Abstract
Genomic disorders result from rearrangement of the human genome. Most genomic disorders are caused by copy number variants (CNV), deletions or duplications of several hundred kilobases. Many CNV loci are associated with autism, schizophrenia, and most commonly, intellectual disability (ID). However, there is little comparison of cognitive ability measures across these CNV disorders. This study aims to understand whether existing data can be leveraged for a cross-comparison of cognitive ability among multiple CNV. We found there is a lack of harmonization among assessment instruments and little standardization for reporting summary data across studies. Despite these limitations, we identified a differential impact of CNV loci on cognitive ability. Our data suggest that future cross-comparisons of CNV disorders will reveal meaningful differences across the phenotypic spectrum, especially if standardized phenotypic assessment is achieved.
Introduction
Genomic disorders, resulting from copy-number variants (CNV) or structural rearrangements of the genome1, 2, account for a significant amount of human morbidity and mortality.3, 4 CNVs include copy number loss (deletions) and copy number gains (duplications).5–7 Phenotypes vary between genomic disorders but often include multi-system impairments, such as cardiac malformations, characteristic craniofacial abnormalities, immune system impairment, metabolic dysregulation, and neurodevelopmental and psychiatric disorders.8–11 There is an increase in the overall burden of large, rare CNV in individuals with these phenotypes, and large, rare CNV are more likely to contain genes with functional roles in neurodevelopmental pathways.12–14 Rare CNV have been implicated in a variety of diseases, and are found to be increased in more severe developmental phenotypes associated with congenital abnormalities.8, 15
A substantial number of CNV are associated with neurodevelopmental and psychiatric phenotypes. For example, there are at least 8 CNV with genome-wide significance for association with schizophrenia, including 1q21.1, 2p16.3, 3q29, 7q11.23, 15q13.3, distal 16p11.2, proximal 16p11.2, and 22q11.2.16–18 Phenotypic heterogeneity has been observed among these CNV; in addition to schizophrenia, each locus may also be associated with autism, epilepsy, and additional neurodevelopmental sequelae.19–21 Many CNV loci cause a degree of cognitive impairment, ranging from mild to severe.22, 23 Few studies have compared the impact on cognitive ability across CNV loci.
There has been a consistent observation that while CNV occur in different parts of the genome and contain non-overlapping genes, they nevertheless are associated with an overlapping set of phenotypes.24, 25 The hypothesis of convergent biology has been advanced as a way to explain these observations. On a genetic level, convergence can reflect a variety of degrees of similarity, ranging from evolutionary changes in the same genes but at different sites, to changes in the same pathways but within different genes.26 Although different genes lie within these CNV loci, they may converge on the same pathways, resulting in shared phenotypes. To study the hypothesis of convergent biology, a first needed step is to have comprehensive, harmonized phenotyping data across a range of CNV, to enable cross-comparison.27
For many CNV, their impact on cognitive function is well-studied. In this review, we sought to ask whether this copious amount of existing data could be leveraged for cross-comparison of cognitive phenotypes. We conducted a thorough literature search across 11 CNV loci, harvesting data from 156 studies. We compared a) the instruments used in these studies and b) estimates of cognitive ability across CNV loci. Our study aims to draw a systematic comparison of cognitive measures across different genomic disorders, and document any barriers that may exist in using existing data to conduct cross-disorder comparisons of cognitive ability.
Methods
Literature Search Strategy
We selected a set of 11 CNVs known to be associated with intellectual disability and conducted a systematic literature search to identify eligible papers with data for analysis. These included 3q29 deletion, 7q11.23 deletion, 15q11.2 deletion (Angelman’s Syndrome (AS)), 15q11.2 deletion (Prader-Willi Syndrome (PWS)), 16p11.2 deletion and duplication, 17p11.2 deletion, and 22q11.2 deletion. (Table 1).
Table 1.
Number of studies and study subjects among CNVs
| CNV | Number of Studies | Total Number of Study Subjects |
|---|---|---|
| 1q21.1 deletion | - | - |
| 2p15p16.1 deletion | - | - |
| 3q29 deletion | 1 | 32 |
| 7q11.23 deletion | 36 | 1,849 |
| 9q34 deletion | - | - |
| 15q11.2 deletion (AS)a | 2 | 45 |
| 15q11.2 deletion (PWS)b | 31 | 1,836 |
| 16p11.2 deletion (proximal) | 6 | 354 |
| 16p11.2 duplication (proximal) | 4 | 198 |
| 17p11.2 deletion | 3 | 134 |
| 22q11.2 deletion | 73 | 4,732 |
Angelman’s Syndrome
Prader-Willi Syndrome
Papers were identified in October 2020 via a PubMed literature search. We identified relevant studies with the search terms “CNV name and cognitive ability”, or “CNV name and IQ”. If a genomic disorder is well-known by an alternate name, we repeated all searches using the alternate name (ie: Williams Syndrome for 7q11.23 deletion). See supplemental table 2 for search details.
Study Selection
Papers matching search criteria were manually reviewed for eligibility. Inclusion criteria were: data measures on human subjects; reporting of cognitive data (Full-Scale IQ (FSIQ), verbal IQ (VIQ), or performance or non-verbal IQ (PIQ or NVIQ)); clear identification of the instrument used for measures of cognitive ability; a sample size > 20; and either open access or reasonable access through a typical University library. When multiple papers reported data using nested, overlapping sample sources, only the paper with the largest sample size was retained. See supplemental table 2 for literature search results.
Data Extraction and Management
For papers that met eligibility criteria, data were extracted from the paper and maintained in a local database. Extracted data included paper title, authors, year published, sample size, assessment instruments, and cognitive ability scores. In this primary literature, data summaries were reported in variable ways across studies. Measures of central tendency (e.g., mean or median) and variance (e.g., standard error or standard deviation) were extracted as reported. Data visualization was performed in R.
Computing Average IQ Scores
For each CNV, we calculated the overall average FSIQ, VIQ, and PIQ across all studies for that CNV, weighting each study by its sample size.
Cognitive Assessment
Measures of cognitive ability, such as IQ tests, have been shrouded in controversy, mainly due to accusations of cultural bias.28 However, they are still considered the most effective form of assessing cognitive ability, and continue to be widely-used.29 Although we compared cognitive ability assessed by different instruments in this study, it is important to note that all instruments are normed on a standard scale (mean = 100, standard deviation (SD) = 15). This means that for almost any of the commonly used instruments, a score that falls two SD below the mean (IQ < 70) indicates the presence of intellectual disability. Additionally, these scores can be used in multiple frameworks. We can use standard scores as a quantitative, dimensional measure to compare cognitive ability across CNVs. Alternatively, we can group people based on whether or not they have intellectual disability. Some common instruments include:
Wechsler Intelligence Scale –
Measures cognitive ability in verbal and non-verbal dimensions. It includes 11 subscales (6 verbal and 5 non-verbal) including general information, numeric memory, vocabulary, computing, comprehension and similarities, verbal intelligence and image completion, image adjustment, cube design, and component insertion30 Different versions of the test may be administered based on the age of the test taker, including Wechsler Preschool and Primary Scale of Intelligence (WPPSI, ages 3–5 years), Wechsler Intelligence Scale for Children (WISC, 6–15 years), and Wechsler Adult Intelligence Scale (WAIS, 16 years and older).31
Kaufman Brief Intelligence Scale –
Assesses verbal and non-verbal ability in people ages 4–90 years. Verbal scale measures verbal knowledge and riddles, while the nonverbal scale measures fluid intelligence (ability to think logically and solve novel problems).32
Differential Ability Scales –
Assesses verbal and non-verbal ability in preschool and school-age children, ages 2.5–18 years. Verbal and non-verbal domains have different subtests depending on the age of test taker. For example, the non-verbal domain for older ages (3.5–7 years) includes matrices and copying, as well as pattern construction, while the younger ages (2.5–3.5 years) are tested on picture similarities.33
Bayley Scales of Infant and Toddler Development –
an assessment for infants aged 1–42 months across three domains: cognitive, language, and motor skills.34
Leiter International Performance Scale –
an assessment for children and adolescents aged 2–20 years.35 Composed of two batteries: attention and memory (AM) and visualization and reasoning (VR). The AM battery is used for the assessment of attention and memory difficulties, while the VR battery is used to measure general intelligence.36
Ammons Quick Test –
an assessment of verbal intelligence using picture identification in children and adults ages 2–90 years.37
Reynolds Intellectual Screening Test –
an assessment of verbal and non-verbal intelligence in people aged 3–99 years.38
Results
Using our search criteria, we identified 156 eligible studies that reported cognitive ability scores. This included 1 paper on 3q29 deletion39, 36 papers on 7q11.23 deletion32, 40–74, 2 papers on 15q11.2 deletion (AS)75, 76, 31 papers on 15q11.2 deletion (PWS)56, 77–106, 6 papers on 16p11.2 deletion (proximal)107–112, 4 papers on 16p11.2 duplication (proximal)107, 109, 110, 112, 3 papers on 17p11.2 deletion113–115, and 73 papers on 22q11.2 deletion syndrome.74, 80, 116–186 Studies reporting cognitive scores for 1q21.1 deletion, 2p15p16.1 deletion, and 9q34 deletion were not identified. The total number of subjects evaluated for a given CNV across all publications for that CNV was variable and ranged from 32 subjects for the 3q29 deletion to 4,732 study subjects for the 22q11.2 deletion. For other CNV included in this review, total sample sizes were: 1,849 (7q11.23 deletion); 45 (15q11.2 deletion, AS); 1,836 (15q11.2 deletion, PWS); 354 (16p11.2 deletion (proximal)); 198 (16p11.2 duplication (proximal)); and 134 (17p11.2 deletion) (Table 1).
We observed significant variability in the assessment instruments used to measure cognitive ability. The Wechsler Intelligence scales (WAIS, WISC, WASI, WPPSI) are the most widely used across studies (Fig. 1, Table S1). However, the Kaufman Brief Intelligence Test has also been administered in a substantial number of study subjects, particularly for the 7q11.23 deletion and 15q11.2 deletion (PWS). Some CNV studies assessed IQ using multiple cognitive assessment instruments, but failed to specify how many subjects were evaluated with each instrument. Across the 8 CNV included in this review, only 2 (15q11.2 deletion (AS) and 17p11.2 deletion) reported cognitive scores with a single instrument (Fig. 1). The variety of instruments used to measure cognitive ability demonstrates the challenges of retrospective data comparison, both within and across disorders.
Figure 1. Cognitive assessment instrument used across CNV studies.
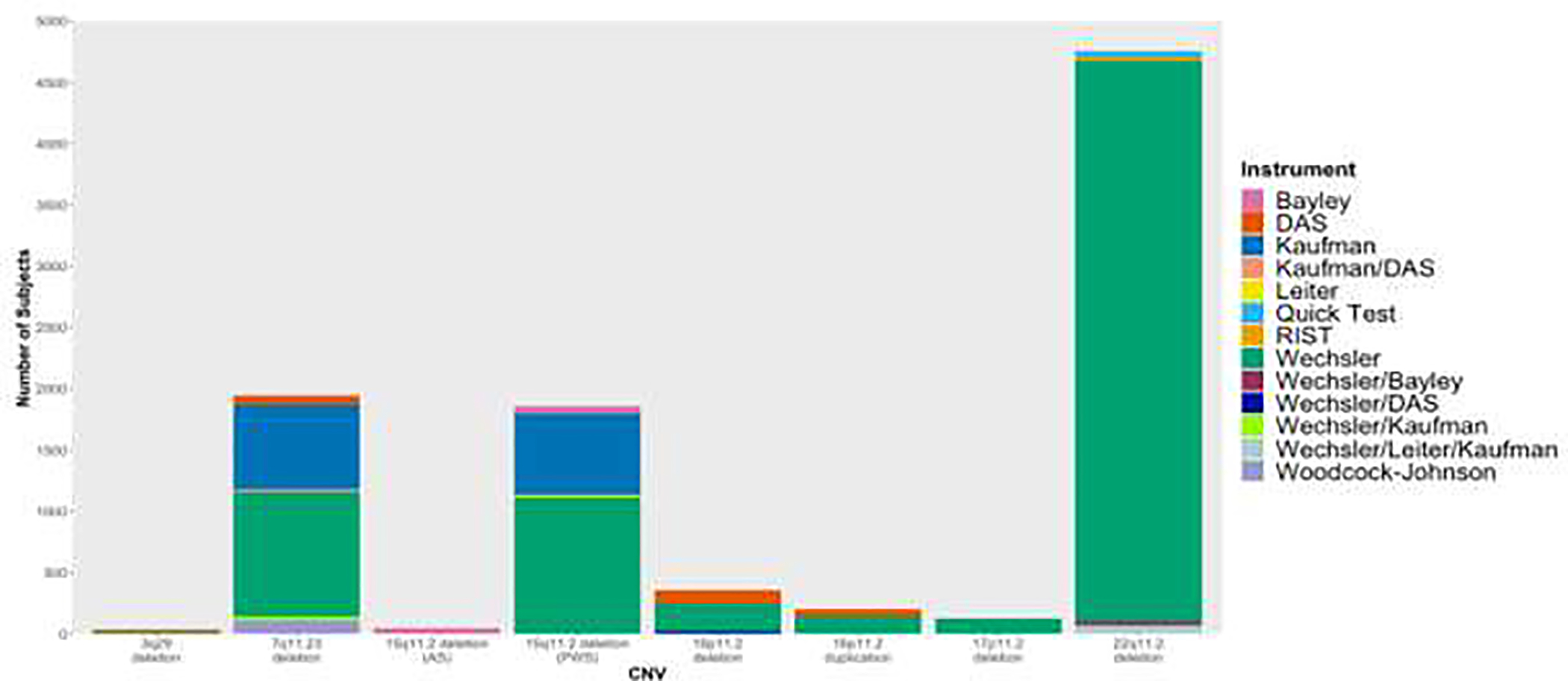
Bars are proportional to number of study subjects that have been evaluated across studies with corresponding instrument. Studies that used any form of the Wechsler Intelligence Scale (WAIS, WASI, WISC, WPPSI) were grouped together under “Wechsler.” Studies that used multiple instruments but did not specify how many subjects were administered each instrument were grouped together with all instruments specified.
aDifferential ability scales
bReynolds Intellectual Screening Test
Figure 2 shows a comparison of FSIQ scores across the CNV included in this study. Within a single CNV, there can be wide variation across studies. For example, FSIQ mean estimates for the 22q11.2 deletion range from a low of 53178 to a high of 88.174 Careful examination of the primary papers does not reveal a clear reason for these differential estimates. Despite this variability, some conclusions can be drawn across CNV. 17p11.2 deletion subjects appear to have the lowest FSIQ estimates among the CNV included in this study, and 16p11.2 deletion and 16p11.2 duplication subjects appear to have the highest. All cognitive data estimates included in this review, across all studies, report FSIQ scores below the population mean IQ of 100 (Fig. 2). As the studies within each CNV are ordered chronologically (from earliest to most recent), we did not notice any temporal changes in FSIQ within the CNV.
Figure 2. FSIQ measures across studies for 8 CNV.
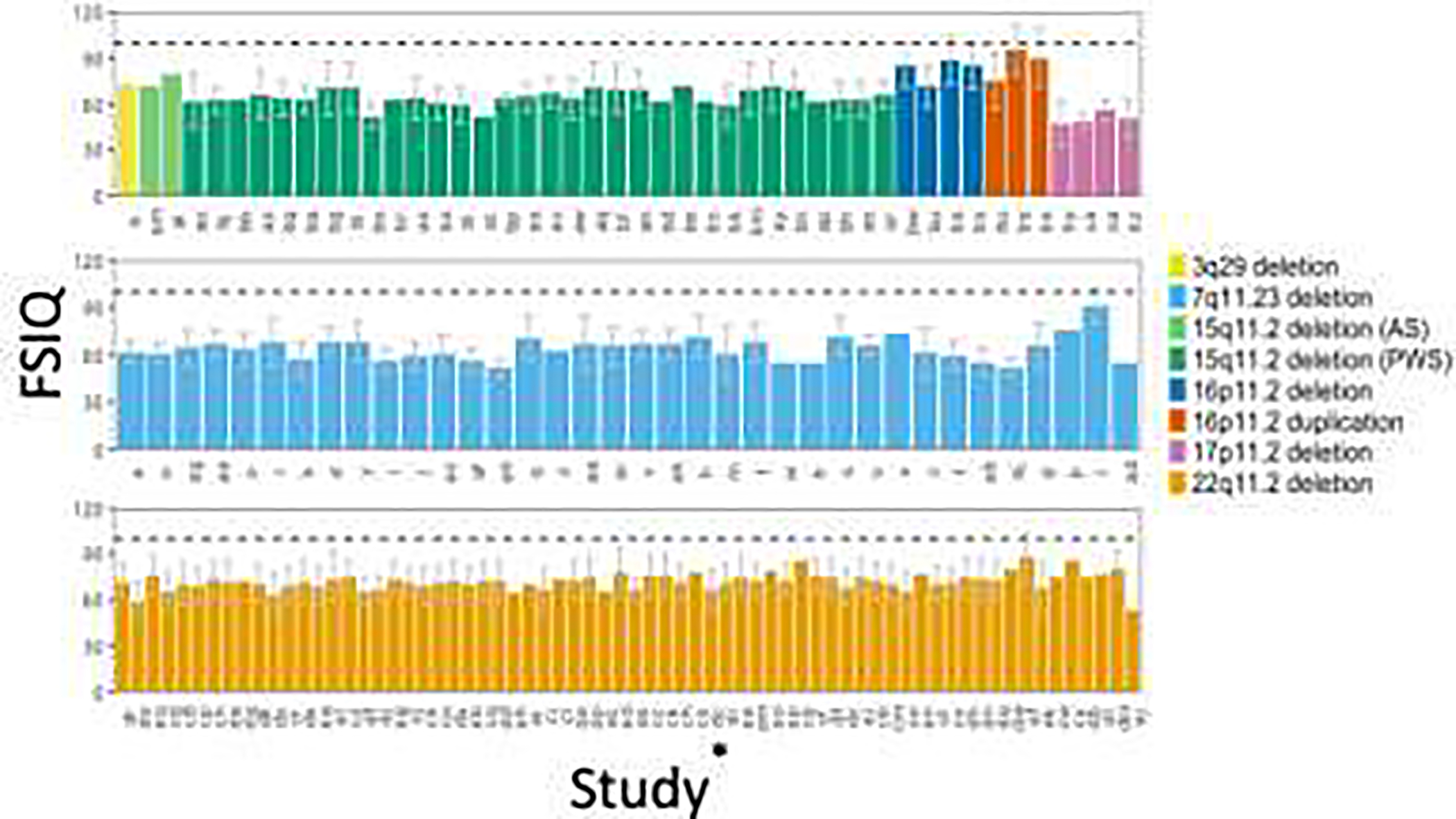
Study indicator on x-axis corresponds to study ID indicator in Table S1. If more than one study for a CNV, studies were listed chronologically from earliest to most recent, or left to right. Dashed black line indicates population mean IQ of 100.
*See Table S1 for study indicators for each study
Though we studied 8 CNV, only 6 contained studies that reported VIQ and PIQ scores (Fig. 3, 4). Inter and intra-CNV VIQ and PIQ scores across studies also showed consistency, but largely mirror the results for FSIQ. 17p11.2 deletion appears to have the lowest PIQ and VIQ, while 16p11.2 deletion and duplication subjects appear to have the highest (Fig. S1, S2). We also did not notice any temporal changes in VIQ or PIQ within CNV across the studies (Fig. S1, S2).
Figure 3. Weighted mean FSIQ, VIQ, and PIQ across CNV studies.
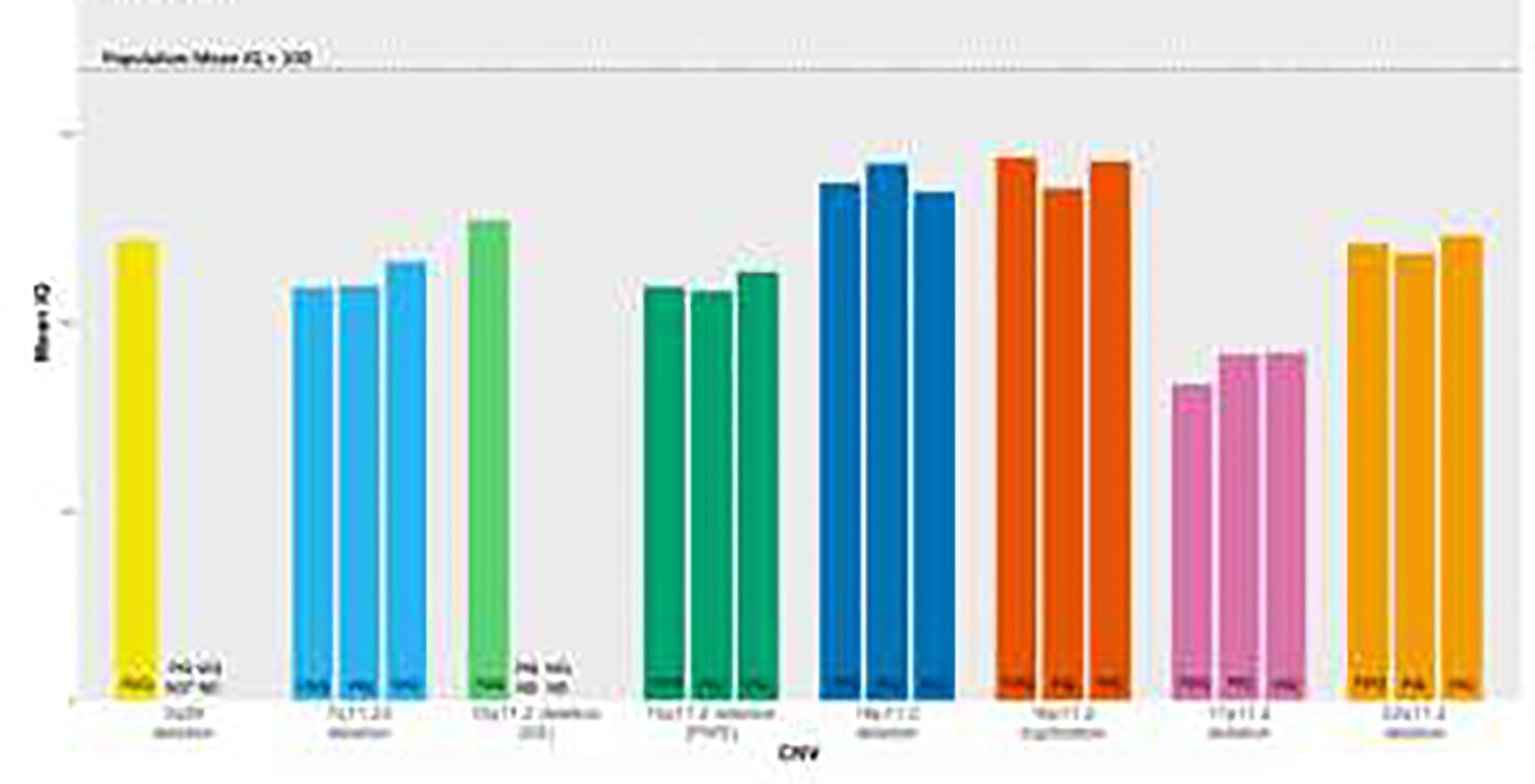
3q29 deletion and 15q11.2 deletion (AS) did not contain studies that reported sample VIQ or PIQ scores. See methods for computation of weighted mean IQ scores.
*Not Determined
Weighted mean FSIQ, VIQ, and PIQ scores across CNV also showed consistency, with all scores falling below population mean (3q29 deletion = 73, 7q11.23 deletion = 66, 15q11.2 deletion (AS) = 76, 15q11.2 (PWS) = 66, 16p11.2 deletion = 82, 16p11.2 duplication = 86, 17p11.2 deletion = 50, 22q11.2 deletion = 71). Mean VIQ scores also fell below population mean (7q11.23 deletion = 70, 15q11.2 deletion (PWS) = 68, 16p11.2 deletion = 81, 16p11.2 duplication = 86, 17p11.2 deletion = 55, 22q11.2 deletion = 74). Mean PIQ scores followed a similar trend (7q11.23 deletion = 66, 15q11.2 deletion (PWS) = 65, 16p11.2 deletion = 85, 16p11.2 duplication = 81, 17p11.2 deletion = 55, 22q11.2 deletion = 71) (Fig. 3).
Discussion
In this study, we sought to compare the impact of a diversity of CNV loci on cognitive ability, using existing data culled from the literature. We document significant challenges in this endeavor. One such challenge is the range of instruments used, with little harmonization within and across loci, as shown in Figure 1. Because many instruments are normed and scored on the same scale, this lack of harmonization presents a minor barrier to cross-comparison. A second challenge is the lack of standardization in data reporting; this is a more significant barrier that obscures our ability to conduct cross-disorder comparisons. Despite these barriers, our analysis reveals a differential impact of CNV loci on cognitive ability.
Our comparisons revealed that the 16p11.2 deletion and 16p11.2 duplication tend to have the smallest effect on FSIQ, VIQ, and PIQ, compared to the other CNV loci included in this analysis (Fig. 2, S1, S2). This was true when examining measures at the individual study level, as well as weighted estimates (Fig. 3). In contrast, we found the 17p11.2 deletion is estimated to depress cognitive ability scores to a great extent than any of the other CNV included in this study. This is consistent with previously reported literature, as 17p11.2 deletion is known to be associated with lower cognitive ability scores.113
The lack of harmonization in cognitive assessment instruments makes a cross comparison difficult, due to instruments using different scales and methods to develop an IQ score.187 CNV cross-comparison would be facilitated if the same instruments were used to measure cognitive ability. Minimally, the instruments used must be explicitly defined. There were multiple studies that we came across in our literature search that were excluded because the cognitive measurement instruments were not identified. It is also important that all studies report the same measures of central tendency and variation when measuring cognitive ability, to allow for a more uniform comparison. An ideal solution is for investigators to make the raw data available upon publication, either in the supplement, or through a public-facing database like NDAR (https://nda.nih.gov/about.html). By making this raw data public, it would allow future studies to download the data for their own analysis, thereby aiding in reproducibility.188
This is one of the first cross-comparisons of cognitive ability in CNV disorders, a paradigm that has previously been suggested as important for study.27, 189 The overlap of these neuropsychiatric phenotypes, within and between these CNV, warrants a cross-disorder analysis, as it may identify biologically defined subcategories of these neuropsychiatric phenotypes.27 Additionally, a cross-comparison like this allows for a global overview of cognitive ability among CNV; despite these CNV originating on different loci, they all share a common phenotype of intellectual disability. Thus, a cross-comparison highlights the fact that thousands of CNV risk factors converge on a limited number of diagnoses190,and that these risk factors overlap with loci that contribute to these disorders.17 Additionally, this type of cross comparison shows evidence of genotype-specific effects on severity of intellectual disability, allowing us to draw a genotype-phenotype association.191
The limitations of this study include, as mentioned earlier, a lack of harmonization among assessment instruments, as well as different measures of central tendency and variation being reported. Ascertainment bias is also a major limitation of this study; carriers of incompletely penetrant CNV may appear phenotypically normal, and hence their cognitive performance may not be captured in the primary literature.192 As a result, only people with severe cognitive and developmental delays are more likely to be referred for a clinical genetics test.191 This would skew our results, as the IQ scores that are reported in the primary papers may over-represent the most severely-affected individuals.
This study presents a promising direction forward into understanding cognitive ability among people with CNV, as it reveals a differential impact of CNV loci on cognitive ability. This study also allows for identification of subcategories of these neuropsychiatric phenotypes, and can be applied to other shared phenotypes aside from cognitive ability. Future studies should ensure consistent, harmonized data collection efforts are being conducted. Additionally, to control for ascertainment bias, studies should conduct population-level analyses of large samples of individuals, such as birth cohorts or health system registries. This will hopefully allow for a more uniform cross-comparison of cognitive ability across CNV to be performed, which will ultimately allow us to better understand the biology and mechanisms of these disorders. This type of study also plays a role into preventative medicine, as it allows us to inform clinicians what to expect when treating patients with these CNV, with the goal of improving the lives of those living with these disorders.
Supplementary Material
Acknowledgements
Funding: This work was supported by the National Institutes of Health (T32 GM0008490–27. R01 MH110701).
Footnotes
Declarations of interest:
none.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eichler E, Copy Number Variation and Human Disease. Nature, 2008. 1. [Google Scholar]
- 2.Harel T and Lupski JR, Genomic disorders 20 years on-mechanisms for clinical manifestations. Clin Genet, 2018. 93(3): p. 439–449. [DOI] [PubMed] [Google Scholar]
- 3.Crawford K, et al. , Medical consequences of pathogenic CNVs in adults: analysis of the UK Biobank. J Med Genet, 2019. 56(3): p. 131–138. [DOI] [PubMed] [Google Scholar]
- 4.Nygaard M, et al. , Copy number variation associates with mortality in long-lived individuals: a genome-wide assessment. Aging Cell, 2016. 15(1): p. 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall CR, et al. , Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet, 2008. 82(2): p. 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin CL, Kirkpatrick BE, and Ledbetter DH, Copy number variants, aneuploidies, and human disease. Clin Perinatol, 2015. 42(2): p. 227–42, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe K, et al. , Delineating the psychiatric and behavioral phenotype of recurrent 2q13 deletions and duplications. Am J Med Genet B Neuropsychiatr Genet, 2018. 177(4): p. 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper GM, et al. , A copy number variation morbidity map of developmental delay. Nat Genet, 2011. 43(9): p. 838–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin CL, et al. , Identification of Neuropsychiatric Copy Number Variants in a Health Care System Population. JAMA Psychiatry, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaikh TH, Copy Number Variation Disorders. Curr Genet Med Rep, 2017. 5(4): p. 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yim SH, et al. , Clinical implications of copy number variations in autoimmune disorders. Korean J Intern Med, 2015. 30(3): p. 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girirajan S, et al. , Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet, 2011. 7(11): p. e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kendall KM, et al. , Association of Rare Copy Number Variants With Risk of Depression. JAMA Psychiatry, 2019. 76(8): p. 818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrow EM, Genomic copy number variation in disorders of cognitive development. J Am Acad Child Adolesc Psychiatry, 2010. 49(11): p. 1091–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coe BP, Girirajan S, and Eichler EE, The genetic variability and commonality of neurodevelopmental disease. Am J Med Genet C Semin Med Genet, 2012. 160C(2): p. 118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirov G, et al. , The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry, 2014. 75(5): p. 378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall CR, et al. , Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet, 2017. 49(1): p. 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan PF, Schizophrenia and the dynamic genome. Genome Med, 2017. 9(1): p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn K, et al. , High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry, 2014. 19(5): p. 568–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders SJ, et al. , Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron, 2011. 70(5): p. 863–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinawi M, et al. , Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet, 2010. 47(5): p. 332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guffanti G, et al. , Increased CNV-region deletions in mild cognitive impairment (MCI) and Alzheimer’s disease (AD) subjects in the ADNI sample. Genomics, 2013. 102(2): p. 112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pizzo L, et al. , Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med, 2019. 21(4): p. 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doelken SC, et al. , Phenotypic overlap in the contribution of individual genes to CNV pathogenicity revealed by cross-species computational analysis of single-gene mutations in humans, mice and zebrafish. Dis Model Mech, 2013. 6(2): p. 358–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vinas-Jornet M, et al. , High Incidence of Copy Number Variants in Adults with Intellectual Disability and Co-morbid Psychiatric Disorders. Behav Genet, 2018. 48(4): p. 323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sackton TB and Clark N, Convergent evolution in the genomics era: new insights and directions. Philos Trans R Soc Lond B Biol Sci, 2019. 374(1777): p. 20190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanders SJ, et al. , A framework for the investigation of rare genetic disorders in neuropsychiatry. Nat Med, 2019. 25(10): p. 1477–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozano-Ruiz A, et al. , Cultural Bias in Intelligence Assessment Using a Culture-Free Test in Moroccan Children. Arch Clin Neuropsychol, 2021. [DOI] [PubMed] [Google Scholar]
- 29.Sansone SM, et al. , Improving IQ measurement in intellectual disabilities using true deviation from population norms. J Neurodev Disord, 2014. 6(1): p. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaman R, et al. , Estimation of Mean Intelligence Quotient with Wechsler Scale in Iran: Systematic Review and Meta-Analysis. Int J Prev Med, 2019. 10: p. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiridigliozzi GA, et al. , Cognitive and academic outcomes in long-term survivors of infantile-onset Pompe disease: A longitudinal follow-up. Mol Genet Metab, 2017. 121(2): p. 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pitts CH and Mervis CB, Performance on the Kaufman Brief Intelligence Test-2 by Children With Williams Syndrome. Am J Intellect Dev Disabil, 2016. 121(1): p. 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farmer C, Golden C, and Thurm A, Concurrent validity of the differential ability scales, second edition with the Mullen Scales of Early Learning in young children with and without neurodevelopmental disorders. Child Neuropsychol, 2016. 22(5): p. 556–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ranjitkar S, et al. , Acceptability and Reliability of the Bayley Scales of Infant and Toddler Development-III Among Children in Bhaktapur, Nepal. Front Psychol, 2018. 9: p. 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Giacomo A, et al. , Can PEP-3 Provide a Cognitive Profile in Children with ASD? A Comparison Between the Developmental Ages of PEP-3 and IQ of Leiter-R. J Appl Res Intellect Disabil, 2016. 29(6): p. 566–573. [DOI] [PubMed] [Google Scholar]
- 36.Scattone D, Raggio DJ, and May W, Brief report: concurrent validity of the Leiter-R and KBIT-2 scales of nonverbal intelligence for children with autism and language impairments. J Autism Dev Disord, 2012. 42(11): p. 2486–90. [DOI] [PubMed] [Google Scholar]
- 37.Zagar RJ, et al. , Ammons quick test validity among randomly selected referrals. Psychol Rep, 2013. 113(3): p. 823–54. [DOI] [PubMed] [Google Scholar]
- 38.Gygi JT, et al. , Measurement Invariance and Latent Mean Differences in the Reynolds Intellectual Assessment Scales (RIAS): Does the German Version of the RIAS Allow a Valid Assessment of Individuals with a Migration Background? PLoS One, 2016. 11(11): p. e0166533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez Russo R, et al. , Deep phenotyping in 3q29 deletion syndrome: recommendations for clinical care. Genet Med, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binelli C, et al. , Facial emotion processing in patients with social anxiety disorder and Williams-Beuren syndrome: an fMRI study. J Psychiatry Neurosci, 2016. 41(3): p. 182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brawn G, et al. , Functional basic reading skills in Williams syndrome. Dev Neuropsychol, 2018. 43(5): p. 454–477. [DOI] [PubMed] [Google Scholar]
- 42.Chiang MC, et al. , 3D pattern of brain abnormalities in Williams syndrome visualized using tensor-based morphometry. Neuroimage, 2007. 36(4): p. 1096–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies RW, et al. , Using common genetic variation to examine phenotypic expression and risk prediction in 22q11.2 deletion syndrome. Nat Med, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Del Cole CG, et al. , Adolescent adaptive behavior profiles in Williams-Beuren syndrome, Down syndrome, and autism spectrum disorder. Child Adolesc Psychiatry Ment Health, 2017. 11: p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dror C, Sinai A, and Gothelf D, Medical, Cognitive, and Psychiatric Characteristics in a Large Israeli Cohort of Individuals with Williams Syndrome. Isr Med Assoc J, 2018. 20(6): p. 373–378. [PubMed] [Google Scholar]
- 46.Dunning BA, Martens MA, and Jungers MK, Music lessons are associated with increased verbal memory in individuals with Williams syndrome. Res Dev Disabil, 2015. 36C: p. 565–578. [DOI] [PubMed] [Google Scholar]
- 47.Elison S, Stinton C, and Howlin P, Health and social outcomes in adults with Williams syndrome: findings from cross-sectional and longitudinal cohorts. Res Dev Disabil, 2010. 31(2): p. 587–99. [DOI] [PubMed] [Google Scholar]
- 48.Farran EK, et al. , How do individuals with Williams syndrome learn a route in a real-world environment? Dev Sci, 2010. 13(3): p. 454–468. [DOI] [PubMed] [Google Scholar]
- 49.Gerard-Desplanches A, et al. , Laterality in persons with intellectual disability II. Hand, foot, ear, and eye laterality in persons with Trisomy 21 and Williams-Beuren syndrome. Dev Psychobiol, 2006. 48(6): p. 482–91. [DOI] [PubMed] [Google Scholar]
- 50.Gregory MD, et al. , Williams syndrome hemideletion and LIMK1 variation both affect dorsal stream functional connectivity. Brain, 2019. 142(12): p. 3963–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hocking DR, Reeve J, and Porter MA, Characterising the Profile of Everyday Executive Functioning and Relation to IQ in Adults with Williams Syndrome: Is the BRIEF Adult Version a Valid Rating Scale? PLoS One, 2015. 10(9): p. e0137628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Howlin P, Davies M, and Udwin O, Cognitive functioning in adults with Williams syndrome. J Child Psychol Psychiatry, 1998. 39(2): p. 183–9. [PubMed] [Google Scholar]
- 53.Jackowski AP and Schultz RT, Foreshortened dorsal extension of the central sulcus in Williams syndrome. Cortex, 2005. 41(3): p. 282–90. [DOI] [PubMed] [Google Scholar]
- 54.Klein-Tasman BP, Li-Barber KT, and Magargee ET, Honing in on the social phenotype in Williams syndrome using multiple measures and multiple raters. J Autism Dev Disord, 2011. 41(3): p. 341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein-Tasman BP and Mervis CB, Autism Spectrum Symptomatology Among Children with Duplication 7q11.23 Syndrome. J Autism Dev Disord, 2018. 48(6): p. 1982–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ly TM and Hodapp RM, Children with Prader-Willi syndrome vs. Williams syndrome: indirect effects on parents during a jigsaw puzzle task. J Intellect Disabil Res, 2005. 49(Pt 12): p. 929–39. [DOI] [PubMed] [Google Scholar]
- 57.McGrath LM, et al. , Attention Bias to Emotional Faces Varies by IQ and Anxiety in Williams Syndrome. J Autism Dev Disord, 2016. 46(6): p. 2174–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mervis CB, et al. , Children with 7q11.23 duplication syndrome: psychological characteristics. Am J Med Genet A, 2015. 167(7): p. 1436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ng R, et al. , Morphological differences in the mirror neuron system in Williams syndrome. Soc Neurosci, 2016. 11(3): p. 277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ng R, Jarvinen A, and Bellugi U, Characterizing associations and dissociations between anxiety, social, and cognitive phenotypes of Williams syndrome. Res Dev Disabil, 2014. 35(10): p. 2403–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Garcia D, et al. , Lateral preference in Williams-Beuren syndrome is associated with cognition and language. Eur Child Adolesc Psychiatry, 2015. 24(9): p. 1025–33. [DOI] [PubMed] [Google Scholar]
- 62.Plesa Skwerer D, et al. , Autonomic responses to dynamic displays of facial expressions in adolescents and adults with Williams syndrome. Soc Cogn Affect Neurosci, 2009. 4(1): p. 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Porter MA, et al. , A role for transcription factor GTF2IRD2 in executive function in Williams-Beuren syndrome. PLoS One, 2012. 7(10): p. e47457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pryweller JR, et al. , The effect of intellectual ability on functional activation in a neurodevelopmental disorder: preliminary evidence from multiple fMRI studies in Williams syndrome. J Neurodev Disord, 2012. 4(1): p. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossi NF and Giacheti CM, Association between speech-language, general cognitive functioning and behaviour problems in individuals with Williams syndrome. J Intellect Disabil Res, 2017. 61(7): p. 707–718. [DOI] [PubMed] [Google Scholar]
- 66.Santos A, et al. , Just another face in the crowd: evidence for decreased detection of angry faces in children with Williams syndrome. Neuropsychologia, 2010. 48(4): p. 1071–8. [DOI] [PubMed] [Google Scholar]
- 67.Sauna-Aho O, et al. , Cognition in adults with Williams syndrome-A 20-year follow-up study. Mol Genet Genomic Med, 2019. 7(6): p. e695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Searcy YM, et al. , The relationship between age and IQ in adults with Williams syndrome. Am J Ment Retard, 2004. 109(3): p. 231–6. [DOI] [PubMed] [Google Scholar]
- 69.Stinton C, Elison S, and Howlin P, Mental health problems in adults with Williams syndrome. Am J Intellect Dev Disabil, 2010. 115(1): p. 3–18. [DOI] [PubMed] [Google Scholar]
- 70.Sullivan K and Tager-Flusberg H, Second-order belief attribution in Williams syndrome: intact or impaired? Am J Ment Retard, 1999. 104(6): p. 523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tavano A, et al. , Neurological soft signs feature a double dissociation within the language system in Williams syndrome. Neuropsychologia, 2010. 48(11): p. 3298–304. [DOI] [PubMed] [Google Scholar]
- 72.van der Fluit F, Gaffrey MS, and Klein-Tasman BP, Social Cognition in Williams Syndrome: Relations between Performance on the Social Attribution Task and Cognitive and Behavioral Characteristics. Front Psychol, 2012. 3: p. 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wuang YP and Tsai HY, Sensorimotor and visual perceptual functioning in school-aged children with Williams syndrome. J Intellect Disabil Res, 2017. 61(4): p. 348–362. [DOI] [PubMed] [Google Scholar]
- 74.Zarchi O, et al. , A comparative study of the neuropsychiatric and neurocognitive phenotype in two microdeletion syndromes: velocardiofacial (22q11.2 deletion) and Williams (7q11.23 deletion) syndromes. Eur Psychiatry, 2014. 29(4): p. 203–10. [DOI] [PubMed] [Google Scholar]
- 75.Grieco JC, et al. , An open-label pilot trial of minocycline in children as a treatment for Angelman syndrome. BMC Neurol, 2014. 14: p. 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peters SU, et al. , Cognitive and adaptive behavior profiles of children with Angelman syndrome. Am J Med Genet A, 2004. 128A(2): p. 110–3. [DOI] [PubMed] [Google Scholar]
- 77.Avrahamy H, et al. , A disease specific questionnaire for assessing behavior in individuals with Prader-Willi syndrome. Compr Psychiatry, 2015. 58: p. 189–97. [DOI] [PubMed] [Google Scholar]
- 78.Azor AM, et al. , Increased brain age in adults with Prader-Willi syndrome. Neuroimage Clin, 2019. 21: p. 101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baker EK, et al. , Exploring autism symptoms in an Australian cohort of patients with Prader-Willi and Angelman syndromes. J Neurodev Disord, 2018. 10(1): p. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bruining H, et al. , Behavioral signatures related to genetic disorders in autism. Mol Autism, 2014. 5(1): p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chevalere J, et al. , Investigation of the relationship between electrodermal and behavioural responses to executive tasks in Prader-Willi syndrome: An event-related experiment. Res Dev Disabil, 2019. 85: p. 229–242. [DOI] [PubMed] [Google Scholar]
- 82.Chevalere J, et al. , Assessment of executive functions in Prader-Willi syndrome and relationship with intellectual level. J Appl Res Intellect Disabil, 2013. 26(4): p. 309–18. [DOI] [PubMed] [Google Scholar]
- 83.Copet P, et al. , Cognitive profile in a large French cohort of adults with Prader-Willi syndrome: differences between genotypes. J Intellect Disabil Res, 2010. 54(3): p. 204–15. [DOI] [PubMed] [Google Scholar]
- 84.Curfs LM, et al. , Strengths and weaknesses in the cognitive profile of youngsters with Prader-Willi syndrome. Clin Genet, 1991. 40(6): p. 430–4. [DOI] [PubMed] [Google Scholar]
- 85.Dimitropoulos A, Ferranti A, and Lemler M, Expressive and receptive language in Prader-Willi syndrome: report on genetic subtype differences. J Commun Disord, 2013. 46(2): p. 193–201. [DOI] [PubMed] [Google Scholar]
- 86.Dimitropoulos A, Ho A, and Feldman B, Social responsiveness and competence in Prader-Willi syndrome: direct comparison to autism spectrum disorder. J Autism Dev Disord, 2013. 43(1): p. 103–13. [DOI] [PubMed] [Google Scholar]
- 87.Dykens EM, Maladaptive and compulsive behavior in Prader-Willi syndrome: new insights from older adults. Am J Ment Retard, 2004. 109(2): p. 142–53. [DOI] [PubMed] [Google Scholar]
- 88.Dykens EM and Roof E, Behavior in Prader-Willi syndrome: relationship to genetic subtypes and age. J Child Psychol Psychiatry, 2008. 49(9): p. 1001–8. [DOI] [PubMed] [Google Scholar]
- 89.Dykens EM, et al. , Diagnoses and characteristics of autism spectrum disorders in children with Prader-Willi syndrome. J Neurodev Disord, 2017. 9: p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hartley SL, et al. , Maladaptive behaviors and risk factors among the genetic subtypes of Prader-Willi syndrome. Am J Med Genet A, 2005. 136(2): p. 140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holland AJ, et al. , Behavioural phenotypes associated with specific genetic disorders: evidence from a population-based study of people with Prader-Willi syndrome. Psychol Med, 2003. 33(1): p. 141–53. [DOI] [PubMed] [Google Scholar]
- 92.Kuppens RJ, et al. , Effect of cessation of GH treatment on cognition during transition phase in Prader-Willi syndrome: results of a 2-year crossover GH trial. Orphanet J Rare Dis, 2016. 11(1): p. 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lo ST, Collin PJ, and Hokken-Koelega AC, Psychiatric disorders in children with Prader-Willi syndrome-Results of a 2-year longitudinal study. Am J Med Genet A, 2015. 167A(5): p. 983–91. [DOI] [PubMed] [Google Scholar]
- 94.Lo ST, et al. , Beneficial Effects of Long-Term Growth Hormone Treatment on Adaptive Functioning in Infants With Prader-Willi Syndrome. Am J Intellect Dev Disabil, 2015. 120(4): p. 315–27. [DOI] [PubMed] [Google Scholar]
- 95.Lukoshe A, et al. , Reduced cortical complexity in children with Prader-Willi Syndrome and its association with cognitive impairment and developmental delay. PLoS One, 2014. 9(9): p. e107320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Manning KE, et al. , Grey matter volume and cortical structure in Prader-Willi syndrome compared to typically developing young adults. Neuroimage Clin, 2018. 17: p. 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milner KM, et al. , Prader-Willi syndrome: intellectual abilities and behavioural features by genetic subtype. J Child Psychol Psychiatry, 2005. 46(10): p. 1089–96. [DOI] [PubMed] [Google Scholar]
- 98.Roof E, et al. , Intellectual characteristics of Prader-Willi syndrome: comparison of genetic subtypes. J Intellect Disabil Res, 2000. 44 (Pt 1): p. 25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salles J, et al. , Deficits in voice and multisensory processing in patients with Prader-Willi syndrome. Neuropsychologia, 2016. 85: p. 137–47. [DOI] [PubMed] [Google Scholar]
- 100.Semenza C, et al. , Genetics and mathematics: evidence from Prader-Willi syndrome. Neuropsychologia, 2008. 46(1): p. 206–12. [DOI] [PubMed] [Google Scholar]
- 101.Shivers CM, Leonczyk CL, and Dykens EM, Life Satisfaction Among Mothers of Individuals with Prader-Willi Syndrome. J Autism Dev Disord, 2016. 46(6): p. 2126–2137. [DOI] [PubMed] [Google Scholar]
- 102.Shriki-Tal L, et al. , Psychiatric disorders in a cohort of individuals with Prader-Willi syndrome. Eur Psychiatry, 2017. 44: p. 47–52. [DOI] [PubMed] [Google Scholar]
- 103.Shu SG, et al. , Anthropometric and intellectual evaluation of individuals with Prader-Willi syndrome. J Formos Med Assoc, 2007. 106(6): p. 509–12. [DOI] [PubMed] [Google Scholar]
- 104.Skokauskas N, et al. , Mental health problems in children with prader-willi syndrome. J Can Acad Child Adolesc Psychiatry, 2012. 21(3): p. 194–203. [PMC free article] [PubMed] [Google Scholar]
- 105.Whittington J, et al. , Cognitive abilities and genotype in a population-based sample of people with Prader-Willi syndrome. J Intellect Disabil Res, 2004. 48(Pt 2): p. 172–87. [DOI] [PubMed] [Google Scholar]
- 106.Zarcone J, et al. , The relationship between compulsive behaviour and academic achievement across the three genetic subtypes of Prader-Willi syndrome. J Intellect Disabil Res, 2007. 51(Pt. 6): p. 478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blackmon K, et al. , Focal Cortical Anomalies and Language Impairment in 16p11.2 Deletion and Duplication Syndrome. Cereb Cortex, 2018. 28(7): p. 2422–2430. [DOI] [PubMed] [Google Scholar]
- 108.Hanson E, et al. , Cognitive and behavioral characterization of 16p11.2 deletion syndrome. J Dev Behav Pediatr, 2010. 31(8): p. 649–57. [DOI] [PubMed] [Google Scholar]
- 109.Hippolyte L, et al. , The Number of Genomic Copies at the 16p11.2 Locus Modulates Language, Verbal Memory, and Inhibition. Biol Psychiatry, 2016. 80(2): p. 129–139. [DOI] [PubMed] [Google Scholar]
- 110.Kim SH, et al. , Language characterization in 16p11.2 deletion and duplication syndromes. Am J Med Genet B Neuropsychiatr Genet, 2020. 183(6): p. 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moreno-De-Luca A, et al. , The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry, 2015. 72(2): p. 119–26. [DOI] [PubMed] [Google Scholar]
- 112.Owen JP, et al. , Brain MR Imaging Findings and Associated Outcomes in Carriers of the Reciprocal Copy Number Variation at 16p11.2. Radiology, 2018. 286(1): p. 217–226. [DOI] [PubMed] [Google Scholar]
- 113.Greenberg F, et al. , Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2). Am J Med Genet, 1996. 62(3): p. 247–54. [DOI] [PubMed] [Google Scholar]
- 114.Madduri N, et al. , Cognitive and adaptive behavior profiles in Smith-Magenis syndrome. J Dev Behav Pediatr, 2006. 27(3): p. 188–92. [DOI] [PubMed] [Google Scholar]
- 115.Udwin O, Webber C, and Horn I, Abilities and attainment in Smith-Magenis syndrome. Dev Med Child Neurol, 2001. 43(12): p. 823–8. [DOI] [PubMed] [Google Scholar]
- 116.Albert AB, et al. , Childhood Executive Functioning Predicts Young Adult Outcomes in 22q11.2 Deletion Syndrome. J Int Neuropsychol Soc, 2018. 24(9): p. 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Angkustsiri K, et al. , Social impairments in chromosome 22q11.2 deletion syndrome (22q11.2DS): autism spectrum disorder or a different endophenotype? J Autism Dev Disord, 2014. 44(4): p. 739–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Antshel KM, et al. , ADHD, major depressive disorder, and simple phobias are prevalent psychiatric conditions in youth with velocardiofacial syndrome. J Am Acad Child Adolesc Psychiatry, 2006. 45(5): p. 596–603. [DOI] [PubMed] [Google Scholar]
- 119.Armando M, et al. , Prevalence and treatment of psychiatric disorders other than psychosis in children and adolescents with 22q11DS: Examining associations with social and role functioning. Psychiatry Res, 2017. 254: p. 238–243. [DOI] [PubMed] [Google Scholar]
- 120.Badoud D, et al. , Understanding others: a pilot investigation of cognitive and affective facets of social cognition in patients with 22q11.2 deletion syndrome (22q11DS). J Neurodev Disord, 2017. 9(1): p. 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baker K, et al. , COMT Val108/158 Met modifies mismatch negativity and cognitive function in 22q11 deletion syndrome. Biol Psychiatry, 2005. 58(1): p. 23–31. [DOI] [PubMed] [Google Scholar]
- 122.Bostelmann M, et al. , Visual memory profile in 22q11.2 microdeletion syndrome: are there differences in performance and neurobiological substrates between tasks linked to ventral and dorsal visual brain structures? A cross-sectional and longitudinal study. J Neurodev Disord, 2016. 8: p. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Butcher NJ, et al. , Functional outcomes of adults with 22q11.2 deletion syndrome. Genet Med, 2012. 14(10): p. 836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Campbell LE, et al. , Social cognition dysfunction in adolescents with 22q11.2 deletion syndrome (velo-cardio-facial syndrome): relationship with executive functioning and social competence/functioning. J Intellect Disabil Res, 2015. 59(9): p. 845–59. [DOI] [PubMed] [Google Scholar]
- 125.Carmel M, et al. , Association of COMT and PRODH gene variants with intelligence quotient (IQ) and executive functions in 22q11.2DS subjects. J Psychiatr Res, 2014. 56: p. 28–35. [DOI] [PubMed] [Google Scholar]
- 126.Chawner S, et al. , Childhood cognitive development in 22q11.2 deletion syndrome: case-control study. Br J Psychiatry, 2017. 211(4): p. 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cunningham AC, et al. , Developmental coordination disorder, psychopathology and IQ in 22q11.2 deletion syndrome. Br J Psychiatry, 2018. 212(1): p. 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.De Smedt B, et al. , Intellectual abilities in a large sample of children with Velo-Cardio-Facial Syndrome: an update. J Intellect Disabil Res, 2007. 51(Pt 9): p. 666–70. [DOI] [PubMed] [Google Scholar]
- 129.de Sonneville LMJ, et al. , [Formula: see text]Executive functioning and its relation to ASD and ADHD symptomatology in 22q11.2 deletion syndrome. Child Neuropsychol, 2018. 24(1): p. 1–19. [DOI] [PubMed] [Google Scholar]
- 130.Debbane M, et al. , Psychotic symptoms in children and adolescents with 22q11.2 deletion syndrome: Neuropsychological and behavioral implications. Schizophr Res, 2006. 84(2–3): p. 187–93. [DOI] [PubMed] [Google Scholar]
- 131.Dewulf D, Noens I, and Swillen A, [Adaptive skills, cognitive functioning and behavioural problems in adolescents with 22q11.2 deletion syndrome]. Tijdschr Psychiatr, 2013. 55(5): p. 369–74. [PubMed] [Google Scholar]
- 132.Dufour F, et al. , Cingulate gyral reductions are related to low executive functioning and psychotic symptoms in 22q 11.2 deletion syndrome. Neuropsychologia, 2008. 46(12): p. 2986–92. [DOI] [PubMed] [Google Scholar]
- 133.Duijff SN, et al. , Cognitive development in children with 22q11.2 deletion syndrome. Br J Psychiatry, 2012. 200(6): p. 462–8. [DOI] [PubMed] [Google Scholar]
- 134.Eaton CB, et al. , Epilepsy and seizures in young people with 22q11.2 deletion syndrome: Prevalence and links with other neurodevelopmental disorders. Epilepsia, 2019. 60(5): p. 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fabbro A, et al. , Depression and anxiety disorders in children and adolescents with velo-cardio-facial syndrome (VCFS). Eur Child Adolesc Psychiatry, 2012. 21(7): p. 379–85. [DOI] [PubMed] [Google Scholar]
- 136.Fiksinski AM, et al. , Neurocognition and adaptive functioning in a genetic high risk model of schizophrenia. Psychol Med, 2019. 49(6): p. 1047–1054. [DOI] [PubMed] [Google Scholar]
- 137.Francisco AA, et al. , Atypical response inhibition and error processing in 22q11.2 Deletion Syndrome and schizophrenia: Towards neuromarkers of disease progression and risk. Neuroimage Clin, 2020. 27: p. 102351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Franconi CP, et al. , IQ and hemizygosity for the Val(158) Met functional polymorphism of COMT in 22q11DS. Am J Med Genet B Neuropsychiatr Genet, 2016. 171(8): p. 1112–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frascarelli M, et al. , Social cognition deficit and genetic vulnerability to schizophrenia in 22q11 deletion syndrome. Ann Ist Super Sanita, 2020. 56(1): p. 107–113. [DOI] [PubMed] [Google Scholar]
- 140.Glaser B, et al. , Eye gaze during face processing in children and adolescents with 22q11.2 deletion syndrome. J Am Acad Child Adolesc Psychiatry, 2010. 49(7): p. 665–74. [DOI] [PubMed] [Google Scholar]
- 141.Gothelf D, et al. , Cognition, psychosocial adjustment and coping in familial cases of velocardiofacial syndrome. J Neural Transm (Vienna), 2007. 114(11): p. 1495–501. [DOI] [PubMed] [Google Scholar]
- 142.Heller C, et al. , Abnormalities in white matter tracts in the fronto-striatal-thalamic circuit are associated with verbal performance in 22q11.2DS. Schizophr Res, 2020. 224: p. 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hidding E, et al. , The role of COMT and plasma proline in the variable penetrance of autistic spectrum symptoms in 22q11.2 deletion syndrome. Clin Genet, 2016. 90(5): p. 420–427. [DOI] [PubMed] [Google Scholar]
- 144.Hooper SR, et al. , A longitudinal examination of the psychoeducational, neurocognitive, and psychiatric functioning in children with 22q11.2 deletion syndrome. Res Dev Disabil, 2013. 34(5): p. 1758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hwang VJ, et al. , Mapping the deletion endpoints in individuals with 22q11.2 deletion syndrome by droplet digital PCR. BMC Med Genet, 2014. 15: p. 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jacobson C, et al. , Core neuropsychological characteristics of children and adolescents with 22q11.2 deletion. J Intellect Disabil Res, 2010. 54(8): p. 701–13. [DOI] [PubMed] [Google Scholar]
- 147.Jensen M, et al. , A higher rare CNV burden in the genetic background potentially contributes to intellectual disability phenotypes in 22q11.2 deletion syndrome. Eur J Med Genet, 2018. 61(4): p. 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kates WR, et al. , Comparing phenotypes in patients with idiopathic autism to patients with velocardiofacial syndrome (22q11 DS) with and without autism. Am J Med Genet A, 2007. 143A(22): p. 2642–50. [DOI] [PubMed] [Google Scholar]
- 149.Kikinis Z, et al. , Abnormalities in gray matter microstructure in young adults with 22q11.2 deletion syndrome. Neuroimage Clin, 2019. 21: p. 101611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Klaassen P, et al. , Explaining the variable penetrance of CNVs: Parental intelligence modulates expression of intellectual impairment caused by the 22q11.2 deletion. Am J Med Genet B Neuropsychiatr Genet, 2016. 171(6): p. 790–6. [DOI] [PubMed] [Google Scholar]
- 151.Kravariti E, et al. , Memory in intellectually matched groups of young participants with 22q11.2 deletion syndrome and those with schizophrenia. Res Dev Disabil, 2010. 31(3): p. 864–8. [DOI] [PubMed] [Google Scholar]
- 152.Lewandowski KE, et al. , Schizophrenic-like neurocognitive deficits in children and adolescents with 22q11 deletion syndrome. Am J Med Genet B Neuropsychiatr Genet, 2007. 144B(1): p. 27–36. [DOI] [PubMed] [Google Scholar]
- 153.Lin A, et al. , Reciprocal Copy Number Variations at 22q11.2 Produce Distinct and Convergent Neurobehavioral Impairments Relevant for Schizophrenia and Autism Spectrum Disorder. Biol Psychiatry, 2020. 88(3): p. 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Linton SR, et al. , Neural and behavioral measures suggest that cognitive and affective functioning interactions mediate risk for psychosis-proneness symptoms in youth with chromosome 22q11.2 deletion syndrome. Am J Med Genet A, 2020. 182(7): p. 1615–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.McCabe KL, et al. , Quantifying the resolution of spatial and temporal representation in children with 22q11.2 deletion syndrome. J Neurodev Disord, 2019. 11(1): p. 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Momma K, et al. , Tetralogy of Fallot associated with chromosome 22q11.2 deletion in adolescents and young adults. Genet Med, 2001. 3(1): p. 56–60. [DOI] [PubMed] [Google Scholar]
- 157.Monks S, et al. , Further evidence for high rates of schizophrenia in 22q11.2 deletion syndrome. Schizophr Res, 2014. 153(1–3): p. 231–6. [DOI] [PubMed] [Google Scholar]
- 158.Mosheva M, et al. , Education and employment trajectories from childhood to adulthood in individuals with 22q11.2 deletion syndrome. Eur Child Adolesc Psychiatry, 2019. 28(1): p. 31–42. [DOI] [PubMed] [Google Scholar]
- 159.Moss EM, et al. , Psychoeducational profile of the 22q11.2 microdeletion: A complex pattern. J Pediatr, 1999. 134(2): p. 193–8. [DOI] [PubMed] [Google Scholar]
- 160.Murphy KC, Jones LA, and Owen MJ, High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch Gen Psychiatry, 1999. 56(10): p. 940–5. [DOI] [PubMed] [Google Scholar]
- 161.Niklasson L, et al. , Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res Dev Disabil, 2009. 30(4): p. 763–73. [DOI] [PubMed] [Google Scholar]
- 162.Olszewski AK, et al. , The social brain network in 22q11.2 deletion syndrome: a diffusion tensor imaging study. Behav Brain Funct, 2017. 13(1): p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ousley O, et al. , Examining the Overlap between Autism Spectrum Disorder and 22q11.2 Deletion Syndrome. Int J Mol Sci, 2017. 18(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Padula MC, et al. , Multimodal investigation of triple network connectivity in patients with 22q11DS and association with executive functions. Hum Brain Mapp, 2017. 38(4): p. 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pontillo M, Menghini D, and Vicari S, Neurocognitive profile and onset of psychosis symptoms in children, adolescents and young adults with 22q11 deletion syndrome: A longitudinal study. Schizophr Res, 2019. 208: p. 76–81. [DOI] [PubMed] [Google Scholar]
- 166.Radoeva PD, et al. , Atlas-based white matter analysis in individuals with velo-cardio-facial syndrome (22q11.2 deletion syndrome) and unaffected siblings. Behav Brain Funct, 2012. 8: p. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Raux G, et al. , Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum Mol Genet, 2007. 16(1): p. 83–91. [DOI] [PubMed] [Google Scholar]
- 168.Roizen NJ, et al. , 22q11.2 deletion syndrome: are motor deficits more than expected for IQ level? J Pediatr, 2010. 157(4): p. 658–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schaer M, et al. , Deviant trajectories of cortical maturation in 22q11.2 deletion syndrome (22q11DS): a cross-sectional and longitudinal study. Schizophr Res, 2009. 115(2–3): p. 182–90. [DOI] [PubMed] [Google Scholar]
- 170.Schneider M, et al. , Preliminary structure and predictive value of attenuated negative symptoms in 22q11.2 deletion syndrome. Psychiatry Res, 2012. 196(2–3): p. 277–84. [DOI] [PubMed] [Google Scholar]
- 171.Schoch K, et al. , Applicability of the nonverbal learning disability paradigm for children with 22q11.2 deletion syndrome. J Learn Disabil, 2014. 47(2): p. 153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schreiner MJ, et al. , Default mode network connectivity and reciprocal social behavior in 22q11.2 deletion syndrome. Soc Cogn Affect Neurosci, 2014. 9(9): p. 1261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Scott JA, et al. , The hippocampi of children with chromosome 22q11.2 deletion syndrome have localized anterior alterations that predict severity of anxiety. J Psychiatry Neurosci, 2016. 41(3): p. 203–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Serur Y, et al. , Psychiatric disorders and autism in young children with 22q11.2 deletion syndrome compared to children with idiopathic autism. Eur Psychiatry, 2019. 55: p. 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shashi V, et al. , Evidence of gray matter reduction and dysfunction in chromosome 22q11.2 deletion syndrome. Psychiatry Res, 2010. 181(1): p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sinderberry B, et al. , Subtypes in 22q11.2 deletion syndrome associated with behaviour and neurofacial morphology. Res Dev Disabil, 2013. 34(1): p. 116–25. [DOI] [PubMed] [Google Scholar]
- 177.Tylee DS, et al. , Machine-learning classification of 22q11.2 deletion syndrome: A diffusion tensor imaging study. Neuroimage Clin, 2017. 15: p. 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ua-Areechit T, et al. , Clinical characteristics and immunological status of patients with 22q11.2 deletion syndrome in Northern Thailand. Asian Pac J Allergy Immunol, 2020. [DOI] [PubMed] [Google Scholar]
- 179.Uljarevic M, et al. , Interrelationship Between Cognitive Control, Anxiety, and Restricted and Repetitive Behaviors in Children with 22q11.2 Deletion Syndrome. Autism Res, 2019. 12(12): p. 1737–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Van Aken K, et al. , The motor profile of primary school-age children with a 22q11.2 deletion syndrome (22q11.2DS) and an age- and IQ-matched control group. Child Neuropsychol, 2009. 15(6): p. 532–42. [DOI] [PubMed] [Google Scholar]
- 181.van Amelsvoort T, et al. , Effects of a functional COMT polymorphism on brain anatomy and cognitive function in adults with velo-cardio-facial syndrome. Psychol Med, 2008. 38(1): p. 89–100. [DOI] [PubMed] [Google Scholar]
- 182.Van Den Heuvel E, et al. , Exploratory study on cognitive abilities and social responsiveness in children with 22q11.2 deletion syndrome (22q11DS) and children with idiopathic intellectual disability (IID). Res Dev Disabil, 2018. 81: p. 89–102. [DOI] [PubMed] [Google Scholar]
- 183.Vangkilde A, et al. , Associations between social cognition, skills, and function and subclinical negative and positive symptoms in 22q11.2 deletion syndrome. J Neurodev Disord, 2016. 8: p. 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Weinberger R, et al. , Neurocognitive profile in psychotic versus nonpsychotic individuals with 22q11.2 deletion syndrome. Eur Neuropsychopharmacol, 2016. 26(10): p. 1610–8. [DOI] [PubMed] [Google Scholar]
- 185.Woodin M, et al. , Neuropsychological profile of children and adolescents with the 22q11.2 microdeletion. Genet Med, 2001. 3(1): p. 34–9. [DOI] [PubMed] [Google Scholar]
- 186.Yuen T, et al. , Premorbid adjustment and schizophrenia in individuals with 22q11.2 deletion syndrome. Schizophr Res, 2013. 151(1–3): p. 221–5. [DOI] [PubMed] [Google Scholar]
- 187.Hays JR, Reas DL, and Shaw JB, Concurrent validity of the Wechsler abbreviated scale of intelligence and the Kaufman brief intelligence test among psychiatric inpatients. Psychol Rep, 2002. 90(2): p. 355–9. [DOI] [PubMed] [Google Scholar]
- 188.Hamburg S, et al. , Assessing general cognitive and adaptive abilities in adults with Down syndrome: a systematic review. J Neurodev Disord, 2019. 11(1): p. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.MacLeod AK, et al. , Genetic copy number variation and general cognitive ability. PLoS One, 2012. 7(12): p. e37385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Moreno-De Luca A, Cubells J, and Sanders S, Cross-Disorder Comparison of Four Neuropsychatric CNV Loci. Curr Genet Med Rep 2014. 2. [Google Scholar]
- 191.Chawner S, et al. , Genotype-phenotype associations in children with copy number variants associated with high neuropsychiatric risk in the UK (IMAGINE-ID): a case-control cohort study. Lancet Psychiatry, 2019. 6(6): p. 493–505. [DOI] [PubMed] [Google Scholar]
- 192.Kendall KM, et al. , Cognitive Performance Among Carriers of Pathogenic Copy Number Variants: Analysis of 152,000 UK Biobank Subjects. Biol Psychiatry, 2017. 82(2): p. 103–110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


