Abstract
Vancomycin-resistant enterococci (VRE), a leading cause of hospital-acquired infections, can occur in wastewater. However, to date, no previous studies have evaluated the occurrence of VRE at wastewater treatment plants (WWTPs) that send their treated effluent to reuse sites. We evaluated the occurrence, concentration, and antimicrobial resistance patterns of VRE at U.S. WWTPs associated with reuse sites. We collected 44 wastewater samples, representing treatment steps from influent to effluent, from two Mid-Atlantic and two Midwest WWTPs between October 2009 and October 2010. Samples were analyzed for total enterococci and VRE using membrane filtration. Isolates were confirmed using biochemical tests and PCR. Antimicrobial susceptibility testing was performed by Sensititre® microbroth dilution. Data were analyzed by two-sample proportion tests and analysis of variance. We detected VRE in 27% (12/44) of all wastewater samples collected and VRE represented 3% of total enterococci detected at all WWTPs. More samples were VRE-positive from the Mid-Atlantic compared to the Midwest WWTPs (p=0.008). VRE concentrations decreased as treatment progressed at all WWTPs, except at Mid-Atlantic WWTP1 where there was an increase in VRE concentrations in activated sludge reactor samples. VRE was not detected in chlorinated effluent, but was detected in one un-chlorinated effluent sample. All unique VRE isolates were multidrug resistant. Fifty-five percent (12/22) of the isolates displayed high-level aminoglycoside resistance. Our findings show that chlorination reduces the occurrence of VRE in wastewater. However, WWTP workers could be exposed to VRE during wastewater treatment. Our data also raise potential concerns about VRE exposure among individuals who come into contact with un-chlorinated reclaimed water.
Keywords: antibiotic-resistant bacteria, enterococci, high-level aminoglycoside resistance, reclaimed water, vancomycin-resistant enterococci, wastewater, wastewater treatment plants
1. Introduction1
The number of hospitalizations associated with antibiotic-resistant bacterial infections in the United States nearly quadrupled between 1997 and 2006 (Mainous et al., 2011). One antibiotic-resistant bacterial pathogen of particular concern is vancomycin-resistant enterococci (VRE), an opportunistic gram-positive bacterium that is resistant to vancomycin – “a drug of last resort” – and can cause urinary tract infections, wound infections, septicemia, and endocarditis (CDC, 2008; Wegener et al., 1999). The first cases of infection with enterococci that expressed high-level vancomycin resistance were reported in the United Kingdom in the 1980s (Uttley et al. 1988). As of 2008, VRE was the third leading cause of hospital-acquired infections in the U.S. (Hidron et al., 2008; Uttley et al., 1988).
In addition to VRE’s ability to cause multiple types of severe infections, this bacterium is a significant public health concern because of its propensity to acquire and transfer mobile resistance genes (Hayakawa et al., 2012). Acquisition of vancomycin resistance genes can take place between strains of enterococci, but these genes can also be transferred from enterococci to other types of bacteria, including Staphylococcus aureus (NIAID, 2009; Sievert et al., 2008). The Michigan Department of Community Health reported the first clinical isolate of vancomycin-resistant S. aureus (VRSA) in 2002 (Sievert et al., 2008). VRSA is cause for concern because of the limited treatment options available for this type of infection. As of 2012, only 13 clinical cases of VRSA had been confirmed in the U.S., but the incidence of VRSA infections could continue to rise (CDC, 2012). Resistance genes can also be transferred between enterococci isolates originating from different settings, including clinical strains released into wastewater and community strains contained in wastewater (Guardabassi and Dalsgaard, 2004; Rizzo et al., 2013).
Previous studies have detected VRE at different stages in the wastewater treatment process, including treated effluent, suggesting that VRE present in wastewater effluent could be partially responsible for the dissemination of VRE into the environment and human communities (Araujo et al., 2010; Caplin et al., 2008; Harwood et al., 2001; Kotzamanidis et al., 2009; Luczkiewicz et al., 2010; Morris et al., 2012; Nagulapally et al., 2009; Poole et al., 2005; Shannon et al., 2007; Talebi et al., 2008). As drought conditions continue to stress freshwater resources in the U.S. and other countries, treated municipal wastewater effluent, or reclaimed water, is increasingly used for applications such as landscape and crop irrigation, groundwater recharge, and snowmaking (EPA, 2012). During these processes, individuals who apply, use, or come in contact with reclaimed water could possibly be exposed to VRE and other bacterial pathogens that may persist in this alternative water source.
To our knowledge, there are no published studies analyzing municipal wastewater intended for reuse for the presence of VRE. In this study, we evaluated the occurrence, concentration and antimicrobial susceptibilities of VRE at four wastewater treatment plants (WWTPs) located in the Mid-Atlantic and Midwest regions of the U.S. from which treated wastewater is reused at spray irrigation sites.
2. Materials and Methods
2.1. Study sites
We sampled four U.S. WWTPs that distribute treated effluent to reuse sites: two WWTPs in the Mid-Atlantic region and two WWTPs in the Midwest region (Rosenberg Goldstein et al., 2012). Sites in the Mid-Atlantic and Midwest were chosen in order to compare WWTPs in two climatically different regions of the United States. Schematics of each WWTP are published in in Rosenberg Goldstein et al. (2012).
Mid-Atlantic WWTP1 is a tertiary WWTP in an urban area that processes 681,390 m3/day of wastewater, with a peak capacity of 1.51 million m3/day. Mid-Atlantic WWTP2 is a tertiary WWTP in a suburban area that processes 7,570 m3/day of wastewater and has a peak capacity of 45,425 m3/day. Tertiary wastewater treatment is defined as any treatment beyond primary treatment (physical removal of solids) and secondary treatment (biological treatment) that provides a final treatment step to further improve effluent quality. Tertiary treatments can include filtration, disinfection and lagooning. The incoming wastewater (influent) at both Mid-Atlantic plants includes domestic and hospital wastewater. At Mid-Atlantic WWTP1, the following treatment steps are employed: screens, primary clarifier, activated sludge reactors, secondary clarifier, sand filters, chlorination, dechlorination and discharge. At Mid-Atlantic WWTP2, the following treatment steps are employed: screens, primary clarifier, primary aeration tank, secondary aeration tank, secondary clarifier, multimedia filter, chlorination, dechlorination and discharge. At both Mid-Atlantic plants, the chlorination dose was 2–3 mg/L, followed by dechlorination with sodium bisulfite such that the chlorine residual in effluent is < 0.1 mg/L. The effluent (discharge) from both Mid-Atlantic plants is piped to landscaping sites for reuse in spray irrigation.
Midwest WWTP1 is a tertiary WWTP in a rural area that processes 1,363 m3/day of wastewater, with a peak capacity of 10,978 m3/day. The incoming wastewater includes domestic wastewater and agriculturally influenced stormwater. At Midwest WWTP1, the following treatment steps are employed: screens, activated sludge lagoons, clarifiers, seasonal chlorination (and dechlorination), and discharge. Seasonal chlorination occurs in June, July, and August, and during these times the chlorination dose is 4 mg/L with a contact time to assure a chlorine residual of 0 mg/L in effluent. This effluent is then piped to a landscaping site for reuse in spray irrigation. Midwest WWTP2 is a tertiary WWTP (with no on-site disinfection) in a rural area that processes 1,439 m3/day and has a peak capacity of 7,571 m3/day. The incoming wastewater at this plant includes domestic wastewater, wastewater from a food production facility, and agriculturally influenced stormwater. At Midwest WWTP2, the following treatment steps are employed: screens, sequencing batch reactor, lagoon cell A, lagoon cell B, lagoon cell C, lagoon cell D, lagoon cell E, and discharge. Unchlorinated effluent from this plant is piped to an agricultural site for crop irrigation.
2.2. Sample collection
Samples were collected throughout the treatment process at all four WWTPs to determine whether certain treatment steps cause the concentration of culturable VRE to increase or decrease, as previous studies have suggested that the concentration of antibiotic-resistant bacteria differs depending on the treatment step sampled (Börjesson et al., 2009; Kim and Aga, 2007; Nakamura and Shirota, 1990). A total of 44 grab samples were collected between October 2009 and October 2010: 12 samples from Mid-Atlantic WWTP1; 8 samples from Mid-Atlantic WWTP2; 12 samples from Midwest WWTP1; and 12 samples from Midwest WWTP2 (Rosenberg Goldstein et al., 2012). The timing of each sampling event was dependent on the availability and schedule of the WWTP operators. Samples were collected in 1‑L sterile polyethylene Nalgene® Wide Mouth Environmental Sample Bottles (Nalgene, Lima, OH), labeled, and transported to the laboratory at 4⁰C within 24 hr for processing.
2.3. Isolation
Standard membrane filtration was used to recover total enterococci and VRE from the water samples (EPA, 2002). Briefly, ten-fold serial dilutions in the range of 1 to 0.001 ml for influent, activated sludge, and post aeration samples; and 100 to 1 ml for secondary clarifier and cell B samples were prepared using sterilized phosphate buffered saline (PBS) and filtered through 0.45 μm, 47 mm mixed cellulose ester filters (Millipore, Billerica, MA). One liter of each non-diluted effluent sample was also filtered in the same fashion. Filters were then plated in duplicate on membrane-Enterococcus Indoxyl-β-D-Glucoside (mEI) agar (EMD Millipore, Billerica, MA) to isolate total enterococci and mEI agar amended with 16 μg/mL of vancomycin to isolate VRE. Plates were incubated at 41°C for 24 hr. Resulting colonies with blue halos were considered presumptive total enterococci and VRE. These colonies were purified on Brain Heart Infusion (BHI) agar (Becton, Dickinson and Company, Franklin Lakes, NJ) and archived in Brucella broth (Becton, Dickinson and Company) with 15% glycerol at −80°C. Enterococcus faecalis ATCC 29212 was used as a positive control and PBS was used as a negative control throughout the isolation process.
2.4. Identification
VRE was confirmed using the Gram stain, the catalase test, and by detection of pyrrolidonyl peptidase activity (Remel, Lenexa, KS). For confirmation, a multiplex PCR assay developed by Micallef et al. (2013) was used. Genomic DNA from VRE was extracted by heat lysis as described previously (Micallef et al., 2013). Briefly, the PCR reaction targeted the D-alanine:D-alanine ligase (ddl) genes of E. faecalis and E. faecium, the vancomycin resistance-encoding vanC1 and vanC2/3 genes of E. gallinarum and E. casseliflavus, respectively, and an internal control targeting a 350 base pair portion of the 16S rRNA gene. PCR amplification consisted of an initial denaturing step of 95°C for 3 min, followed by 35 cycles of denaturing at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 5 min. Positive controls used for PCR amplification were E. faecalis ATCC 51299, E. faecium ATCC 51559, E. casseliflavus ATCC 25788, and E. gallinarum ATCC 49573.
2.5. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the Sensititre® microbroth dilution system in accordance with the manufacturer’s instructions on all confirmed VRE (n=34) and vancomycin-intermediate enterococci (VIE) (enterococci that express intermediate, and not complete, resistance to vancomycin) (n=22) isolates (Trek Diagnostic Systems Inc., Cleveland, OH). Overnight cultures were transferred to sterile demineralized water (Trek Diagnostic Systems) to achieve a 0.5 McFarland standard. Then, 50 μL of each suspension was transferred to sterile cation-adjusted Mueller Hinton broth (Trek Diagnostic Systems), and 50 μL of the broth solution was then dispensed into GPN3F or custom designed CMV5ACDC minimal inhibitory concentration plates (Trek Diagnostic Systems) that included the following antibiotics (range of concentrations in μg/ml): erythromycin (0.25–8), quinupristin/dalfopristin (synercid) (0.12–32), vancomycin (1–128), tetracycline (2–32), gentamicin (2–16, 128–1024), linezolid (0.5–8), streptomycin (512–2048), penicillin (0.06–16), and ciprofloxacin (0.5–4). E. faecalis ATCC 29212 and S. aureus ATCC 29213 were used as quality control strains. Minimal inhibitory concentrations were recorded as the lowest concentration of an antimicrobial that completely inhibited bacterial growth (CLSI, 2013). Resistance breakpoints published by the Clinical and Laboratory Standards Institute were used (CLSI, 2013). Multidrug resistance was defined as resistance to two or more classes of antibiotics.
2.6. Statistical analyses
Descriptive statistics included the percentage of wastewater samples that were positive for VRE and the percentage of VRE out of total enterococci by WWTP. A two-sample test of binomial proportions was used to compare the percentage of positive VRE samples between the Mid-Atlantic and the Midwest WWTPs. One-way analysis of variance (ANOVA) was then used to compare the average log concentration of presumptive VRE by treatment step for each WWTP. ANOVA was followed by linear contrasts as a post-hoc test to compare concentrations of presumptive VRE between each specific treatment step. In all cases, p-values of ≤ 0.05 were defined as statistically significant. All statistical analyses were performed using Stata/IC 10 (StatCorp LP, College Station, TX).
3. Results
3.1. Presence and concentration of VRE
VRE were detected at all WWTPs in this study and made up 3% of the total enterococci recovered (Table 1). Total enterococci were detected at all WWTPs in all treatment steps, including chlorinated effluent. Across all treatment plants sampled, 27% (12/44) of wastewater samples were positive for VRE: 45% (9/20) of samples from the Mid-Atlantic WWTPs; and 13% (3/24) of samples from the Midwest WWTPs (p=0.008). Thirty-three percent (4/12) of influent samples from all WWTPs were VRE-positive; 60% (3/5) from the Mid-Atlantic WWTPs and 14% (1/7) from the Midwest WWTPs. The percentage of VRE out of total enterococci increased in the activated sludge reactor step at both Mid-Atlantic WWTP1 and Mid-Atlantic WWTP2 but decreased to undetectable levels in the effluent (Table 1). At Midwest WWTP1, the percentage of VRE out of total enterococci increased as treatment progressed, with the highest percentage of VRE present in the effluent (Table 1). No confirmed VRE were detected in any tertiary-treated (chlorinated) effluent samples. However, VRE were detected in one effluent sample from Midwest WWTP1 in October 2010 when chlorination was not being used – the same sample that methicillin-resistant Staphylococcus aureus (MRSA) was isolated from as described in Rosenberg Goldstein et al. (2012).
Table 1.
Average concentration of total enterococci and vancomycin-resistant enterococci (VRE) by wastewater treatment plant and treatment step across all sample collection dates.
| Total Enterococci (CFU/100 mls) | VRE (CFU/100 mls) | Percentage of VRE | |
|---|---|---|---|
| Mid-Atlantic WWTP1 | |||
| Influent (n=3) | 7.17×106 | 1.93×104 | 0.3 |
| Activated Sludge Reactor (n=3) | 4.64×105 | 1.89×105 | 40.7 |
| Secondary Clarifier (n=3) | 1.49×103 | 9.60×101 | 6.5 |
| Effluent (n=3) | 1.04 | 0 | 0.0 |
| Mid-Atlantic WWTP2 | |||
| Influent (n=2) | 1.73×106 | 8.56×104 | 4.9 |
| Activated Sludge Reactor (n=2) | 2.19×105 | 1.32×104 | 6.0 |
| Secondary Clarifier (n=2) | 1.86×104 | 8.46×102 | 4.6 |
| Effluent (n=2) | 5.00×10−2 | 0 | 0.0 |
| Midwest WWTP1 | |||
| Influent (n=3) | 1.21×106 | 4.03×104 | 3.3 |
| Post Aeration (n=3) | 6.34×104 | 8.55×102 | 1.3 |
| Secondary Clarifier (n=3) | 6.63×103 | 1.03×103 | 15.5 |
| Effluent (n=3) | 1.56×101 | 3.29a | 21.1 |
| Midwest WWTP2 | |||
| Influent (n=4) | 5.30×105 | 2.52×103 | 0.5 |
| Cell B (n=4) | 5.08×102 | 0 | 0.0 |
| Effluent (n=4) | 6.05×101 | 0 | 0.0 |
| Total | 1.14×107 | 3.52×105 | 3.1 |
Sample collected in October 2010 when chlorination was not taking place.
In general, the average concentration of presumptive VRE at each WWTP decreased as treatment progressed (Figure 1). Specifically, the ANOVA showed that there were significant differences in VRE concentrations between treatment steps at all WWTPs, except at Midwest WWTP 2 where VRE was only detected in one influent sample (Mid-Atlantic WWTP1 p≤0.001; Mid-Atlantic WWTP2 p=0.001; Midwest WWTP1 p≤0.001). At Mid-Atlantic WWTP1 and Mid-Atlantic WWTP2, this statistical significance was sustained when comparing each treatment step to one another using linear contrasts (all p-values were less than 0.05). At Midwest WWTP1, the statistical significance achieved by ANOVA was sustained for all linear contrasts between all treatment steps (all p-values were less than 0.05), except when comparing the activated sludge reactor step with post aeration (p=0.83). At Mid-Atlantic WWTP1, there was a slight increase in presumptive VRE concentrations in the activated sludge reactor from 1.9 × 104 CFU/100 ml in influent to 1.9 × 105 CFU/100 ml in the activated sludge reactor. At all WWTPs, the lowest concentration of VRE was detected in the effluent samples, with no VRE identified in the effluent at Mid-Atlantic WWTP1, Mid-Atlantic WWTP 2, or Midwest WWTP2.
Figure 1:
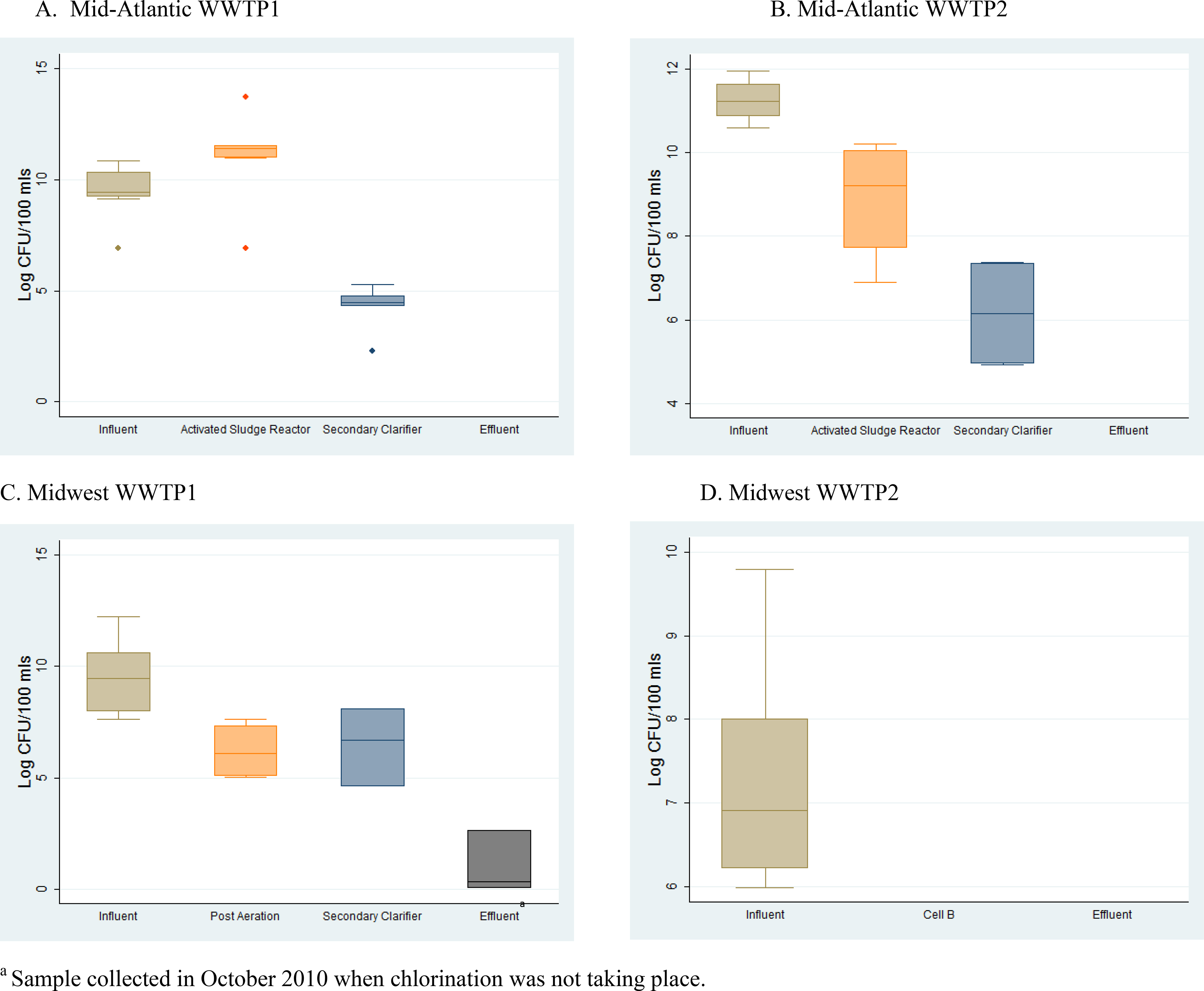
Average concentration of presumptive vancomycin-resistant enterococci (VRE) by wastewater treatment plant and sampling location. Whiskers are drawn from the 75th percentile to the upper adjacent value and from the 25th percentile to the lower adjacent value, the mid-line is the median, and the points represent outliers.
The majority of unique VRE isolates from all WWTPs were identified as E. faecium (78.26%), followed by E. faecalis (17.39%), and E. gallinarum (4.35%) (Table 2).
Table 2.
Number and percentage of vancomycin-resistant enterococci (VRE) isolates by species and wastewater treatment plant
| Number of Isolates(%) | |||||
|---|---|---|---|---|---|
| Enterococcus species | Mid-Atlantic WWTP1 | Mid-Atlantic WWTP2 | Midwest WWTP1 | Midwest WWTP2 | Total |
| E. faecium | 14(77.8) | 0(0) | 3(100) | 1(100) | 18(78.3) |
| E. faecalis | 4(22.2) | 0(0) | 0(0) | 0(0) | 4(17.4) |
| E. gallinarum | 0(0) | 1(100) | 0(0) | 0(0) | 1(4.4) |
3.2. Presence of VIE
VIE were detected at all WWTPs in this study except for Mid-Atlantic WWTP1. No VIE were detected in any of the tertiary-treated (chlorinated) effluent samples. However, similar to VRE, VIE was detected in one effluent sample from Midwest WWTP1 in October 2010 when chlorination was not performed. VIE isolates from all of the WWTPs were identified as E. gallinarum (78.95%) and E. casseliflavus (21.05%).
3.3. Antibiotic resistance patterns
In total, 34 VRE isolates were recovered from the four WWTPs. However, only 23 isolates that could be identified as unique using phenotypic analyses were included in the analysis of antibiotic resistance patterns in order to eliminate bias that would have resulted from including clones. All phenotypically unique VRE isolates, of all species, from all WWTPs were multidrug resistant. The resistance patterns among unique isolates varied by WWTP.
The VR E. faecium isolates (n=14) from Mid-Atlantic WWTP1 were resistant to multiple antibiotics used to treated enterococci infections including penicillin, ciprofloxacin, and streptomycin (Figure 2a). The percentage of resistant VR E. faecium isolates were mostly constant throughout the treatment process at this WWTP, except for an increasing percentage of isolates displaying resistance to streptomycin as the treatment process progressed and a spike in the percentage of isolates resistant to tetracycline in the activated sludge reactor (Figure 2a). Four VR E. faecalis isolates were detected at Mid-Atlantic WWTP1 (one from influent, one from the activated sludge reactor, and two from the secondary clarifier) (Figure 2b). All of these VR E. faecalis isolates were resistant to erythromycin, quinupristin/dalfopristin (synercid), tetracycline, vancomycin, and ciprofloxacin. Quinupristin/dalfopristin (synercid) resistance observed among VR E. faecalis represents intrinsic antibiotic resistance. Three out of four VR E. faecalis isolates from Mid-Atlantic WWTP1 were resistant to gentamicin (one from the activated sludge reactor and two from the secondary clarifier), one of the isolates from the secondary clarifier was resistant to streptomycin, and the one isolate from the influent was resistant to penicillin (Figure 2b).
Figure 2:
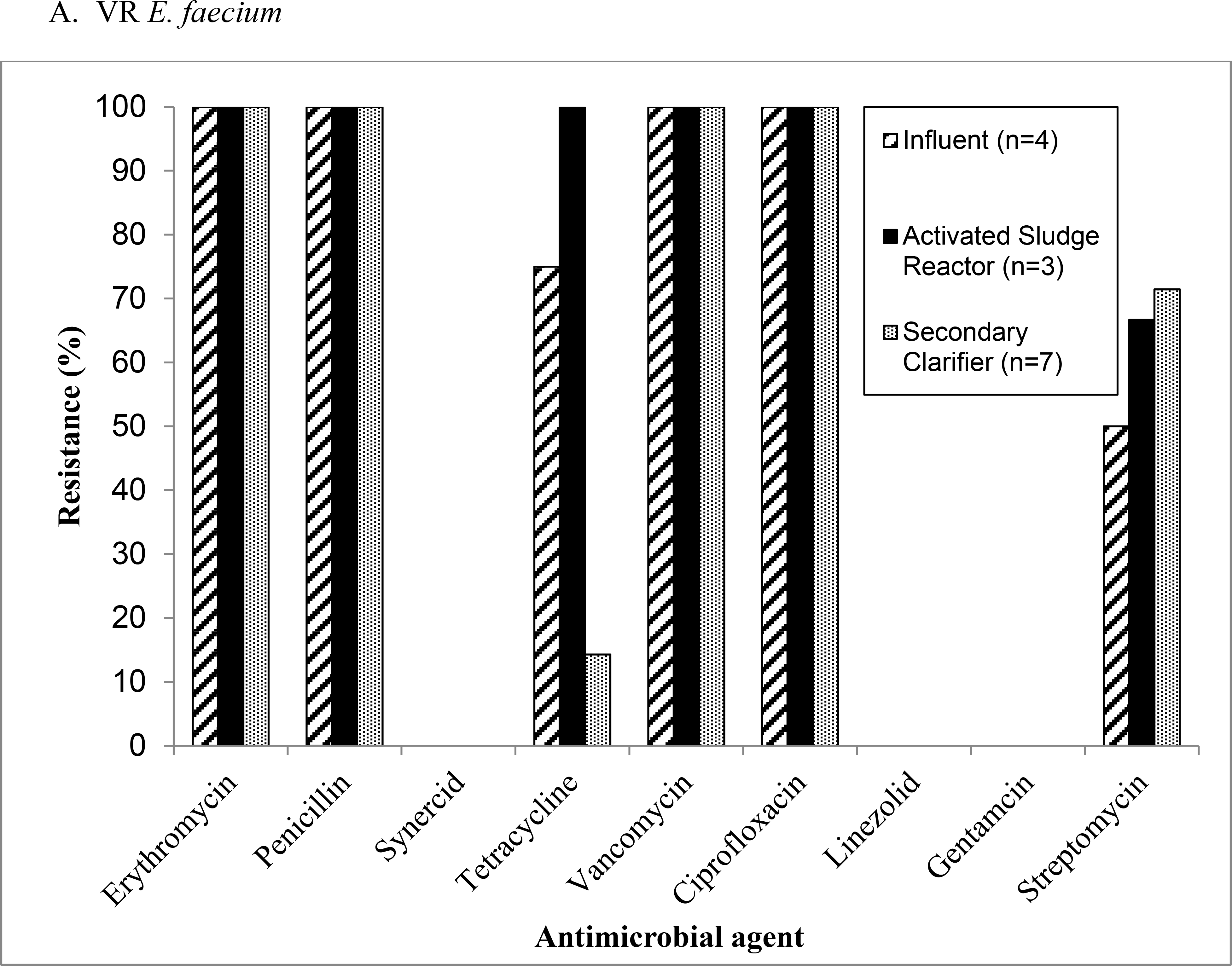
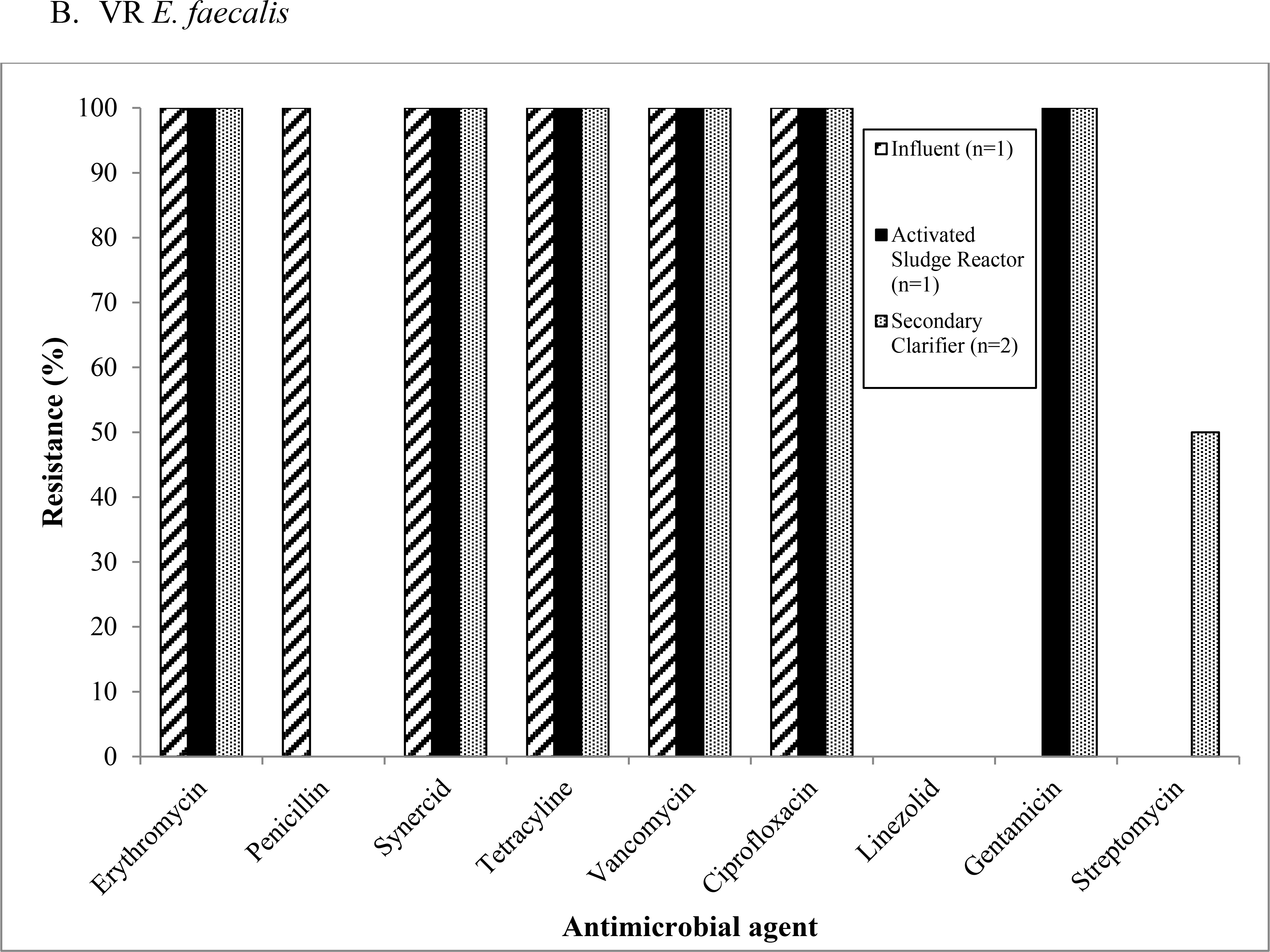
Antimicrobial resistance patterns among (a) vancomycin-resistant Enterococcus (VRE) faecium and (b) VR E. faecalis isolates recovered from Mid-Atlantic WWTP1.
At Mid-Atlantic WWTP2 the one unique VR E. gallinarum isolate was resistant to quinupristin/dalfopristin (synercid) in addition to vancomycin. The three unique VR E. faecium isolates from Midwest WWTP 1 (one from the secondary clarifier, two from the effluent) were all resistant to erythromycin, penicillin, vancomycin, and ciprofloxacin. From Midwest WWTP2, the one VR E. faecium isolate from the influent expressed resistance to erythromycin, penicillin, tetracycline, vancomycin, ciprofloxacin, and streptomycin.
In total, 22 VIE isolates were isolated from Mid-Atlantic WWTP2, Midwest WWTP1, and Midwest WWTP2, however only 19 isolates that could be identified as phenotypically unique were included in the analysis of antibiotic resistance patterns. The isolates were resistant or intermediately resistant to a number of clinically relevant antibiotics including erythromycin, tetracycline, and ciprofloxacin, although patterns differed by treatment plant (Figure 3). At Mid-Atlantic WWTP2 (Figure 3a) and Midwest WWTP1 (Figure 3b) the percentage of resistant or intermediately resistant isolates generally remained the same or increased as treatment progressed. At Midwest WWTP2 (Figure 3c), VIE were only isolated from influent samples.
Figure 3:
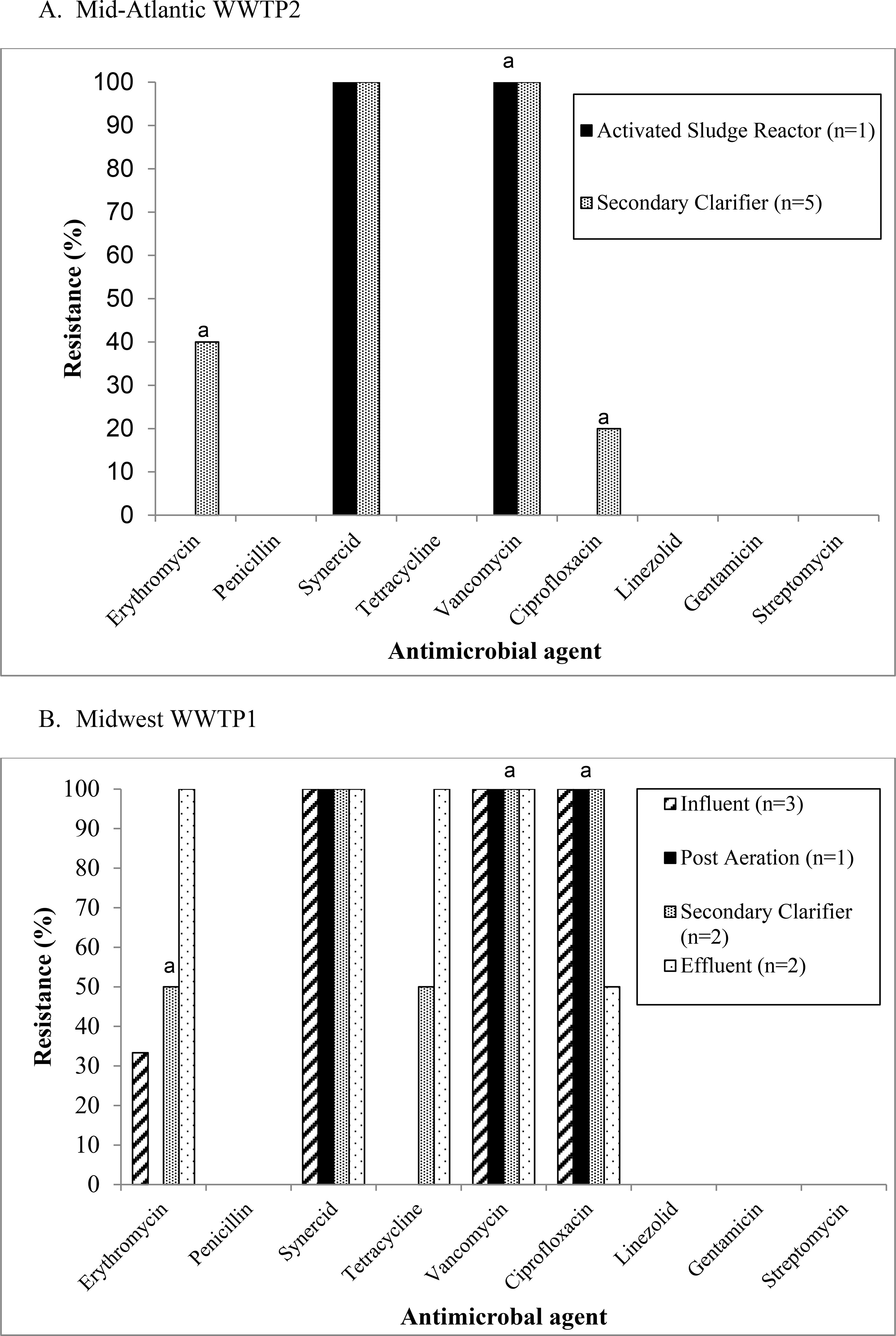
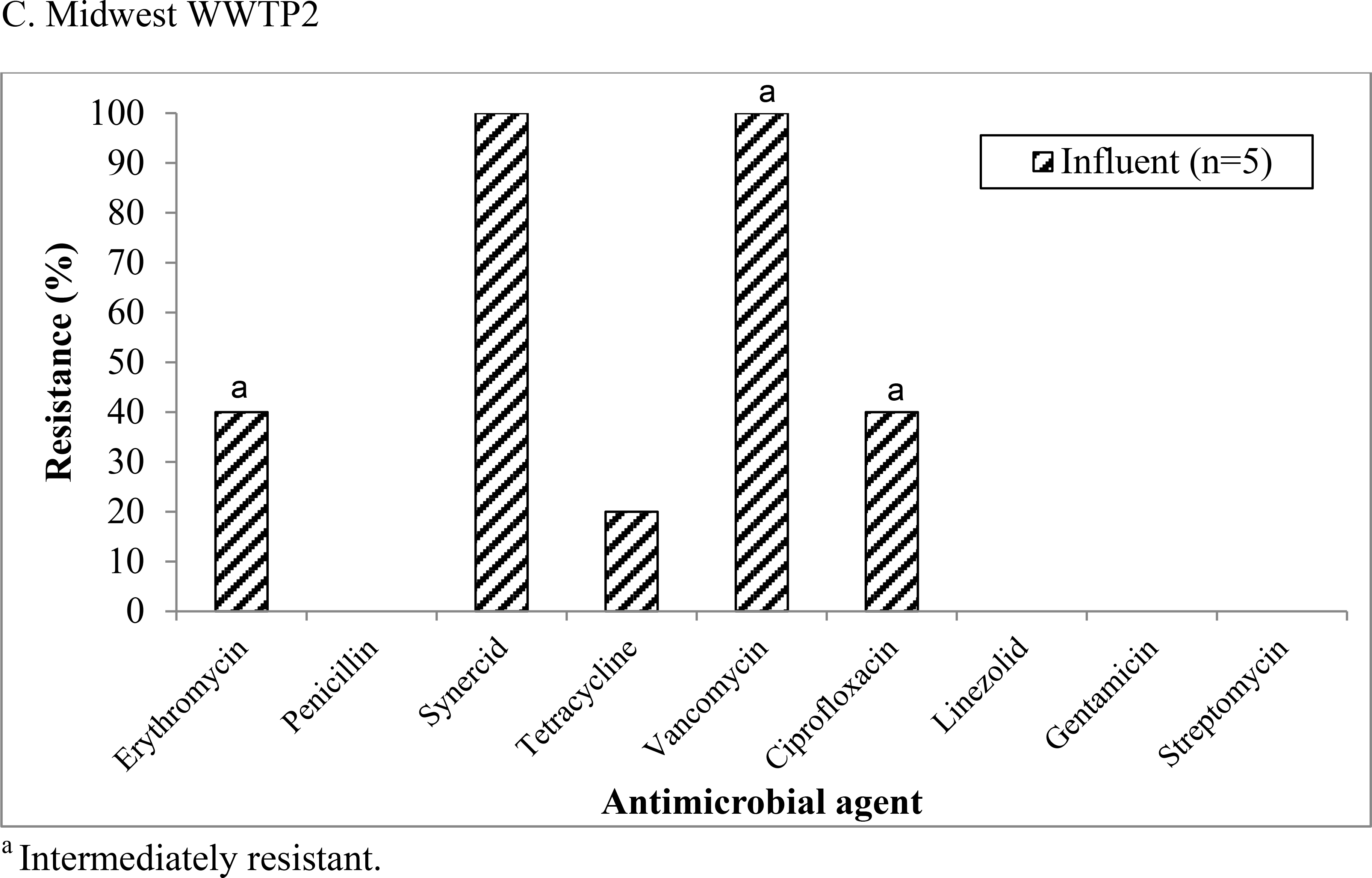
Antimicrobial resistance patterns among vancomycin-intermediate enterococci (VIE) isolates recovered from (a) Mid-Atlantic WWTP2, (b) Midwest WWTP1, and (c) Midwest WWTP2.
4. Discussion
4.1. Occurrence of VRE
Previous studies have detected VRE in wastewater, including effluent samples (Araujo et al., 2010; Caplin et al., 2008; Kotzamanidis et al., 2009; Morris et al., 2012). However, to our knowledge, this is the first study to evaluate the occurrence of VRE at U.S. WWTPs from which treated effluent is reclaimed and reused at spray irrigation sites. We identified VRE in 27% of all samples from all WWTPs, including an un-chlorinated effluent sample from Midwest WWTP1 (a WWTP that chlorinates only in summer). On average, the concentration of VRE at each WWTP decreased as treatment progressed, indicating that wastewater treatment seems to be effective in reducing the numbers of VRE released in final effluent.
The majority of studies assessing the presence of VRE in wastewater have been conducted in Europe and have found VRE prevalence ranging from 2–52% (Kotzamanidis et al., 2009; Luczkiewicz et al., 2010; Morris et al., 2012). Interestingly, VRE was isolated in 2–3% of samples from European secondary WWTPs (Luczkiewicz et al., 2010; Morris et al., 2012), and in 52% of wastewater samples from a tertiary WWTP that uses chlorination (Kotzamanidis et al., 2009). To our knowledge, Nagulapally et al. (2009) is the only published study to have detected VRE at a U.S. municipal WWTP. Nagulapally et al. (2009) isolated VRE from influent and secondary clarifier samples, but not from UV-disinfected effluent samples (Nagulapally et al., 2009). The lack of VRE in UV-disinfected effluent samples in the Nagulapally et al. (2009) study is interesting given the findings of Luczkiewicz et al. (2011) that showed that UV irradiation actually increased levels of antibiotic-resistant enterococci in treated water (Luczkiewicz et al., 2011). In our study we also identified VRE in influent and biologically treated samples (from the activated sludge reactors, post aeration, and secondary clarifiers), but not in tertiary-treated effluent. These findings provide evidence that tertiary treatments involving chlorination (Mid-Atlantic WWTP 1 and Mid-Atlantic WWTP2) or lagooning (Midwest WWTP2) are effective in reducing numbers of VRE to undetectable levels.
Prior studies have also shown that VRE concentrations decrease as wastewater treatment progresses (Kotzamanidis et al., 2009; Morris et al., 2012; Nagulapally et al., 2009). In the current study, we also found that the concentration of VRE decreased as treatment progressed at most WWTPs where samples were collected (Figure 1). Yet, at Mid-Atlantic WWTP1, there was a slight increase in the concentration of VRE in the activated sludge reactor step. Also, at Mid-Atlantic WWTP1 and Mid-Atlantic WWTP2 the proportion of VRE out of total enterococci was highest in the activated sludge reactor step (Table 1). Previous studies have identified an increase in the concentration or percentage of antibiotic-resistant bacteria in the secondary treatment step, and specifically in activated sludge reactors, which decreased with tertiary treatments (Börjesson et al., 2009; Luczkiewicz et al., 2010; Nakamura and Shirota, 1990). The combination of high concentrations of microorganisms, nutrients, oxygen, and antibiotics found in the activated sludge reactor step could both encourage bacterial growth and select for antibiotic-resistant bacteria.
Interestingly, the un-chlorinated effluent sample from Midwest WWTP1 that contained a confirmed VRE isolate is from the same WWTP and sampling date (October 2010) where MRSA was detected in a previous study by our group (Rosenberg Goldstein et al., 2012), suggesting that chlorination plays an important role in reducing the presence of multiple types of antibiotic-resistant bacteria in wastewater. On the other hand, it is important to note that both VRE and MRSA (Rosenberg Goldstein et al., 2012) were only isolated from influent samples at Midwest WWTP2. Although this plant does not utilize chlorination or filtration, the sequencing batch reactor at this plant, along with its series of five lagoons seem to effectively eliminate both VRE and MRSA without the use of disinfection or filtration (Figure 1).
Beyond VRE specifically, it is important to note that vancomycin-susceptible enterococci were isolated from the effluent of all tested treatment plants (Table 1), including those that used chlorination. The survival of enterococci after chlorination during wastewater treatment can be explained by previous studies that have shown enterococci to be extremely hardy microorganisms that not only can survive for long periods of time on environmental fomites but also can tolerate excessive heat, chlorine and certain alcohol concentrations (Arias and Murray, 2012;Bradley and Fraise, 1996). The presence of vancomcyin-susceptible enterococci in treated wastewater effluent has both public health implications for users of reclaimed water and ecological implications for receiving waters.
4.2. Regional differences
The different VRE prevalence rates between WWTPs in the Mid-Atlantic (45%) and Midwest (13%) (p=0.008) observed in this study could be a reflection of geographic differences in human VRE infection rates (Bouchillon et al., 2005; CDDEP, 2013). Since 2004, more VRE has been isolated from humans in the Mid-Atlantic region of the U.S. compared to the Midwest (Bouchillon et al., 2005; CDDEP, 2013). The regional differences in clinically confirmed VRE in the U.S. could be due, in part, to aggressive intervention and control efforts undertaken in the late 1990s at acute and long-term care facilities in Iowa, Nebraska, and South Dakota, including the isolation of patients colonized with VRE and a greater emphasis on handwashing and cleaning (Low et al., 2001; Ostrowsky et al., 2001). The prevalence of VRE in facilities that participated in the intervention program in this region decreased from 2.2% to 0.5% between 1997 and 1999 (Ostrowsky et al., 2001). Beyond regional differences in human VRE infection rates, the Mid-Atlantic and Midwest WWTPs differed by surrounding land uses and types of influent received, which also could help explain the varying VRE prevalence rates observed in this study. The Mid-Atlantic WWTPs are located in urban and suburban settings and receive domestic and hospital influent, whereas both Midwest WWTPs are located in rural areas and receive agriculturally influenced stormwater in addition to domestic wastewater (Rosenberg Goldstein et al., 2012).
4.3. Species diversity
E. faecium was the dominant species of VRE identified in wastewater samples in this study (78.26%). Because wastewater contains human excreta, and E. faecium and E. faecalis are the two species of enterococci that most commonly cause human infections, our findings on species diversity verified our expectations (Table 1) (Hayakawa et al., 2012). Several previous studies in Europe and Iran that have analyzed species diversity among VRE isolated from wastewater also found a greater percentage of VR E. faecium than VR E. faecalis in wastewater (Araujo et al., 2010; Kotzamanidis et al., 2009; Luczkiewicz et al., 2010; Morris et al., 2012; Talebi et al., 2008). In a study of clinical VRE species diversity, Deshpande et al. (2007) also found that 92.8% of VRE isolates from 26 U.S. sites were E. faecium (Deshpande et al., 2007). However, the study by Nagulapally et al. (2009) that detected VRE at one U.S. municipal WWTP in the Midwest region found a greater percentage of E. faecalis among VRE isolates than any other species (Nagulapally et al., 2009).
4.4. Antibiotic resistance patterns
The presence of high-level aminoglycoside resistance in more than half (54.5%) of the enterococci isolated in this study is cause for concern given that the current international guidelines for treatment of Enterococcus infections recommend a combination of ampicillin and an aminoglycoside antibiotic (including gentamicin, streptomycin, and kanamycin) (Fernández-Hidalgo et al., 2013). Fifty percent (9/18) of VR E. faecium isolates from all WWTPs sampled in this study were resistant to streptomycin, and none were resistant to gentamicin. Among the four VR E. faecalis isolates detected at Mid-Atlantic WWTP1, 25% (1/4) were streptomycin-resistant and 75% (3/4) were gentamicin-resistant. Gentamicin-resistance has previously been found to be more prevalent among clinical VR E. faecalis isolates compared to VR E. faecium (Deshpande et al., 2007). Previous studies have also isolated high-level aminoglycoside-resistant enterococci and VRE from wastewater samples and found a wide range of the prevalence of VRE displaying high-level aminoglycoside resistance (Kotzamanidis et al., 2009; Luczkiewicz et al., 2010; Rice et al., 1995; Talebi et al., 2008; Tejedor Junco et al., 2001). Among VR E. faecalis isolates collected from municipal wastewater, Kotzamanidis et al. (2009) reported that 5% had high-level gentamicin resistance and 95% had high-level streptomycin-resistance (Kotzamanidis et al., 2009). Twenty-five percent and 75% of VR E. faecium isolates, respectively, from wastewater and sludge from Portuguese WWTPs were resistant to gentamicin and streptomycin (Araujo et al., 2010).
4.5. Multidrug resistance
Our finding that 100% of the VRE isolates detected in this study were multidrug resistant is consistent with findings from previous studies analyzing VRE from municipal wastewater (Araujo et al., 2010; Kotzamanidis et al., 2009; Talebi et al., 2008). Because multidrug resistance further limits treatment options for infections, this finding is concerning considering potential occupational exposures to VRE in wastewater among WWTP workers.
4.6. Public health and ecological implications
Our study raises possible public health concerns for WWTP workers and for individuals exposed to treated wastewater during reuse. Based on data from numerous studies, WWTP workers report higher levels of gastrointestinal (GI) and respiratory disease symptoms compared to the general population, although there is conflicting evidence about increased infection risks due to specific pathogens identified in wastewater (Glas et al., 2001; Khuder et al., 1998; Seuri et al., 2005; Thorn and Kerekes, 2001). Because VRE was identified at all WWTPs in the current study, WWTP workers could be exposed to both enterococci and VRE through inhalation, dermal, and accidental ingestion exposure routes. The same workers could be exposed to MRSA (Rosenberg Goldstein et al., 2012), as well as Legionella spp. and Aeromonas spp., two important human pathogens more commonly associated with waterborne disease that have previously been isolated from reclaimed water (Brissaud et al., 2008; Jjemba et al., 2010; Palmer et al., 1995). These exposures could potentially result in infections among these workers.
Moreover, because both VRE and MRSA (Rosenberg Goldstein et al. 2012) were identified in the final effluent at Midwest WWTP1, and the effluent from this WWTP is sent to a reuse site for irrigation, individuals exposed to reclaimed water could also be at risk for exposure to both of these microorganisms, as well as other human pathogens that survive the distribution process and persist in reclaimed water. Interventions aimed at reducing VRE exposures and infections in clinical facilities including education and increased handwashing have been found to be effective (Low et al., 2001; Ostrowsky et al., 2001). Improving awareness about VRE, and other human pathogens, at WWTPs and reuse sites and encouraging handwashing could be important interventions to explore to reduce potential exposures among WWTP workers and spray irrigation workers.
Beyond human health impacts, our study findings raise ecological concerns with regard to surface waters that receive treated wastewater effluent (Lata et al., 2009). The release of treated wastewater containing antibiotic-resistant enterococci can result in the dissemination and persistence of bacterial antimicrobial resistance in water and the surrounding ecosystem (Lata et al., 2009). Specifically, antibiotic-resistant enterococci released into the water column can be transmitted to aquatic macrophytes, sediment and soil (Mundt, 1961;Mundt, 1963), subsequently impacting the surrounding ecosystem through the transmission of these microorganisms by animals, fish, insects, wind and rain (Lata et al., 2009). This resulting environmental reservoir of antimicrobial resistance could then serve as a medium for the cycling of antibiotic-resistant strains, such as VRE, between the environment, the food chain and humans (Blanch et al., 2003).
4.7. Limitations
As with all field studies, there are some limitations to this study. If VRE were injured or otherwise present in a viable but nonculturable state, they would not have grown on the vancomycin-amended mEI plates, which are a less hospitable media, possibly resulting in underestimations of VRE occurrence and concentrations (Lleò et al., 2005; Lleò et al., 2001). The concentrations of VRE could also have been underestimated because of the use of grab samples in this study. Moreover, because we sought to generate data on concentrations of enterococci and VRE throughout the wastewater treatment process we did not use enrichment methods to culture the isolates. As a result, the total number of isolates that were recovered in this study was relatively low. Finally, the generalizability of our results is limited because we sampled only four WWTPs in two U.S. regions.
5. Conclusions
To our knowledge, our study is the first to demonstrate the occurrence of VRE at U.S. WWTPs from which effluent is reused at spray irrigation sites. Although the concentration of VRE decreased as treatment progressed at the WWTPs sampled, VRE was isolated in one secondary-treated (un-chlorinated) effluent sample. All unique VRE isolates from all WWTPs sampled in this study were multidrug resistant, and were specifically resistant to a number of antibiotics used to treat VRE infections, including the aminoglycosides gentamicin and streptomycin. WWTP workers and those exposed to secondary-treated (un-chlorinated) reclaimed water, a growing alternative freshwater source, could potentially be exposed to VRE that persist in this water.
Acknowledgements:
We thank the operators at the wastewater treatment plants for their participation and assistance. We thank D. Jack, V. Long, and B. Zappe for performing laboratory analyses. This work was supported by R03 Small Grants Program, Grant #1-R03-OH009598-01 from the National Institute for Occupational Safety and Health, Centers for Disease Control and Prevention (CDC). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of CDC. The Maryland Water Resources Research Center also provided a summer fellowship to R.E.R.G. that supported this work.
Footnotes
Abbreviations: Vancomycin-intermediate enterococci (VIE); vancomycin-resistant enterococci (VRE); vancomycin-resistant S. aureus (VRSA); wastewater treatment plant (WWTP)
References
- Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 2012; 10: 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo C, Torres C, Silva N, Carneiro C, Gonclaves A, Radhouani H, et al. Vancomycin-resistant enterococci from Portugese wastewater treatment plants. J Basic Microbiol 2010; 50. [DOI] [PubMed] [Google Scholar]
- Blanch AR, Caplin JL, Iversen A, Kuhn I, Manero A, Taylor HD, Vilanova X. Comparison of enterococcal populations related to urban and hospital wastewater in various climatic and geographic European regions. J Appl Microbiol 2003; 94: 994–1002. [DOI] [PubMed] [Google Scholar]
- Bouchillon SK, Hoban DJ, Johnson BM, Johnson JL, Hsiung A, Dowzicky MJ. In vitro activity of tigecycline against 3989 Gram-negative and Gram-positive clinical isolates from the United States Tigecycline Evaluation and Surveillance Trial (TEST Program; 2004). Diagn Microbiol Infect Dis 2005; 52: 173–179. [DOI] [PubMed] [Google Scholar]
- Bradley CR, Fraise AP. Heat and chemical resistance of enterococci. J Hosp Infect 1996; 34: 191–196. [DOI] [PubMed] [Google Scholar]
- Brissaud F, Blin E, Hemous S, L G. Water reuse for urban landscape irrigation: aspersion and health related regulations. Water Sci Technol 2008; 57: 781–787. [DOI] [PubMed] [Google Scholar]
- Börjesson S, Melin S, Matussek A, Lindgren P-E. A seasonal study of the mecA gene and Staphylococcus aureus including methicillin-resistant S. aureus in a municipal wastewater treatment plant. Water Res 2009; 43: 925–932. [DOI] [PubMed] [Google Scholar]
- Caplin JL, Hanlon GW, Taylor HD. Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ Microbiol 2008; 10: 885–892. [DOI] [PubMed] [Google Scholar]
- CDC. CDC Reminds Clinical Laboratories and Healthcare Infection Preventionists of their Role in the Search and Containment of Vancomycin-Resistant Staphylococcus aureus (VRSA). Centers for Disease Control and Prevention, Division of Healthcare Quality Promotion (DHQP), Healthcare-associated Infections (HAIs), Atlanta, GA, 2012. http://www.cdc.gov/HAI/settings/lab/vrsa_lab_search_containment.html [Accessed 24 October 2012]. [Google Scholar]
- CDC. Information for the public about VRE, 2008. http://www.cdc.gov/ncidod/dhqp/ar_VRE_publicFAQ.html [Accessed 8 September 2009].
- CDDEP. Vancomycin-Resistant Enterococcus. Resistance Map. 2013. The Center for Disease Dynamics, Economics & Policy, Washington, D.C., 2013. http://www.cddep.org/ResistanceMap/bug-drug/VRE#.US4_pVcp4iP [Accessed 25 February 2013]. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA, 2013. [Google Scholar]
- Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 2007; 58: 163–170. [DOI] [PubMed] [Google Scholar]
- EPA. Method 1600: Enterococci in Water by Membrane Filtration Using membrane-Enterococcus Indoxyl-B-D-Glucoside Agar (mEI). 2002.
- EPA . 2012 guidelines for water reuse. EPA 600-R-12–618. Washington, DC. 2012. [Google Scholar]
- Fernández-Hidalgo N, Almirante B, Gavaldà J, Gurgui M, Peña C, de Alarcón A, et al. Ampicillin plus ceftriaxone is as effective as ampicillin plus gentamicin for treating Enterococcus faecalis infective endocarditis. Clin Infect Dis 2013. [DOI] [PubMed] [Google Scholar]
- Glas C, Hotz P, Steffen R. Hepatitis A in workers exposed to sewage: a systematic review. Occup Environ Med 2001; 58: 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardabassi L, Dalsgaard A. Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl Environ Microbiol. 2004; 70: 984–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood VJ, Brownell M, Perusek W, Whitlock JE. Vancomycin-resistant Enterococcus spp. isolated from wastewater and chicken feces in the United States. Appl Environ Microbiol. 2001; 67: 4930–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa K, Marchaim D, Martin ET, Tiwari N, Yousuf A, Sunkara B, et al. Comparison of the clinical characteristics and outcomes associated with vancomycin-resistant Enterococcus faecalis and vancomycin-resistant E. faecium bacteremia. Antimicrob Agents Chemother 2012; 56: 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: Antimicrobial‐resistant pathogens associated with healthcare‐associated infections: Annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol 2008; 29: 996–1011. [DOI] [PubMed] [Google Scholar]
- Jjemba PK, Weinrich LA, Cheng W, Giraldo E, LeChevallier MW. Regrowth of potential opportunistic pathogens and algae in reclaimed-water distribution systems. Appl Environ Microbiol 2010; 76: 4169–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuder SA, Arthur T, Bisesi MS, Schaub EA. Prevalence of infectious diseases and associated symptoms in wastewater treatment workers. Am J Ind Med 1998; 33: 571–577. [DOI] [PubMed] [Google Scholar]
- Kim S, Aga DS. Potential ecological and human health impacts of antibiotics and antibiotic-resistant bacteria from wastewater treatment plants. J Toxicol Environ Health B Crit Rev 2007; 10: 559–573. [DOI] [PubMed] [Google Scholar]
- Kotzamanidis C, Zdragas A, Kourelis A, Moraitou E, Papa A, Yiantzi V, et al. Characterization of vanA-type Enterococcus faecium isolates from urban and hospital wastewater and pigs. J Appl Microbiol 2009; 107: 997–1005. [DOI] [PubMed] [Google Scholar]
- Lata P, Ram S, Agrawal M, Shanker R. Real time PCR for the rapid detection of vanA gene in surface waters and aquatic macrophyte by molecular beacon probe. Environ Sci Technol 2009; 43: 3343–3348. [DOI] [PubMed] [Google Scholar]
- Lleò MdM, Bonato B, Benedetti D, Canepari P. Survival of enterococcal species in aquatic environments. FEMS Microbiol Ecol 2005; 54: 189–196. [DOI] [PubMed] [Google Scholar]
- Lleò MM, Bonato B, Tafi MC, Signoretto C, Boaretti M, Canepari P. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J Appl Microbiol 2001; 91: 1095–1102. [DOI] [PubMed] [Google Scholar]
- Low DE, Keller N, Barth A, Jones RN. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: Results from the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis 2001; 32: S133–S145. [DOI] [PubMed] [Google Scholar]
- Luczkiewicz A, Jankowska K, Fudala-Ksiazek S, Ola nczuk-Neyman K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Wat Res 2010; 44: 5089–5097. [DOI] [PubMed] [Google Scholar]
- Luczkiewicz A, Jankowska K, Bray R, Kulbat E, Quant B, Sokolowska A, Olanczuk-Neyman K. Antimicrobial resistance of fecal indicators in disinfected wastewater. Water Sci Technol 2011; 64: 2352–2361. [DOI] [PubMed] [Google Scholar]
- Mainous I, Arch G., Diaz VA, Matheson EM, Gregorie SH, Hueston WJ. Trends in hospitalizations with antibiotic-resistant infections: U.S., 1997–2006. Public Health Rep 2011; 126: 354–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef SA, Rosenberg Goldstein RE, George A, Ewing L, Tall BD, Boyer MS, et al. Diversity, distribution and antibiotic resistance of Enterococcus spp. recovered from tomatoes, leaves, water and soil on Mid-Atlantic farms. Food Microbiol 2013; 10.1016/j.fm.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Morris D, Galvin S, Boyle F, Hickey P, Mulligan M, Cormican M. Enterococcus faecium of the vanA genotype in rural drinking water, effluent, and the aqueous environment. Appl Environ Microbiol 2012; 78: 596–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt JO. Occurrence of enterococci: Bud, blossom, and soil studies. Appl Microbiol 1961; 9: 541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt JO. Occurrence of enterococci on plants in a wild environment. Appl Microbiol 1963; 11: 141–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagulapally SR, Ahmad A, Henry A, Marchin GL, Zurek L, Bhandari A. Occurence of ciprofloxacin-, trimethoprim-sulfamethoxazole-, and vancomycin-resistant bacteria in a municipal wastewater treatment plant. Water Environ Res 2009; 81: 82–90. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Shirota H. Behavior of drug resistant fecal coliforms and R plasmids in a wastewater treatment plant. Nippon Koshu Eisei Zasshi 1990; 37: 83–90. [PubMed] [Google Scholar]
- NIAID. Vancomycin-Resistant Enterococci (VRE). 2009. http://www.niaid.nih.gov/topics/antimicrobialresistance/examples/vre/Pages/overview.aspx [Accessed 24 October 2012]
- Ostrowsky BE, Trick WE, Sohn AH, Quirk SB, Holt S, Carson LA, et al. Control of vancomycin-resistant Enterococcus in health care facilities in a region. N Engl J Med 2001; 344: 1427–1433. [DOI] [PubMed] [Google Scholar]
- Palmer CJ, Bonilla GF, Roll B, Paszko-Kolva C, Sangermano LR, Fujioka RS. Detection of Legionella species in reclaimed water and air with the EnviroAmp Legionella PCR kit and direct fluorescent antibody staining. Appl Environ Microbiol 1995; 61: 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole TL, Hume ME, Campbell LD, Scott HM, Alali WQ, Harvey RB. Vancomycin-resistant Enterococcus faecium strains isolated from community wastewater from a semiclosed agri-food system in Texas. Antimicrob Agents Chemother. 2005; 49: 4382–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice EW, Messer JW, Johnson CH, Reasoner DJ. Occurrence of high-level aminoglycoside resistance in environmental isolates of enterococci. Appl Environ Microbiol 1995; 61: 374–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg Goldstein RE, Micallef SA, Gibbs SG, Davis JA, He X, George A, et al. Methicillin-resistant Staphylococcus aureus (MRSA) detected at four U.S. wastewater treatment plants. Environ Health Perspect 2012; 120: 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuri M, Koivunen J, Granfors K, Heinonen-Tanski H. Work-related symptoms and Salmonella antibodies among wastewater treatment plant workers. Epidemiol Infect. 2005; 133: 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon KE, Lee DY, Trevors JT, Beaudette LA. Application of real-time quantitative PCR for the detection of selected bacterial pathogens during municipal wastewater treatment. Sci Total Environ 2007; 382: 121–129. [DOI] [PubMed] [Google Scholar]
- Sievert DM, Rudrik JT, Patel JB, McDonald LC, Wilkins MJ, Hageman JC. Vancomycin-resistant Staphylococcus aureus in the United States, 2002–2006. Clin Infect Dis 2008; 46: 668–674. [DOI] [PubMed] [Google Scholar]
- Talebi M, Rahimi F, Katouli M, Mollby R, Pourshafie MR. Epidemiological link between wastewater and human vancomycin-resistant Enterococcus faecium Isolates. Curr Microbiol 2008; 56: 468–473. [DOI] [PubMed] [Google Scholar]
- Tejedor Junco MT, González Martín M, Pita Toledo ML, Lupiola Gómez P, Martín Barrasa JL. Identification and antibiotic resistance of faecal enterococci isolated from water samples. Int J Hyg Environ Health 2001; 203: 363–368. [DOI] [PubMed] [Google Scholar]
- Thorn J, Kerekes E. Health effects among employees in sewage treatment plants: A literature survey. Am J Ind Med 2001; 40: 170–179. [DOI] [PubMed] [Google Scholar]
- Uttley A, Collins C, Naidoo J, George R. Vancomycin-resistant enterococci [letter]. Lancet 1988; 1: 57–58. [DOI] [PubMed] [Google Scholar]
- Wegener H, Aarestrup FM, Jensen L, Hammerum A, Bager F. Use of antimicrobial growth promoters in food animals and Enterococcus faecium resistance to therapeutic antimicrobial drugs in Europe. Emerg Infect Dis 1999; 5: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]


