Abstract
Background
COVID-19, as a novel coronavirus disease caused by new coronavirus SARS-CoV-2, spreads all over the world, and brings harm to human in many countries. Humans suffered a lot from both SARS-CoV-2 now and by SARS-CoV in the year 2003. It is important to understand the differences and the relationships between these two types of viruses.
Objective
To compare relative synonymous codon usage of ORF1ab gene in SARS-CoV-2 and SARS-CoV, relative synonymous codon usage of their genomes are studied in this paper from the bioinformatics perspective.
Methods
The ORF1ab gene, which is an important non-structural polyprotein coding gene and now used for nucleic acid detection markers in many measurement method, in both SARS-CoV-2 (30 strains) and SARS-CoV (20 strains) are considered to be the research object in the present paper. The relative synonymous codon usage values of the ORF1ab gene are calculated to characterize the differences and the evolutionary characteristics among 50 strains.
Results
There is a significant difference between SARS-CoV and SARS-CoV-2 when the relative synonymous codon usage value of ORF1ab genes is concerned. The results suggest that codon usage pattern of SARS-CoV is more similar to human than that of the SARS-CoV-2, and that the inner difference in SARS-CoV-2 strains is larger than that of SARS-CoV, which denote the larger diversity exits in the SARS-CoV-2 virus.
Conclusion
These results show that the relative synonymous codon usage values in the coronavirus could be used for further research on their evolutionary phenomenon.
Keywords: COVID-19, SARS-CoV-2, SARS-CoV, Coronavirus, Relative synonymous codon usage, Codon usage pattern, Gene evolution
Introduction
Coronavirus often cause cold and upper respiratory tract infections in human body. A new kind of coronavirus named as COVID-19 by the World Health of Organization (WHO) on February, 11, 2020 (World Health Organization 2019, 2020), has rapidly expanded over a short time. At present, the COVID-19 cases have been reported in many parts of the world (Harapan et al. 2020; Hussin and Byrareddy, 2020). According to the latest data, as of May 28th, 2021, more than 156 million people were infected by SARS-CoV-2. Among them, more than 3.2 million cases were dead. The global incidence of COVID-19 has grown dramatically recently, meanwhile, more and more population is now at risk for there is no specific treatment for COVID-19 (Serafin et al. 2020).
A good knowledge of COVID-19 infection prevention is of utmost importance (Na et al. 2020). Patients infected by SARS-CoV-2 show clinical manifestations of pneumonia (Xiaoping et al. 2020; Asadi-Pooya and Simani 2019; Chen et al. 2020), which are similar to the symptoms of SARS-CoV (Huang et al. 2020; Peiris et al. 2004). Learning from previous experiences with SARS is much better, and many current understanding of COVID-19 is compared with the outbreak of SARS in 2003 by many scientists (Gorbalenya et al. 2020; Wei-Jie et al. 2020; Tort et al. 2020), especially the clinical methods for dealing with the COVID-19 (Mahmoud et al. 2020) and the genetic diversity and evolution (Li et al. 2020a). All previous researches have neglected the extent of how relative synonymous codon usage (RSCU) values of SARS-CoV-2 parallel to hosts. How to evaluate the distance between the RSCU value of virus and the host becomes a key issue. SARS-CoV-2 is an enveloped virus with an about 29 kb positive sense single-stranded RNA genome (Shuo et al. 2016). The genome of SARS-CoV-2, like other coronavirus, has a long ORF1ab polyprotein coding gene. The polyprotein ORF1ab in SARS-CoV-2 is split into 16 putative proteins based on its alignment with the human SARS-CoV polyprotein (Srinivasan et al. 2020). Since the translated ORF1ab polyprotein regions are very long, the sequence similarity is sufficient to classify them as homologs (Muhamad et al. 2020). Many factors may affect the RSCU in viruses, such as mutation pressure, gene length, natural selection, etc. That is the hosts can affect the RSCU of viruses via affecting their suitability and immune escape, so the RSCU of ORF1ab may be useful for understanding of their molecular evolution (He et al. 2020). Almost all previous researches have not mentioned the RSCU of ORF1ab in coronavirus. In this study, to investigate the hypothesis that different coronavirus leads to the RSCU varies in coronavirus, we calculated the RSCU values for ORF1ab in 30 SARS-CoV-2 virus strains and 20 SARS-CoV strains. Comprehensive analysis on the differences between the RSCU values of them and their evolution characteristics are all performed.
Material and methods
In order to study the coronavirus, completed genomes of SARS-CoV and SARS-CoV-2 were obtained from the NCBI (http://www.ncbi.nlm.nih.gov) database. Genomes in the NCBI database, including 23 SARS-CoV and 31 SARS-CoV-2 respectively, were downloaded on September, 10th, 2020. The sequences with correct start and stop codons, and having multiple of three bases were considered as effective sequences. The detailed information of effective gene sequences including accession numbers in the NCBI database and their classification are as shown in the Table 1. The ORF1ab gene sequences were then extracted from these initial sequences for calculating their relative synonymous codon usages.
Table 1.
Basic sequence information of SARS-CoV and SARS-CoV-2 genes
| Number | Accession no. | Sequence name | Classification |
|---|---|---|---|
| 1 | AY291451.1 | SARS coronavirus TW1 | SARS-CoV |
| 2 | AY502925.1 | SARS coronavirus TW2 | |
| 3 | AY502926.1 | SARS coronavirus TW3 | |
| 4 | AY502927.1 | SARS coronavirus TW4 | |
| 5 | AY502928.1 | SARS coronavirus TW5 | |
| 6 | AY502929.1 | SARS coronavirus TW6 | |
| 7 | AY502930.1 | SARS coronavirus TW7 | |
| 8 | AY502931.1 | SARS coronavirus TW8 | |
| 9 | AY502932.1 | SARS coronavirus TW9 | |
| 10 | AY502923.1 | SARS coronavirus TW10 | |
| 11 | AY502924.1 | SARS coronavirus TW11 | |
| 12 | AY338174.1 | SARS coronavirus TC1 | |
| 13 | AY338175.1 | SARS coronavirus TC2 | |
| 14 | AY348314.1 | SARS coronavirus TC3 | |
| 15 | AP006557.1 | SARS coronavirus TWH | |
| 16 | AP006559.1 | SARS coronavirus TWK | |
| 17 | AY502927.1 | SARS coronavirus TWC2 | |
| 18 | AP006561.1 | SARS coronavirus TWY | |
| 19 | AP006560.1 | SARS coronavirus TWS | |
| 20 | AP006558.1 | SARS coronavirus TWJ | |
| 21 | MT370516.1 | SARS-CoV-2/human/TWN/CGMH-CGU-03/2020 | SARS-CoV-2 |
| 22 | MT374115.1 | SARS-CoV-2/human/TWN/CGMH-CGU-19/2020 | |
| 23 | MT479225.1 | SARS-CoV-2/human/TWN/CGMH-CGU-23/2020 | |
| 24 | MT374111.1 | SARS-CoV-2/human/TWN/CGMH-CGU-15/2020 | |
| 25 | MT374112.1 | SARS-CoV-2/human/TWN/CGMH-CGU-16/2020 | |
| 26 | MT479226.1 | SARS-CoV-2/human/TWN/CGMH-CGU-24/2020 | |
| 27 | MT479227.1 | SARS-CoV-2/human/TWN/CGMH-CGU-25/2020 | |
| 28 | MT517437.1 | SARS-CoV-2/human/TWN/CGMH-CGU-27/2020 | |
| 29 | MT517436.1 | SARS-CoV-2/human/TWN/CGMH-CGU-26/2020 | |
| 30 | MT374101.1 | SARS-CoV-2/human/TWN/CGMH-CGU-02/2020 | |
| 31 | MT374102.1 | SARS-CoV-2/human/TWN/CGMH-CGU-06/2020 | |
| 32 | MT374103.1 | SARS-CoV-2/human/TWN/CGMH-CGU-07/2020 | |
| 33 | MT374104.1 | SARS-CoV-2/human/TWN/CGMH-CGU-08/2020 | |
| 34 | MT374105.1 | SARS-CoV-2/human/TWN/CGMH-CGU-09/2020 | |
| 35 | MT374106.1 | SARS-CoV-2/human/TWN/CGMH-CGU-10/2020 | |
| 36 | MT374107.1 | SARS-CoV-2/human/TWN/CGMH-CGU-11/2020 | |
| 37 | MT374108.1 | SARS-CoV-2/human/TWN/CGMH-CGU-12/2020 | |
| 38 | MT374109.1 | SARS-CoV-2/human/TWN/CGMH-CGU-13/2020 | |
| 39 | MT374110.1 | SARS-CoV-2/human/TWN/CGMH-CGU-14/2020 | |
| 40 | MT374113.1 | SARS-CoV-2/human/TWN/CGMH-CGU-17/2020 | |
| 41 | MT374114.1 | SARS-CoV-2/human/TWN/CGMH-CGU-18/2020 | |
| 42 | MT374116.1 | SARS-CoV-2/human/TWN/CGMH-CGU-20/2020 | |
| 43 | MT479223.1 | SARS-CoV-2/human/TWN/CGMH-CGU-21/2020 | |
| 44 | MT479224.1 | SARS-CoV-2/human/TWN/CGMH-CGU-22/2020 | |
| 45 | MT590598.1 | SARS-CoV-2/human/TWN/CGMH-CGU-28/2020 | |
| 46 | MT590600.1 | SARS-CoV-2/human/TWN/CGMH-CGU-30/2020 | |
| 47 | MT590599.1 | SARS-CoV-2/human/TWN/CGMH-CGU-29/2020 | |
| 48 | MT066176.1 | SARS-CoV-2/NTU02/TWN/human/2020 | |
| 49 | MT066175.1 | SARS-CoV-2/NTU01/TWN/human/2020 | |
| 50 | MT192759.1 | SARS-CoV-2/human/TWN/CGMH-CGU-01/2020 |
The RSCU values of ORF1ab genes for all coronavirus were calculated to determine their codon usage pattern. The quantitative difference between SARS-CoV and SARS-CoV-2 of all codons were calculated. The RSCU values were calculated as follows:
| 1 |
Here gij is the observed number of the ith codon for the jth amino acid, which has ni kinds of different synonymous codons. The codons, whose RSCU values greater than 1.0, are usually regarded as abundant codons, whereas those with the RSCU values less than 1.0 are defined as less-abundant codons. Based on the RSCU values, it is easily to calculate the distance between coronavirus and their hosts theoretically. The formula of D(A, B) was established to evaluate the potential role of the overall codon usage pattern of the host in the formation of the overall codon usage pattern of viruses.
| 2 |
| 3 |
where R(A,B) is used to evaluate the similarity distance between coronavirus and human from the aspect of the RSCU. Here aiis defined as the RSCU value for a specific codon in 59 synonymous codons of coronavirus, bi is the RSCU value for the same codon of the human host (Siddiq et al. 2017).
D(A,B) represents the potential effect of the overall codon usage of the host on that of coronavirus, and this value ranges from zero to 1.0. Further, based on the RSCU values, the evolutionary distance of all 50 coronavirus was calculated without considering the confounding influence of stop codons. The Euclidean distances among all observations of RSCU values were used to analysis the divergence among the coronavirus.
Results and discussion
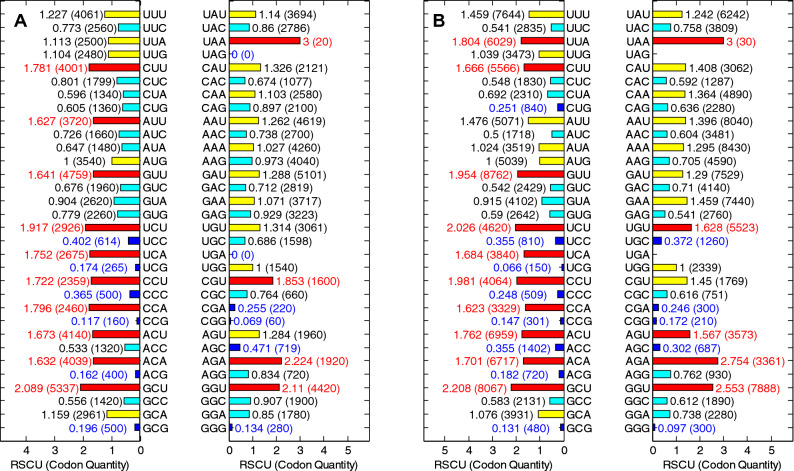
As an important parameter, the RSCU value, which represents the ratio occurrence frequency of one codon and the expected usage frequency, is usually used for evaluating the bias of the synonymous codon (Qi et al. 2020; Prajna et al. 2018). If the RSCU value of a codon is more than 1.0, it would be regarded as a positive codon usage pattern. On the contrary, the codon with less RSCU value (less than 0.5) could be regarded as a less-abundant codon. The overall RSCU values of ORF1ab gene in 20 SARS-CoV and 30 SARS-CoV-2 genomes were calculated, and the result is shown in Fig. 1 From the result, G and C-ended codons are obviously less than A and U-ended codons for two kinds of coronavirus.
Fig. 1.
A Overall RSCU for ORF1ab of 20 SARS-CoV genomes, and B Overall RSCU for ORF1ab of 30 SARS-CoV-2 genomes. Red bars denote the codons, such as the GUG, UCA, CCA, ACA, UAA, AGA, AGG and GGA, with high RSCU values (more than 1.5) and could be regarded as the abundant codons, and the blue bars in the figure denote the less-abundant codons. All ORF1ab genes of 50 coronavirus select UAA as the stop codon, so the RSCU value of UAA is 3 (color figure online)
The RSCU values shown in the Fig. 1 can reflect the overall characteristics of codon usage in 50 coronavirus ORF1ab genes. For each particular codons, the RSCU values of 50 coronavirus ORF1ab genes are separately shown in the Fig. 1A and the Fig. 1B. It can be seen that there are significant differences among codon usage pattern of all codons except particular ones (AUG and UGG). Among 3 terminal codons, all sequences, within both SARS-CoV and SARS-CoV-2, select UAA as terminal codons, so the RSCU values for UAG and UGA all equal to zero. The RSCU value of some codons such as UUU, UUA, AUA, GUU, CCU, CAA, AAA, GAA, UGU, AGA and GGU of SARS-CoV-2 are all greater than that of SARS-CoV. Another recent research showed that the RSCU pattern of SARS-CoV-2 resembles to human to some extent, while bat-SL-CoVZC45 has similar synonymous codon usage pattern to its host-the bat. The distance between the SARS-CoV-2 and other animals is greater than it to human (Ji et al. 2020).
Interestingly, From the Fig. 1, compared to SARS-CoV, we could see that the frequencies of the codons with the U-ending and A-ending in SARS-CoV-2 tend to be higher. And the average usage frequency for ended codons is shown in the Fig. 2. It can clearly show the composition of ORF1ab gene sequences (for both SARS-CoV-2 and SARS-CoV). The G and the C ended codons in both SARS-CoV-2 and SARS-CoV are less frequently used compared to A and T ended codon. More interestingly, the calculated results show that the G and the C ended codons used in the SARS-CoV-2 are less than that in SARS-CoV, the p values are less than 0.001. The results show the codon usage has a stronger bias in SARS-CoV-2.
Fig. 2.
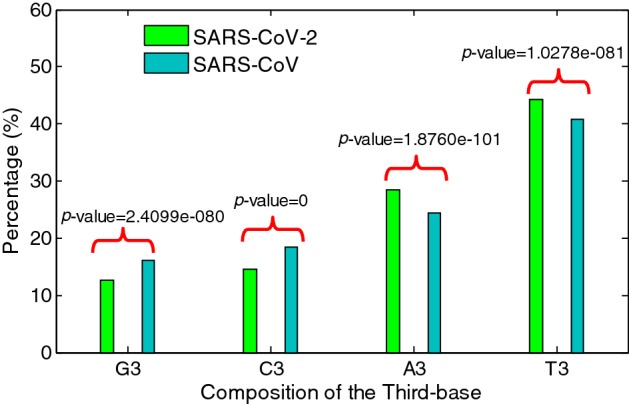
Third-base usage frequency for ORF1ab of SARS-CoV-2 and SARS-CoV
When other genes in conorovirus, such as the M and the S coding sequences are concerned, the G and the C ended codons also have higher usage compared to the A and the C ended codons (see Table 2). For the same gene, the G and the C were preferred to be used as the third codon base in SARS-CoV-2.
Table 2.
Mean value of the composition for ended bases in conorovirus
| Gene name | Conorovirus | G3 | C3 | A3 | T3 |
|---|---|---|---|---|---|
| ORF1ab | SARS-CoV-2 | 0.127 | 0.145 | 0.284 | 0.443 |
| SARS-CoV | 0.162 | 0.184 | 0.244 | 0.409 | |
| M | SARS-CoV-2 | 0.161 | 0.230 | 0.242 | 0.368 |
| SARS-CoV | 0.212 | 0.248 | 0.23 | 0.311 | |
| S | SARS-CoV-2 | 0.108 | 0.159 | 0.270 | 0.463 |
| SARS-CoV | 0.119 | 0.182 | 0.233 | 0.466 |
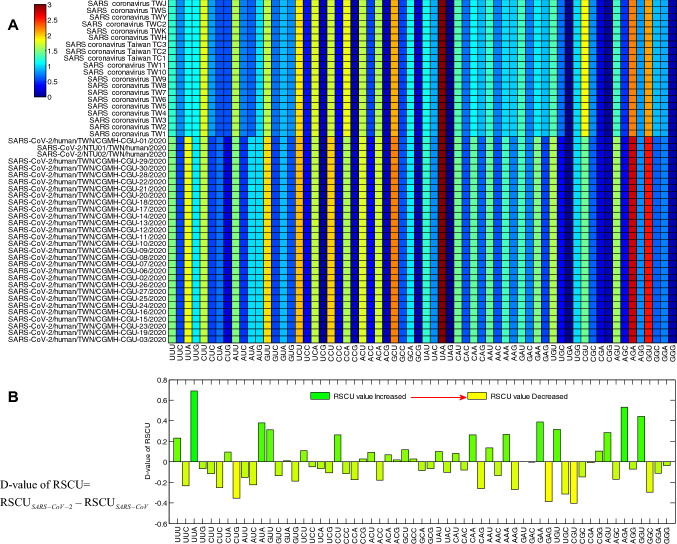
Codon links to nucleic acids and proteins, so, the RSCU values sometimes can be used to describe the evolutionary of genomes via calculating their distance (Xiaoyue et al. 2019). The heat map of the RSCU value for 59 codons of coronavirus is shown in Fig. 3A separately. The differences of the RSCU values between the coronavirus were calculated and the result is shown in the Fig. 3B. These results showed that in coronavirus genomes, the codon usage preference showed that the SARS-CoV-2 tend to use more UUA, AUA, GUU, CAA, AAA, GAA, UGU, AGU, AGA and GUU. This is consistent with the conclusion expressed in the Fig. 2. In the Fig. 3B, compared to the SARS-CoV, the results show thatthe T and the A ended codons are more than the GC ended ones in SARS-CoV-2.
Fig. 3.
A Heat map of RSCU values of 50 ORF1ab coronavirus genes separately, and B The difference the mean RSCU values between SARS-CoV and SARS-CoV-2
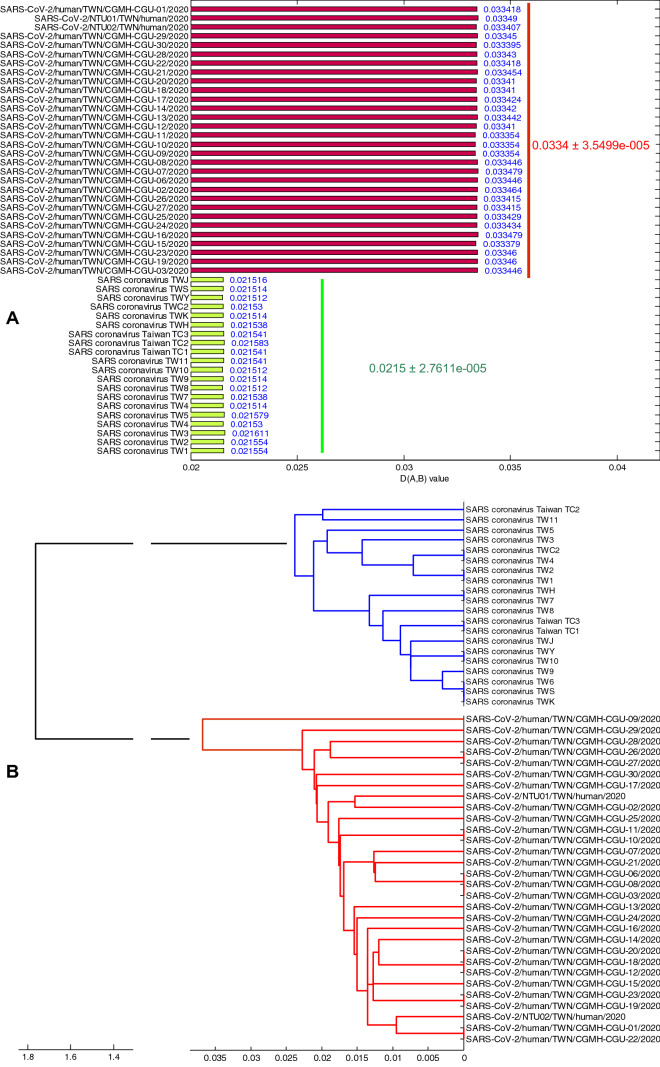
In order to facilitate comparison with the evolutionary characteristics of SARS-CoV-2 and to fully understand the virus evolutionary divergence, we selected SARS-CoV as a contrast. Evolutionary distance—the values of D(A,B), which denote the RSCU distance between the viruses and the hosts, of SARS-CoV-2 (= 0.0334) and SARS-CoV (= 0.0215) are shown in the Fig. 4A. Here, the mean value is used to evaluate the evolutionary distance, while their standard deviation is used to evaluate the degree of evolutionary divergence. The results in the Fig. 4A show that the standard deviations of SARS-CoV (= 2.7611e−5) is less than that of SARS-CoV-2 (= 3.5499e−5), revealing the broader evolutionary divergence existing in the SARS-CoV-2 genomes. Small evolutionary distance may cause the little rate of variation, consequently, little rate of variation may cause the little degree of evolutionary divergence. We speculate that if SARS-CoV-2 continues to exist in human hosts, its evolutionary divergence degree in the future would be larger than it is now. The RSCU analysis is very important for exploring the evolution of virus from the molecular level. Differences of the RSCU values among genomes can be used to describe the evolutionary characteristics (Paraskevis et al. 2020). Evolutionary characteristics of 50 coronavirus genomes, their phylogenetic tree, have subtle differences, and the result is shown in the Fig. 4B. The maximum intraspecies difference is about 0.03, while the interspecific difference is about 1.8, which denote that there is no genetic relationship between SARS-CoV and SARS-CoV-2.
Fig. 4.
A Distance between overall codon usage pattern of coronavirus and human, and B phylogenetic tree of coronavirus genomes
MERS-CoV (Jiang et al. 2020; Rokni et al. 2020), another important coronavirus, emerged in 2012, is compared to the SARS-CoV-2 by many researchers recently (Singh et al. 2020). But the present study does not consider the MERS-CoV for it is a virus which had been already exited in humans for about 8 years even until now, as well as it has not spread all over the world. Unlike the MERS-CoV, the SARS-CoV and the SARS-CoV-2 are all controllable viruses for some certain countries. It showed that the coronavirus strains of the same classification are more similar to each other, indicating that the virus show similar codon usage pattern. On the other hand, the small D(A,B) value also reflect the greater adaption of the SARS strains to their hosts, or a longer time exist in their hosts. In terms of evolutionary distance, the differences between strains of SARS-CoV are smaller, while the differences between SARS-CoV-2 strains are larger (Fig. 4B).
Novel SARS-CoV-2 lies behind the seriously ongoing outbreak of COVID-19 (Li et al. 2020b). The genome of SARS-CoV-2 has a long ORF1ab gene which coding the polyprotein. Although there are growing researches on SARS-CoV-2 from the perspective of virology and clinical strategies (Lai et al. 2020; Koyama et al. 2020), recent researches revealed its attractive mechanisms, even the content, the adaption to human hosts and evolutionary pressures of the SARS-CoV-2 are studied (Dittadi et al. 2020; Dilucca et al. 2020), no bioinformatics method is used for exploring the ORF1ab gene in the coronavirus.
Genetic analysis on SARS-CoV-2 was studied by other researches recently by using eighty-six complete or near-complete genomes of SARS-CoV-2 (Phan 2020), the results conclude the evidence of the genetic diversity and rapid evolution of SARS-CoV-2. In the present study, the certain ORF1ab gene, which is usually used for nucleic acid testing of the SARS-CoV-2 (Mathuria et al. 2020), is used for exploring the diversity, evolution of SARS-CoV-2 from the RSCU values.
Although many methods have been used for exploring the gene evolution of SARS-CoV-2 (Shi et al. 2020; Yin 2020; Bartolini et al. 2020), the samples are so complex, for instance, big numbers and many kinds of sequences (Yoshimoto 2020; Devaux et al. 2020; Pfefferle et al. 2020; Yadav et al. 2020) from too many areas that they may be hardly to get a comprehensive result. In the present study, the scope of research objects was defined and all suited samples of SARS-CoV-2 and SARS-CoV were downloaded and considered.
Conclusions
COVID-19, now, spreads all over the world, and exists in most countries. Exploring its codon usage pattern is useful for understanding genetic characteristics and geographical differences. Coronaviruses caught our attention when they caused more and more human diseases recently. The RSCU value has a very broad significance for exploring the evolution characteristics of coronavirus. It is critical to determine the differences between them understand the molecular mechanism of transmission. Information obtained from the RSCU analysis in ORF1ab coronaviruses will provide some insights to this question and will be helpful for investigation of its recombination. In this paper, ORF1ab genes from samples of SARS-CoV-2 and SARS-CoV are collected for research. The results show that there is significant difference between SARS-CoV and SARS-CoV-2 when the RSCU values of ORF1ab are concerned. Most coronavirus tend to use A and U as their third base. Interestingly, in SARS-CoV-2, this phenomenon becomes more pronounced. Most important, the differences between strains of SARS-CoV-2 are larger than that in SARS-CoV, probably for the longer time existing period in human being of the SARS-CoV-2. The unique RSCU features of ORF1ab in SARS-CoV-2 reveal there is no close genetic relationship with SARS-CoV. New information obtained from present analysis is highly significant for effective control of SARS-CoV-2 induced pneumonia of the whole world.
Acknowledgements
This research work was supported by Special Scientific Research Plan Project of Education Department of Shaanxi Province (No. 18JK0377).
Declarations
Conflict of interest
None.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Asadi-Pooya AA, Simani L. Central nervous system manifestations of COVID-19: a systematic review. J Neurol Sci. 2019 doi: 10.1016/j.jns.2020.116832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini B, Rueca M, Gruber CEM, Messina F, Carletti F, Giombini E, Lalle E, Bordi L, Matusali G, Colavita F, Castilletti C, Vairo F, Ippolito G, Capobianchi MR, Di Caro A. SARS-CoV-2 phylogenetic analysis, Lazio Region, Italy, February–March 2020. Emerg Infect Dis. 2020;26(8):1842–1845. doi: 10.3201/eid2608.201525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53(3):425–435. doi: 10.1016/j.jmii.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilucca M, Forcelloni S, Georgakilas AG, Giansanti A, Pavlopoulou A. Codon usage and phenotypic divergences of SARS-CoV-2 genes. Viruses. 2020;12(5):498. doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittadi R, Afshar H, Carraro P. The early antibody response to SARS-Cov-2 infection. Clin Chem Lab Med. 2020;58(10):e201–e203. doi: 10.1515/cclm-2020-0617. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J. Severe acute respiratory syndrome-related coronavirus: classifying 2019-n CoV and naming it SARS-CoV-2. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H, Itoh N, Yufika A, Winardi W, Keam S, Te H, Megawati D, Hayati Z, Wagner AL, Mudatsir M. Coronavirus disease 2019 (COVID-19): a literature review. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tao H, Yan Y, et al. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin AR, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Wang W, Zhao X, et al. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Rayner S, Luo MH. Does SARS-CoV-2 has a longer incubation period than SARS and MERS? J Med Virol. 2020;92(5):476–478. doi: 10.1002/jmv.25708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrobial Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou Y, Zhang M, et al. Updated approaches against SARS-CoV-2. Antimicrob Agents Chemother. 2020;64(6):e00483–e520. doi: 10.1128/AAC.00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liang Y, Fengyu Hu, et al. Molecular and serological characterization of SARS-CoV-2 infection among COVID-19 patients. Virology. 2020;551:26–35. doi: 10.1016/j.virol.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud K, Ibrahim A, Fayez M, Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020 doi: 10.1002/jmv.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathuria JP, Yadav R. Laboratory diagnosis of SARS-CoV-2—a review of current methods. J Infect Public Health. 2020;13(7):901–905. doi: 10.1016/j.jiph.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhamad F, Kubota Y, Ito M. Nonstructural proteins NS7b and NS8 are likely to be phylogenetically associated with evolution of 2019-nCoV. Infect Genet Evolut. 2020;81:104272. doi: 10.1016/j.meegid.2020.104272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Z, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D, Kostaki EG, Magiorkinis G, et al. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evolut. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Guan Y, Yuen KY. Severe acute respiratory syndrome. Nat Med. 2004;10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S, Reucher S, Nörz D, Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Eurosurveillance. 2020;25(9):2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Genetic diversity and evolution of SARS-CoV-2. Infect Genet Evolut. 2020;81:104260. doi: 10.1016/j.meegid.2020.104260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajna M, Suneetha S, Grace R. Genome-wide codon usage bias analysis in Beauveria bassiana. Bioinformation. 2018;14(9):580–586. doi: 10.6026/97320630014580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Wei C, Li Y, et al. The characteristic of the synonymous codon usage and phylogenetic analysis of hepatitis B virus. Genes Genom. 2020;42(7):805–815. doi: 10.1007/s13258-020-00932-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokni M, Ghasemi V, Tavakoli Z. Immune responses and pathogenesis of SARS-CoV-2 during an outbreak in Iran: comparison with SARS and MERS. Rev Med Virol. 2020;30(3):e2107. doi: 10.1002/rmv.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafin MB, Bottega A, Foletto VS, et al. Drug repositioning an alternative for the treatment of coronavirus COVID-19. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Han D, Zhang R, Li J, Zhang R. Molecular and serological assays for SARS-CoV-2: insights from genome and clinical characteristics. Clin Chem. 2020;66(8):1030–1046. doi: 10.1093/clinchem/hvaa122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuo Su, Gary W, Weifeng S, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq UR, Mao Y, Tao S. Codon usage bias and evolutionary analyses of Zika virus genomes. Genes Genom. 2017;39:855–866. doi: 10.1007/s13258-017-0549-0. [DOI] [Google Scholar]
- Singh A, Singh RS, Sarma P, Batra G, Joshi R, Kaur H, Sharma AR, Prakash A, Medhi B. A comprehensive review of animal models for coronaviruses: SARS-CoV-2, SARS-CoV, and MERS-CoV. Virol Sin. 2020;35(3):290–304. doi: 10.1007/s12250-020-00252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Cui H, Gao Z, et al. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses. 2020;12(4):360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tort FL, Castells M, Cristina J. A comprehensive analysis of genome composition and codon usage patterns of emerging coronaviruses. Virus Res. 2020 doi: 10.1016/j.virusres.2020.197976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei-Jie G, Zheng-Yi N, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2019 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2019) Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- World Health Organization (2020) Novel coronavirus (2019-nCoV) technical guidance: infection prevention and control. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/infection-prevention-and-control
- Xiaoping Y, Dong L, Zhang Y, et al. A mild type of childhood Covid-19—a case report. Radiol Infect Dis. 2020 doi: 10.1016/j.jrid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaoyue Yu, Zuo L, Dandan Lu, Bin Lu, et al. Comparative analysis of chloroplast genomes of five Robinia species: genome comparative and evolution analysis. Gene. 2019;689(20):141–151. doi: 10.1016/j.gene.2018.12.023. [DOI] [PubMed] [Google Scholar]
- Yadav PD, Potdar VA, Choudhary ML, Nyayanit DA, Agrawal M, Jadhav SM, Majumdar TD, Shete-Aich A, Basu A, Abraham P, Cherian SS. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J Med Res. 2020;151(2–3):200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. Genomics. 2020;112(5):3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]