Abstract
Background & Aims
DJ-1 is universally expressed in various tissues and organs and is involved in the physiological processes in various liver diseases. However, the role of DJ-1 in liver ischemia-reperfusion (I/R) injury is largely unknown.
Methods
In this study, we first examined the DJ-1 expression changes in the liver tissues of mice and clinical donor after hepatic I/R by both quantitative polymerase chain reaction and Western blotting assays. Then we investigated the role of DJ-1 in I/R injury by using a murine liver I/R model.
Results
We demonstrated that DJ-1 down-regulation in both human and mouse liver tissues in response to I/R injury and Dj-1 deficiency in hepatocytes but not in myeloid cells could significantly ameliorate I/R induced liver injury and inflammatory responses. This hepatoprotective effect was dependent on enhanced autophagy in Dj-1 knockout mice, because inhibition of autophagy by 3-methyladenine and chloroquine could reverse the protective effect on hepatic I/R injury in Dj-1 knockout mice.
Conclusions
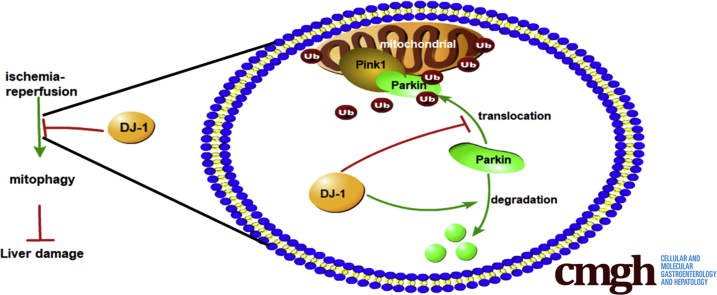
Dj-1 deficiency in hepatocytes significantly enhanced mitochondrial accumulation and protein stability of PARKIN, which in turn promotes the onset of mitophagy resulting in elevated clearance of damaged mitochondria during I/R injury.
Keywords: DJ-1, Liver I/R Injury, PARKIN, Mitophagy
Abbreviations used in this paper: ALT, alanine aminotransferase; AST, aspartate aminotransferase; CQ, chloroquine; Dj-1-/-, global Dj-1 knockout; GEO, Gene Expression Omnibus; H/R, hypoxia/reoxygenation; IHC, immunohistochemical; IL, interleukin; I/R, ischemia-reperfusion; KO, knockout; LDH, lactate dehydrogenase; 3-MA, 3-methyladenine; MPO, myeloperoxidase; mtDNA, mitochondrial DNA; ΔΨm, mitochondrial membrane potential; OMM, outer mitochondrial membrane; PD, Parkinson’s disease; PINK1, PTEN-induced kinase-1; qRT-PCR, quantitative real-time polymerase chain reaction; TEM, transmission electron microscope; TNF, tumor necrosis factor; WT, wild-type
Graphical abstract
Summary.
DJ-1 is significantly down-regulated in human donor liver tissues and the mouse liver subjected to I/R injury. In a mouse model of liver I/R injury, Dj-1 deficiency in hepatocytes but not in myeloid cells ameliorates liver damage and inflammatory responses, probably by promoting mitochondrial accumulation of PARKIN and stabilizing PARKIN protein, which consequently result in enhanced mitophagy in response to I/R injury
Liver ischemia/reperfusion (I/R) injury remains a major problem in clinical practice and occurs in multiple clinical settings, including hemorrhagic shock, partial hepatectomy, and liver transplantation.1 Although the mechanisms of liver I/R injury are complex, loss of mitochondrial membrane potential (ΔΨm) is the key event to trigger I/R injury, mainly because the dysfunctional mitochondria can produce excessive reactive oxygen species, which in turn cause DNA damage, protein oxidation, and lipid peroxidation and ultimately result in cell death.2, 3, 4, 5 Failure to remove damaged mitochondria has been implicated in many pathophysiological processes, especially in neurodegenerative diseases and aging.6 Therefore, it is generally accepted that prompt and sufficient removal of dysfunctional mitochondria improves cell death.7
Mitophagy is a form of selective autophagy for clearance of damaged mitochondria and is essential in maintaining mitochondrial homeostasis.8 The serine/threonine kinase PTEN-induced kinase-1 (PINK1) and the E3 ubiquitin ligase PARKIN are the 2 key regulators of mitophagy working in concert to remove dysfunctional mitochondria. PINK1 is encoded in nuclear DNA and translocated from cytosol into mitochondria through the translocases localized in the outer and inner mitochondrial membranes.9 In healthy mitochondria, full-length PINK1 is first cleaved by a mitochondrial protease, presenilin-associated rhomboid-like (PARL), to form a truncated PINK1 protein (52KD),10 and the truncated PINK1 is subsequently retro-translocated into cytosol and subjected to rapid degradation in a proteasome-dependent manner.11,12 In response to ΔΨm loss, full-length PINK1 is stabilized and accumulated on the outer mitochondrial membrane (OMM) and recruits PARKIN from cytosol to damaged mitochondria.13, 14, 15, 16 PARKIN then ubiquitinates a number of proteins at the OMM to consign damaged mitochondria to autophagic turnover.17
DJ-1 is a highly conserved and homodimeric protein18 that participates in multiple biological functions including cell survival,19 apoptosis,20 transcriptional regulation,21 mitochondrial function, and oxidative stress.22 Some DJ-1 mutations cause early-onset familial recessive Parkinson’s disease (PD).23 Interestingly, DJ-1 has been reported to interact with PINK1 and PARKIN, the other 2 PD-related genes, to form a trimer complex,24 which may be associated with the function and homeostasis of mitochondria.25 Previous studies have demonstrated the involvement of DJ-1 in the pathogenesis of various liver diseases and conditions including liver fibrosis,26 nonalcoholic fatty liver disease,27,28 liver cancer,29 and liver injury and repair.30 However, the role of DJ-1 in hepatic I/R injury remains largely unknown.
In this study we show that DJ-1 is significantly down-regulated in the human donor liver tissues and the mouse liver subjected to I/R injury. In a mouse model of liver I/R injury, we demonstrate that Dj-1 deficiency in hepatocytes but not in myeloid cells ameliorates liver damage and inflammatory responses, probably by promoting mitochondrial accumulation of PARKIN and stabilizing PARKIN protein, which consequently result in enhanced mitophagy in response to I/R injury.
Results
DJ-1 Down-Regulation in Liver I/R Injury
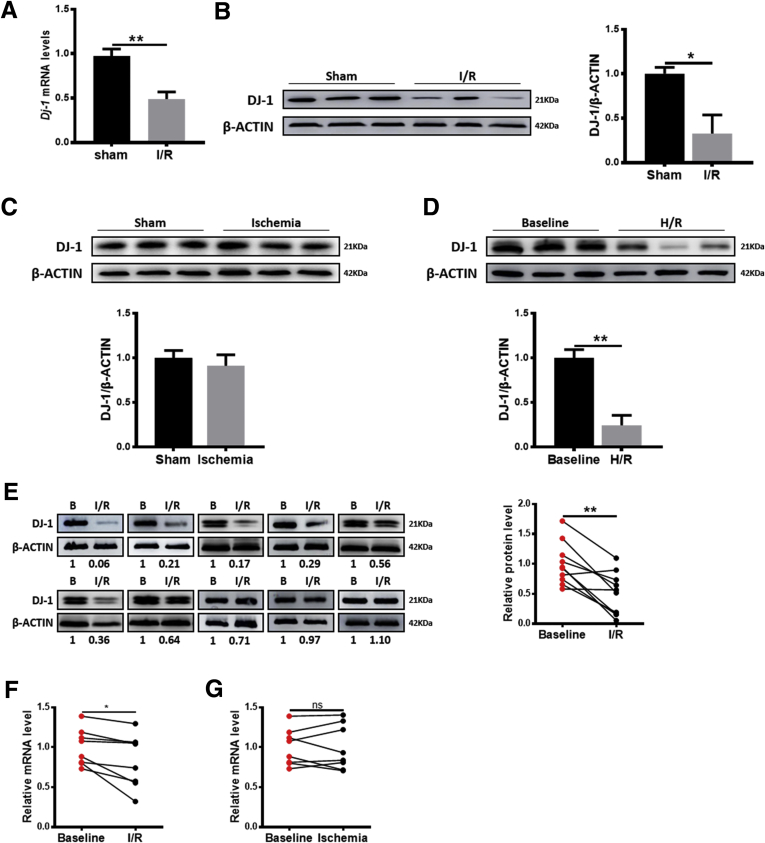
To examine whether DJ-1 is involved in liver I/R injury, the DJ-1 expression changes were first examined in the liver tissues in a mouse hepatic I/R model. Compared with the sham group, both quantitative real-time polymerase chain reaction (qRT-PCR) and Western blotting assays showed that DJ-1 expression was significantly decreased in the I/R group (Figure 1A and B) but not in the ischemia group (Figure 1C). Meanwhile, to detect the influence of I/R in the expression of DJ-1 on primary hepatocytes, we adopted the hypoxia/reoxygenation (H/R) model (hypoxia for 4 hours and reoxygenation for 6 hours) to treat primary hepatocytes from wild-type (WT) mice in vitro to mimic in vivo liver I/R injury. Compared with baseline controls, the primary hepatocytes treated with H/R showed lower DJ-1 expression levels (Figure 1D). More importantly, analyses of clinical living donor liver tissues that were subjected to I/R injury confirmed the decrease in DJ-1 expression (Figure 1E). In addition, bioinformatic analyses in a Gene Expression Omnibus (GEO) data set (GSE12720) also indicated a hepatic DJ-1 decrease in response to I/R but not ischemia injury in living donor liver grafts (Figure 1F and G). These findings indicate that the expression of DJ-1 in liver tissues is down-regulated in response to I/R injury.
Figure 1.
Liver DJ-1 expression is significantly decreased during hepatic I/R injury. (A) mRNA levels of DJ-1 in mouse liver tissues subjected to hepatic I/R injury. β-actin served as a control. (B–E) Representative Western blotting and quantification showing DJ-1 expression in liver tissues in mice subjected to I/R injury (B), in liver tissues in mice subjected to ischemia injury (C), in primary hepatocytes from WT mice in response to H/R treatment in vitro mimic of I/R injury (D), and in human liver biopsies either before any manipulation of the liver or after reperfusion (E). (F and G) Dj-1 mRNA levels in the sham group and the reperfused group (F) or the ischemia group (G) in GEO data sets (GSE12720). (n = 3 per group for A–C, n = 3 independent experiments for D, n = 10 per group for E, n = 8 per group for F and G). ∗P < .05, ∗∗P < .01.
Ablation of DJ-1 Ameliorates Hepatic I/R Injury
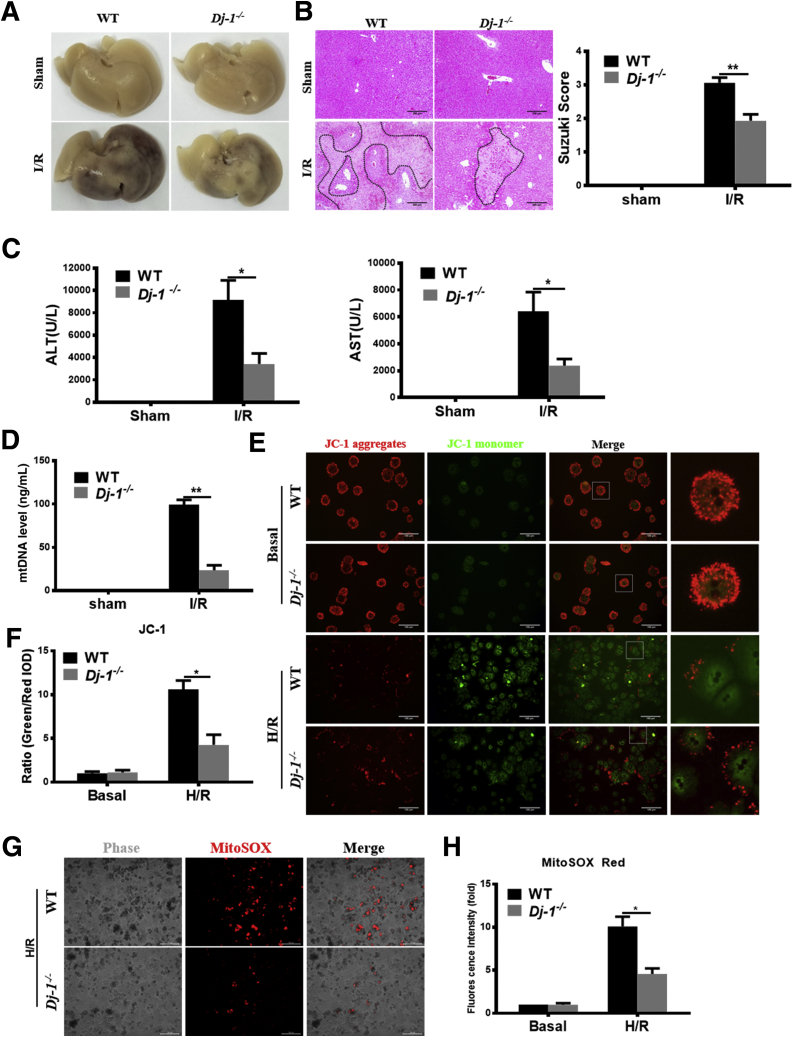
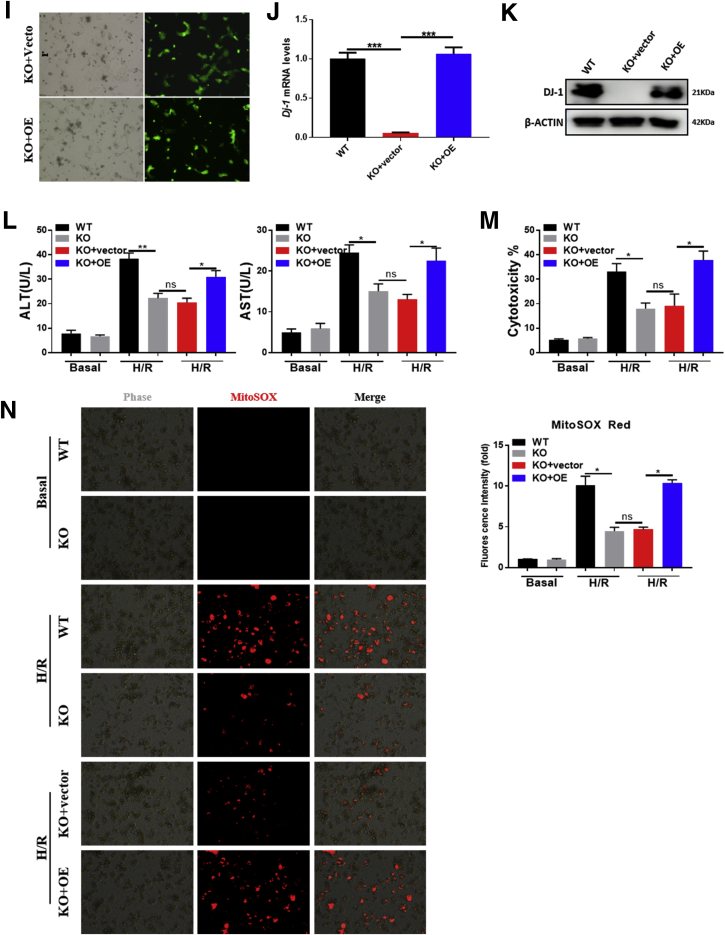
To determine the role of DJ-1 in I/R injury, the hepatic I/R injury model was performed in Dj-1 knockout (Dj-1-/-) mice. Compared with WT mice, Dj-1-/- mice showed significantly less hepatic necrosis (Figure 2A and B), serum alanine aminotransferase (ALT)/aspartate aminotransferase (AST) (Figure 2C), and mitochondrial DNA (mtDNA) (Figure 2D) levels at 6 hours after reperfusion, suggesting that Dj-1 deficiency improves I/R injury. Because mitochondrial dysfunction is a key event to trigger the onset of I/R injury, we then used JC-1, a membrane potential–sensitive dye, and MitoSox, a mitochondrial superoxide indicator, to evaluate ΔΨm loss and mitochondrial superoxide, respectively, in primary hepatocytes after in vitro H/R treatment. JC-1 dye aggregates in healthy mitochondria fluorescing red or exports from damaged mitochondria into cytosol fluorescing green. The ratio of green/red signals could present the severity of mitochondrial damage. As shown in Figure 2E–H, compared with hepatocytes isolated from WT mice, the hepatocytes isolated from Dj-1-/- mice showed less ΔΨm loss and mitochondrial superoxide in response to H/R treatment. These findings indicate that Dj-1 deficiency improves mitochondrial dysfunction and hepatic I/R injury. To further confirm the pathophysiological functions of DJ-1, we have set up experiments to investigate whether Dj-1 overexpression by adenovirus vector in Dj-1 knockout (KO) primary hepatocytes and observed whether rescue DJ-1 could counteract the protective effect of Dj-1 deficiency in vitro. As shown in Figure 2I, the expression levels of green fluorescent protein in primary hepatocytes were detected to assay the transfection efficiency under fluorescence microscope after transfection for 48 hours. The expressions of DJ-1 in primary hepatocytes were detected by Western blotting and qRT-PCR after transfection for 48 hours (Figure 2J and K). As shown in Figure 2L–N, the ALT and AST levels were significantly increased after DJ-1 overexpressed in DJ-1 KO primary hepatocytes compared with DJ-1 KO hepatocytes after H/R treatment. Meanwhile, cytotoxicity and mitochondrial superoxide levels were also reversed by DJ-1 overexpression in DJ-1 KO primary hepatocytes.
Figure 2.
DJ-1 KO improves I/R-induced liver injury. (A) Representative morphology of the whole liver organ from WT and Dj-1-/- mice in both sham and I/R groups. (B) Representative image of H&E staining of liver sections from Dj-1-/- and WT mice in both sham and I/R groups. (C) Serum ALT and AST levels of WT and Dj-1-/- mice in both sham and I/R groups. (D) Serum mtDNA levels of WT and Dj-1-/- mice in both sham and I/R groups. (E and F) Representative fluorescent images (E) and quantification (F) of loss of mitochondrial membrane potential in primary hepatocytes isolated from either WT or Dj-1-/- mice subjected to H/R injury. (G and H) Representative images (G) and quantification (H) of mitochondrial superoxide levels in primary hepatocytes isolated from either WT or Dj-1-/- mice subjected to H/R injury. (I–N) Primary hepatocytes were isolated from WT and Dj-1-/- mice, and Dj-1 was overexpressed in primary hepatocytes of Dj-1-/- mice by adenovirus vector in vitro. (I) Expression levels of green fluorescent protein in primary hepatocytes were captured to assay the transfection efficiency by using bright field (left panel) and fluorescence (right panel) microscopy after transfection for 48 hours. (J) mRNA and (K) proteins levels of DJ-1 in primary hepatocytes after transfection for 48 hours. (L) ALT and AST levels of medium supernatant in both sham and H/R groups. (M) Quantification of cytotoxicity was assayed by measuring levels of LDH in the medium supernatant. (N) Representative images (left) and quantification (right) of mitochondrial superoxide levels in primary hepatocytes. (KO, Dj-1 knockout; OE, Dj-1 overexpression; n = 3–5 per group for A–D; n = 3 independent experiments for E–N.)
DJ-1 Deficiency Improves I/R-Related Inflammatory Responses
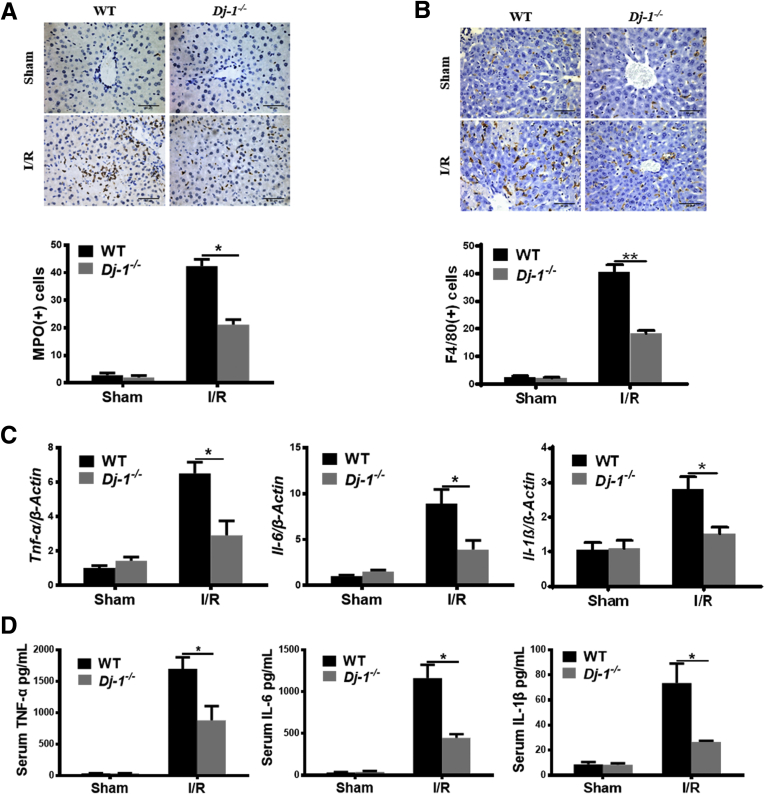
Because hepatic I/R injury frequently associates with the surge of sterile inflammatory responses,31 we then performed immunohistochemical (IHC) staining of myeloperoxidase (MPO) (a marker of neutrophils) and F4/80 (a marker of macrophages) to determine the hepatic infiltration of those immune cells. Significantly attenuated hepatic infiltration of both cell types was observed in Dj-1-/- mice compared with WT mice (Figure 3A and B). Correspondingly, the induction of proinflammatory cytokines such as interleukin (IL) 6, IL1β, and tumor necrosis factor (TNF) α was also decreased in Dj-1-/- mice compared with WT mice (Figure 3C and D). Collectively, these results demonstrate that Dj-1 deficiency improves the inflammatory responses after hepatic I/R injury.
Figure 3.
DJ-1 deficiency improves I/R-induced inflammation responses. (A) Representative images of MPO and quantification of MPO positive cells showing hepatic infiltration of neutrophils in response to I/R surgery in both genotypes. (B) Representative images of F4/80 and quantification of F4/80 positive cells showing hepatic infiltration of macrophages in response to I/R surgery in both genotypes. (C) Expression levels of Tnf-α, Il-6, and Il-1β in liver tissues in both WT and Dj-1-/- mice after hepatic I/R injury. (D) Serum levels of TNF-α, IL6, and IL1β in both WT and Dj-1-/- mice after hepatic I/R injury. (n = 3–5 per group for A–D).
Hepatocyte-Specific but not Myeloid-Specific Dj-1 KO Ameliorates Hepatic I/R Injury
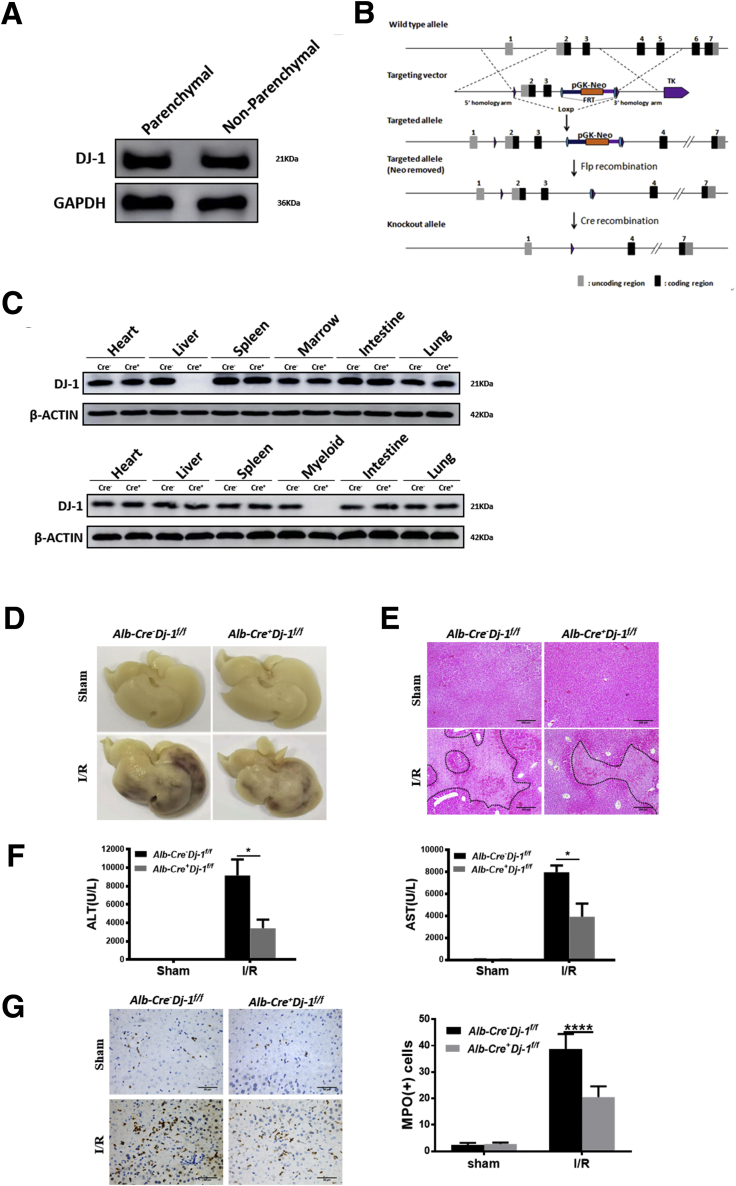
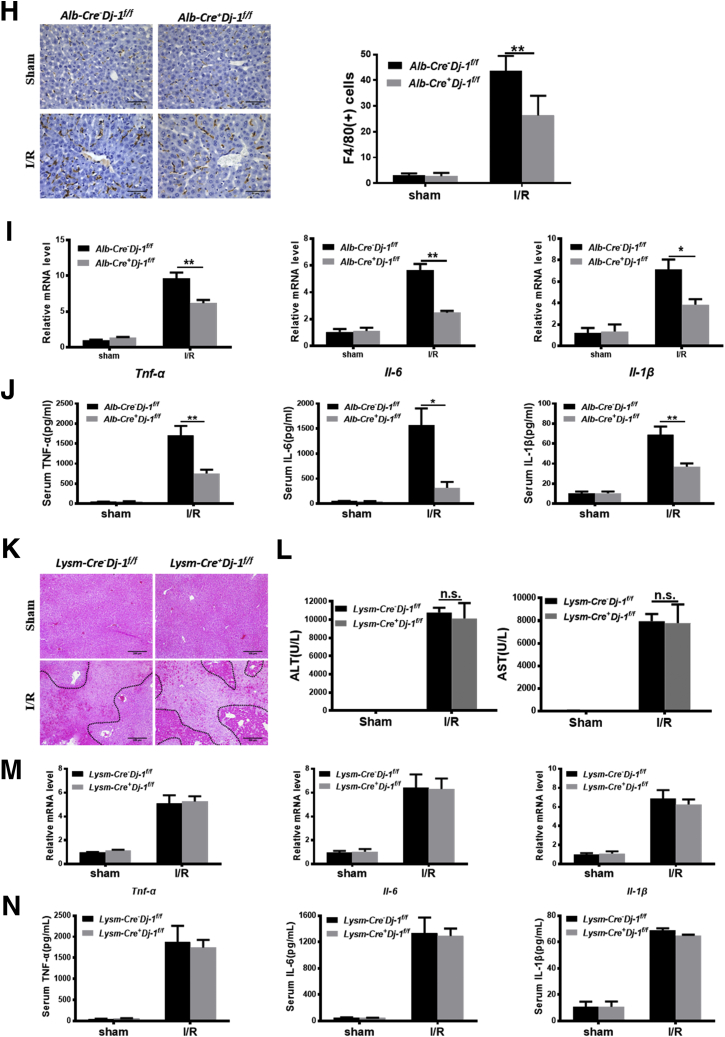
Both hepatocytes and myeloid cells have been recognized to play important roles in the progression of liver I/R injury.32, 33, 34 We then started to investigate which type of cells are important for I/R protection in Dj-1-/- mice. We first examined the DJ-1 expression in both parenchymal and non-parenchymal cells in mouse liver. As shown in Figure 4A, DJ-1 levels were comparable in both cell types. We generated Dj-1f/f mice by inserting loxp sites flanking the second and third exons of Dj-1 (Figure 4B). We then used Alb-Cre+Dj-1f/f mice and Lysm-Cre+Dj-1f/f mice, which possess specific Dj-1 depletion in hepatocytes and myeloid cells, respectively, to perform liver I/R injury. Tissue-specific Dj-1 depletion was confirmed by Western blotting assays, the upper blots were for Alb-Cre+Dj-1f/f mice, and the lower blots were for Lysm-Cre+Dj-1f/f mice (Figure 4C). Interestingly, hepatocyte-specific but not myeloid-specific Dj-1 KO improved liver I/R injury as demonstrated by reduced levels of liver necrosis, serum ALT/AST, hepatic infiltration of both neutrophils and macrophages, and proinflammatory cytokines in Alb-Cre+Dj-1f/f mice (Figure 4D–J) but not in Lysm-Cre+Dj-1f/f mice (Figure 4K–N). Collectively, these findings indicate that Dj-1 depletion in hepatocytes but not in myeloid cells improves liver I/R injury.
Figure 4.
Hepatocyte-specific DJ-1 KO improves I/R-induced liver injury and inflammatory responses. (A) Protein levels of DJ-1 in both parenchymal and non-parenchymal cells in mouse liver. (B) Schematic diagram of conditional Dj-1 KO mice. (C) Expression levels of DJ-1 in various tissues of Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice (upper blots) or Lysm-Cre-Dj-1f/f and Lysm-Cre+Dj-1f/f mice (lower blots). (D) Representative morphology of the whole liver organ from Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in both sham and I/R groups. (E) Representative image of H&E staining of liver sections from Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in both sham and I/R groups. (F) Serum levels of ALT and AST of Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in both sham and I/R groups. (G and H) Representative IHC image of MPO (G) and F4/80 (H), respectively, showing the hepatic infiltration of neutrophils and macrophages in Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in both sham and I/R groups. (I and J) Hepatic mRNA levels (I) and serum levels (J) of TNF-α, IL6, and IL1β in Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in response to I/R injury. (K) Representative image of H&E staining of liver sections from Lysm-Cre-Dj-1f/f and Lysm-Cre+Dj-1f/f mice in response to I/R injury. (L) Serum levels of ALT and AST of Lysm-Cre-Dj-1f/f and Lysm-Cre+Dj-1f/f mice in response to I/R injury. (M) Relative mRNA levels of Tnf-α, Il-6, and Il-1β in liver tissues of Lysm-Cre-Dj-1f/f and Lysm-Cre+Dj-1f/f mice after I/R injury. (N) Serum levels of TNF-α, IL6, and IL1β in Lysm-Cre-Dj-1f/f and Lysm-Cre+Dj-1f/f mice after hepatic I/R injury. (n = 3–5 per group for A–N.) ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001.
DJ-1 Deficiency Enhances Hepatic Autophagy in Response to I/R Injury
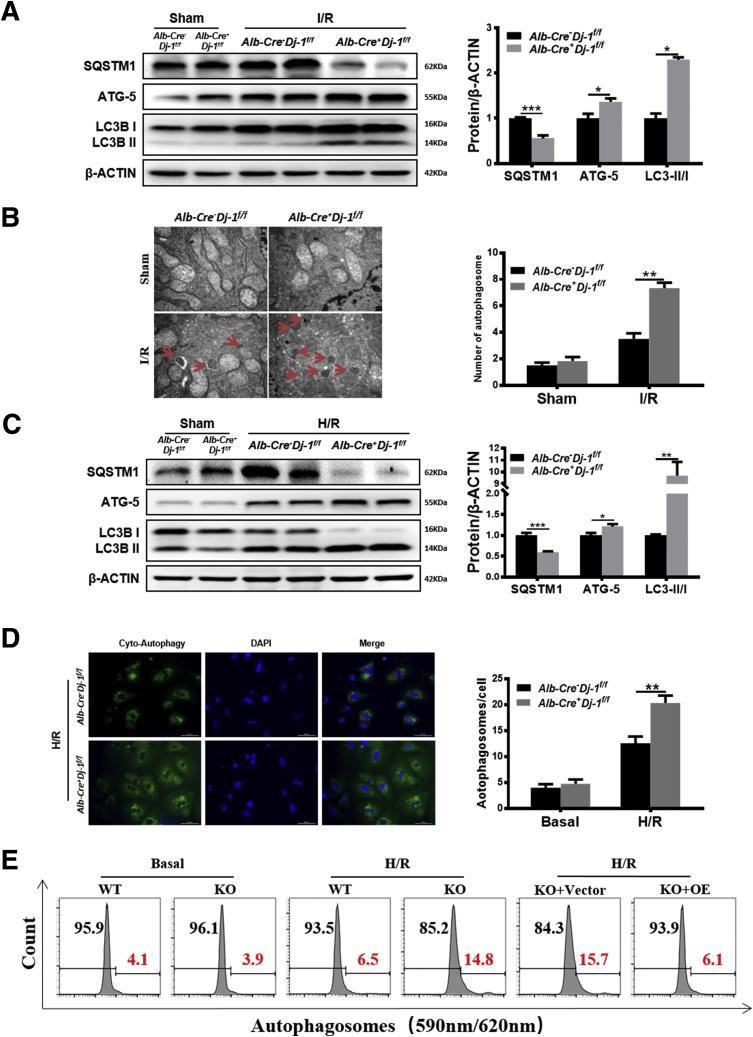
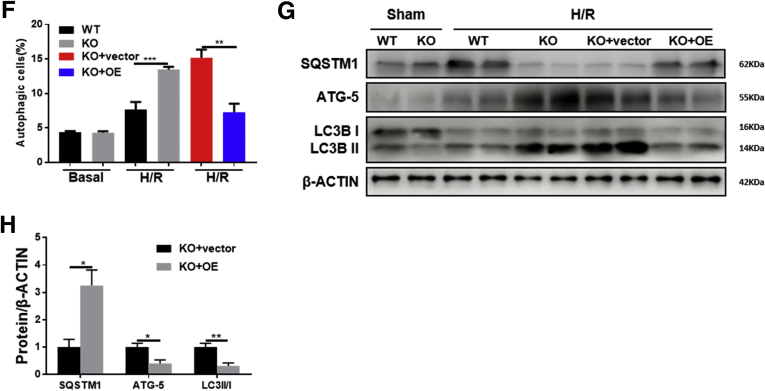
Autophagy is an evolutionarily conserved self-degradation cellular process and enables cells to recycle intracellular proteins and organelles in response to various extracellular stimuli such as depletion of nutrients, energy, and growth factors.35 Although the exact role of autophagy on I/R injury is still under debate,36 our and some other groups have demonstrated the protective role of autophagy on liver I/R injury.37 To determine whether the hepatoprotective phenomenon in Alb-Cre+Dj-1f/f mice is related to autophagy, we examined the levels of autophagy markers, LC3 ratio (LC3-II/I), ATG5, and SQSTM1, in liver tissues after I/R injury. Western blotting assays showed increased LC3 ratio and ATG5 levels and decreased SQSTM1 expression in Alb-Cre+Dj-1f/f mice compared with control mice after I/R injury, suggesting enhanced autophagy in Alb-Cre+Dj-1f/f mice (Figure 5A). More importantly, transmission electron microscopy (TEM) confirmed that I/R induced a greater number of autophagosomes in the liver tissues of Alb-Cre+Dj-1f/f mice compared with control mice (Figure 5B). In addition, autophagy levels were also measured in primary hepatocytes isolated from both genotypes after in vitro H/R treatment. As shown in Figure 5C, compared with hepatocytes isolated from control mice, H/R treatment caused greater LC3 ratio and ATG5 expression and less SQSTM1 expression in the primary hepatocytes isolated from Alb-Cre+Dj-1f/f mice. To further confirm the enhancement of autophagy in hepatocytes with DJ-1 depletion, a novel amphiphilic tracer that stains lysosomes minimally while maximizing the fluorescence in autophagic vesicles such as pre-autophagosomes, autophagosomes, and autolysosomes38 was used to measure the autophagy flux. H/R treatment induced more abundant autophagic vesicles (green signals) in the primary hepatocytes of Alb-Cre+Dj-1f/f mice than in control hepatocytes (Figure 5D). We also found that adenovirus virus-mediated overexpression of Dj-1 in DJ-1 KO primary hepatocytes that were isolated from Alb-Cre+Dj-1f/f mice showed a marked decreased autophagosome (Figure 5E–H). These findings suggest that hepatic-specific Dj-1 deficiency enhances autophagy in liver tissues in I/R injury.
Figure 5.
DJ-1 deficiency enhances hepatic autophagy in response to I/R injury. (A) Representative Western blotting and quantification for expression of SQSTM1, ATG-5, and LC3 in the liver tissues of Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice after hepatic I/R injury. β-ACTIN was used as the loading control. (B) Representative transmission electron images showing autophagosomes in the liver sections of both Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice after hepatic I/R injury. Autophagosomes are indicated by arrows. (C) Representative Western blotting and quantification for expression of SQSTM1, ATG-5, and LC3 in the primary hepatocytes of the Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice after H/R treatment. β-ACTIN was used as the loading control. (D) Representative fluorescence images of autophagosomes in the primary hepatocytes of the Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice after H/R treatment. (E–H) Primary hepatocytes were isolated from Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice, and Dj-1 was overexpressed in the primary hepatocytes by adenovirus vector in vitro. (E and F) Levels of autophagy were measured by flow cytometry using Autophagy Probe. (G and H) Representative Western blotting and quantification for expression of SQSTM1, ATG-5, and LC3 in the primary hepatocytes after H/R treatment. β-ACTIN was used as the loading control. KO, Dj-1 knockout; OE, Dj-1 overexpression; n = 3–6 per group for A and B, n = 3 independent experiments for C–H. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Autophagy Is Essential for the Protective Effect of DJ-1 Depletion
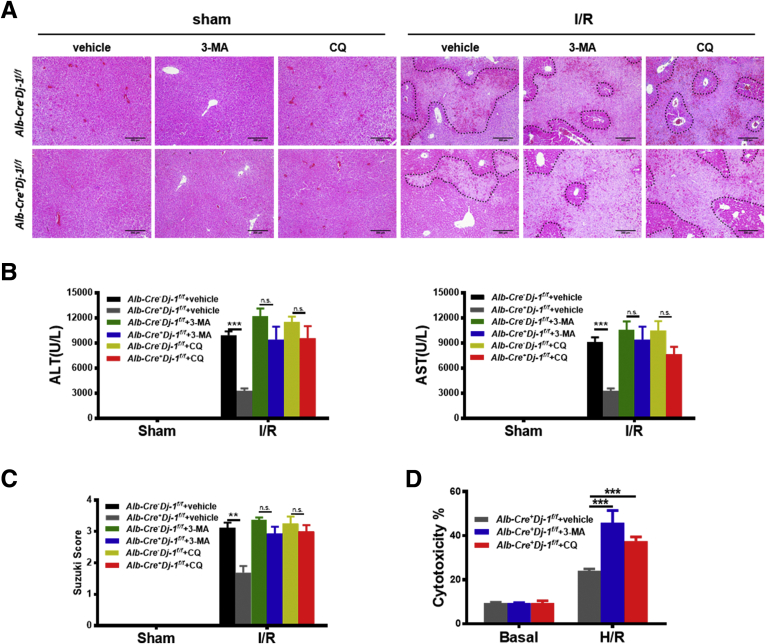
To test whether autophagy is required for the hepatoprotective effect in Alb-Cre+Dj-1f/f mice in response to liver I/R injury, we used 2 autophagy inhibitors, 3-methyladenine (3-MA) and chloroquine (CQ), to block autophagy in both genotypes. Autophagy inhibition significantly eliminated the hepatoprotective effect in Alb-Cre+Dj-1f/f mice as evidenced by substantially increased hepatic necrosis and serum ALT/AST levels in response to I/R injury (Figure 6A–C). In line with these in vivo results, 3-MA and CQ pretreatment significantly counteracted the protective effect of Dj-1 deficiency in hepatocytes after in vitro H/R treatment (Figure 6D). Together, these observations suggest that Dj-1 deficiency protects hepatic I/R injury in an autophagy-dependent manner.
Figure 6.
Autophagy inhibition attenuates the protective effect on hepatic I/R injury in DJ-1 deficiency mice. (A–C) Hepatic necrosis levels (A) and Suzuki score (C) and the serum ALT and AST (B) levels were examined in Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice pretreated with autophagy inhibitors 3-MA and CQ, followed by hepatic I/R injury. (D) Quantification of cytotoxicity (% cell death) from the primary hepatocytes isolated from Alb-Cre+Dj-1f/f mice, which were pretreated with autophagy inhibitors 3-MA and CQ, followed by H/R treatment. (n = 4 per group for A–C, n = 3 independent experiments for D). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
DJ-1 Loss Increases Mitophagy by Enhanced Mitochondrial Translocation of Parkin
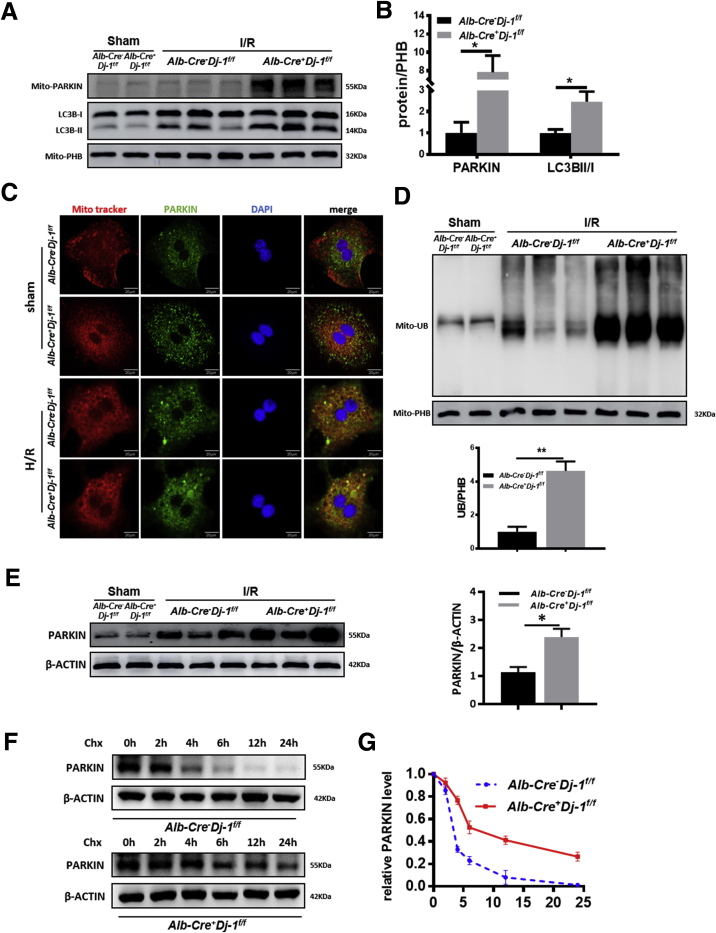
As a form of selective autophagy, mitophagy has been indicated to play a protective role in I/R injury in various organs by selectively removing damaged mitochondria.39, 40, 41 A previous study has demonstrated that Dj-1 deficiency could promote mitochondrial translocation of PARKIN, which is a key step of the onset of mitophagy.42 To examine whether Dj-1 deficiency promotes mitophagy during liver I/R injury, we measured the mitochondrial accumulation of PARKIN in the liver tissues of both genotypes in response to I/R injury. The mitochondrial accumulation of PARKIN and LC3BI/II ratios were significantly higher in Alb-Cre+Dj-1f/f mice compared with control mice (Figure 7A and B), suggesting that hepatic-specific Dj-1 deficiency may enhance mitophagy levels in liver tissues in response to I/R injury. To further confirm this point, we performed immunofluorescent co-staining of mitochondria via MitoTracker Red and PARKIN in primary hepatocytes after H/R treatment. Co-localization of PARKIN and mitochondria was more apparent in Dj-1 depleted hepatocytes than in WT hepatocytes (Figure 7C). Moreover, compared with control mice, mitochondria isolated from the liver of Alb-Cre+Dj-1f/f mice possessed significantly higher ubiquitin levels in response to I/R injury (Figure 7D). Interestingly, Western blotting assays showed that compared with control mice, Dj-1 deficiency could significantly increase PARKIN protein levels in the whole liver tissue lysates after I/R injury (Figure 7E). Meanwhile, the steady-state levels of PARKIN were comparable in both genotypes (Figure 7E). In addition, cycloheximide chase assays showed that PARKIN was more stable in Dj-1 depleted hepatocytes than in WT hepatocytes in response to H/R treatment (Figure 7F and G). This result is consistent with a previous study showing increased PARKIN accumulation in Dj-1-null cells.24 Therefore, in response to I/R injury, Dj-1 deficiency not only enhanced mitochondrial recruitment of PARKIN but also increased PARKIN stability, which contribute to elevated mitophagy levels.
Figure 7.
DJ-1 deficiency increases mitophagy by enhanced mitochondrial translocation and protein stability of Parkin. (A and B) Representative Western blotting images (A) and quantification (B) of the mitochondrial PARKIN and LC3 in the liver tissues in the Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in response to I/R injury. PHB was used as a loading control. (C) Representative confocal images of the co-staining of mitochondria and PARKIN in the primary hepatocytes isolated from either Alb-Cre-Dj-1f/f or Alb-Cre+Dj-1f/f mice after in vitro H/R treatment. (D) Representative Western blotting images and quantification of the ubiquitin levels on the mitochondria isolated from liver tissues of both Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in response to I/R injury. PHB was used as a loading control. (E) Representative Western blotting images and quantification for expression of PARKIN in the whole tissue lysates of liver tissues of both Alb-Cre-Dj-1f/f and Alb-Cre+Dj-1f/f mice in response to I/R injury. (F and G) Representative Western blotting and quantification of PARKIN in the primary hepatocytes isolated from either Alb-Cre-Dj-1f/f or Alb-Cre+Dj-1f/f mice subjected to in vitro H/R treatment followed with CHX treatment. β-ACTIN was used as a loading control. (n = 3 per group for A, B, D, and E; n = 3 independent experiments for C, F, and G). CHX, cycloheximide. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Discussion
DJ-1 is abundantly expressed in various tissues and organs and regulates many biological functions.43 Our previous studies revealed that DJ-1 is a stress sensor in the pathogenic progression of various liver diseases.26,28,29 However, the functions and molecular mechanisms of DJ-1 in hepatic I/R injury are still unknown and required for further investigation. In this study, we investigate the functions and mechanisms of DJ-1 in hepatic I/R injury.
We demonstrated that I/R injury leads to DJ-1 down-regulation in both human and mouse liver tissues. Meanwhile, in vitro H/R treatment also decreased DJ-1 expression in hepatocytes. These results suggest that DJ-1 may play a role in hepatic I/R injury. Consistent with our previous studies showing that Dj-1 deficiency improves CCL4- and DEN-induced liver injury,26,29 our current study has demonstrated that ablation of Dj-1 ameliorates liver I/R injury. We further demonstrated that the specific Dj-1 depletion in hepatocytes but not in myeloid cells contributes to the improvement of I/R injury.
Recently, many groups have demonstrated the involvement of autophagy in I/R injury in various organs. Although the exact role of autophagy in I/R injury is still under debate, we and other groups have demonstrated that autophagy plays a positive role in protecting I/R injury.37,44, 45, 46, 47 In this study, we showed that elevated autophagy levels are required for the protective effect in Dj-1 deficiency hepatocytes, because inhibition of autophagy by 3-MA and CQ counteracted the hepatoprotective function of Dj-1 deficiency during hepatic I/R injury. Interestingly, like PINK1 and PARKIN, which are frequently mutated in many cases of early-onset familial PD, some DJ-1 mutations also lead to early-onset PD. Moreover, a previous study has identified a trimer complex consisted of PINK1-Parkin-DJ-1, suggesting that DJ-1 may play a role in PINK1-PARKIN mediated mitophagy. In this study, we demonstrate that Dj-1 deficiency enhances mitophagy by either increasing mitochondrial recruitment of Parkin or elevating the protein stability of Parkin.
In fact, the association of DJ-1 and autophagy has been demonstrated controversially in many studies. It seems that DJ-1 could either positively48, 49, 50 or negatively25,51, 52, 53 regulate the autophagy levels. This discrepancy may be due to the different role of DJ-1 in various cell types. The results in current study support that DJ-1 negatively regulates autophagy in hepatocytes.
In summary, the present work demonstrates that DJ-1 is a novel cellular regulator of hepatic I/R injury. Dj-1 depletion protects against hepatocyte damage and inflammatory responses through argument of mitophagy during hepatic I/R injury. Our study provides experimental evidence supporting that DJ-1 could serve as a potential therapeutic target for the amelioration of hepatocyte damage caused by I/R injury.
Materials and Methods
Patient Samples
The human liver biopsies used in this study were obtained from Renji Hospital. The baseline biopsies were taken in the living donor before manipulation of the liver. The I/R biopsies were taken at 2–3 hours after reperfusion of the donor liver with recipient blood during transplant. Informed consent forms of all participants were signed by all donors or their families. All human samples were used only for scientific research, and the human tissues experiment was approved by the institutional review board at the Renji Hospital and performed according to the principles outlined by the Declaration of Helsinki.
Mice
DJ-1 KO (Dj-1-/-) mice (JAX stock# 006577) were purchased from the Jackson Laboratory (Bar Harbor, ME). The experimental Dj-1-/- and WT littermate controls were generated by Dj-1+/- × Dj-1+/-. The conditional Dj-1 KO mice were generated by the Shanghai Model Organisms by inserting loxp sites flanking the second and third exons of Dj-1 (Figure 4B). Mice with conditional ablation of the Dj-1 gene were generated by crossing Alb-Cre+/- and Lysm-Cre+/- with Dj-1f/f mice. In all experiments, Dj-1f/f littermates were used as controls. The sources of all the above mice are C57BL/6 background. Colonies were housed in the virus/antigen-free Animal Core Facility of the ShuGuang Hospital Affiliated to Shanghai University of Chinese Traditional Medicine, with a 12-hour light/dark cycle and controlled temperature and humidity and were provided with water and food ad libitum.
Murine Model of Warm Liver Ischemia-Reperfusion Injury
Eight- to 10-week-old male mice (weight 23–27 g) were subjected to 70% warm hepatic I/R injury as we previously described.37 In brief, after the mice were anesthetized by sodium pentobarbital (40 mg/kg, intraperitoneally), midline laparotomy was performed to expose the liver. An atraumatic microvascular clamp was used to occlude the blood supply to the left and median lobes of the liver for 90 minutes, and then the clamp was removed for 6-hour reperfusion. Mice were euthanized at each indicated time point after reperfusion, and the liver and serum samples were collected immediately for subsequent examination. Sham controls underwent the same surgical procedure without vasculature occlusion. To examine the role of autophagy on liver I/R injury, the mice were injected intraperitoneally with CQ (60 mg/kg) or 3-MA (30 mg/kg) (Sigma-Aldrich, St Louis, MO) at 1 hour before ischemia. A same volume of 0.5% dimethyl sulfoxide or blank solution was used as the vehicle control. All procedures involving animals were reviewed and approved by the criteria outlined in Guide for the Care and Use of Laboratory Animals, which was approved by the Institutional Animal Care and Use Committee of the ShuGuang Hospital Affiliated to Shanghai University of Chinese Traditional Medicine.
Liver Function Measurement
The liver function of mice was assessed by measuring the concentrations of serum ALT and AST by standard clinical automatic analyzer (Dimension Xpand; Siemens Dade Behring, Munich, Germany) according to the manufacturer’s instructions.
Liver Histologic and IHC Staining
Liver issues were fixed in 4% paraformaldehyde for 24 hours and then paraffin-embedded through standard procedures. The paraffin-embedded tissue sections were sliced by a rotary microtome to give a thickness of 5 μm. For liver histopathology, in brief, the slides were stained with H&E to visualize the pattern in necrotic areas of the liver, and the Suzuki criteria were used to evaluate the histologic severity of hepatic I/R injury. For IHC staining, the hepatic infiltration of macrophages and neutrophils was determined by incubating the liver tissue slides with MPO (rabbit polyclonal, 1:300 dilution; Cell Signaling Technology, Danvers, MA) and F4/80 (rat monoclonal, 1:250 dilution; AbD Serotec, Oxford, UK) primary antibodies, respectively. After overnight incubation at 4°C, the slides were incubated with horseradish peroxidase–conjugated secondary antibodies (1:500 dilution; Jackson Laboratory), and immunoreactive cells were visualized using DAB.
Primary Hepatocyte Isolation, Culture, and Treatment
Primary hepatocytes were isolated from 6- to 8-week male mice and then plated on plates as previously described.54 The primary hepatocytes were challenged with 4-hour hypoxia at 37°C in a modular incubator chamber (Biospherix, Lacona, NY) gassed with 5% CO2 and 95% N2 and followed by 6-hour reoxygenation to mimic in vivo liver I/R injury. Autophagy inhibitors, 3-MA (5 mmol/L) and CQ (20 μmol/L), were added to the cell culture medium of primary hepatocytes, and autophagy inhibition was confirmed by Western blotting. To assess the H/R induced cytotoxicity in primary hepatocytes, the lactate dehydrogenase (LDH) levels in cell culture supernatant were measured by LDH Release Assay Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. TheΔΨm assay kit with JC-1 (Beyotime, Shanghai, China) was used to measuring mitochondrial transmembrane potential in hepatocytes according to the manufacturer’s instructions. Mitochondrial superoxide was stained with MitoSOX Red (Invitrogen, Waltham, MA). The autophagosomes of the hepatocytes were identified by the Cyto-ID Autophagy Detection Kit (Enzo Life Sciences, Farmington, NY) and Autophagy Assay Red kit (ImmunoChemistry Technologies, Bloomington, MN). The PARKIN stability was measured in primary hepatocytes after in vitro H/R treatment by incubating cells with cycloheximide at 20 μg/mL for indicated time points.
Plasmids Construction and Adenovirus Package
Mouse full-length DJ-1 cDNA was inserted into pAdEasy-EF1-MCS-CMV-EGFP (EcoRI MCS) vector to obtain Dj-1 expression plasmid (pAdEasy-DJ-1). Furthermore, pAdEasy-DJ-1 or its control vector plasmid (pAdEasy-GFP) was co-transfected with packaging plasmids pHBAd-BHG into HEK293A cells to produce Ad-DJ-1 or Ad-GFP adenovirus particles, respectively. Adenovirus supernatant was collected 46 hours after transfection. The recombinant DJ-1 overexpression and its control adenovirus were prepared by Hanbio (Shanghai, China).
Cell Transfection and Evaluation of DJ-1
Primary hepatocytes were cultured in a 24-well or 6-well plate and infected with Ad-DJ-1 or its control Ad-GFP by using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) according to the protocol provided by Ribobio Corporation. The expression levels of green fluorescent protein in primary hepatocytes were detected to assay the transfection efficiency under fluorescence microscope after transfection for 48 hours (Figure 2I). The expressions of DJ-1 in primary hepatocytes were detected by Western blotting and qRT-PCR after transfection for 48 hours (Figure 2J and K).
qPCR and Western Blotting Analysis
Total RNA of liver tissues was isolated with the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Eight hundred nanograms total RNA was reverse-transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Tokyo, Japan) following the manufacturer’s instructions. The PCR amplification products were quantified by SYBR-green based qRT-PCR (Takara) following a standard procedure. The mRNA expression levels of target genes were normalized by β-Actin. Primer sequences are provided in Table 1.
Table 1.
Primers Used for qRT-PCR
| Gene | Sequence 5′—3′ forward | Sequence 5′—3′ reverse |
|---|---|---|
| β-Actin | GTGACGTTGACATCCGTAAAGA | GCCGGACTCATCGTACTCC |
| Il-6 | GCTACCAAACTGGATATAATCAGGA | CCAGGTAGCTATGGTACTCCAGAA |
| Il-1β | TGTAATGAAAGACGGCACACC | TCTTCTTTGGGTATTGCTTGG |
| Tnfα | TTCTATGGCCCAGACCCTCA | TTTGCTACGACGTGGGCTAC |
| Dj-1 | GTGCAGTGTAGCCGTGATGT | CCTCCTGGAAGAACCACCAC |
Liver tissue or cell samples were processed to Western blotting analysis as we previously described.28 Primary antibodies were rabbit anti-DJ-1 (Abcam, Cambridge, MA), mouse anti-PARKIN (Santa Cruz Technology, Santa Cruz, CA), mouse anti-Ub (Santa Cruz Technology), rabbit anti-LC3B (Proteintech, Wuhan, China), rabbit anti-PHB (Proteintech), rabbit anti-SQSTM1 (Cell Signaling Technology), rabbit anti-ATG5 (Cell Signaling Technology), and anti-β-actin (Sigma-Aldrich).
TEM analysis
The fresh liver samples were fixed with 2.5% glutaraldehyde, then cut into ultrathin sections, stained with uranyl acetate and lead citrate, and then observed on an electron microscope. Ten visual fields were randomly taken from each sample, and the total amount of autophagic vacuoles was counted.
mtDNA Detection
Absolute qRT-PCR was used to measure serum levels of mtDNA as we previously described.37 Total DNA from serum samples was isolated with the QIAamp Blood and Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions and diluted to the appropriate concentration. The same amount of total DNA was added to each reaction on each plate. mtDNA was determined using a pair of mouse primers of cytochrome c oxidase subunit III (5′-ACCAAGGCCACCACA CTCCT-3′ and 5′-ACGCTCAGAAGAATCCTGCAAAGAA-3′). To construct standard curves, mitochondria were isolated from mouse liver by Tissue Mitochondria Isolation Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Real-time PCR was used to verify the purity of mtDNA standards using 2 pairs of mouse primers, one for mitochondrial genes and other for nuclear-encoded β-actin. Dilutions of these purified mtDNA samples and mtDNA standards were included on each PCR plate for each gene tested. The limit of detection for the assay was determined to be <0.05 ng/mL.
Cytoplasmic and Mitochondrial Fractionation
A Tissue Mitochondria Isolation Kit was used to isolate intact mitochondria according to the manufacturer’s instructions. Briefly, 50–100 mg liver tissues were homogenized with 2 mL Mito-Cyto Isolation Buffer on ice for 10 passes, and the homogenate was then centrifuged at 1000g for 5 minutes to remove the sediments. The supernatant was further centrifuged at 3500g for 10 minutes to collect the mitochondrial pellet.
Immunofluorescence and Confocal Microscopy
To analyze the co-localization of PARKIN with mitochondria, immunofluorescence co-staining against mitochondria and PARKIN was performed by MitoTracker Red (Beyotime, Shanghai, China) and the PARKIN antibody (Santa Cruz Technology). The mitochondrial accumulation of PARKIN was detected by Olympus FV3000 confocal laser scanning microscope.
Serum Concentration of Inflammatory Cytokines
Serum levels of inflammatory cytokines including IL6, TNF-α, and IL1β were measured using commercial enzyme-linked immunosorbent assay kits (NeoBioscience Technology, Shenzhen, China) according to the manufacturer’s protocols.
Dj-1 Expression Analysis in Public Data Sets
Affymetrix microarray data in GSE1272055 were downloaded from the GEO (https://www.ncbi.nlm.nih.gov/geo/) database, which contains 63 biopsies in 13 deceased donor and 8 living donor liver grafts done at serial time points (baseline, no manipulation; ischemia, cold preservation; and I/R, 1 hour after reperfusion). Dj-1 expression was extracted and analyzed in GraphPad Prism 7 (San Diego, CA).
Statistical Analysis
Paired and unpaired data were evaluated with the Student t test in this study. All data are presented as mean ± standard error of the mean of at least 3 independent biological replicated experiments, except where otherwise specifically noted. P < .05 was considered to be statistically significant (∗P < .05, ∗∗P < .01, ∗∗∗P < .001).
Acknowledgments
The authors thank Dr Zhang Ming (Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China) for the gifts of Alb-Cre and Lysm-Cre mice.
CRediT Authorship Contributions
Min Xu (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Lead)
Jinyang Gu (Funding acquisition: Equal; Writing – original draft: Equal)
Jichang Li (Data curation: Supporting; Investigation: Supporting)
Dongwei Xu (Data curation: Supporting; Investigation: Supporting)
Miao Huang (Investigation: Equal; Formal analysis: Equal; Writing - original draft: Equal)
Fang Wang (Investigation: Supporting)
Hailong Wu (Funding acquisition: Equal; Supervision: Equal; Writing – review & editing: Supporting)
Xuehua Sun (Funding acquisition: Equal; Supervision: Equal)
Yueqiu Gao (Funding acquisition: Equal; Supervision: Equal)
Hualian Hang (Investigation: Equal; Methodology: Equal; Writing – original draft: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the National Natural Science Foundation of China (81873582 and 82070633 to X. Kong, 81874436 to X. Sun, 31870905 to H. Wu, 81874436 to Y. Gao, 82072645 and 81772507 to J. Gu), grants from the Committee of Science and Technology of Shanghai Municipal Government (Z.Y. (2018-2020)-CCCX-2003-01 to Y Gao), Shanghai Municipal Education Commission–Gaofeng Clinical Medicine Grant Support (No. 20191910), Foundation for Shanghai Jiao Tong University for SMC-Morning Star Youth Scholars Program, Clinical Research Plan of SHDC (No. SHDC2020CR3005A) to J. Gu, and the National Key Sci-Tech Special Project of China (2018ZX10725-504) to Y. Gao.
Contributor Information
Jinyang Gu, Email: gjynyd@126.com.
Xiaoni Kong, Email: xiaoni-kong@126.com.
Yueqiu Gao, Email: gaoyueqiu@hotmail.com.
References
- 1.Zhai Y., Busuttil R.W., Kupiec-Weglinski J.W. Liver ischemia and reperfusion injury: new insights into mechanisms of innate-adaptive immune-mediated tissue inflammation. Am J Transplant. 2001;11:1563–1569. doi: 10.1111/j.1600-6143.2011.03579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luan S., Yun X., RaoW, Xiao C., Xu Z., Lang J., Huang Q. Emamectin benzoate induces ROS-mediated DNA damage and apoptosis in Trichoplusia Tn5B1-4 cells. Chem Biol Interact. 2017;273:90–98. doi: 10.1016/j.cbi.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Sun S., Xie F., Xu X., Cai Q., Zhang Q., Cui Z., Zheng Y., Zhou J. Advanced oxidation protein products induce S-phase arrest of hepatocytes via the ROS-dependent, beta-catenin-CDK2-mediated pathway. Redox Biol. 2018;14:338–353. doi: 10.1016/j.redox.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farmer E.E., Mueller M.J. ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol. 2013;64:429–450. doi: 10.1146/annurev-arplant-050312-120132. [DOI] [PubMed] [Google Scholar]
- 5.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 6.Wang K., Klionsky D.J. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang Y.C., Tian H.X., Yi T., Chen H.B. The critical roles of mitophagy in cerebral ischemia. Protein Cell. 2016;7:699–713. doi: 10.1007/s13238-016-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Um J.H., Yun J. Emerging role of mitophagy in human diseases and physiology. BMB Rep. 2017;50:299–307. doi: 10.5483/BMBRep.2017.50.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 10.Spinazzi M., De Strooper B. PARL: the mitochondrial rhomboid protease. Semin Cell Dev Biol. 2016;60:19–28. doi: 10.1016/j.semcdb.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 11.Takatori S., Ito G., Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci Lett. 2008;430:13–17. doi: 10.1016/j.neulet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Lin W., Kang U.J. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S., Holmstrom K.M., Treis A., Skujat D., Weber S.S., Fiesel F.C., Kahle P.J., Springer W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 16.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., Magrane J., Moore D.J., Dawson V.L., Grailhe R., Dawson T.M., Li C., Tieu K., Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J., Wang K.Z., Chu C.T. After the banquet: mitochondrial biogenesis, mitophagy, and cell survival. Autophagy. 2013;9:1663–1676. doi: 10.4161/auto.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonifati V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J.C., Brice A., van Duijn C.M., Oostra B., Meco G., Heutink P. DJ-1 (PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci. 2003;24:159–160. doi: 10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- 19.Aron L., Klein P., Pham T.T., Kramer E.R., Wurst W., Klein R. Pro-survival role for Parkinson’s associated gene DJ-1 revealed in trophically impaired dopaminergic neurons. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cookson M.R. Pathways to parkinsonism. Neuron. 2003;37:7–10. doi: 10.1016/s0896-6273(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 21.Yamane T., Suzui S., Kitaura H., Takahashi-Niki K., Iguchi-Ariga S.M., Ariga H. Transcriptional activation of the cholecystokinin gene by DJ-1 through interaction of DJ-1 with RREB1 and the effect of DJ-1 on the cholecystokinin level in mice. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy M.K., Cookson M.R. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy. 2011;7:531–532. doi: 10.4161/auto.7.5.14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., van Dongen J.W., Vanacore N., van Swieten J.C., Brice A., Meco G., van Duijn C.M., Oostra B.A., Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 24.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X., Zhang Z. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J Clin Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas K.J., McCoy M.K., Blackinton J., Beilina A., van der Brug M., Sandebring A., Miller D., Maric D., Cedazo-Minguez A., Cookson M.R. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Y., SunX, Gu J., Yu C., Wen Y., Gao Y., Xia Q., Kong X. Deficiency of DJ-1 ameliorates liver fibrosis through inhibition of hepatic ROS production and inflammation. Int J Biol Sci. 2016;12:1225–1235. doi: 10.7150/ijbs.15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu R., Liu X.M., Sun J.G., Chen H., Ma J., Dong M., Peng S., Wang J.Q., Ding J.Q., Li D.H., Speakman J.R., Ning G., Jin W., Yuan Z. DJ-1 maintains energy and glucose homeostasis by regulating the function of brown adipose tissue. Cell Discov. 2017;3:16054. doi: 10.1038/celldisc.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M., Wu H., Li M., Wen Y., Yu C., Xia L., Xia Q., Kong X. DJ-1 deficiency protects hepatic steatosis by enhancing fatty acid oxidation in mice. Int J Biol Sci. 2018;14:1892–1900. doi: 10.7150/ijbs.28620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu B., Wang J., Yu Y., Zhen C., Gu J., Liu W., Wen Y., Chen L., Gao Y., Xia Q., Kong X. DJ-1 promotes development of DEN-induced hepatocellular carcinoma and proliferation of liver cancer cells. Oncotarget. 2017;8:8499–8511. doi: 10.18632/oncotarget.14293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L., Luo M., Sun X., Qin J., Yu C., Wen Y., Zhang Q., Gu J., Xia Q., Kong X. DJ-1 deficiency attenuates expansion of liver progenitor cells through modulating the inflammatory and fibrogenic niches. Cell Death Dis. 2016;7:e2257. doi: 10.1038/cddis.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Golen R.F., Reiniers M.J., Olthof P.B., van Gulik T.M., Heger M. Sterile inflammation in hepatic ischemia/reperfusion injury: present concepts and potential therapeutics. J Gastroenterol Hepatol. 2013;28:394–400. doi: 10.1111/jgh.12072. [DOI] [PubMed] [Google Scholar]
- 32.Loi P., Yuan Q., Torres D., Delbauve S., Laute M.A., Lalmand M.C., Petein M., Goriely S., Goldman M., Flamand V. Interferon regulatory factor 3 deficiency leads to interleukin-17-mediated liver ischemia-reperfusion injury. Hepatology. 2013;57:351–361. doi: 10.1002/hep.26022. [DOI] [PubMed] [Google Scholar]
- 33.Wang P.X., Zhang R., Huang L., Zhu L.H., Jiang D.S., Chen H.Z., Zhang Y., Tian S., Zhang X.F., Zhang X.D., Liu D.P., Li H. Interferon regulatory factor 9 is a key mediator of hepatic ischemia/reperfusion injury. J Hepatol. 2015;62:111–120. doi: 10.1016/j.jhep.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Huang H., Nace G.W., McDonald K.A., Tai S., Klune J.R., Rosborough B.R., Ding Q., Loughran P., Zhu X., Beer-Stolz D., Chang E.B., Billiar T., Tsung A. Hepatocyte-specific high-mobility group box 1 deletion worsens the injury in liver ischemia/reperfusion: a role for intracellular high-mobility group box 1 in cellular protection. Hepatology. 2014;59:1984–1997. doi: 10.1002/hep.26976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cursio R., Colosetti P., Gugenheim J. Autophagy and liver ischemia-reperfusion injury. Biomed Res Int. 2015;2015:417590. doi: 10.1155/2015/417590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu D., Chen L., Chen X., Wen Y., Yu C., Yao J., Wu H., Wang X., Xia Q., Kong X. The triterpenoid CDDO-imidazolide ameliorates mouse liver ischemia-reperfusion injury through activating the Nrf2/HO-1 pathway enhanced autophagy. Cell Death Dis. 2017;8:e2983. doi: 10.1038/cddis.2017.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marx V. Autophagy: eat thyself, sustain thyself. Nature Methods. 2015;12:1121–1125. doi: 10.1038/nmeth.3661. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X., Yan H., Yuan Y., Gao J., Shen Z., Cheng Y., Shen Y., Wang R.R., Wang X., Hu W.W., Wang G., Chen Z. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013;9:1321–1333. doi: 10.4161/auto.25132. [DOI] [PubMed] [Google Scholar]
- 40.Tang C., Han H., Yan M., Zhu S., Liu J., Liu Z., He L., Tan J., Liu Y., Liu H., Sun L., Duan S., Peng Y., Liu F., Yin X.M., Zhang Z., Dong Z. PINK1-PRKN/PARK2 pathway of mitophagy is activated to protect against renal ischemia-reperfusion injury. Autophagy. 2018;14:880–897. doi: 10.1080/15548627.2017.1405880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W., Chen C., Wang J., Liu L., He Y., Chen Q. Mitophagy in cardiomyocytes and in platelets: a major mechanism of cardioprotection against ischemia/reperfusion injury. Physiology (Bethesda) 2018;33:86–98. doi: 10.1152/physiol.00030.2017. [DOI] [PubMed] [Google Scholar]
- 42.Joselin A.P., Hewitt S.J., Callaghan S.M., Kim R.H., Chung Y.H., Mak T.W., Shen J., Slack R.S., Park D.S. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- 43.Xu M., Chen L., Li J., Wu H., Xia Q., Kong X. Emerging roles of DJ-1 in liver diseases through regulation of oxidative stress and immune response. Liver Research. 2018 [Google Scholar]
- 44.Wang J.H., Ahn I.S., Fischer T.D., Byeon J.I., Dunn W.A., Jr., Behrns K.E., Leeuwenburgh C., Kim J.S. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. 2011;141:2188–2199 e2186. doi: 10.1053/j.gastro.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J.S., Nitta T., Mohuczy D., O’Malley K.A., Moldawer L.L., Dunn W.A., Jr., Behrns K.E. Impaired autophagy: a mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725–1736. doi: 10.1002/hep.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khader A., Yang W.L., Godwin A., Prince J.M., Nicastro J.M., Coppa G.F., Wang P. Sirtuin 1 stimulation attenuates ischemic liver injury and enhances mitochondrial recovery and autophagy. Crit Care Med. 2016;44:e651–e663. doi: 10.1097/CCM.0000000000001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li M., Xu M., Li J., Chen L., Xu D., Tong Y., Zhang J., Wu H., Kong X., Xia Q. Alda-1 ameliorates liver ischemia-reperfusion injury by activating aldehyde dehydrogenase 2 and enhancing autophagy in mice. J Immunol Res. 2018;2018:9807139. doi: 10.1155/2018/9807139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nash Y., Schmukler E., Trudler D., Pinkas-Kramarski R., Frenkel D. DJ-1 deficiency impairs autophagy and reduces alpha-synuclein phagocytosis by microglia. J Neurochem. 2017;143:584–594. doi: 10.1111/jnc.14222. [DOI] [PubMed] [Google Scholar]
- 49.Xue R., Jiang J., Dong B., Tan W., Sun Y., Zhao J., Chen Y., Dong Y., Liu C. DJ-1 activates autophagy in the repression of cardiac hypertrophy. Arch Biochem Biophys. 2017;633:124–132. doi: 10.1016/j.abb.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Zhou B., Lei S., Xue R., Leng Y., Xia Z., Xia Z.Y. DJ-1 overexpression restores ischaemic post-conditioning-mediated cardioprotection in diabetic rats: role of autophagy. Clin Sci (Lond) 2017;131:1161–1178. doi: 10.1042/CS20170052. [DOI] [PubMed] [Google Scholar]
- 51.Ren H., Fu K., Mu C., Li B., Wang D., Wang G. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett. 2010;297:101–108. doi: 10.1016/j.canlet.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Jaramillo-Gomez J., Nino A., Arboleda H., Arboleda G. Overexpression of DJ-1 protects against C2-ceramide-induced neuronal death through activation of the PI3K/AKT pathway and inhibition of autophagy. Neurosci Lett. 2015;603:71–76. doi: 10.1016/j.neulet.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 53.Qin X., Lu A., Ke M., Zhu W., Ye X., Wang G., Weng G. DJ-1 inhibits autophagy activity of prostate cancer cells by repressing JNK-Bcl2-Beclin1 signaling. Cell Biol Int. 2020;44:937–946. doi: 10.1002/cbin.11290. [DOI] [PubMed] [Google Scholar]
- 54.Wen Y., Feng D., Wu H., Liu W., Li H., Wang F., Xia Q., Gao W.Q., Kong X. Defective initiation of liver regeneration in osteopontin-deficient mice after partial hepatectomy due to insufficient activation of IL-6/Stat3 pathway. Int J Biol Sci. 2015;11:1236–1247. doi: 10.7150/ijbs.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Jonge J., Kurian S., Shaked A., Reddy K.R., Hancock W., Salomon D.R., Olthoff K.M. Unique early gene expression patterns in human adult-to-adult living donor liver grafts compared to deceased donor grafts. Am J Transplant Surgeons. 2009;9:758–772. doi: 10.1111/j.1600-6143.2009.02557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]