig. 2 C). Ubiquitous expression of TIR-1 caused a complete disappearance of fluorescence in the worm, including at the nerve cords (Fig. 2, F and G). Specific degradation of SDN-1-AID-mNG either in neurons (Fig. 2 F, H) or in the epidermis (Fig. 2, F and I) roughly caused a 25% reduction of the SDN-1-AID-mNG synaptic fluorescence, while degradation of SDN-1-AID-mNG in body-wall muscles caused a 70% decrease of this fluorescence (Fig. 2, F and J). Therefore, SDN-1 is expressed by pre-, post- and perisynaptic cells, yet the vast majority of SDN-1 present at synaptic sites is contributed by postsynaptic muscle cells.
Figure S4.
The AID leads to tissue-specific degradation. Confocal detection of AID-GFP expressed in all tissues by the ubiquitous promoter Peft-3 from animals grown on regular NGM plates (control; left panels) or auxin plates (right panels). Tissue-specific degradation was achieved by coexpressing Peft-3::AID-GFP and Peft-3::tir-1-bfp (ubiquitous), Pdpy-7::tir-1-bfp (epidermis), Pmyo-3::tir-1-bfp (body-wall muscles), or Prab-3::tir-1-bfp (neurons). Arrowheads indicate the position of the nerve ring. Scale bars, 10 µm.
HS chains modulate syndecan synaptic content but are dispensable for its synaptic localization
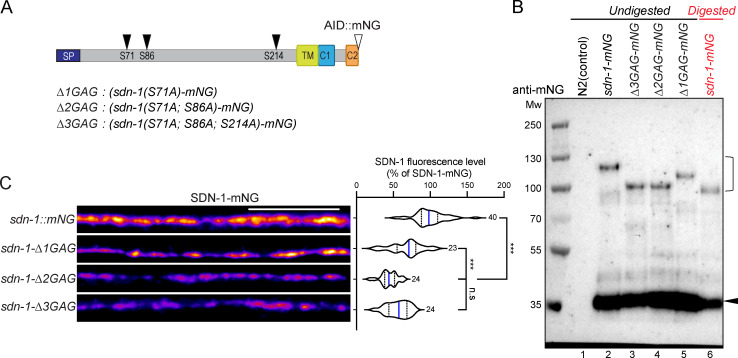
SDN-1 contains two putative HS attachment sites in its N-terminal region, and a third putative site closer to the transmembrane region (Minniti et al., 2004). We mutated these sites by CRISPR-Cas9 in a C-terminally tagged version of SDN-1 in order to avoid any potential interaction of the fluorescent protein with the extracellular matrix (Fig. 3 A). Immunoprecipitation followed by Western blot analysis first indicated that SDN-1 likely form SDS-resistant dimers, as previously described for mammalian syndecans (Fig. 3 B; Asundi and Carey, 1995; Choi et al., 2005). Mutation of one or both N-terminal attachment sites each induced a size shift, indicating the loss of corresponding GAG modifications (Fig. 3 B). Because we did not detect any additional weight difference between the double and triple mutants, the third putative GAG attachment site is most probably not modified (Fig. 3 B). Consistently, when SDN-1-mNG precipitates were treated with heparinase (I and III), the apparent molecular weight was close to SDN-1(Δ2GAG)-mNG (Fig. 3 B). Noteworthy, SDN-1(Δ3GAG)-mNG still localized at synapses, but the overall fluorescence level was decreased by 30% in Δ1GAG and by ∼50% in Δ2GAG and Δ3GAG as compared with the wild-type SDN-1-mNG (Fig. 3 C). Although GAG chains likely stabilize SDN-1 at the synapse, these data indicate that the SDN-1 core protein carries sufficient information for its synaptic localization.
Figure 3.
HS chains modulate SDN-1 synaptic content.(A) Black arrowheads indicate the position of putative GAG chain anchoring serines in SDN-1. Serines were mutated to alanines to generate ΔGAG mutants. Open arrowhead indicates the insertion site of AID::mNG fluorescence tag. (B) Western blot analysis (anti-mNG) of nonmutated SDN-1-mNG (lane 2), SDN-1(Δ3GAG)-mNG (lane 3), SDN-1(Δ2GAG)-mNG (lane 4), or SDN-1(Δ1GAG)-mNG (lane 5) proteins precipitated from worm lysate using mNG beads. Expected SDN-1-mNG molecular weight is 58 kD. Bracket likely indicates the SDS-resistant dimerization form of SDN-1. Arrowhead indicates potential cleaved C terminus–mNG band. Protein samples from lane 6 (shown in red) were digested by heparinase I and III. Protein lysate from N2 wild-type animals was used as control in lane 1. (C) Confocal detection and quantification of SDN-1 levels from SDN-1-mNG and ΔGAG mutants. Scale bar, 10 µm.
Ce-Punctin/MADD-4 localizes syndecan at synapses
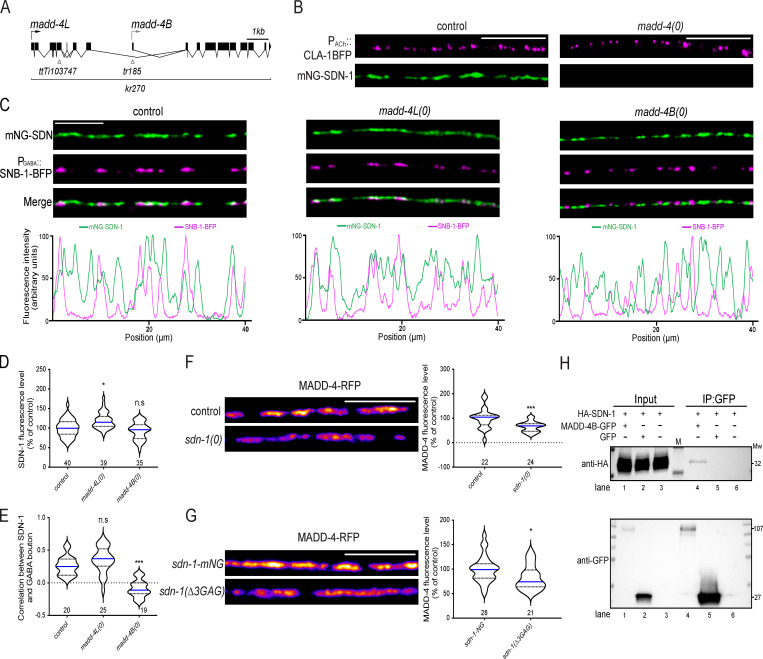
AChR localization at cholinergic NMJs is controlled by the anterograde synaptic organizer Ce-Punctin/MADD-4 (Pinan-Lucarré et al., 2014). madd-4 encodes madd-4L (long) and madd-4B (short) isoforms that share a common C-terminal moiety (Fig. 4 A). MADD-4L is present at cholinergic NMJs and is required for retaining both L- and N-AChR clusters at synapses. MADD-4B is present at GABAergic NMJs, where it promotes GABAAR clustering, and at cholinergic NMJs, where it prevents MADD-4L from recruiting GABAARs (Pinan-Lucarré et al., 2014).
Figure 4.
Ce-Punctin/MADD-4 localizes SDN-1 at synapses.(A) Structure of the madd-4 locus. Boxes represent exons. Arrows indicate the beginning of the ORF of madd-4L and madd-4B. Localization of isoform specific mutations (ttTi103747 and tr185) and full knockout (kr270), referred to as madd-4(0), are indicated. (B) Confocal detection of mNG-SDN-1 and cholinergic active zones (CLA-1-BFP). (C) Confocal detection and fluorescence profiles of mNG-SDN-1 and GABAergic boutons using the SNB-1-BFP marker driven by the Punc-47 promoter. (D) Quantification of mNG-SDN-1 fluorescence levels. (E) Pearson’s correlation coefficient between mNG-SDN-1 and GABA boutons. (F and G) Confocal detection and quantification of MADD-4-RFP fluorescence. (H) Coimmunoprecipitation of SDN-1 and MADD-4B expressed in HEK293 cells. Cells were transfected with HA-SDN-1 alone (lanes 3 and 6), MADD-4B-GFP (lanes 1 and 4), or GFP (lanes 2 and 5). Cell lysates were precipitated by GFP nanobody beads and probed with anti-HA antibody (upper). The expression of GFP was detected using anti-GFP antibody (lower). M, protein ladder. Molecular weight (Mw) is shown on the right. Scale bars, 10 µm.
We therefore tested if MADD-4 regulates the localization of SDN-1. Strikingly, mNG-SDN-1 was almost undetectable at the nerve cords of madd-4(0) null mutants (Fig. 4 B). Genetic removal of MADD-4L alone had no impact on the localization nor the level of SDN-1 (Fig. 4, C–E). In madd-4B mutants, the overall content of mNG-SDN1 did not change; mNG-SDN-1 was no longer detectable at GABAergic synapses but relocalized to cholinergic junctions, where MADD-4L was still present (Fig. 4, C–E). Hence, these results show that SDN-1 localization is controlled by the neurally secreted ECM protein MADD-4 and that this localization can be instructed by any of the two MADD-4 isoforms, likely through its C-terminal moiety.
We then asked whether SDN-1 would, in turn, regulate MADD-4. In sdn-1(0) mutants, the synaptic content of MADD-4 was reduced by 30%, based on the expression of an RFP C-terminal knock-in that labels both MADD-4 isoforms (Fig. 4 F). In sdn-1(Δ3GAG)-mNG, MADD-4-RFP was also decreased, albeit to a lesser extent (17%; Fig. 4 G). The interpretation of these results is complicated because removing the GAG chains causes a partial loss of SDN-1, yet they suggest that the GAG chains carried by SDN-1 might partly stabilize MADD-4 at the synapse.
To test if MADD-4 and SDN-1 might physically interact, we expressed MADD-4B-GFP and HA-SDN-1 in HEK293 cells. Both proteins coimmunoprecipitated, suggesting that SDN-1 and MADD-4 can directly interact (Fig. 4 H).
Ce-Punctin/MADD-4 recruits UNC-40/DCC to stabilize the α7-like N-AChR ACR-16 at the synapse
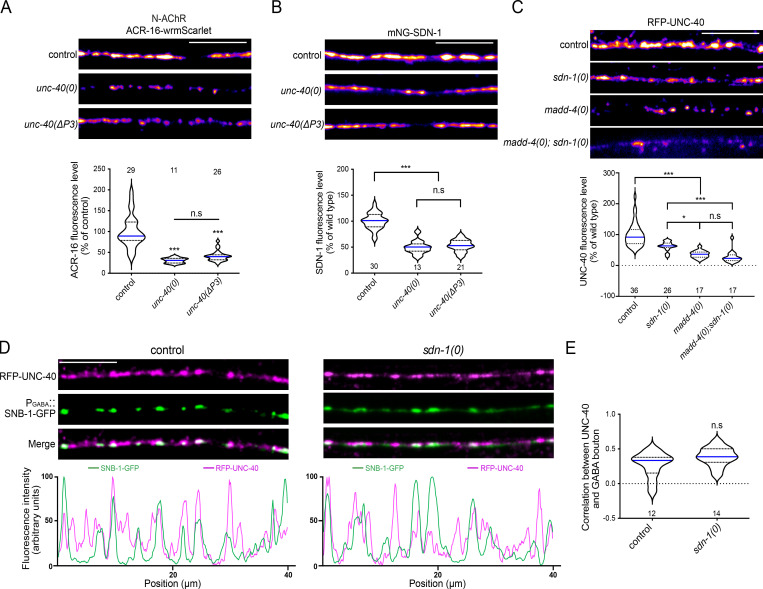
At GABA synapses, MADD-4 recruits GABAARs through an UNC-40/DCC-dependent pathway (Tu et al., 2015). We therefore wondered whether UNC-40 might also be required for the synaptic localization of N-AChRs. In unc-40(0) mutants, ACR-16-wrmScarlet fluorescence was indeed reduced by ∼70%, but the residual signal had a punctate pattern (Fig. 5 A). We then analyzed the SDN-1 content at the nerve cord of unc-40(0) and observed a 50% loss of mNG-SDN-1 fluorescence (Fig. 5 B). To test for a putative reverse interaction between SDN-1 and UNC-40, as previously reported in other systems (Bennett et al., 1997; Blanchette et al., 2015), we analyzed the synaptic localization of RFP-UNC-40 in sdn-1(0) mutants and observed a 38% decrease of fluorescence (Fig. 5 C). Interestingly, UNC-40 remained present at both cholinergic and GABAergic NMJs (Fig. 5, D and E). These results support the existence of a crosstalk between SDN-1 and UNC-40 to maintain the proper level of either protein at NMJs.
Figure 5.
UNC-40 promotes the synaptic localization of ACR-16 and SDN-1.(A and B) Confocal detection and quantification of ACR-16-wrmScarlet (A) and mNG-SDN-1 (B) fluorescence. (C) Confocal detection and quantification of RFP-UNC-40 driven by the muscle-specific Pmyo-3 promoter. (D) Confocal detection and fluorescent profiles of RFP-UNC-40 and GABA boutons using the SNB-1-GFP marker driven by the Punc-25 promoter. (E) Pearson’s correlation coefficient between RFP-UNC-40 and GABA boutons. Scale bars, 10 µm.
The localization of UNC-40 at NMJs depends on MADD-4 (Tu et al., 2015). Since MADD-4 also localizes SDN-1 at synapses, the localization of UNC-40 might depend on SDN-1. Alternatively, MADD-4 might localize UNC-40 at synapses independently from SDN-1, because it can directly bind UNC-40 (Tu et al., 2015). To test these hypotheses, we analyzed RFP-UNC-40 expression in madd-4(0) mutants and observed a stronger decrease than in sdn-1(0) mutants (Fig. 5 C), suggesting that MADD-4 has the capability to localize UNC-40 at NMJs independently from SDN-1. Consistently, the loss of UNC-40 was not aggravated in a madd-4(0); sdn-1(0) double mutant as compared with madd-4(0) single mutants (Fig. 5 C).
At GABA synapses, the positive regulation of GABAAR synaptic content by UNC-40 requires its C-terminal region, called the P3 domain, which physically interacts with FERM-3, a cytosolic protein orthologous to FARP1/2 that in turn recruits LIN-2, the orthologue of CASK, and form a subsynaptic scaffold (Zhou et al., 2020). To test the role of the P3 domain at cholinergic synapses, we quantified ACR-16-wrmScarlet and mNG-SDN-1 fluorescence in unc-40(ΔP3) mutant, which carries only a deletion of the P3 domain, and observed the same phenotypes as in unc-40(0) (Fig. 5, A and B). These results indicated that the control of N-AChRs by UNC-40 requires the P3 domain and thus might involve the LIN-2/FRM-3 intracellular scaffold.
SDN-1 recruits LIN-2/CASK at cholinergic synapses to cluster α7-like N-AChRs
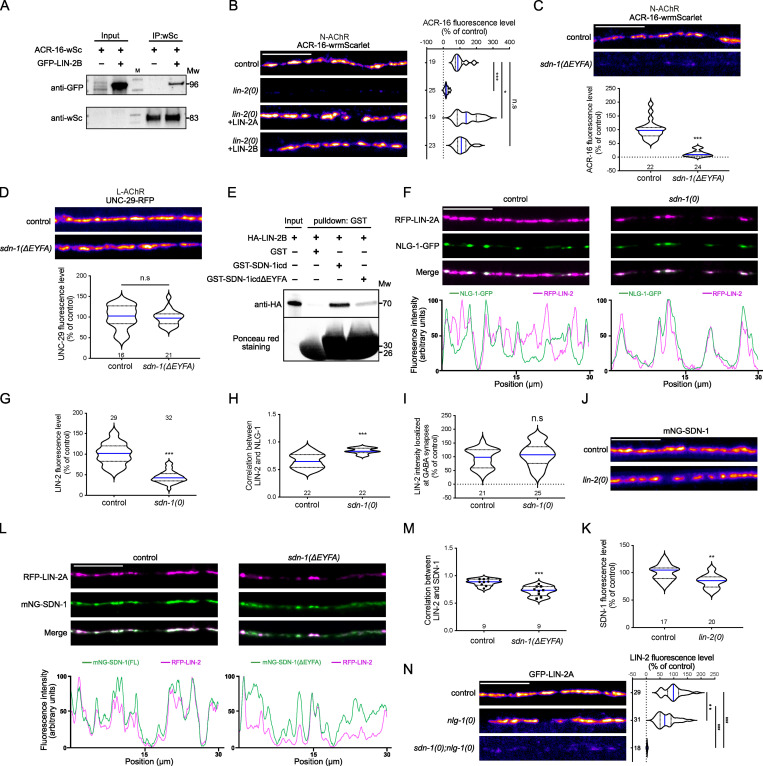
Syndecan contains in its C terminus an evolutionarily conserved PDZ domain–binding site (EFYA), which binds the scaffolding protein CASK in the mammalian nervous system (Ethell and Yamaguchi, 1999; Hsueh et al., 1998). We showed in a previous study that LIN-2/CASK was present at cholinergic synapses (Zhou et al., 2020). We therefore hypothesized that LIN-2 might be involved in N-AChR clustering by interacting with the SDN-1 PDZ-binding site. Coimmunoprecipitation showed that LIN-2 and ACR-16 are able to interact in vivo (Fig. 6 A). Moreover, acr-16-wrmScarlet was reduced by 95% in lin-2(0) null mutants (Fig. 6 B). This defect could be rescued by muscle-specific expression of either the long isoform LIN-2A or the short isoform LIN-2B that lacks the N-terminal CaM-kinase domain but retains the PDZ, SH3, and the MAGUK domains (Fig. 6 B). Finally, we used CRISPR-Cas9 to delete the last four residues of mNG-SDN-1; mNG-SDN-1(ΔEFYA) properly localized at synapses, although its content was slightly decreased (Fig. S5 A). Strikingly, N-AChRs were almost undetectable at the nerve cords in sdn-1(ΔEFYA), while L-AChRs remained expressed and clustered at synapses indistinguishably from the wild type (Fig. 6, C and D).
Figure 6.
SDN-1 recruits LIN-2/CASK at cholinergic synapses to cluster α7-like N-AChRs.(A) Coimmunoprecipitation analysis of the interaction between LIN-2B and ACR-16. The worm lysates of animals expressing ACR-16-wrmScarlet with or without coexpression of GFP-LIN-2B in muscle were precipitated by wrmScarlet nanobody beads and detected by anti-GFP or anti-wrmScarlet antibodies. (B) Confocal detection and quantification of ACR-16-wrmScarlet in control, lin-2(0), and lin-2(0) animals expressing GFP-LIN-2A or GFP-LIN-2B driven by the muscle-specific Pmyo-3 promoter. (C and D) Confocal detection and quantification of ACR-16-wrmScarlet (A) or UNC-29-RFP (B) fluorescence levels in control and sdn-1(ΔEYFA) animals that carry a mutation deleting the SDN-1 C-terminal PDZ-binding motif. (E) GST pull-down analysis of LIN-2B interaction with the entire intracellular part of SDN-1 (SDN-1ICD) or after deleting the PDZ-binding motif (SDN-1ICD ΔEFYA). Samples were analyzed using an anti-HA antibody. The same membrane was stained with Ponceau S red to show GST expression. Molecular weights (Mw) are shown on the right. (F) Confocal detection and fluorescence profiles of RFP-LIN-2A and NLG-1-GFP in control and sdn-1(0) animals expressing rfp-lin-2a and nlg-1-gfp driven by the muscle-specific promoter Pmyo-3. (G) Quantification of the fluorescence levels of RFP-LIN-2A at the dorsal nerve cord. (H) Pearson’s correlation coefficient between RFP-LIN-2A and NLG-1-GFP fluorescence. (I) Fluorescence levels of RFP-LIN-2A at GABA synapses. RFP-LIN-2A fluorescence level was quantified in GABA synaptic regions as defined by the presence of the NLG-1-GFP marker (see Material and methods for details). (J and K) Confocal detection (J) and quantification (K) of mNG-SDN-1. (L) Confocal detection and fluorescence profiles of RFP-LIN-2A and mNG-SDN-1 full length (control) or lacking the PDZ-binding motif (sdn-1(ΔEYFA)) in animals expressing rfp-lin-2a driven by the muscle-specific promoter Pmyo-3. (M) Pearson’s correlation coefficient between RFP-LIN-2A and mNG-SDN-1 full length or mNG-SDN-1 (ΔEYFA). Individual values are shown as dots. (N) Confocal detection and quantification of LIN-2A-GFP. Scale bars, 10 µm.
Figure S5.
LIN-2 is concentrated at GABA synapses in animals lacking SDN-1 PDZ-binding motif.(A) Confocal detection and quantification of mNG-SDN-1 fluorescence levels in control (mNG-sdn-1) and mNG-sdn-1(ΔEYFA) animals. (B) Confocal detection and fluorescence profiles of RFP-LIN-2A, expressed under the muscle-specific Pmyo-3 promoter, and GABAergic presynaptic active zones, using the CLA-1-BFP marker under Punc-47 promoter, in control and sdn-1(ΔEYFA) animals. (C) Pearson’s correlation coefficient between RFP-LIN-2A and GABAergic active zones in control and sdn-1(ΔEYFA) animals. (D) Fluorescence levels of RFP-LIN-2A along the dorsal nerve cord (left) or at GABA synapses (right) in control and sdn-1(ΔEYFA) animals. Total fluorescence (left panel) was measured as for other figures. To measure fluorescence specifically at GABA synapses, RFP-LIN-2A fluorescence was measured within GABA regions, which were defined by the presence of the Punc-47::cla-1-bfp marker. Scale bars, 10 µm.
We then tested if LIN-2/CASK and SDN-1 were able to physically interact. Using in vitro translated and purified recombinant proteins, we showed that the intracellular domain of SDN-1 (SDN-1ICD) could efficiently pull down LIN-2B. This interaction was strongly reduced after deletion of the SDN-1 PDZ-binding site (Fig. 6 E). The relevance of this interaction was further tested in vivo. Analysis of LIN-2 expression in sdn-1(0) null mutants revealed a 55% decrease of LIN-2 at the nerve cord (Fig. 6, F and G). Strikingly, the remaining LIN-2 was exclusively detected at GABAergic synapses, as shown by its perfect colocalization with neuroligin/NLG-1 that is only present at GABA synapses (Maro et al., 2015; Tu et al., 2015; Fig. 6, F–I). These results suggest that SDN-1 is the limiting factor that controls the localization of LIN-2 at cholinergic synapses. By contrast, SDN-1 was only decreased by 16% in lin-2(0) (Fig. 6, J and K). Next, we analyzed the distribution of LIN-2 and SDN-1. The two proteins perfectly overlap in controls, while they were less colocalized in the absence of SDN-1 EFYA motif (Fig. 6, L and M), due to a GABAergic relocalization of LIN-2 (Fig. S5, B–D). Because neuroligin is necessary for LIN-2 localization at GABAergic synapses, this predicted that removing both syndecan and neuroligin would cause a complete loss of LIN-2 at NMJs. Accordingly, GFP-LIN-2 was almost undetectable at the nerve cord of nlg-1(0); sdn-1(0) double mutants (Fig. 6 N).
Altogether, these data indicate that LIN-2/CASK is positioned in the postsynaptic domains of excitatory and inhibitory synapses by two distinct transmembrane proteins, namely syndecan SDN-1 and neuroligin NLG-1, respectively, that each provide binding sites for LIN-2, which in turn participates in the recruitment of AChR and GABAAR.
FRM-3/FARP bridges α7-like N-AChRs with LIN-2/CASK and SDN-1
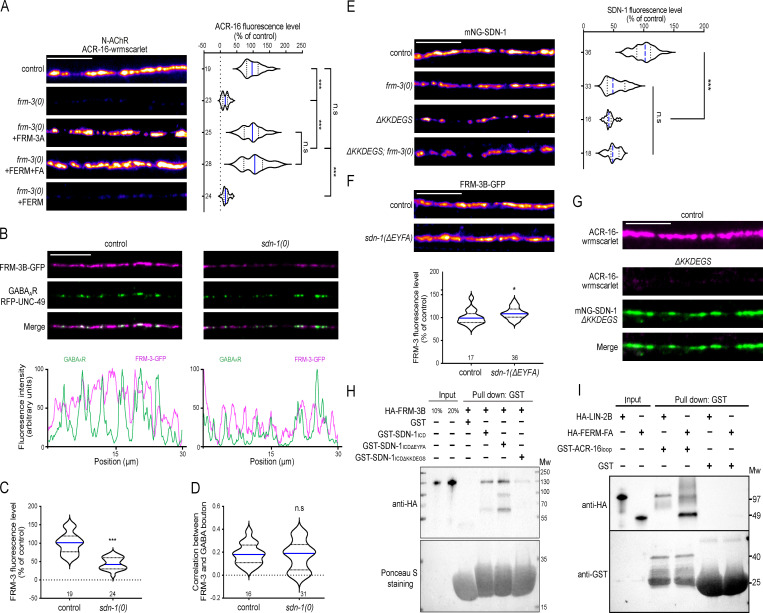
Because the LIN-2 partner FRM-3 is present at cholinergic NMJs (Zhou et al., 2020), we tested if FRM-3 might also be involved in the clustering of N-AChRs. Strikingly, ACR-16-wrmScarlet was almost undetectable in a frm-3(0) null mutant (Fig. 7 A). This phenotype could be rescued by muscle-specific expression of either full-length FRM-3A or its FERM-FA (FERM adjacent) domain. Interestingly, the FERM domain alone was not able to rescue N-AChR clustering defects, as also observed for GABA receptors at inhibitory NMJs (Fig. 7 A; Zhou et al., 2020).
Figure 7.
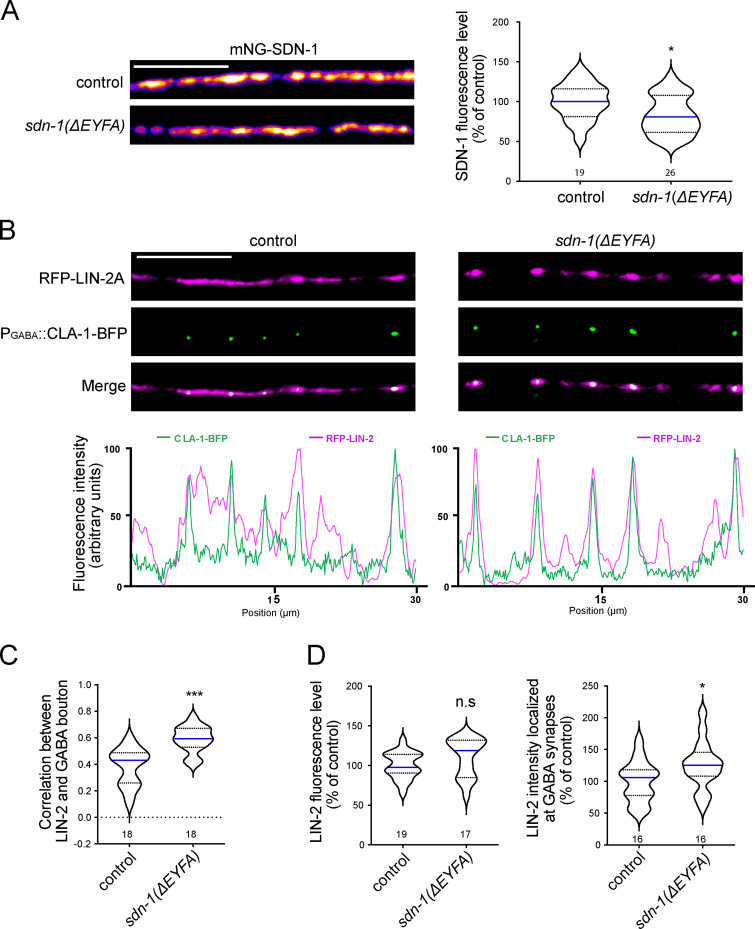
FRM-3/FARP bridges α7-like N-AChRs with LIN-2/CASK and SDN-1.(A) Confocal detection and quantification of ACR-16-wrmScarlet in control, frm-3(0) and frm-3(0) animals expressing full length FRM-3A-GFP, FERM-FA-GFP or FERM-GFP truncations driven by the muscle-specific Pmyo-3 promoter. (B) Confocal detection and fluorescent profiles of GABAARs (RFP-UNC-49) and FRM-3B-GFP in control and sdn-1(0) animals expressing frm-3b-gfp driven by the muscle-specific promoter Pmyo-3. (C) Quantification of FRM-3B-GFP fluorescence level. (D) Pearson’s correlation coefficient between FRM-3B-GFP and GABA bouton. (E) Confocal detection and quantification of mNG-SDN-1 full length or lacking the KKDEGS motif (ΔKKDEGS). (F) Confocal detection and quantification of FRM-3B-GFP in control and sdn-1(ΔEYFA) animals expressing frm-3b-gfp under the control of the muscle-specific promoter Pmyo-3. (G) Confocal detection of ACR-16-wrmScarlet in control or in mNG-SDN-1 knock-in animals lacking the KKDEGS motif. (H) GST pull-down analysis of the binding between HA-tagged FRM-3B and SDN-1ICD, PDZ-binding motif deletion (SDN-1ICDΔEYFA), C1 motif deletion (SDN-1ICDΔKKDEGS) by immunoblotting using anti-HA antibody. The same membrane was stained by Ponceau red to show GST expression. Molecular weights (Mw) are shown on the right. (I) GST pull-down analysis of ACR-16 TM3-TM4 cytosolic loop binding with HA tagged LIN-2B or FERM-FA domain of FRM-3 by immunoblotting using anti-HA antibody. The same membrane was probed by anti-GST antibody to show GST expression. Molecular weight (Mw) is shown on the right. Scale bars, 10 µm.
We then analyzed the expression of FRM-3-GFP in a sdn-1(0) mutant and observed a 55% decrease of fluorescence at the nerve cord (Fig. 7, B and C). However, in contrast to LIN-2, the FRM-3 distribution remained unchanged (Fig. 7, B–D). Hence, despite the loss of LIN-2 at cholinergic synapses in sdn-1(0) mutants, FRM-3 remained partially concentrated in postsynaptic domains of cholinergic synapses. In a reciprocal experiment, we measured a 47% decrease of SDN-1 content at the nerve cord of frm-3(0) mutants (Fig. 7 E), much more than in lin-2(0) mutants. This suggested that the interactions between FRM-3 and LIN-2 with SDN-1 were different. Indeed, in contrast to LIN-2, FRM-3 content was not reduced by deleting the PDZ-binding site of SDN-1 (Fig. 7 F). An early study reported direct binding of the FERM domain of ezrin and syndecan-2 through the DEGS motif present in its C1 intracellular region (Granés et al., 2003). This sequence is conserved in SDN-1 (Fig. 2 A). CRISPR-Cas9 deletion of the KKDEGS sequence caused a 50% decrease of mNG-SDN-1 fluorescence level, similar to what was observed in frm-3(0) mutants (Fig. 7 E). The deletion of the SDN-1 KKDEGS sequence in frm-3(0) animals did not cause an additional decrease of mNG-SDN-1 fluorescence, suggesting that the C1 domain of SDN-1 is the site of interaction with FRM-3. Accordingly, in vitro pull-down experiments demonstrated interactions of the FERM-FA domain of FRM-3 with SDN-1ICD, and this interaction was weakened when the KKDEGS sequence was deleted, while the removal of the PDZ-binding motif had no effect (Fig. 7 H).
Our data show that FRM-3 and LIN-2 are both critical for SDN-1–dependent clustering of N-AChR and might form a complex connecting N-AChR and SDN-1. Accordingly, in vivo coimmunoprecipitation indicates that ACR-16 and LIN-2 are in a complex (Fig. 6 A). Using in vitro pull-down experiments, we could confirm this interaction and also detect an interaction between the FERM-FA domain of FRM-3 and the TM3-TM4 cytoplasmic loop of ACR-16 (Fig. 7 I). Altogether, these data suggest that a macromolecular scaffold assembles at cholinergic synapses to localize N-AChRs.
SDN-1ICD is sufficient to relocalize N-AChRs at GABAergic synapses independently of L-AChRs
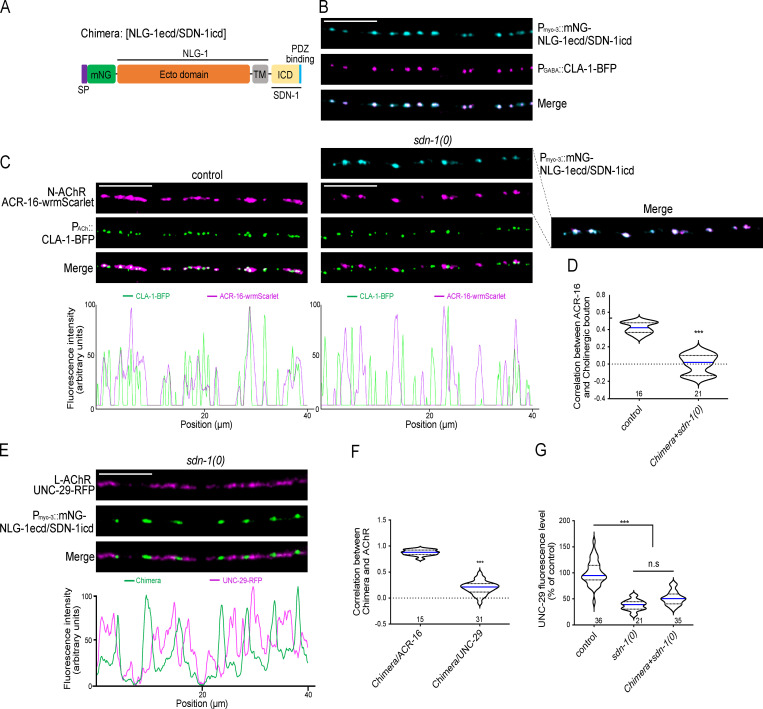
At cholinergic synapses, SDN-1 is required for the localization of both L-AChRs and N-AChRs. Yet, the interaction between the intracellular C terminus of SDN-1 and the PDZ domain of LIN-2/CASK is specifically involved in the clustering of homomeric N-AChRs, but not heteromeric L-AChRs. We therefore asked whether SDN-1 was only a permissive factor requiring additional components to specify N-AChR localization or if it was carrying sufficient information to dictate N-AChR localization. We expressed in the muscle cells of sdn-1(0) mutants a mNG-tagged chimeric protein containing the extracellular and transmembrane regions of NLG-1 and SDN-1ICD (Fig. 8 A). As predicted, this NLG-1 chimera localized at GABA synapses (Fig. 8 B). Strikingly, as opposed to sdn-1(0) mutants, ACR-16-wrmScarlet were readily detected in transgenic animals, but they relocalized at GABAergic NMJs together with the chimera (Fig. 8, C and D). In contrast, the decreased level of L-AChRs was not rescued and the receptors remained localized at cholinergic synapses (Fig. 8, E–G). These results demonstrate that SDN-1ICD is necessary and sufficient to recruit N-AChR at synaptic sites. Furthermore, despite the fact that homomeric and heteromeric AChRs colocalize at cholinergic NMJs, their clustering relies on two distinct machineries (see Fig. 9 and discussion).
Figure 8.
SDN-1ICD recruits N-AChR to GABA synapses.(A) Structure of the chimeric protein containing the NLG-1 ecto- and transmembrane (TM) domains and the SDN-1ICD. mNG tag was inserted immediately after the signal peptide (SP). The PDZ-binding motif of SDN-1 is shown in blue. (B) Confocal detection of mNG-NLG-1ECD/SDN-1ICD expression driven by the muscle-specific promoter Pmyo-3 and of the presynaptic marker CLA-1-BFP driven by the GABAergic neuron-specific promoter Punc-47. (C) Confocal detection and fluorescent profiles of ACR-16-wrmScarlet and cholinergic active zone marker CLA-1-BFP. NLG-1ECD/SDN-1ICD was expressed under the muscle-specific promoter Pmyo-3 in sdn-1(0) mutant animals. (D) Pearson’s correlation coefficient between N-AChRs (ACR-16-wrmScarlet) and cholinergic boutons. (E) Confocal detection and fluorescent profiles of L-AChRs (UNC-29-RFP) and mNG-NLG-1ECD/SDN-1ICD driven by the muscle-specific promoter Pmyo-3 in sdn-1(0) mutants. (F) Pearson’s correlation coefficient between the chimera and N-AChRs (ACR-16-wrmScarlet) or L-AChRs (UNC-29-RFP) in sdn-1(0) mutants. (G) Quantification of UNC-29-RFP in control, sdn-1(0), and sdn-1(0) animals expressing the chimera specifically in muscle. Scale bars, 10 µm.
Figure 9.
Working model for N-AChR clustering at NMJs. See Discussion for details.
To test if the intracellular domain of NLG-1 might be sufficient to recruit GABAARs, we fused the intracellular domain of NLG-1 to the extracellular domain of LEV-10, a protein involved in L-AChR clustering and located only at cholinergic synapses. Unfortunately, this protein was unstable, which prevented us from drawing any conclusions.
Discussion
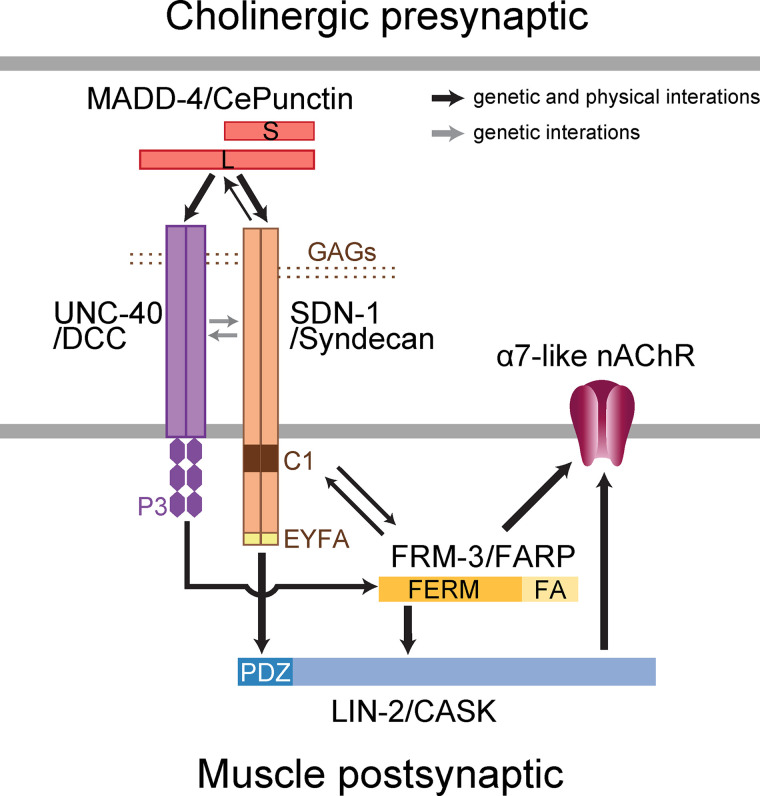
Our results identified the syndecan SDN-1 as a core organizer of cholinergic and, to a lesser extent, GABAergic neuromuscular junctions in C. elegans. SDN-1 is absolutely required for the synaptic clustering of homomeric α7-like N-AChRs and is necessary for achieving proper synaptic levels of heteromeric L-AChRs and GABAARs. We therefore uncovered a previously unknown function of syndecan in the direct control of synaptic receptor composition. Syndecan acts by bridging extracellular matrix components with intracellular scaffolding proteins (Fig. 9). In our model, the anterograde organizer Punctin is secreted by cholinergic and GABAergic motoneurons in the synaptic cleft and triggers appropriate postsynaptic differentiation. At cholinergic synapses, Punctin has at least two parallel functions. First, it localizes the syndecan produced by muscle cells at postsynaptic sites. Second, it concentrates and likely activates the transmembrane receptor UNC-40/DCC. Syndecan, in turn, stabilizes Punctin and UNC-40 at synaptic sites. Coincident clustering of SDN-1 and UNC-40 at cholinergic NMJs triggers the intracellular recruitment of the scaffolding molecules LIN-2/CASK and FRM-3/FARP by direct interaction with the PDZ domain–binding site of SDN-1 and the C-terminal P3 domain of UNC-40, respectively. The FERM-FA domain of FRM-3/FARP also engages direct interaction with SDN-1, likely with its submembrane C1 domain. The resulting CASK–FARP complex then triggers synaptic clustering of the α7-like N-AChR.
The synaptic localization of syndecan is controlled by the ECM protein Punctin
The molecular mechanisms that control the synaptic localization of syndecan at Drosophila NMJs or in rodent synapses remain elusive. At the C. elegans NMJ, our genetic analyses demonstrate the instructive role of Punctin for synaptic localization of SDN-1/syndecan. Strikingly, removing Punctin from GABAergic NMJs relocalizes SDN-1 exclusively at cholinergic NMJs, while in a Punctin null mutant, SDN-1 is almost undetectable. Our data are consistent with a direct interaction of the C-terminal half of Punctin, shared by all Punctin isoforms, and SDN-1. This interaction must involve the SDN-1 core protein, since ΔGAG SDN-1 mutants still localize at the synapse. Accordingly, the human ADAMTS1 protease, which contains structural domains similar to Punctin, is able to cleave syndecan 4 independently of the presence of GAG chains (Rodríguez-Manzaneque et al., 2009). However, HS chains are important to regulate the amount of SDN-1 at synapses. HSs form long, highly negatively charged molecules able to engage versatile electrostatic interactions (Xu and Esko, 2014). Punctin is a good candidate for synaptic stabilization of SDN-1 through HS interaction, because it contains an immunoglobulin-like domain with a predicted highly electropositive surface pocket, which we demonstrated to bind heparin in vitro (Platsaki et al., 2020). SDN-1 is the first identified component able to regulate the amount of Punctin. Although Punctin might interact with a yet-unidentified presynaptic receptor, an alternative hypothesis is that it is locally deposited at synaptic terminals and its interaction with the ECM would prevent its diffusion and trigger the local differentiation of postsynaptic domains (Pinan-Lucarré et al., 2014). The Punctin/SDN-1 crosstalk identified at the C. elegans NMJ provides an interesting example of a positive feedback loop between a synaptic organizer and its effector, which likely contributes to synaptic stabilization.
Cooperation between syndecan- and UNC-40/DCC-dependent signaling to localize α7-like N-AChRs
Syndecan impacts the second signaling output of Punctin, which involves the transmembrane receptor UNC-40/DCC. During development, Punctin promotes the growth of muscle arms toward nerve cords by positioning and activating UNC-40–dependent remodeling of the actin cytoskeleton (Alexander et al., 2009). In the adult, Punctin binds, recruits, and likely activates UNC-40/DCC at the postsynaptic sites of NMJs (Tu et al., 2015). At GABAergic NMJs, UNC-40 activation sets the amount of synaptic GABAARs by promoting their recruitment to NLG-1/neuroligin (Zhou et al., 2020). Here, we show that in addition to its dependence on Punctin, the synaptic content of UNC-40 is regulated by SDN-1. DCC is known to bind heparin (Bennett et al., 1997), and numerous studies have documented the interplay between GAGs and DCC-signaling for cellular and axonal migration in various organisms (Blanchette et al., 2015, 2017; Bülow et al., 2002; Matsumoto et al., 2007; Rhiner et al., 2005). An elegant structural study demonstrated the critical role of HS for the binding of netrin to DCC by providing sulfate ions that bridge the positively charged surfaces that interact which each other on both proteins (Finci et al., 2014). Similarly, syndecan might serve as a coreceptor to strengthen the binding of Punctin on UNC-40 or stabilize UNC-40 at the plasma membrane by favoring its interaction with the ECM.
Syndecan- and DCC-dependent signaling then converge to build an intracellular scaffold containing FRM-3/FARP and LIN-2/CASK at cholinergic NMJs. This configuration parallels what we recently described at GABAergic NMJs for the adhesion molecule NLG-1/neuroligin and the transmembrane receptor UNC-40/DCC, where (1) UNC-40 recruits FRM-3 through its C-terminal P3 domain, (2) FRM-3 recruits LIN-2, (3) LIN-2 binds the C-terminal tail of neuroligin through its PDZ domain, and (4) LIN-2 interacts with the intracellular loop of the GABAAR UNC-49 and stabilizes the receptors at the synapse by bridging them with Punctin-dependent neuroligin clusters (Zhou et al., 2020). At a first glance, syndecan replaces neuroligin at cholinergic synapses to eventually achieve N-AChR clustering. SDN-1 interacts with LIN-2/CASK via its C-terminal PDZ domain–binding site, and this interaction is necessary for the localization of LIN-2/CASK at cholinergic synapses. The PDZ-dependent interaction between syndecans and CASK is evolutionarily conserved (Hsueh et al., 1998). Interestingly, CASK is mostly found in axons during development, together with SDC3, and redistributes to somatodendritic compartments in the adult, where it is coenriched with SDC2 at various synapses (Hsueh and Sheng, 1999; Hsueh et al., 1998). Our results in C. elegans suggest that syndecan localization might instruct the subcellular localization of CASK in mammalian neurons.
The α7-like N-AChR clustering relies on an evolutionarily conserved intracellular scaffold
The intracellular moiety of SDN-1 syndecan is the core component that controls the synaptic clustering of the α7-like N-AChR by nucleating the assembly of the LIN-2–CASK-FRM-3–FARP complex at cholinergic synapses. Punctin therefore achieves N-AChR clustering by positioning SDN-1 in register with cholinergic boutons, and SDN-1 subsequently recruits N-AChRs via the LIN-2–FRM-3 cytoplasmic complex. This mechanism is fundamentally different from the clustering mechanisms of the heteromeric L-AChRs, where a set of extracellular proteins associates with L-AChRs to form small clusters that are subsequently localized at cholinergic synapses by the long isoform of Punctin (Gally et al., 2004; Gendrel et al., 2009; Pinan-Lucarré et al., 2014; Rapti et al., 2011). However, SDN-1 also modulates the synaptic content of L-AChRs, likely through extracellular interactions, since the disruption of the SDN-1/LIN-2 association or the ectopic expression of SDN-1ICD have no impact on L-AChRs. SDN-1 might control L-AChR content by stabilizing Punctin at synapses or by enhancing the interactions between Punctin L and the L-AChR clustering machinery, which have still not been solved at the molecular level.
The central and specific role of SDN-1 for N-AChR clustering at cholinergic synapses raises questions about the mechanisms underlying this specificity. Not only is SDN-1 equally present at GABAergic NMJs, but its associated proteins UNC-40/DCC, FRM-3/FARP, and LIN-2/CASK are used at inhibitory NMJs to cluster GABAARs. Yet, ACR-16 is exclusively detected at cholinergic synapses. Several scenarios can be envisioned. First, NLG-1 could outcompete SDN-1 for LIN-2/CASK binding and stabilize a complex that preferentially binds GABAARs. Second, Punctin L, which is exclusively present at cholinergic NMJs, might induce distinct oligomerization of SDN-1 at the nanoscale, which is known to impact its signaling output (Choi et al., 2005; Jang et al., 2018). Third, SDN-1 might be differentially phosphorylated (Ethell et al., 2001; Horowitz and Simons, 1998; Murakami et al., 2002; Oh et al., 1997), modified, or associated with a nonidentified cofactor to promote the formation of an ACR-16–binding complex. The insertion of ACR-16 in the muscle membrane was shown to be regulated by Wnt ligands possibly secreted from cholinergic neurons (Babu et al., 2011; Francis et al., 2005; Jensen et al., 2012; Tikiyani et al., 2018). A crosstalk between Wnt’s and syndecan has been documented in many systems (Pataki et al., 2015), and SDN-1 might serve as a coreceptor at cholinergic NMJs. Yet, this putative Wnt–syndecan crosstalk can be bypassed by overexpression of the SDN-1 tail at GABAergic synapses, which emphasizes the central role of SDN-1 as a postsynaptic organizer.
The C. elegans ACR-16 is strikingly close to the mammalian α7 N-AChR subunit (Ballivet et al., 1996; Touroutine et al., 2005). These receptors have attracted attention, because they have been implicated in cognition and memory and their dysfunction was associated to neuropsychiatric disorders (for review, see Dineley et al., 2015). In some instances, they mediate fast synaptic transmission, as in the hippocampus (Alkondon et al., 1997; Bürli et al., 2010; Fabian-Fine et al., 2001; Gray et al., 1996; Wonnacott, 1997; Zarei et al., 1999), and in many cases, they localize presynaptically on GABAergic or glutamatergic terminals, where they enhance neurotransmitter release (Alkondon et al., 1997; Bürli et al., 2010; Fabian-Fine et al., 2001; Gray et al., 1996; Liu et al., 2001). Surprisingly, the mechanisms that regulate these localizations remain mostly unknown. Among many factors, synaptic activity, neurotrophin, anti-apoptotic Bcl-2 proteins, src kinase, Wnt and neuroregulin signaling pathways, or plasma membrane lipid composition, have been shown to impact α7 receptor localization, yet always partially (Brusés et al., 2001; Kawai et al., 2002; Charpantier et al., 2005; Farías et al., 2007; Hancock et al., 2008; Dawe et al., 2019). Colocalization between α7 receptor clusters and PSD-95 family members has been described, yet direct interaction was not documented (Conroy et al., 2003; Farías et al., 2007; Parker et al., 2004). Because all the components we identified in C. elegans are evolutionarily conserved and expressed in neurons, it would be worth testing if syndecan regulates the localization of nicotinic receptors in the mammalian brain.
Materials and methods
Strains and genetics
All C. elegans strains were originally derived from the wild-type Bristol N2 strain. Worm cultivation, genetic crosses, and manipulation of C. elegans were performed according to standard protocols (Brenner, 1974). All strains were maintained on nematode growth medium (NGM) agar plates with Escherichia coli OP50 as a food source at 20°C. A complete list of strains used in this study can be found in Table 1.
Table 1. List of strains.
| Figure | Strain | Genotype |
|---|---|---|
| 1 | EN7849 | kr440 [acr-16-wrmScarlet] (I); krSi144 [Punc-17::cla-1-TagBFP] (I); sdn-1(zh20; X) |
| 1 | EN7850 | kr440 [acr-16-wrmScarlet] (I); krSi144 [Punc-17::cla-1-TagBFP] (I) |
| 1, S2 | EN208 | kr208 [unc-29-TagRFP-T] (I) |
| 1 | EN2649 | kr208 [unc-29-TagRFP-T] (I); sdn-1(zh20; X) |
| 1 | EN7244 | zxIs6 [Punc-17::ChR2(H134R)-YFP+lin-15(+)] (V) |
| 1 | EN7243 | zxIs6 [Punc-17::ChR2(H134R)-YFP+lin-15(+)] (V); sdn-1(zh20; X) |
| S1 | EN3886 | kr296 [unc-49-TagRFP-T] (III); sdn-1(zh20; X); krIs67 [Punc-47::snb-1-TagBFP] |
| S1 | EN3796 | kr296 [unc-49-TagRFP-T] (III); krIs67 [Punc-47::snb-1-TagBFP] |
| S1 | EN3513 | kr296 [unc-49-TagRFP-T] (III); sdn-1(zh20; X) |
| S1, S2 | EN296 | kr296 [unc-49-TagRFP-T] (III) |
| 2 | EN387 | kr387 [sdn-1::SL2-wrmScarlet] (X) |
| 2 | EN7094 | kr208 [unc-29-TagRFP-T] (I); krSi50 [Peft-3::TIR1-TagBFP] (IV); kr383 [sdn-1::AID-mNG] (X) |
| 2 | EN7158 | kr208 [unc-29-TagRFP-T] (I); krSi55 [Pmyo-3::TIR1-TagBFP] (V); kr383 [sdn-1::AID-mNG] (X) |
| 2 | EN7029 | kr208 [unc-29-TagRFP-T] (I); krSi36 [Prab-3::TIR1-TagBFP] (V); kr383 [sdn-1::AID-mNG] (X) |
| 2 | EN7235 | kr208 [unc-29-TagRFP-T] (I); krSi63 [Pdpy-7::TIR1-TagBFP] (II); kr383 [sdn-1::AID-mNG] (X) |
| 2, 5, 6, 7, S5 | EN437 | kr437 [mNG-sdn-1] (X) |
| 2, S2 | EN7839 | kr208 [unc-29-TagRFP-T] (I); kr437 [mNG-sdn-1] (X) |
| S2, S3 | EN7838 | kr296 [unc-49-TagRFP-T] (III); kr437 [mNG-sdn-1] (X) |
| S2, S3, 4 | EN7860 | krSi144 [Punc-17::cla-1-TagBFP] (I); kr437 [mNG-sdn-1] (X) |
| S2, S3 | EN7915 | krSi143 [Punc-47::cla-1-TagBFP] (V); kr437 [mNG-sdn-1] (X) |
| S2 | EN7808 | krSi144 [Punc-17::cla-1-TagBFP] (I) |
| S2 | EN7809 | krSi143 [Punc-47::cla-1-TagBFP] (V) |
| S4 | EN7131 | ieSi58 [Peft-3::AID-GFP::unc-54 3′UTR + Cbr-unc-119(+)] (IV); krSi50 [Peft-3::TIR1-TagBFP] (IV) |
| S4 | EN7157 | ieSi58 [Peft-3::AID-GFP::unc-54 3′UTR + Cbr-unc-119(+)] (IV); krSi55 [Pmyo-3::TIR1-TagBFP] (V) |
| S4 | EN7028 | ieSi58 [Peft-3::AID-GFP::unc-54 3′UTR + Cbr-unc-119(+)] (IV); krSi36 [Prab-3::TIR1-TagBFP] (V) |
| S4 | EN7236 | krsi63 [Pdpy-7::TIR1-TagBFP] (II); ieSi58 [Peft-3::AID-GFP::unc-54 3′UTR + Cbr-unc-119(+)] (IV) |
| 3 | EN383 | kr383 [sdn-1-AID-mNG] (X) |
| 3 | EN441 | kr441 [sdn-1(S71A)-AID-mNG] |
| 3 | EN388 | kr388 [sdn-1(S71A,S86A)-AID-mNG] |
| 3 | EN475 | kr475 [sdn-1(S71A,S86A,S214A)-AID-mNG] |
| 4 | EN7937 | krSi145 [Punc-17::cla-1-TagBFP] (IV); kr437 [mNG-sdn-1] (X) |
| 4 | EN7938 | madd-4(kr270; I); krSi145 [Punc-17::cla-1-TagBFP] (IV); kr437 [mNG-sdn-1] (X) |
| 4 | EN7747 | kr373 [madd-4-TagRFP-T] (I); kr437 [mNG-sdn-1] (X); krIs67 [Punc-47::snb-1-TagBFP] |
| 4 | EN7754 | madd-4(tr185; I); kr437 [mNG-sdn-1] (X); krSi67 [Punc-47::snb-1-TagBFP] |
| 4 | EN7755 | madd-4(ttTi103747; I); kr437 [mNG-sdn-1] (X); krSi67 [Punc-47::snb-1-TagBFP] |
| 4 | EN373 | kr373 [madd-4-TagRFP-T] (I) |
| 4 | EN7899 | kr373 [madd-4-TagRFP-T] (I); sdn-1(zh20; X) |
| 4 | EN7631 | kr373 [madd-4-TagRFP-T] (I); kr383 [sdn-1-AID-mNG] (X) |
| 4 | EN7758 | kr373 [madd-4-TagRFP-T] (I); kr475 [sdn-1(S71A,S86A,S214A)-AID-mNG] (X) |
| 5–7 | EN440 | kr440 [acr-16-wrmScarlet] (I) |
| 5 | EN7512 | unc-40(e1430; I); kr440 [acr-16-wrmScarlet] (I) |
| 5 | EN7910 | krSi145 [Punc-17::cla-1-TagBFP]; kr457[unc-40(ΔP3)] (I); kr440 [acr-16-wrmScarlet] (I) |
| 5 | EN7733 | unc-40(e1430; I); kr437 [mNG-sdn-1] (X) |
| 5 | EN7833 | krSi11 [Pmyo-3::TagRFP-T-unc-40] (II); juIs1 [Punc-25::snb-1-GFP]; sdn-1(zh20; X) |
| 5 | EN3333 | krSi11 [Pmyo-3::RFP-unc-40] (II) |
| 5 | EN3361 | krSi11 [Pmyo-3::RFP-unc-40] (II); juIs1 [Punc-25::snb-1-GFP] |
| 5 | EN7845 | krSi11 [Pmyo-3::TagRFP-T-unc-40] (II); madd-4(kr270; I) |
| 5 | EN7831 | krSi11 [Pmyo-3::Tag-RFP-T-unc-40] (II); madd-4(kr270; I); sdn-1(zh20; X) |
| 6 | EN7779 | krSi35[Pmyo-3::lin-2a-RFP]; krSi10 [Pmyo-3::nlg-1-GFP]; sdn-1(zh20; X) |
| 6 | EN7720 | kr437 [mNG-sdn-1] (X); lin-2(n397; X) |
| 6 | EN7858 | krSi35 [Pmyo-3::lin-2a-RFP]; krSi143 [Punc-47::cla-1-TagBFP]; kr437 [mNG-sdn-1] (X) |
| 6 | EN7859 | krSi35 [Pmyo-3::lin-2a-RFP]; krSi143 [Punc-47::cla-1-TagBFP]; kr437 [mNG-sdn-1] (X); kr496 [mNG-sdn-1(ΔEYFA)] (X) |
| 6 | EN3921 | krSi30[Pmyo-3::lin-2a-GFP]; krIs67 [Punc-47::snb-1-TagBFP]; kr296 [unc-49-TagRFP-T] (III) |
| 6 | EN7013 | krSi30[Pmyo-3::lin-2a-GFP]; krIs67 [Punc-47::snb-1-TagBFP]; kr296 [unc-49-TagRFP-T] (III); nlg-1 (ok259; X) |
| 6 | EN7832 | krSi30[Pmyo-3::lin-2a-GFP]; krIs67 [Punc-47::snb-1-TagBFP]; kr296 [unc-49-TagRFP-T] (III); nlg-1 (ok259)] (X); sdn-1(zh20; X) |
| S5 | EN496 | kr496 [mNG-sdn-1(ΔEYFA)] (X) |
| S5 | EN7858 | krSi143 [Punc-47::cla-1-TagBFP] V; krSi35 [Pmyo-3::lin-2a-RFP] |
| S5 | EN7859 | krSi143 [Punc-47::cla-1-TagBFP] V; krSi35 [Pmyo-3::lin-2a-RFP]; kr496 [mNG-sdn-1(ΔEYFA)] (X) |
| 7 | EN7489 | kr440 [acr-16-wrmScarlet] (I); frm-3(gk585; X) |
| 7 | EN7959 | krSi31 [Pmyo-3::GFP-frm-3a]; kr440 [acr-16-wrmScarlet] (I); frm-3(gk585; X) |
| 7 | EN8055 | krSi32 [Pmyo-3::GFP-frm-3a(1-412aa)]; kr440 [acr-16-wrmScarlet] (I); frm-3(gk585; X) |
| 7 | EN7912 | krSi39 [Pmyo-3::GFP-frm-3a(1-318aa)]; kr440 [acr-16-wrmScarlet] (I); frm-3(gk585; X) |
| 7 | EN8048 | kr440 [acr-16-wrmScarlet] (I); kr437 [mNG-sdn-1] (X); frm-3(gk585; X) |
| 7 | EN50519 | sdn-1 [mNG-kr519(ΔKKDEGS)] (X) |
| 7 | EN8057 | sdn-1 [mNG-kr519(ΔKKDEGS)] (X); frm-3(gk585; X) |
| 7 | EN8082 | kr440 [acr-16-wrmScarlet] (I); sdn-1 [mNG-kr519(ΔKKDEGS)] (X) |
| 7 | EN7177 | krSi61 [Pmyo-3::frm-3-GFP]; kr296 [unc-49-TagRFP-T] (III) |
| 7 | EN8046 | krSi144 [Punc-17::cla-1-TagBFP]; krSi61 [Pmyo-3::frm-3-GFP]; kr499 [sdn-1(ΔEYFA)] (X) |
| 7 | EN7208 | krSi61 [Pmyo-3::frm-3-GFP]; krIs67 [Punc-47::snb-1-TagBFP]; kr296 [unc-49-TagRFP-T] (III) |
| 7 | EN7209 | krSi61 [Pmyo-3::frm-3-GFP]; krIs67 [Punc-47::snb-1-TagBFP]; kr296 [unc-49-TagRFP-T] (III); sdn-1(zh20; X) |
| 8 | EN8107 | krSi157[Pmyo-3::mNG-NLG-1 ECD /SDN-1 ICD ]; krSi143 [Punc-47::cla-1-TagBFP]; kr296 [unc-49-TagRFP-T] (III) |
| 8 | EN7926 | krSi157[Pmyo-3::mNG-NLG-1 ECD /SDN-1 ICD ]; krSi145 [Punc-17::cla-1-TagBFP]; kr440 [acr-16-wrmScarlet] (I); sdn-1(zh20; X) |
| 8 | EN7851 | krSi144 [Punc-17::cla-1-TagBFP]; kr208 [unc-29-TagRFP-T] (I); sdn-1(zh20; X) |
| 8 | EN8040 | krSi160[Pmyo-3::mNG-NLG-1 ECD /SDN-1 ICD ]; kr208 [unc-29-TagRFP-T] (I); sdn-1(zh20; X) |
Plasmids
The constructs created in this study are described in Table 2. The full open reading frames of all constructs were verified by Sanger sequencing from GATC Company. The other plasmids used to create single-copy insertion alleles by the miniMos method are described in a previous study (Zhou et al., 2020).
Table 2. List of generated plasmids.
| Plasmid | Description | Usage |
|---|---|---|
| pUA111 | TIR-1-TagBFP | MiniMos plasmid to build tissue-specific expression of TIR1 alleles |
| pDD268 | mNG-3xFLAG | Template used to insert homology arms to create sdn-1::AID-mNG (EN383) strain |
| pAV52 | TagRFP-T | Repair template to create EN373 CRISPR knock-in strain |
| pCV01 | AID-mNG | Repair template to create EN383 CRISPR knock-in strain |
| pCV02 | SL2::wrmScarlet | Repair template to create EN387 CRISPR knock-in strain |
| pCV03 | PU6pol(III)::crCV02 | Vector expressing guide RNA to create EN387 strain |
| pCV04 | Pmyo-3::TIR1-TagBFP::unc-54 3′UTR | Minimal Mos 1 vector for Pmyo-3::TIR1-TagBFP insertion; this plasmid was used to create krSi55 allele; contains a neomycin-resistant cassette |
| pCV05 | Prab-3::TIR1-TagBFP::unc-54 3′UTR | Minimal Mos 1 vector for Prab-3::TIR1-TagBFP insertion; this plasmid was used to create krSi36 allele; contains a neomycin-resistant cassette |
| pCV08 | Pdpy-7::TIR1-TagBFP::unc-54 3′UTR | Minimal Mos 1 vector for Pdpy-7::TIR1-TagBFP insertion; this plasmid was used to create krSi63 allele; contains a neomycin-resistant cassette |
| pCV09 | Peft-3::TIR1-TagBFP::unc-54 3′UTR | Minimal Mos 1 vector for Peft-3::TIR1-TagBFP insertion; this plasmid was used to create krSi50 allele; contains a neomycin-resistant cassette |
| pXZ48 | pcDNA3.1(+)-HA-lin-2b | To synthesize HA-LIN-2B in vitro by TnT kit |
| pXZ64 | pcDNA3.1(+)-frm-3_FERM-FA | To synthesize HA-FERM-FA domain of FRM-3 in vitro by TnT kit |
| pXZ92 | pGEX-3X-SDN-1ICD | To express GST-SDN-1 intracellular domain in ArcticExpress E. coli |
| pXZ94 | pGEX-3X-SDN-1ICDΔEYFA | To express GST-SDN-1 intracellular domain lacking EYFA PDZ-binding motif in ArcticExpress E. coli |
| pXZ97 | pGEX-3X-ACR-16_loop | To express GST-ACR-16 TM3-4 loop in ArcticExpress E. coli |
| pXZ102 | Pmyo-3::sp-mNG-nlg-1 ECD +TM/sdn-1 ICD | Minimal Mos 1 vector to express NLG-1 ecto-transmembrane domain and SDN-1ICD in body-wall muscle |
| pXZ119 | pcDNA3.1(+)::SP-HA-SDN-1 | To express HA-SDN-1 in HEK293 cell |
| pBP40 | Pcmv::MADD-4B-egfp | To express MADD-4B-egfp in HEK293 cell |
TIR1, transport inhibitor response 1.
Generation of single-copy insertion alleles
The single-copy insertion alleles generated by the miniMos method (Frøkjær-Jensen et al., 2014) are listed in Table 3. Genes encoding fluorescent-tagged proteins are driven by myo-3– (body-wall muscle), unc-47– (GABAergic motoneuron), unc-17– (cholinergic motoneuron), rab-3– (pan-neuronal), dpy-7– (epidermis), or eft-3–specific (ubiquitous) promoters. N2 animals were injected with 15 ng/µl plasmid of interest containing the promoter and the open reading frame, 50 ng/µl pCFJ601 (Mos1 transposase), 10 ng/µl pMA122 (negative selective marker Phsp16.2::peel-1), and 2.5 ng/µl pCFJ90 (Pmyo-2::mCherry). Neomycin (G418) was added to plates 24 h after injection at 1.5 µg/µl final concentration. Candidate plates were heat shocked for 2 h at 34°C. Selected lines were then bred to homozygosity.
Table 3. List of miniMos single-copy insertion alleles.
| Allele name | Construct |
|---|---|
| krSi143 | Punc-47::cla-1-TagBFP V |
| krSi144 | Punc-17::cla-1-TagBFP I |
| krSi145 | Punc-17::cla-1-TagBFP IV |
| krSi50 | Peft-3::TIR1-TagBFP IV |
| krSi55 | Pmyo-3::TIR1-TagBFP V |
| krSi36 | Prab-3::TIR1-TagBFP V |
| krSi63 | Pdpy-7::TIR1-TagBFP II |
| krSi157 | Pmyo-3::mNG-NLG-1 ECD /SDN-1 ICD , line1 |
| krSi160 | Pmyo-3::mNG-NLG-1 ECD /SDN-1 ICD , line2 |
| krSi30 | Pmyo-3::sfGFP-lin-2a |
| krSi31 | Pmyo-3::frm-3-sfGFP |
| krSi32 | Pmyo-3::frm-3(1-412aa)-sfGFP |
| krSi35 | Pmyo-3::tagRFP-T-lin-2a |
| krSi39 | Pmyo-3::frm-3(1-318aa)-sfGFP |
| krSi60 | Pmyo-3::sfGFP-lin-2b |
| krSi61 | Pmyo-3::sfGFP-frm-3b |
Modifications of the endogenous loci
To generate the kr383 allele, the AID sequence (Zhang et al., 2015) and mNG were inserted tandemly into the sdn-1 locus, just before the stop codon. To generate the kr373 and kr387 alleles, the TagRFP-T and the wrmScarlet (Bindels et al., 2017; El Mouridi et al., 2017) sequences were inserted in the madd-4 and in the sdn-1 loci, respectively, just before the stop codon. The detailed protocol was described previously (Dickinson et al., 2015). CRISPR RNA (CrRNA) sequences targeting the insertion region were designed and synthesized from IDT (Integrated DNA Technologies). CrRNA and tracer RNA were mixed as 1:1 ratio to form a short guidance RNA (sgRNA) duplex. Adult worms were microinjected in the gonads with a mix that contained Cas9 nuclease 0.5 µl of 10 µg/µl (Integrated DNA Technologies), sgRNA duplex 3 µl (100 µM), repair templates (containing the DNA fragment to insert, a hygromycin resistance selection cassette surrounded by loxP sites, Phsp16.41::Cre, and homology arms to target the genomic region) 50 ng/µl, pCFJ90 [Pmyo-2::mCherry] coinjection marker 2.5 ng/µl, and RNase/DNase-free water up to 10 µl. Injected animals were then grown at 25°C, and after 48 h, a positive selection was performed by adding hygromycin on plates at 0.2 µg/µl. Resistant animals were then heat shocked for 2 h at 34°C by water bath to excise the hygromycin selection cassette from the genome. Worms were then screened by sequencing PCR products from the region of interest. Candidates were outcrossed once with N2 to remove unspecific background mutations.
To generate the kr437 and kr440 alleles, PCR amplified sequences of mNG or wrmScarlet with or without homology arms were used as repair templates, respectively (Dokshin et al., 2018). CrRNA was designed and synthesized by IDT (Integrated DNA Technologies). The injection mix contained annealed double-stranded DNA donor cocktail as repair template 200 ng/µl (total 4 µg), Cas9 nuclease 0.5 µl of 10 µg/µl (Integrated DNA Technologies), and sgRNA duplex 3 µl (100 µM). The candidate F1 animals were isolated by tracking the initial fluorescence knock-in. The F2 progenies were then isolated and homozygosed. The insertion was then confirmed by PCR and sequencing.
To generate kr388, kr441, kr475, kr496, kr499, and kr519, ultramer DNA repair templates were produced by Integrated DNA Technologies. The injection mix contained Cas9 nuclease 0.5 µl of 10 µg/µl (Integrated DNA Technologies), sgRNA duplex 3 µl (100 µM), ultramer repair template 1.25 µl (100 µM), pRF4 [Peft-3::rol-6] coinjection marker 2.5 ng/µl, and RNase/DNase-free water filled up to 10 µl. Insertions in F1 roller progenies were checked by PCR. Homozygotes were isolated, and the PCR product was then sent for sequencing. Candidates were outcrossed once with N2 to remove unspecific background mutations.
An extensive list of modified genomic loci can be found in Table 4.
Table 4. List of modified genomic loci.
| Allele name | Modification |
|---|---|
| kr208 | TagRFP-T inserted into TM3-4 loop of UNC-29 |
| kr270 | Full deletion of madd-4 locus |
| kr296 | TagRFP-T inserted at the N terminus of UNC-49 |
| kr373 | TagRFP inserted at the C terminus of MADD-4L/B |
| kr383 | AID-mNG fusion tag inserted right after PDZ-binding motif of SDN-1 |
| kr387 | SL2::wrmScarlet tag inserted right after PDZ-binding motif of SDN-1 |
| kr441 | S71A in sdn-1-aid-mNG (sdn-1(Δ1GAG)) |
| kr388 | S71A and S86A in sdn-1-aid-mNG (sdn-1(Δ2GAG)) |
| kr475 | S71A, S86A, and S214A in sdn-1-aid-mNG (sdn-1(Δ3GAG)) |
| kr437 | mNG inserted at the N terminus (after signal peptide) of SDN-1 |
| kr440 | wrmScarlet inserted into TM3-4 loop of ACR-16 |
| kr457 | Deletion of the P3 motif in UNC-40 |
| kr496 | Deletion of the PDZ-binding motif (EYFA) in mNG-SDN-1 |
| kr499 | Deletion of the PDZ-binding motif (EYFA) in N2 wild type |
| kr519 | Deletion of the ezrin-binding motif (KKDEGS) in mNG-SDN-1 |
Auxin-induced degradation
Auxin plates were prepared by adding auxin indole-3-acetic acid (Sigma-Aldrich) from a 400-mM stock solution in ethanol into NGM at the final concentration of 1 mM (Zhang et al., 2015). Tissue-specific degradation efficiency was monitored on an animal expressing GFP fused to the AID under the ubiquitous promoter Peft-3 and Prab-3::TIR1-TagBFP, Peft-3::TIR1-TagBFP, Pdpy-7::TIR1-TagBFP, or Pmyo-3::TIR1-TagBFP. Animals were grown on regular or auxin plates for 3, 36, 96, and 120 h, respectively. SDN-1-AID-mNG fluorescence was scored after tissue-specific degradation on young adult animals that had hatched on regular or auxin plates.
Microscopy imaging and quantification
For confocal imaging, young live adult hermaphrodites (24 h after L4 larval stage) were mounted on 2% agarose dry pads with 5% poly-lysine beads in M9 buffer (3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl, 0.25 g MgSO4·7 H2O, and distilled water up to 1 liter). Fluorescence images were captured at 20°C using an Andor spinning disk system (Oxford Instruments) installed on a Nikon-IX86 microscope (Olympus) equipped with a 60×/NA 1.42 oil-immersion objective and an Evolve electron-multiplying charge-coupled device camera. Each animal was imaged with IQ software (APIS Informationstechnologien) as a stack of optical sections (0.2 µm apart), and 41 planes were projected along the z axis.
Quantification of images was performed using ImageJ (v1.48 by National Institutes of Health) with Fiji plugin add-ons. For fluorescence intensity measurement, 50 µm (wide) × 3 µm (high) regions along the dorsal cord near the midbody were cropped and analyzed. Acquisition settings were the same across genotypes for quantitative analysis. Each strain was imaged on at least three different days, and the data were pooled together. Data are presented as a percentage of the average fluorescence relative to that of the control strain. For colocalization quantification, images were captured along 40 µm of the dorsal nerve cord anterior to the vulva. The fluorescence intensity along the cord was evaluated with the Plot Profile plugin. For each channel, the values along the x axis were normalized to the value of maximal intensity. Data are presented as minimum to maximum values for animals of each genotype. The colocalization between two channels were analyzed by Pearson’s correlation coefficient as described previously (Pinan-Lucarré et al., 2014).
To quantify the intensity of mNG-SDN-1 at ACh vs GABA NMJs (Fig. S3 E), images of the dorsal cord of animals coexpressing mNG-sdn-1 and Punc-17::cla-1-BFP or Punc-47::cla-1-BFP were acquired and cropped for quantification. In both channels, the background was subtracted using a rolling ball radius of 10 pixels in Fiji (ImageJ). The CLA-1–BFP channel of cropped images was converted to binary using the Ostu threshold. For Punc-17::CLA-1-BFP, noise was removed from the binary mask using the despeckle function (twice). The thresholded image was then multiplied with the binary mask to generate the preprocessed image for further analysis. To generate the plot profiles, the intensities of each pixel along the x axis (nerve cord was converted to a two-dimensional axis) were recorded for each channel in csv files. To determine the level of mNG-SDN-1 at cholinergic synapses, the csv files were then read by an R script to define the cholinergic CLA-1-BFP domains based on values of Punc-17::CLA-1-BFP signals detected along the nerve cord. Next, the mean intensity of SDN-1 was calculated by measuring the area under the curve of SDN-1 profiles within cholinergic CLA-1-BFP domains and then dividing by the cholinergic CLA-1–BFP domain area. The same procedure was applied to determine the level of mNG-SDN-1 at GABAergic synapses and was also used in Fig. S5 D to determine the level of RFP-LIN-2 level at GABA synapses. To quantify the percentage of L-AChRs and GABA receptors colocalized with SDN-1 (Fig. S3 B), the mNG-SDN-1 domains were defined by the same processing as described above for CLA-1-BFP. Next, the percentage of colocalized receptor was calculated by determining the ratio between the area under the curve (in receptor channel) within SDN-1 domains and the area under the curve (in receptor channel) out of the SDN-1 domains. The Plot Profile fluorescence signals were measured and processed by Manders’ overlapping correlation analysis modified from previous studies (Bolte and Cordelières, 2006; Dunn et al., 2011).
To quantify RFP-LIN-2 levels at GABA synapses in Fig. 6 I, GABA domains were defined by the same processing as described for CLA-1-BFP above using NLG-1-GFP as GABAergic synapse marker. To analyze colocalization between N-AChR and cholinergic boutons in Fig. 8, C and D, images were processed as described in the previous paragraph.
GST pull-down assay
The GST pull-down experiment was performed based on previous protocol (Zhou et al., 2016). DNA fragments encoding the ACR-16 TM3-4 loop or SDN-1ICD full length or lacking the EYFA PDZ-binding motif were cloned to pGEX-3X expression vector and then transfected into ArcticExpress E. coli strain (Agilent). Bacterial clones were first cultured overnight in 5 ml LB medium supplemented with 100 mg/ml ampicillin. The volume of overnight-grown bacteria was enlarged in 100 ml LB/ampicillin medium by culturing 2 h at 37°C with shaking until OD600 reached 0.6–0.8. IPTG was added to medium at 1 mM final concentration to induce protein expression at 12°C for 24 h. The induced bacteria were collected by centrifugation and lysed by 20 mW pulsed sonication for 30 min in cold bacteria lysis buffer (50 mM Hepes, pH 7.5, 400 mM NaCl, 1 mM DTT, 1 mM PMSF, and one tablet of Roche-Merck protease inhibitor cocktail/50 ml; 11697498001). Protein lysate was centrifuged at 15,000 ×g for 45 min at 4°C. Clear supernatants were incubated with 100 µl Glutathione sepharose-4B beads (Sigma-Aldrich; GE17-0756-01) at 4°C for 12h. To express HA (human influenza)–tagged LIN-2B, FRM-3B, or FERM-FA domain, coding sequences with an HA N-terminal tag were cloned into pcDNA3.1(+) expression vector. Fusion proteins were expressed by TnT Quick Coupled system (Promega; L1170). 15 µl HA-tagged protein was then incubated with candidate GST-coated beads at 4°C for 2 h. The beads were washed five times with STE (10 mM Tris-Cl, pH8.0, 150 mM NaCl, 1 mM EDTA, and 0.1% Tween-20) buffer. Proteins were eluted by adding equal volume of 2xSDS sampling buffer (Bio-Rad; #1610737) and boiled for 10 min. Samples were electrophoresed in 12% SDS/PAGE (Bio-Rad; #4561043). The separated proteins were incubated with an anti-HA antibody (Cell Signaling Technology; #3724) at 1:1,000 dilution. The GST fusion loading amount was detected by stain-free gel imaging or Ponceau S red staining immediately after membrane transfer. The membrane or gels were imaged by Chemidoc system (Bio-Rad) using autoexposure settings.
Immunoprecipitation from worm lysate and heparinase treatment
Worm lysis and immunoprecipitation were performed as previous published (Zhou et al., 2020). In brief, mixed-stage worms of each genetic background were collected from 40 10-cm dishes with 0.1 mM NaCl. After washing three times at 4°C, worm slurry was frozen in liquid nitrogen to form worm beads ∼15 ml in volume. Five milliliters of worm beads was grinded in liquid nitrogen and thawed in 7.5 ml ice-cold worm lysis buffer (50 mM Hepes, 50 mM KCl, 100 mM NaCl, 1 mM EDTA, 2% Triton X-100, 2 mM PMSF and one tablet of Roche-Merck cOmplete Protease inhibitor cocktail in 25 ml). Worm lysis was incubated at 4°C for 2 h with rotation and then centrifuged at 15,000 ×g for 20 min at 4°C. Supernatant was collected and diluted to a final concentration of 0.2% Triton-X100 by worm lysis buffer. Precleaned 100 µl Trap-A beads coupled with anti-RFP (Chromotek; rta-100) were incubated overnight at 4°C with the samples. The samples were eluted and analyzed by Western blot using anti-RFP(Chromotek; 6G6, 6g6-100) or anti-GFP (Roche; #11814460001) antibodies at 1:1,000 dilution, respectively. To digest HS chains of SDN-1 and compare with GAG-binding site mutant animals, mNG-SDN-1 nonmutated or Δ1, 2, and 3 GAGs fusion proteins were first concentrated by immunoprecipitation of ∼20,000 nematodes per genotype using Trap-A beads coupled with anti-mNG nanobody (Chromotek; nta-100). 1.5 U heparinase I and III (Sigma-Aldrich; H3917-50UN) was added to nonmutated tube containing 500 µl washing buffer (50 mM Hepes, pH 7.7, and 50 mM NaCl) and 100 µl Trap-A beads. The tube was then incubated at 37°C for 2 h. The proteins of each group were eluted by adding 100 µl 2x sampling buffer and boiled for 10 min. Samples were separated by 12% SDS/PAGE (Bio-Rad) and blotted with anti-mNG antibody (Cell Signaling Technology; #53061S) at 1:1,000 dilution. N2 wild-type animals were used as negative control.
HEK293 EBNA cell culture and coimmunoprecipitation
HEK293 EBNA cells were cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen) in a humidified atmosphere of 5% CO2 in air at 37°C. The cells were plated onto six-well plates overnight, and plasmids pBP40 (expressing MADD-4B-egfp), pXZ119 (expressing HA-SDN-1), and pcDND3.1::egfp were cotransfected or transfected alone the next day using Lipofectamine 3000 according to the manufacturer’s instruction (Invitrogen). Cells were collected 48 h after transfection. Cells were lysed in 500 µl RIPA lysis buffer (25 mM Tris•HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS) for 30 min on ice. The cell lysates were then incubated with 30 µl Trap-A beads coupled with anti-GFP nanobody (Chromotek; gta-100). The samples were eluted and analyzed by Western blot. Anti-HA antibody (Cell Signaling Technology; C29F4) at 1:1,000 dilution was used to detect the HA-SDN-1 in the precipitates. The membranes were imaged by Chemidoc system (Bio-Rad) using auto exposure settings.
Electrophysiology
Adult nematodes were glued (B. Braun Medical; Histoacryl Blue) along the dorsal side of the body to the surface of a plastic culture dish (Corning). A sharpened tungsten rod (A-M Systems) was used to perform a lateral incision before the vulva and to remove the viscera. The cuticle flap was glued back to expose the ventral body-wall muscles, and the preparation was treated by collagenase type IV for 20 s at a concentration of 0.5 mg/ml. Recordings were performed on the two first ventromedial muscle cells before the vulva from the right muscle quadrant. For all recordings, strains express ChR2 in cholinergic motoneurons (zxIs6; Liewald et al., 2008). Animals were grown on NGM plates freshly seeded with 300 µl OP50 culture supplemented by 0.25 µl 100 mM all-trans retinal stock (dissolved in ethanol). Membrane currents were recorded in the whole-cell configuration using a MultiClamp 700B amplifier (Molecular Devices). Acquisition and command voltage were done using the Clampex 10 software driving an Axon Digidata 1550 (Molecular Devices). The resistance of recording pipettes ranged between 3 and 4 MΩ. Data were analyzed with Clampfit 10 (Molecular Devices) and graphed with Origin software (OriginLab). The bath solution contained (in mM) 150 NaCl, 5 KCl, 5 CaCl2, 1 MgCl2, 10 glucose, 15 Hepes, and sucrose to 320 mOsm/liter (pH 7.2), and the pipette solution contained (in mM) 120 KCl, 4 NaCl, 5 EGTA, 10 TES (N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid), 4 MgATP, and sucrose to 310 mOsm/liter (pH 7.2).
For currents evoked by cholinergic motoneuron stimulation, the slit worm preparations were exposed to 10-ms light stimulation performed with the pE2 system (CoolLED) at a wavelength of 460 nm driven by Clampex 10 software. The protocol was the following: a first stimulation triggered the activation of L-AChR and N-AChR; the peak current value was measured and named total peak current. The preparation was then perfused with bath solution supplemented with dihydro-β-erythroidine (DHβE) at the concentration of 0.01 mM leading to the specific inhibition of N-AChR. In these conditions, a second stimulation was performed activating only L-AChR. A third stimulation was then applied to check if the current was stable. and the value of the peak current was measured and named L-AChR evoked current. Finally, for each cell, the L-AChR evoked current was subtracted from the total peak current to obtain the N-AChR evoked current values.
For agonist-evoked currents, nicotine and levamisole diluted at the concentration of 0.1 mM in the bath solution were pressured-ejected during 100 ms from a pipette similar in size to the patch pipette at 10 psi using a PDES-2DX system (NPI Electronic). The slit worm preparations were continuously perfused with fresh bath solution via a gravity flow delivery system. All chemicals were obtained from Sigma-Aldrich, except DHβE, which was provided by Tocris. All experiments were done at 20°C.
Statistical analysis
Data were analyzed by nonparametric tests (Mann–Whitney tests for Figs. 1, S1, 2, S2, and S3; Fig. 4, F and G; Fig. 5 E; Fig. 6, A, B, G–I, K, and M; Fig. S5; Fig. 7, C, D, and F; and Fig. 8, D and F or Kruskal–Wallis tests followed by Dunn’s multiple comparison tests for Figs. 4 E and 8 G). One-way ANOVA followed by Tukey’s multiple comparison tests were performed for Figs. 3 and 4 D; Fig. 5, A–C; Fig. 6, B and N; and Fig. 7, A and E; data distribution was assumed to be normal, but this was not formally tested. For all tests, ***, P < 0.001; **, P < 0.01; *, P < 0.05.
Online supplemental material
Fig. S1 (complementary to Fig. 1) shows that SDN-1 has a moderate effect on GABAAR synaptic content. Figs. S2 and S3 (complementary to Fig. 2) illustrate how mNG insertion in the sdn-1 locus has no effect on NMJ organization and how SDN-1 is equally abundant at excitatory and inhibitory synapses. Fig. S4 (complementary to Fig. 2) show the efficiency of the AID in different tissues. Fig. S5 (complementary to Fig. 6) reveals that LIN-2 is concentrated at GABA synapses in animals lacking the SDN-1 PDZ-binding motif.
Supplementary Material
Acknowledgments
We thank the Hobert laboratory for plasmids, the Caenorhabditis Genetic Center for strains, Alexis Weinreb for image analysis scripts, Laure Granger for HEK293 EBNA cell culture, and Le Centre d'Imagerie Quantitative Lyon-Est (LyMIC-CIQLE, Lyon, France) imaging facility for support and access to equipment.
The Caenorhabditis Genetic Center is funded by the National Institutes of Health Office of Research Infrastructure Programs (grant P40 OD010440). This work was supported by the European Research Council (ERC_Adg C.NAPSE 695295) within the framework of the Université de Lyon LABEX CORTEX (ANR-11-LABX-0042) within the program “Investissements d’Avenir” operated by the French National Research Agency (grant ANR-11-IDEX-0007). Work in the Bülow laboratory is supported by the National Institutes of Health (grants R21NS111145, R01NS096672, and U01CA241981).
The authors declare no competing financial interests.
Author contributions: conceptualization, J.-L. Bessereau, X. Zhou, C. Vachon, and H.E. Bülow; methodology, X. Zhou, C. Vachon, M. Cizeron, O. Romatif, and M. Jospin; investigation, X. Zhou, C. Vachon, M. Cizeron, O. Romatif, and M. Jospin; writing – original draft, J.-L. Bessereau, X. Zhou, C. Vachon, and M. Jospin; writing – review & editing, M. Cizeron and H.E. Bülow; funding acquisition, J.-L. Bessereau and H.E. Bülow.
References
- Alexander, M., Chan K.K.M., Byrne A.B., Selman G., Lee T., Ono J., Wong E., Puckrin R., Dixon S.J., and Roy P.J.. 2009. An UNC-40 pathway directs postsynaptic membrane extension in Caenorhabditis elegans. Development. 136:911–922. 10.1242/dev.030759 [DOI] [PubMed] [Google Scholar]
- Alkondon, M., Pereira E.F., Barbosa C.T., and Albuquerque E.X.. 1997. Neuronal nicotinic acetylcholine receptor activation modulates gamma-aminobutyric acid release from CA1 neurons of rat hippocampal slices. J. Pharmacol. Exp. Ther. 283:1396–1411. [PubMed] [Google Scholar]
- Allen, N.J., Bennett M.L., Foo L.C., Wang G.X., Chakraborty C., Smith S.J., and Barres B.A.. 2012. Astrocyte glypicans 4 and 6 promote formation of excitatory synapses via GluA1 AMPA receptors. Nature. 486:410–414. 10.1038/nature11059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun, Z.F., Crocker C., and Hall D.H. 2021. Handbook of C. Elegans Anatomy. In WormAtlas. [Google Scholar]
- Apte, S.S.2009. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J. Biol. Chem. 284:31493–31497. 10.1074/jbc.R109.052340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araque, A., Parpura V., Sanzgiri R.P., and Haydon P.G.. 1999. Tripartite synapses: glia, the unacknowledged partner. Trends Neurosci. 22:208–215. 10.1016/S0166-2236(98)01349-6 [DOI] [PubMed] [Google Scholar]
- Asundi, V.K., and Carey D.J.. 1995. Self-association of N-syndecan (syndecan-3) core protein is mediated by a novel structural motif in the transmembrane domain and ectodomain flanking region. J. Biol. Chem. 270:26404–26410. 10.1074/jbc.270.44.26404 [DOI] [PubMed] [Google Scholar]
- Attreed, M., Desbois M., van Kuppevelt T.H., and Bülow H.E.. 2012. Direct visualization of specifically modified extracellular glycans in living animals. Nat. Methods. 9:477–479. 10.1038/nmeth.1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu, K., Hu Z., Chien S.-C., Garriga G., and Kaplan J.M.. 2011. The immunoglobulin super family protein RIG-3 prevents synaptic potentiation and regulates Wnt signaling. Neuron. 71:103–116. 10.1016/j.neuron.2011.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballivet, M., Alliod C., Bertrand S., and Bertrand D.. 1996. Nicotinic acetylcholine receptors in the nematode Caenorhabditis elegans. J. Mol. Biol. 258:261–269. 10.1006/jmbi.1996.0248 [DOI] [PubMed] [Google Scholar]
- Bennett, K.L., Bradshaw J., Youngman T., Rodgers J., Greenfield B., Aruffo A., and Linsley P.S.. 1997. Deleted in colorectal carcinoma (DCC) binds heparin via its fifth fibronectin type III domain. J. Biol. Chem. 272:26940–26946. 10.1074/jbc.272.43.26940 [DOI] [PubMed] [Google Scholar]
- Bindels, D.S., Haarbosch L., van Weeren L., Postma M., Wiese K.E., Mastop M., Aumonier S., Gotthard G., Royant A., Hink M.A., et al. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 14:53–56. 10.1038/nmeth.4074 [DOI] [PubMed] [Google Scholar]
- Blanchette, C.R., Perrat P.N., Thackeray A., and Bénard C.Y.. 2015. Glypican Is a Modulator of Netrin-Mediated Axon Guidance. PLoS Biol. 13:e1002183. 10.1371/journal.pbio.1002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette, C.R., Thackeray A., Perrat P.N., Hekimi S., and Bénard C.Y.. 2017. Functional Requirements for Heparan Sulfate Biosynthesis in Morphogenesis and Nervous System Development in C. elegans. PLoS Genet. 13:e1006525. 10.1371/journal.pgen.1006525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte, S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Brenner, Sydney. 1974. The Genetics of Caenorhabditis Elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusés, J.L., Chauvet N., and Rutishauser U.. 2001. Membrane lipid rafts are necessary for the maintenance of the (alpha)7 nicotinic acetylcholine receptor in somatic spines of ciliary neurons. J. Neurosci. 21:504–512. 10.1523/JNEUROSCI.21-02-00504.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bülow, H.E., Berry K.L., Topper L.H., Peles E., and Hobert O.. 2002. Heparan sulfate proteoglycan-dependent induction of axon branching and axon misrouting by the Kallmann syndrome gene kal-1. Proc. Natl. Acad. Sci. USA. 99:6346–6351. 10.1073/pnas.092128099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burden, S.J., Huijbers M.G., and Remedio L.. 2018. Fundamental Molecules and Mechanisms for Forming and Maintaining Neuromuscular Synapses. Int. J. Mol. Sci. 19:490. 10.3390/ijms19020490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürli, T., Baer K., Ewers H., Sidler C., Fuhrer C., and Fritschy J.-M.. 2010. Single particle tracking of alpha7 nicotinic AChR in hippocampal neurons reveals regulated confinement at glutamatergic and GABAergic perisynaptic sites. PLoS One. 5:e11507. 10.1371/journal.pone.0011507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpantier, E., Wiesner A., Huh K.-H., Ogier R., Hoda J.-C., Allaman G., Raggenbass M., Feuerbach D., Bertrand D., and Fuhrer C.. 2005. α7 neuronal nicotinic acetylcholine receptors are negatively regulated by tyrosine phosphorylation and Src-family kinases. J. Neurosci. 25:9836–9849. 10.1523/JNEUROSCI.3497-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., Lee E., Kwon S., Park H., Yi J.Y., Kim S., Han I.-O., Yun Y., and Oh E.-S.. 2005. Transmembrane domain-induced oligomerization is crucial for the functions of syndecan-2 and syndecan-4. J. Biol. Chem. 280:42573–42579. 10.1074/jbc.M509238200 [DOI] [PubMed] [Google Scholar]
- Condomitti, G., and de Wit J.. 2018. Heparan Sulfate Proteoglycans as Emerging Players in Synaptic Specificity. Front. Mol. Neurosci. 11:14. 10.3389/fnmol.2018.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condomitti, G., Wierda K.D., Schroeder A., Rubio S.E., Vennekens K.M., Orlandi C., Martemyanov K.A., Gounko N.V., Savas J.N., and de Wit J.. 2018. An Input-Specific Orphan Receptor GPR158-HSPG Interaction Organizes Hippocampal Mossy Fiber-CA3 Synapses. Neuron. 100:201–215.e9. 10.1016/j.neuron.2018.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy, W.G., Liu Z., Nai Q., Coggan J.S., and Berg D.K.. 2003. PDZ-containing proteins provide a functional postsynaptic scaffold for nicotinic receptors in neurons. Neuron. 38:759–771. 10.1016/S0896-6273(03)00324-6 [DOI] [PubMed] [Google Scholar]
- Dani, N., and Broadie K.. 2012. Glycosylated synaptomatrix regulation of trans-synaptic signaling. Dev. Neurobiol. 72:2–21. 10.1002/dneu.20891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, G.B., Yu H., Gu S., Blackler A.N., Matta J.A., Siuda E.R., Rex E.B., and Bredt D.S.. 2019. α7 nicotinic acetylcholine receptor upregulation by anti-apoptotic Bcl-2 proteins. Nat. Commun. 10:2746. 10.1038/s41467-019-10723-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D.J., Pani A.M., Heppert J.K., Higgins C.D., and Goldstein B.. 2015. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 200:1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineley, K.T., Pandya A.A., and Yakel J.L.. 2015. Nicotinic ACh receptors as therapeutic targets in CNS disorders. Trends Pharmacol. Sci. 36:96–108. 10.1016/j.tips.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev, A., and Rusakov D.A.. 2011. Molecular signals of plasticity at the tetrapartite synapse. Curr. Opin. Neurobiol. 21:353–359. 10.1016/j.conb.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dityatev, A., Frischknecht R., and Seidenbecher C.I.. 2006. Extracellular matrix and synaptic functions. Results Probl. Cell Differ. 43:69–97. 10.1007/400_025 [DOI] [PubMed] [Google Scholar]
- Dokshin, G.A., Ghanta K.S., Piscopo K.M., and Mello C.C.. 2018. Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics. 210:781–787. 10.1534/genetics.118.301532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow, D.J., Huxley-Jones J., Hall J.M., Francks C., Maycox P.R., Kew J.N.C., Gloger I.S., Mehta N.A.L., Kelly F.M., Muglia P., et al. 2011. ADAMTSL3 as a candidate gene for schizophrenia: gene sequencing and ultra-high density association analysis by imputation. Schizophr. Res. 127:28–34. 10.1016/j.schres.2010.12.009 [DOI] [PubMed] [Google Scholar]
- Dunn, K.W., Kamocka M.M., and McDonald J.H.. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300:C723–C742. 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mouridi, S., Lecroisey C., Tardy P., Mercier M., Leclercq-Blondel A., Zariohi N., and Boulin T.. 2017. Reliable CRISPR/Cas9 Genome Engineering in Caenorhabditis elegans Using a Single Efficient sgRNA and an Easily Recognizable Phenotype. G3 (Bethesda). 7:1429–1437. 10.1534/g3.117.040824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell, I.M., and Yamaguchi Y.. 1999. Cell surface heparan sulfate proteoglycan syndecan-2 induces the maturation of dendritic spines in rat hippocampal neurons. J. Cell Biol. 144:575–586. 10.1083/jcb.144.3.575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell, I.M., Hagihara K., Miura Y., Irie F., and Yamaguchi Y.. 2000. Synbindin, A novel syndecan-2-binding protein in neuronal dendritic spines. J. Cell Biol. 151:53–68. 10.1083/jcb.151.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell, I.M., Irie F., Kalo M.S., Couchman J.R., Pasquale E.B., and Yamaguchi Y.. 2001. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 31:1001–1013. 10.1016/S0896-6273(01)00440-8 [DOI] [PubMed] [Google Scholar]
- Fabian-Fine, R., Skehel P., Errington M.L., Davies H.A., Sher E., Stewart M.G., and Fine A.. 2001. Ultrastructural distribution of the alpha7 nicotinic acetylcholine receptor subunit in rat hippocampus. J. Neurosci. 21:7993–8003. 10.1523/JNEUROSCI.21-20-07993.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farías, G.G., Vallés A.S., Colombres M., Godoy J.A., Toledo E.M., Lukas R.J., Barrantes F.J., and Inestrosa N.C.. 2007. Wnt-7a induces presynaptic colocalization of alpha 7-nicotinic acetylcholine receptors and adenomatous polyposis coli in hippocampal neurons. J. Neurosci. 27:5313–5325. 10.1523/JNEUROSCI.3934-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Ferrer, M., and Dityatev A.. 2018. Shaping Synapses by the Neural Extracellular Matrix. Front. Neuroanat. 12:40. 10.3389/fnana.2018.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finci, L.I., Krüger N., Sun X., Zhang J., Chegkazi M., Wu Y., Schenk G., Mertens H.D.T., Svergun D.I., Zhang Y., et al. 2014. The crystal structure of netrin-1 in complex with DCC reveals the bifunctionality of netrin-1 as a guidance cue. Neuron. 83:839–849. 10.1016/j.neuron.2014.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, M.M., Evans S.P., Jensen M., Madsen D.M., Mancuso J., Norman K.R., and Maricq A.V.. 2005. The Ror receptor tyrosine kinase CAM-1 is required for ACR-16-mediated synaptic transmission at the C. elegans neuromuscular junction. Neuron. 46:581–594. 10.1016/j.neuron.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Frøkjær-Jensen, C., Davis M.W., Sarov M., Taylor J., Flibotte S., LaBella M., Pozniakovsky A., Moerman D.G., and Jorgensen E.M.. 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods. 11:529–534. . 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gally, C., Eimer S., Richmond J.E., and Bessereau J.-L.. 2004. A transmembrane protein required for acetylcholine receptor clustering in Caenorhabditis elegans. Nature. 431:578–582. 10.1038/nature02893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel, M., Rapti G., Richmond J.E., and Bessereau J.-L.. 2009. A secreted complement-control-related protein ensures acetylcholine receptor clustering. Nature. 461:992–996. 10.1038/nature08430 [DOI] [PubMed] [Google Scholar]
- Granés, F., Berndt C., Roy C., Mangeat P., Reina M., and Vilaró S.. 2003. Identification of a novel Ezrin-binding site in syndecan-2 cytoplasmic domain. FEBS Lett. 547:212–216. 10.1016/S0014-5793(03)00712-9 [DOI] [PubMed] [Google Scholar]
- Gray, R., Rajan A.S., Radcliffe K.A., Yakehiro M., and Dani J.A.. 1996. Hippocampal synaptic transmission enhanced by low concentrations of nicotine. Nature. 383:713–716. 10.1038/383713a0 [DOI] [PubMed] [Google Scholar]
- Halassa, M.M., Fellin T., and Haydon P.G.. 2007. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol. Med. 13:54–63. 10.1016/j.molmed.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Hancock, M.L., Canetta S.E., Role L.W., and Talmage D.A.. 2008. Presynaptic type III neuregulin1-ErbB signaling targets α7 nicotinic acetylcholine receptors to axons. J. Cell Biol. 181:511–521. 10.1083/jcb.200710037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen, A., Pihlajaniemi T., Faissner A., and Yuzaki M.. 2014. Neural ECM and synaptogenesis. Prog. Brain Res. 214:29–51. 10.1016/B978-0-444-63486-3.00002-5 [DOI] [PubMed] [Google Scholar]
- Hienola, A., Tumova S., Kulesskiy E., and Rauvala H.. 2006. N-syndecan deficiency impairs neural migration in brain. J. Cell Biol. 174:569–580. 10.1083/jcb.200602043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz, A., and Simons M.. 1998. Phosphorylation of the cytoplasmic tail of syndecan-4 regulates activation of protein kinase Calpha. J. Biol. Chem. 273:25548–25551. 10.1074/jbc.273.40.25548 [DOI] [PubMed] [Google Scholar]
- Hsueh, Y.-P., and Sheng M.. 1999. Regulated expression and subcellular localization of syndecan heparan sulfate proteoglycans and the syndecan-binding protein CASK/LIN-2 during rat brain development. J. Neurosci. 19:7415–7425. 10.1523/JNEUROSCI.19-17-07415.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh, Y.-P., Yang F.-C., Kharazia V., Naisbitt S., Cohen A.R., Weinberg R.J., and Sheng M.. 1998. Direct interaction of CASK/LIN-2 and syndecan heparan sulfate proteoglycan and their overlapping distribution in neuronal synapses. J. Cell Biol. 142:139–151. 10.1083/jcb.142.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H.-T., Umemori H., and Hsueh Y.-P.. 2016. Postsynaptic SDC2 induces transsynaptic signaling via FGF22 for bidirectional synaptic formation. Sci. Rep. 6:33592. 10.1038/srep33592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, B., Jung H., Hong H., and Oh E.-S.. 2018. Syndecan transmembrane domain modulates intracellular signaling by regulating the oligomeric status of the cytoplasmic domain. Cell. Signal. 52:121–126. 10.1016/j.cellsig.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Jensen, M., Hoerndli F.J., Brockie P.J., Wang R., Johnson E., Maxfield D., Francis M.M., Madsen D.M., and Maricq A.V.. 2012. Wnt signaling regulates acetylcholine receptor translocation and synaptic plasticity in the adult nervous system. Cell. 149:173–187. 10.1016/j.cell.2011.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, K.G., Tenney A.P., Ghose A., Duckworth A.M., Higashi M.E., Parfitt K., Marcu O., Heslip T.R., Marsh J.L., Schwarz T.L., et al. 2006. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron. 49:517–531. 10.1016/j.neuron.2006.01.026 [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Pavlov I., Võikar V., Lauri S.E., Hienola A., Riekki R., Lakso M., Taira T., and Rauvala H.. 2002. Syndecan-3-deficient mice exhibit enhanced LTP and impaired hippocampus-dependent memory. Mol. Cell. Neurosci. 21:158–172. 10.1006/mcne.2002.1167 [DOI] [PubMed] [Google Scholar]
- Kawai, H., Zago W., and Berg D.K.. 2002. Nicotinic α 7 receptor clusters on hippocampal GABAergic neurons: regulation by synaptic activity and neurotrophins. J. Neurosci. 22:7903–7912. 10.1523/JNEUROSCI.22-18-07903.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshan, P.T., Phan A.Q., Wang G.J., Crane M.M., Lu H., and Shen K.. 2014. Regulation of synaptic extracellular matrix composition is critical for proper synapse morphology. J. Neurosci. 34:12678–12689. 10.1523/JNEUROSCI.1183-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Xiong W.-C., and Mei L.. 2018. Neuromuscular Junction Formation, Aging, and Disorders. Annu. Rev. Physiol. 80:159–188. 10.1146/annurev-physiol-022516-034255 [DOI] [PubMed] [Google Scholar]
- Liewald, J.F., Brauner M., Stephens G.J., Bouhours M., Schultheis C., Zhen M., and Gottschalk A.. 2008. Optogenetic analysis of synaptic function. Nat. Methods. 5:895–902. 10.1038/nmeth.1252 [DOI] [PubMed] [Google Scholar]
- Lin, Y.-L., Lei Y.-T., Hong C.-J., and Hsueh Y.-P.. 2007. Syndecan-2 induces filopodia and dendritic spine formation via the neurofibromin-PKA-Ena/VASP pathway. J. Cell Biol. 177:829–841. 10.1083/jcb.200608121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Ford B., Mann M.A., and Fischbach G.D.. 2001. Neuregulins increase alpha7 nicotinic acetylcholine receptors and enhance excitatory synaptic transmission in GABAergic interneurons of the hippocampus. J. Neurosci. 21:5660–5669. 10.1523/JNEUROSCI.21-15-05660.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maro, G.S., Gao S., Olechwier A.M., Hung W.L., Liu M., Özkan E., Zhen M., and Shen K.. 2015. MADD-4/Punctin and Neurexin Organize C. elegans GABAergic Postsynapses through Neuroligin. Neuron. 86:1420–1432. 10.1016/j.neuron.2015.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, Y., Irie F., Inatani M., Tessier-Lavigne M., and Yamaguchi Y.. 2007. Netrin-1/DCC signaling in commissural axon guidance requires cell-autonomous expression of heparan sulfate. J. Neurosci. 27:4342–4350. 10.1523/JNEUROSCI.0700-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minniti, A.N., Labarca M., Hurtado C., and Brandan E.. 2004. Caenorhabditis elegans syndecan (SDN-1) is required for normal egg laying and associates with the nervous system and the vulva. J. Cell Sci. 117:5179–5190. 10.1242/jcs.01394 [DOI] [PubMed] [Google Scholar]
- Murakami, M., Horowitz A., Tang S., Ware J.A., and Simons M.. 2002. Protein kinase C (PKC) δ regulates PKCalpha activity in a Syndecan-4-dependent manner. J. Biol. Chem. 277:20367–20371. 10.1074/jbc.M202501200 [DOI] [PubMed] [Google Scholar]
- Nguyen, M.U., Kwong J., Chang J., Gillet V.G., Lee R.M., and Johnson K.G.. 2016. The Extracellular and Cytoplasmic Domains of Syndecan Cooperate Postsynaptically to Promote Synapse Growth at the Drosophila Neuromuscular Junction. PLoS One. 11:e0151621. 10.1371/journal.pone.0151621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E.-S., Couchman J.R., and Woods A.. 1997. Serine phosphorylation of syndecan-2 proteoglycan cytoplasmic domain. Arch. Biochem. Biophys. 344:67–74. 10.1006/abbi.1997.0180 [DOI] [PubMed] [Google Scholar]
- Parker, M.J., Zhao S., Bredt D.S., Sanes J.R., and Feng G.. 2004. PSD93 regulates synaptic stability at neuronal cholinergic synapses. J. Neurosci. 24:378–388. 10.1523/JNEUROSCI.3865-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataki, C.A., Couchman J.R., and Brábek J.. 2015. Wnt Signaling Cascades and the Roles of Syndecan Proteoglycans. J. Histochem. Cytochem. 63:465–480. 10.1369/0022155415586961 [DOI] [PubMed] [Google Scholar]
- Pinan-Lucarré, B., Tu H., Pierron M., Cruceyra P.I., Zhan H., Stigloher C., Richmond J.E., and Bessereau J.-L.. 2014. C. elegans Punctin specifies cholinergic versus GABAergic identity of postsynaptic domains. Nature. 511:466–470. 10.1038/nature13313 [DOI] [PubMed] [Google Scholar]
- Platsaki, S., Zhou X., Pinan-Lucarré B., Delauzun V., Tu H., Mansuelle P., Fourquet P., Bourne Y., Bessereau J.-L., and Marchot P.. 2020. The Ig-like domain of Punctin/MADD-4 is the primary determinant for interaction with the ectodomain of neuroligin NLG-1. J. Biol. Chem. 295:16267–16279. 10.1074/jbc.RA120.014591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti, G., Richmond J., and Bessereau J.-L.. 2011. A single immunoglobulin-domain protein required for clustering acetylcholine receptors in C. elegans. EMBO J. 30:706–718. 10.1038/emboj.2010.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhiner, C., Gysi S., Fröhli E., Hengartner M.O., and Hajnal A.. 2005. Syndecan regulates cell migration and axon guidance in C. elegans. Development. 132:4621–4633. 10.1242/dev.02042 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Manzaneque, J.C., Carpizo D., Plaza-Calonge M. del C., Torres-Collado A.X., Thai S.N., Simons M., Horowitz A., and Iruela-Arispe M.L.. 2009. Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. Int. J. Biochem. Cell Biol. 41:800–810. 10.1016/j.biocel.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saied-Santiago, K., and Bülow H.E.. 2018. Diverse roles for glycosaminoglycans in neural patterning. Dev. Dyn. 247:54–74. 10.1002/dvdy.24555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin, S., Lamanna W.C., and Esko J.D.. 2011. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3:a004952. 10.1101/cshperspect.a004952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y.S., and Kim E.. 2013. Presynaptic proteoglycans: sweet organizers of synapse development. Neuron. 79:609–611. 10.1016/j.neuron.2013.07.048 [DOI] [PubMed] [Google Scholar]
- Swenarchuk, L.E.2019. Nerve, Muscle, and Synaptogenesis. Cells. 8:1448. 10.3390/cells8111448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikiyani, V., Li L., Sharma P., Liu H., Hu Z., and Babu K.. 2018. Wnt Secretion Is Regulated by the Tetraspan Protein HIC-1 through Its Interaction with Neurabin/NAB-1. Cell Rep. 25:1856–1871.e6. 10.1016/j.celrep.2018.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touroutine, D., Fox R.M., Von Stetina S.E., Burdina A., Miller D.M. III, and Richmond J.E.. 2005. acr-16 encodes an essential subunit of the levamisole-resistant nicotinic receptor at the Caenorhabditis elegans neuromuscular junction. J. Biol. Chem. 280:27013–27021. 10.1074/jbc.M502818200 [DOI] [PubMed] [Google Scholar]
- Tu, H., Pinan-Lucarré B., Ji T., Jospin M., and Bessereau J.-L.. 2015. C. elegans Punctin Clusters GABA(A) Receptors via Neuroligin Binding and UNC-40/DCC Recruitment. Neuron. 86:1407–1419. 10.1016/j.neuron.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Wonnacott, S.1997. Presynaptic nicotinic ACh receptors. Trends Neurosci. 20:92–98. 10.1016/S0166-2236(96)10073-4 [DOI] [PubMed] [Google Scholar]
- Xu, D., and Esko J.D.. 2014. Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83:129–157. 10.1146/annurev-biochem-060713-035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, Z., Manning L., Nelson J., Richmond J.E., Colón-Ramos D.A., Shen K., and Kurshan P.T.. 2017. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. eLife. 6:e29276. 10.7554/eLife.29276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki, M.2018. Two Classes of Secreted Synaptic Organizers in the Central Nervous System. Annu. Rev. Physiol. 80:243–262. 10.1146/annurev-physiol-021317-121322 [DOI] [PubMed] [Google Scholar]
- Zarei, M.M., Radcliffe K.A., Chen D., Patrick J.W., and Dani J.A.. 1999. Distributions of nicotinic acetylcholine receptor α7 and β2 subunits on cultured hippocampal neurons. Neuroscience. 88:755–764. 10.1016/S0306-4522(98)00246-2 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Ward J.D., Cheng Z., and Dernburg A.F.. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 142:4374–4384. 10.1242/dev.129635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., and Bessereau J.-L.. 2019. Molecular Architecture of Genetically-Tractable GABA Synapses in C. elegans. Front. Mol. Neurosci. 12:304. 10.3389/fnmol.2019.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X., Zeng J., Ouyang C., Luo Q., Yu M., Yang Z., Wang H., Shen K., and Shi A.. 2016. A novel bipartite UNC-101/AP-1 μ1 binding signal mediates KVS-4/Kv2.1 somatodendritic distribution in Caenorhabditis elegans. FEBS Lett. 590:76–92. 10.1002/1873-3468.12043 [DOI] [PubMed] [Google Scholar]
- Zhou, X., Gueydan M., Jospin M., Ji T., Valfort A., Pinan-Lucarré B., and Bessereau J.-L.. 2020. The netrin receptor UNC-40/DCC assembles a postsynaptic scaffold and sets the synaptic content of GABAA receptors. Nat. Commun. 11:2674. 10.1038/s41467-020-16473-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.