Abstract
Growing evidence suggests that imbalances in resident microbes (dysbiosis) can promote chronic inflammation, immune-subversion, and production of carcinogenic metabolites, thus leading to neoplasia. Yet, evidence to support a direct link of individual bacteria species to human sporadic cancer is still limited. This chapter focuses on several emerging bacterial toxins that have recently been characterized for their potential oncogenic properties toward human orodigestive cancer and the presence of which in human tissue samples has been documented. These include cytolethal distending toxins produced by various members of gamma and epsilon Proteobacteria, Dentilisin from mammalian oral Treponema, Pasteurella multocida toxin, two Fusobacterial toxins, FadA and Fap2, Bacteroides fragilis toxin, colibactin, cytotoxic necrotizing factors and α-hemolysin from Escherichia coli, and Salmonella enterica AvrA. It was clear that these bacterial toxins have biological activities to induce several hallmarks of cancer. Some toxins directly interact with DNA or chromosomes leading to their breakdowns, causing mutations and genome instability, and others modulate cell proliferation, replication and death and facilitate immune evasion and tumor invasion, prying specific oncogene and tumor suppressor pathways, such as p53 and β-catenin/Wnt. In addition, most bacterial toxins control tumor-promoting inflammation in complex and diverse mechanisms. Despite growing laboratory evidence to support oncogenic potential of selected bacterial toxins, we need more direct evidence from human studies and mechanistic data from physiologically relevant experimental animal models, which can reflect chronic infection in vivo, as well as take bacterial-bacterial interactions among microbiome into consideration.
1. Introduction
1.1. Overview of association between infectious agents, microbiome and cancer
Infections have been recognized as a major preventable cause of human cancer.1 Plummer et al.2 estimate that 15.4% of the worldwide cancer cases are attributable to infection, amounting to approximately 2.2 million cases per year. A variety of infectious agents have been linked to pathogenesis of human cancers, including parasites, such as Schistosoma haematobium (urinary bladder cancer) and Opisthorchis viverrini (cholangiocarcinoma), bacteria, such as, Helicobacter (H) pylori (stomach cancer and gastric lymphoma) and viruses, such as hepatitis virus types B and C (liver cancer), human papillomavirus (HPV) (anogenital and oropharyngeal cancer), Epstein-Barr (EB) virus (Burkitt lymphoma and nasopharyngeal cancer), human herpes virus type 8 (Kaposi’s sarcoma and non-Hodgkin’s lymphoma) and human T cell leukemia/lymphoma virus type 1.2,3 Etiological roles of these oncoviruses have been appreciated more unequivocally, as viral genomes can be integrated to the host genome and transcribed, producing own oncoproteins and manipulating host signaling pathways critical in oncogenesis. On the other hand, precise mechanisms of bacteria-induced carcinogenesis have been less clear, except a well characterized oncoprotein from H. pylori, CagA,4–9 despite the fact that bacteria are the most ubiquitous microorganisms that collectively make up to 100 trillion cells in our bodies, 10-fold the number of human cells.10 Bacteria are primarily extracellular organisms and thus their oncogenic effects may be rather indirect. Growing evidence also suggests that imbalances in resident microbes (dysbiosis) can promote chronic inflammation, immune-subversion and production of carcinogenic metabolites, thus leading to neoplasia.1,11–13 Yet, evidence to support a direct link of individual bacteria species to human sporadic cancer is still limited, except the aforementioned H. pylori producing a cytotoxin, CagA. There are other bacterial toxins that have recently been characterized for their potential oncogenic properties and the presence in human tissue samples. This book chapter focuses on those emerging bacterial toxins in order to expand the repertoire of bacteria to be interrogated further to establish a causal association.
1.2. Overview of bacterial toxins
Toxins are a wide range of substances biologically produced within living cells or organisms that are harmful to other organisms while benefiting own survival. They are inanimate and not capable of reproducing themselves. Toxins can be small molecules, peptides, or proteins that are capable of causing disease on contact with or absorption by body tissues interacting with biological macromolecules such as enzymes or cellular receptors. Bacterial toxins are broadly grouped into two main types, lipopolysaccharides (LPS), which are associated with the cell wall of Gram-negative bacteria, and proteins, which are released from bacterial cells and may act at tissue sites removed from the site of bacterial growth. The cell-associated toxins are referred to endotoxins and the extracellular diffusible toxins are referred to as exotoxins, or protein toxins.14 This book chapter focuses on exotoxins produced by distinctive groups of bacteria, as LPS is not bacteria specific, produced by a virtually all Gram-negative bacteria and has been known for a long time causing sepsis (endotoxemia) and a myriad of innate inflammatory responses in infected hosts.15–17 While exotoxins can be named after organisms generating the toxin or after susceptible tissue/organ types, protein toxins can be further divided according to modes of actions; (1) targeting host membrane receptors; (2) targeting host membrane lipid bilayer, and (3) targeting host cytosolic proteins, either through uptake by host endocytosis of paired bacterial proteins called AB toxins, or by direct delivery through molecular syringe of a bacterial injection apparatus.18 Protein toxins are also characterized based in their secretion systems.19 To exert virulence, bacterial toxins produced in cytosol must be transported across the bacterial membranes through co-translational and post-translational mechanisms to reach their targets. These transports are mediated through various secretion systems, which now include nine different mechanisms, types I–IX.20,21 Gram-positive bacteria utilize only types I and VII,19 Gram-negative bacteria exploit all but type VII,20 while one bacterial species can possess multiple systems. Finally, toxins are also classified based on virulence mechanisms, i.e., targeting cytoskeleton components, targeting ubiquitin/proteasomal system, targeting cell translational machinery, targeting neurotransmission system, targeting cAMP production and MAP kinase signaling, and targeting DNA and inducing endoplasmic reticulum stress responses,18 some of which may be highly relevant to carcinogenesis. Subsequent sections provide more details about individual bacterial toxins, with which potential links to human cancers have been increasingly recognized.
2. Cytolethal distending toxins (CDTs)
2.1. Biochemical properties of CDTs
CDTs belong to the AB toxin family produced by several groups of Gram-negative bacteria and is composed of an active subunit (CdtB) and two binding subunits (CdtA and CdtC) (AB2 toxin).22 The crystal structure solved by Nesic et al. indicates that this holotoxin is a tripartite complex,23 and thus it is distinguished from binary CDT toxin secreted by Clostridium difficile in its structure and functions. CDT consists of an enzyme of the DNase-I family (CdtB) bound to two ricin-like lectin domains (CdtA and CdtC), which are required for host cell attachment and translocation of CdtB into host cells.22 It forms a ternary complex with three interdependent molecular interfaces, characterized by globular, as well as non-globular extensions from CdtA and CdtC that facilitate interactions among the three subunits. CdtA and CdtB have also been reported to contain disordered amino acid sequences pointing to dynamic properties of this molecule.23,24 An overall surface area of interaction is estimated to be over 10,700 Å2 for the entire holotoxin, with relative molecular mass ~70,000 (Mr 70 K).23
The CdtA and CdtC subunits are lectin-type molecules, sharing structural homology with the B-chain repeats of the plant toxin ricin and form a deeply grooved, highly aromatic surface that is critical for toxicity.23 The CdtB subunit is functionally and structurally homologous to mammalian DNase I, is translocated into the nucleus, and acts as a genotoxin leading to DNA damage.25 After cell surface binding via CdtA and CdtC, CdtB has to cross the plasma membrane through endocytosis to reach the nucleus, traveling through the trans-Golgi, where sulfation occurs, and then it is retrograde translocated via the endoplasmic reticulum (ER), where it is glycosylated.26 Further confocal microscopic analysis indicates that most likely the active subunit of CDT is directly translocated from the ER to the nucleus.27 CdtB is a highly potent DNase, inducing DNA at an extremely low concentration (50 pg/mL), leading to both single-strand and double-strand breaks.25 CdtB also possesses lipid phosphatase activity, converting PI3,4,5,P3 to PI,4,5P2.22,28,29 This leads to the blockade of PI-3K signaling, which is required for an extremely diverse array of cellular functions, most notably cellular proliferation and survival and frequently dysregulated in many cancers.22,30 These biochemical characteristics of this toxin are indicative of its potential involvement in human cancers.
2.2. Microbiological characteristics of CDT-producing bacteria
CDTs are produced by a small but diverse group of bacterial pathogens that primarily belong to gamma and epsilon Proteobacteria.28 To date, CDT have been documented in Aggregatibacter actinomycetemcomitans (Aa), Campylobacter coli/fetus/hyointestinalis/jejuni (Cj)/lari/upsaliensis, Escherichia albertii/coli (Ea/Ec), Haemophilus ducreyi (Hd), Helicobacter canis/cinaedi/fennelliae/hepaticus/pullorum/winghamensis, Salmonella enterica (Se) and Shigella dysenteriae/boydii,5,25,28,31 which were isolated from human samples. However, it is important to note that this toxin is not necessarily produced by all subspecies/strains that belong to these specific bacterial groups. The higher carriage rates (>80%) have been reported for Aa, Hd, Se and enterohepatic Campylobacter and Helicobacter species, while it is generally low in Ec and Shigella.28,31–33
In nearly all bacteria, CDTs are encoded by the cdtABC gene cluster, a constitutively expressed operon on the chromosome, which yields molecular masses of 23–30, 28–29 and 19–21 kDa of CdtA, CdtB and CdtC subunits, respectively, according to bacterial species.28 Among these three genes, cdtB is the most conserved with homology across species of 50% or higher, while that of cdtA and cdtC is generally less than 30%.22,31,34 Nonsynonymous mutation in any conserved residue that is critical for the catalytic activity or the Mg2+ binding abolishes the ability of CdtB to cleave DNA.22,23,35 Single nucleotide polymorphisms at amino acid 281 (H/R) in Aa cdtB and at 95 (P/S) in Cj cdtB have been shown to change its biological activity by several log orders.36,37 Strains carrying the high activity allele were reported to be dominant in clinical isolates.36,37 A notable exception for the structure of this gene cluster is Se, which is an intracellular pathogen and expresses CDT only after bacterial internalization into host cells.22,38 Se does not carry any homologous genes to cdtA and cdtC, but possesses two other genes called, pltA and pltb (persussis-like toxin A and B), located upstream of cdtB in the same pathogenicity island, the products of which form with CdtB a tripartite.39,40 These two subunits have been projected to facilitate the transport of CdtB from its production site within infected host cells to extracellular medium, from where CdtB can intoxicate other non-infected cells.22,39
Each of the translated CdtA, CdtB and CdtC subunits is independently translocated across the cytoplasmic membrane, through a standard mechanism for the movement of secreted proteins across the cytoplasmic membrane of bacteria, where precursor proteins are exported by a common sec-dependent pathway that involves both signal peptidase I and signal peptidase II.41,42 The CdtB and CdtC proteins processed by signal peptidase I and the CdtA processed by signal peptidase II accumulate in the periplasmic space and all three proteins self-assemble at the outer membrane. Catalyzed by an unidentified processing enzyme to remove the hydrophobic glycerolipid of CdtA, the holotoxin is translocated across the outer membrane.41,42 The hydrophilic holotoxin accumulates in the aqueous environment outside the bacterium until it recognizes a specific receptor on the target cell surface.
An alternative mode of bacterial toxin secretion is the release of holotoxins into enclosed outer membrane vesicles (OMVs). Bacterial OMVs are nano-sized compartments consisting of a lipid bilayer that encapsulates periplasm-derived, luminal content. OMVs produced by of Gram-negative bacteria are now recognized as a generalized secretion pathway that provides a means to transfer cargo to the host cells.43 In the context of intestinal colonization, packaging and release of CDT into outer membrane vesicles may provide the toxin with extra protection against enzymatic digestion, thus increasing the likelihood of the uptake of intact toxin by host cells. Although a mechanism of delivering the toxin from the vesicles to the target cell has not been well understood, several potential routes, including clathrin or caveolin mediated or non-mediated endocytosis and membrane fusion, have been demonstrated.43 OMVs-like vesicles have been also found to be synthesized by internalized Se, transported and released to the extracellular medium through host microtubule and actin tracks.44
2.3. In vitro cellular responses to CDTs
Biological consequences from CDT intoxication in mammalian cells have been evaluated in vitro using various normal and tumor cell lines that are exposed to bacterial lysates or purified toxins or are transfected with cdtB or CdtB, of different bacterial origins. DNA damage caused by CDT has been shown to induce the activation of DNA damage response (DDR), leading to an ATM (Ataxia telangiectasia mutated)-dependent cell cycle arrest at G2/M and/or G1/S transition, and initiation of multiple DNA repair pathways. However, the DDR system sometimes fails to properly repair DNA damage, leading to cell death by apoptosis or to a long-term cell cycle arrest (senescence).25 This cellular senescence may alternatively lead to a senescence-associated secretory phenotype (SASP), which is characterized by secretion of a large number of growth factors and pro-inflammatory cytokines and can promote survival and proliferation of transformed cells.45
Cell fate following CDT-induced DNA damage depends on types of cells.46,47 Hematopoietic lineages, particularly lymphocyte, are known to be greatly more sensitive to CDT than most other cells, inducing rapid move toward apoptosis after a brief cell cycle arrest.29,46 Accordingly, CDT is considered an immunotoxin. It is capable of inhibiting phagocytosis in murine macrophages48 as well of inhibiting functions of both T- and B-cells.29,46 Furthermore, CDT has other immunomodulatory properties as it has been documented to induce the expression of pro-inflammatory cytokines/chemokines (such as IL-1β, IL-6, IL-8) in human macrophages and peripheral mononuclear cells.25,49,50
Epithelial and mesenchymal lineages mainly undergo cell cycle arrest accompanied by cytoplasmic elongation and distension due to accumulation of F-actin assemblies.51 DNA double-strand breaks induced by HdCDT led to the formation of actin stress fibers and the rearrangement of the actin cytoskeleton in epithelial cells and fibroblasts, via activation of the small GTPase RhoA. Activation of RhoA was part of the ATM-induced cellular responses to genotoxic stresses to support cell survival.52 Moreover, in colorectal cancer cell lines, exposure to Hd CDT stimulated a marker of autophagy, a pro-survival pathway, in an ATM and p53-dependent manner.53 CDT also induced remodeling of adherens junctions in gingival tissue and normal epithelial cells and compromised mucosal barrier function, which was accompanied by a pronounced increase in the expression and cytosolic distribution of E-cadherin and β-catenin.54
Investigators have demonstrated in both fibroblast and normal and cancer cell lines the enhanced senescence-associated β-galactosidase activity, expansion of promyelocytic leukemia nuclear compartments and induced expression of several cytokines (especially interleukins IL-6, IL-8 and IL-24), which were overall features shared by cells undergoing replicative or premature cellular senescence.55 Furthermore, a long-term (30 weeks) of exposure of fibroblast and intestinal epithelial cells to Hd CDT led to increased frequency of mutations (mainly transversions), accumulation of chromosomal aberrations and enhanced anchorage-independent growth.56 These observations suggest that CDT-induced DNA damage promotes genomic instability, favoring tumor initiation and progression.57 Graillot et al. postulate that CDT does not initiate colorectal cancer by itself, but may have promoting effects in premalignant epithelial cells. They found that chronic exposure (3 weeks) to Ec CDT resulted in enhanced anchorage-independent growth and genetic instability measured by the micronucleus formation in colonic epithelial cells defective in APC and p53 in comparison to normal cells, which were considered hallmarks of malignant transformation.58 Frisan et al. also postulate that CDT may act synergistically with an oncovirus, specifically EBV. They found that exposure of gastric cancer cell lines to Aa CDT reactivated latent EBV infection and promoted latent infection in non-infected cells via genomic instability.59
2.4. CDT toxicity in animal models
Potential carcinogenic effects of CDTs in vivo have been evaluated in a few animal models. The vast majority of the studies were based on inoculation of murine enterohepatic Helicobacter species, either H. hepaticus or H. bilis, while a minority used enteric Cj. In these studies, natural or engineered CDT-deficient strains were often used for comparison.60,61 In both liver and colon carcinogenesis, a progressive pathological sequence from chronic active inflammation (hepatitis/colitis), dysplasia, benign tumor to cancer has been acknowledged.60,61 Hh inoculation to A/J, C57BL/6 and their F1 mice induced hepatitis, some of which further progressed to hepatocellular carcinomas or dysplastic liver lesions, in a gender-specific manner, resulting in 69% incidence in F1 male mice. Hh was isolated from hepatic tissues of all F1 mice with liver tumors, which were developed in the presence of hepatic steatosis and active hepatitis.62 Likewise, perinatal intraperitoneal inoculation of Hh to A/J female mice resulted in increased incidence of hepatitis and hepatocellular tumors in their male (not female) offspring.63 However, these experiments did not rule out the effects of other virulence factors co-expressed in CDT-positive Hh. Thus, others have tried to address the independent effects of CDT. While infection of A/JCr mice with wild-type (WT) Hh or an isogenic mutant lacking CDT activity induced comparable chronic hepatitis, the CDT mutant-infected mice did not develop any hepatic dysplastic nodules in contrast to the WT-infected male (not female) mice that developed dysplasia. Male mice infected with WT also exhibited significantly enhanced hepatic transcription of pro-inflammatory TNF-α, IFN-γ and Cox-2, growth mediators IL-6 and TGF-α, anti-apoptotic Bcl-2 and Bcl-XL, and increased hepatocyte proliferation compared with the CDT mutant-infected male mice, confirming that CDT plays a key role in promoting the dysplastic changes in the Hh-infected mouse livers.64 Fox et al. further investigated the potential interactions of Hh with chemical or virial liver carcinogens. In both models, Hh intestinal colonization was sufficient to promote the development of hepatocellular carcinoma (HCC), via transactivation of the nuclear factor kappa N (NF-κB) network in the liver. Thus bacterial translocation to the liver was not a requisite and, in the presence of a chemical carcinogen, aflatoxin, both male female mice equally developed HCC.65 Tumor promoting effects of Hh on hepatitis C virus transgenic mice shown in this study also suggest potential bacterial involvement in viral activation as in the case of Aa CDT and EBV.59
Orogastric administration of culture supernatants of CDT-producing isolates to suckling mice induced overt inflammation in stomach, small intestine and large intestine, which was not seen in the experiments with CDT-negative Cj strains,66 demonstrating that CDT plays a crucial role in gastrointestinal pathology in this model. In NF-κB-deficient mice, inoculation of the cdtB mutant strain did not induce gastroenteritis and gastric hyperplasia/dysplasia comparable to that developed in the WT-infected mice.67 Furthermore, inoculation of CDT-negative Hh mutant showed a significantly diminished capacity to induce colitis lesions in an interleukin-10 deficient mouse model for inflammatory bowel disease.68 Shen et al. tested mutant and WT CDT from zoonotic H. cinaedi using the same mouse model. Both mutant and WT colonized to a similar extent, but the mice infected with the mutant showed attenuated colitis and hyperplasia but no dysplasia or intra-mucosal carcinoma, compared to those infected with WT, which exhibited elevated mRNA expression of tumor necrosis factor alpha, inducible nitric oxide synthase, and gamma interferon in the cecum.69 Another newly found CDT-positive H. japonicum induced the histopathology characterized by moderate to severe inflammation, mild edema, epithelial defects, mild to severe hyperplasia, dysplasia and carcinoma in the lower bowel of the same mouse model.70 In an immunocompromised mouse model, inoculation of CDT-deficient Hh mutant led to less severe cecal pathology without inducing pre-neoplastic lesions, while WT-infected animals developed pre-neoplastic dysplasia and cancer, with concomitant upregulation of cecal Il-6 and Tnfα.71 In a mouse xenograft model with Hh cdtB-transfected human hepatic and intestinal cancer cell lines, both types of the CdtB-derived tumors showed increased apoptosis, senescence, p21 and Ki-67 nuclear antigen expression, an overexpression of cytokeratins at the invasive front of the tumor and an increase in ploidy, while no difference in proliferating cells undergoing mitosis. All these features were considered hallmarks of endoreplication and aggressiveness in cancer.72 More recently, oral infection of the ApcMin/+ mice with CDT-producing Cj together with dextran sulfate sodium administration has been reported to induce significantly larger number and size of tumors when compared with a ctdB mutant Cj. WT Cj infection also induced expression of hundreds of colonic genes, with 22 genes dependent on the presence of cdtB, as well as altered microbial gene expression, which could also modulate colorectal carcinogenesis.73
2.5. Clinical and epidemiological studies concerning CDT
The direct evidence to link CDTs or CDT-positive bacteria to human cancer are still limited. To our knowledge, the presence of CDT itself in human tumor tissue or in surrounding mucosa has not been demonstrated. Alternatively, CDT-positive (by PCR) Ec has been reported to be more commonly present in colorectal cancer tissue than in colorectal mucosa from diverticulosis patients (16% vs 0%).33 However, these CDT-positive strains also harbored other known virulence genes and thus it was difficult to disentangle the effect of CDT itself. Another routinely used method to assess the past exposure is serological assays to detect specific antibodies. Although anti-CDT antibody assays have been available, demonstrating increased titers in diarrhea type of irritable bowel syndrome (IRS) compared with control subjects,74,75 their association with cancer or premalignant pathologies has not been studied. More consistent evidence from human populations has been complied with regard to hepatobiliary tract cancers. It should be noted that these studies evaluated the exposure to bacterial species with known high carriage rates of CDT, without confirmation of CDT status. The best studied is gallbladder cancer and its predisposing condition, cholelithiasis, in relation to infection with Se serovar Typhi or Hh. Se serovar Typhi status has been most often assessed by antibodies against a capsular polysaccharide (Vi). The summary odds ratio (OR) from a meta-analysis based on such serological studies was 4.6, consistent with the summary ORs based on stool or bile culture (4.7 and 5.5, respectively). A recent study in India also reported an equivalent difference in serology using the same assay between cancer and benign conditions, but more positive samples (74%) were detected using PCR for S. typhi fliC genes, whereas PCR-positivity in healthy gallbladder samples was very rare.76 Serological assays against a Hh antigen have revealed a significantly higher antibody prevalence in bile duct/gallbladder cancer patients (39%) and even higher prevalence in patients with viral hepatitis (60%) or viral liver cirrhosis (68%), compared with cholelithiasis (13%) or healthy donors (27%), respectively.77,78 Given low antibody prevalence in non-viral hepatitis and cirrhosis reported at the same time,78 these findings are also be indicative of bacterial-viral interaction. Alternatively, using PCR, in situ hybridization for16S rRNA and Western blot for Hh antigens Hamada et al. reported high Hh carriage rates (40%) in cholelithiasis patients with or without gastric cancer, compared to the patients with other benign conditions (13%).79
3. Dentilisin (chymotrypsin-like protease (CTLP))
3.1. Biochemical properties of CTLP
Dentilisin belongs to only two known lipoproteins in the subtilase family of subtilisin-like serine proteases. Its activity is suggested to be based on an active seryl residue, on an active imidazole group, and on an active carboxyl group but not on metal cations.80 The enzyme hydrolyzes specifically N-succinyl-L-Ala-L-Ala-L-Pro-L-Phe-p-nitroaniline (SAAPFNA, a typical chymotrypsin substrate) and various host proteins and peptides, including basement membrane components (type IV collagen, laminin, and fibronectin), serum proteins (transferrin, fibrinogen, IgG, IgA, and α1-antitrypsin), and bioactive peptides (cytokines, and chemokines).81,82 These biochemical properties are considered to be instrumental in bacterial invasion, immune evasion and cytotoxicity.81,82 The matured dentilisin is present as a complex, composed of an active enzyme, PrtP (~65 kDa), and two auxiliary proteins, PrcA (~70 kDa) and PrcB (~22 kDa).83 The two auxiliary proteins are postulated to be required for stabilization of the active enzyme unit.84,85 PrtP protease activity facilitates maturation of PrtP and PrcA, by releasing a 16 kDa N-terminal of the premature PrtP and cleaving PrcA to A1 (~30 kDa) and A2 (~40 kDa).85,86
3.2. Microbiological characteristics of dentilisin synthesis
Dentilisin is produced primarily by mammalian oral Treponema species,83,87,88 represented by T. denticola (Td), a key pathogen of periodontitis, which are Gram-negative obligate anaerobic bacteria, spirochaetes uniquely lacking LPS. It is important to note that substantial inter-strain variability exist in this enzymatic activity, which is not entirely explained by the presence or sequence variabilities of encoding genes.83,87,88 Typically, the dentilisin complex is encoded by a three-gene operon, consisting of prcB, prcA and prtP, with a putative promoter upstream of prcB.85,86 These gene sequences are recently annotated,83 demonstrating that prcB is the most conserved (89% out of 202 amino acids identical among the strains). prcA encoding 639 amino acids was 77% identical among the strains, and much of the variability was in three regions (151–180, 328–362 and 512–545). In prtP that transcribes 766 amino acids, the predicted catalytic triad (Asp203, His258, Ser447) was conserved in all strains, and most prtP inter-strain variations were concentrated in the C-terminal 270 residues (up to 20%).83 However, variability in proteolytic activities was not correlated with overall homology of prtP, suggesting that other unidentified sensing and signaling mechanisms modulate expression of these operons.
In Td, the dentilisin complex is located on bacterial outer membrane and extracellularly.89 However, the secretion system by which the three subunits are transported through inner and outer membranes has not been well understood, Yet, studies have demonstrated that this complex is co-localized with another major Td virulence factor, Msp (major outer sheath protein),83,85 encoded by msp and playing a major role in host adhesion.90 MSP locates within the outer membrane, but is predominantly periplasmic, with only limited surface exposure.91 Importantly, there is close interplay between PrtP and outer membrane segment of Msp,92 and, as a result, msp mutants lack CTLP activity while prtP mutants show aberrant Msp expression.93 In addition to cell surface expression, a recent mass-spectrometry-based study revealed that the three dentilisin subunits were enriched in outer membrane vesicles,89 which enable remote delivery of the toxin.
3.3. Dentilisin in vitro activities
Cytopathic effects of isolated dentilisin, including membrane blebbing, vacuolization, inhibition of motility, loss of epithelial cell contacts and release of a cytosolic enzyme have been demonstrated in multi-layer epithelial cell culture system.94,95 In this culture model, Td did not invade the cells of epithelial multilayers, but evidently shed dentilisin, which rapidly penetrated the cell layers, was transported into large intracellular vacuoles and led to increased epithelial permeability.94 Using reconstructed basement membrane (Matrigel),96 Grenier et al. also reported the migration of Td through the basement membrane, which was facilitated by protease activity of dentilisin to degrade the basement membrane components. Other investigators employed dentilisin mutant strains and confirmed that dentilisin was responsible for the increase in epithelial permeability and also found an evidence that dentilisin facilitated degradation of the tight junction protein, ZO-1.97,98
In addition, higher production from polymorphonuclear leukocytes (PMNs) was observed in a dentilisin-positive wild-type strain in comparison to a dentilisin-negative mutant, suggesting another possible mechanism mediating the cytotoxicity of this enzyme.99 The production was medicated through C3 complement activation, which was associated with matrix metalloproteinase (MMP) 9 release from PMNs. On the other hand, gingival epithelial cells co-cultured with wild-type or dentilisin mutant Td showed dentilisin-dependent preferential degradation of tumor necrosis factor α, which resulted in substantial reduction in both IL-8 protein and mRNA levels.100 These results were indicative of host immune evasion.
An additional potentially oncogenic property of dentilisin is its ability to induce MMP-2, a key enzyme both in tissue homeostasis and tissue destruction, which has been linked to tumor invasion, progression as well as tumorigenesis.101–103 In periodontal ligament cell (PDL) culture, purified dentilisin protease induced MMP-2 activation, mediated through fragmentation of fibronectin in extracellular matrix.104 Subsequently, it was found that Td plays a key role in the transcriptional regulation of MMP-2 and its activating complex MT1-MMP/TIMP-2 through downregulation of several chromatin modification enzymes, specifically histone phosphorylases (aurora kinases) and histone deacetylase. Inhibition of these enzymes that mediated epigenetic modifications prevented Td-mediated increases in MMP-2, MT1-MMP, and TIMP-2 in PDL cells.105 While specific mechanisms by which dentilisin modulates the expression of these chromatin modification enzymes remain to be studied, this has substantial implication in transcriptional control of a wide range of tumor suppressor and oncogenes. Finally it is intriguing that a low dose exposure to Td (5×107 cells/mL) increased DNA synthesis (3H-thymidine uptake) via activation of MAP kinase signal pathways in PDL cells, while infection at higher concentrations reduced DNA synthesis, exerting cytotoxic effects. Western blot analysis showed that Td strongly but transiently activated ERK1 and ERK2, signals mediating cell proliferation, and JNK and p38, kinases mediating apoptosis.106 These results underscore the ability of Td to modulate cell proliferation and cell survival.
3.4. Dentilisin activities in animal models and clinical studies
To our knowledge, there have been no animal carcinogenesis models using dentilisin itself or Td inoculation, as the primary endpoint in animal models have been periodontal disease. However the effects of wild-type and dentilisin mutant strains have been tested in a mouse subcutaneous abscess model, which showed a significantly reduction in size of the lesion (abscess) with the mutant.107 Despite the lack of in vivo data in animals, several recent clinical studies suggest potential carcinogenic effects of dentilisin in humans. IgG antibodies against CTLP have been shown to be detectable in human sera,108 but no seroepidemiological studies have examined the associations between orodigestive cancers and Td or dentilisin. In saliva of periodontitis patients and healthy controls, the concentration of Td was correlated with activated forms of MMP-8, which was cleaved from its inactive precursor form and which exerts both tumorigenic and anti-tumorigenic effects.109 In 2004, based on 16S rRNA gene sequencing, Narikiyo et al. reported that Td was significantly enriched in esophageal cancer tissue compared with adjacent normal mucosa and it was absent in saliva from healthy volunteers.110 More recently, Nieminen et al. demonstrated using immunehitochemical staining the presence of CTLP in several types of orodigestive cancer tissue. CTLP was positive in 19 out of 29 tongue, 20 out of 25 tonsillar, 3 out of 3 esophageal, 21 out of 32 gastric, 6 out of 6 pancreatic and 25 out of 54 colon cancer cases.111 The investigators also reported that CTLP degraded several MMP inhibitors, including TIMP-1, TIMP-2, hypothesizing that CTLP may contribute to carcinogenesis through immunomodulation.111 Subsequently, the same group of investigators analyzed more tissue samples of mobile tongue squamous cell carcinoma. CTLP was present in 95% out of 141 tumors, of which many (40.4%) showed high immunopositivity. CTLP positivity was significantly associated with invasion depth, tumor size and the expression of TLR-7, TLR-9 and c-Myc. Furthermore high CTLP immunopositivity in younger patients (≤60 years old) predicted early relapse.112 Kylmä et al. also evaluated the presence of CTLP by immunohistochemistry in a series of 201 unselected consecutive oropharyngeal squamous cell carcinoma patients and reported 81% overall positivity, which was significantly more pronounced in HPV-negative than positive tumors. Among those HPV-negative, higher TLR 5 and lower TLR 7 expression associated with high CTLP expression, which resulted in poor disease-specific survival. No similar association emerged among HPV-positive subgroup.113 In a prospective cohort study that characterized microbial compositions of oral rinse samples of cancer-free individuals, the risk of subsequent colorectal cancer statistically significantly increased in those who were Td-positive, yielding the OR of 1.8, which was highest among ORs for all other periodontal pathogens.114 In order to substantiate oncogenic potentials of Td-CTLP, intrigued by the recent clinical observations, more mechanistic studies are warranted to delineate specific molecular pathways.
4. Pasteurella multocida toxin (PMT)
4.1. Biochemical properties of PMT
PMT belongs to a large group of deamidating toxins/effectors, deamidases, which remove the amide functional group from a key glutamine residue of the protein substrate and provoke significant pathophysiological changes of target cells. Other bacterial toxins structurally and functionally related to PMT include the cytotoxic necrotizing factors from E. coli and Yersinia pseudotuberculosis and the dermonecrotic toxin from Bordetella spp.115 PMT selectively targets the α-subunit of host heterotrimeric G proteins and stimulate its substrates, Gαq, Gα13 and the Gαi-family proteins, leading to activation of various host signal transduction pathways.116,117 As a result, it acts as a highly potent mitogen at picomolar concentrations that stimulates quiescent cells to grow and divide.118,119
PMT is an AB toxin, consisting of a receptor binding and translocation domain (B) and a biologically active (A) domain. This AB toxin binds to host cell receptors through their binding B domains and facilitate the cellular uptake and delivery (translocation) of their toxic activity A domains into the host cell cytosol, where the A domains then interact with and modify their cellular G protein targets to cause cellular toxicity.120 PMT consists of 1285 amino acid residues resulting in a mass of 146 kDa. The receptor binding domain is located in the N-terminal part of the protein, including the amino acid residues 1–580. Within this domain a putative translocation domain is located between residues 402–457 in the C-terminus.116
The C-terminal part of PMT further contains three domains designated C1, C2 and C3. Of these, the C3 is the major biologically active domain which harbors the catalytic activity of the toxin to modify intracellular targets. The C1-domain (feet), encompassing amino acid residues 575–719, consists of seven helices, four of which serve as a plasma membrane targeting signal in toxin B. Because the primary target proteins of PMT are plasma membrane-bound heterotrimeric G proteins, dysfunctional C1 impairs localization to the substrate and diminishes PMT toxicity. The largest domain in the C-terminal part of PMT is the so-called body or C2 domain (amino acid residues 720–1104), consisting of 18 helices and nine beta-strands and exhibit a structure typical of nucleotide-binding proteins. This C3 domain (amino acid residues 1105–1285) is separated into two subdomains, providing the catalytic cleft for the enzymatic function of the toxin. It has been further clarified that Cys-1165, His-1205, His-1223, Asp-1220 and Gln-1225 are essential for PMT activity.116
4.2. Microbiological characteristics of PMT
PMT is produced by toxinogenic strains of Pasteurella multocida, mostly serogroups D and some A, which are classified based on capsular polysaccharides. These strains contain a unique 18-kbp region carrying 14 genes (PMCN06_2106 to PMCN06_2119), including the toxA gene for PMT flanked with several phage-related genes. Pm is a small, pleomorphic, Gram-negative, nonflagellated facultatively anaerobic coccobacillus and a multihost animal, zoonotic and opportunistic pathogen that is capable of causing respiratory and multisystemic diseases, bacteremia, and bite wound infections, while it is part of the normal flora in many animals.121 The sequence analysis of toxA, indicating lower GC content than the rest of the genome, suggests its acquisition through horizontal transmission. Further sequence analysis has revealed that PMT is encoded within a lysogenic bacteriophage and that phage regulatory elements are present upstream and downstream of toxA, which control toxA expression.122
Specific secretion systems that translocate PMT from cytoplasm through bacterial external membrane surface have not been identified in Pm. Instead, it has been speculated that stress response induced by environmental factors encountered during host infections leads to induction of the phage lytic cycle, with resultant bacterial cell lysis and toxin release.122 Knowledge of the uptake of PMT into eukaryotic cells has been still limited. Three major steps are generally acknowledged: (i) Binding of the toxin to a host cell membrane receptor; (ii) Internalization by endocytosis; and (iii) Release of the biologically active toxin into the host cytosol. To date specific host PMT receptors have not been well characterized. However, surface plasmon resonance analysis indicates that PMT initially binds with low affinity to a wide range of abundant membrane lipid components, which is followed by a more specific binding to sphingomyelin and possibly additional putative proteinaceous receptors, which would induce endocytosis.116,120 Further trafficking within host cells is facilitated by interaction with transferrin receptors and a small regulatory G protein Art6.116,120 It has been also clarified that PMT N-terminal mediates cytosolic delivery of its native C-terminal cargo as a single polypeptide (C1-C2-C3), without cleavage between subdomains.123 However, cellular uptake process may be cell-type specific. While no endocytic uptake was detectable in enterocytes, mast cells in the lamina propria distinctly accumulated PMT in their secretory granules.124
4.3. Cellular activities of PMT
The incubation of mammalian cells with purified recombinant PMT induces strong mitogenic and anti-apoptotic effects in the cell-type specific manner. Myogenic effects have been reported in fibroblasts, preadipocytes, osteoblasts and embryonic kidney cells, while cytopathic responses have been noted in lung cells and cardiomyocytes.115 Mitogenic responses are mediated through increased intracellular Ca2+ and inositol phosphate levels as a result of activation of phospholipase Cβ (PLCβ) and Rho-dependent cytoskeletal signaling.115 PMT induces Ras-dependent activation of extracellular signal-regulated serine/threonine protein kinase125 mitogen-activated protein kinase (MAPK) via Gq-dependent, protein kinase C (PKC)-independent transactivation of the epidermal growth factor receptor in a human embryonic kidney cell line.118,126 The MAPK pathway, also known as the RAS-RAF-MEK-ERK cascade, is often activated in many human cancers.127 Additionally, PMT has been shown in fibroblast culture to stimulate the mammalian target of rapamycin complex 1, which is also known to be often activated in human cancer,128 through the Gαq/11/PLCβ/PKC pathway.129 PMT induces anchorage-independent growth in rat fibroblast culture with greater potency than that achieved by epidermal growth factor or platelet-derived growth factor.119 PMT also activates anti-apoptotic pathways, e.g., protein kinase D signaling in both cardiac fibroblasts and cardiomyocytes, which leads to the phosphorylation of the transcription factor, cAMP response element binding protein (CREB), and upregulation of CREB target genes, including the anti-apoptotic Bcl-2 protein.130,131 These cells exposed to PMT also exhibit increased cell migration capacity,130,131 which may be instrumental for tumor progression. Additional evidence pointing to the oncogenic potential of PMT is the finding that PMT stimulates Janus kinases (JAK)132/signal transducer and activator of transcription (STAT) signaling in embryonic kidney cells and fibroblasts,133,134 through Gq-dependent phosphorylation and activation of the JAK1 and JAK2 and increased expression of proto-oncogene serine/threonine kinase Pim-1. This is followed by activation of STAT1, STAT3 and STAT5 transcriptional factors, the upregulation of cyclooxygenase 2, a pro-inflammatory protein that is upregulated in many cancers and downregulation of the transcription factor suppressor of cytokine signaling-3 (SOCS-3).115 Similarly to NF-κB, it has been acknowledged that STAT3 can act as a non-classical oncogene and plays a role in inflammation-associated cancer.135 Through the STAT activation, PMT exposure in vitro induces proliferation of native T cells and the differentiation of Th17 cells/IL-17 production,136,137 which is linked to excessive inflammation such as inflammatory bowel disease. PMT also initiates cytoskeletal rearrangements, including focal adhesion assembly and actin stress fiber development in the RhoA-dependent manner,115 which may underlie increased motility and tumor-aggressive behavior.138 In summary, these in vitro data support PMT’s ability to manipulate host signaling pathways just like oncoviruses.
4.4. Animal, epidemiological and clinical data of PMT and Pm
There have been no animal carcinogenesis models using PMT to our knowledge. However, a limited number of experimental animal studies for PMT-induced atrophic rhinitis have reported the development of epithelial squamous metaplasia in some of the PMT-injected animals.139,140 This seems compatible to H. pylori-induced gastric carcinogenesis where atrophic gastritis and intestinal metaplasia precede gastric cancer.141,142
Likewise, a few studies have reported carriage of Pm in human populations using serology or oropharyngeal swab culture. Serological studies were designed to target capsular antigen types A and D or non-specific non-PMT somatic antigens. These studies have revealed that Pm carriage is quite common in people routinely exposed to animals, such as meat industry workers and pet owners and the highest rate was found in pig breeders (~77%, serology and culture combined).143–145 Nevertheless, information concerning prevalence of toxigenic Pm has been limited, but it is estimated to be very low in human populations (<10%).146 Because toxigenic activities were tested biochemically in earlier studies, it was not clear whether strains negative to PMT did not carry the toxA gene or whether its expression was transcriptionally repressed. There have also been case reports of Pm infection diagnosed in various types of cancer patients.147,148 Yet, as Pm was primarily isolated from non-cancerous infectious lesions or blood, it is difficult to infer any causal associations from these reports.
5. Fusobacterium nucleatum toxins
5.1. Fusobacterium nucleatum background
The oral microbiota contains one of the highest diversities of bacteria in the human body. Many of these bacteria are believed to contribute to the development of cancer. Fusobacterium nucleatum (F. nucleatum) is one such bacteria and is considered an oncobacterium.149 F. nucleatum is a Gram-negative anaerobic bacteria that is found primarily in the oral mucosa under normal conditions. However, F. nucleatum has been implicated in several disease states including colorectal cancer.150 F. nucleatum is more abundantly found in diseased lesions.150 Multiple studies have shown that F. nucleatum is found to be present in potentially high numbers in cancerous tissues.149 There has been a renewed interest in studying the pathogenicity of F. nucleatum given its prominent placement in diseased states. F. nucleatum featured prominently in a review of the bacteria associated with the oral microbiome and their potential contribution to systemic disease and cancer.16 Two F. nucleatum exotoxins with potential links to carcinogenesis are FadA and Fap2. In this review we will focus on the FadA and Fap2 adhesins which may play significant roles in tumor formation and cancer.
5.2. Fusobacterium adhesin A (FadA)
5.2.1. FadA molecular and chemical characteristics
Fusobacterium adhesin A (FadA) was discovered and found to be conserved among Fusobacterium genera that inhabit the oral mucosa and is important for cell binding.151 FadA is a 129 amino acid protein with an 18 amino acid signaling peptide.151 It has a secreted form that was shown to cause an upregulation of the β-catenin pathway and Wnt gene expression, both important developmental pathways that when dysregulated are leading causes of carcinogenesis.152 The crystal structure of FadA revealed a unique “leucine chain” structure which, when mutated, abrogated the binding of host cells.153 Although they did not identify the receptor binding site for FadA they suggest that FadA functions as a filament and therapeutic targets in this region might be suitable for future clinical importance.153
5.2.2. Microbial characteristics of FadA
FadA is coded by the fadA gene and shown to be expressed on the cell surface.151,153 FadA contains a signaling peptide that would be used for its secretory pathway out of the cell and studies have shown that it also has a secreted form.151
5.2.3. FadA in vitro studies
Colorectal cancer cell lines HCT116, DLD1, SW480 and HT29 exposed to wild-type F. nucleatum demonstrated proliferation but those exposed to deleted FadA mutants did not have the same result.152 FadA was shown to promote the activation of the β-catenin pathway and Wnt signaling seen by Western blotting suggesting a pathway that potentially contributes to carcinogenesis.152 When F. nucleatum is introduced to E-cadherin expressing CRC cells there is an increase of cell proliferation but not in CRC cells that do not express E-cadherin indicating that FadA promotes tumor growth.152 Thus, FadA acts like a ligand of E-cadherin and when it binds E-cadherin it reduces its tumor suppressor activity.152 Work by this same group showed that Annexin A1, a modulator of the Wnt/β-catenin molecular pathway, is also upregulated by FadA when acting on the E-cadherin molecular pathway.154 This upregulation enters into a positive feedback loop thereby promoting cancer progression by this microbe.154
FadA was also shown to stimulate inflammatory responses in these cells to the same extent as F. nucleatum pointing to its significance as an oncogenic microorganism.152 FadA might represent a pathway whereby F. nucleatum might attach and invade. When FadA was deleted it was shown that F. nucleatum was unable to attach and invade but FadA was shown to trigger a signaling cascade inside HCT116 cell lines promoting lymphoid enhancing factor expression.152 Collectively, these results suggest a strong contribution to colorectal cancer progression by FadA.
5.2.4. FadA activities in animal models
Both FadA and F. nucleatum were studied in vivo by subcutaneously injecting them into nude mice.152 When HCT116 cells with FadA were injected subcutaneously in nude mice this promoted growth of tumors.152 Tumor growth increased in nude mice 20% when compared to controls after treatment with FadA protein.152 Rubinstein et al. also reported positive correlation between fadA gene copy numbers and ANXA1 mRNA levels in a F. nucleatum-induced Apcmin/+ mouse model.154 Further studies in mice also conclusively demonstrated that F. nucleatum, resulted in increased tumor growth and enriched populations in cancer tissues.155
5.2.5. FadA clinical and epidemiological results
Expression of the FadA gene was statistically significantly higher in colorectal cancer tissues than in normal mucosa or adenoma tissues.152 Presence of FadA in colon tissue from patients with colorectal cancer showed that it was 10–100 times the normal level.152 However, we are just starting to understand the mechanistic roles that F. nucleatum may be contributing to the progression of cancer or if it is merely a “hitch hiker” found at tumor tissues. Rubinstein et al. also demonstrated co-localization of FadA and Annexin A1 proteins as well as positive correlation between fadA gene copy numbers and ANXA1 mRNA levels in human colon cancer tissue.154 This group suggests that Annexin A1 presents as a novel marker of cancer prognosis based on the results of disease-free survival of 466 CRC cancer patients.154 On the other hand, a recent study measured the antibody responses to 11 F. nucleatum antigens, including FadA, in pre-diagnostic serum samples of colorectal cancer patients and did not find that FadA was statistically significant as an indication of colorectal cancer risk.156
5.3. Fusobacterium apoptosis protein (Fap2)
5.3.1. Fap2 molecular and chemical characteristics
Fap2 was shown to have a predicted molecular mass of 389.8 kDa and a size of 11.3 kb.157 This corresponds to a 3692 amino acid protein with no cysteine residues.157 Using site-directed mutagenesis it was shown that Fap2 contains three domains: the C-terminal autotransporter domain, the central domain containing the repeats seen in other filamentous hemagglutinins and adhesins, and the N-terminal domain.158 The 3596th to 3606th amino acids have been identified to be an immunogenic epitope of the Fap2 protein.159 Using an ELISA-based analysis; the immunogenic region: “TELAYKHYFGT,” in the Fap2 protein was identified as the specific region.159
5.3.2. Microbial characteristics of Fap2
Fap2 was previously identified as a potential apoptosis inducing protein that was treated with proteases and heat which suggested that Fap2 (unnamed at that time) was probably a membrane-bound protein.160 Further studies by the same group elucidated that the gene product of fap2 codes for the large outer membrane protein with similar parallel β-helix structures and highly conserved C-terminal barrel autotransporter domains seen in other homologues.161 The fap2 gene codes for the membrane-bound Fap2 protein and was identified as an important binding and galactose sensitive adhesin that was not able to hemagglutinate.157
5.3.3. Fap2 in vitro studies
Fusobacterium apoptosis protein (Fap2) was initially isolated as a potentially important membrane-bound protein that could play a role in apoptosis.160 F. nucleatum was shown to activate apoptosis in human peripheral blood mononuclear cells (PBMCs) and polymorphonuclear cells (PMNs).160 More recently it was shown that Fap2 has the capacity to bind to galactose residues and fap2 mutants failed to coaggregate with human embryonic kidney92 293 T cells presumably due to lack of galactose binding.157 Fap2 is able to bind to TIGIT, an inhibitory receptor present on all human natural killer cells that is then able to abrogate the immune cell action against F. nucleatum.162 Human cell lines HCT116, RKO and HT29 were used to demonstrate the binding of F. nucleatum to colorectal cancer cells via Fap2, while the binding was absent with Fap2 deletion mutants.163 When colorectal cancer cells (CRC) were treated with O-glycanase, the binding of F. nucleatum and CRC cells was reduced.163 This could be as a result of loss of sugar residues Gal-Gal-NAc which are needed to bind the Fap2 lectin expressed by F. nucleatum. It was also shown that F. nucleatum strains that lacked Fap2 or had Fap2 mutant proteins showed reduced binding to Gal-Gal-NAc expressing CRC cells.163 The binding between Fap2 and Gal-Gal-NAc provides an important mechanism by which F. nucleatum is able to alter the tumor microenvironment to promote cancer. Given the capacity for binding to a widely present sugar residue, Fap2 presents an important target of studies aimed at uncovering its contribution to cancer.159,163
5.3.4. Fap2 activities in animal models
Use of Fap2 mutants that lack Fap2 show decreased colorectal cancer cells in mice.163 The mice model showed that established colorectal cancer tumors when inoculated with F. nucleatum had more colonization than with the Fap2 deletion mutant and F. nucleatum localized to mouse tumor tissues in a Fap2-dependent manner.163 Consistent with the observations in human tissue described below, Gal-GalNAc (measured using FITC-labeled PNA) was overexpressed in the mouse CRC sections compared with sections prepared from adjacent normal colon tissues.163
5.3.5. Fap2 clinical and epidemiological studies
Fap2 and Gal-GalNAc was shown to potentially give rise to the presence of F. nucleatum in cancer and its precursor lesions and metastasis in human.163 When Gal-GalNAc levels on healthy human colorectal tissues, human colonic adenomas, and human colorectal adenocarcinomas was assessed, its levels were significantly higher in adenocarcinomas compared with adenomas.163 Within the adenoma group, there were statistically significant trends by histology, i.e., the highest levels of Gal-GalNAc expression on villous adenomas, followed by tubulous villous adenomas.163
When 37 human patient colorectal cancer cases were tested immuno- and seroreactivity to Fap2 protein, investigators found a 100% seroreactivity with the colorectal cancer samples and a 32% seroreactivity with healthy controls.159 The Fap2 peptide mimotope proved to be immunogenic in both animal and human models of colorectal cancer giving some indication of its potential use as a biomarker for cancer progression. However, in a study using 11 F. nucleatum antigens (including Fap2 and FadA) in pre-diagnostic serum samples from 485 colorectal cancer patients the results indicated that these proteins did not implicate a statistically significant risk to contract colorectal cancer.156 The approach used differs from other studies in that they had access to pre-diagnostic serum samples, which were not taken before cancer diagnosis and matched to controls who did not develop colorectal cancer.156
5.4. FadA and Fap2 potential mechanistic roles in carcinogenesis
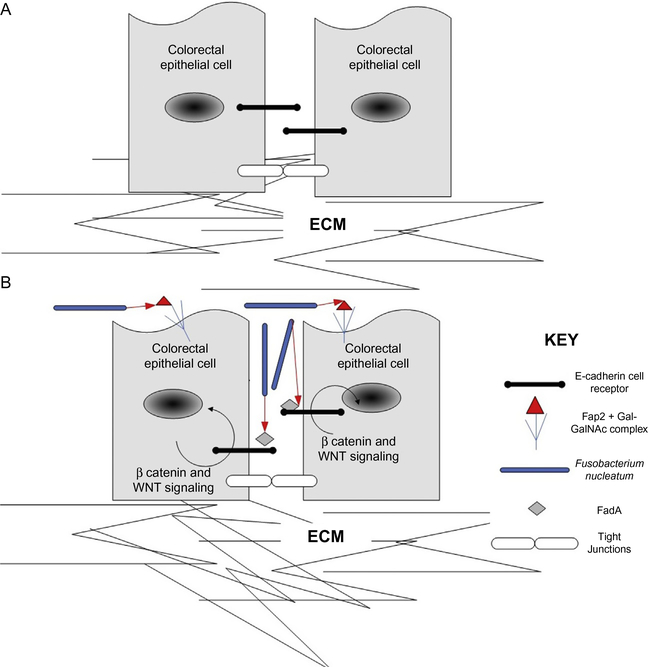
FadA and Fap2 are two adhesins produced by F. nucleatum that are of significant interest to cancer researchers. F. nucleatum has the ability to adhere to multiple cell types creating a platform for invasion and persistence of other cell types. F. nucleatum is considered an important “bridge” organism paving the way for early colonizers of the oral microbiome such as Streptococcus.132,149 In this regard F. nucleatum might be looked at as an ecosystem engineer, a term first coined by Jones.164 An ecosystem engineer has the ability to transform the environment to suit its need for survival. Studies looking at cancer formation has already implicated cancer cells as being ecosystem engineers.165 This understanding could help form future research decisions and possible medical interventions. The human oral microbiome contains a very diverse collection of bacteria in addition to other constituents such as viruses and fungi.16 The mouth has been described as an island with many biogeographical features which result in multiple unique ways for bacteria to colonize the different micro niches.132 These adaptations may include the ability for F. nucleatum to signal to other microbial and host cells (as in the case of FadA which acts as a ligand to E-cadherin-expressing colorectal cancer cell types) and adhere to other cells (as Fap2 can bind to sugar residues found on colorectal cancer cells). Under dysbiosis, bacteria, including their toxins, can make their way to other parts of the body and result in many diseased states.16 These same adaptations might serve to disrupt homeostasis leading to severe conditions such as cancer. The role of F. nucleatum in adhering to other cells and creating a stable habitat for other colonizers has already been shown but we are suggesting that it plays an additional role as an ecosystem engineer in altering the tumor environment to facilitate proliferation and colonization. In Fig. 1B F. nucleatum functions as a putative ecosystem engineer by signaling to other cell types such as colorectal cancer cells via FadA which in turns leads to proliferation and tumor growth. This would have significant impacts to the tumor microenvironment such as invasion and increase in size. F. nucleatum also has the ability to adhere to other cell types via Fap2 binding to sugar residues on the cell surface and this would result in an increased recruitment of F. nucleatum. This contributes to the modification of the tumor microenvironment thus implicating F. nucleatum as an ecosystem engineer.
Fig. 1.
F. nucleatum’s putative role as an ecosystem engineer. (A) Normal state without the presence of F. nucleatum. ECM: extracellular matrix. (B) The presence of rod shaped F. nucleatum releases FadA which triggers a signaling cascade in colorectal epithelial cells leading to proliferation.152,154 Fap2 binds to sugar residues on colorectal epithelial cells which leads to co-aggregation and increased recruitment of F. nucleatum thus modifying the tumor microenvironment and behaving as an ecosystem engineer.163
6. Bacteroides fragilis toxin (Bft)
6.1. Chemical and molecular characteristics of Bft
Bacteroides fragilis toxin (Bft) is a zinc-dependent metalloprotease toxin with three isotypes (BFT-1, BFT-2, and BFT-3).166 While this enterotoxin has been known for several decades, it was not until the early 1990s that Bft was purified and characterized extensively. At this point it was established through the use of SDS-PAGE that Bft had a molecular weight of approximately 19,000,167 a value similar to previous estimates that predicted a value of 19,500.168 Bft was further characterized to have an isoelectric point in the range of 4.4–4.6 and to be stable within a pH range of 5–10.167 Interestingly, Bft appears to be stable at temperatures ranging from −20 °C to 20 °C, but shows marked inactivation at higher temperatures. Specifically when incubated for 1 h at 55 °C over 90% of the toxin activity was lost, and when incubated for 1 h at 65 °C over 99% of the toxin activity was lost.167
At this time the enzyme susceptibility of Bft was also tested, revealing that the toxin is resistant to trypsin, chymotrypsin, dispase, bacterial amylase and bacterial lipase, yet sensitive to proteinase K and Streptomyces protease.167 Antiserum specific to the toxigenic strain of B. fragilis has been shown to neutralize the activity of the toxin, while exposure to antiserum specific to nontoxigenic strains does not produce the same effect.167 In addition to these physical characteristics, the molecular structure of Bft was also established at this time, with the first 20 amino acids from the N-terminal known to be Ala-Val-Pro-Ser-Glu-Pro-Lys-Thr-Val-Tyr-Val-Ile-Xxx-Leu-Arg-Glu-Asn-Gly-Ser-Thr.167
6.2. Microbial characteristics of Bft
Bacteroides Fragilis is an obligate anaerobe, Gram-negative bacteria that is a part of the normal biome of the human colon. Functionally, there are two distinct groups of Bacteroides fragilis. Strains that secrete a zinc-dependent metalloprotease toxin (Bft) which is encoded from the B. fragilis pathogenicity island are referred to as enterotoxigenic B. fragilis (ETBF). Strains that lack this ability are called nontoxigenic B. fragilis (NTBF).169 While Bacteroides fragilis normally comprises only 1–2% of colon flora,170 a value 10–100 times smaller than some other species of Bacteroides found in human intestines, it is the species most frequently isolated from clinical specimens, and as such is often viewed as the most virulent species of Bacteroides.171 B. fragilis is so clinically prevalent due to its propensity to cause abscesses, soft-tissue infections, and bacteremias.172
Moncrief et al. generated a Bft recombinant PCR product that, when cloned into pUC19 and sequenced was determined to be 538 nucleotides in length,173 providing insight into B. fragilis’ enterotoxin encoding gene. This PCR product, which they designated as rBF107, was further analyzed and found to contain sequence identity to a number of metalloproteases, including Pseudomonas aeruginosa alkaline protease, and matrix metalloproteases.173 Additionally, the sequence HELGHILGAEH, found in rBF107, exhibits consistency with the zinc-binding motif HEXXHXXGXXH, which is characteristic of the metzincin family.173 This discovery gave strong support to the theory that Bft is a zinc-dependent metalloprotease.
Kling et al. did additional analysis of the encoding gene for the enterotoxin belonging to B. fragilis. This gene, termed BftP was found to be 1191 nucleotides long and code for a 44.4 kDa protein that is comprised of 397 amino acids.174 BftP has a signal peptide 18 amino acids in length that is immediately succeeded by a protoxin that is 379 amino acids in length, while the protoxin found in stool samples begins at the 212th amino acid.174 Three potential AUG start codons have been identified in the promoter region of the B. fragilis enterotoxin, with the middle start codon being followed by the amino acid sequence typical of a signal peptide. The two additional start codons are located 24 nucleotides upstream and 25 nucleotides downstream relative to the middle start codon. BftP has a signal I peptidase cleavage site located between two alanine residues at amino acid 17 and lacks an obvious ribosome-binding site.174
B. fragilis does not appear to utilize types III, IV, autotransporter or two-partner secretion systems,175 yet does employ the type VI secretion system.176 More specifically, B. fragilis expresses the GA3 subtype of T6SS (type VI secretion system), which is not highly conserved among other bacterial species.177 The type VI secretion system exports the Bft exotoxin directly into eukaryotic host cells or other co-growing bacterial cells. This is accomplished through the T6 apparatus, a multiprotein, cell envelope spanning complex comprised of a core of Tss proteins.177 This complex is characterized by a needle-like structure that becomes inundated with toxins in the cytoplasm.178 The needle is then driven from the cell via contraction of the surrounding sheath in order to implant the toxins into extracellular matter.177 Additionally, B. fragilis also appears to use type I secretory system, as it was determined to have three Tolc-like proteins that are encoded adjacent to genes that have previously been shown to be associated with type I secretion systems.175
6.3. In vitro toxicity of Bft
In vitro experiments have helped shed light on the potential mechanisms of action used by Bft. On a biochemical level Bft has been shown to utilize its zinc-dependent protease activity to cleave the extracellular domain of E-cadherin, which is followed by further intracellular degradation.179 This is relevant because E-cadherin is not only the primary protein of zonula adherens, which function to maintain tight epithelial junctions, but also has tumor suppressor functions. Bft has been shown to provoke cell rounding, increase in volume, and effacement of microvilli and apical junctional complexes in intestinal and renal cells in culture.180 This supports the idea that cadherins are functionally involved. This is thought to lead to actin rearrangement, and consequently pro-inflammatory cytokines, which diminish epithelial barrier function.180 In addition, this leads to an increase in cellular proliferation mediated by elevated expression of the c-Myc oncogene.181 Interestingly, these consequences appear to be reversible, as cells appear normal 2–3 days after toxin treatment.180
Additional experiments have shown that exposure to 15 ng of Bft per mL resulted in formation of blebs on the cell surface and complete disappearance of stress fibers along with F-actin condensation at the cell periphery.182 These results were more pronounced at increased doses.182 Cell organelles (including mitochondria and nucleus) appear to be unaffected by exposure to Bft,182 as the toxin’s effects seem to be localized to the actin cytoskeleton and associated structures. Additionally, Bft has been shown to stimulate intestinal epithelial shedding, which is dependent, in part, on E-cadherin degradation and β-catenin–T cell-factor nuclear signaling183 and to stimulate proliferation and migration of human colon cancer cells in vitro.181
Incubation of human colorectal cancer cells with BFT led to upregulation of spermine oxidase, a polyamine catabolic, resulting in increased reactive oxygen species (ROS) and DNA damage measured by γ-H2A induction.184 Exposure to Bft has also been shown to induce changes in gene expression and chromatin availability in colonic epithelial cells.185 Eight genes have been identified in colonic epithelial cells that are significantly differentially expressed 24 h post-Bft exposure, but not 48 h post-exposure. It has also been demonstrated that while Bft does induce changes in chromatin accessibility in colonic epithelial cells, these changes are transient, and greatly decline by 48 h after Bft treatment.185 Specifically, there is a noted increase in chromatin accessibility at transcription factor binding sites, particularly those belonging to the AP-1/ATF family, many of which function downstream of mitogen-activated protein kinase (MAPK) pathways. Critically, while an increase in chromatin accessibility has been shown to have an association with differential DNA methylation and DNA mutation, these differences are not statistically significant.185
6.4. Toxicity of Bft in animal models
Mouse models have been used to demonstrate a link between Bft and colorectal cancer.186 Mice colonized by the enterotoxigenic B. fragilis experienced a rapid onset colitis followed by tumors of the distal colon.186 This was in contrast to mice exposed to nontoxigenic B. fragilis, which did not induce tumor formation at a higher rate than the control. Mice inoculated with the toxigenic strain developed observable tumors as early as 4 weeks after exposure, and microadenomas as early as 1 week after exposure.186 Bft also induced a prominent lamina propria intrusion of IL-17-producing CD4+T cells (Th17) and γδ-T cells with a dominant signal transducer activator of transcription-3 (STAT-3) pathway.186 The importance of IL-17 in the tumor-inducing response to Bft was demonstrated by the decrease in colonic tumors when the mice were treated with IL-17 blocking antibodies.186 IL-17 has been shown to induce tumor growth both in vitro and in vivo via an IL-6-stat3 signaling pathway.187 B. fragilis has also been found in increased quantity in the intestinal lumen of the 1,2-dimethylhydrazine (DMH)-induced rat colorectal cancer model.188
6.5. Clinical and epidemiological evidence to support in vivo toxicity in humans
In addition to animal models, there is now substantial indication that Bft is involved in the development of colorectal cancer in humans. In addition to being the most commonly isolated Bacteroides species found in clinical specimens,171 enterotoxigenic B. fragilis has been shown to have an increased prevalence in samples from cases involving colorectal cancer when compared to a control group.166 There has also been shown to be a difference in Bft detection depending on the stage in the course of the disease, with late-stage colorectal cancer patients having an increased rate of Bft detection in comparison to colorectal cancer patients at earlier stages of the disease.166 This observation becomes more pronounced as the disease progresses, to the point where Bft was identified in 100% of late-stage colorectal cancer patients in one study.166 It should be noted that this study still found Bft in approximately 54% of the controls when one-sided colon samples were obtained, yet this number was notably lower that the approximately 87% of colorectal cancer cases that tested positive for Bft.166
This link is reinforced by additional studies, which have demonstrated that stools from colorectal cancer patients were shown to test positive for Bft at significantly higher rates than controls.189 Interestingly, B. fragilis has been reported to be found in more abundance in mucosal as opposed to luminal samples taken from colorectal cancer patients.190 Here it is once again important to differentiate between toxigenic B. fragilis and nontoxigenic B. fragilis. One study determined that B. fragilis as a whole was found in 77% of colorectal cancer patients yet 68% of healthy controls, a difference that was not significant.189 However, the Bft gene was detected in 38% of the B. fragilis isolated from the cancer patients, but only 12% from the control, a statistically significant difference.189 Additionally, individuals with familial adenomatous polyposis, a disease that leads to digestive tract polyp formation at a young age due to mutation of tumor suppression gene APC, has been shown to be associated with enterotoxic B. fragilis.191 In one study 60% of patients with familial adenomatous polyposis had gut mucosa with enterotoxic B. fragilis present, compared to only 30% of controls.191
7. Escherichia (E) coli toxins
E. coli secretes a number of protein exotoxins, some of them are lineage-specific, and among those, three toxins: colibactin, cytotoxic necrotizing factors (CNF) and α-hemolysin, which are pertinent to human cancer, are reviewed here in Sections 7.1–7.3.
7.1. Colibactin
7.1.1. Chemical and molecular characteristics
Colibactin is a genotoxin causing DNA double strand breaks in a contact-dependent manner. Colibactin is synthesized by a hybrid non-ribosomal peptide synthetase-polyketide synthase (NRPS-PKS) assembly line,192 a sequence of tailoring and editing enzymes, and a critical phosphopantetheinyl transferase that mediates NRPS and PKS activation.193 Despite multiple attempts, colibactin had not been able to be isolated successfully due to its’ extreme instability.194 Finally, in September 2019, through the interdisciplinary approach encompassing chemical synthesis, metabolomics, and probe-mediated natural product capture, a group of scientists from Yale University claimed a complete elucidation of colibactin structure.195 It has a nearly symmetrical structure that contains two electrophilic spirocyclopropyldihydro-2-pyrrolone and the hydrolytically labile C36–C37 α-dicarbonyl.195 These two electrophilic cyclopropane residues undergoes ring-opening through nucleotide addition,195 consistent with earlier observation that colibactin-producing bacteria cross-link DNA.196–198 Using genome-editing techniques, the group also showed the production of colibactin’s precursor, precolibactin 1489, that requires every biosynthetic gene in the colibactin gene cluster, demonstrating it as being the first reported product derived from the long-elusive and completed biosynthetic pathway.195 However, precolibactin 1489 was considered a stable product arising from oxidation and macrocyclization of a putative linear direct precursor of colibactin, precolibactin 1491.195
7.1.2. Microbial characteristics of colibactin
Colibactin is encoded by the pks genomic island, and is often found on E. coli strains belonging to the B2 phylogenetic group.194 The bacteria Klebsiella pneumoniae is also known to carry the colibactin-producing pks island, with one study finding 16.7% of K. pneumoniae isolates were pks+.190 It is estimated that roughly 20% of healthy individuals are colonized by pks+ E. coli.199 In terms of E. coli prevalence, the pks genomic island is found in 30–40% of all B2 strains.200 Colibactin-producing E. coli has also been shown to be relatively resistant to human macrophages and to not influence COX-2 expression.201
The pks genomic island is 54-kb in length and directs the synthesis of colibactin via specific cellular machinery.202 It consists of 19 protein producing genes named clbA to clbS, which are arranged in sequence as clbA, clbR, clbC to clbQ and clbS.192
The first step in the assembly line of colibactin synthesis consists of the ClbA protein, a phosphopantetheinyl (PPant) transferase, activating the NRPS and PKS enzymes via the addition of a PPant onto the NRPS and PKS carrier protein domains.192 ClbN and ClbB serve as the NRPS enzymes, with ClbN producing N-myristoyl-D-Asn, which is then accepted by ClbB. ClbB then proceeds to add either Ala or Val and incorporate a malonyl-CoA. ClbH and ClbI have been shown to be essential for the formation of cyclopropane, which is responsible for colibactin-induced DNA alkylation.192 ClbsD-G are required for the synthesis of the unique PKS extender aminomalonyl unit (AM), which is necessary for genotoxicity. ClbG transfers AM to ClbK, which then incorporates AM into colibactin. While less is knows about the off-loading process, it is thought that ClbQ is involved in this process, and therefore controlling the flux of colibactin production.192 Finally, ClbM (a MATE transporter) is responsible for precolibactin release into the periplasmic space, where ClbP generates the mature colibactin via removal of the N-myristoyl-D-Asn side chain.192 ClbS is also produced by pks+ E. coli where it is thought to sequester colibactin and therefore protect the E. coli from damage from its own product.192 Additionally, clbM has been shown to play a role in the transport of colibactin,203 but to date the precise mechanism of how colibactin is secreted and transported to host cell cytosol is poorly understood.
7.1.3. In vitro toxicity data of colibactin
Colibactin induces cellular damage in eukaryotic cells via introduction of a double-stranded break into the DNA of the host cell.193 This action requires the presence of live bacteria, not being observed with bacteria culture supernatants or cell lysates. This damage serves to activate a signaling pathway that eventually leads to cell cycle arrest, enlargement of cell bodies, and ultimately death of the cell.193 Cells exposed to pks+ E. coli strains have been shown to demonstrate many of the defining characteristics of cell death, including chronic DNA double strand breaks, enhanced senescence-associated β-galactosidase (SA-β-Gal) activity, expansion of promyelocytic leukemia nuclear foci and senescence-associated heterochromatin foci.204 Additionally, exposed cells increase their reactive oxygen species production and pro-inflammatory cytokines, chemokines and proteases secretion.204 Importantly, the presence of colibactin-producing E. coli strains in fibroblasts in vitro has been shown to promote the growth of epithelial human lung adenocarcinoma and colon carcinoma tumor cells.204 Pks+ E. coli infection has also been associated with the induction of IL-6 production, which is known to induce epithelial cell invasion, and MMP production, which has been shown to have direct tumor-promoting effects.204 Taken together this suggests a mechanism for pks+ E. coli reprogramming fibroblast expression profiles in a way that promotes growth of cancer cells.204
Brief exposure to low doses of pks+ E. coli strains has been shown to transiently damage DNA in mammalian epithelial cells, followed by cellular division with incomplete DNA repair.205 Increased rates of chromosomal aberrations, instability, and gene mutations were also observed in cells post-exposure.205 Due to the difficulty in isolating colibactin, alternative methods have been turned to in order to study this toxin’s exact mechanism of action. Production and observation of synthetic adenine-colibactin adducts mimicking the results of pks+ E. coli incubation in human cells demonstrated that colibactin breaks DNA strands by utilizing alkylation via a cyclopropane “warhead.”197 The degradation products observed in this experiment consisted of two diastereomeric adducts that both contain a 5-hyfroxypyrrolidin-2-one ring system with an attached N3-substituted adenine ring. Due to the small size of these adducts, it is likely that they are actually derived from a larger, yet unstable colibactin-DNA interstrand cross-link.197
7.1.4. Colibactin toxicity in animal models
In vivo experimentation has shown that pks+ E. coli strains induced the formation of phosphorylated H2AX foci in mouse enterocytes and indicates DNA damage to colonic epithelial cells.205 Pks+ E. coli strains have also been found to induce a higher degree of lymphopenia in septicemic mice as compared to strain that did not produce colibactin.206 Perhaps most notably, Pks+ E. coli has been observed to promote tumorigenesis in the ApcMin/+; Il10−/− mouse model in a colibactin-dependent manner, suggesting colibactin is a driver of carcinogenesis.207 A rat model has also demonstrated the link between pks+ E. coli strains and DNA damage, with observable signs of genotoxin stress including anaphase bridges, higher occurrence of crypt fission and accelerated renewal of the mature epithelium detected in adult rats.208 This same model revealed that colibactin-producing E. coli strains increased intestinal epithelial cell apoptosis and proliferation, as well as amplified goblet, enteroendocrine and paneth cells.208 This serves to support the connection between Pks+ E. coli and intestinal tumorigenesis.208
Notably, even transient exposure to Pks+ E. coli has been shown to increase tumor growth.209 This was shown when intestinal epithelial cells HCT116 that had been mixed with Pks+ E. coli were injected into nude mice. 3 h post injection the mice were given a broad-spectrum antibiotic (imipenem) in order to kill the bacteria. This short exposure resulted in significantly increased tumor growth, while injection of epithelial cells that had been mixed with Pks − E. coli had no effect on tumor growth, similarly to uninfected cells.209 This tumor-promoting effects was mediated by induction of a senescence-associated secretory phenotype via overexpression of microRNA 20a-5p, which led to accumulation of SUMO conjugated p53.209 The previously mentioned adenosine-colibactin adducts that was shown in vitro mechanistic study of colibactin action have also been observed in the colonic epithelial cells of mice that have been monocolonized with pks+ E. coli.197
7.1.5. Clinical and epidemiological data to support in vivo toxicity in humans
Among B2 E. coli isolated from colorectal cancer and diverticulosis patients, colibactin-producing variants were found to be prevalent at a high rate in the colorectal cancer patients (55.3%), especially when compared with the 19.3% seen among the diverticulosis patients.33 This observation corroborates additional data showing colibactin-producing E. coli strains are associated with colorectal tumors, with 55–66.7% of cancer patient biopsies testing positive while only 19–21% of intestinal control tissue did.199 Recently, PCR analysis has been used to demonstrate that the clbA gene is found at an increased prevalence in the stools of patients with colorectal cancer.210 In this study 56.4% of stool from colorectal cancer patients was found to have clbA, as compared to only 18.5% seen in the controls.210
The presence of the pks island has also been shown to enhance the ability of E. coli to persist in the human gut microbiota.211 One study on this subject showed that long-term colonizers of the gut were significantly more likely to have the pks island than either intermediate-term colonizers or transient strains.211 This may help explain why B2 E. coli strains are represented to a large degree in the gut microbiome.211 Additionally, individuals with familial adenomatous polyposis, a disease that leads to digestive tract polyp formation at a young age due to mutation of tumor suppression gene APC, has been shown to be associated with pks+ E. coli.191 In one study 68% of patients with familial adenomatous polyposis had gut mucosa with pks+E. coli present, compared to only 22% of controls.191
7.2. CNFs (cytotoxic necrotizing factors)
7.2.1. Chemical and molecular characteristics of CNFs
CNFs belong to a family of deamidating toxins that deamidate glutamine 63/61 in the switch II region of Rho GTPases, leading to constitutive activation of Rho GTPases.212 There are four identified variants of cytotoxic necrotizing factor (CNF), CNF1, CNF2 and CNF3, which are secreted by E. coli and CNFY, which is secreted by Yersinia. CNF1 is the best characterized of all CNFs, while CNF2 shares 85% sequence identity with CNF1, CNF3 shares 70% sequence identity with CNF1 and CNFY shares 61% sequence identity with CNF1.212 While there is a great deal of similarity among the CNFs, they differ in terms of their substrate specificity. CNFY exclusively acts on RhoA, B and C and has no activity for Rac or Cdc42.213 CNF2 preferentially modifies RhoA and Rac,214 whereas CNF1 and CNF3 deamidate RhoA, Rac and Cdc42.212
All CNFs are identical in length at 1013/1014 amino acids long and posses an N-terminal receptor binding domain.212 Amino acids 710–1014 in CNF1 have been shown to be responsible for the catalytic activity of this protein, while amino acids 53–190 are responsible for cellular uptake.212
CNF1 is a protein with a molecular weight of 115 kDa.215 Experimentation has determined that CNF is stable within a pH range of 6–10.5.216 The toxic activity of CNF was destroyed when exposed both to a temperature of 75 °C for 15 min and 60 °C for 1 h.216 As CNF is primarily a protein, it has a low carbohydrate content of no more than 25 μg.216
7.2.2. Microbial characteristics of CNFs
Compared to the pks island, CNF1 is relatively less common, with one study determining that, of 78 E. coli strains isolated from adults with urinary tract infections, 13% were positive for the CNF gene.217 The nucleotide sequence of the CNF gene has been identified, and notably the start codon is located at nucleotide 856 while the TGA termination codon is located at nucleotide 3900.218 Interestingly, the C+G content of the CNF1 is lower than that of the E. coli genome as a whole at 36.5%.218
For CNF1 specifically, ferredoxin is required for secretion at the step of crossing the cytoplasmic membrane.219 CNF1 is transported in its active form by outer membrane vesicles.220 This is supported by the fact that the histone-like nucleoid structuring protein H-NS has been shown to down regulate CNF1 production by affecting outer membrane vesicle release.220 CNFY utilizes the type III secretory pathway, which supports CNF1 using the same pathway.221
7.2.3. In vitro toxicity data of CNFs
CNF induces actin assembly and multinucleation in mouse embryonic fibroblasts that were incubated with the purified toxin.222 CNF has been shown to inhibit the intrinsic GTPase activity of RhoA and completely blocks GTPase activity stimulated by the Rho-GTPase-activating protein (rhoGAP).222 CNF has been observed to reduce function at the basolateral membrane of intercellular epithelial junctions.125 This is accomplished through displacement of the proteins occludin and zonula occludens-1 (ZO-1), and reorganization of junction adhesion molecule-1 (JAM-1).125 CNF1 has also been shown to increase F-actin content in cytoskeletons of polymorphonuclear leukocytes.223 CNF1 also increases superoxide generation and adherence on epithelial monolayers, but significantly decreases their phagocytic function.223 This implies a possible role of CNF1 in intestinal infections by way of by stimulating polymorphonuclear leukocyte cytotoxicity.223
It has also been shown that treatment with CNF1 initially inhibits mitosis and cytokinesis and elicits endoreplication and polyploidization in human colon cancer cells in culture.224 These outcomes were found to be followed by depolyploidization-associated survival of cells. An important finding from this study was the discovery that progeny derived from cells treated with CNF1 exhibited marked genomic instability, which was clearly seen by an increased rate of aneuploidy.224 These progeny cells were noted to be more resistant to CNF1, but not to commonly used chemotherapy agents 5-fluorouracil and oxaliplatin.224 There is also evidence that CNF1 induces COX2 and NF-κB expression in mouse fibroblasts and a human squamous cancer cell line, respectively.225,226 CNF1 has also been shown to act in endothelial cells to both inhibit apoptosis and promote pro-inflammatory cytokine release, with the later effect observed in both epithelial and endothelial cells.227
7.2.4. CNF toxicity in animal models
The majority of CNF-related research using animal models to date has focused primarily on inflammation rather than cancer, yet is still relevant for the purposes of this chapter. CNF positive E. coli has been shown to cause more inflammation-mediated morphological and histological tissue damage in a rat prostate model than an isogenic mutant.228 Interestingly, this occurred despite the fact that total bacterial counts for each strain were nearly identical. A mouse bladder model demonstrated that mice infected with CNF1-positive strains consistently showed deeper and more extensive inflammation than those infected with the isogenic mutants.229
CNF1 has also been shown to have an association with spread of prostate cancer in a mouse model.230 In this experiment induced overexpression of CNF1 was observed to promote both cellular invasion and migration.230 Injection of CNF1 expressing cells into the tails of mice was observed to lead to a higher level of lung metastasis when compared to mice receiving a control vector.230
7.2.5. Clinical and epidemiological data to support carcinogenic potential in humans
Similar to colibactin, cnf+ B2 E. coli strains have been shown to have an association with colorectal cancer.33 Of total E. coli strains isolated from colorectal cancer patients, cnf+ B2 E. coli strains comprised 18%.231 Once again, this is more prevalent that what is seen in patients with diverticulitis.231 One study found that 39.5% of biopsies from patients with colorectal cancer contained CNF1-producing E. coli, whereas 12.9% of biopsies from diverticulitis patients were positive for CNF1-producing E. coli.33 Notably, All cnf1-harboring strains belonged to the B2 phylogroup.33
It is important to note that an individual E. coli strain is not limited to producing only one exotoxin. In fact, the previous study found that 33% of B2 strains possessed both the pks island and the cnf1 gene.33 Additional studies have shown that both the cnf1 gene and the hlyC (discussed below) are detectable in significantly lager numbers from E. coli blood isolates from cancer patients than in fecal isolates from healthy volunteers.33 Cnf1-producing E. coli has also been associated with pathological conditions in other body systems. Strains of E. coli that possess the cnf1 gene have been shown to be associated with urosepsis,232 possible indicating that the cnf1 gene is indicative of higher bacterial virulence. This higher virulence may lead to more severe inflammation and increases risk of carcinogenesis.
7.3. α-Hemolysin
7.3.1. Chemical and molecular characteristics of α-hemolysin
α-Hemolysin is a cytotoxic protein that is a member of the repeats in toxin (RTX) group. This group is characterized by glycine and aspartate-rich repeats located at the C-terminus of these toxin proteins.233 This nonapeptide sequence is GGXGXDXUX, where U represents a large, hydrophobic residue.233 α-Hemolysin requires calcium to function optimally, and it is this repeating domain that is responsible for calcium binding.233 Specifically, Asp-863 has been shown to be of special importance in calcium binding, and therefore HlyA conformational change and ability to breakdown the bilayer permeability layer.233 HlyA possesses 15 of these nonapeptide repeats, implying the presence of a large number of calcium binding sites.233
HlyA consists of both protein (95% of weight) and carbohydrates, but lipids have not been detected.234 The majority of hlyA is not heat-stable as it is inactivated at 56 °C,234 but when exposed to temperatures of 37 °C inactivation did not occur for upwards of 18 h.235 HlyA appears to be generally more stable at a lower pH, as it is stable at pH 3.0 for 6 h, but inactivates at pH 7.0–10.0 within 6 h.236 This is not an unexpected result due to the acidity of the gut, a frequent location of E. coli colonization. Functionally, α-hemolysin of E. coli is a pore-forming toxin that acts by generating small cation-permeable channels in the membrane of a host cell.237
7.3.2. Microbial characteristics of α-hemolysin
The hemolysin gene (hly) was identified on 23% of 78 E. coli strains taken from adults with UTI’s.217 This study also concluded that hly was more prevalent than CNF in this population, with hly being present on 18 E. coli strains while CNF was present on 10.217 Four defined genes encode for α-hemolysin. HlyA codes for a 107,000-kDa protein that is thought to be the inactive precursor of hemolysin.238 HlyC, codes for a 20,000 polypeptide that is crucial for post-translational modification of the protein encoded by hlyA, rendering this protein hemolytically active. HlyB and hlyD are necessary for hemolysin secretion.239 The gene for all four hly citrons has been localized to an 8211-base-pair region.240 Of note, there is an ATG codon preceded by a potential ribosome-binding site (GCGG) at base-pair 5317.240
The secretory pathway of hly is well known, as it is both the prototypical and best characterized type I secretion system.241 HlyA reaches the extracellular space in a soluble form without the formation of periplasmic intermediates.242 Interestingly, a fraction of hlyA appears to be very tightly associated with outer membrane vesicles.242 This corresponds with the observation that hlyA localized in outer membrane vesicles is frequently found among hemolytic E. coli strains.242
7.3.3. In vitro toxicity data of α-hemolysin
E. coli hemolysin has a pronounced effect on human monocytes, with nanomolar concentrations (250–2000 ng/mL) resulting in rapid and irreversible depletion of cellular ATP to levels below 20% of controls within 60 min.223 Exposure of human monocytes to lower doses (10–200 ng/mL) of hemolysin results in the release of large amounts of interleukin 1 beta within 1–2 h.223 50% of cellular ATP is observed to be depleted within 90 min when incubated with only 0.3–3.0 colonyforming units of E. coli per monocyte.223 In neutrophils, hemolysin causes defects in membrane permeability, resulting in ATP efflux and propidium iodide influx.243 These cellular membrane lesions do not appear to be repaired.243 Human serum albumin, lipoproteins and IgG protect erythrocytes and platelets, but not neutrophils, from the effects of hemolysin.243 The binding of hemolysin to neutrophils results in granule exocytosis and loss of cellular phagocytic killing ability.243 This is expected to impair or destroy the typical immunoprotective properties of these cells.
HlyA has also been shown to play an important role in giving E. coli the ability to translocate across cellular monolayers.237 In this case, translocation was associated with decreased epithelial integrity of colonic cell cultures, which exhibited increased ion permeability. Experimentation has demonstrated that hemolysin secreting E. coli strains were able to translocate, whereas non-hemolytic mutants were unable to do so, and did not compromise the epithelial barrier.237 In human uroepithelial cell lines, incubation with α-hemolysin substantially reduced IL-6 secretion by LPS through degradation of β-catenin and NF-κB subunit RelA.244 Co-culture of a normal human colon mucosal epithelial cell line with hly+ E. coli activated expression of HIF1α and the glucose transporter GLUT1, which often upregulated in many types of cancer as an indication of reprogrammed energy metabolism, and repressed expression of the tumor suppressor BIM,245 suggesting undelaying mechanisms of tumor promotion.
7.3.4. α-Hemolysin toxicity in animal models
Much like the case of CNF, there has been relatively little cancer-specific research pertaining to animal models and hemolysin exposure. However, some relevant studies on hemolysin toxicity are discussed in this section. In a rat model, Hly+ strains provoked higher urinary shedding of leukocytes and erythrocytes, indicating an affect of hemolysin on inflammatory reactions.246 Rat models have revealed that hemolysin mediates neutrophil toxicity, resulting in in vivo necrosis and lysis.247 In the same study mentioned above that showed the importance of hemolysin in E. coli translocation ability in vitro, hemolysin was also demonstrated to induce focal leaks in native rat colon.237 The rat colons exposed to hemolytic E. coli in this experiment were observed to have both absent nuclei and notable bacterial adherence to the epithelial surface in addition to decreased epithelial integrity.237
7.3.5. Clinical and epidemiological data to support carcinogenic potential in humans
The effect of hemolysin has also been examined using E. coli strains taken directly from human patients. α-Hemolysin synthesized by E. coli strains isolated from ileal mucosa of patients with Crohn’s disease has been shown to adhere to differentiated intestinal cells, and may disrupt the intestinal barrier.248 Hemolysin has also been shown to be associated with bacteremia in hospitalized cancer patients.249 One study found that 26% of blood samples from cancer patients with bacteremia were positive for hemolysin-producing E. coli, while only 6% of fecal samples from healthy patients were positive for hemolysin-producing E. coli.249
In a very interesting outcome, hly+ E. coli has been implicated in adenomagenesis and colorectal cancer in females but not males.245 Hly+ E. coli has been found to be relatively more prevalent in stools from females with adenoma and colorectal cancer, and to be correlated with poor survival in female but not male colorectal cancer patients.245 In this experiment, hly+ E. coli was detected in 36.1% of female colorectal cancer patients, 39.3% of female adenoma patients as compared to just 6.7% of female healthy controls.245 The association of hly+ E. coli with colorectal cancer was shown to be independent of both cancer stage as well as patient age.245 It is hypothesized that the reason hly+ E. coli appears to be correlated to increased negative outcomes in female as opposed to males is due to immunological, hormonal or behavioral differences.245 It is unlikely that genetic factors are causative of this discrepancy due to the fact that in vitro experimentation demonstrated both cytotoxic and carcinogenic effects on male derived cellular lines.245
8. Salmonella enterica avirulence protein A (AvrA)
8.1. Background of Salmonella enterica infection in humans
Salmonella enterica is a Gram-negative, facultative anaerobe, not a symbiotic commensal, but an intracellular pathogen to both humans and animals. Salmonella infection poses a major public health concern worldwide as foodborne illness.250 There has been also a concern that long-standing Salmonella infection may increase the risk of colorectal cancer in humans as increased risk of colorectal tumor has been reported in several populations worldwide.251,252 The Salmonella genome contains a remarkably large number (up to 23) of pathogenicity islands (SPI),253,254 synthesizing over 60 effectors.255 We describe here Avirulence protein A (AvrA), one of the bacterial type three secretion system effectors as there have been recent significant findings to support its oncogenicity.
8.2. Microbial characteristics of AvrA
AvrA represents the least characterized effector from the SPI1 which is originally thought to be non-virulent.256 AvrA is directly injected into host cytoplasm by type 3 secretion system (T3SS) with a filament like-needle that is encoded by the same SPI1 in a finely coordinated manner.257,258 The AvrA gene is present in most Salmonella enterica isolates from humans and animals. This is particularly the case for non-typhoid Salmonella as its prevalence was reported to be 98–100% (514/523 and 185/185),259,260 while it was absent in typhoid strains, which has been linked to gallbladder cancer. Natural AvrA genetic variants have been reported for some Salmonella typhimurium strains, which lacks an internal leucine residue (L140) and has a profound effect on AvrA function.261,262 Among non-typhoidal strains, AvrA protein expression is known to vary markedly with clinical presentation. AvrA protein is not often produced by clinical isolates from systemic disease, but it is often detectable in those from limited enteritis.263 Interestingly, whereas expressions of most effectors from SPI1 are controlled at the transcriptional level,264 it is now clear that expression of AvrA is regulated in a post-transcriptional manner and that mRNA transcription takes place constitutively in all AvrA-positive strains,265 as AvrA remains silent when cloned into E. coli.266 In fact, a number of Proteobacteria have evolved global regulation systems of post-transcriptional gene expression for their various virulence factors to be responsive to changes in the environment and to flourish in specialized host niches. Salmonella enterica is equipped with the Csr (carbon storage regulator) system, consisting of a small RNA binding protein (CrsA) and non-coding RNAs (CsrB/CsrC).267 CsrA binding to the 5′ untranslated and/or early coding regions of mRNAs alters mRNA translation, turnover and/or transcript elongation, leading to either decay or stabilization of specific mRNA targets, while CsrB/CsrC containing multiple CsrA binding sites bind and sequester CsrA, thereby antagonizing CsrA activities.267,268 Overexpression of either CsrA or CsrB shuts down AvrA expression, but that constitutional CsrA expression was required for AvrA expression.265
8.3. Chemical and molecular characteristics of AvrA
AvrA belong to a superfamily of the Yersinia outer protein J (YopJ) acetyltransferases, consisting of 287 amino acids.262,269,270 The regulatory regions are located at flanking N- and C-terminal segments (residues 1–45 and 220–287, respectively) and the intervening residues (46–219) form the catalytic domain, which is structurally homologous to the CE clan peptidases.262,270 In addition, AvrA possesses ubiquitin-like protein protease and deubiquitination activities. These enzymatic activities have been considered crucial for bacterial survival and virulence through post-translational modifications (PTMs) of host proteins to modulate the activity of key proteins on oncogenic and inflammatory pathways. Specific host targets for deubiquitination include IκBα and β-catenin and those for O (threonine)- and Nε(lysine)-acetylation include MAPKK271 and p53.272,273 p53 acetylation has been shown to increase its stability and transcriptional activities.274
8.4. In vitro toxicity data of AvrA
AvrA transfection to HeLa cells led to inhibition of activation of NF-κB, a key pro-inflammatory transcription factor, and augmented epithelial apoptosis.275 AvrA transfection to human 293 T cells induced repression of c-Jun N-terminal kinase and NF-κB signaling pathways271 as acetylation of critical amino acid residues in MAPKK blocks MAPKK phosphorylation activities.272 Colonization of AvrA-positive Salmonella with human colon cancer cells corroborated this finding showing inhibition of IκBα degradation.276 The ubiquitination of β-catenin decreased in the presence of AvrA expression, indicating that the expression of the Salmonella AvrA effector stabilizes β-catenin by decreasing the ubiquitination of β-catenin.276 Further colonization experiments with human colon cancer cell lines demonstrated modulation of several members of the Wnt pathway by AvrA expression, i.e., upregulation of Wnt11 and Wnt2 and downregulation of Wnt1,277–279 which was also confirmed in vivo with murine models.
8.5. AvrA toxicity in animal models
In both transgenic Drosophila and murine models, AvrA was shown to potently inhibit c-Jun N-terminal kinase (JNK) and NF-κB signaling pathways and, in the mouse intestinal mucosa, infection with AvrA-positive Salmonella dampened the proapoptotic innate immune response to Salmonella, suggesting a survival strategy for intracellular pathogen, where the bacteria elicit transient inflammation but do not destroy epithelial cells.271 In a murine short-term infection model, epithelial cell proliferation of the large intestine was increased in mice infected with AvrA-positive Salmonella than those infected with AvrA-negative Salmonella.276,278 Further studies have revealed that infection with AvrA-expressing Salmonella increased the Wnt/β-catenin activity, intestinal stem cell population, and cell proliferation in the infected intestinal mucosa.280 In their murine chronic infection model, Lu et al. found that AvrA persistently regulated β-catenin post-translational modifications, including phosphorylation and acetylation. Moreover, the upstream regulator Akt, transcription factors, T cell factors, nuclear β-catenin, and β-catenin target genes were enhanced in mice infected with Salmonella-expressing AvrA, suggesting its functional role in promoting intestinal renewal.281 The subsequent studies using mouse chemical carcinogenesis models have demonstrated that colorectal tumor incidence indeed markedly increased (almost doubled) in the AvrA+ Salmonella infected mice, compared with mice without bacterial gavage or mice infected with AvrA− Salmonella.282 While the Wnt1 protein level in the mouse intestine was decreased in the AvrA+ infected mice compared with those infected with an AvrA− strain,279 Wint11 expression was enhanced in animals infected with AvrA+ strains.277 In the same animal model colonized with Salmonella AvrA-sufficient or AvrA-deficient bacterial strains, AvrA-expressing bacteria activated the STAT3 pathway, which is predicted to enhance proliferation and promote tumorigenesis, providing new insights regarding a STAT3-dependent mechanism by which the specific bacterial product AvrA enhances the development of infection-associated colon cancer.283
8.6. Clinical and epidemiological data to support carcinogenic potential in humans
There have been sparse data from epidemiological and clinical studies to directly support oncogenicity of AvrA in humans. However, Lu et al. repotted that AvrA-gene positive Salmonella was detectable in human feces and demonstrated that AvrA protein was immunohistochemically detectable in human colorectal mucosa and that its expression in tumor adjacent mucosa in colorectal cancer cases was significantly higher than in normal mucosa of non-cancer patients.284 Consistent with the results from in vitro and animal studies, decreased Wint1 expression was observed in human colorectal cancer tissue.279
9. Concluding remarks
We have conducted an in-depth review on several emerging bacterial toxins that may be involved in pathogenesis of human orodigestive tract cancer. It was clear that these bacterial toxins have biological activities to induce several hallmarks of cancer.285 Some toxins (CDT, colibactin and CNF) can directly interact with DNA or chromosomes leading to their breakdowns, causing mutations and genome instability, which is considered a crucial step for tumor initiation. Other cancer hallmarks commonly manipulated by bacterial toxins are cell proliferation, replication and death. While sustained proliferation and unlimited replication of epithelial cells are essential for tumor growth, enhanced cell apoptosis and senescence can lead to evasion of immune surveillance if they occur preferentially in hematologic cells, and to tumor stimulatory SASP45 if they primarily involve epithelial and stromal cells. Manipulation of cell proliferation and death by bacterial toxins often are achieved by induction or degradation of oncogenes or tumor suppressors. The two most often modulated pathways were p53286 and β-catenin/Wnt pathways,287,288 while others, such as JAK-STAT, MAPK/ERK, Ras and Myc pathways are also exploited by PMT, Bft and AvrA as discussed above.
Wnt/β-catenin signaling pathway controls several crucial biological processes, such as cell motility, proliferation and the induction of differentiation. The activation of its canonical pathway is dependent on the binding of Wnt glycoprotein ligands to Frizzled (Fzd) receptors and its co-receptor, low-density lipoprotein receptor-related protein 5/6 (LRP5/6) on the plasma membrane, forming the ternary complex Wnt–Fzd–LRP5/6. Subsequent recruitment of a scaffolding protein disheveled (Dvl) leads to the inhibition of Axin-mediated β-catenin phosphorylation, and thus β-catenin stabilization, cytoplasmic accumulation, and nuclear translocation. Once in the nucleus, β-catenin activates Wnt-dependent gene expression.154 Wnt/β-catenin has recently emerged as an important target of several virulence factors produced by bacteria. The mechanisms are diverse and include the repression of Wnt inhibitors’ expression by the epigenetic modification of histones, blocking Wnt–Fzd ligand binding, manipulating β-catenin nuclear translocation, down- or upregulation of Wnt family members, post-translational modification of β-catenin, and inhibition of Axin-1 expression, which promotes β-catenin activity,154,280,287 while others, such as HlyA directly degrades β-catenin through its protease activity and suppresses its activity.244,288 Accordingly, outcomes of manipulations of the Wnt/β-catenin pathway by these bacterial toxins are diverse. Both Salmonella AvrA that deubiquitinates β-catenin to prevent its degradation and Fusobacterium FadA that binds E-cadherin to stimulate β-catenin lead to increased cell proliferation and tumor formation,154,282,288 while E. coli HlyA results in immunosuppression.244,288
Many bacteria as well as oncoviruses, such as HPV and EBV, are known to repress p53 through direct cleavage or proteasome degradation.286 However, as we discussed above, other bacterial toxins, such as CDT and colibactin, upregulate P53 due to their genotoxic effects, while AvrA introduces a post-translational modification by acetylation to stabilize and stimulate this pathway.273 Resulting sustained apoptosis has been speculated to lead eventually to the development of SASP, which in turn promotes tumorigenesis.45 An additional way to resist cell death has been recognized to be enhancing autophagy, which is considered a pro-survival signal in challenging cellular environment.285 CDT exploits this strategy.
An emerging hallmark of cancer is reprograming of cellular metabolism and energetics from mitochondrial respiration to aerobic glycolysis, so-called the Warburg effect.285 While this metabolic shift caused by exposure to LPS/endotoxin has been well documented,289,290 the effects of the exotoxins reviewed here on cellular energetics have been less well delineated. Yet, there are ample laboratory data to support this possibility. CDT from Campylobacter and Helicobacter species and HlyA from E coli have been shown to damage mitochondria, which may lead to reduced respiratory function.291–293 On the other hand, activation of key signaling molecules, such as HIF1α and GLUT1 by HlyA245 and c-Myc by Bft,181 on the glycolysis pathway has been noted. Tan et al. directly assessed glycolysis and mitochondria biogenesis in mouse melanoma cells exposed to E. coli CNF, reporting an increase in glycolysis and a decrease in mitochondrial biogenesis.294 However, conflicting data exist, suggesting that explore to CNF enhances mitochondrial volume and function in other cell culture systems,295,296 and thus the net effect on cellular energetics in human non-transformed cells is unknown.
Many bacterial toxins, such as CDT, CTLP, PMT, FadA, Bft, CFA and HlyA, also exert biological activates to degrade intercellular junction proteins, including tight junction ZO-1 and E-cadherin, as well as extracellular matrix, partly through activation of matrix metalloproteinases, which leads to increased mucosal permeability, decreased mucosal defense and disruption of cellular communications, and activates tumor invasion.285 On the other hand, bacteria effectors, such as AvrA, stabilizes tight junction to accommodate their own survival in the host.297,298
However, by far, the cancer hallmark “tumor-promoting inflammation” is most often controlled by bacterial toxins. In fact, all but Fap2 reviewed here act on this pathway, through diverse mechanisms and with distinct outcomes. A common aspect of infection-related cancer is the induction of chronic inflammation through various mechanisms.1,299 Whereas most bacteria toxins reviewed here and others induce inflammation and often produce tumor-promoting cytokines, such as IL-17 and IL-6,187 some pathogenic bacteria have evolved to synthesize bacterial toxins, such as AvrA, HlyA and CTLP, with the ability to temper the inflammatory response to create a suitable niche for their survival and proliferation in the host, not killing the host or host cells they infected. Acute excessive inflammation is a short-term response that either clears the pathogenic bacteria from the host through recruitment of immunological cells, or results in fatal consequences (host death). Thus, it does not lead to the development cancer. Accordingly, anti-inflammatory properties are particularly important and common in many T3SS pathogens with numerous virulence factors that induce strong pro-inflammatory reactions, such as Salmonella and E coli.272 The NF-κB network is often manipulated by bacterial toxins to both up- and downregulate inflammatory responses.300
Table 1 summarizes the strength of evidence from human studies and mechanistic studies to support carcinogenicity of each selected toxin. Causal association has not been established for any of the toxins or bacteria reviewed here and the strength in human studies simply reflect consistency in statistical associations. It is important to note that the vast majority of in vitro studies with pathogens are short-term infections that induce a range of host cell responses. Besides, intestinal cells have short turnover rate of 3–4 days. Thus, the interpretation of transitory cellular signaling in the context of in vitro infection as tumorigenic may not be necessarily warranted. The tumor-promoting effects seen in animal models thus far have been delivered from genetically susceptible or chemically induced carcinogenesis models, and no transgenic animal models for the genes encoding these toxins have been tested. Overall, despite growing laboratory evidence to support oncogenic potential of selected bacterial toxins reviewed here,3,301,302 evidence from human studies has been rather limited. This is partly because most microbiome research to date have focused on global community composition based on 16S rRNA gene sequencing. More targeted research will certainly help advance establishing the bacterial etiology of orodigestive tract cancer, which includes studies testing various human tissue samples, including not only tumors, but also surrounding mucosa, precursor lesions and healthy mucosa, as well as fecal, oral rinse/plaque and pre-diagnostic blood samples to directly detect bacterial toxins and their molecular signatures and to assess antibody responses. In addition, we need more physiologically relevant experimental animal models, which can reflect chronic infection in vivo in humans as well as take bacterial-bacterial interactions among microbiome into consideration. Such effort may lead to the development of new probiotic, prebiotic or antibiotic approaches or immunotherapies to prevent cancer development and progression.
Table 1.
Summary of existing evidence to support carcinogenicity of selected bacterial toxins.
| Toxins | Mechanistic (in vitro/animal) | Human (clinical/epidemiological) | Target organs |
|---|---|---|---|
| CDT | a | b | Oral, enterohepatic, genital |
| CTLP | c | a | Oro-digestive tract/organ |
| PMT | b | c | Ororespiratory tract |
| FadA | a | c | Oral, colorectum |
| Fap2 | c | c | Oral, colorectum |
| Bft | a | a | Colorectum |
| Colibactin | a | a | Colorectum |
| CNF | c | b | Colorectum |
| α-Hemolysin | c | b | Colorectum |
| AvrA | a | b | Colorectum |
Strong mechanisms with animal model/association with cancer in multiple studies.
Strong mechanisms only/association with cancer from single reports.
Suggestive mechanisms only/presence in target organs or in circulation.
Acknowledgments
A.A.V. is a post-doctoral researcher with Healthy UrbanWaters (HUW) supported by the Fred A. and Barbara M. Erb Family Foundation. A.A.V. was also supported by a National Institutes of Health (NIH) Post-doctoral fellowship from the NIH Common Fund and Office of Scientific Workforce Diversity under three linked awards RL5GM118981, TL4GM118983, 1UL1GM118982 administered by the National Institute of General Medical Sciences.
References
- 1.Kuper H, Adami H-O, Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248(3):171–183. [DOI] [PubMed] [Google Scholar]
- 2.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4(9):e609–e616. [DOI] [PubMed] [Google Scholar]
- 3.van Elsland D, Neefjes J. Bacterial infections and cancer. EMBO Rep. 2018;19(11): e46632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert S, Tegtmeyer N. Type IV secretion and signal transduction of Helicobacter pylori CagA through interactions with host cell receptors. Toxins. 2017;9(4):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatakeyama M Helicobacter pylori CagA—a bacterial intruder conspiring gastric carcinogenesis. Int J Cancer. 2006;119(6):1217–1223. [DOI] [PubMed] [Google Scholar]
- 6.Tegtmeyer N, Neddermann M, Asche CI, Backert S. Subversion of host kinases: a key network in cellular signaling hijacked by Helicobacter pylori CagA. Mol Microbiol. 2017;105:358–372. [DOI] [PubMed] [Google Scholar]
- 7.El-Etr SH, Mueller A, Tompkins LS, Falkow S, Merrell DS. Phosphorylation-independent effects of CagA during interaction between Helicobacter pylori and T84 polarized monolayers. J Infect Dis. 2004;190(8):1516–1523. [DOI] [PubMed] [Google Scholar]
- 8.Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4CagA-independent mechanisms. Infect Immun. 2002;70(2):665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohnishi N, Yuasa H, Tanaka S, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci U S A. 2008;105(3):1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60(5):631–637. [DOI] [PubMed] [Google Scholar]
- 12.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One. 2011;6(5):e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todar K Online Textbook of Bacteriology. Madison, WI: University of Wisconsin; 2008–2012. [Google Scholar]
- 15.Moreira APB, Texeira TFS, Ferreira AB, do Carmo Gouveia Peluzio M, de Cássia Gonçalves Alfenas R. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br J Nutr. 2012;108(5):801–809. [DOI] [PubMed] [Google Scholar]
- 16.Vasquez AA, Ram JL, Qazazi MS, Sun J, Kato I. Oral microbiome: potential link to systemic diseases and oral cancer. In: Sun J, Dudeja PK, eds. Mechanisms Underlying Host-Microbiome Interactions in Pathophysiology of Human Diseases. Boston, MA: Springer US; 2018:195–246. [Google Scholar]
- 17.Trent MS, Stead CM, Tran AX, Hankins JV. Invited review: diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res. 2006;12(4):205–223. [DOI] [PubMed] [Google Scholar]
- 18.Lemichez E, Barbieri JT. General aspects and recent advances on bacterial protein toxins. Cold Spring Harb Perspect Med. 2013;3(2):a013573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkel JS, Baldwin MR, Barbieri JT. Toxins from bacteria. EXS. 2010;100:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veith PD, Glew MD, Gorasia DG, Reynolds EC. Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol. 2017;106(1):35–53. [DOI] [PubMed] [Google Scholar]
- 21.Abby SS, Cury J, Guglielmini J, Néron B, Touchon M, Rocha EPC. Identification of protein secretion systems in bacterial genomes. Sci Rep. 2016;6:23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerra L, Cortes-Bratti X, Guidi R, Frisan T. The biology of the cytolethal distending toxins. Toxins. 2011;3(3):172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesic D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature. 2004;429(6990):429–433. [DOI] [PubMed] [Google Scholar]
- 24.Hontz JS, Villar-Lecumberri MT, Potter BM, Yoder MD, Dreyfus LA, Laity JH. Differences in crystal and solution structures of the cytolethal distending toxin B subunit: relevance to nuclear translocation and functional activation. J Biol Chem. 2006;281(35):25365–25372. [DOI] [PubMed] [Google Scholar]
- 25.Faïs T, Delmas J, Serres A, Bonnet R, Dalmasso G. Impact of CDT toxin on human diseases. Toxins. 2016;8(7):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerra L, Teter K, Lilley BN, et al. Cellular internalization of cytolethal distending toxin: a new end to a known pathway. Cell Microbiol. 2005;7(7):921–934. [DOI] [PubMed] [Google Scholar]
- 27.Guerra L, Nemec KN, Massey S, et al. A novel mode of translocation for cytolethal distending toxin. Biochim Biophys Acta (BBA)—Mol Cell Res. 2009;1793(3):489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology. 2011;157(7):1851–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shenker BJ, Dlakić M, Walker LP, et al. A novel mode of action for a microbial-derived immunotoxin: the cytolethal distending toxin subunit B exhibits phosphatidylinositol 3,4,5-triphosphate phosphatase activity. J Immunol. 2007;178(8):5099–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boesze-Battaglia K, Alexander D, Dlakić M, Shenker BJ. A journey of cytolethal distending toxins through cell membranes. Front Cell Infect Microbiol. 2016;6:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pickett CL, Whitehouse CA. The cytolethal distending toxin family. Trends Microbiol. 1999;7(7):292–297. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed HJ, Svensson LA, Cope LD, et al. Prevalence of cdtABC genes encoding cytolethal distending toxin among Haemophilus ducreyi and Actinobacillus actinomycetemcomitans strains. J Med Microbiol. 2001;50(10):860–864. [DOI] [PubMed] [Google Scholar]
- 33.Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013;8(2):e56964–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamasaki S, Asakura M, Tsukamoto T, Faruque SM, Deb R, Ramamurthy T. Cytolethal distending toxin (cdt): genetic diversity, structure and role in diarrheal disease. Toxin Rev. 2006;25(1):61–88. [Google Scholar]
- 35.Lara-Tejero M, Galán JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290(5490):354–357. [DOI] [PubMed] [Google Scholar]
- 36.Nishikubo S, Ohara M, Ikura M, et al. Single nucleotide polymorphism in the cytolethal distending toxin B gene confers heterogeneity in the cytotoxicity of actinobacillus actinomycetemcomitans. Infect Immun. 2006;74(12):7014–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.AbuOun M, Manning G, Cawthraw SA, et al. Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect Immun. 2005;73(5):3053–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haghjoo E, Galán JE. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A. 2004;101(13):4614–4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spanò S, Ugalde JE, Galán JE. Delivery of a Salmonella typhi exotoxin from a host intracellular compartment. Cell Host Microbe. 2008;3(1):30–38. [DOI] [PubMed] [Google Scholar]
- 40.Miller R, Wiedmann M. Dynamic duo—the salmonella cytolethal distending toxin combines ADP-ribosyltransferase and nuclease activities in a novel form of the cytolethal distending toxin. Toxins. 2016;8(5):121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiRienzo JM. Uptake and processing of the cytolethal distending toxin by mammalian cells. Toxins. 2014;6(11):3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueno Y, Ohara M, Kawamoto T, et al. Biogenesis of the Actinobacillus actinomycetemcomitans cytolethal distending toxin holotoxin. Infect Immun. 2006;74(6): 3480–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Donoghue EJ, Krachler AM. Mechanisms of outer membrane vesicle entry into host cells. Cell Microbiol. 2016;18(11):1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guidi R, Levi L, Rouf SF, Puiac S, Rhen M, Frisan T. Salmonella enterica delivers its genotoxin through outer membrane vesicles secreted from infected cells. Cell Microbiol. 2013;15(12):2034–2050. [DOI] [PubMed] [Google Scholar]
- 45.Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shenker BJ, McKay T, Datar S, Miller M, Chowhan R, Demuth D. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2 arrest in human T cells. J Immunol. 1999;162(8):4773–4780. [PubMed] [Google Scholar]
- 47.Cortes-Bratti X, Karlsson C, Lagergård T, Thelestam M, Frisan T. The Haemophilus ducreyi cytolethal distending toxin induces cell cycle arrest and apoptosis via the DNA damage checkpoint pathways. J Biol Chem. 2001;276(7):5296–5302. [DOI] [PubMed] [Google Scholar]
- 48.Ando-Suguimoto ES, da Silva MP, Kawamoto D, Chen C, DiRienzo JM, Mayer MPA. The cytolethal distending toxin of Aggregatibacter actinomycetemcomitans inhibits macrophage phagocytosis and subverts cytokine production. Cytokine. 2014;66(1):46–53. [DOI] [PubMed] [Google Scholar]
- 49.Shenker BJ, Walker LP, Zekavat A, Dlakić M, Boesze-Battaglia K. Blockade of the PI-3K signalling pathway by the Aggregatibacter actinomycetemcomitans cytolethal distending toxin induces macrophages to synthesize and secrete pro-inflammatory cytokines. Cell Microbiol. 2014;16(9):1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akifusa S, Poole S, Lewthwaite J, Henderson B, Nair SP. Recombinant Actinobacillus actinomycetemcomitans cytolethal distending toxin proteins are required to interact to inhibit human cell cycle progression and to stimulate human leukocyte cytokine synthesis. Infect Immun. 2001;69(9):5925–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aragon V, Chao K, Dreyfus LA. Effect of cytolethal distending toxin on F-actin assembly and cell division in chinese hamster ovary cells. Infect Immun. 1997;65(9): 3774–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frisan T, Cortes-Bratti X, Chaves-Olarte E, Stenerlöw B, Thelestam M. The Haemophilus ducreyi cytolethal distending toxin induces DNA double-strand breaks and promotes ATM-dependent activation of RhoA. Cell Microbiol. 2003;5(10):695–707. [DOI] [PubMed] [Google Scholar]
- 53.Seiwert N, Neitzel C, Stroh S, et al. AKT2 suppresses pro-survival autophagy triggered by DNA double-strand breaks in colorectal cancer cells. Cell Death Dis. 2017;8:e3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Damek-Poprawa M, Korostoff J, Gill R, DiRienzo JM. Cell junction remodeling in gingival tissue exposed to a microbial toxin. J Dent Res. 2013;92(6):518–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blazkova H, Krejcikova K, Moudry P, Frisan T, Hodny Z, Bartek J. Bacterial intoxication evokes cellular senescence with persistent DNA damage and cytokine signalling. J Cell Mol Med. 2010;14(1–2):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guidi R, Guerra L, Levi L, et al. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell Microbiol. 2013;15(1):98–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guerra L, Guidi R, Frisan T. Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? FEBS J. 2011;278(23): 4577–4588. [DOI] [PubMed] [Google Scholar]
- 58.Graillot V, Dormoy I, Dupuy J, et al. Genotoxicity of cytolethal distending toxin (CDT) on isogenic human colorectal cell lines: potential promoting effects for colorectal carcinogenesis. Front Cell Infect Microbiol. 2016;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frisan T, Nagy N, Chioureas D, Terol M, Grasso F, Masucci MG. A bacterial genotoxin causes virus reactivation and genomic instability in Epstein–Barr virus infected epithelial cells pointing to a role of co-infection in viral oncogenesis. Int J Cancer. 2019;144(1):98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell Microbiol. 2008;10(8):1599–1607. [DOI] [PubMed] [Google Scholar]
- 61.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2010;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.García A, Ihrig MM, Fry RC, et al. Genetic susceptibility to chronic hepatitis is inherited codominantly in Helicobacter hepaticus-infected AB6F1 and B6AF1 hybrid male mice, and progression to hepatocellular carcinoma is linked to hepatic expression of lipogenic genes and immune function-associated networks. Infect Immun. 2008;76(5):1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diwan BA, Sipowicz M, Logsdon D, et al. Marked liver tumorigenesis by Helicobacter hepaticus requires perinatal exposure. Environ Health Perspect. 2008;116(10):1352–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ge Z, Rogers AB, Feng Y, et al. Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell Microbiol. 2007;9(8):2070–2080. [DOI] [PubMed] [Google Scholar]
- 65.Fox JG, Feng Y, Theve EJ, et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut. 2010;59(1):88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jain D, Prasad KN, Sinha S, Husain N. Differences in virulence attributes between cytolethal distending toxin positive and negative Campylobacter jejuni strains. J Med Microbiol. 2008;57(3):267–272. [DOI] [PubMed] [Google Scholar]
- 67.Fox JG, Rogers AB, Whary MT, et al. Gastroenteritis in NF-κB-deficient mice is produced with wild-type Camplyobacter jejuni but not with C. jejuni lacking cytolethal distending toxin despite persistent colonization with both strains. Infect Immun. 2004;72(2):1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young VB, Knox KA, Pratt JS, et al. In vitro and in vivo characterization of Helicobacter hepaticus cytolethal distending toxin mutants. Infect Immun. 2004;72(5):2521–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen Z, Feng Y, Rogers AB, et al. Cytolethal distending toxin promotes Helicobacter cinaedi-associated typhlocolitis in interleukin-10-deficient mice. Infect Immun. 2009; 77(6):2508–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen Z, Feng Y, Muthupalani S, et al. Novel Helicobacter species H. japonicum isolated from laboratory mice from Japan induces typhlocolitis and lower bowel carcinoma in C57BL/129 IL10−/− mice. Carcinogenesis. 2016;37(12):1190–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ge Z, Feng Y, Ge L, Parry N, Muthupalani S, Fox JG. Helicobacter hepaticus cytolethal distending toxin promotes intestinal carcinogenesis in 129Rag2-deficient mice. Cell Microbiol. 2017;19(7):e12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Péré-Védrenne C, Prochazkova-Carlotti M, Rousseau B, et al. The cytolethal distending toxin subunit CdtB of Helicobacter hepaticus promotes senescence and endoreplication in xenograft mouse models of hepatic and intestinal cell lines. Front Cell Infect Microbiol. 2017;7:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He Z, Gharaibeh RZ, Newsome RC, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2): 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pimentel M, Morales W, Rezaie A, et al. Development and validation of a biomarker for diarrhea-predominant irritable bowel syndrome in human subjects. PLoS One. 2015;10(5):e0126438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rezaie A, Park SC, Morales W, et al. Assessment of anti-vinculin and anti-cytolethal distending toxin B antibodies in subtypes of irritable bowel syndrome. Dig Dis Sci. 2017;62(6):1480–1485. [DOI] [PubMed] [Google Scholar]
- 76.Scanu T, Spaapen Robbert M, Bakker Jeroen M, et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe. 2015;17(6):763–774. [DOI] [PubMed] [Google Scholar]
- 77.Shimoyama T, Takahashi R, Abe D, Mizuki I, Endo T, Fukuda S. Serological analysis of Helicobacter hepaticus infection in patients with biliary and pancreatic diseases. J Gastroenterol Hepatol. 2010;25(s1):S86–S89. [DOI] [PubMed] [Google Scholar]
- 78.Murakami K, Takahashi R, Ono M, et al. Serodiagnosis of Helicobacter hepaticus infection in patients with liver and gastrointestinal diseases: western blot analysis and ELISA using a highly specific monoclonal antibody for H. hepaticus antigen. J Gastroenterol. 2011;46(9):1120–1126. [DOI] [PubMed] [Google Scholar]
- 79.Hamada T, Yokota K, Ayada K, et al. Detection of Helicobacter hepaticus in human bile samples of patients with biliary disease. Helicobacter. 2009;14(6):545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mäkinen PL, Mäkinen KK, Syed SA. Role of the chymotrypsin-like membrane-associated proteinase from Treponema denticola ATCC 35405 in inactivation of bioactive peptides. Infect Immun. 1995;63(9):3567–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ishihara K Virulence factors of Treponema denticola. Periodontol 2000. 2010;54(1): 117–135. [DOI] [PubMed] [Google Scholar]
- 82.Fenno JC. Treponema denticola interactions with host proteins. J Oral Microbiol. 2012;4. 10.3402/jom.v4i0.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goetting-Minesky MP, Godovikova V, Li JJ, et al. Conservation and revised annotation of the Treponema denticola prcB-prcA-prtP locus encoding the dentilisin (CTLP) protease complex. Mol Oral Microbiol. 2013;28(3):181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee SY, Bian X-L, Wong GWK, Hannam PM, McBride BC, Fenno JC. Cleavage of Treponema denticola PrcA polypeptide to yield protease complex-associated proteins Prca1 and Prca2 is dependent on PrtP. J Bacteriol. 2002;184(14):3864–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Godovikova V, Goetting-Minesky MP, Fenno JC. Composition and localization of Treponema denticola outer membrane complexes. Infect Immun. 2011;79(12):4868–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Godovikova V, Wang H-T, Goetting-Minesky MP, et al. Treponema denticola PrcB is required for expression and activity of the PrcA-PrtP (Dentilisin) complex. J Bacteriol. 2010;192(13):3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heuner K, Bergmann I, Heckenbach K, Göbel UB. Proteolytic activity among various oral Treponema species and cloning of a prtP-like gene of Treponema socranskii subsp. socranskii. FEMS Microbiol Lett. 2001;201(2):169–176. [DOI] [PubMed] [Google Scholar]
- 88.Correia FF, Plummer AR, Ellen RP, et al. Two paralogous families of a two-gene subtilisin operon are widely distributed in oral treponemes. J Bacteriol. 2003;185(23): 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Veith PD, Glew MD, Gorasia DG, Chen D, O’Brien-Simpson NM, Reynolds EC. Localization of outer membrane proteins in Treponema denticola by quantitative proteome analyses of outer membrane vesicles and cellular fractions. J Proteome Res. 2019;18:1567–1581. [DOI] [PubMed] [Google Scholar]
- 90.Fenno JC, Müller KH, McBride BC. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178(9):2489–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Caimano MJ, Bourell KW, Bannister TD, Cox DL, Radolf JD. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect Immun. 1999;67(8):4072–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Puthenveetil R, Kumar S, Caimano MJ, et al. The major outer sheath protein forms distinct conformers and multimeric complexes in the outer membrane and periplasm of Treponema denticola. Sci Rep. 2017;7(1):13260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fenno JC, Wong GWK, Hannam PM, McBride BC. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol Lett. 1998;163(2):209–215. [DOI] [PubMed] [Google Scholar]
- 94.Uitto VJ, Pan YM, Leung WK, et al. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect Immun. 1995;63(9):3401–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto V-J, McBride BC. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun. 1998;66(5):1869–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grenier D, Uitto VJ, McBride BC. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect Immun. 1990;58(2):347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chi B, Qi M, Kuramitsu HK. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol. 2003;154(9):637–643. [DOI] [PubMed] [Google Scholar]
- 98.Ellen RP, Ko KS, Lo CM, Grove DA, Ishihara K. Insertional inactivation of the prtP gene of Treponema denticola confirms dentilisin’s disruption of epithelial junctions. J Mol Microbiol Biotechnol. 2000;2(4):581–586. [PubMed] [Google Scholar]
- 99.Yamazaki T, Miyamoto M, Yamada S, Okuda K, Ishihara K. Surface protease of Treponema denticola hydrolyzes C3 and influences function of polymorphonuclear leukocytes. Microbes Infect. 2006;8(7):1758–1763. [DOI] [PubMed] [Google Scholar]
- 100.AR J, Baek KJ, Shin JE, Choi Y. Mechanisms of IL-8 suppression by Treponema denticola in gingival epithelial cells. Immunol Cell Biol. 2014;92(2):139–147. [DOI] [PubMed] [Google Scholar]
- 101.Li H, Lu S, Chen Y, et al. AKT2 phosphorylation of hexokinase 2 at T473 promotes tumorigenesis and metastasis in colon cancer cells via NF-κB, HIF1α, MMP2, and MMP9 upregulation. Cell Signal. 2019;58:99–110. [DOI] [PubMed] [Google Scholar]
- 102.Pietruszewska W, Bojanowska-Pozniak K, Kobos J. Matrix metalloproteinases MMP1, MMP2, MMP9 and their tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: an immunohistochemical study. Otolaryngol Pol. 2016;70(3):32–43. [DOI] [PubMed] [Google Scholar]
- 103.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miao D, Fenno JC, Timm JC, Joo NE, Kapila YL. The Treponema denticola chymotrypsin-like protease dentilisin induces matrix metalloproteinase-2-dependent fibronectin fragmentation in periodontal ligament cells. Infect Immun. 2011;79(2): 806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ateia IM, Sutthiboonyapan P, Kamarajan P, et al. Treponema denticola increases MMP-2 expression and activation in the periodontium via reversible DNA and histone modifications. Cell Microbiol. 2018;20(4):e12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leung WK, Wu Q, Hannam PM, McBride BC, Uitto V-J. Treponema denticola may stimulate both epithelial proliferation and apoptosis through MAP kinase signal pathways. J Periodontal Res. 2002;37(6):445–455. [DOI] [PubMed] [Google Scholar]
- 107.Ishihara K, Kuramitsu HK, Miura T, Okuda K. Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J Bacteriol. 1998;180(15): 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Capone R, Wang HT, Ning Y, Sweier DG, Lopatin DE, Fenno JC. Human serum antibodies recognize Treponema denticola Msp and PrtP protease complex proteins. Oral Microbiol Immunol. 2008;23(2):165–169. [DOI] [PubMed] [Google Scholar]
- 109.Dejonckheere E, Vandenbroucke RE, Libert C. Matrix metalloproteinase8 has a central role in inflammatory disorders and cancer progression. Cytokine Growth Factor Rev. 2011;22(2):73–81. [DOI] [PubMed] [Google Scholar]
- 110.Narikiyo M, Tanabe C, Yamada Y, et al. Frequent and preferential infection of Treponema denticola, Streptococcus mitis, and Streptococcus anginosus in esophageal cancers. Cancer Sci. 2004;95(7):569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nieminen MT, Listyarifah D, Hagström J, et al. Treponema denticola chymotrypsin-like proteinase may contribute to orodigestive carcinogenesis through immunomodulation. Br J Cancer. 2017;118:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Listyarifah D, Nieminen MT, Mäkinen LK, et al. Treponema denticola chymotrypsin-like proteinase is present in early-stage mobile tongue squamous cell carcinoma and related to the clinicopathological features. J Oral Pathol Med. 2018;47(8):764–772. [DOI] [PubMed] [Google Scholar]
- 113.Kylmä AK, Jouhi L, Listyarifah D, et al. Treponema denticola chymotrypsin-like protease as associated with HPV-negative oropharyngeal squamous cell carcinoma. Br J Cancer. 2018;119(1):89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Y, Cai Q, Shu X-O, et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer. 2019;144(10):2381–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wilson BA, Ho M. Cellular and molecular action of the mitogenic proteindeamidating toxin from Pasteurella multocida. FEBS J. 2011;278(23):4616–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Orth JHC, Aktories K. Molecular biology of Pasteurella multocida toxin. In: Aktories K, JHC O, Adler B, eds. Pasteurella multocida: Molecular Biology, Toxins and Infection. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012:73–92. [Google Scholar]
- 117.Orth JHC, Aktories K. Pasteurella multocida toxin activates various heterotrimeric G proteins by deamidation. Toxins. 2010;2(2):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lax AJ, Grigoriadis AE. Pasteurella multocida toxin: the mitogenic toxin that stimulates signalling cascades to regulate growth and differentiation. Int J Med Microbiol. 2001;291(4):261–268. [DOI] [PubMed] [Google Scholar]
- 119.Higgins TE, Murphy AC, Staddon JM, Lax AJ, Rozengurt E. Pasteurella multocida toxin is a potent inducer of anchorage-independent cell growth. Proc Natl Acad Sci U S A. 1992;89(10):4240–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wilson BA, Ho M. Pasteurella multocida toxin interaction with host cells: entry and cellular effects. Curr Top Microbiol Immunol. 2012;361:93–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wilson BA, Ho M. Pasteurella multocida: from zoonosis to cellular microbiology. Clin Microbiol Rev. 2013;26(3):631–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pullinger GD, Bevir T, Lax AJ. The Pasteurella multocida toxin is encoded within a lysogenic bacteriophage. Mol Microbiol. 2004;51(1):255–269. [DOI] [PubMed] [Google Scholar]
- 123.Clemons NC, Bannai Y, Haywood EE, et al. Cytosolic delivery of multidomain cargos by the N terminus of Pasteurella multocida toxin. Infect Immun. 2018;86(8): e00248–e00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Danielsen EM, Christiansen N, Danielsen EM. Pasteurella multocida toxin: targeting mast cell secretory granules during kiss-and-run secretion. Tissue Cell. 2016; 48(1):1–9. [DOI] [PubMed] [Google Scholar]
- 125.Hopkins AM, Walsh SV, Verkade P, Boquet P, Nusrat A. Constitutive activation of Rho proteins by CNF-1 influences tight junction structure and epithelial barrier function. J Cell Sci. 2003;116(Pt. 4):725–742. [DOI] [PubMed] [Google Scholar]
- 126.Seo B, Choy EW, Maudsley S, Miller WE, Wilson BA, Luttrell LM. Pasteurella multocida toxin stimulates mitogen-activated protein kinase via Gq/11-dependent transactivation of the epidermal growth factor receptor. J Biol Chem. 2000;275(3): 2239–2245. [DOI] [PubMed] [Google Scholar]
- 127.Liu F, Yang X, Geng M, Huang M. Targeting ERK, an Achilles’ Heel of the MAPK pathway, in cancer therapy. Acta Pharm Sin B. 2018;8(4):552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci. 2012;13(2):1886–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oubrahim H, Wong A, Wilson BA, Chock PB. Mammalian target of rapamycin complex 1 (mTORC1) plays a role in Pasteurella multocida toxin (PMT)-induced protein synthesis and proliferation in Swiss 3T3 cells. J Biol Chem. 2013;288(4):2805–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ozgen N, Obreztchikova M, Guo J, et al. Protein kinase D Links Gq-coupled receptors to cAMP response element-binding protein (CREB)-Ser133 phosphorylation in the heart. J Biol Chem. 2008;283(25):17009–17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Preuβ I, Hildebrand D, Orth JHC, Aktories K, Kubatzky KF. Pasteurella multocida toxin is a potent activator of anti-apoptotic signalling pathways. Cell Microbiol. 2010;12(8):1174–1185. [DOI] [PubMed] [Google Scholar]
- 132.Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. [DOI] [PubMed] [Google Scholar]
- 133.Orth JHC, Aktories K, Kubatzky KF. Modulation of host cell gene expression through activation of STAT transcription factors by Pasteurella multocida toxin. J Biol Chem. 2007;282(5):3050–3057. [DOI] [PubMed] [Google Scholar]
- 134.Hildebrand D, Walker P, Dalpke A, Heeg K, Kubatzky KF. Pasteurella multocida toxininduced Pim-1 expression disrupts suppressor of cytokine signalling (SOCS)-1 activity. Cell Microbiol. 2010;12(12):1732–1745. [DOI] [PubMed] [Google Scholar]
- 135.Kubatzky K, Kloos B, Hildebrand D. Signaling cascades of Pasteurella multocida toxin in immune evasion. Toxins. 2013;5(9):1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bagley KC, Abdelwahab SF, Tuskan RG, Lewis GK. Pasteurella multocida toxin activates human monocyte-derived and murine bone marrow-derived dendritic cells in vitro but suppresses antibody production in vivo. Infect Immun. 2005;73(1):413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hildebrand D, Heeg K, Kubatzky K. Pasteurella multocida toxin manipulates T cell differentiation. Front Microbiol. 2015;6:1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pani G, Galeotti T, Chiarugi P. Metastasis: cancer cell’s escape from oxidative stress. Cancer Metastasis Rev. 2010;29(2):351–378. [DOI] [PubMed] [Google Scholar]
- 139.Elias B, Boros G, Albert M, et al. Clinical and pathological effects of the dermonecrotic toxin of Bordetella bronchiseptica and Pasteurella multocida in specific-pathogen-free piglets. Nihon Juigaku Zasshi. 1990;52(4):677–688. [DOI] [PubMed] [Google Scholar]
- 140.Balur MB, Koçak HE, Altınay S, Özdamar K, Taşkın Ü, Oktay MF. Is submucosal fat injection effective in atrophic rhinitis? An experimental animal study. Eur Arch Otorhinolaryngol. 2017;274(10):3637–3642. [DOI] [PubMed] [Google Scholar]
- 141.Correa P, Haenszel W, Cuello C, et al. Gastric precancerous process in a high risk population: cross-sectional studies. Cancer Res. 1990;50(15):4731–4736. [PubMed] [Google Scholar]
- 142.Correa P, Haenszel W, Cuello C, et al. Gastric precancerous process in a high risk population: cohort follow-up. Cancer Res. 1990;50(15):4737–4740. [PubMed] [Google Scholar]
- 143.Choudat D, Le Goff C, Delemotte B, et al. Occupational exposure to animals and antibodies against Pasteurella multocida. Br J Ind Med. 1987;44(12):829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Donnio PY, Le Goff C, Avril JL, Pouedras P, Gras-Rouzet S. Pasteurella multocida: oropharyngeal carriage and antibody response in breeders. Vet Res. 1994;25(1):8–15. [PubMed] [Google Scholar]
- 145.Avril JL, Donnio PY, Pouedras P. Selective medium for Pasteurella multocida and its use to detect oropharyngeal carriage in pig breeders. J Clin Microbiol. 1990;28(6): 1438–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Donnio PY, Avril JL, Andre PM, Vaucel J. Dermonecrotic toxin production by strains of Pasteurella multocida isolated from man. J Med Microbiol. 1991;34(6):333–337. [DOI] [PubMed] [Google Scholar]
- 147.Harris PJ, Osswald MB. Pasteurella multocida epiglottitis: a review and report of a new case with associated chronic lymphocytic leukemia. Ear Nose Throat J. 2010;89(12):E4–E7. [DOI] [PubMed] [Google Scholar]
- 148.Véleza M, Casanas B, Greene JN. Pasteurella multocida infections in cancer patients. Asian Biomed. 2010;4(3):449. [Google Scholar]
- 149.Brennan CA, Garrett WS. Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Han YW, Wang X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J Dent Res. 2013;92(6):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Han YW, Ikegami A, Rajanna C, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187(15):5330–5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nithianantham S, Xu M, Yamada M, Ikegami A, Shoham M, Han YW. Crystal structure of FadA adhesin from Fusobacterium nucleatum reveals a novel oligomerization motif, the leucine chain. J Biol Chem. 2009;284(6):3865–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rubinstein MR, Baik JE, Lagana SM, et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/β-catenin modulator Annexin A1. EMBO Rep. 2019;20(4): e47638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology. 2017;152(4):851–866.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Butt J, Jenab M, Pawlita M, et al. Antibody responses to Fusobacterium nucleatum proteins in prediagnostic blood samples are not associated with risk of developing colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2019;28(9):1552–1555. [DOI] [PubMed] [Google Scholar]
- 157.Coppenhagen-Glazer S, Sol A, Abed J, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect Immun. 2015;83(3):1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, Haake SK. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J Dent Res. 2005;84(8):700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Guevarra LA Jr, Afable ACF, Belza PJO, et al. Immunogenicity of a Fap2 peptide mimotope of Fusobacterium nucleatum and its potential use in the diagnosis of colorectal cancer. Infect Agent Cancer. 2018;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jewett A, Hume WR, Le H, et al. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun. 2000;68(4):1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaplan CW, Ma X, Paranjpe A, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010;78(11):4773–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Abed J, Emgard JE, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jones CG, Lawton JH, Shachak M. Organisms as ecosystem engineers. Oikos. 1994;69(3):373–386. [Google Scholar]
- 165.Yang KR, Mooney SM, Zarif JC, Coffey DS, Taichman RS, Pienta KJ. Niche inheritance: a cooperative pathway to enhance cancer cell fitness through ecosystem engineering. J Cell Biochem. 2014;115(9):1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60(2):208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Van Tassell RL, Lyerly DM, Wilkins TD. Purification and characterization of an enterotoxin from Bacteroides fragilis. Infect Immun. 1992;60(4):1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Myers LL, Shoop DS, Stackhouse LL, et al. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J Clin Microbiol. 1987;25(12):2330–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sears CL, Geis AL, Housseau F. Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J Clin Invest. 2014;124(10):4166–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Moore WE, Cato EP, Holdeman LV. Some current concepts in intestinal bacteriology. Am J Clin Nutr. 1978;31(10 suppl):S33–S42. [DOI] [PubMed] [Google Scholar]
- 171.Wexler HM. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev. 2007;20(4):593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Simon GL, Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984;86(1):174–193. [PubMed] [Google Scholar]
- 173.Moncrief JS, Obiso R, Barroso LA, et al. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect Immun. 1995;63(1):175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kling JJ, Wright RL, Moncrief JS, Wilkins TD. Cloning and characterization of the gene for the metalloprotease enterotoxin of Bacteroides fragilis. FEMS Microbiol Lett. 1997;146(2):279–284. [DOI] [PubMed] [Google Scholar]
- 175.Wilson MM, Anderson DE, Bernstein HD. Analysis of the outer membrane proteome and secretome of Bacteroides fragilis reveals a multiplicity of secretion mechanisms. PLoS One. 2015;10(2):e0117732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A. 2016;113(13):3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Silverman JM, Agnello DM, Zheng H, et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013; 51(5):584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Obiso RJ, Azghani AO, Wilkins TD. The Bacteroides fragilis toxin fragilysin disrupts the paracellular barrier of epithelial cells. Infect Immun. 1997;65(4):1431–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Popoff M Concepts in bacterial virulence. In: Russell W, Herwald H, eds. Contributions to Microbiology. Switzerland: Reinhardt Druck, Basel, Karger; 2004:31–34. [Google Scholar]
- 181.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400. [DOI] [PubMed] [Google Scholar]
- 182.Donelli G, Fabbri A, Fiorentini C. Bacteroides fragilis enterotoxin induces cytoskeletal changes and surface blebbing in HT-29 cells. Infect Immun. 1996;64(1):113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci. 2007;120(pt 11):1944–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Goodwin AC, Shields CED, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(37):15354–15359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Allen J, Hao S, Sears CL, Timp W. Epigenetic changes induced by Bacteroides fragilis toxin. Infect Immun. 2019;87(6):pii: e00447–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Housseau F, Sears CL. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/−) mice: a human commensal-based murine model of colon carcinogenesis. Cell Cycle. 2010;9(1):3–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009;206(7): 1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhu Q, Jin Z, Wu W, et al. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One. 2014;9(6):e90849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8): 782–786. [DOI] [PubMed] [Google Scholar]
- 190.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359(6375): 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Faïs T, Delmas J, Barnich N, Bonnet R, Dalmasso G. Colibactin: more than a new bacterial toxin. Toxins (Basel). 2018;10(4):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nougayrède JP, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. [DOI] [PubMed] [Google Scholar]
- 194.Healy AR, Vizcaino MI, Crawford JM, Herzon SB. Convergent and modular synthesis of candidate precolibactins. Structural revision of precolibactin A. J Am Chem Soc. 2016;138(16):5426–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Xue M, Kim CS, Healy AR, et al. Structure elucidation of colibactin and its DNA cross-links. Science. 2019;365(6457):eaax2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vizcaino MI, Crawford JM. The colibactin warhead crosslinks DNA. Nat Chem. 2015;7(5):411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wilson MR, Jiang Y, Villalta PW, et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science. 2019;363(6428):pii: eaar7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li Z-R, Li J, Cai W, et al. Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage. Nat Chem. 2019;11(10):880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.McCarthy AJ, Martin P, Cloup E, Stabler RA, Oswald E, Taylor PW. The genotoxin colibactin is a determinant of virulence in Escherichia coli K1 experimental neonatal systemic infection. Infect Immun. 2015;83(9):3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Raisch J, Rolhion N, Dubois A, Darfeuille-Michaud A, Bringer MA. Intracellular colon cancer-associated Escherichia coli promote protumoral activities of human macrophages by inducing sustained COX-2 expression. Lab Invest. 2015;95(3):296–307. [DOI] [PubMed] [Google Scholar]
- 202.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol. 2008;46(12):3906–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Mousa JJ, Yang Y, Tomkovich S, et al. MATE transport of the E. coli-derived genotoxin colibactin. Nat Microbiol. 2016;1:15009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Secher T, Samba-Louaka A, Oswald E, Nougayrède JP. Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PLoS One. 2013;8(10):e77157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. 2010;107(25):11537–11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marcq I, Martin P, Payros D, et al. The genotoxin colibactin exacerbates lymphopenia and decreases survival rate in mice infected with septicemic Escherichia coli. J Infect Dis. 2014;210(2):285–294. [DOI] [PubMed] [Google Scholar]
- 207.Tomkovich S, Yang Y, Winglee K, et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017;77(10):2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Payros D, Secher T, Boury M, et al. Maternally acquired genotoxic Escherichia coli alters offspring’s intestinal homeostasis. Gut Microbes. 2014;5(3):313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cougnoux A, Dalmasso G, Martinez R, et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63(12):1932–1942. [DOI] [PubMed] [Google Scholar]
- 210.Eklöf V, Löfgren-Burström A, Zingmark C, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141(12):2528–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nowrouzian FL, Oswald E. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb Pathog. 2012;53(3–4):180–182. [DOI] [PubMed] [Google Scholar]
- 212.Knust Z, Schmidt G. Cytotoxic necrotizing factors (CNFs)—a growing toxin family. Toxins (Basel). 2010;2(1):116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Hoffmann C, Pop M, Leemhuis J, Schirmer J, Aktories K, Schmidt G. The Yersinia pseudotuberculosis cytotoxic necrotizing factor (CNFY) selectively activates RhoA. J Biol Chem. 2004;279(16):16026–16032. [DOI] [PubMed] [Google Scholar]
- 214.Sugai M, Hatazaki K, Mogami A, et al. Cytotoxic necrotizing factor type 2 produced by pathogenic Escherichia coli deamidates a gln residue in the conserved G-3 domain of the rho family and preferentially inhibits the GTPase activity of RhoA and rac1. Infect Immun. 1999;67(12):6550–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.De Rycke J, Phan-Thanh L, Bernard S. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J Clin Microbiol. 1989;27(5):983–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Caprioli A, Falbo V, Roda LG, Ruggeri FM, Zona C. Partial purification and characterization of an Escherichia coli toxic factor that induces morphological cell alterations. Infect Immun. 1983;39(3):1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Usein CR, Damian M, Tatu-Chitoiu D, et al. Prevalence of virulence genes in Escherichia coli strains isolated from Romanian adult urinary tract infection cases. J Cell Mol Med. 2001;5(3):303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993; 61(11):4909–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yu H, Kim KS. Ferredoxin is involved in secretion of cytotoxic necrotizing factor 1 across the cytoplasmic membrane in Escherichia coli K1. Infect Immun. 2010; 78(2):838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kouokam JC, Wai SN, Fällman M, Dobrindt U, Hacker J, Uhlin BE. Active cytotoxic necrotizing factor 1 associated with outer membrane vesicles from uropathogenic Escherichia coli. Infect Immun. 2006;74(4):2022–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Blumenthal B, Hoffmann C, Aktories K, Backert S, Schmidt G. The cytotoxic necrotizing factors from Yersinia pseudotuberculosis and from Escherichia coli bind to different cellular receptors but take the same route to the cytosol. Infect Immun. 2007;75(7):3344–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387(6634): 725–729. [DOI] [PubMed] [Google Scholar]
- 223.Bhakdi S, Muhly M, Korom S, Schmidt G. Effects of Escherichia coli hemolysin on human monocytes. Cytocidal action and stimulation of interleukin 1 release. J Clin Invest. 1990;85(6):1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhang Z, Aung KM, Uhlin BE, Wai SN. Reversible senescence of human colon cancer cells after blockage of mitosis/cytokinesis caused by the CNF1 cyclomodulin from Escherichia coli. Sci Rep. 2018;8(1):17780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Thomas W, Ascott ZK, Harmey D, Slice LW, Rozengurt E, Lax AJ. Cytotoxic necrotizing factor from Escherichia coli induces RhoA-dependent expression of the cyclooxygenase-2 Gene. Infect Immun. 2001;69(11):6839–6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Boyer L, Travaglione S, Falzano L, et al. Rac GTPase instructs nuclear factor-kappaB activation by conveying the SCF complex and IkBalpha to the ruffling membranes. Mol Biol Cell. 2004;15(3):1124–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Travaglione S, Fabbri A, Fiorentini C. The Rho-activating CNF1 toxin from pathogenic E. coli: a risk factor for human cancer development? Infect Agent Cancer. 2008;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Rippere-Lampe KE, Lang M, Ceri H, Olson M, Lockman HA, O’Brien AD. Cytotoxic necrotizing factor type 1-positive Escherichia coli causes increased inflammation and tissue damage to the prostate in a rat prostatitis model. Infect Immun. 2001;69(10):6515–6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rippere-Lampe KE, O’Brien AD, Conran R, Lockman HA. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf(1)) attenuates the virulence of uropathogenic Escherichia coli. Infect Immun. 2001;69(6):3954–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Guo Y, Zhang Z, Wei H, et al. Cytotoxic necrotizing factor 1 promotes prostate cancer progression through activating the Cdc42-PAK1 axis. J Pathol. 2017;243(2):208–219. [DOI] [PubMed] [Google Scholar]
- 231.Raisch J, Buc E, Bonnet M, et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol. 2014;20(21): 6560–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Dubois D, Delmas J, Cady A, et al. Cyclomodulins in urosepsis strains of Escherichia coli. J Clin Microbiol. 2010;48(6):2122–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cortajarena AL, Goñi FM, Ostolaza H. Asp-863 is a key residue for calcium-dependent activity of Escherichia coli RTX toxin alpha-haemolysin. FEBS Lett. 2003;546(2–3): 271–275. [DOI] [PubMed] [Google Scholar]
- 234.Jorgensen SE, Short EC, Kurtz HJ, Mussen HK, Wu GK. Studies on the origin of the alpha-haemolysin produced by Escherichia coli. J Med Microbiol. 1976;9(2):173–189. [DOI] [PubMed] [Google Scholar]
- 235.LovelL R, Rees TA. A filterable haemolysin from Escherichia coli. Nature. 1960; 188:755–756. [DOI] [PubMed] [Google Scholar]
- 236.Smith HW. The haemolysins of Escherichia coli. J Pathol Bacteriol. 1963;85:197–211. [DOI] [PubMed] [Google Scholar]
- 237.Troeger H, Richter JF, Beutin L, et al. Escherichia coli alpha-haemolysin induces focal leaks in colonic epithelium: a novel mechanism of bacterial translocation. Cell Microbiol. 2007;9(10):2530–2540. [DOI] [PubMed] [Google Scholar]
- 238.Goebel W, Hedgpeth J. Cloning and functional characterization of the plasmidencoded hemolysin determinant of Escherichia coli. J Bacteriol. 1982;151(3):1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Bhakdi S, Mackman N, Nicaud JM, Holland IB. Escherichia coli hemolysin may damage target cell membranes by generating transmembrane pores. Infect Immun. 1986;52(1):63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Felmlee T, Pellett S, Welch RA. Nucleotide sequence of an Escherichia coli chromosomal hemolysin. J Bacteriol. 1985;163(1):94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Gentschev I, Dietrich G, Goebel W. The E. coli alpha-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 2002;10(1):39–45. [DOI] [PubMed] [Google Scholar]
- 242.Balsalobre C, Silván JM, Berglund S, Mizunoe Y, Uhlin BE, Wai SN. Release of the type I secreted alpha-haemolysin via outer membrane vesicles from Escherichia coli. Mol Microbiol. 2006;59(1):99–112. [DOI] [PubMed] [Google Scholar]
- 243.Bhakdi S, Greulich S, Muhly M, et al. Potent leukocidal action of Escherichia coli hemolysin mediated by permeabilization of target cell membranes. J Exp Med. 1989;169(3):737–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Dhakal BK, Mulvey MA. The UPEC pore-forming toxin α-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe. 2012;11(1):58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Jin Y, Tang S, Li W, et al. Hemolytic E. coli promotes colonic tumorigenesis in females. Cancer Res. 2016;76(10):2891–2900. [DOI] [PubMed] [Google Scholar]
- 246.Marre R, Hacker J, Henkel W, Goebel W. Contribution of cloned virulence factors from uropathogenic Escherichia coli strains to nephropathogenicity in an experimental rat pyelonephritis model. Infect Immun. 1986;54(3):761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Russo TA, Davidson BA, Genagon SA, et al. E. coli virulence factor hemolysin induces neutrophil apoptosis and necrosis/lysis in vitro and necrosis/lysis and lung injury in a rat pneumonia model. Am J Physiol Lung Cell Mol Physiol. 2005;289(2):L207–L216. [DOI] [PubMed] [Google Scholar]
- 248.Darfeuille-Michaud A, Neut C, Barnich N, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998; 115(6):1405–1413. [DOI] [PubMed] [Google Scholar]
- 249.Hilali F, Ruimy R, Saulnier P, et al. Prevalence of virulence genes and clonality in Escherichia coli strains that cause bacteremia in cancer patients. Infect Immun. 2000;68(7):3983–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Cianflone NFC. Salmonellosis and the GI tract: more than just peanut butter. Curr Gastroenterol Rep. 2008;10(4):424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kato I, Boleij A, Kortman GA, et al. Partial associations of dietary iron, smoking and intestinal bacteria with colorectal cancer risk. Nutr Cancer. 2013;65(2):169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mughini-Gras L, Schaapveld M, Kramers J, et al. Increased colon cancer risk after severe Salmonella infection. PLoS One. 2018;13(1):e0189721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hayward MR, AbuOun M, La Ragione RM, et al. SPI-23 of S. Derby: role in adherence and invasion of porcine tissues. PLoS One. 2014;9(9):e107857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Sabbagh SC, Forest CG, Lepage C, Leclerc J-M, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010;305(1):1–13. [DOI] [PubMed] [Google Scholar]
- 255.Van Engelenburg SB, Palmer AE. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat Methods. 2010;7(4):325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Phoebe Lostroh C, Lee CA. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 2001;3(14–15):1281–1291. [DOI] [PubMed] [Google Scholar]
- 257.Costa TRD, Felisberto-Rodrigues C, Meir A, et al. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13(6):343–359. [DOI] [PubMed] [Google Scholar]
- 258.Gaytan MO, Martinez-Santos VI, Soto E, Gonzalez-Pedrajo B. Type three secretion system in attaching and effacing pathogens. Front Cell Infect Microbiol. 2016;6:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Huehn S, La Ragione RM, Anjum M, et al. Virulotyping and antimicrobial resistance typing of Salmonella enterica serovars relevant to human health in Europe. Foodborne Pathog Dis. 2009;7(5):523–535. [DOI] [PubMed] [Google Scholar]
- 260.Prager R, Mirold S, Tietze E, et al. Prevalence and polymorphism of genes encoding translocated effector proteins among clinical isolates of Salmonella enterica. Int J Med Microbiol. 2000;290(7):605–617. [DOI] [PubMed] [Google Scholar]
- 261.Du F, Galán JE. Selective inhibition of Type III secretion activated signaling by the salmonella effector AvrA. PLoS Pathog. 2009;5(9):e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Labriola JM, Zhou Y, Nagar B. Structural analysis of the bacterial effector AvrA identifies a critical helix involved in substrate recognition. Biochemistry. 2018;57(33): 4985–4996. [DOI] [PubMed] [Google Scholar]
- 263.Streckel W, Wolff A-C, Prager R, Tietze E, Tschäpe H. Expression profiles of effector proteins SopB, SopD1, SopE1, and AvrA differ with systemic, enteric, and epidemic strains of Salmonella enterica. Mol Nutr Food Res. 2004;48(7):496–503. [DOI] [PubMed] [Google Scholar]
- 264.Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10(1):24–29. [DOI] [PubMed] [Google Scholar]
- 265.Kerrinnes T, Zelas ZB-B, Streckel W, et al. CsrA and CsrB are required for the post-transcriptional control of the virulence-associated effector protein AvrA of Salmonella enterica. Int J Med Microbiol. 2009;299(5):333–341. [DOI] [PubMed] [Google Scholar]
- 266.Ben-Barak Z, Streckel W, Yaron S, Cohen S, Prager R, Tschäpe H. The expression of the virulence-associated effector protein gene avrA is dependent on a Salmonella enterica-specific regulatory function. Int J Med Microbiol. 2006;296(1):25–38. [DOI] [PubMed] [Google Scholar]
- 267.Timmermans J, Van Melderen L. Post-transcriptional global regulation by CsrA in bacteria. Cell Mol Life Sci. 2010;67(17):2897–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Vakulskas CA, Potts AH, Babitzke P, Ahmer BMM, Romeo T. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev. 2015;79(2):193–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ma K-W, Ma W. YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiol Mol Biol Rev. 2016;80(4):1011–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zhang Z-M, Ma K-W, Yuan S, et al. Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat Struct Mol Biol. 2016;23:847. [DOI] [PubMed] [Google Scholar]
- 271.Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 2008;3(4):233–244. [DOI] [PubMed] [Google Scholar]
- 272.Sun J Pathogenic bacterial proteins and their anti-inflammatory effects in the eukaryotic host. Anti-Inflammatory Anti-Allergy Agents Med Chem. 2009;8(3):214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Wu S, Ye Z, Liu X, et al. Salmonella typhimurium infection increases p53 acetylation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;298(5): G784–G794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Reed SM, Quelle DE. p53 acetylation: regulation and consequences. Cancer. 2015; 7(1):30–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Collier-Hyams LS, Zeng H, Sun J, et al. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-κB pathway. J Immunol. 2002;169(6): 2846–2850. [DOI] [PubMed] [Google Scholar]
- 276.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007; 171(3):882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Liu X, Wu S, Xia Y, et al. Wingless homolog Wnt11 suppresses bacterial invasion and inflammation in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2011;301(6):G992–G1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Liu X, Lu R, Wu S, et al. Wnt2 inhibits enteric bacterial-induced inflammation in intestinal epithelial cells. Inflamm Bowel Dis. 2012;18(3):418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wang J, Lu R, Fu X, et al. Novel regulatory roles of Wnt1 in infection-associated colorectal cancer. Neoplasia. 2018;20(5):499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Liu X, Lu R, Wu S, Sun J. Salmonella regulation of intestinal stem cells through the Wnt/β-catenin pathway. FEBS Lett. 2010;584(5):911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Lu R, Liu X, Wu S, et al. Consistent activation of the β-catenin pathway by Salmonella type-three secretion effector protein AvrA in chronically infected intestine. Am J Physiol Gastrointest Liver Physiol. 2012;303(10):G1113–G1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lu R, Wu S, Zhang YG, et al. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis. 2014;3:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Lu R, Wu S, Zhang Y-G, et al. Salmonella protein AvrA activates the STAT3 signaling pathway in colon cancer. Neoplasia. 2016;18(5):307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Lu R, Bosland M, Xia Y, Zhang Y-G, Kato I, Sun J. Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget. 2017;8(33):55104–55115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 286.Siegl C, Rudel T. Modulation of p53 during bacterial infections. Nat Rev Microbiol. 2015;13:741. [DOI] [PubMed] [Google Scholar]
- 287.Silva-García O, Valdez-Alarcón JJ, Baizabal-Aguirre VM. Wnt/β-catenin signaling as a molecular target by pathogenic bacteria. Front Immunol. 2019;10:2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Rogan MR, Patterson LL, Wang JY, JW MB. Bacterial manipulation of Wnt signaling: a host-pathogen Tug-of-Wnt. Front Immunol. 2019;10:2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Escoll P, Buchrieser C. Metabolic reprogramming of host cells upon bacterial infection: why shift to a Warburg-like metabolism? FEBS J. 2018;285(12):2146–2160. [DOI] [PubMed] [Google Scholar]
- 290.Ramond E, Jamet A, Coureuil M, Charbit A. Pivotal role of mitochondria in macrophage response to bacterial pathogens. Front Immunol. 2019;10:2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Canonico B, Campana R, Luchetti F, et al. Campylobacter jejuni cell lysates differently target mitochondria and lysosomes on HeLa cells. Apoptosis. 2014;19(8): 1225–1242. [DOI] [PubMed] [Google Scholar]
- 292.Liyanage NPM, Manthey KC, Dassanayake RP, Kuszynski CA, Oakley GG, Duhamel GE. Helicobacter hepaticus cytolethal distending toxin causes cell death in intestinal epithelial cells via mitochondrial apoptotic pathway. Helicobacter. 2010;15(2): 98–107. [DOI] [PubMed] [Google Scholar]
- 293.Bielaszewska M, Rüter C, Kunsmann L, et al. Enterohemorrhagic Escherichia coli hemolysin employs outer membrane vesicles to target mitochondria and cause endothelial and epithelial apoptosis. PLoS Pathog. 2013;9(12):e1003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Tan X, Xu A, Zhao T, et al. Simulated microgravity inhibits cell focal adhesions leading to reduced melanoma cell proliferation and metastasis via FAK/RhoA-regulated mTORC1 and AMPK pathways. Sci Rep. 2018;8(1):3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Travaglione S, Loizzo S, Rizza T, et al. Enhancement of mitochondrial ATP production by the Escherichia coli cytotoxic necrotizing factor 1. FEBS J. 2014;281(15): 3473–3488. [DOI] [PubMed] [Google Scholar]
- 296.Fiorentini C, Matarrese P, Straface E, et al. Toxin-induced activation of Rho GTP-binding protein increases Bcl-2 expression and influences mitochondrial homeostasis. Exp Cell Res. 1998;242(1):341–350. [DOI] [PubMed] [Google Scholar]
- 297.Lin Z, Zhang Y-G, Xia Y, Xu X, Jiao X, Sun J. Salmonella enteritidis effector AvrA stabilizes intestinal tight junctions via the JNK pathway. J Biol Chem. 2016;291(52): 26837–26849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Liao AP, Petrof EO, Kuppireddi S, et al. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS One. 2008;3(6):e2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Merchant JL. Inflammation, atrophy, gastric cancer: connecting the molecular dots. Gastroenterology. 2005;129(3):1079–1082. [DOI] [PubMed] [Google Scholar]
- 300.Gambhir S, Vyas D, Hollis M, Aekka A, Vyas A. Nuclear factor kappa B role in inflammation associated gastrointestinal malignancies. World J Gastroenterol. 2015;21(11): 3174–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Gagnaire A, Nadel B, Raoult D, Neefjes J, Gorvel J-P. Collateral damage: insights into bacterial mechanisms that predispose host cells to cancer. Nat Rev Microbiol. 2017;15:109. [DOI] [PubMed] [Google Scholar]
- 302.Rosadi F, Fiorentini C, Fabbri A. Bacterial protein toxins in human cancers. Pathog Dis. 2016;74(1):ftv105. [DOI] [PubMed] [Google Scholar]