Abstract
Background
Oral squamous cell carcinoma (OSCC) is a common tumor of the head and neck. Its treatment usually requires multiple modalities. Currently, there are no molecular biomarkers to guide these treatment strategies. Studies have shown that microfibril-associated protein 4 (MFAP4) is potentially useful for non-invasive assessment of various diseases; however, its biological function in tumors is still unknown. In this study, we propose that MFAP4 is a new prognostic target for OSCC.
Material/Methods
First, we collected OSCC data (GSE25099 and GSE30784 datasets) from the Gene Expression Omnibus (GEO) database and compared the differential expression of MFAP4 gene between the patients (tumor) and normal (control) groups. The comparison was done with University of California Santa Cruz Xena (https://xenabrowser.net/Datapages/), and we calculated the difference in MFAP4 gene expression between normal and tumor tissues in a pan-cancer analysis. Then, we compared the 2 groups with high and low expression of MFAP4 gene in terms of tumor mutation burden (TMB), miRNA regulation, and immune cell infiltration.
Results
We found that the expression of MFAP4 gene was significantly decreased in tumors. Our research also showed that high expression of MFAP4 was related to better prognosis of patients and may be related to tumor gene mutation, miRNA regulation, and infiltration of different immune cells.
Conclusions
Our work provides evidence that expression of MFAP4 can be used as a prognostic biomarker for risk stratification of OSCC patients and elaborates on its relation with the regulation of TMB, miRNAs, and immune cell infiltration.
Keywords: MFAP4 Protein, Human; Mouth Neoplasms; Prognosis
Background
Oral squamous cell carcinoma (OSCC) is one of the most common tumors of the head and neck. There are more than 300 000 new confirmed cases worldwide each year [1,2]. It is estimated that OSCC accounts for about 3% of all malignant tumors, and more than 13 000 patients with OSCC die in the United States each year [3]. The histological grade of OSCC can range from highly differentiated keratinizing carcinoma to undifferentiated non-keratinizing carcinoma with high metastatic tendency. Risk factors for OSCC include smoking, drinking, viral infections (eg, Epstein-Barr virus, human papillomavirus, and herpes simplex virus), chewing betel nuts, occupational exposure to carcinogens, immune deficiency, radiation, diet, and genetic susceptibility [4,5]. The treatment of OSCC is generally multimodal, including surgery followed by chemoradiotherapy (CRT). The epidermal growth factor receptor (EGFR) monoclonal antibody cetuximab in combination with radiation can be used in HPV-negative OSCC where comorbidities prevent the use of cytotoxic chemotherapy. Immune checkpoint inhibitors such as pembrolizumab and nivolumab also play an important role in the treatment of OSCC and can be used for treatment of recurrent or metastatic OSCC and pembrolizumab is used as a primary treatment for unresectable tumors [6]. Currently, there are no molecular biomarkers to guide these treatment strategies.
Microfibril-associated protein 4 (MFAP4) is an extracellular matrix protein belonging to the fibrinogen-related domain family. This family includes several proteins involved in tissue homeostasis and innate immunity, such as fibrinogen C domain-containing protein 1 (FIBCD1), fibrin, and angiogenin [7–9]. MFAP4 was initially identified as a gene that is commonly missing in patients with Smith-Magenis syndrome [10]. Studies have shown that MFAP4 can be useful for non-invasive assessment of various diseases, such as chronic obstructive pulmonary disease [11,12], liver cirrhosis [13,14], diabetic neuropathy [15], and cardiovascular complications [16]. However, the biological function of MFAP4 in tumors is still unknown. It is worth noting that a recent study reported that high expression of MFAP4 mRNA was significantly associated with decreased overall survival of patients with neuroblastoma. However, the prognostic value of MFAP4 expression in OSCC patients remains unclear.
In this study, we propose MFAP4 as a new prognostic target for OSCC. We also studied its relationship with the regulation of tumor mutation burden (TMB), miRNAs, and immune cell infiltration.
Material and Methods
Differences in Expression of MFAP4
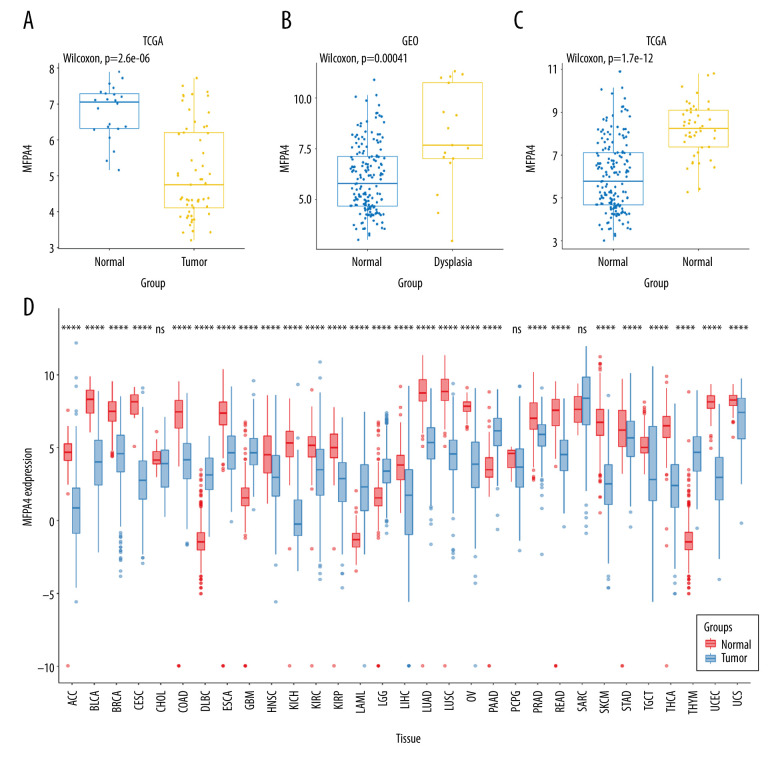
We used the GEOquery package to download 2 oral cancer datasets – GSE25099 and GSE30784 – from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo). The GSE25099 dataset included 57 tumor samples and 22 normal samples, and the GSE30784 dataset included 17 samples of dysplasia, 45 normal samples, and 167 tumor samples. We compared the differences in MFAP4 expression between normal and tumor samples. In addition, we downloaded the transcripts per kilobase million values from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression databases, which were uniformly processed with the Toil process of the University of California Santa Cruz (UCSC) Xena tool (https://xenabrowser.net/datapages/), and we compared the differences between groups (Figure 1). A Wilcoxon test was used to compare the difference between MFAP4 expression in the tumor and normal groups, and the ggplot2 package was used for drawing graphs.
Figure 1.
Expression of MFAP4 in tumors. The GEO and TCGA datasets showed that the expression of MFAP4 gene in tumors was lower than that in normal samples (A–C). Pan-cancer analysis showed that MFAP4 gene had significantly lower expression in multiple tumors (D).
Survival analysis
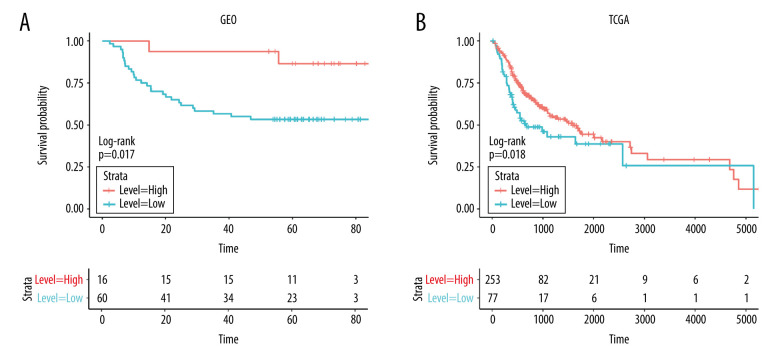
We directly downloaded the head and neck squamous cell carcinoma (HNSC) head and neck cancer expression matrix (HTSeq-Counts and HTSeq-FPKM files) and clinical information from UCSC Xena (https://xenabrowser.net/datapages/). After we eliminated non-oral cancer and incomplete OS samples, 330 samples were included in the study as the experimental group. At the same time, we downloaded 76 samples of oral cancer patients from the GSE41613 dataset as a verification group. GEOquery package was used for data download. We performed survival analysis on the 2 groups of samples (Figure 2) to assess the relationship between difference in survival and expression of MFAP4 between normal and tumor samples. Finally, we used tableone package to compare the differences between clinical phenotypes of TCGA after grouping (Supplementary Table 1). Survival and survminer packages were used for best separation grouping and survival analysis.
Figure 2.
(A, B) The prognostic relationship between MFAP4 and OSCC. In the TCGA and GEO datasets, high expression of MFAP4 was related to better prognosis of patients.
Difference Analysis and Enrichment Analysis
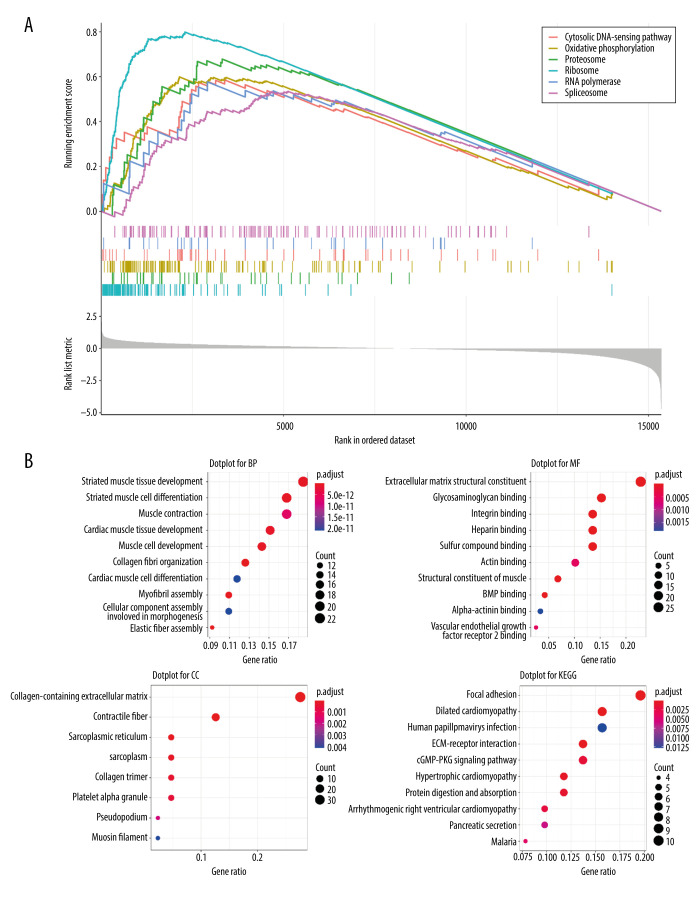
We divided the 330 samples from the TCGA dataset into 2 groups based on high or low expression of MFAP4 (high: 253, low: 77). Then, we performed gene set enrichment analysis (GSEA), gene ontology (GO) enrichment analysis, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. EdgeR package was used for differential analysis, and clusterProfiler package was used for enrichment analysis.
Relationship Between MFAP4 Expression and TMB
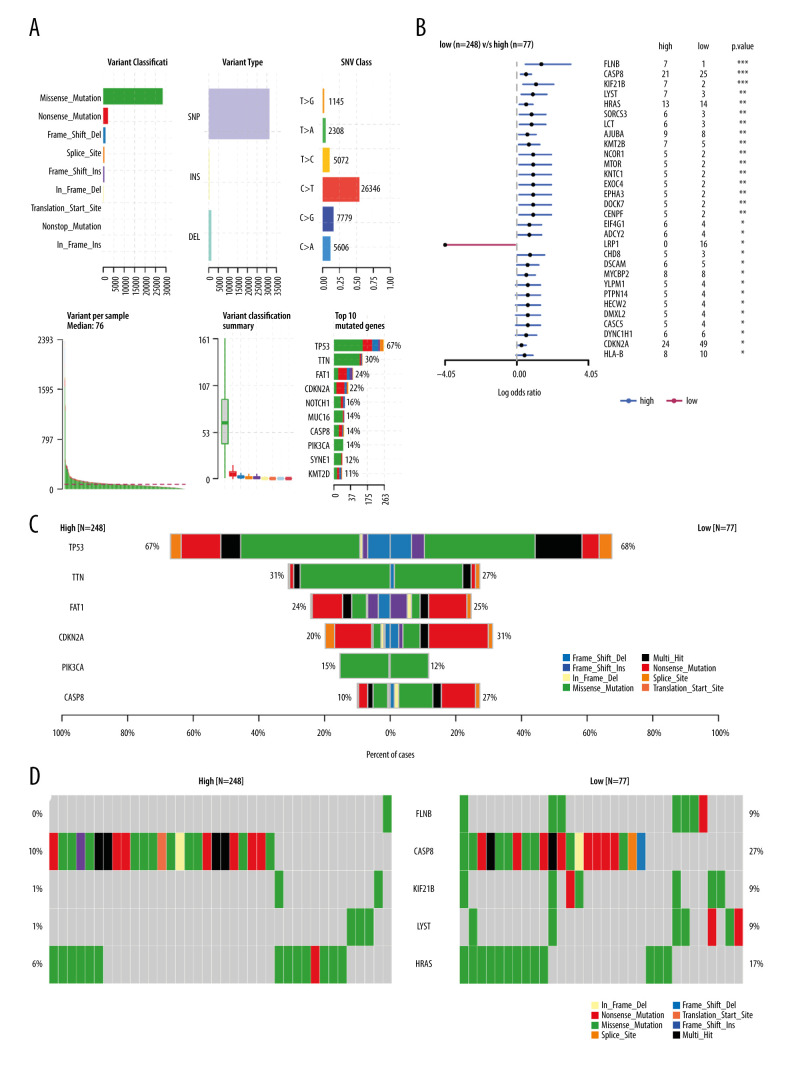
TMB is becoming a new predictive biomarker for selecting patients who will benefit from immune checkpoint inhibitor therapy [17–19]. It is usually defined as the total number of somatically encoded mutations, and it is related to the emergence of new antigens that trigger anti-tumor immunity [20–22]. We downloaded the Mutation Annotation Format file for storing somatic variant information of HNSC patients from the TCGA database, and we screened tumor mutation data from 330 OSCC samples. We compared the mutation data of the high and low MFAP4 expression groups and mapped the mutation data of the top 6 genes. The highly mutated genes in the high MFAP4 expression group were screened, and the first 5 mutated genes were compared and mapped. Maftools package was used to analyze mutation data [23].
Relationship Between MFAP4 Expression and MicroRNA
We used the TCGAbiolinks package to download the miRNA expression profiles of OSCC samples and divided them into 2 groups (high MFAP4 expression: 251, low MFAP4 expression: 75). Then, edgeR package was used for differential analysis to identify miRNAs that were related to MFAP4, and to analyze the correlation between them.
Relationship Between MFAP4 and Immune Cells
Single-sample GSEA (ssGSEA) is a method for quantifying infiltrating immune cells [24]. We analyzed data on immune cell infiltration of OSCC samples in TCGA and GEO databases, based on the most commonly used immune cell marker genes [25]. Differences in immune cell infiltration between the low- and high-expression groups were identified. Gene Set Variation Analysis package was used to quantify 24 types of immune cells. We compared the differences in quantified immune cells between the 2 groups and selected different immune cells in TCGA and GEO databases, including eosinophils, macrophages, mast cells, natural killer (NK) cells, T cells, T helper cells, and effector memory T (Tem) cells.
Correlation of MFAP4 and Immune Checkpoints
Immune checkpoints were reported in previous research, and we evaluated the relationship between MFAP4 and immune checkpoints to assess the importance of MFAP4. The relationship was evaluated with Pearson correlation.
Ethics Statement
The data of this study are from GEO and TCGA datasets, and did not involve animal experiments or human specimens. There were no ethics-related issues.
Results
MFAP4 Expression Was Low in Tumor Samples
We performed Wilcoxon testing to assess the differential expression of MFAP4 between tumor samples and normal samples in the GSE25099 and GSE30784 datasets. We found that the expression of MFAP4 was lower in tumor samples, and box plots were drawn to show the difference. TCGA network researchers described the molecular characteristics of tumors in 11 160 patients with 33 cancer types and identified many molecular subtypes. In these cancers, we calculated the difference in expression of MFAP4 between tumor and normal samples, and the results are shown in Figure 1.
Analysis of Relationship Between survival and MFAP4 Expression
In addition to the 330 samples included in the study as the experimental group, we downloaded 76 samples of oral cancer patients from the GSE41613 dataset as a verification group. We drew the survival curve of MFAP4 expression between the 2 groups and found that high expression of MFAP4 gene was related to better prognosis of patients (Figure 2).
GSEA, GO terms, and KEGG Pathway Analysis
After performing GSEA on both the high and low MFAP4 expression groups, we selected the first 6 enriched pathways for mapping (Figure 3A). The cytosolic DNA-sensing, oxidative phosphorylation, proteasome, ribosome, RNA polymerase, and spliceosome pathways were enriched in GSEA. Finally, we performed functional enrichment GO and KEGG analyses on the genes that differed (criteria were |logFC| >2 and P<0.05; a total of 121 differentially expressed genes were included) (Figure 3B). In GO analysis, genes that were involved in cell functions such as striated muscle tissue development, extracellular matrix structural constituents, and collagen-containing extracellular matrix were enriched. In KEGG pathway analysis, genes mainly involved in focal adhesion, dilated cardiomyopathy, and human papillomavirus infection were enriched.
Figure 3.
Various enrichment analysis results. (A) The cytosolic DNA-sensing, oxidative phosphorylation, proteasome, ribosome, RNA polymerase, and spliceosome pathways were enriched in GSEA. (B) GO was enriched in cell functions such as striated muscle tissue development, extracellular matrix structural constituent, and collagen-containing extracellular matrix. KEGG was mainly enriched in focal adhesion, dilated cardiomyopathy, and human papillomavirus infection.
Relationship Between MFAP4 and TMB
We downloaded 330 OSCC samples and data on OSCC tumor mutations from the TCGA database (Figure 4A). Maftools package was used to calculate the TMB between the high and low MFAP4 expression groups, and to compare the differences between the groups. We also compared and plotted the graph of the top 6 mutation groups (Figure 4B). Forest plots were used to show the differential mutation genes between the high and low MFAP4 expression groups (Figure 4C). Compared with the low expression group, the top 10 highly mutated genes in the high MFAP4 expression group were FLNB, CASP8, KIF21B, LYST, HRAS, LCT, SORCS3, AJUBA, KMT2B, and CENPF. Finally, we compared the data of the first 5 mutant genes between the 2 groups. The results are shown in Figure 4D.
Figure 4.
The relationship between MFAP4 and TMB. (A) Overview of mutation data of OSCC tumors in the TCGA database. (B) Group comparison of the first 6 mutant genes. (C) Forest plot of genes that were highly mutated in the high MFAP4 expression group compared to the low expression group. (D) Comparison of mutation data of the top 5 mutant genes between the high and low MFAP4 expression groups.
Identification of miRNAs Related to MFAP4
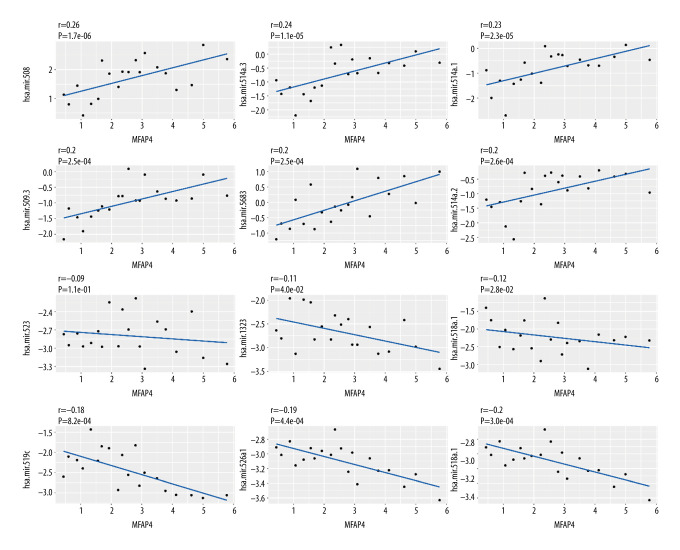
With the criteria set as |logFC| >2 and P value <0.05, 23 differential miRNAs were included in the Pearson correlation analysis of the relationship between miRNA and MFAP4 in both the high- and low-MFAP4 expression groups. We determined the positive correlation and negative correlation of the top 6 miRNAs, and the results are shown in Figure 5.
Figure 5.
Correlation between miRNA and MFAP4. The top 6 positively correlated miRNA and top 6 negatively correlated miRNA showed good linear relationships with MFAP4.
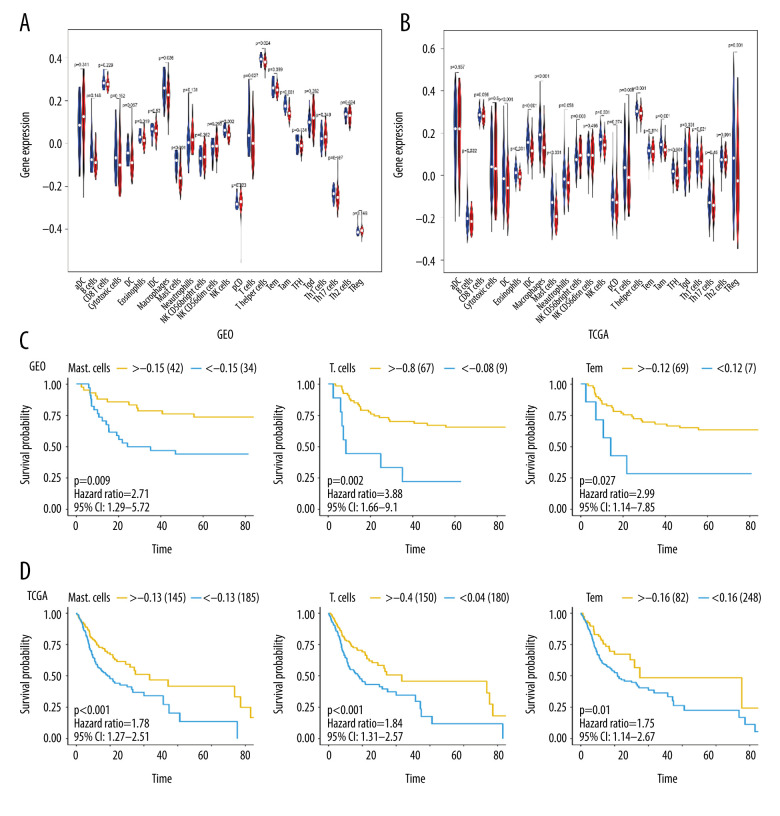
Relationship Between MFAP4 and Immune Infiltration
We used ssGSEA to quantify immune cell infiltration in OSCC samples from TCGA and GEO databases, and compared the high- and low-MFAP4 expression groups (Figure 6A, 6B). After grouping the samples according to best separation and analyzing the survival information of each patient with respect to the type of infiltrating immune cell, we found that 3 immune cell types (mast cells, T cells, and Tem cells) were related to survival (Figure 6C, 6D).
Figure 6.
The relationship between MFAP4 gene expression and tumor immune cell infiltration. (A, B) Differences in immune cell infiltration between samples from TCGA and GEO databases. Seven immune cells showed statistical difference in the MFAP4 expression group. (C, D) The infiltration of 3 immune cells was related to the prognosis of the patients.
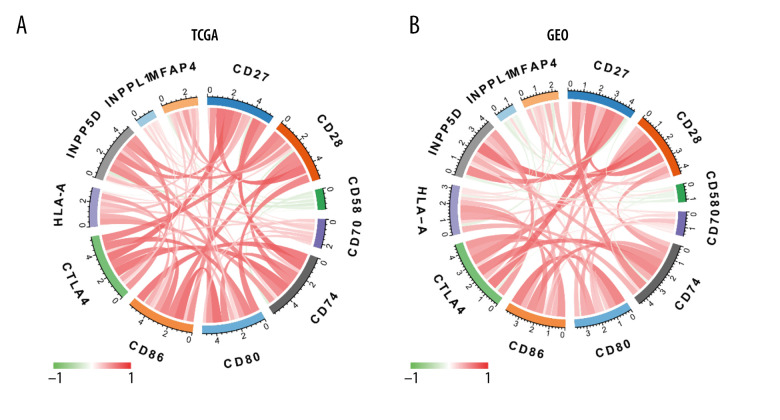
Correlation of MFAP4 and Immune Checkpoints
With Spearman’s rank correlation analysis, we used the most commonly recognized and used immune cell markers to calculate the correlation of immune checkpoints with the MFAP4 gene [25]. These immune checkpoints included CD28 (GEO, r=0.332; TCGA, r=0.532), CD80 (GEO, r=−0.111; TCGA, r=0.221), CD86 (GEO, r=0.030; TCGA, r=0.291), CTLA4 (GEO, r=0.235; TCGA, r=0.221), INPP5D (GEO, r=0.296; TCGA, r=0.376), INPPL1 (GEO, r=0.213; TCGA, r=0.266), CD58 (GEO, r=−0.140; TCGA, r=−0.198), CD27 (GEO, r=0.399; TCGA, r=0.403), CD70 (GEO, r=0.045; TCGA, r=0.057), HLA-A (GEO, r=−0.198; TCGA, r=−0.079), and CD74 (GEO, r=0.352; TCGA, r=0.235) (Figure 7). We drew a graph to show the correlation of the relationship of each 2 genes; the color and width of each line represent the correlation coefficient.
Figure 7.
(A, B) Diagram of the relationship between MFAP4 and immune targets.
Discussion
OSCC is one of the most common tumors of the head and neck and seriously affects health. Its treatment usually requires multiple modalities, including surgery followed by chemoradiotherapy (CRT), epidermal growth factor receptor (EGFR) monoclonal antibody combined with radiation and immune checkpoint inhibitors, but patients often have a poor prognosis nonetheless. Therefore, it is very important to determine new therapeutic targets for the treatment of OSCC patients. This study proposed that MFAP4 is a new prognostic target for OSCC and also studied its relation with the regulation of TMB, miRNAs, and immune cell infiltration.
MFAP4 is a stromal cell protein belonging to the fibrinogen-associated protein superfamily. This family also includes ficolins, fibroleukin, FIBCD1, angiopoietins, and tenascins, which play multifaceted roles in innate immunity, development of the cardiovascular system, and normal functioning of the endothelium [26,27]. However, there are no relevant reports on the role of MFAP4 in OSCC. In this study, we found that the MFAP4 gene showed low expression in oral cancer tumor samples and other tumors, and high expression of MFAP4 was related to better prognosis.
After establishing this connection between MFAP4 gene and OSCC, we then further explored the mechanism of influence of MFAP4 on OSCC and found that the cytosolic DNA-sensing, oxidative phosphorylation, proteasome, ribosome, RNA polymerase, and spliceosome pathways were enriched on GSEA. At the same time, we assessed differences in enrichment with GO and KEGG functions. GO was enriched in cell functions such as striated muscle tissue development, extracellular matrix structural constituents, and collagen-containing extracellular matrix. KEGG analysis showed enrichment in focal adhesion, dilated cardiomyopathy, and human papillomavirus infection. Therefore, studying these pathways will help to clarify the underlying mechanisms of the development, proliferation, and invasion of oral cancer cells and also help to predict the progression of cancer.
TMB is becoming a new predictive biomarker for selecting patients who will benefit from immune checkpoint inhibitor therapy [17–19]. It is usually defined as the total number of somatically encoded mutations and is related to the emergence of new antigens that trigger anti-tumor immunity [20–22]. When we compared the mutation data of the high- and low-MFAP4 expression groups, we found that the Nonsense_Mutation and Missense_Mutation of FLNB mutant genes in the low MFAP4 expression group samples increased significantly. The Frame_Shift_Ins and Multi_Hit mutations of CASP8 mutant genes in the low-MFAP4 expression group were significantly different from those in the high-expression group. Our results therefore indicate that the MFAP4 gene may affect the occurrence of oral cancer by affecting the mutations of these genes.
MicroRNAs are small non-coding RNAs that have been recently identified as regulators of gene expression through binding to the 3′ untranslated regions (UTRs) of target mRNA [28]. This results in mRNA degradation or translational inhibition. Emerging evidence has shown that microRNAs are abnormally expressed in various cancers [29], and deregulated microRNA expression is strongly associated with tumor initiation, promotion, and progression [30,31]. Each microRNA can have multiple target genes, and several microRNAs can also regulate the same gene. Therefore, this complex regulatory network not only regulates the expression of multiple genes through one microRNA but also finely regulates the expression of a certain gene through the combination of several microRNAs. In view of the important relationship between MFAP4 gene and oral cancer, we further explored the relationship between it and microRNA. We obtained 23 differentially expressed microRNAs, calculated the correlation between these differentially expressed microRNAs and MFAP4 expression, and finally selected the first 6 positively correlated and the last 6 negatively correlated microRNAs, and then drew the correlation diagram. Previous studies have shown that NFKB1 may be negatively regulated by miR-508-3p and plays an important role in the occurrence of gastric cancer [32]. The positive regulatory relationship between MFAP4 and miR-508-3p was discovered for the first time in the present study, and it may be helpful to understand the mechanism of oral cancer. Only a few studies have found relationships of other positive regulations, such as with hsa.mir.514a.3, hsa.mir.514a.1, hsa.mir.514a.2, hsa.mir.509.3, and hsa.mir.5683, with cancer. Therefore, the relationship between microRNAs and oral cancer needs further exploration. Hsa-miR-519c decreases endogenous ABCG2 mRNA and protein levels by acting through a putative miR-519c binding site located within the longer 3UTR region found only in the parental cells [33]. There are relatively few studies on the relationship between other negative regulatory microRNAs and cancer, and the regulatory relationship of microRNA with MFAP4 gene and the mechanism of oral cancer development still needs further research and verification.
The process of tumor immune cell infiltration has been well explained in a previous article [24]. In this study, we screened 7 common differentially infiltrating immune cells (eosinophils, macrophages, mast cells, NK cells, T cells, T helper cells, Tem) from the data of GEO and TCGA. After grouping by best separation, we found that high infiltration of tumors by mast cells, T cells, and Tem cells was related to better prognosis. Mast cells are immune cells that are present in all classes of vertebrates [34]. They have a widespread distribution in close proximity to epithelia, fibroblasts, blood, lymphatic vessels, and nerves [35]. Mast cell progenitors enter the circulation and complete their maturation in tissues. These cells are involved in several physiological and inflammatory processes, including organ development [36], wound healing [45], heart function [37], and tumorigenesis [38–42]. Encouragingly, in some models, T cell-mediated immune response can efficiently control tumors. In the last 2 decades, a large number of antigens have been identified in tumors that can be recognized by T cells [43–46], suggesting a potential role of T lymphocytes in anti-tumor immune response. Functional cytolytic T cells have been shown to develop action against tumor cells that express self-tumor antigens in some patients with melanoma [47], pancreatic cancer [48], and Epstein-Barr virus-associated malignancies [49]. We therefore analyzed the correlation between the immune targets of these immune cells and MFAP4, and found that MFAP4 was related to the immune targets of CD27, CD28, CD58, CD70, CD74, CD80, CTLA4, HLA-A, INPP5D, and INPPL1. This result provides a new prospect for further studying the immune infiltration mechanism in relation to the MFAP4 gene and OSCC.
Some studies have shown miR-24-3-p, CD163, and CD204 have predictive value in OSCC [50,55. He found miR-24-3p can maintain the proliferation of OSCC cells through targeting PER1 [50]. Kubota suggested that CD 163+ CD204+ tumor-associated macrophages (TAMs) could play an important role in the invasion and metastasis of OSCC by T cell regulation via IL10 and PD-L1 production [55]. In the present study, the prognosis of patients was significantly different with different expression levels of MFAP4, which indicated that MFAP4, like miR-24-3-p, CD163, and CD204, has potential as a biomarker of oral squamous cell carcinoma. Similarly, we also know that it is difficult to accurately predict the prognosis of patients based on expression of a single gene, so a single gene could be combined with other prognostic genes, TMB, immune infiltration, and so on to study which combination most improves the prediction effect. In this study, we found that the top 6 positively correlated miRNA and the top 6 negatively correlated miRNA showed good linear relationships with MFAP4, 7 immune cells showed a significant difference in the MFAP4 expression group, and the infiltration of 3 immune cells was related to the prognosis of the patients. Thus, the prognostic value of MFAP4 lies not only in its use as a separate predictive marker, but in its combination with other factors.
Conclusions
Our research clarified the relationship between MFAP4 and oral cancer (particularly OSCC). We found that the MFAP4 gene is related to the prognosis of OSCC and is also closely related to TMB. We also identified some miRNAs that are related to MFAP4, which is of great significance for exploring the impact of OSCC mutations. Finally, we found a connection between mast cells, T cells, and Tem cells and MFAP4 in terms of immune infiltration and their relationship to the prognosis of tumors. These findings suggest that MFAP4 may be a promising therapeutic target for OSCC.
Supplementary Data
Supplementary Table 1.
Comparision of clinical phenotypes in diffient groups.
| High | Low | p | |
|---|---|---|---|
| n | 144 | 186 | |
| Anatomic neoplasm subdivision (%) | NA | ||
| Alveolar ridge | 6 (4.2) | 12 (6.5) | |
| Base of tongue | 11 (7.6) | 12 (6.5) | |
| Buccal mucosa | 9 (6.2) | 13 (7.0) | |
| Floor of mouth | 33 (22.9) | 27 (14.5) | |
| Hard Palate | 4 (2.8) | 3 (1.6) | |
| Lip | 1 (0.7) | 2 (1.1) | |
| Oral cavity | 38 (26.4) | 34 (18.3) | |
| Oral tongue | 42 (29.2) | 83 (44.6) | |
| Clinical M (%) | 0.709 | ||
| M0 | 138 (95.8) | 173 (94.5) | |
| M1 | 0 (0.0) | 2 (1.1) | |
| MX | 6 (4.2) | 8 (4.4) | |
| Clinical N (%) | 0.800 | ||
| N0 | 73 (52.1) | 94 (53.4) | |
| N1 | 24 (17.1) | 34 (19.3) | |
| N2 | 41 (29.3) | 47 (26.7) | |
| N3 | 2 (1.4) | 1 (0.6) | |
| Clinical T (%) | 0.028 | ||
| T1 | 4 (2.8) | 16 (8.9) | |
| T2 | 41 (29.1) | 63 (35.2) | |
| T3 | 36 (25.5) | 46 (25.7) | |
| T4 | 60 (42.6) | 54 (30.2) | |
| Clinical stage (%) | 0.066 | ||
| Stage I | 1 (0.7) | 10 (5.6) | |
| Stage II | 31 (22.0) | 47 (26.3) | |
| Stage III | 32 (22.7) | 36 (20.1) | |
| Stage IV | 77 (54.6) | 86 (48.0) | |
| OS=1 (%) | 81 (56.2) | 69 (37.1) | 0.001 |
| DSS=1 (%) | 45 (33.6) | 47 (26.3) | 0.170 |
| DFI=1 (%) | 10 (29.4) | 13 (21.3) | 0.456 |
| PFI=1 (%) | 64 (44.4) | 70 (37.6) | 0.216 |
Acknowledgements
We thank the researchers who gave their data for this analysis. We gratefully acknowledge their contributions.
Abbreviations
- TCGA
The Cancer Atlas Genome
- GEO
Gene Expression Omnibus
- MFAP4
microfibril-associated protein 4
- Tem
effector memory T cell
- NK
natural killer
- GSEA
gene set enrichment analysis
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TMB
tumor mutation burden
- OSCC
oral squamous cell carcinoma
Footnotes
Ethical Statement
The data of this study are from the GEO and TCGA databases, and do not involve animal experiments or human specimens, and there are no ethics-related issues.
Availability of Data and Material
The data of this study are from the GEO and TCGA databases.
Conflict of Interest
None.
Source of support: Departmental sources
References
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359(11):1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Argiris A, Karamouzis MV, Raben D, et al. Head and neck cancer. Lancet. 2008;371(9625):1695–709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashibe M, Brennan P, Chuang SC, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18(2):541–50. doi: 10.1158/1055-9965.EPI-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DE, Burtness B, Leemans CR, et al. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6(1):92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlosser A, Thomsen T, Moeller JB, et al. Characterization of FIBCD1 as an acetyl group-binding receptor that binds chitin. J Immunol. 2009;183(6):3800–9. doi: 10.4049/jimmunol.0901526. [DOI] [PubMed] [Google Scholar]
- 8.Thomsen T, Schlosser A, Holmskov U, et al. Ficolins and FIBCD1: Soluble and membrane bound pattern recognition molecules with acetyl group selectivity. Mol Immunol. 2011;48(4):369–81. doi: 10.1016/j.molimm.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Procopio WN, Pelavin PI, Lee WM, et al. Angiopoietin-1 and -2 coiled coil domains mediate distinct homo-oligomerization patterns, but fibrinogen-like domains mediate ligand activity. J Biol Chem. 1999;274(42):30196–201. doi: 10.1074/jbc.274.42.30196. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Z, Lee CC, Jiralerspong S, et al. The gene for a human microfibril-associated glycoprotein is commonly deleted in Smith-Magenis syndrome patients. Hum Mol Genet. 1995;4(4):589–97. doi: 10.1093/hmg/4.4.589. [DOI] [PubMed] [Google Scholar]
- 11.Brandsma CA, van den Berge M, Postma DS, et al. A large lung gene expression study identifying fibulin-5 as a novel player in tissue repair in COPD. Thorax. 2015;70(1):21–32. doi: 10.1136/thoraxjnl-2014-205091. [DOI] [PubMed] [Google Scholar]
- 12.Johansson SL, Roberts NB, Schlosser A, et al. Microfibrillar-associated protein 4: A potential biomarker of chronic obstructive pulmonary disease. Respir Med. 2014;108(9):1336–44. doi: 10.1016/j.rmed.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Molleken C, Sitek B, Henkel C, et al. Detection of novel biomarkers of liver cirrhosis by proteomic analysis. Hepatology. 2009;49(4):1257–66. doi: 10.1002/hep.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bracht T, Schweinsberg V, Trippler M, et al. Analysis of disease-associated protein expression using quantitative proteomics-fibulin-5 is expressed in association with hepatic fibrosis. J Proteome Res. 2015;14(5):2278–86. doi: 10.1021/acs.jproteome.5b00053. [DOI] [PubMed] [Google Scholar]
- 15.Blindbaek SL, Schlosser A, Green A, et al. Association between microfibrillar-associated protein 4 (MFAP4) and micro- and macrovascular complications in long-term type 1 diabetes mellitus. Acta Diabetol. 2017;54(4):367–72. doi: 10.1007/s00592-016-0953-y. [DOI] [PubMed] [Google Scholar]
- 16.Wulf-Johansson H, Lock Johansson S, Schlosser A, et al. Localization of microfibrillar-associated protein 4 (MFAP4) in human tissues: Clinical evaluation of serum MFAP4 and its association with various cardiovascular conditions. PLoS One. 2013;8(12):e82243. doi: 10.1371/journal.pone.0082243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topalian SL, Taube JM, Anders RA, et al. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–87. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–28. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubin MM, Artyomov MN, Mardis ER, et al. Tumor neoantigens: Building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–21. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher TN, Kesmir C, van Buuren MM. Biomarkers in cancer immunotherapy. Cancer Cell. 2015;27(1):12–14. doi: 10.1016/j.ccell.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Grizzi G, Caccese M, Gkountakos A, et al. Putative predictors of efficacy for immune checkpoint inhibitors in non-small-cell lung cancer: Facing the complexity of the immune system. Expert Rev Mol Diagn. 2017;17(12):1055–69. doi: 10.1080/14737159.2017.1393333. [DOI] [PubMed] [Google Scholar]
- 23.Mayakonda A, Lin DC, Assenov Y, et al. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–56. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 2018;67(7):1031–40. doi: 10.1007/s00262-018-2150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39(4):782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Kasamatsu S, Hachiya A, Fujimura T, et al. Essential role of microfibrillar-associated protein 4 in human cutaneous homeostasis and in its photoprotection. Sci Rep. 2011;1:164. doi: 10.1038/srep00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlosser A, Pilecki B, Hemstra LE, et al. MFAP4 promotes vascular smooth muscle migration, proliferation and accelerates neointima formation. Arterioscler Thromb Vasc Biol. 2016;36(1):122–33. doi: 10.1161/ATVBAHA.115.306672. [DOI] [PubMed] [Google Scholar]
- 28.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–38. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 30.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang T, Kang W, Zhang B, et al. miR-508-3p concordantly silences NFKB1 and RELA to inactivate canonical NF-kappaB signaling in gastric carcinogenesis. Mol Cancer. 2016;15:9. doi: 10.1186/s12943-016-0493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.To KK, Zhan Z, Litman T, et al. Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol. 2008;28(17):5147–61. doi: 10.1128/MCB.00331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulero I, Sepulcre MP, Meseguer J, et al. Histamine is stored in mast cells of most evolutionarily advanced fish and regulates the fish inflammatory response. Proc Natl Acad Sci USA. 2007;104(49):19434–39. doi: 10.1073/pnas.0704535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marone G, Galli SJ, Kitamura Y. Probing the roles of mast cells and basophils in natural and acquired immunity, physiology and disease. Trends Immunol. 2002;23(9):425–27. doi: 10.1016/s1471-4906(02)02274-3. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Fu T, Song F, et al. Mast cells participate in corneal development in mice. Sci Rep. 2015;5:17569. doi: 10.1038/srep17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid AC, Silver RB, Levi R. Renin: At the heart of the mast cell. Immunol Rev. 2007;217:123–40. doi: 10.1111/j.1600-065X.2007.00514.x. [DOI] [PubMed] [Google Scholar]
- 38.Varricchi G, Galdiero MR, Marone G, et al. Controversial role of mast cells in skin cancers. Exp Dermatol. 2017;26(1):11–17. doi: 10.1111/exd.13107. [DOI] [PubMed] [Google Scholar]
- 39.Galdiero MR, Varricchi G, Marone G. The immune network in thyroid cancer. Oncoimmunology. 2016;5(6):e1168556. doi: 10.1080/2162402X.2016.1168556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varricchi G, Galdiero MR, Loffredo S, et al. Are mast cells MASTers in cancer? Front Immunol. 2017;8:424. doi: 10.3389/fimmu.2017.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oskeritzian CA. Mast cell plasticity and sphingosine-1-phosphate in immunity, inflammation and cancer. Mol Immunol. 2015;63(1):104–12. doi: 10.1016/j.molimm.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sammarco G, Varricchi G, Ferraro V, et al. Mast cells, angiogenesis and lymphangiogenesis in human gastric cancer. Int J Mol Sci. 2019;20(9):2106. doi: 10.3390/ijms20092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulie PG, Hanagiri T, Takenoyama M. From tumor antigens to immunotherapy. Int J Clin Oncol. 2001;6(4):163–70. doi: 10.1007/pl00012101. [DOI] [PubMed] [Google Scholar]
- 44.Van Der Bruggen P, Zhang Y, Chaux P, et al. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol Rev. 2002;188:51–64. doi: 10.1034/j.1600-065x.2002.18806.x. [DOI] [PubMed] [Google Scholar]
- 45.Coggin JH, Jr, Barsoum AL, Rohrer JW, et al. Contemporary definitions of tumor specific antigens, immunogens and markers as related to the adaptive responses of the cancer-bearing host. Anticancer Res. 2005;25(3c):2345–55. [PubMed] [Google Scholar]
- 46.Kazemi T, Younesi V, Jadidi-Niaragh F, et al. Immunotherapeutic approaches for cancer therapy: An updated review. Artif Cells Nanomed Biotechnol. 2016;44(3):769–79. doi: 10.3109/21691401.2015.1019669. [DOI] [PubMed] [Google Scholar]
- 47.Dudley ME, Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nat Rev Cancer. 2003;3(9):666–75. doi: 10.1038/nrc1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryschich E, Notzel T, Hinz U, et al. Control of T-cell-mediated immune response by HLA class I in human pancreatic carcinoma. Clin Cancer Res. 2005;11(2 Pt 1):498–504. [PubMed] [Google Scholar]
- 49.Gottschalk S, Heslop HE, Rooney CM. Adoptive immunotherapy for EBV-associated malignancies. Leuk Lymphoma. 2005;46(1):1–10. doi: 10.1080/10428190400002202. [DOI] [PubMed] [Google Scholar]
- 50.He L, Ping F, Fan Z, et al. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed Pharmacother. 2020;121:109553. doi: 10.1016/j.biopha.2019.109553. [DOI] [PubMed] [Google Scholar]
- 51.Lindholt JS, Madsen M, Kirketerp-Moller KL, et al. High plasma microfibrillar-associated protein 4 is associated with reduced surgical repair in abdominal aortic aneurysms. J Vasc Surg. 2020;71(6):1921–29. doi: 10.1016/j.jvs.2019.08.253. [DOI] [PubMed] [Google Scholar]
- 52.Zwadlo-Klarwasser G, Neubert R, Stahlmann R, et al. Influence of dexamethasone on the RM 3/1-positive macrophages in the peripheral blood and tissues of a New World monkey (the marmoset Callithrix jacchus) Int Arch Allergy Immunol. 1992;97(2):178–80. doi: 10.1159/000236115. [DOI] [PubMed] [Google Scholar]
- 53.Platt N, Gordon S. Is the class A macrophage scavenger receptor (SR-A) multifunctional? – The mouse’s tale. J Clin Invest. 2001;108(5):649–54. doi: 10.1172/JCI13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu S, Ye L, Bennett S, et al. Molecular structure and function of microfibrillar-associated proteins in skeletal and metabolic disorders and cancers. J Cell Physiol. 2021;236(1):41–48. doi: 10.1002/jcp.29893. [DOI] [PubMed] [Google Scholar]
- 55.Kubota K, Moriyama M, Furukawa S, et al. CD163(+)CD204(+) tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci Rep. 2017;7(1):1755. doi: 10.1038/s41598-017-01661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Comparision of clinical phenotypes in diffient groups.
| High | Low | p | |
|---|---|---|---|
| n | 144 | 186 | |
| Anatomic neoplasm subdivision (%) | NA | ||
| Alveolar ridge | 6 (4.2) | 12 (6.5) | |
| Base of tongue | 11 (7.6) | 12 (6.5) | |
| Buccal mucosa | 9 (6.2) | 13 (7.0) | |
| Floor of mouth | 33 (22.9) | 27 (14.5) | |
| Hard Palate | 4 (2.8) | 3 (1.6) | |
| Lip | 1 (0.7) | 2 (1.1) | |
| Oral cavity | 38 (26.4) | 34 (18.3) | |
| Oral tongue | 42 (29.2) | 83 (44.6) | |
| Clinical M (%) | 0.709 | ||
| M0 | 138 (95.8) | 173 (94.5) | |
| M1 | 0 (0.0) | 2 (1.1) | |
| MX | 6 (4.2) | 8 (4.4) | |
| Clinical N (%) | 0.800 | ||
| N0 | 73 (52.1) | 94 (53.4) | |
| N1 | 24 (17.1) | 34 (19.3) | |
| N2 | 41 (29.3) | 47 (26.7) | |
| N3 | 2 (1.4) | 1 (0.6) | |
| Clinical T (%) | 0.028 | ||
| T1 | 4 (2.8) | 16 (8.9) | |
| T2 | 41 (29.1) | 63 (35.2) | |
| T3 | 36 (25.5) | 46 (25.7) | |
| T4 | 60 (42.6) | 54 (30.2) | |
| Clinical stage (%) | 0.066 | ||
| Stage I | 1 (0.7) | 10 (5.6) | |
| Stage II | 31 (22.0) | 47 (26.3) | |
| Stage III | 32 (22.7) | 36 (20.1) | |
| Stage IV | 77 (54.6) | 86 (48.0) | |
| OS=1 (%) | 81 (56.2) | 69 (37.1) | 0.001 |
| DSS=1 (%) | 45 (33.6) | 47 (26.3) | 0.170 |
| DFI=1 (%) | 10 (29.4) | 13 (21.3) | 0.456 |
| PFI=1 (%) | 64 (44.4) | 70 (37.6) | 0.216 |