Abstract
Colorectal cancer (CRC) is the fourth most lethal cancer in the world. Heat shock protein 60 (HSP60), a mitochondrial chaperone that maintains mitochondrial proteostasis, is highly expressed in tumors compared with in paracancerous tissues, suggesting that high HSP60 expression benefits tumor growth. To determine the effects of HSP60 expression on tumor progression, stable HSP60-knockdown HCT116 cells were constructed in the present study, revealing that knockdown of HSP60 inhibited cell proliferation. Proteomic analysis demonstrated that mitochondrial proteins were downregulated, indicating that knockdown of HSP60 disrupted mitochondrial homeostasis. Metabolomic analysis demonstrated that cellular adenine levels were >30-fold higher in HSP60-knockdown cells than in control cells. It was further confirmed that elevated adenine activated the AMPK signaling pathway, which inhibited mTOR-regulated protein synthesis to slow down cell proliferation. Overall, the current results provide a valuable resource for understanding mitochondrial function in CRC, suggesting that HSP60 may be a potential target for CRC intervention.
Keywords: adenine, colorectal cancer, heat shock protein 60, metabolomics, mTOR pathway, quantitative proteomics
Introduction
Colorectal cancer (CRC) is one of the most common types of cancer, with >1.8 million new cases diagnosed worldwide every year, resulting in 881,000 fatalities (1). The occurrence of CRC is driven by mutations of tumor suppressor genes, oncogenes and genes associated with DNA repair (2), and it can be classified as sporadic, hereditary or familial. Colorectal carcinogenesis is characterized as a multi-step, multi-mechanism process in which tumor initiation and progression occur via a progressive accumulation of genetic mutations (3). Understanding tumor progression in CRC at the molecular level is important for facilitating diagnosis and therapeutic intervention.
Heat shock protein 60 (HSP60) is a major ATP-dependent mitochondrial chaperone and is well-conserved from bacteria to mammals. Previous studies have revealed that HSP60 serves an important role in the pathology of complex diseases, such as neurodegenerative disorders, atherosclerosis and heart disease, as well as multiple inflammatory diseases (4–6). The effects of HSP60 expression on cancer progression have been extensively studied and suggest that HSP60 is either pro-survival or pro-apoptotic in different types of tumor (7–12). Our previous study has indicated that HSP60 expression is downregulated in clear cell renal cell carcinoma (ccRCC) tissues compared with in paracancerous tissues (13). HSP60 overexpression in ccRCC cells inhibits proliferation, while HSP60-knockdown promotes cell proliferation in both cell culture and nude mouse xenografts. Additionally, HSP60-knockdown activates the AMPK signaling pathway, which promotes acquisition of the Warburg phenotype in ccRCC cells (14). In addition, HSP60 promotes tumor cell proliferation and metastasis (8–10) and is highly expressed in colorectal, ovarian and prostate cancer (12,15,16). However, it is unclear whether HSP60-knockdown activates the AMPK signaling pathway and inhibits proliferation in CRC cells.
To determine the effects of HSP60 expression on CRC tumor progression, stable HCT116 cells with HSP60-knockdown were constructed in the present study. The current study demonstrated how HSP60-knockdown affected proliferation of HCT116 cells via proteomic and metabolic analysis, and validated the regulation of the adenine-AMPK-mTOR signaling pathway in HSP60-knockdown CRC cells. The current results suggested that HSP60 may be a potential target for CRC therapy.
Materials and methods
Cell lines and culture
The human colon cancer cell line (HCT116) and 293T cell line were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, and the cells were grown in McCoy's 5A medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Wisent, Inc.) and 1% penicillin/streptomycin (Wisent, Inc.). The cells were cultured at 37°C with 5% CO2. Mycoplasma testing was performed for the cell lines used.
Establishment of a stable cell line with HSP60-knockdown
The HSP60-knockdown stable cell line was established based on a previously published protocol (17). Two short hairpin (sh)RNAs (KD1 and KD2) specifically targeting HSPD1 were chosen (17), and a scramble shRNA was used as negative control (sequences shown in Table SI). Restriction sites were created on the double-strand shRNAs and pLL3.7 vector (Addgene, Inc.; cat. no. 11795) and the shRNAs were inserted into plasmid pLL3.7 lentivirus vectors with T4 DNA ligase (cat. no. EL0011; Thermo Fisher Scientific, Inc,). pLL3.7-shRNAs (20 µg; 1.33 µg/ml) were co-transfected with pMD2.G, pMDLg/p RRE and pREV-Rev (preserved in our laboratory) into 293T cells using polyethyleneimine (Sigma-Aldrich; Merck KGaA) when cells reached 60–70% confluence. Cells were incubated at 37°C with 5% CO2. After 72 h, supernatants were harvested with PEG6000 (Tokyo Chemical Industry Co., Ltd.) and were used to infect HCT116 cells with 6 µg/ml polybrene at 37°C for 6 h when cells reached 30–40% confluence. After 48 h, infected cells were sorted by a flow cytometer (FACSCalibur; BD Biosciences) to generate the monoclonal stable cell line. The clones with the uniform GFP expression were selected in the present study (data not shown).
Detection of cell proliferation by cell counting Kit-8 (CCK-8) assay
HCT116 cells were seeded in a 96-well plate (2,000 cells/well). The proliferation rate was determined using the CCK-8 assay according to the manufacturer's instructions (Dojindo Molecular Technologies, Inc.). Briefly, the CCK-8 reagent was added into each well after 0, 12, 24, 36, 48, 72, 84, 96 and 108 h of cell culture. Absorbance at 450 nm was measured 2 h after CCK-8 addition.
Xenograft experiments
All animal studies were approved by the Animal Research Ethics Committee of the Tsinghua University (Beijing, China). The mice were housed with free access to food and water in a temperature- and light-regulated pathogen-free room at Tsinghua University Animal Facilities (temperature, 24±1°C; humidity, 60±5%; 12-h light/dark cycle). The protocols used in the present study were approved by the Institutional Animal Care and Use Committee (IACUC) of Tsinghua University and were performed in accordance with the IACUC guidelines. The humane endpoint was tumor burden >15 mm at the largest dimension. Carbon Dioxide Euthanasia for Mice (BU ASC Guidelines) (17) was used to provide a rapid, painless, stress-free death. CO2 overdose causes rapid unconsciousness followed by death. A gradual CO2 fill rate of 10–30% of the chamber volume per minute was used when euthanizing mice. A total of 5×106 control cells (transfected with the negative control shRNA) or HSP60-KD1 or HSP60-KD2 cells were resuspended with PBS and subcutaneously injected into five 5-week-old nude male mice for each group (15 mice in total; weight, 19.9–23.4 g; Beijing Vital River Laboratory Animal Technology Co., Ltd.). Tumor size was quantified by fluorescence imaging using an in vivo imaging system (PerkinElmer, Inc.) to measure the GFP fluorescence intensity of tumors in mice. 20 days after injection, The tumor volume was calculated as π/6 × length (mm) × width (mm) × height (mm).
Quantitative proteomic analysis and metabolomic analysis
Quantitative proteomic analysis was performed as previously described (3). Tandem Mass Tag (TMT) reagents were purchased from Thermo Fisher Scientific, Inc., and peptides were labeled according to the manufacturer's instructions (Thermo Fisher Scientific, Inc.). The labeled peptides were mixed and desalted on a SEP Pak C18 column (Waters Corporation) and separated by reverse phase chromatography. Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific, Inc.) and Xcalibur 3.0 software (Thermo Fisher Scientific, Inc.) were applied with data-dependent acquisition mode. Proteomic analysis of cell lines was performed in four biological replicates. The MS/MS spectra were searched using the search engine SEQUEST from Proteome Discoverer Software (version 2.1; Thermo Fisher Scientific, Inc.) against UniProt human database. Metabolomic analysis was performed according to a previous study (7). TSQ Quantiva™ Triple Quadrupole Mass Spectrometer (Thermo Fisher Scientific, Inc.) with positive/negative ion switching for targeted quantitation with selective reaction monitoring was used. C18 based reverse phase chromatography was utilized with 10 mM tributylamine, 15 mM acetate in water and 100% methanol as mobile phase A and B, respectively. The source voltage was 3,500 V for positive and 2,500 V for negative ion mode. The source parameters were as follows: Spray voltage, 3,000 V; capillary temperature, 320°C; heater temperature, 300°C; sheath gas flow rate, 35 units; auxiliary gas flow rate, 10 units. Metabolite identification was based on TraceFinder software (Version 3.2; Thermo Fisher Scientific, Inc.) with a home-built database containing ~300 compounds. Ingenuity Pathway Analysis (IPA) software (version 2.0; Qiagen, Inc.) was used to perform pathway analysis.
Western blotting
Cells were lysed with lysis buffer (20 mmol/l Tris-HCl pH 7.5, 150 mmol/l NaCl, 1% Triton X-100, 1% sodium pyrophosphate and Protease Inhibitor Cocktail) for 30 min on ice. The supernatant was collected by centrifugation at 13,800 × g for 20 min at 4°C. Protein concentrations were determined using the BCA protein assay kit. Proteins (40 µg protein/lane) were separated via 12% SDS-PAGE and transferred onto a PVDF membrane. The membrane was blocked with 5% skim milk powder in TBST solution at room temperature for 1 h. Anti-β-actin (cat. no. AC038; 1:5,000), anti-HSP60 (cat. no. A0564), anti-AMPKα (cat. no. A1229), anti-phospho (p)-AMPKα (Thr172; cat. no. AP0116), anti-Raptor (cat. no. A8992), anti-p-Raptor (Ser792; cat. no. AP0928), anti-eukaryotic translation initiation factor 4E binding protein 1 (4EBP1; cat. no. A19045) and anti-p-4EBP1 (Thr37/46) primary antibodies were obtained from ABclonal Biotech Co., Ltd. Antibodies against mTOR (cat. no. 2983), p-mTOR (Ser2448; cat. no. 5536), Bcl-2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3; cat. no. 44060) and Parkin (cat. no. 2132), as well as HRP-conjugated anti-rabbit (cat. no. 7074) and anti-mouse IgG (cat. no. 7076) secondary antibodies were obtained from Cell Signaling Technology, Inc. Antibodies against p70S6 kinase (p70S6K; cat. no. S4047) and p-p70S6K (T421/S424; cat. no. S6436) were obtained from Sigma-Aldrich. Dilution rates for all antibodies, except for the anti-β-actin antibody, were 1:1,000. Primary antibodies were incubated at 4°C overnight, while secondary antibodies were incubated at room temperature for 1 h. The visualization reagent was part of an enhanced chemiluminescent kit (Beijing Solarbio Science & Technology Co., Ltd.).
Statistical analysis
At least three biological replicates were performed for all in vitro assays, and the data were presented as the mean ± SEM. Statistical analysis was performed using GraphPad Prism 7.0 software (GraphPad Software, Inc.). One-way or two-way ANOVA was used for the comparison of multiple groups followed by Bonferroni's correction, Benjamini-Hochberg method or Dunnett's test. Wilcoxon rank-sum test was used to determine the significance of the differences in Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/). Fisher's exact test was used for pathway enrichment analysis. The cut-off values for downregulated proteins were fold-change ≤0.76 and P<0.05; and those for upregulated proteins were fold-change ≥1.3 and P<0.05. P<0.05 was considered to indicate a statistically significant difference.
Results
HSP60-knockdown inhibits the proliferation of CRC cells
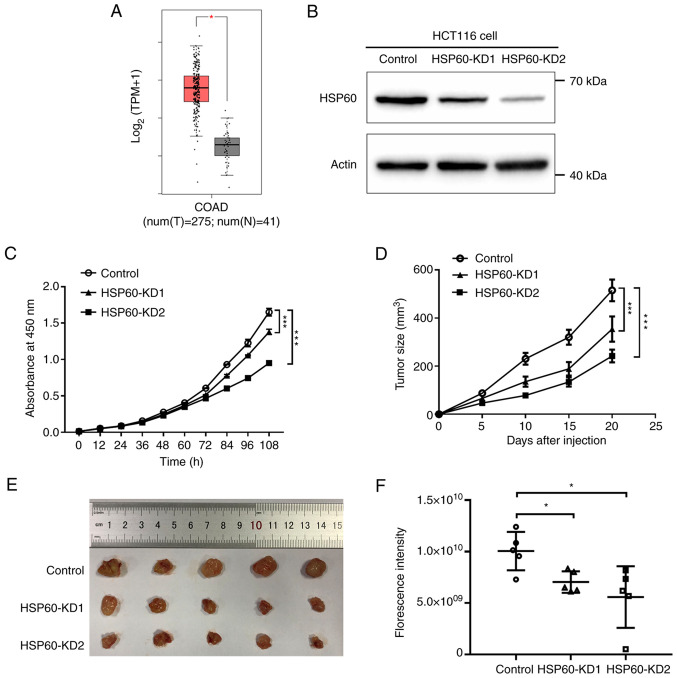
GEPIA (18) revealed that HSP60 expression in tumor samples was significantly higher than that in paired normal tissues from patients with CRC, suggesting that HSP60 may be required for tumor progression (Fig. 1A). To explore the effects of HSP60-knockdown on the progression of CRC, stable HSP60-knockdown cells were established using HSP60-directed shRNAs in human CRC HCT116 cells. Cells transfected with scrambled shRNA that had no homology in the human genome were used as the negative control. HSP60 expression in these cells was examined by western blotting, confirming that HSP60 expression was significantly silenced in HSP60-KD1 and HSP60-KD2 cells (Fig. 1B). HSP60 expression in HSP60-KD2 cells was markedly lower than in HSP60-KD1 cells (Fig. 1B).
Figure 1.
HSP60-knockdown suppresses the proliferation of colorectal cancer cells in vitro and in vivo. (A) HSP60 was highly expressed in colorectal cancer tissues (red) compared with in paired normal tissues (gray) from patients with colorectal cancer. The log2-transformed values of HSP60 gene expression of the COAD dataset in GEPIA were used to compare the difference between cancer and normal samples. Significance was calculated using the Wilcoxon rank-sum test. (B) HSP60 was knocked down in HSP60-KD1 and -KD2 cells. (C) Cell Counting Kit-8 assay indicated that cell proliferation was significantly decreased in HSP60-KD cells than in control cells. (D and E) HSP60-knockdown inhibited tumor growth in nude mice. The volumes of xenograft tumors of HCT HSP60-KD and control cells were monitored for 20 days after injection. Data were analyzed using ordinary two-way ANOVA followed by Bonferroni's test as multiple comparisons test. (F) Florescent intensities of tumors on the 20th day after injection indicated the relative size of xenograft tumors generated from HSP60-KD and control cells in nude mice. Data were analyzed using ordinary one-way ANOVA followed by Benjamini-Hochberg method as multiple comparisons test. *P<0.05; ***P<0.001. HSP60, heat shock protein 60; KD, knockdown; T, tumor; N, normal; TPM, transcripts per million; COAD, colon adenocarcinoma; GEPIA, Gene Expression Profiling Interactive Analysis.
The proliferation rates of HSP60-knockdown cells were then measured using CCK-8 assay. HSP60-knockdown significantly inhibited the proliferation rate of HCT116 cells (Fig. 1C). Additionally, the proliferation rate of HSP60-KD2 cells was slower than that in HSP60-KD1 cells, indicating that HSP60 expression was positively associated with the proliferation rate. Subsequently, HSP60-KD1 and -KD2 cells were subcutaneously injected into 5-week-old immunodeficient mice to verify whether HSP60-knockdown inhibited tumor growth. Vernier calipers were used to calculate tumor volume 20 days after injection via measuring the major and minor diameters of tumors. The results indicated that HSP60-knockdown significantly decreased tumor size compared with the control cells (Fig. 1D and E). Since HSP60-KD1 and -KD2 cells stably expressed GFP, the tumor volume of subcutaneous mouse xenografts was measured using fluorescence imaging. The fluorescence intensity of HSP60-KD1 and -KD2 tumors was significantly lower than that in control tumors (Figs. 1F and S1). These results indicated that HSP60-knockdown slowed the proliferation of CRC cells in vitro and in vivo, and that HSP60 expression was positively associated with tumor growth.
HSP60-knockdown disrupts mitochondrial proteostasis
To explore how HSP60-knockdown inhibited the proliferation of CRC cells, a TMT-based quantitative proteomic analysis was performed to identify differentially expressed proteins between HSP60-knockdown and control cells. The analysis quantified a total of 5,590 proteins. Based on the cut-off threshold for determining the differentially expressed proteins, 575 proteins were considered to be downregulated (fold change ≤0.76; P<0.05), while 53 proteins were upregulated (fold change ≥1.3; P<0.05) (Fig. 2A).
Figure 2.
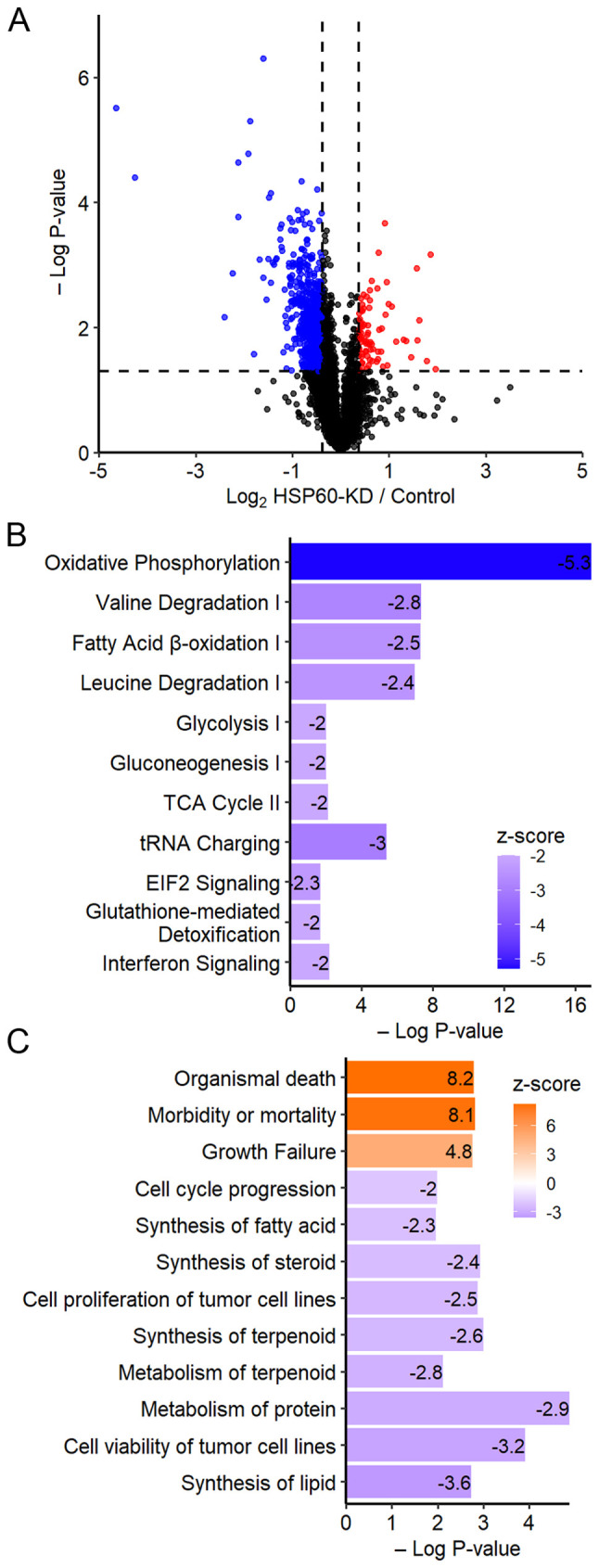
Proteomic analysis identified deferentially expressed proteins between HSP60-KD cells and control cells. (A) A total of 575 proteins and 53 proteins were found to be significantly upregulated and downregulated, respectively, in HSP60-KD HCT116 cells by proteomic analysis. (B) Ingenuity Pathway Analysis indicated that 11 cancer-associated pathways were significantly inhibited in HSP60-KD HCT116 cells, and ‘oxidative phosphorylation’ was the most inhibited pathway based on z-scores. (C) Downstream functional analysis by Ingenuity Pathway Analysis identified significantly activated (z-score >2; P<0.05) and inhibited (z-score <-2; P<0.05) functions in HSP60-KD HCT116 cells. HSP60, heat shock protein 60; KD, knockdown.
IPA indicated that 11 cancer-associated pathways were significantly inhibited in HSP60-knockdown cells (Fig. 2B). Inhibition of ‘oxidative phosphorylation’ suggested that HSP60-knockdown may cause mitochondrial dysfunction (19), while inhibition of ‘tRNA charging’ and ‘EIF2 signaling’ implied that HSP60-knockdown may inhibit protein synthesis (20). Other signaling pathways inhibited in HSP60-knockdown cells included ‘valine degradation’, ‘fatty acid b-oxidation’, ‘leucine degradation’, ‘glycolysis’, ‘gluconeogenesis’, ‘TCA cycle’, ‘glutathione-mediated detoxification’ and ‘interferon signaling’ (Fig. 2B). From the Disease and Function Analysis in IPA, it was revealed that the functions associated with ‘organismal death’, ‘morbidity or mortality’, and ‘growth failure’ were activated in HSP60-knockdown cells (z-score >2; P<0.05; Fig. 2C). Other disease-associated pathways that were inactivated included ‘synthesis of lipid’ and ‘cell viability of tumor cell lines’, ‘metabolism of protein’, ‘metabolism of terpenoid’, ‘cell proliferation of tumor cell lines’, ‘synthesis of steroid’, ‘synthesis of fatty acid’ and ‘cell cycle progression’ (z-score <-2; P<0.05; Fig. 2C).
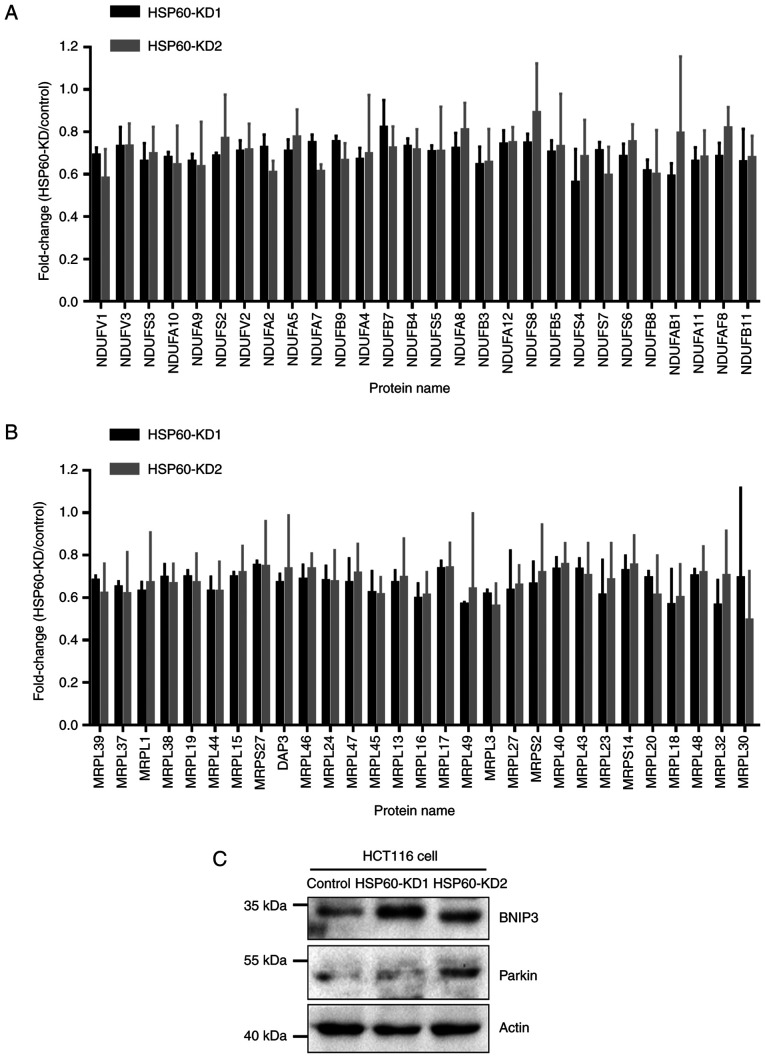
Quantitative proteomic analysis results revealed that proteins associated with oxidative phosphorylation were downregulated in HSP60-knockdown cells, in which 28 subunits of mitochondrial complex I were downregulated in HSP60-KD1 and -KD2 cells compared with in control cells (Fig. 3A). Moreover, quantitative proteomic analysis results indicated that HSP60-knockdown downregulated 29 mitochondrial ribosomal proteins (MRPs) in HSP60-KD1 and -KD2 HCT116 cells compared with in control cells (Fig. 3B). These results indicated that HSP60-knockdown disrupted mitochondrial proteostasis and inhibited oxidative phosphorylation in HCT116 cells. Immunoblotting experiments confirmed the upregulation of mitochondrial autophagy-associated proteins, such as BNIP3 and Parkin, following HSP60-knockdown (Fig. 3C).
Figure 3.
Mitochondrial-associated proteins are significantly downregulated in HSP60-KD HCT116 cells. (A) A total of 28 proteins associated with mitochondrial complex I were downregulated in HSP60-KD1 and -KD2 cells compared with in control cells. (B) A total of 29 MRPs were decreased in HSP60-KD1 and -KD2 HCT116 cells compared with in control cells. (C) Mitochondrial autophagy-associated proteins BNIP3 and Parkin were increased in HSP60-KD cells compared with in control cells. HSP60, heat shock protein 60; KD, knockdown; MRP, mitochondrial ribosomal protein; NDUF, NADH:ubiquinone oxidoreductase core subunit; BNIP3, Bcl-2/adenovirus E1B 19-kDa interacting protein 3.
HSP60-knockdown in HCT116 cells inhibits the mTOR signaling pathway through the adenine/AMPK/mTOR signaling pathway
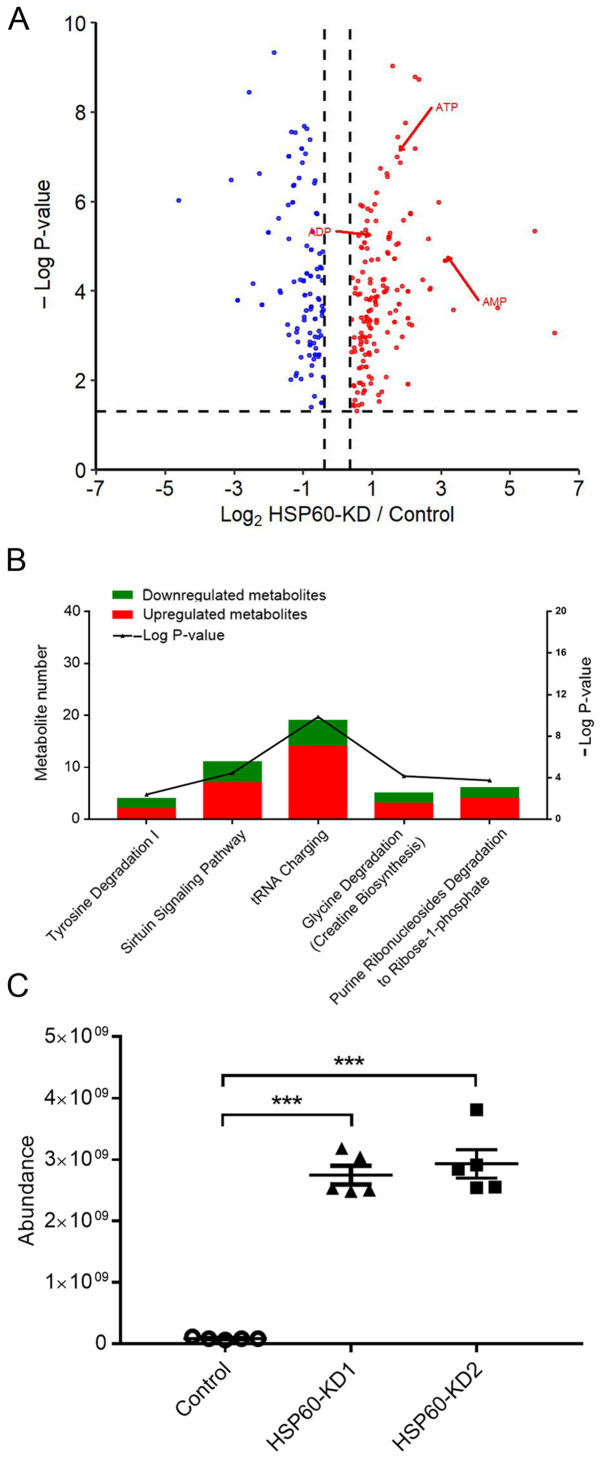
The aforementioned results indicated that HSP60-knockdown may reprogram cell metabolism. To determine the metabolic changes in HSP60-knockdown cells and control cells, a metabolomics analysis was performed in five biological replicates. Notably, it was revealed that HSP60-knockdown significantly changed the levels of 297 metabolites in HSP60-KD1 cells (Fig. 4A), supporting the hypothesis that HSP60-knockdown reprogramed cellular metabolic pathways. IPA analysis indicated that the following pathways were associated with the changed metabolites: ‘Tyrosine degradation’, ‘sirtuin signaling pathway’, ‘tRNA charging’, ‘glycine degradation’ and ‘purine ribonucleoside degradation to ribose-1-phosphate’ (Fig. 4B). Additionally, the metabolomics analysis revealed that the adenine level was increased by >30-fold (Fig. 4C), and cellular AMP, ADP and ATP were also increased by 6.2-, 2.8- and 2.4-fold, respectively (Fig. 4A).
Figure 4.
Metabolomic analysis of HSP60-KD cells. (A) Significantly changed metabolites in HSP60-knockdown cells compared with in control cells (P<0.05; fold-change ≥1.3 or ≤0.75). (B) Metabolic pathway analysis and downstream function analysis using Ingenuity Pathway Analysis showed significantly altered pathways (Fisher's exact test; P<0.05) with the number of identified metabolites in associated pathways reported in the literature. (C) Adenine was increased in HSP60-KD cells by >30-fold. Data were analyzed using ordinary one-way ANOVA followed by Dunnett's test. ***P<0.001. HSP60, heat shock protein 60; KD, knockdown.
It is known that excess adenine can increase AMP, which promotes AMPK signaling (21,22). To confirm that HSP60-knockdown activated the AMPK signaling pathway, western blotting experiments were performed, revealing that HSP60-knockdown markedly increased AMPKα phosphorylation at T172 (Fig. 5A), while it decreased phosphorylation of p70S6k, 4EBP1 and mTOR (Fig. 5B). Additionally, western blotting revealed an increase in Raptor phosphorylation (Fig. 5B). These results demonstrated that HSP60-knockdown inactivated the mTOR signaling pathway to inhibit protein translation.
Figure 5.
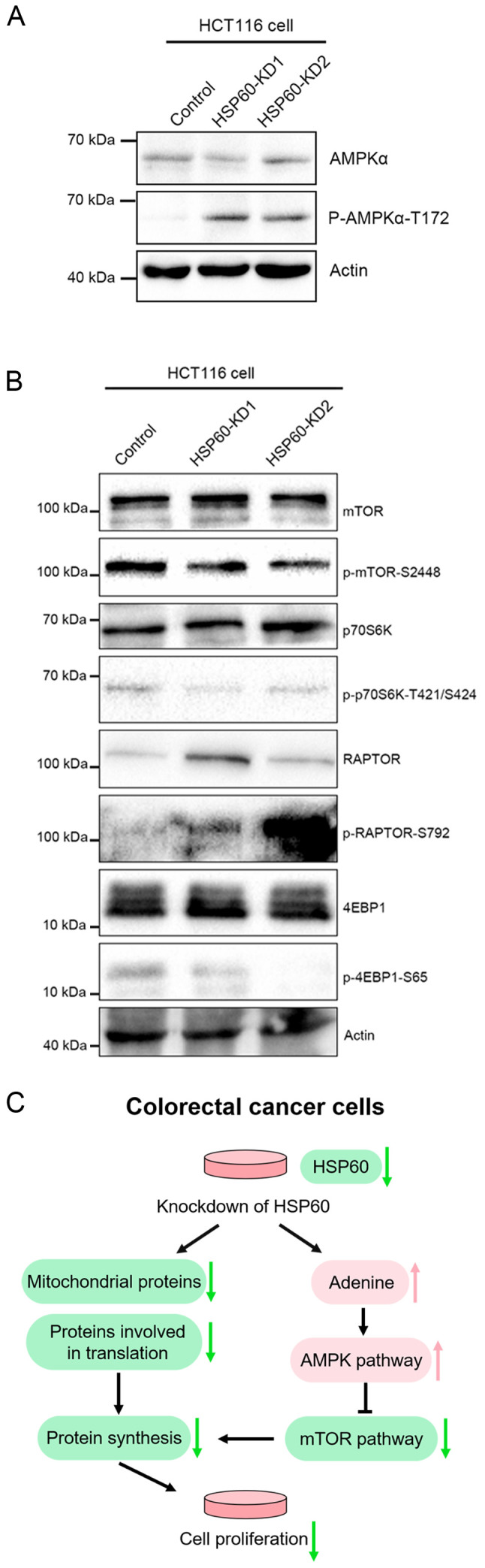
HSP60-knockdown in HCT116 cells activates the AMPK signaling pathway and inhibits mTOR and protein synthesis. (A) Western blotting indicated that phosphorylation of AMPK (T172) in HSP60-KD HCT116 cells was increased compared with in control cells. (B) Western blotting indicated that the mTOR signaling pathway was inhibited in HSP60-KD cells compared with in control cells. (C) Diagram illustrating the effects of HSP60-knockdown on the adenine/AMPK/mTOR signaling pathway and proliferation of HCT116 cells. HSP60, heat shock protein 60; KD, knockdown; p, phosphorylated; 4EBP1, eukaryotic translation initiation factor 4E binding protein 1; p70S6K, p70S6 kinase.
Discussion
Previous studies have demonstrated that HSP60 suppresses or promotes cell proliferation in a context-dependent manner. HSP60-knockdown decreases the proliferation rates of pancreatic cancer cells (23) and glioblastoma cells (24), while it increases the proliferation rates of ccRCC cells (13,14). HSP60-knockdown attenuates pancreatic ductal cancer cell proliferation and migration/invasion via decreasing Erk1/2 phosphorylation (23), while it switches the mitochondrial function from ATP production to biosynthesis in ccRCC cells (14). Analysis of HSP60 mRNA expression in cancer and normal tissues in GEPIA revealed that HSP60 was highly expressed in CRC, suggesting that HSP60 expression may be positively associated with tumor progression. Consistently, a previous study has demonstrated that the serum levels of HSP60 protein are significantly higher in patients with CRC compared with in healthy controls, and serum HSP60 exhibited the same sensitivity and specificity compared with carcinoembryonic antigen (25). Until now, it was unclear whether HSP60-knockdown inhibited the proliferation of CRC cells. The present study demonstrated that HSP60-knockdown inhibited the proliferation of CRC cells.
The present study conducted proteomics analysis and revealed the global effects of HSP60-knockdown on cellular processes. MRPs and respiratory complex I subunits were identified to be downregulated following HSP60-knockdown. Metabolomics analysis indicated that HSP60-knockdown significantly increased cellular adenine levels and adenine-associated molecules, such as AMP and ADP, resulting in activation of the AMPK signaling pathway. The current results revealed that HSP60-knockdown in HCT116 cells significantly activated AMPKα and inhibited mTOR and its downstream targets p70S6 kinase and 4EBP1 (Fig. 5C), providing information on the mechanisms underlying HSP60-knockdown causing adenine accumulation, which requires further exploration. In our future study, stable cell lines in which HSP60 is knocked down or overexpressed will be generated from patient-derived colon cancer cells.
In conclusion, the present results indicated that high HSP60 expression was important for CRC progression. HSP60-knockdown inhibited cell proliferation in vitro and in nude mouse xenografts. Additionally, HSP60-knockdown interrupted mitochondrial protein homeostasis and caused adenine accumulation, which activated the AMPK signaling pathway to inhibit protein translation and cell proliferation. These data provide a valuable resource for understanding mitochondrial homeostasis in CRC progression, and HSP60 may be used as a potential therapeutic target for CRC treatment.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by Projects of Shandong Science and Technology Committee (grant no. 2018GSF118159), Natural Science Foundation of Shandong Province (grant no. ZR2016CL08), Medical and Health Technology Development Program of Shandong Province (grant no. 2016WS0013) and the Taishan Scholar Program Foundation of China (grant no. tshw20110515).
Funding
The present study was supported by Projects of Shandong Science and Technology Committee (grant no. 2018GSF118159), Natural Science Foundation of Shandong Province (grant no. ZR2016CL08), Medical and Health Technology Development Program of Shandong Province (grant no. 2016WS0013) and the Taishan Scholar Program Foundation of China (grant no. tshw20110515).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JG, HD and RX conceived and designed the study. JG performed the experiments. JG, HD and RX wrote the paper. SZ analyzed the data, reviewed and edited the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors are responsible for confirming the authenticity of the raw data.
Ethics approval and consent to participate
All experimental protocols were approved (approval no. 17-DHT1) by the Animal Research Ethics Committee of Tsinghua University (Beijing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 3.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 4.Knowlton AA, Srivatsa U. Heat-shock protein 60 and cardiovascular disease: A paradoxical role. Future Cardiol. 2008;4:151–161. doi: 10.2217/14796678.4.2.151. [DOI] [PubMed] [Google Scholar]
- 5.Hansen JJ, Durr A, Cournu-Rebeix I, Georgopoulos C, Ang D, Nielsen MN, Davoine CS, Brice A, Fontaine B, Gregersen N, Bross P. Hereditary spastic paraplegia SPG13 is associated with a mutation in the gene encoding the mitochondrial chaperonin Hsp60. Am J Hum Genet. 2002;70:1328–1332. doi: 10.1086/339935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundtman C, Wick G. The autoimmune concept of atherosclerosis. Curr Opin Lipidol. 2011;22:327–334. doi: 10.1097/MOL.0b013e32834aa0c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo J, Li X, Zhang W, Chen Y, Zhu S, Chen L, Xu R, Lv Y, Wu D, Guo M, et al. HSP60-regulated mitochondrial proteostasis and protein translation promote tumor growth of ovarian cancer. Sci Rep. 2019;9:12628. doi: 10.1038/s41598-019-48992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappello F, Conway DME, Marasa L, Zummo G, Macario AJ. Hsp60 expression, new locations, functions and perspectives for cancer diagnosis and therapy. Cancer Biol Ther. 2008;7:801–809. doi: 10.4161/cbt.7.6.6281. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 10.Tsai YP, Yang MH, Huang CH, Chang SY, Chen PM, Liu CJ, Teng SC, Wu KJ. Interaction between HSP60 and beta-catenin promotes metastasis. Carcinogenesis. 2009;30:1049–1057. doi: 10.1093/carcin/bgp087. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh JC, Siegelin MD, Dohi T, Altieri DC. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010;70:8988–8993. doi: 10.1158/0008-5472.CAN-10-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappello F, Bellafiore M, Palma A, David S, Marciano V, Bartolotta T, Sciume C, Modica G, Farina F, Zummo G, Bucchieri F. 60KDa chaperonin (HSP60) is over-expressed during colorectal carcinogenesis. Eur J Histochem. 2003;47:105–110. doi: 10.4081/814. [DOI] [PubMed] [Google Scholar]
- 13.Tang H, Chen Y, Liu X, Wang S, Lv Y, Wu D, Wang Q, Luo M, Deng H. Downregulation of HSP60 disrupts mitochondrial proteostasis to promote tumorigenesis and progression in clear cell renal cell carcinoma. Oncotarget. 2016;7:38822–38834. doi: 10.18632/oncotarget.9615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng R, Liu Z, Tang H, Zhang W, Chen Y, Xu R, Chen L, Song J, Liu X, Deng H. HSP60 silencing promotes Warburg-like phenotypes and switches the mitochondrial function from ATP production to biosynthesis in ccRCC cells. Redox Biol. 2019;24:101218. doi: 10.1016/j.redox.2019.101218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hjerpe E, Egyhazi S, Carlson J, Stolt MF, Schedvins K, Johansson H, Shoshan M, Avall-Lundqvist E. HSP60 predicts survival in advanced serous ovarian cancer. Int J Gynecol Cancer. 2013;23:448–455. doi: 10.1097/IGC.0b013e318284308b. [DOI] [PubMed] [Google Scholar]
- 16.Cappello F, Rappa F, David S, Anzalone R, Zummo G. Immunohistochemical evaluation of PCNA, p53, HSP60, HSP10 and MUC-2 presence and expression in prostate carcinogenesis. Anticancer Res. 2003;23:1325–1331. [PubMed] [Google Scholar]
- 17.Boston University Research Support, corp-author. Boston University Research Support; Boston, MA: 2019. Carbon Dioxide Euthanasia for Rats and Mice (BU ASC Guidelines) Revised May 2019. [Google Scholar]
- 18.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-González C, Nuevo-Tapioles C, Herrero Martín JC, Pereira MP, Serrano Sanz S, Ramírez de Molina A, Cuezva JM, Formentini L. Dysfunctional oxidative phosphorylation shunts branched-chain amino acid catabolism onto lipogenesis in skeletal muscle. EMBO J. 2020;39:e103812. doi: 10.15252/embj.2019103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaborske JM, Narasimhan J, Jiang L, Wek SA, Dittmar KA, Freimoser F, Pan T, Wek RC. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J Biol Chem. 2009;284:25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills GC, Schmalstieg FC, Trimmer KB, Goldman AS, Goldblum RM. Purine metabolism in adenosine deaminase deficiency. Proc Natl Acad Sci USA. 1976;73:2867–2871. doi: 10.1073/pnas.73.8.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeon SM. Regulation and function of AMPK in physiology and diseases. Exp Mol Med. 2016;48:e245. doi: 10.1038/emm.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou C, Sun H, Zheng C, Gao J, Fu Q, Hu N, Shao X, Zhou Y, Xiong J, Nie K, et al. Oncogenic HSP60 regulates mitochondrial oxidative phosphorylation to support Erk1/2 activation during pancreatic cancer cell growth. Cell Death Dis. 2018;9:161. doi: 10.1038/s41419-017-0196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang H, Li J, Liu X, Wang G, Luo M, Deng H. Down-regulation of HSP60 suppresses the proliferation of Glioblastoma cells via the ROS/AMPK/mTOR pathway. Sci Rep. 2016;6:28388. doi: 10.1038/srep28388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vocka M, Langer D, Fryba V, Petrtyl J, Hanus T, Kalousova M, Zima T, Petruzelka L. Novel serum markers HSP60, CHI3L1, and IGFBP-2 in metastatic colorectal cancer. Oncol Lett. 2019;18:6284–6292. doi: 10.3892/ol.2019.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.