Abstract
Synaptosomal-associated protein of 23 kDa (SNAP-23) plays an important role during regulated exocytosis of various inflammatory mediators, stored in secretory granules, from mast cells in response to physiological triggers. It is however synthesized as a soluble protein, and the mechanisms by which free SNAP-23 gets peripherally associated with membrane for the regulation of exocytosis, are poorly defined. SNAP-23 contains a hydrophobic domain with five closely spaced cysteines which get palmitoylated, and we show that SNAP-23 cysteine mutants show differential membrane association when transfected in rat basophilic leukemia (RBL) mast cells. SNAP-23 Cys− mutant, devoid of all five cysteines, and SNAP-23 P119A (proline to alanine) mutant, that likely interferes with palmitoylation of SNAP-23 by palmitoyl transferases are completely cytosolic. Mutating specific cysteines (Cys; C) to leucine or phenylalanine (L or F; retains hydrophobicity but lacks palmitoylation) partially decreases the membrane association of SNAP-23 which is further hampered by alanine (A; has lesser hydrophobicity, and lacks palmitoylation) mutation at C79, C80 or C83 position. Cloning a transmembrane domain MDR31–145 from multidrug resistance protein into SNAP-23 Cys− mutant is able to partially restore its membrane association. Regulated exocytosis studies using co-transfected human growth hormone (hGH) secretion reporter plasmid revealed that over expression of SNAP-23 Cys− and P119A mutants significantly inhibits the overall extent of exocytosis from RBL mast cells, whereas expression of SNAP-23 Cys−-MDR31–145 fusion protein is able to restore exocytosis. These results establish that the cysteine-rich domain of SNAP-23 regulates its membrane association and thereby also regulates exocytosis from mast cells.
Keywords: Exocytosis, Mast cells, SNAP-23, Cysteine-rich domain, Membrane association, Protein palmitoylation
1. Introduction
Regulated exocytosis is critical for the functioning of mast cells which are important immune effector cells mediating various inflammatory and immunoregulatory responses. During regulated exocytosis, the pre-formed secretory granules in the cells move towards and fuse with the plasma membrane only upon receiving a physiological trigger, thereby releasing the granular contents into extracellular space. Soluble N-ethylmaleimide-sensitive-factor attachment protein receptors (SNAREs) mediated secretory granule-plasma membrane fusion is the essence of regulated exocytosis. SNAREs are a large family of membrane associated proteins essential for membrane-membrane fusion in all eukaryotes [1]. They have been categorized into members of the vesicle-associated membrane protein (VAMP) family of v-SNAREs and members of the syntaxin and SNAP-23/25 families of target membrane t-SNAREs [2]. One copy of v-SNAREs binds with cognate t-SNAREs, one copy each of syntaxin and SNAP-23 or the neuronal SNAP-25 to form a trimolecular complex, whose formation drives the energetically demanding process of membrane fusion [3]. Since SNAREs regulate the membrane fusion process, their functioning must also be tightly regulated. Post-translational modifications of SNAREs and the binding of regulatory/accessory proteins are thought to be the main mechanisms which can temporally and spatially regulate the formation of trans-SNARE complexes [3] and research is underway to understand the molecular mechanisms regulating SNAREs so as to identify ways to modulate exocytosis.
Our results predict similar canonical cysteine motif with five cysteines along with downstream proline residue conserved in linker region of SNAP-23. SNAP-23/25 is an essential component of ternary SNARE complex. While SNAP-25 is a neuronal t-SNARE, SNAP-23 is it’s 60% identical homologue which is ubiquitously expressed in all nonneuronal cells [4,5]. Previous studies have shown critical role of canonical cysteine motif with four cysteines along with downstream proline conserved in the linker region of SNAP-25 in membrane targeting and fusion [6]. SNAP-23 is a peripheral membrane protein [7–9] which is however, synthesized as a soluble protein lacking any transmembrane domain or membrane-targeting signal and the mechanism of its membrane association is poorly defined.
SNAP-23 contains two coiled-coil SNARE motifs linked by a hydrophobic cysteine-rich domain containing five closely spaced cysteines which get palmitoylated [10]. Palmitoylation is a post-translational protein modification that involves the attachment of hydrophobic palmitate group to cysteine residues of proteins via a labile thioester linkage and helps in targeting otherwise soluble proteins to cell membranes [11–14]. Interestingly, palmitate linkage is reversible and palmitoylation dynamics of proteins is regulated by membrane bound DHHC (aspartic acid-histidine-histidine-cysteine) palmitoyl transferases (PATs) and cytosolic protein acyl thioesterases [14–18]. It has been shown that cysteine residues and other surrounding amino acids of SNAP-25 are important in initial membrane binding via hydrophobic interactions [6]. Initial access to membrane helps in interacting with membrane bound DHHC proteins, which palmitoylate SNAP-25 and promote stable membrane attachment. Thus, palmitoylation of cysteine residues is thought to regulate the intracellular localization and hence functional activity of palmitoylated proteins [14].
Mutations within the hydrophobic cysteine-rich domain have been reported to perturb palmitoylation, hence, the targeting and stable membrane association of neuronal SNAP-25 [6,19]; and membrane binding of syndet, i.e., mouse SNAP-23 [20]. Previous studies [9] have found two induced phosphorylation sites, serines S95 and S120 in close proximity to the cysteine residues in the hydrophobic linker region of SNAP-23. These sites get phosphorylated during mast cells stimulation and overexpression of SNAP-23 phosphorylation mutants inhibits mast cell exocytosis [9,21,22]. Recently we have also seen that at least the basal phosphorylation site T102 present in the linker region close to conserved cysteines is also important for membrane association of SNAP-23 in mast cells [22]. Another study claims that the initial membrane association of SNAP-23 and SNAP-25 is due to electrostatic interactions that help its palmitoylation at plasma membrane [23]. In neither of these reports, functional role of the hydrophobic cysteine-rich domain of SNAP-23 in regulated exocytosis in mast cells has been investigated.
In the present study, we have investigated the role of the five conserved cysteines in regulating its membrane association and their physiological importance in regulating exocytosis from RBL mast cells. RBL-2H3 (mentioned in short as RBL) cell line has been extensively studied as a model for mast cell biology [24]. Important aspects of mast cell physiology like degranulation of various pro-inflammatory mediators following cross-linking of their IgE-bound FcεRI receptors by multivalent antigens are observed in RBL cells. We first used the bioinformatic approach to predict the important cysteines and proline residues in the linker region of SNAP-23, and mutated those to investigate their roles. Wild-type SNAP-23 and its mutants were transfected and expressed as EGFP-fusion proteins in RBL cells for studying their membrane localization and their effect on regulated exocytosis from mast cells. Our data suggest that the conserved cysteine and proline residues in the linker region of SNAP-23 are essential for proper trafficking and membrane association of free SNAP-23 due to hydrophobic nature and dynamic palmitoylation chemistry of cysteines; and thereby also regulate exocytosis from mast cells.
2. Materials and methods
2.1. Recombinant plasmids, site-directed mutagenesis, bioinformatics and cloning
Mammalian expression vector EGFPC2 was from Clontech. cDNA encoding full-length human SNAP-23 and SNAP-23 Cys− mutant cloned in EGFPC2 [10], pCMV-FLAG-rat Syntaxin-4 and pCMV-FLAG rat Syntaxin-4 ΔCT mutant, and human growth hormone cDNA cloned in pcDNA3 mammalian expression vector have been described before [25]. Individual cysteine to leucine, cysteine to alanine mutations; C79F and P119A mutants were made in the coding sequence of wild type human SNAP-23 using Quick Change Site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions. SNAP-23 transmembrane fusion protein was designed using bioinformatics-tool. Multi-pass transmembrane proteins suitable for cloning into SNAP-23 were searched in Universal protein resource Knowledgebase (Uniprot KB). Individual transmembrane domains with appropriate topology were further narrowed down using transmembrane topology prediction model TMMOD and literature mining [26–29]. To make SNAP-23 Cys−-MDR31–145 fusion construct, total RNA was isolated from LS180, a human colon adenocarcinoma cell line (a kind gift from Prof. Rakesh K. Tyagi, SCMM, JNU, New Delhi) using Trizol reagent (Sigma). Coding region for MDR31–145 domain was amplified by One-step RT-PCR kit (Qiagen) according to manufacturer’s instructions using sequence specific forward primer 5′GCGCGACAAATGGTCAGATGGATCTTGAGGC3′ and reverse primer 5′GCGCGACCATTTGTCTGTCGACCAGCTGC3′ incorporating AhdI (Eam 1105I) site, unique restriction enzyme site in the linker region of EGFPC2-SNAP-23 Cys− plasmid (Restriction mapper version 3). The amplified sequence was digested by AhdI enzyme (NEB) and ligated into AhdI digested EGFPC2-SNAP-23 Cys− plasmid using Quick Ligation kit (NEB). All the mutants and the fusion construct were confirmed by sequencing. FLAG-rat Syntaxin-4 and FLAG rat Syntaxin-4 ΔCT mutant were subcloned into EGFP-C2 (Clontech) plasmid by using EcoRI and ApaI restriction sites. The integrity of subcloned plasmids was confirmed by sequencing from GCC Biotech and SciGenom Labs Pvt. Ltd., India.
2.2. Multiple sequence alignment (MSA) and generation of the sequence logo
SNARE protein sequences belonging to SNAP-23 and SNAP-25 isoform b subfamilies were extracted from Uniprot database (http://www.uniprot.org). The resultant dataset thus included 46 members of SNAP-23 and 67 members of SNAP-25b after removal of redundant sequences. The sequences were then aligned using the multiple sequence alignment programme MAFFT [30] with default parameters and the linker region corresponding to SNAP 23 and SNAP-25b sequences were excised from the alignment. The sequence logo was generated from the multiple sequence alignment for aligned linker region of SNAP-23 and SNAP-25 isoform b using weblogo 3 sequence logo generator (http://weblogo.berkeley.edu/logo.cgi).
2.3. Antibodies
Rabbit polyclonal anti-serum recognizing the SNAP-23 carboxyl terminus has been described previously [7,9]. Anti–DNP IgE (clone TIB 142) was obtained from the American Type Culture Collection (Manassas, VA, USA). Anti-GFP rabbit mAb antibody is from Clontech (Living Colors), CA, USA. For Western blotting anti-mouse IgG-HRP conjugated antibody (Southern Biotech, Birmingham, AL, USA) and anti-Rabbit Protein A-HRP conjugated antibody (Southern Biotech, Birmingham, AL, USA) were used. Alexa dye-conjugated secondary antibodies were obtained from Molecular Probes (Eugene, OR, USA).
2.4. Cell culture and transfections
Rat basophilic leukemia mast cells (RBL—2H3), were grown in equal parts of minimal essential medium and Iscove’s medium containing 20% fetal bovine serum (Gibco, life technologies, Grand Island, NY, USA), 25 mM N-2-Hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) (Sigma, MO, USA) (pH 7.2), and 120 μg/ml gentamicin (RBL medium). Cells were maintained as subconfluent monolayers at 37 °C in a humidified atmosphere containing 5% CO2 and passaged with trypsin. Exponentially growing RBL cells were harvested and re-suspended in serum-free, antibiotic-free RBL medium (10 million cells per 0.5 ml medium) and were transfected by electroporation (320 V, 950 μF, Bio-Rad Gene Pulser) using total 20 μg of plasmid DNA as described previously [25]. Transfected cells were immediately plated in antibiotic-free RBL medium in tissue-culture dishes and processed for all experiments 24 h post-transfection unless otherwise indicated. Percentage transfection efficiencies and mean fluorescence intensities (Supplementary Fig. S1) of all plasmids were checked by flowcytometry and RBL cell recovery was also calculated in each case (Supplementary Fig. S2).
2.5. Subcellular fractionation, SDS-PAGE and immunoblotting
Membrane/cytosol enrichment analysis of RBL cells was done as described previously [9,22]. Briefly, 10 million cells were suspended in 1 ml of hypotonic buffer containing 1 mg/ml BSA and protease inhibitors [31], and were disrupted by repeated passage through a 30-gauge syringe. Nuclei and unbroken cells were removed by centrifugation and the post-nuclear supernatant was collected. A portion of it was analyzed by immunoblotting with anti-SNAP-23 c-terminus antibody [32] to compare the expression of transfected versus endogenous SNAP-23. The remaining supernatant was subjected to centrifugation at 100,000 g for 1 h at 4 °C to isolate membranes (pellet) and cytosol (supernatant) fractions. Each fraction was brought to the same volume in hypotonic buffer and was adjusted to a final concentration of 1% Triton X-100. Equivalent portions of each fraction were separated on 10.5% SDS-PAGE gels, transferred to Sequi-Blot PVDF membranes (Bio-Rad, USA) and analyzed by immunoblotting [25] with anti-GFP monoclonal antibody clone A.v. (Clontech, CA, USA) or anti-SNAP-23 c-terminus antibody. We have shown representative equivalent membrane/cytosol enrichment fraction blots out of three independent transfection-membrane/cytosol enrichment fractionation experiments for each SNAP-23 mutant. All blots for specific mutants were developed, and quantitated similarly. Each time for one PVDF membrane/blot, several films were developed with different exposure times (5 s to 1 min) for best visualization of the bands. The representative blots are shown with the molecular weight markers. Protein bands were visualized with Immobilon Western Chemiluminescent HRP substrate (Millipore, MA, USA). Band intensities were quantified by densitometry using AlphaImager gel documentation system and AlphaEaseFC 4.0 software as mentioned previously in [22]. Briefly, SNAP-23 membrane localization is calculated as [(Band intensity of SNAP-23 in membrane fraction/(Band intensity of SNAP-23 in membrane fraction + Band intensity of SNAP-23 in cytosolic fraction)) × 100], that is, we took the percent of SNAP-23 membrane association with respect to total SNAP-23 (Band intensity of SNAP-23 in membrane fraction + Band intensity of SNAP-23 in cytosolic fraction).
2.6. Confocal microscopy
RBL cells transfected with EGFP (Enhanced green fluorescent protein) tagged constructs were seeded on coverslips of 18 mm diameter and 0.15 mm thickness. Cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 30 min and excess paraformaldehyde was quenched with 50 mM NH4Cl in PBS as described earlier [25]. Briefly, after washing, the fixed cells were permeabilized with 1% IGEPAL (Sigma, MO, USA) in the presence of 3% normal goat serum (Sigma, MO, USA) and 0.05% saponin (SD Fine Chem. Limited, Boisar, India) in PBS. The cells were then incubated with 3% normal goat serum and 0.05% saponin in PBS for 1 h at room temperature to prevent nonspecific protein binding. Primary Abs diluted in the same buffer were added to the cells, and incubation was conducted for 1 h at room temperature. After washing, the cells were incubated for 30 min in the presence of secondary goat Abs conjugated to Alexa Fluor 546 (red) (Molecular Probes, Eugene, OR, USA). As a control, samples were stained with an irrelevant antibody and no staining was observed in the respective channel for all confocal fluorescence microscopy experiments. Coverslips were mounted in Fluoromount G (Southern Biotech, USA) and images having optical slice thickness of 1.0 μm were collected with Olympus Fluoview FV1000 laser scanning microscope or Nikon Ti2 microscope in X-Y direction using 60× or 100× magnification (sometimes with 2× zoom). Z-stacks of the images were collected with the depth of the cell using 0.4 μm step size. In each experiment, images were acquired using identical settings for all groups. Co-localization studies were carried out as described before [22]. Briefly, individual channels were thresholded by using the co-localization analysis feature of the NIS-Elements AR 5.01.00. The region of interest was considered for each cell, and a scatter plot was generated and co-localization coefficient value was calculated from it. The colocalization coefficient describes the co-localization in the green channel with respect to the red channel. All experiments were repeated three times. Intensity and contrast of images were adjusted without disturbing the gamma settings using the look-up table of Olympus Fluoview version 1.7a software. Single images were then saved as JPEG files and organized into figures using Microsoft Office PowerPoint 2007 and above. For some experiments Nikon Ti2 (Software-NIS-Elements AR 5.01.00,64-bit) was also used.
For quantitation of number of cells showing some plasma membrane association of transfected SNAP-23 Cys−-MDR31–145 fusion protein at 24 or 48 h after transfection in RBL cells, blinded observation (by an expert in mast cell degranulation studies, unaware of conditions under which experiments were done) was carried out. The DIC and green fluorescence channel images of mast cells were captured (Confocal microscope, Olympus, 100×) at 24 or 48 h after transfection with EGFP-tagged SNAP-23 Cys−-MDR31–145 fusion construct. The plasma membrane or completely cytosolic localization of green fluorescence was scored for at least 30 cells for each condition from at least three independent transfection experiments.
2.7. Mast cell exocytosis assay
RBL cells were co-transfected with 2 μg of human growth hormone (hGH) secretion reporter plasmid [25] together with 18 μg of empty vector or various test plasmids (EGFPC2 vector alone or EGFP-tagged SNAP-23 cysteine mutants, SNAP-23 P119A mutant or SNAP-23 Cys−-MDR31–145 fusion construct). Transfected cells were triggered for exocytosis by cross-linking their high affinity FcεRI receptor using previously described method [7]. Briefly, subconfluent cells were sensitized overnight with DNP (dinitrophenol)-specific IgE (TIB-142 hybridoma culture supernatant) and IgE sensitized cells were either mock-stimulated (resting) with media alone or stimulated with 100 ng/ml DNP-BSA (Bovine serum albumin) for 45 min. The amount of hGH released into the culture supernatant and remaining in the cell lysates was determined using hGH ELISA kit (Roche Diagnostics) and expressed as a percentage of net hGH (stimulated-resting) released in the supernatant relative to total hGH in the cells.
2.8. Statistical analysis
All results were expressed as mean ± SEM values of at least three independent experiments. Statistical calculations were done using a one-tailed distribution in a two-sample equal variance Student’s t-test. Results were considered significant when a p value of < 0.05 was obtained.
3. Results
3.1. Multiple sequence alignment predicts canonical cysteine and proline motifs in linker region of SNAP-23
Sequence logo generated from the Multiple sequence alignment of linker region corresponding to SNAP-23 and SNAP-25b predicts canonical cysteine motif along with downstream residues conserved in the linker region of SNAP-23 which have been also found to be conserved in SNAP-25b. The only difference observed in the cysteine motif is the first cysteine in SNAP-23 being replaced by phenylalanine in SNAP-25b. Multiple sequence alignment also revealed a proline residue present in QPARV motif in SNAP-25b also found to be conserved in QPXXV in SNAP-23 (Fig. 1A).
Fig. 1.
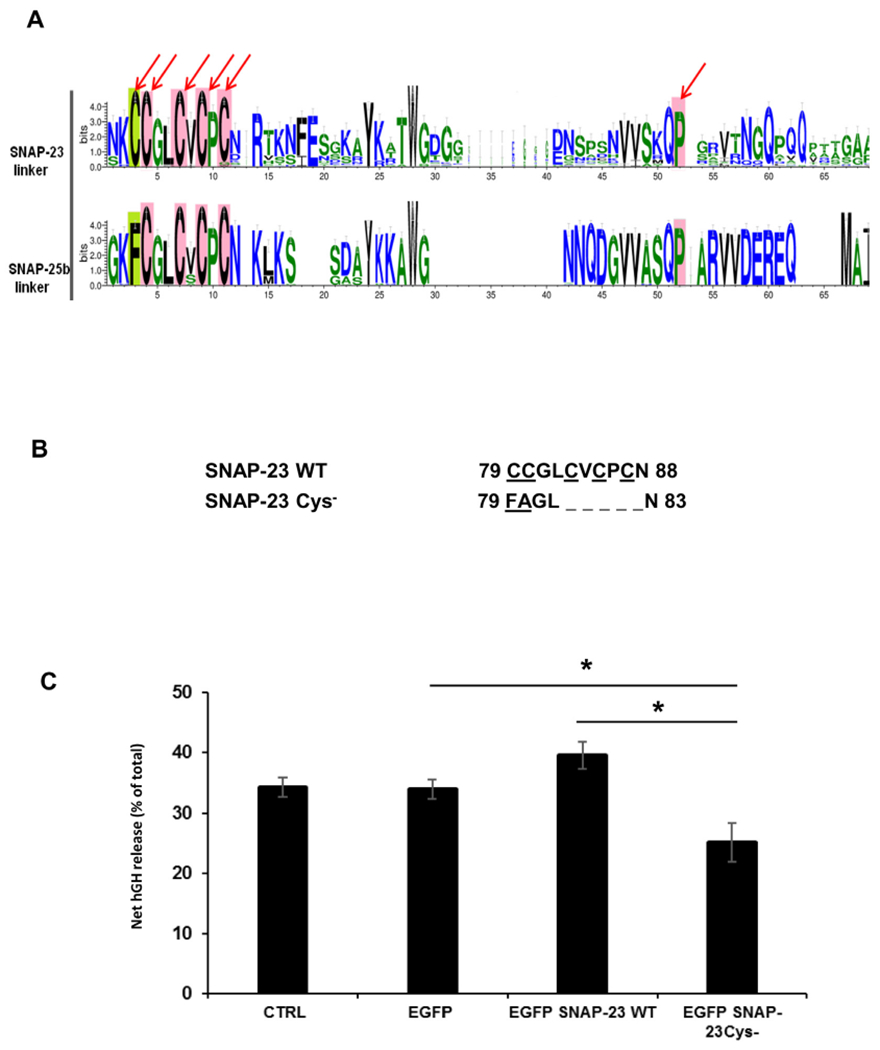
Importance of five conserved cysteines in linker region of SNAP-23 in RBL mast cell exocytosis. A. Comparison of canonical cysteine rich motif and downstream conserved residues in the minimal membrane binding sequence of linker region of SNAP-23 and SNAP-25b proteins. The figure compares the motif logo representation generated from aligned sequences for SNAP-25b and SNAP-23. The residues highlighted with pink blocks are globally conserved in SNAP-23 and SNAP25b while the first cysteine highlighted in green block is differentially conserved, that is, cysteine in SNAP-23 being replaced with phenylalanine in SNAP-25b. The residues pointed with arrows were selected and further subjected to site directed mutagenesis in human SNAP-23. B. Amino acid sequence of the cysteine-rich domain of SNAP-23 and of the corresponding region of SNAP-23 Cys− mutant. C. Inhibition of RBL mast cell exocytosis on transfection of SNAP-23 Cys− mutant. RBL cells were co-transfected with hGH secretion reporter plasmid together with either empty vector (control), EGFPC2 vector alone or EGFP-tagged SNAP-23 WT, SNAP-23 Cys− mutants construct. The transfected cells were sensitized with DNP-specific IgE and either mock-stimulated (resting) or stimulated by IgE cross-linking with DNP-BSA for 45 min. The extent of degranulation was determined by measuring the amount of hGH released from the cells and was expressed as percent of net hGH (stimulated - resting) released in the supernatant relative to total hGH in the cells after stimulation. Each data point represents mean ± SEM of three independent experiments. Asterisks indicate statistically significant differences in net hGH release from mast cells caused by overexpressing SNAP-23 variants as compared to wild-type SNAP-23 overexpression (*, p < 0.05).
3.2. Overexpression of SNAP-23 Cys− mutant inhibits regulated exocytosis from transiently transfected RBL mast cells
The t-SNARE SNAP-23 is important for regulated exocytosis in different cell types including mast cells [9,10,33–35]. In mast cells, we and others have previously shown that SNAP-23 is peripherally associated with the plasma membrane for the regulation of exocytosis, even though it lacks a transmembrane domain or membrane-targeting signal [7–9]. SNAP-23 contains five conserved cysteine residues in its hydrophobic linker region, and these cysteine residues have been shown to undergo palmitoylation, and thus have been thought to be the main anchors holding SNAP-23 at the membrane for the regulation of exocytosis [10]. In order to study the importance of these cysteine residues in the process of regulated exocytosis, we first obtained cDNA encoding wild type SNAP-23 and a mutant lacking all five cysteines in the linker region of SNAP-23 (Fig. 1B) [10], and cloned these into EGFP vector, for easier tracking of expression of these proteins in RBL mast cells. RBL mast cells were transiently transfected with these EGFP-tagged SNAP-23 WT and SNAP-23 Cys− mutant and percentage transfection efficiency and mean fluorescence intensity were checked by flowcytometry (Supplementary Fig. S1) to monitor the expression of transfected protein. Post-transfection cell recoveries were also calculated in each case (Supplementary Fig. S2) as further experiments to be done were based on exact cell numbers. Transfection efficiency was 50–60% in each case (Supplementary Fig. S1), but cell recovery was less than one-fourth (0.4 million) in case of SNAP-23 Cys− mutant transfection as compared to SNAP-23 WT transfected cells (1.8 million; Supplementary Fig. S2). Numbers of transfections were done and cells were seeded accordingly for further experiments. To check if these cysteine residues were important for regulated exocytosis from mast cells, RBL cells were transiently co-transfected with either wild type or the various cysteine mutants of SNAP-23 or the SNAP-23 transmembrane fusion construct together with hGH secretion reporter plasmid as standardized earlier [25]. Transfected cells were sensitized with DNP-specific IgE and either mock-stimulated (resting) or stimulated by IgE cross-linking with DNP-BSA for 45 min as described in materials and methods. The percentage of net hGH secretion (stimulated-resting) was calculated from the cells overexpressing each construct to determine their effect on mast cell regulated exocytosis. As shown in Fig. 1C, overexpression of SNAP-23 Cys− mutant significantly inhibited exocytosis of hGH from the transfected RBL cells (only 25% net hGH release after 45 min of stimulation as compared to 39% in case of SNAP-23 WT overexpression). These results confirm that cysteine-rich domain of SNAP-23 regulates mast cell exocytosis. There were no significant differences between total hGH in these mast cell transfectants (data not shown). These results suggest that certain cysteine residues in the linker region of SNAP-23 play an important role in regulating mast cell degranulation.
3.3. SNAP-23 Cys− mutant fails to traffic to the plasma membrane and remains stuck in the cytosol of RBL mast cells
Having established the importance of cysteine residues in the linker region of SNAP-23 in regulating mast cell degranulation, we decided to study the importance of these cysteine residues in membrane localization of SNAP-23 in mast cells. Confocal microscopy was done to observe the location of transfected SNAP-23 at different time points after transfection in RBL cells. It was found that SNAP-23 WT could be visualized in the RBL cells after 6 h of transfection, it could be seen associated partially with the plasma membrane. After 12 h of transfection, almost all SNAP-23 WT was associated with plasma membrane. SNAP-23 Cys− mutant could also be detected 6 h after transfection, but it failed to traffic to the plasma membrane at all and remained stuck in the cytosol of RBL cells at all tested time points (Fig. 2A).
Fig. 2.
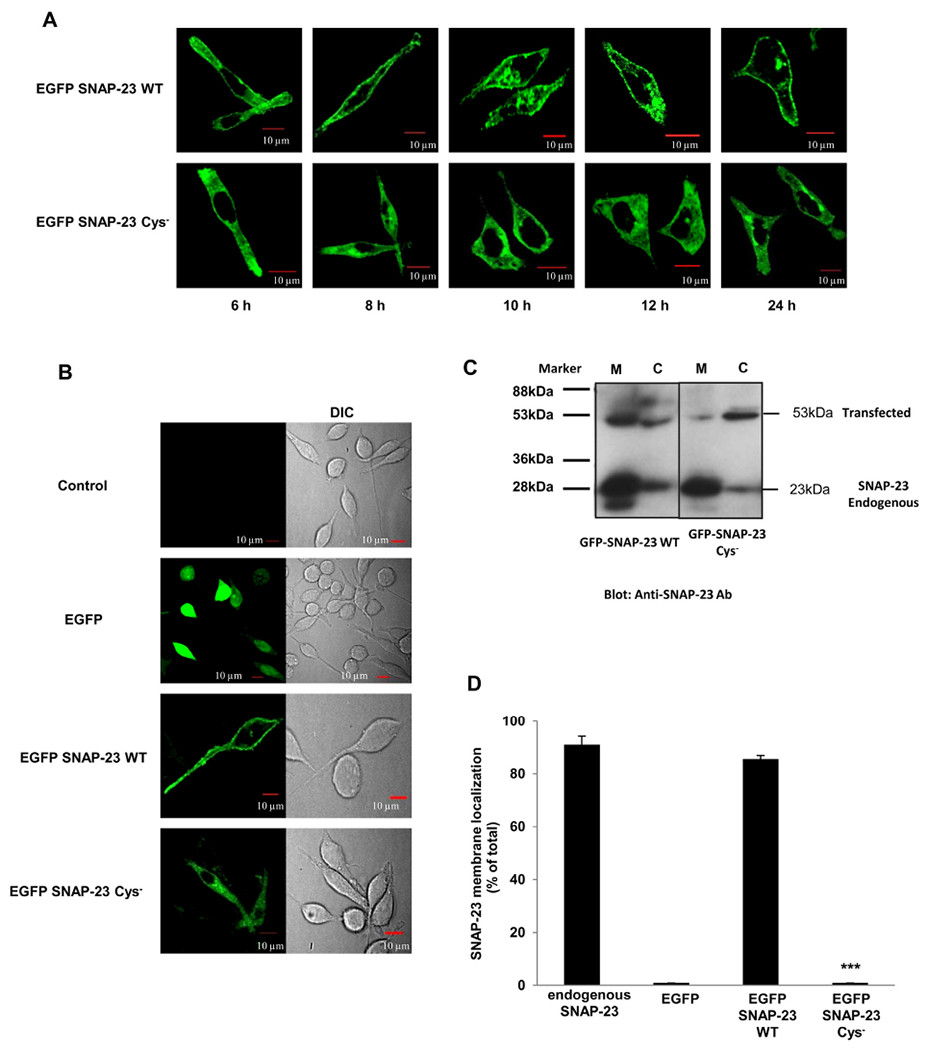
SNAP-23 Cys− mutant fails to traffic to the plasma membrane and remains stuck in the cytosol of RBL mast cells. A. RBL cells were transfected with EGFP-tagged SNAP-23 WT and SNAP-23 Cys− mutant. Transfected cells were plated on coverslips and analyzed by confocal microscopy after 6, 8, 10, 12 and 24 h post-transfection. Shown here are representative images. Scale = 10 μm. B. RBL cells were either mock transfected (control) or transfected with EGFPC2 vector, EGFP-tagged SNAP-23 WT or SNAP-23 Cys− mutant. The cells were analyzed by confocal microscopy after 24 h of transfection. Shown here are representative images. Scale = 10 μm. C. Transfected cells were also analyzed by membrane/cytosol enrichment analysis followed by immunoblotting with anti-SNAP-23 c-terminus or anti-GFP antibody. The representative immunoblots are shown here along with the molecular weight markers. The image is of a single blot having membrane/cytosol enrichment analysis for both SNAP-23 WT and Cys− mutant transfected cells. The black line in between the lanes represents an empty lane. EGFP and endogenous SNAP-23 served as cytosolic and membrane marker proteins respectively. D. Band intensities from three independent experiments were quantified by densitometry and expressed as percentage of SNAP-23 membrane localization with respect to total SNAP-23 (mean ± SEM). *** indicate that displacement of SNAP-23 Cys− mutant from the membrane is statistically significant as compared to membrane association of wild-type SNAP-23 (p < 0.0005).
To test the membrane association of SNAP-23 WT and SNAP-23 Cys− mutant, we transfected the EGFP-tagged constructs into RBL cells and the transfected cells were either analyzed by confocal microscopy (Fig. 2A, B & Supplementary Fig. S3) or subjected to membrane/cytosol enrichment fractionation followed by immunoblotting (Fig. 2C & D). As expected, EGFP alone was expressed in the cytosol and EGFP-tagged SNAP-23 WT was found to localize mainly to the plasma membrane (86% by immunoblotting; 78% by microscopic analysis) of RBL cells (Fig. 2D & Supplementary Fig. S3). Vogel et al. [10] have shown that SNAP-23 Cys− mutant does not get palmitoylated. Not to our surprise, we found that SNAP-23 Cys− mutant completely failed to anchor to the plasma membrane and was seen stuck in the cytosol of RBL cells (Fig. 2A, last panel; Fig. 2B, C, D & Supplementary Fig. S3). The inability of SNAP-23 Cys− mutant to localize to the plasma membrane suggests that either some or all cysteine residues in the linker region of SNAP-23 are required for anchoring of SNAP-23 to the plasma membrane in mast cells.
3.4. SNAP-23 P119A mutant behaves like SNAP-23 Cys− mutant in terms of its trafficking, membrane association, and regulated exocytosis in RBL mast cells
It has been previously reported that SNAP-25 P117A mutant which inhibits palmitoylation of SNAP-25 by DHHC17, also significantly inhibits the membrane binding of SNAP-25 [6,36]. So, we were interested in studying the effect of corresponding P119A mutant of SNAP-23 which seems to reside in a similarly conserved motif (Fig. 3A) on its trafficking and membrane association. RBL cells were successfully transfected with EGFP-tagged SNAP-23 P119A mutant with 55.3% transfection efficiency (Supplementary Fig. S1). However, post-transfection cell recovery was significantly low (0.7 million) as compared to SNAP-23 WT transfection (1.8 million; Supplementary Fig. S2). Confocal microscopy was done to observe the intracellular location of the mutant protein at different time points after transfection in RBL cells. As in case of SNAP-23 Cys− mutant, the SNAP-23 P119A mutant could also be detected 6 h after transfection, but it failed to traffic to the plasma membrane and remained stuck in the cytosol of RBL cells at all-time points tested (Fig. 3B). To test the membrane association of SNAP-23 P119A mutant, the transfected RBL cells were either analyzed by confocal microscopy (Fig. 3B, C & Supplementary Fig. S3) or subjected to membrane/cytosol enrichment fractionation followed by immunoblotting (Fig. 3D & E) after 24 h of transfection. The P119A mutation completely shifted localization of SNAP-23 to cytosol [96% decrease in membrane association by immunoblotting, Fig. 3E; the microscopy data showed 86% reduction in membrane association (Supplementary Fig. S3)], unlike the similar P117A mutant of SNAP-25 that showed only 50% decrease in membrane binding as compared to wild-type SNAP-25 [6]. Since, SNAP-23 P119A mutant is likely to interfere with palmitoylation of SNAP-23 by DHHC palmitoyl transferases as in case of SNAP-25 P117A mutant, our results strongly suggest that palmitoylation of the cysteine-rich linker is essential for the membrane association of SNAP-23. Further, just like the SNAP-23 Cys− mutant, overexpression of SNAP-23 P119A mutant also significantly inhibited exocytosis of hGH from the transfected RBL cells (only 23% net hGH release after 45 min of stimulation as compared to 39% in case of SNAP-23 WT overexpression) (Fig. 3F).
Fig. 3.
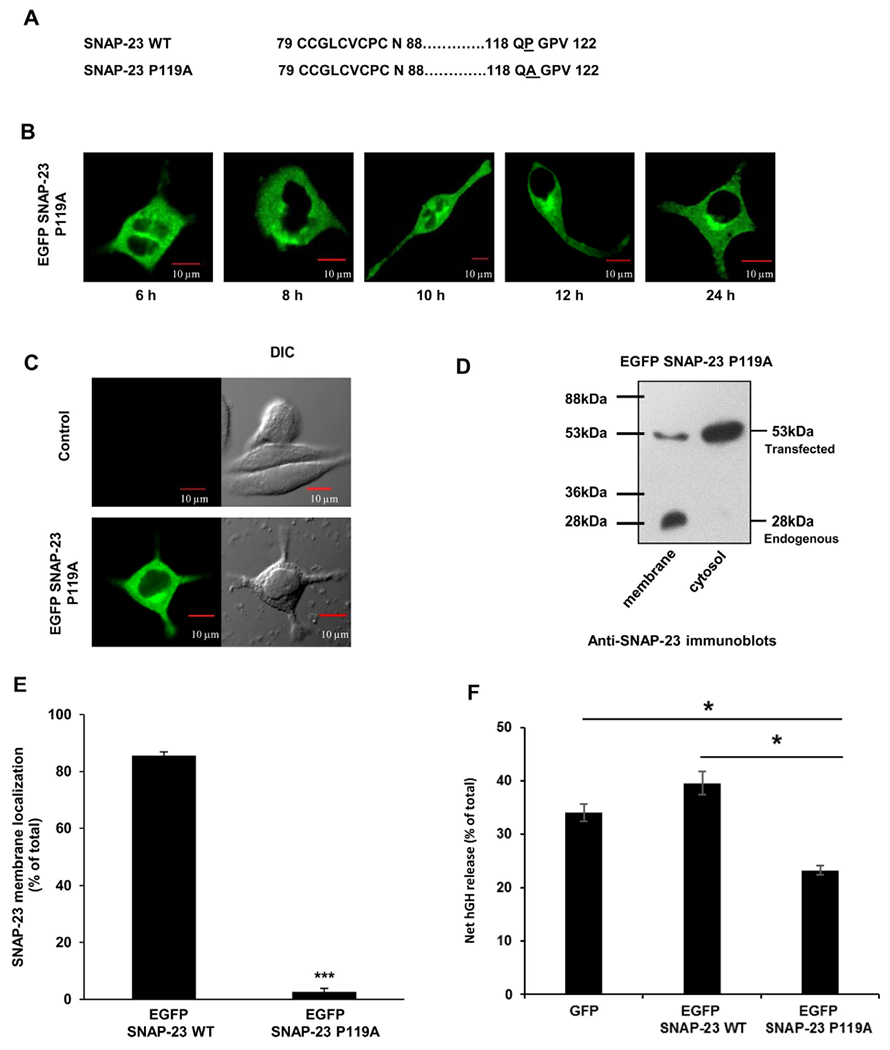
SNAP-23 P119A mutant fails to traffic to the plasma membrane and affects RBL mast cell exocytosis. A. Amino acid sequence of the cysteine-rich hydrophobic domain of SNAP-23 WT and of the corresponding region of SNAP-23 P119A mutant. B. RBL cells were transfected with EGFP-tagged SNAP-23 P119A mutant. Transfected cells were plated on coverslips and analyzed by confocal microscopy after 6, 8, 10, 12 and 24 h post-transfection. Shown here are representative images. Scale = 10 μm. C. RBL cells were either mock transfected (control) or transfected with EGFP-tagged SNAP-23 P119A mutant. Twenty-four hours post-transfection the cells were analyzed by confocal microscopy. Shown here are representative images. Scale = 10 μm. D. Transfected cells were also analyzed by membrane/cytosol enrichment followed by immunoblotting with anti-SNAP-23 antibody. Here the representative immunoblots are showing the transfected as well as the endogenous protein along with the position of the molecular weight markers. E. Band intensities from three independent experiments were quantified by densitometry and expressed as SNAP-23% membrane localization with respect to total SNAP-23 (mean ± SEM), (***, p < 0.0005). F. The extent of hGH secretion is assayed by using hGH kit (discussed in materials and methods) in EGFP, EGFP SNAP-23 WT and EGFP SNAP-23 P119A mutant transfected RBL cells at 45 min after allergen cross linking. Each data point is a mean ± SEM of three independent experiments (*, p > 0.005).
3.5. Importance of individual cysteine residues in the linker region of SNAP-23 in its membrane association in RBL mast cells
After having established the importance of cysteine-rich linker region of SNAP-23 in its membrane localization in RBL mast cells, we wanted to study the contribution of individual cysteine residues towards SNAP-23 membrane localization. Cysteine residues could assist in membrane localization due to their hydrophobicity and/or may be responsible for stable anchoring of SNAP-23 into plasma membrane due to palmitoylation of their free sulfhydryl group. To distinguish between these two possibilities, two types of individual cysteine mutants of SNAP-23 were made. In one set, individual cysteine residues in the linker region were separately mutated to leucine or phenylalanine (Fig. 4A, left panel), so as to retain hydrophobicity [37] but loose palmitoylation. In the other set of mutants, individual cysteines were separately mutated to alanine (Fig. 4A, right panel), so that the residues lost hydrophobicity [37] as well as palmitoylation. EGFP-tagged plasmid constructs encoding all these mutants were individually transfected into RBL mast cells, and percentage transfection efficiencies, mean fluorescence intensities and post-transfection cell recoveries were checked in each case as previously described. Transfection efficiencies were 50–65% in each case (Supplementary Fig. S1), while cell recoveries were about one-third to half after transfecting cysteine single mutants as compared to recovery after transfecting SNAP-23 WT. However, SNAP-23 C85L and C83A mutants gave cell recovery similar to SNAP-23 WT transfection (Supplementary Fig. S2). Membrane/cytosol enrichment fractions were then isolated from the transfected cells and analyzed by immunoblotting with anti-GFP antibody (Fig. 4B). SNAP-23 C79F mutant shows ~57% membrane association. Apart from that we found that mutating four cysteine residues C79, C80, C83 and C87 to leucine individually led to a significant inhibition in membrane localization of SNAP-23. SNAP-23 C79L, C83L and C87L mutants showed about 50–60% loss of membrane association, whereas SNAP-23 C80L mutant showed almost 70% loss of membrane association in comparison to SNAP-23 WT. SNAP-23 C85L mutant showed membrane association comparable to SNAP-23 WT (Fig. 4B & C). These data suggest that palmitoylation of C80 may be most important, followed by palmitoylation of C83, C79 and C87 for stable membrane localization of SNAP-23 in RBL mast cells. Palmitoylation of C85 does not seem to be required for membrane association of SNAP-23 in mast cells.
Fig. 4.
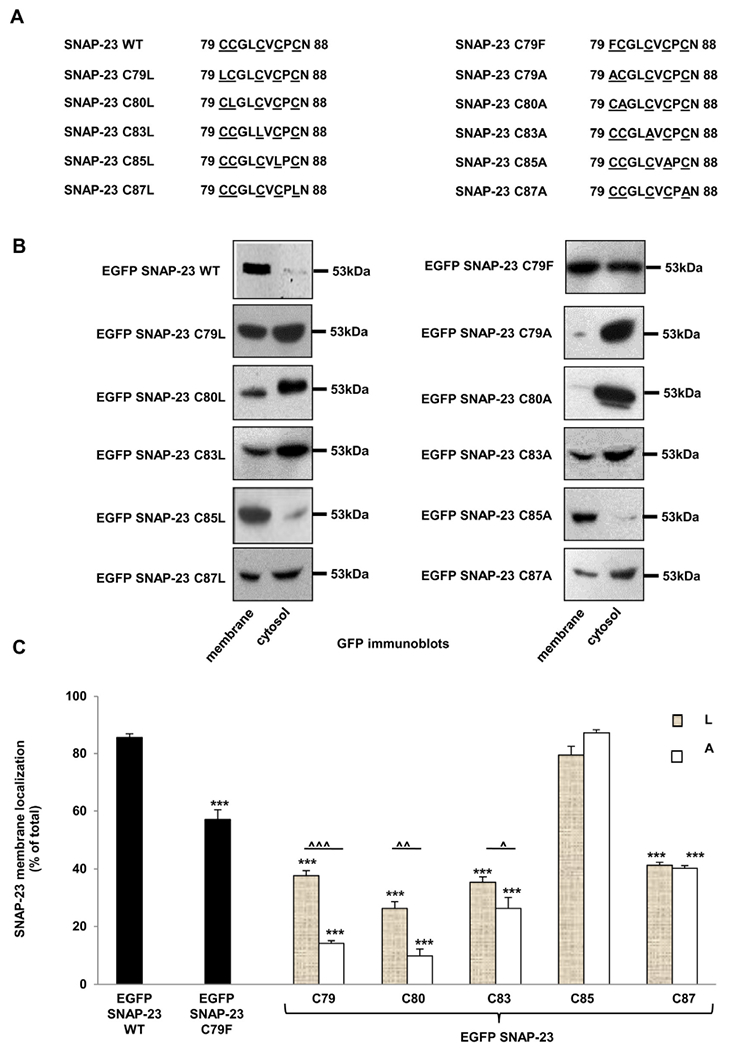
SNAP-23 single cysteine mutants show differential membrane association in RBL mast cells. A. Amino acid sequence of the cysteine-rich domain of SNAP-23 and of the corresponding region of SNAP-23 single cysteine mutants. B. RBL cells were transfected with EGFP-tagged SNAP-23 WT and various SNAP-23 single cysteine mutants. Twenty-four hours post-transfection the cells were fractionated for membrane/cytosol enrichment analysis Equivalent portions of each fraction were analyzed by immunoblotting with anti-SNAP-23 and anti-GFP antibody. Shown here are representative equivalent membrane/cytosol enrichment fraction blots out of three independent transfection- membrane/cytosol enrichment fractionation experiments for each SNAP-23 mutant. All blots for specific mutants were developed, and quantitated similarly. Each time for one PVDF membrane/blot, several films were developed with different exposure times (5 s to 1 min) for best visualization of the bands. Here the representative blots are shown with the molecular weight markers. C. Band intensities from three independent experiments were quantified by densitometry and expressed as percentage of SNAP-23 membrane localization with respect to total SNAP-23 (mean ± SEM). *** indicate statistically significant decrease in membrane binding of the mutants compared with wild-type SNAP-23 (p < 0.0005). Carets indicate statistically significant differences in membrane binding of SNAP-23 cysteine to leucine (L) and cysteine to alanine (A) mutants (^ ^ ^, p < 0.0005; ^ ^, p < 0.005 and ^, p < 0.05).
SNAP-23 C85A mutant still showed comparable membrane localization just like SNAP-23 WT and SNAP-23 C85L mutant (Fig. 4B–C). This confirms that hydrophobicity as well as palmitoylation of this residue is not important for membrane association of SNAP-23 in mast cells. SNAP-23 C83A and C87A mutants showed membrane localization to almost similar extent as SNAP-23 C83L and C87L mutants respectively (Fig. 4B–C). This indicates that probably palmitoylation alone, rather than hydrophobicity of these two residues is important for their role in membrane localization of SNAP-23 in mast cells. Whereas when residues C79 and C80 in the linker region of SNAP-23 were mutated to alanine, the membrane localization of SNAP-23 was almost completely abolished (Fig. 4B–C). This implies that hydrophobicity as well as palmitoylation of these two cysteine residues plays an important role in membrane localization of SNAP-23 in RBL mast cells. Also, these two residues are the most important residues involved in membrane localization of SNAP-23, as mutating either one is sufficient to almost completely move SNAP-23 to cytosolic localization. Our results demonstrate that both palmitoylation and hydrophobicity of various cysteine residues in the linker region of SNAP-23 regulate its membrane association.
Our immunoblotting data in Fig. 4B and C, showed a strong signal for GFP SNAP-23 C79A and GFP SNAP-23 C80A present exclusively in the cytosolic fraction, hence we decided to investigate the localization of these two mutants in RBL cells after transfection by confocal immunofluorescence microscopy. After 24 h of transfection in the RBL mast cells with EGFP SNAP-23 C79A and C80A constructs (separately), the cells were fixed and images acquired and analyzed by confocal microscope. These mutants accumulated in the cytosol (Fig. 5 & Supplementary Fig. S3) and the percent membrane association of GFP SNAP-23 C79A and GFP SNAP-23 C80A was also analyzed from the confocal microscopy data. Their membrane associations were only 10% and 9% respectively (the plasma membrane association decreased by 88% and 89% respectively in comparison to WT SNAP-23, that is, 78% membrane association) (Supplementary Fig. S3). These results confirm our Western blot data, and suggest that these two residues are critical for the palmitoylation and targeting of SNAP-23 to membrane.
Fig. 5.
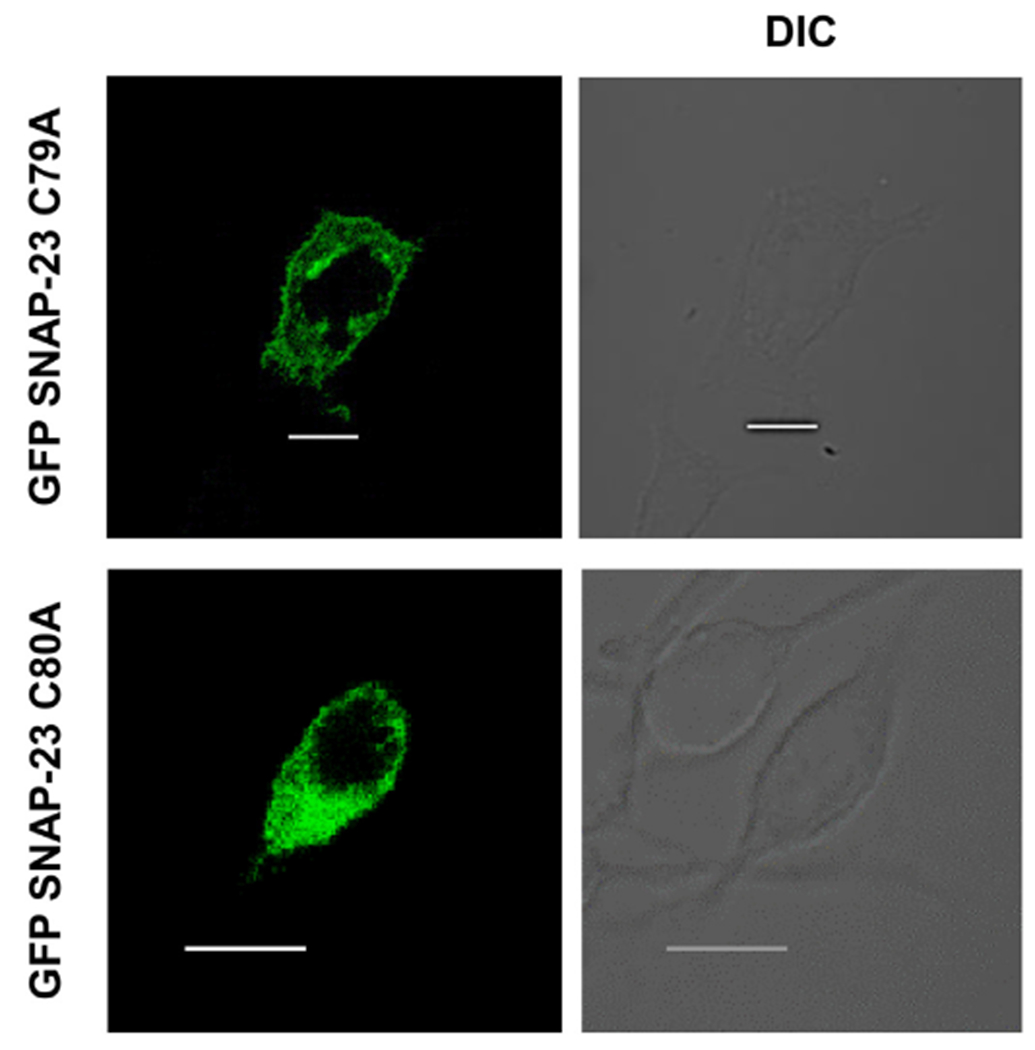
SNAP-23 C79A and C80A mutants show cytosolic localization in RBL mast cells. RBL cells were transfected with EGFP-tagged SNAP-23 C79A and C80A constructs. Transfected cells were plated on coverslips and analyzed by confocal microscopy 24 h post-transfection. Shown here are representative images. Scale = 10 μm.
3.6. Generation and membrane localization of a transmembrane domain containing SNAP-23 Cys− mutant fusion protein
We have seen above that cysteine-rich domain is essential for targeting SNAP-23 to membrane, which otherwise lacks any transmembrane domain or membrane-targeting signal. So, we were interested in studying the effect of replacing this cysteine-rich domain with a transmembrane domain from some other protein, on membrane trafficking of SNAP-23. We set out to genetically engineer a SNAP-23 fusion construct in which the cysteine-rich domain of SNAP-23 would be replaced by a transmembrane domain. The fusion construct was designed following a bioinformatics approach described in Fig. 6. Briefly, through searching in UniprotKB and extensive database mining, we narrowed down to three multi-pass transmembrane proteins (MDR3, NaKATPase alpha-2 subunit and CLP24) that express in human or rat cells, localize to the plasma membrane, have known sequence, exhibit constitutive expression, and that would not likely affect the function of SNAP-23, and were thus suitable for cloning. Further for all individual transmembrane domains from these three proteins, topology prediction was done using TMMOD model [26], which led to narrowing down of five suitable transmembrane domains (Fig. 6). MDR3 protein is constitutively expressed in various human cancer cell lines [27] and topology mapping of individual transmembrane domains of MDR3 has been done in great detail [28,29]. Based on these studies we chose MDR31–145 transmembrane domain for cloning. TMMOD model was first used to predict the transmembrane topology of fusion protein SNAP-23 Cys−-MDR31–145. Fig. 6 shows the topology prediction showing two transmembrane regions and the correct expected orientation of the fusion protein with N and C terminals of SNAP-23 facing inside the cytosol. Fusion construct SNAP-23 Cys−-MDR31–145 was then made by cloning MDR31–145 domain into the linker region of SNAP-23 Cys− mutant (Fig. 7A) using a unique restriction enzyme site AhdI as described in materials and methods.
Fig. 6.
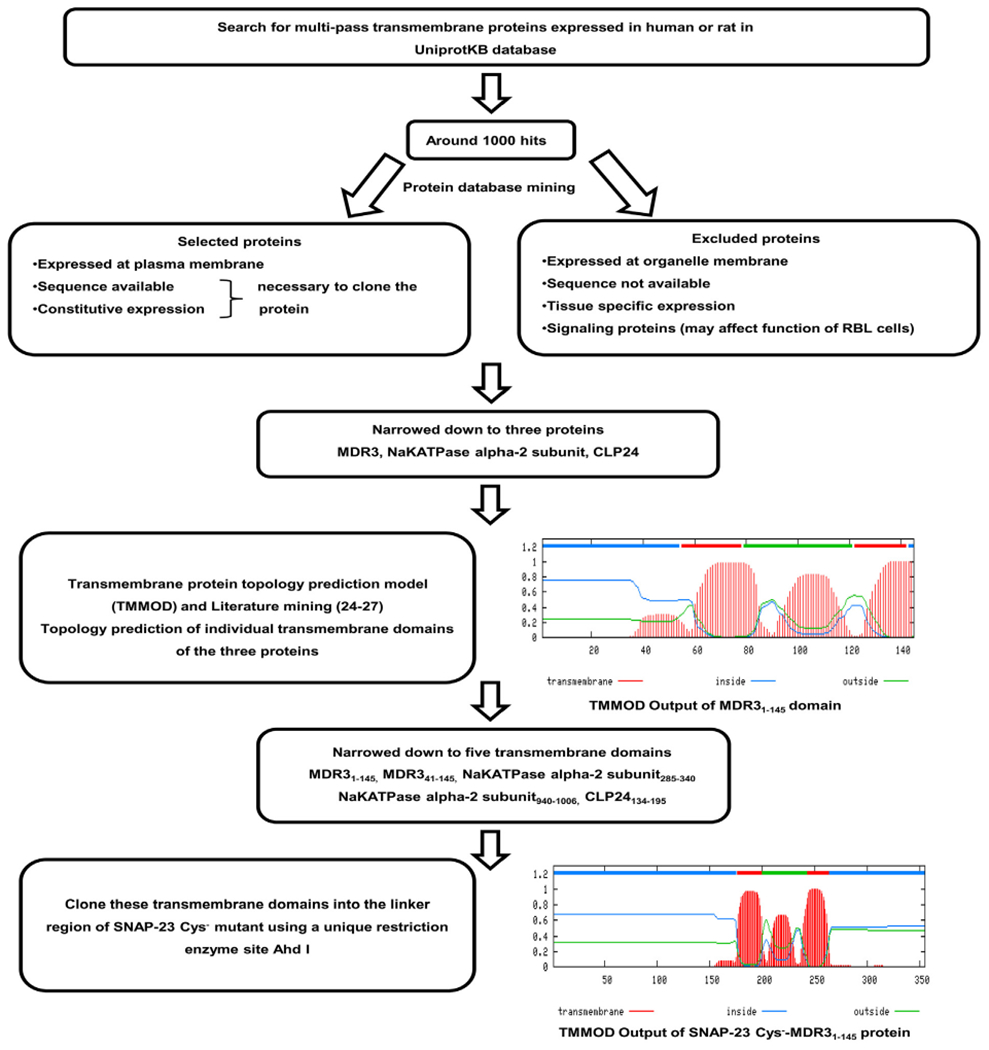
Rationale of designing a SNAP-23 transmembrane fusion protein SNAP-23 Cys−-MDR31–145, Multi-pass transmembrane proteins expressed in human or rat cells were searched in UniprotKB database. Out of around 1000 hits obtained, three proteins namely MDR3, NaKATPase alpha-2 subunit and CLP24 were narrowed down after extensive database mining. Based on the database information, these proteins localize to the plasma membrane, have known sequence, exhibit constitutive expression, and would not likely affect the function of SNAP-23, and were thus suitable for cloning into SNAP-23. Other proteins showing tissue specific or organelle specific expression; or with unknown sequence; or signaling functions were excluded. Further for all individual transmembrane domains from these three proteins, topology prediction was done using TMMOD model [26], which led to narrowing down of five suitable transmembrane domains for cloning (MDR31–145, MDR341–145, NaKATPase alpha-2 subunit285–340, NaKATPase alpha-2 subunit940–1006 and CLP24134–195). Shown here is a representative TMMOD output of MDR31–145 domain with two transmembrane regions; and correct orientation of N and C terminals facing inside the cytosol. MDR3 protein is constitutively expressed in various human cancer cell lines [27] and topology mapping of individual transmembrane domains of MDR3 has been done in great detail [28,29]. Based on these studies we chose MDR31–145 transmembrane domain for cloning. TMMOD model was first used to predict the transmembrane topology of fusion protein SNAP-23 Cys−-MDR31–145. Shown here is the topology prediction showing two transmembrane regions and the correct expected orientation of the fusion protein with N and C terminals of SNAP-23 facing inside the cytosol. Fusion construct SNAP-23 Cys−-MDR31–145 was then made by cloning MDR31–145 domain into the linker region of SNAP-23 Cys− mutant using a unique restriction enzyme site Ahd I.
Fig. 7.
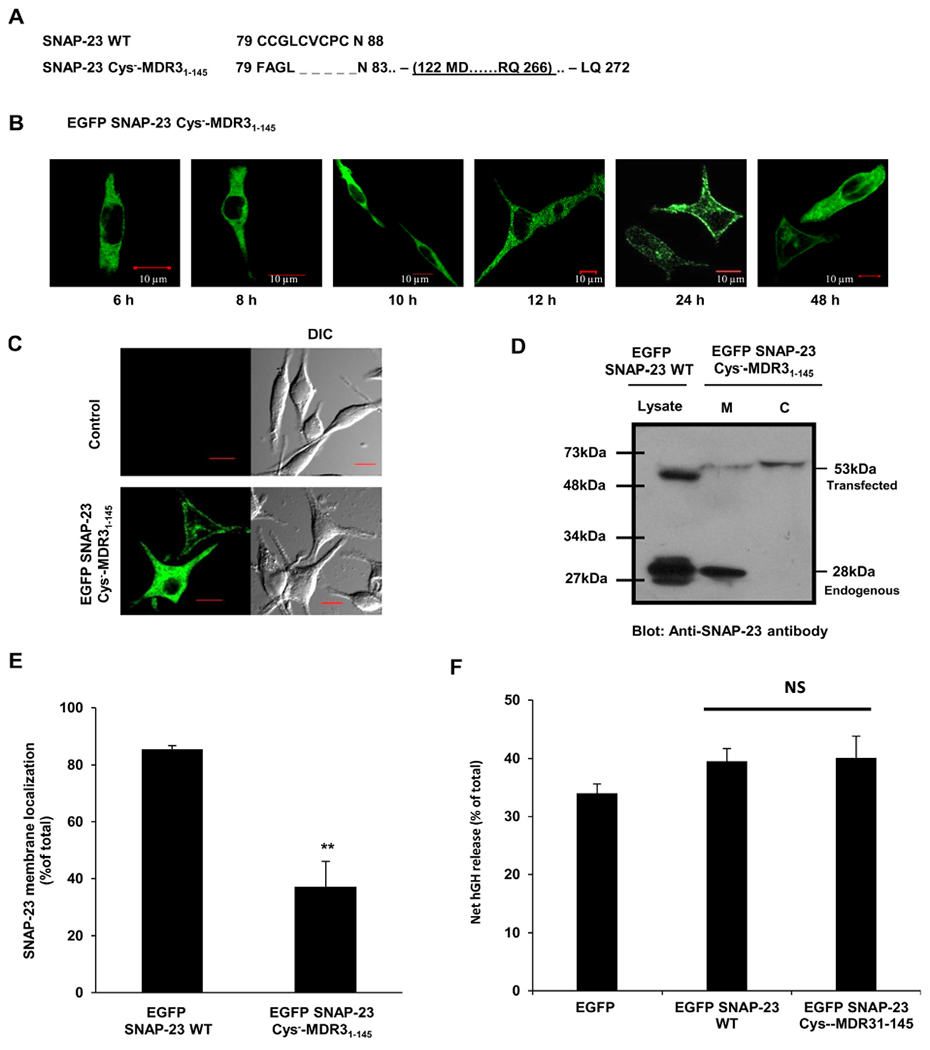
SNAP-23 Cys−-MDR31–145 transmembrane fusion protein shows delayed and only partial trafficking to the plasma membrane of RBL mast cells. A. Amino acid sequence of the cysteine-rich domain of SNAP-23 WT and of the corresponding region of SNAP-23 Cys−-MDR31–145 fusion construct. Also shown (underlined) is the cloning site of MDR31–145 domain in SNAP-23 Cys− mutant. B. RBL cells were transfected with EGFP-tagged SNAP-23 Cys−-MDR31–145 fusion construct. Transfected cells were plated on coverslips and analyzed by confocal microscopy after 6, 8, 10, 12, 24 and 48 h post-transfection. Shown here are representative images. Scale = 10 μm. C. RBL cells were either mock transfected (control) or transfected with EGFP-tagged SNAP-23 Cys−-MDR31–145 fusion construct. 48 h post-transfection the cells were analyzed by confocal microscopy. Shown here are representative images. Scale = 10 μm. D. Lysates and membrane/cytosol enrichment fractions from EGFP-tagged SNAP-23 Cys−-MDR31–145 fusion construct transfected RBL cells were analyzed by immunoblotting with anti-SNAP-23 antibody. Shown here is a representative immunoblot with position of molecular weight markers. E. Band intensities from three independent experiments were quantified by densitometry and expressed as SNAP-23% membrane localization with respect to total SNAP-23 (mean ± SEM). ** indicate statistically significant difference in membrane association of SNAP-23 Cys−-MDR31–145 fusion protein as compared to wild-type SNAP-23 (p < 0.005). F. Effect of SNAP-23 Cys−-MDR31–145 fusion protein in mast cell exocytosis was detected by hGH reporter assay. Each data point is a mean ± SEM of three independent experiments (ns = not significant).
RBL mast cells transfected with this EGFP-tagged SNAP-23 Cys−-MDR31–145 fusion construct expressed the fusion protein at a significantly lower level (60%) as compared to the expression of endogenous SNAP-23 (Fig. 7D). The transfection efficiency of fusion construct and post-transfection recovery of the transfected RBL cells were comparable to that obtained in case of SNAP-23 WT transfection (Supplementary Fig. S1 & 2). Confocal microscopy was done to observe the expression and trafficking of fusion protein at different time points after transfection in RBL cells. As in case of SNAP-23 WT, the expression of fusion protein was detectable at 6 h after transfection. At 12 h post-transfection, most of the fusion protein showed cytosolic or internal organelle localization. But it was only until 24–48 h post-transfection, that it was seen partially localized to the plasma membrane of RBL cells (Fig. 7B, & Supplementary Fig. S3). Even after 48 h, a significant proportion of fusion protein was seen localized to cytosol or internal compartments (Fig. 7B–C, Supplementary Fig. S3). This observation was further confirmed by membrane/cytosol enrichment fractionation of the RBL mast cells transfected with fusion protein (Fig. 7D–E). Only 37% of fusion protein was found associated with membrane even 48 h post-transfection as compared to 86% in case of SNAP-23 WT (Fig. 7D–E). From membrane/cytosol enrichment analysis from microscopy data it was found that after 48 h of transfection SNAP-23 Cys−-MDR31–145 fusion protein shows 26% membrane association (Supplementary Fig. S3). After 24 h only 36% cells showed some membrane targeting of the fusion protein by microscopic analysis. After 48 h, 74% cells showed some plasma membrane targeting of the fusion protein, and only in 26% cells the fusion protein seems to be completely cytosolic or not trafficked to plasma membrane at all. These results further reiterate that for optimal and stable plasma membrane association of SNAP-23, the cysteine residues in the linker region of SNAP-23 are required.
3.7. Overexpression of SNAP-23 Cys−-MDR31–145 fusion protein restores regulated exocytosis from transiendy transfected RBL mast cells
Having established the importance of cysteine residues in the linker region of SNAP-23 in its membrane localization and observing the partial and delayed membrane association of SNAP-23 Cys−-MDR31–145 fusion protein, we decided to check if SNAP-23 Cys−-MDR31–145 fusion protein could support regulated exocytosis from mast cells. To determine this, RBL cells were transiently co-transfected with either wild type SNAP-23 or the SNAP-23 transmembrane fusion construct together with hGH secretion reporter plasmid as standardized earlier [25]. Transfected cells were sensitized with DNP-specific IgE and either mock-stimulated (resting) or stimulated by IgE cross-linking with DNP-BSA for 45 min as described in materials and methods. The percentage of net hGH secretion (stimulated-resting) was calculated from the cells overexpressing each construct to determine their effect on mast cell regulated exocytosis. The overexpression of SNAP-23 Cys−-MDR31–145 fusion protein which partially localized to the membrane, showed mast cell degranulation just like in case of SNAP-23 WT transfected cells (40% net hGH release after 45 min of stimulation) (Fig. 7F).
As Cys−-MDR31–145 fusion protein partially localized to the plasma membrane, and was able to restore exocytosis when expressed in RBL mast cells, we decided to investigate if this fusion protein and wildtype SNAP-23 follow same mechanism to regulate exocytosis. Previously [22], we have shown that after mast cell stimulation the SNAP-23 relocates to LAMP-3 positive late endosomal compartments and mediates homotypic fusions of granules. So, here we have tried to do similar colocalization studies with RBL cell transfected with Cys−-MDR31–145 fusion protein. The results are shown in Supplementary Fig. S4. The results indicated that in resting state itself the GFP tagged Cys−-MDR31–145 fusion protein showed slightly higher association with LAMP-3 positive compartments (P. Coefficient 0.32), but like the wildtype SNAP-23, on stimulation the association of Cys−-MDR31–145 fusion protein with LAMP-3 positive compartments increased (P. coefficient 0.55), indicating that it partially relocated to the lysosomes. Hence the mechanism of action may be similar to that of wildtype SNAP-23.
3.8. SNAP-23 initial membrane association does not depend on Syntaxin 4 in mast cells
From literature it is known that SNAP-25 initial membrane association depends on Syntaxin 1 in neurons [38]. To check whether SNAP-23 membrane association in mast cells depends on Syntaxin 4, GFP Syntaxin 4 WT and GFP Syntaxin 4 ΔCT constructs were transfected into RBL mast cells and at 6 h and 24 h their trafficking to plasma membrane was observed under confocal microscope. It was found that Syntaxin 4 WT is associated with plasma membrane at 6h but the mutant is unable to associate with the plasma membrane even after 24 h of transfection in RBL mast cells 97% association for Syntaxin 4 WT whereas for the Syntaxin 4 ΔCT the plasma membrane association was only 10% (Fig. 8A&B, Supplementary Fig. S3). Now to check whether during membrane association Syntaxin 4 carries SNAP-23 for its initial membrane association, GFP Syntaxin 4 WT and GFP Syntaxin 4 ΔCT mutants transfected RBL mast cells were counterstained with SNAP-23 specific antibody (in red) after 6 h of transfection. It was seen that Syntaxin 4 associated with plasma membrane completely after 6 h, in case of SNAP-23 was still seen in cytosol. Here we found partial colocalization of Syntaxin 4 with endogenous SNAP-23 (P. coefficient 0.63 ± 0.02). But after 6 h, Syntaxin 4 ΔCT was found stuck in cytosol and there were hardly any association with SNAP-23 as calculated from its colocalization coefficient (P. coefficient 0.35 ± 0.2). So it can be concluded that endogenous SNAP-23 is not associated with Syntaxin 4 during its initial membrane association as its mutant Syntaxin 4 ΔCT cannot drag behind SNAP-23 from plasma membrane association (Fig. 8C).
Fig. 8.
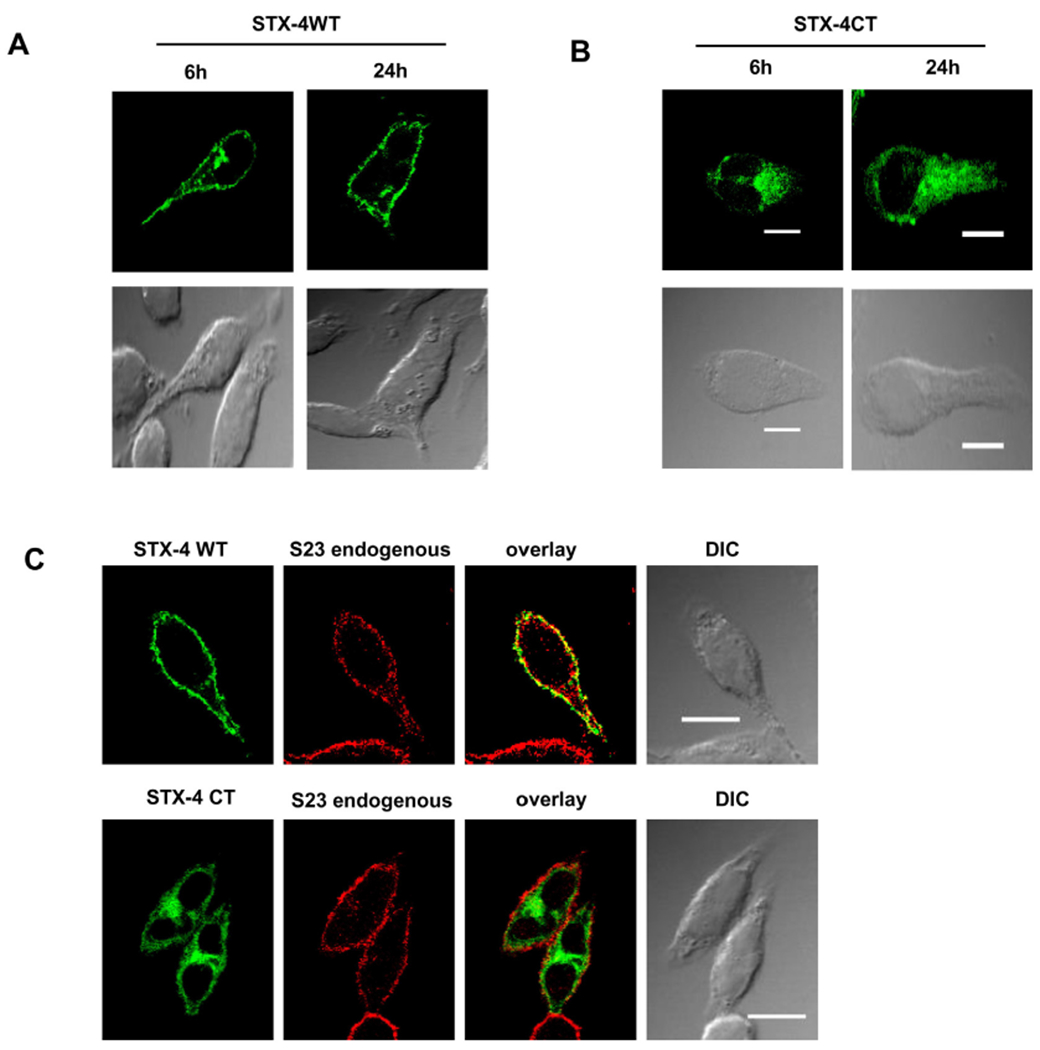
SNAP-23 initial plasma membrane association is independent of Syntaxin 4. A–B. RBL mast cells were transfected with GFP Syntaxin 4 (STX-4) WT and GFP Syntaxin 4 ΔCT (STX-4 CT) mutant and plated on coverslip. After 6 h and 24 h of transfection their membrane association dynamics were observed under confocal microscope after PFA fixation. C. GFP Syntaxin 4 (STX-4) WT and GFP Syntaxin 4 ΔCT (STX-4 CT) transfected RBL cells were fixed with 4% PFA after 6 h and then counterstained with SNAP-23 specific antibody (red), separately. Then they were observed under confocal microscope. Scale bar is 10 μm. No significant staining was observed in the green or red channel when using an irrelevant Ab.
4. Discussion
Mast cells are important immune effector cells whose function depends on regulated exocytosis of various inflammatory mediators stored in their secretory granules. [3]. Thus it is very important to investigate the molecular mechanisms of regulated exocytosis from mast cells in order to devise new interventions for immune disorders involving mast cell mediator release. The protein trafficking mechanisms involved in mediator release from mast cells have been studied to some extent in the past [39,40]. We and others have identified some specific v and t SNAREs as being important for regulation of mast cell exocytosis [3,41,42]. SNAREs may also be regulated through specific signaling or orchestrated events in resting as well as activated states of mast cells, which is important for mast cell exocytosis [9,31]. We have also shown that in mast cells, in response to a physiological trigger like cross-linking of their high affinity cell surface receptor FcεRI, the signaling causes a transient phosphorylation of SNAP-23. This transient phosphorylation followed by dephosphorylation is a very important sequence of events required for mast cell degranulation as blocking either of these events leads to significant inhibition in mast cell exocytosis [9]. Along with phosphorylation, another important post-translational modification of SNAP-23 which may regulate SNAP-23 function is palmitoylation. It is known that though SNAP-23 is a membrane-associated SNARE, it lacks a transmembrane domain just like its neuronal homologue SNAP-25 [6,9]. Nonetheless, SNAP-23 does contain five highly conserved cysteine residues in its linker region at positions 79, 80, 83, 85 and 87 which get palmitoylated and have been thought to be the main anchors holding SNAP-23 at the membrane for the regulation of exocytosis [10]. A linker connecting two SNARE domains is a unique feature of SNAP-25 subfamily including SNAP-23 and SNAP-25 proteins. On the basis of our results for Multiple sequence alignment, we predicted canonical cysteine motif with five cysteines conserved in the membrane binding domain sequence (79–122) of the linker region of SNAP-23. Notably the predicted canonical cysteine motif in SNAP-23 has been already found to be conserved in minimal membrane binding sequence (85–120) of SNAP-25 [36] with four residues, the first cysteine being replaced with phenylalanine in case of SNAP-25b. Further the proline residue present in downstream QPARV motif in SNAP-25 also seems to be conserved in QPXXV region in SNAP-23. The proline residue has been already known to be critical in determining the specificity of interaction with DHHC palmitoyl transferases in SNAP-25 [6].
So, we set out to study the importance of these cysteine residues in trafficking of free newly synthesized SNAP-23 to its functionally relevant destination, that is, plasma membrane in mast cells. Not surprisingly, when all the cysteine residues in the linker region of SNAP-23 were mutated/deleted, the SNAP-23 Cys− mutant failed to associate with the membrane and seemed stuck in the cytosol which may be its site of synthesis. This result was in agreement to earlier studies [20] which have shown that deletion of the entire cysteine-rich domain of syndet, i.e., mouse SNAP-23 shifted its immunofluorescence from plasma membrane to a diffuse intracellular localization.
Deletion or mutation of cysteine residues in SNAP-23 will not only prevent palmitoylation, but will also change hydrophobicity of the linker region, maybe even preventing the initial association with membrane. In such a scenario, mutant SNAP-23 may fail to traffic to its destination, fail to fold properly and may even fail to associate with any chaperones or other interacting partners which may be important for its initial trafficking. Such mutant proteins may show inappropriate interactions or actions that may cause toxicity to the expressing cells. It may also interfere with the functioning of endogenous SNAP-23, therefore behaving as a dominant negative mutant. We also found very low cell recoveries in mast cells transfected with SNAP-23 Cys− mutant. Also the transfected mutant may be targeted for degradation as it may misfold or fail to bind appropriate targets (membranes/chaperones).
Though SNAP-23 has five conserved cysteines in the linker region, SNAP-25 has only four cysteines and it still targets to membrane in neuronal cells [6]. So, we decided to investigate whether all the cysteine residues in the linker region of SNAP-23 contribute to its membrane targeting, and also to dissect out the importance of hydrophobicity and palmitoylation of cysteines for membrane association of SNAP-23 in mast cells. Two kinds of mutants for all individual cysteine residues in the linker region of SNAP-23 were made; (i) cysteine to leucine (which retain hydrophobicity but lose palmitoylation), and (ii) cysteine to alanine (which have much reduced hydrophobicity besides lacking palmitoylation) [37]. All cysteine residues except for C85 seemed to be important for membrane association of SNAP-23 in mast cells, as SNAP-23 C85L and C85A mutants, both showed membrane association similar to wild-type SNAP-23. Two residues which seemed to be the most crucial ones for membrane association of SNAP-23 were C79 and C80 as mutation of either one of these to alanine led to an almost complete mistargeting of SNAP-23 to cytosol (with only 10–14% remaining associated with membrane). This was further validated by the microscopy images and their quantitation. Mutating individual cysteine residues led to a significant decrease in membrane association of SNAP-23, the decrease generally being more pronounced in C to A mutant than in C to L mutants. Our results were a bit contrary to the finding of Greaves et al. [6] where replacement with leucine almost completely reversed the decrease in membrane binding of SNAP-25B caused by cysteine to alanine single mutants. However, just like the corresponding C90L and C90A mutants of SNAP-25B in their study, our C85L and C85A mutants of SNAP-23 showed percentage membrane association similar to the wild-type protein. Our results are in agreement to previous reports where the hydrophobic cysteine-rich domain of SNAP-25 undergoes dynamic palmitoylation to regulate intracellular patterning and membrane interactions of SNAP-25 [6,19]. Our study has highlighted the importance of palmitoylation as well as hydrophobicity of cysteine residues in the linker region of SNAP-23 in it’s membrane association. A previous study by Weber et al. [23] has shown that electrostatic interactions between basic amino acid residues close to cysteine residues in the linker region of SNAP-25/SNAP-23 and acidic lipids precede stable membrane association of these t-SNAREs. Actually, many studies have established that for peripheral membrane proteins lacking a trans-membrane domain, both hydrophobic as well as electrostatic interactions may be important for initial membrane recruitment. For some proteins electrostatic interactions recruit proteins to membrane non-specifically, whereas the hydrophobic amino acids facilitate partitioning into the hydrophobic core of the lipid bilayer [43]. In certain other proteins hydrophobic residues drive initial recruitment to the membrane, but the electrostatic interactions involving basic residues may increase its residency time at the membrane for further modifications [44]. Overall, it seems that the hydrophobic and basic amino acids may both in optimum numbers and locations; contribute to the secondary structure of the protein for facilitation of initial membrane association.
In order to study the contribution of palmitoylation alone of the cysteines in membrane targeting of SNAP-23, a P119A mutant of SNAP-23 was made. It had been shown earlier that a corresponding P117A mutation in SNAP-25 blocked the palmitoylation of cysteine residues in its linker region thereby significantly inhibiting its membrane targeting [6,36]. In our present study, the SNAP-23 P119A mutant behaved exactly as the SNAP-23 Cys− mutant in terms of post-transfection cell recovery, as well as lack of membrane targeting, reiterating that palmitoylation of cysteines in linker region is essential for targeting of SNAP-23 to its plasma membrane location in mast cells.
Further, to investigate the biological role of cysteine-rich domain of SNAP-23 in regulation of mast cell exocytosis, we overexpressed SNAP-23 mutants in RBL mast cells together with human growth hormone secretion reporter. Not to our surprise, we found that overexpression of SNAP-23 Cys− mutant (which could not at all attach to the plasma membrane for fusion events required during exocytosis) significantly inhibited regulated exocytosis from mast cells. The P119A mutant also showed similar effect causing inhibition of MC exocytosis. Hence, our results clearly demonstrate that the cysteine-rich domain of SNAP-23, or palmitoylation of these conserved cys residues in the linker region of SNAP-23, regulates its membrane association and thereby also the extent of mast cell exocytosis.
We were also interested in studying the effect of replacing the cysteine-rich domain of SNAP-23 with a transmembrane domain from some other protein, on membrane targeting of SNAP-23 and on regulated exocytosis from mast cells. Attachment of SNAP-23 to membrane via cysteine-rich linker is thought to be dynamic because of the dynamic chemistry of palmitoylation [14–18]. On the contrary, membrane attachment of SNAP-23 via a permanent transmembrane domain would be stable. So, we made SNAP-23 Cys−-MDR31–145 fusion construct and transfected it into RBL cells. The SNAP-23 Cys−-MDR31–145 fusion protein showed delayed trafficking and only partial association with the plasma membrane of RBL mast cells. The possible reasons for this could be the large size of fusion protein as compared to SNAP-23 WT, or the fact that it is attached to membrane via transmembrane domain and not via palmitoylation, or its membrane trafficking pathway might have changed. It might be following transmembrane transport to endoplasmic reticulum and then vesicular transport to plasma membrane. It might not be getting correctly folded by chaperones and thus might be binding to ubiquitinases for degradation, as was suggested by its significant low expression (60%) in comparison to endogenous SNAP-23. It was interesting to find out that although SNAP-23 Cys−-MDR31–145 fusion protein showed delayed and that too only partial membrane association, its overexpression did not at all affect the regulated exocytosis from mast cells. And this fusion protein was able to partially relocate to the lysosomal compartments (LAMP-3 positive) during mast cell exocytosis. May be 37% membrane association as shown by fusion protein is sufficient to carry out exocytosis. But, there may be discrepancy between membrane association and exocytosis results due to the presence of endogenous SNAP-23. It may also be possible that the fusion protein is not at all competing with endogenous SNAP-23 for exocytosis and thereby does not show any effect on the process. Endogenous SNAP-23 knockout or knockdown should be done to see the exact functional consequences of the mutants. But the challenge is that 64% maximum knockdown of endogenous SNAP-23 that we could achieve using siRNA in RBL mast cells was not found to be sufficient to knockdown all the function of endogenous SNAP-23 (data not shown).
Interestingly, palmitate linkage (which is responsible for stable membrane attachment of SNAP-23) is reversible. Moreover, there is evidence that palmitoylation/depalmitoylation dynamics can also be modulated by distinct posttranslational modifications present on the target protein. Interplay between palmitoylation and phosphorylation has been much studied [17]. SNAP-23 also contains induced and basal phosphorylation sites near the hydrophobic cysteine-rich domain that gets palmitoylated [9,10]. We have also recently found that the SNAP-23 membrane association and thus its function in regulated exocytosis from mast cells may be regulated by various physiological triggers and intracellular signaling pathways that are induced by those triggers, which ultimately regulate SNAP-23 palmitoylation and phosphorylation dynamics and thereby its function. Much work has been done to study the role of phosphorylation of SNAP-23 in regulating exocytosis from mast cells [9,22,35] and platelets [33,45]. However, the role of palmitoylation of the hydrophobic cysteine-rich domain of SNAP-23 in regulating exocytosis has not been studied so far. In this study, we have shown for the first time that overexpression of SNAP-23 Cys− mutant, which does not get palmitoylated [10] inhibits mast cell exocytosis, thus, strongly suggesting that palmitoylation of SNAP-23 regulates mast cell exocytosis. Based on our study of SNAP-23 palmitoylation and phosphorylation a model is proposed (Fig. 9) that shows that SNAP-23 initial plasma membrane association is mediated by the constitutively phosphorylated threonine residue at 102 position and hydrophobic interaction. At the plasma membrane P119 residue may play an important role in SNAP- 23 interaction with the PAT. Here in this representation the overview of SNAP-23 palmitoylation and phosphorylation [22] is shown. SNAP-23 gets phosphorylated at S95 and S120 residures duning mast cell exocytosis and this transient phosphorylation mediates its relocation to internal granule membranes (LAMP-3 positive and Serotonin positive granules). This phosphorylated SNAP-23 also interacts with Syntaxin 3 (on the granule membrane), and subsequently these interactions accomplish the homotypic fusions of granules that lead to compound exocytosis of the granular contents. And the subseiquent dephosphorylation of SNAP-23 mediates the final fusion with plasma membrane [21,22]. Although further study is required to determine the precise mechanistic details, it is clear that the cysteine-rich domain of SNAP-23 plays an important role in regulating its membrane association and regulated exocytosis from mast cells.
Fig. 9.
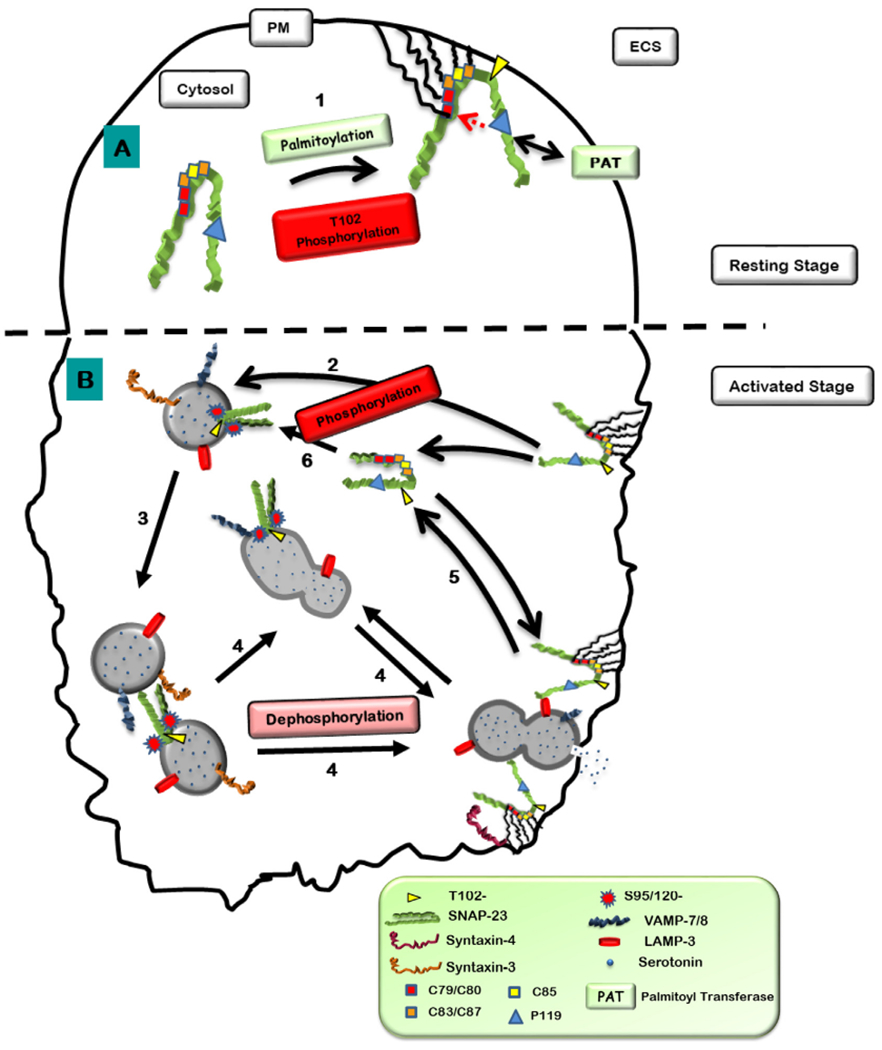
Hypothetical model showing hydrophobic interactions and palmitoylations of cysteine in the linker region of SNAP-23 regulate its membrane association. A. After synthesis in the cytosol, 1. SNAP-23 may initially associate with membrane (internal or plasma membrane) through some hydrophobic interactions due to presence of hydrophobic amino acids like cysteines in the linker region. Since DHHC palmitoyl transferases (PAT) which palmitoylate cysteine residues in proteins are localized on membranes, initial hydrophobic and electrostatic interaction with membrane may bring SNAP-23 close to these DHHC proteins which can now palmitoylate the conserved cysteine residues in SNAP-23 and thereby confine it to membrane. All cysteine residues except for C85 seemed to be important for membrane association of SNAP-23 in mast cells. P119 residue of SNAP-23 (corresponding P117 residue in SNAP-25 involved in the palmitoylation of cysteine residues in its linker region thereby in its membrane targeting) seems to be important for membrane association of SNAP-23 in mast cells and may be required for interaction with DHHC palmitoyl transferases. Apart from that it has also shown that the basal phosphorylation at T102 site also plays an important role in SNAP-23 initial plasma membrane association [22]. B. After Crosslinking of mast cells, 2. The phosphorylation of SNAP-23 at S95 and S120 sites mediates its relocation to internal LAMP-3 positive granule membranes, 3–4. There may be the phosphorylation helps in homotypic fusions (granule-granule fusions) as Syntaxin-3 present there (may play the cognitive t-SNARE of SNARE-tri-molecular complex, [22]). Then the dephosphorylation of these residues mediate the plasma membrane fusions of these granules (compound exocytosis) [21]. 5–6. Then SNAP-23 relocates to cytosol and phosphorylated and associates to LAMP-3 positive granule membrane.
Supplementary Material
Transparency document.
The Transparency document associated with this article can be found, in online version.
Acknowledgements
We thank confocal fluorescent Microscopy Facility, AIRF (Advanced instrument research facility), JNU, for help with image analysis. We are grateful to Professor Rakesh K. Tyagi [Special Centre for Molecular Medicine (SCMM), JNU, New Delhi] for his kind gift of LS180 cell line. This work was supported by research grants from Department of Science and Technology (DST) Govt. of India (SR/SO/HS-0122/2009; and DST-PURSE), and University Grants Commission (UGC), India (UPE-II Project ID-54; and UGC-resource networking) to NP. VA and GKK were supported by a grant from DBT, India; PN was supported by a grant from UGC, India; SA was supported by a grant from CSIR, India.
Abbreviations:
- MC
mast cell
- SNAP-23/25
synaptosomal-associated protein of 23/25 kDa
- RBL
rat basophilic leukemia
- PBS
phosphate buffered saline
- MDR3
multidrug resistance protein 3
- DNP
dinitrophenol
- BSA
Bovine serum albumin
- hGH
human Growth Hormone
- SNAREs
soluble N-ethylmaleimide-sensitive-factor attachment protein receptors
- VAMP
vesicle-associated membrane protein
- DHHC
aspartic acid-histidine-histidine-cysteine
- PATs
palmitoyl transferases
- UniprotKB
Universal protein resource Knowledgebase
- EGFP
enhanced green fluorescent protein
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbamcr.2019.06.015.
References
- [1].Jahn R, Sudhof TC, Membrane fusion and exocytosis, Annu. Rev. Biochem. 68 (1999) 863–911. [DOI] [PubMed] [Google Scholar]
- [2].Rothman JE, Mechanisms of intracellular protein transport, Nature 372 (1994) 55–63. [DOI] [PubMed] [Google Scholar]
- [3].Stow JL, Manderson AP, Murray RZ, SNAREing immunity: the role of SNAREs in the immune system, Nat Rev Immunol 6 (2006) 919–929. [DOI] [PubMed] [Google Scholar]
- [4].Ravichandran V, Chawla A, Roche PA, Identification of a novel syntaxin- and synaptobrevin/VAMP-binding protein, SNAP-23, expressed in non-neuronal tissues, J. Biol. Chem. 271 (1996) 13300–13303. [DOI] [PubMed] [Google Scholar]
- [5].Wang G, Witkin JW, Hao G, Bankaitis VA, Scherer PE, Baldini G, Syndet is a novel SNAP-25 related protein expressed in many tissues, J. Cell Sci. 110 (1997) 505–513 Pt 4. [DOI] [PubMed] [Google Scholar]
- [6].Greaves J, Prescott GR, Fukata Y, Fukata M, Salaun C, Chamberlain LH, The hydrophobic cysteine-rich domain of SNAP25 couples with downstream residues to mediate membrane interactions and recognition by DHHC palmitoyl transferases, Mol. Biol. Cell 20 (2009) 1845–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Puri N, Roche PA, Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis, Traffic 7 (2006) 1482–1494. [DOI] [PubMed] [Google Scholar]
- [8].Guo Z, Turner C, Castle D, Relocation of the t-SNARE SNAP-23 from lamellipodia-like cell surface projections regulates compound exocytosis in mast cells, Cell 94 (1998) 537–548. [DOI] [PubMed] [Google Scholar]
- [9].Hepp R, Puri N, Hohenstein AC, Crawford GL, Whiteheart SW, Roche PA, Phosphorylation of SNAP-23 regulates exocytosis from mast cells, J. Biol. Chem. 280 (2005) 6610–6620. [DOI] [PubMed] [Google Scholar]
- [10].Vogel K, Roche PA, SNAP-23 and SNAP-25 are palmitoylated in vivo, Biochem. Biophys. Res. Commun. 258 (1999) 407–410. [DOI] [PubMed] [Google Scholar]
- [11].Greaves J, Chamberlain LH, Palmitoylation-dependent protein sorting J. Cell Biol. 176 (2007) 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Greaves J, Prescott GR, Gorleku OA, Chamberlain LH, The fat controller: roles of palmitoylation in intracellular protein trafficking and targeting to membrane microdomains (review), Mol. Membr. Biol. 26 (2009) 67–79. [DOI] [PubMed] [Google Scholar]
- [13].Linder ME, Deschenes RJ, Palmitoylation: policing protein stability and traffic, Nat Rev Mol Cell Biol 8 (2007) 74–84. [DOI] [PubMed] [Google Scholar]
- [14].Prescott GR, Gorleku OA, Greaves J, Chamberlain LH, Palmitoylation of the synaptic vesicle fusion machinery, J. Neurochem. 110 (2009) 1135–1149. [DOI] [PubMed] [Google Scholar]
- [15].Greaves J, Gorleku OA, Salaun C, Chamberlain LH, Palmitoylation of the SNAP25 protein family: specificity and regulation by DHHC palmitoyl transferases, J. Biol. Chem. 285 (2010) 24629–24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Greaves J, Chamberlain LH, DHHC palmitoyl transferases: substrate interactions and (patho)physiology, Trends Biochem. Sci. 36 (2011) 245–253. [DOI] [PubMed] [Google Scholar]
- [17].Salaun C, Greaves J, Chamberlain LH, The intracellular dynamic of protein palmitoylation, J. Cell Biol. 191 (2010) 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zeidman R, Jackson CS, Magee AI, Protein acyl thioesterases (review), Mol. Membr. Biol. 26 (2009) 32–41. [DOI] [PubMed] [Google Scholar]
- [19].Greaves J, Chamberlain LH, Differential palmitoylation regulates intracellular patterning of SNAP25, J. Cell Sci. 124 (2011) 1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Koticha DK, Huddleston SJ, Witkin JW, Baldini G, Role of the cysteine-rich domain of the t-SNARE component, SYNDET, in membrane binding and subcellular localization, J. Biol. Chem. 274 (1999) 9053–9060. [DOI] [PubMed] [Google Scholar]
- [21].Naskar P, Naqvi N, Puri N, Blocking dephosphorylation at serine 120 residue in t-SNARE SNAP-23 leads to massive inhibition in exocytosis from mast cells, J. Biosci. 43 (2018) 127–138. [PubMed] [Google Scholar]
- [22].Naskar P, Puri N, Phosphorylation of SNAP-23 regulates its dynamic membrane association during mast cell exocytosis, Biol Open 6 (2017) 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Weber P, Batoulis H, Rink KM, Dahlhoff S, Pinkwart K, Sollner TH, Lang T, Electrostatic anchoring precedes stable membrane attachment of SNAP25/SNAP23 to the plasma membrane, eLife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Passante E, Frankish N, The RBL-2H3 cell line: its provenance and suitability as a model for the mast cell, Inflammation research : official journal of the European Histamine Research Society … [et al. ] 58 (2009) 737–745. [DOI] [PubMed] [Google Scholar]
- [25].Puri N, Kruhlak MJ, Whiteheart SW, Roche PA, Mast cell degranulation requires N-ethylmaleimide-sensitive factor-mediated SNARE disassembly, J. Immunol. 171 (2003) 5345–5352. [DOI] [PubMed] [Google Scholar]
- [26].Kahsay RY, Gao G, Liao L, An improved hidden Markov model for transmembrane protein detection and topology prediction and its applications to complete genomes, Bioinformatics 21 (2005) 1853–1858. [DOI] [PubMed] [Google Scholar]
- [27].Orlowski S, Martin S, Escargueil A, P-glycoprotein and ‘lipid rafts’: some ambiguous mutual relationships (floating on them, building them or meeting them by chance?), Cellular and molecular life sciences : CMLS 63 (2006) 1038–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kast C, Gros P, Topology mapping of the amino-terminal half of multidrug resistance-associated protein by epitope insertion and immunofluorescence, J. Biol. Chem. 272 (1997) 26479–26487. [DOI] [PubMed] [Google Scholar]
- [29].Zhang JT, Sequence requirements for membrane assembly of polytopic membrane proteins: molecular dissection of the membrane insertion process and topogenesis of the human MDR3 P-glycoprotein, Mol. Biol. Cell 7 (1996) 1709–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Katoh K, Asimenos G, Toh H, Multiple alignment of DNA sequences with MAFFT, Methods Mol. Biol. 537 (2009) 39–64. [DOI] [PubMed] [Google Scholar]
- [31].Vaidyanathan VV, Puri N, Roche PA, The last exon of SNAP-23 regulates granule exocytosis from mast cells, J. Biol. Chem. 276 (2001) 25101–25106. [DOI] [PubMed] [Google Scholar]
- [32].Low SH, Roche PA, Anderson HA, van Ijzendoorn SC, Zhang M, Mostov KE, Weimbs T, Targeting of SNAP-23 and SNAP-25 in polarized epithelial cells, J. Biol. Chem. 273 (1998) 3422–3430. [DOI] [PubMed] [Google Scholar]
- [33].Karim ZA, Zhang J, Banerjee M, Chicka MC, Al Hawas R, Hamilton TR, Roche PA, Whiteheart SW, IkappaB kinase phosphorylation of SNAP-23 controls platelet secretion, Blood 121 (2013) 4567–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Martin-Martin B, Nabokina SM, Blasi J, Lazo PA, Mollinedo F, Involvement of SNAP-23 and syntaxin 6 in human neutrophil exocytosis, Blood 96 (2000) 2574–2583. [PubMed] [Google Scholar]
- [35].Suzuki K, Verma IM, Phosphorylation of SNAP-23 by IkappaB kinase 2 regulates mast cell degranulation, Cell 134 (2008) 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gonzalo S, Greentree WK, Linder ME, SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain, J. Biol. Chem. 274 (1999) 21313–21318. [DOI] [PubMed] [Google Scholar]
- [37].Kyte J, Doolittle RF, A simple method for displaying the hydropathic character of a protein, J. Mol. Biol. 157 (1982) 105–132. [DOI] [PubMed] [Google Scholar]
- [38].Vogel K, Cabaniols JP, Roche PA, Targeting of SNAP-25 to membranes is mediated by its association with the target SNARE syntaxin, J. Biol. Chem. 275 (2000) 2959–2965. [DOI] [PubMed] [Google Scholar]
- [39].Lorentz A, Baumann A, Vitte J, Blank U, The SNARE machinery in mast cell secretion, Front. Immunol. 3 (2012) 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Woska JR Jr., Gillespie ME,SNARE complex-mediated degranulation in mast cells, J. Cell. Mol. Med. 16 (2012) 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Puri N, Roche PA, Mast cells possess distinct secretory granule subsets whose exocytosis is regulated by different SNARE isoforms, Proc. Natl. Acad. Sci. U. S. A. 105 (2008) 2580–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Paumet F, Le Mao J, Martin S, Galli T, David B, Blank U, Roa M, Soluble NSF attachment protein receptors (SNAREs) in RBL-2H3 mast cells: functional role of syntaxin 4 in exocytosis and identification of a vesicle-associated membrane protein 8-containing secretory compartment, J. Immunol. 164 (2000) 5850–5857. [DOI] [PubMed] [Google Scholar]
- [43].Whited AM, Johs A, The interactions of peripheral membrane proteins with biological membranes, Chem. Phys. Lipids 192 (2015) 51–59. [DOI] [PubMed] [Google Scholar]
- [44].Scott AM, Antal CE, Newton AC, Electrostatic and hydrophobic interactions differentially tune membrane binding kinetics of the C2 domain of protein kinase Calpha, J. Biol. Chem. 288 (2013) 16905–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Polgar J, Lane WS, Chung SH, Houng AK, Reed GL, Phosphorylation of SNAP-23 in activated human platelets, J. Biol. Chem. 278 (2003) 44369–44376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


