Summary
In the thymus, T cell progenitors differentiate in order to generate naive T lymphocytes which migrate in the periphery where they will fulfill their function in the adaptive immune response. During thymopoiesis, genomic alterations in thymocytes can promote leukemia development. Among recurrent alteration is PTEN inactivation, which is associated to MYC overexpression. Herein, we used conditional Pten and Myc knockout mouse models and single-cell RNA-sequencing approach, to investigate the impact of MYC loss on physio-pathological development of PTEN-proficient or PTEN-deficient T lymphocytes. First, our results confirm that MYC is mandatory for PTEN loss-mediated leukemogenesis, while it is not required for terminal steps of thymopoiesis. In contrast, we uncovered that Myc ablation in CD4+CD8+ thymocytes disrupts T lymphocytes homeostasis in the spleen, notably by drastically reducing the number of MYC-deficient effector/memory T cells. Collectively, our data show that besides naive T cells proliferation, MYC is essential for effector/memory differentiation.
Subject areas: Molecular genetics, Molecular biology, Immunology
Graphical abstract
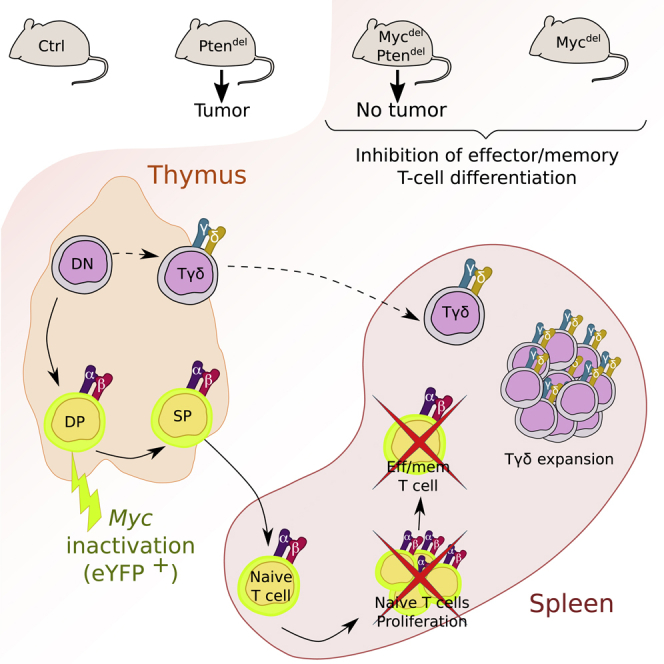
Highlights
-
•
MYC is essential for PTEN loss-mediated T cell leukemogenesis
-
•
MYC is required for effector/memory T cell differentiation
-
•
Expansion of splenic CD8+TCRγδ+ cells in MYC-deficient background
Molecular genetics; Molecular biology; Immunology
Introduction
T lymphocyte development is rhythmed by both proliferation and cell death phases. The first ones take place in the thymus during T lymphocyte maturation. Indeed, the aim of thymopoiesis is to produce T lymphocytes expressing a T cell receptor (TCR) able to induce an immune response against foreign antigens. For this purpose, TCR loci are rearranged through V(D)J recombination and thymocytes undergo various TCR-dependent quality controls. The first occurs at CD4−CD8− (DN) stage and corresponds to the selection of thymocytes that have successfully rearranged their TCRβ locus; this β selection process leads to an intense proliferation wave. Then, at the CD4+CD8+ double-positive (DP) stage, thymocytes harboring a TCR displaying too low or too high affinity for self-peptide-major histocompatibility complex (p-MHC) are eliminated by death-by-neglect or negative selection respectively. Only thymocytes with adequate affinity for p-MHC survive and differentiate into CD4 or CD8 single-positive (SP) thymocyte (Klein et al., 2014). Following thymopoiesis, mature naive T cells migrate in the secondary lymphoid organs, where they can encounter their specific antigen for the first time. Then, the activated T lymphocyte undergoes clonal expansion, which corresponds to an intense phase of proliferation and initiates a program of differentiation into effector and memory T lymphocytes. After antigen clearance, most effector T lymphocytes die by apoptosis (known as clonal contraction), while a surviving subset persists as long-lived memory cells (Kaech et al., 2002).
Disruption of the normal control of cell growth, proliferation, and thymocyte differentiation through genetic alterations may lead to T cell acute lymphoblastic leukemias (T-ALL). T-ALL is a malignant proliferation of T cell progenitors abnormally arrested at various stages of thymopoiesis. Numerous oncogenes and tumor suppressors (TS) have been reported in T-ALL, leading to complex oncogenic networks (Belver and Ferrando, 2016; Girardi et al., 2017). One of the most potent, PTEN which is deleted in around 15–20% of T-ALL cases, represents the main negative regulator of PI3K/AKT signaling pathway and is a well-known TS involved in numerous type of cancers (Milella et al., 2015). In human T-ALL, PTEN loss is correlated with unfavorable prognostic (Trinquand et al., 2013) and with the accumulation of MYC protein in leukemic blasts (Bonnet et al., 2011). In addition, T-ALL from the subgroup harboring TCR-MYC translocations are recurrently associated with PTEN loss-of-function mutations (La Starza et al., 2014; Milani et al., 2019). The functional interaction between MYC and PTEN is also sustained by mouse models, showing that inactivation of PTEN in thymocytes leads to T-ALL over-expressing MYC due to Myc-TCR translocations or NOTCH1 hyperactivation (Guo et al., 2008; Liu et al., 2010). Indeed, MYC is a master transcription factor and an oncogene known to be activated through multiple mechanisms in various human cancers (Dang, 2013).
Herein, using conditional gene inactivation in developing thymocytes, we assess the impact of the absence of MYC on physio-pathological development of PTEN-proficient or PTEN-deficient T lymphocytes. We showed that, as expected, Myc deletion at the DP stage prevents PTEN loss-mediated leukemogenesis, and has a limited impact on thymocytes differentiation. Yet, it strongly affects splenic T lymphocytes homeostasis, notably by impeding effector/memory T cell development.
Results
Myc deletion impedes T cell leukemogenesis mediated by PTEN loss
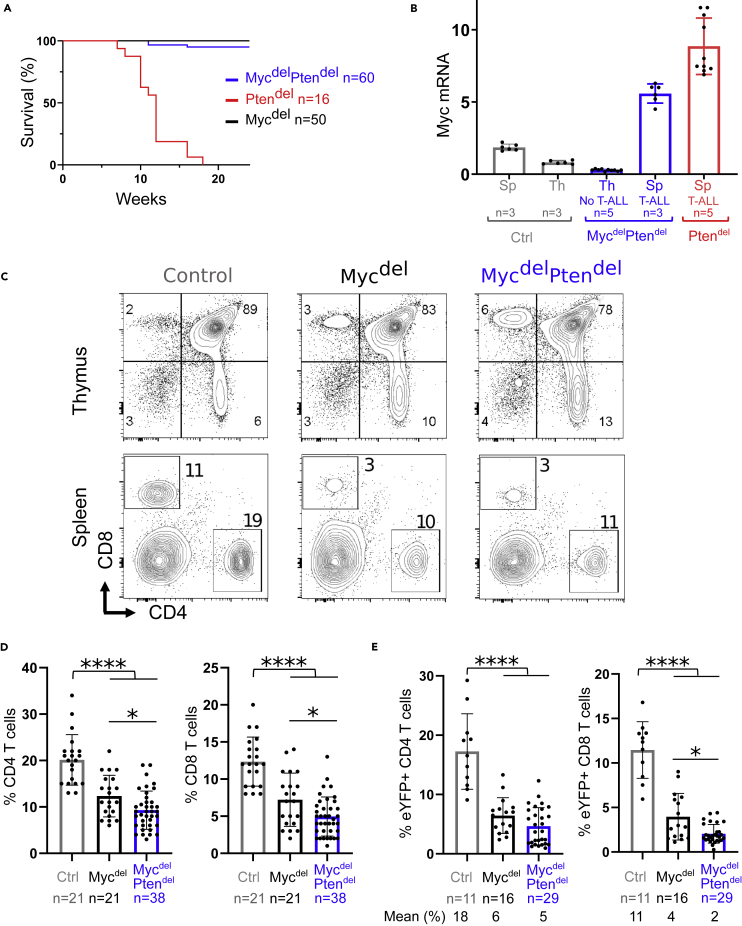
We used CD4-Cre mice to inactivate Pten and/or Myc genes at the DP stage of thymocyte differentiation. Thus, besides PTEN and MYC proficient mice (control), 3 models were generated: CD4-cre x PtenFlox/Flox (Ptendel), CD4-cre x MycFlox/Flox (Mycdel), and the double knockout CD4-cre x MycFlox/Flox x PtenFlox/Flox (MycdelPtendel). As previously described (Gon et al., 2018; Guo et al., 2008; Liu et al., 2010), Ptendel mice developed T-ALL in around 11 week (Figure 1A). Conversely, of 60 MycdelPtendel mice monitored for up to 1 year, only 3 mice developed T-ALL. Those, similarly to Ptendel T-ALL, arose in less than 4 months and were characterized by malignant proliferation of αβTCR+ T cells in the spleen (Figure S1A). However, Myc mRNA analysis of T-ALL cells of these 3 mice shows that Myc transcript level is similar in both Ptendel and MycdelPtendel T-ALL cells (Figure 1B). Thus, the few T-ALL arising in MycdelPtendel models expressed Myc, indicating that Myc gene escaped Cre-mediated inactivation. In PTEN-deficient T-ALL mouse models, oncogenic Myc activation occurs through Myc-TCR translocations, or alternatively, when translocation is impaired (for instance in RAG-deficient or in Tcra−/− backgrounds), through NOTCH1 hyperactivation (Gon et al., 2018; Liu et al., 2010). T-ALL developed by MycdelPtendel mice do not display NOTCH1 hyperactivation, suggesting that MYC activation is likely due to the translocation of one Myc allele (Figures S1B and S1C).
Figure 1.
Myc is required for Pten-loss mediated leukemogenesis and for splenic T cell homeostasis
(A) Survival curves of Ptendel, Mycdel and MycdelPtendel mice.
(B) Quantitative PCR for Myc mRNA expression in thymus (Th) or spleen (Sp) from Control mice, MycdelPtendel mice (disease-free and leukemic) and leukemic Ptendel mice. Transcripts levels were normalized to ABL. The analysis was performed in duplicate. Error bars represent means with standard deviation (SD).
(C) Representative FACS contour plots showing CD4 and CD8 expression on thymocytes and splenocytes from the indicated genotypes. Percentages of cells in depicted gates are indicated.
(D) Percentages of CD4 and CD8 lymphocytes in spleens.
(E) Percentages of eYFP positive CD4 and CD8 lymphocytes.
(A, B, D, and E) Numbers of mice that were analyzed are indicated. (D and E) Each dot represents a distinct mouse. Error bars represent means with SD. Statistical significant differences were assessed using Mann-Whitney test: ∗p < 0.05; ∗∗∗∗p < 0.0001.
In conclusion, our data show that MYC is required for PTEN loss-mediated leukemogenesis.
Disruption of T lymphocyte homeostasis upon Myc deletion
As MycdelPtendel mice do not develop leukemia, we undertook to analyze the impact of this double knockout on T lymphocyte development. Compared to control mice, thymocytes number has a tendency to decrease in aging Mycdel and MycdelPtendel mice (Figures S1D and S1E), and this is mainly due to reduced number of DP cells (Figure S1E). Typical FACS plots of thymocytes show that Myc deletion or Myc/Pten double deletion from DP stage do not strongly impact conventional thymocytes differentiation (Figure 1C).
In the spleen, the most obvious phenotype of Mycdel and MycdelPtendel mice is a significant reduction of CD4 and CD8 T cells, both of them affected in the same extent (Figures 1C and 1D). We crossed our mice models with ROSA26-LSL-eYFP reporter mice in which Cre-expressing cells express the enhanced yellow fluorescent protein (eYFP) (Srinivas et al., 2001), allowing us to monitor Myc and Pten-deleted cells. Compared to control, Mycdel and MycdelPtendel spleens displayed more eYFP negative T cells (not shown) indicating that in these cells, Cre recombinase was not expressed and thus Myc and/or Pten were not inactivated (Figure S1F).
In fine, eYFP+ T cells (CD4+ plus CD8+) from control mice account for 29% of total splenocytes while eYFP+ T cells from Mycdel and MycdelPtendel mice account for 10% and 7% of splenic cells, respectively (Figure 1E).
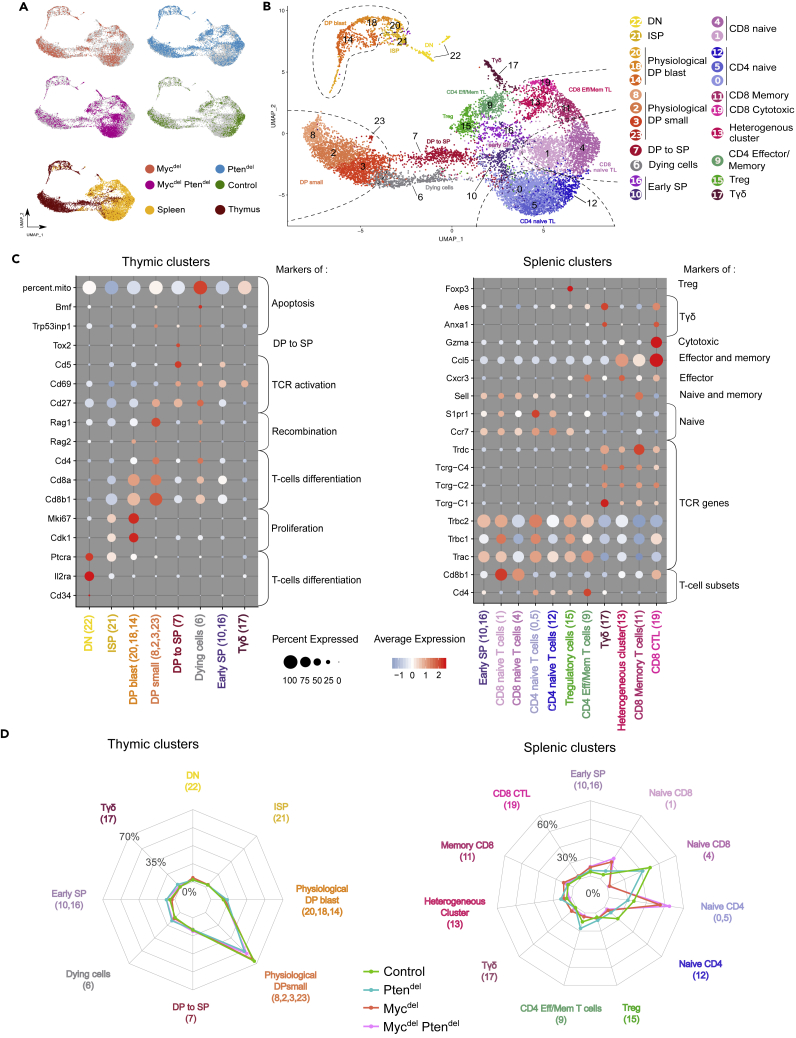
We used a single-cell RNA sequencing (scRNAseq) approach to investigate thymus and T lymphocytes from spleens of control, Mycdel, MycdelPtendel and Ptendel disease-free mice. Then, we applied the UMAP non-linear dimensionality reduction method to visualize cell transcriptome heterogeneity (Butler et al., 2018). Sample demultiplexing allowed us to visualize sample of origin for each cell on the UMAP plot (Figure 2A). According to various gene markers (Chopp et al., 2020; Mingueneau et al., 2013; Park et al., 2020), we assigned cell type to the 23 identified clusters (Figures 2B and 2C & Table S1); also Myc expression is displayed in Figure S2. We notably distinguished the main cell subsets of thymopoiesis (DN to early SP cells). We clearly identified in the spleen, naive (clusters 0, 5, & 12) and effector/memory (cluster 9) CD4 T lymphocytes, as well as naive (clusters 1 and 4), memory (cluster 11), and cytotoxic (cluster 19) CD8 T lymphocytes.
Figure 2.
Single cell RNAseq profiling reveals a disruption of splenic T cell composition in Myc-deficient mice
The scRNAseq assays were performed in duplicate with thymus and spleens from 4 to 6 weeks-old Control, Ptendel, Mycdel and MycdelPtendel mice (n = 2 for each genotype).
(A) UMAP plot of the analyzed samples after integration of the two datasets and exclusion of non T cells. The four upper plots are colored according to their sample of origin, and the lower plot according to tissue of origin.
(B) Same UMAP plot colored by cluster. Each cluster is annotated with their corresponding cell type.
(C) Seurat Dotplot showing the expression level of principal marker genes used to identify thymic and splenic clusters. Color represents the scaled average expression of the gene of interest across the various clusters, while dot size indicates the proportion of cells expressing the gene of interest.
(D) Radar plot showing the percentage of cells by clusters for each genotypes in the thymus and in the spleen.
For each genotype, we defined the percentage of cells in each cluster (Figure 2D). For the thymus, radar plots of the various genotypes superpose indicating that there is no major impact of Myc and/or Pten deletion on cellular composition of the thymus. Conversely, in the spleen, cell distribution differs according to genotype. We distinguish 2 types of radar plots, one corresponding to Mycdel and MycdelPtendel mice and the other one to control and Ptendel mice. Notably, we clearly observe that naive CD8 T lymphocytes split into 2 clusters: cluster 1, predominantly composed of MYC-deficient cells, while cluster 4 mainly comprises control and Ptendel cells. To better understand the difference between MYC-proficient versus MYC-deficient naive T lymphocytes, we performed a differential gene expression analysis. This reveals that MYC-deficient naive T cells downregulate genes related to ribosomes and protein synthesis (Figure S3).
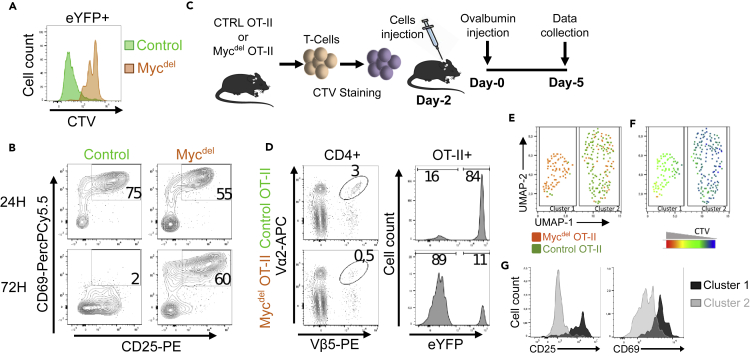
MYC deficiency blocks stimulated T lymphocytes in an early activation stage
Next, we undertook to assess the ability of MYC-deficient naive T cells to proliferate. To do so, we performed ex vivo TCR-stimulation assays using anti-CD3/CD28 beads and showed that after 72 hr MYC-deficient T lymphocytes retained CellTrace Violet (CTV) staining indicating that they do not proliferate (Figure 3A). Yet, at 24 hr those cells were activated as they expressed, similarly to control cells, early activation markers, CD25 and CD69. However, at 72 hr, MYC-deficient T cells retain the expression of CD25 and CD69 while control T cells lose CD69 and downregulate CD25 (Figure 3B).
Figure 3.
Stimulated MYC-deficient T cells are blocked in an activated state
(A and B) Ex vivo stimulation assays. T lymphocytes from control and Mycdel spleens were labeled with CTV, stimulated with anti-CD3/CD28 beads for 24 and 72 hr and analyzed by flow cytometry. CTV staining at 72 H (A) and CD25/CD69 expression at 24H and 72H (B) of eYFP+ cells from Control and Mycdel spleens.
(C–G) In vivo stimulation assays. (C) Overview of the procedure used for monitoring OT-II T lymphocytes proliferation in vivo. T lymphocytes from Control OT-II or Mycdel OT-II spleens were labeled with CTV and injected in recipient C57BL/6 mice, which were then immunized with Ovalbumin and finally euthanized 5 days later for lymph nodes analysis (D–G). (D) CD4+ cells from mouse injected with control OT-II or MycdelOT-II cells were analyzed for OT-II expression, then eYFP expression was assessed in OT-II+ cells (right histograms). (E-G) All Myc-deficient CD4+OT-II+YFP+ cells (113 cells) were concatenated with 120 control CD4+OT-II+YFP+ T cells and then analyzed using the UMAP tool from FlowJo software. The resulting UMAP graph is colored according to the origin of the cells (E) and to CTV labeling (F). (G) Expression of CD25 and CD69 in clusters 1 and 2 that were defined in the UMAP (panel E).
For in vivo assays, Mycdel mice were crossed with the OT-II mouse model, which expresses a transgenic Vα2/Vβ5.1 TCR recognizing the chicken Ovalbumin antigen in the context of MHC-II molecules (Barnden et al., 1998). We obtained MycdelOT-II mice (OT-II x Mycdel x ROSA26-LSL-eYFP) and control OT-II mice (OT-II x CD4-Cre x ROSA26-LSL-eYFP) that were used to perform in vivo stimulation assays (Figure 3C). In mice injected with MycdelOT-II cells, we detect only 0.5% of OT-II+ cells, among them only 10% are eYFP+ (while at the time of injection 99% of OT-II+ cells were eYFP+). Therefore, conversely to control OT-II cells, TCR-stimulation did not give rise to clonal expansion of MYC-deficient OT-II+ T cells (Figure 3D). Following FACS acquisition, T cells of interest (CD4+OT-II+eYFP+) from control and MycdelOT-II genotypes were concatenated to facilitate the comparison between genotypes. We distinguish two main clusters: cluster 1 composed of cells originating from MycdelOT-II mice, and cluster 2 comprising mainly cells originating from control OT-II mice (Figure 3E). Similarly to in vitro stimulation assays, Myc-deleted cells (cluster 1) retained CTV staining indicating that those cells do not proliferate (Figure 3F). Moreover, comparison of clusters 1 and 2 reveals that Myc-deleted OT-II+ cells retained the expression of early activation markers (CD25 and CD69) (Figure 3G).
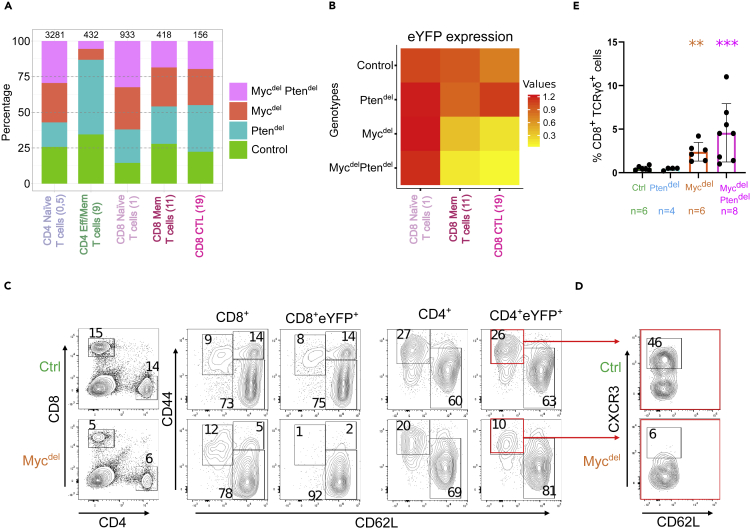
MYC is required for effector/memory T lymphocytes differentiation
We then reasoned that the blockade of naive T cells in an “activated” state may impede their differentiation. Thus, we next analyzed effector/memory T-cells. In the UMAP plot, effector/memory CD4 T cells cluster (cluster 9) is mainly composed of Control and Ptendel cells, while cells from both Mycdel and MycdelPtendel mice represent less than 20% of the cells (Figure 4A). This observation suggests that Myc-deletion impairs the expansion of effector/memory CD4 T cells. Conversely, memory/effector CD8 T lymphocytes clusters comprise more than 50% of cells from Mycdel and MycdelPtendel mice. Yet, a deeper analysis through the monitoring of eYFP expression shows that most cells originating from Mycdel and MycdelPtendel mice are eYFP negative (Figure 4B), indicating that effector/memory CD8 T cells found in Myc-deleted genetic backgrounds, have not inactivated Myc gene. Altogether, this transcriptomic analysis suggests that effector/memory T lymphocytes fail to develop in MYC-deficient background. We then undertook to validate this observation using flow cytometry analysis. First, the analysis of CD8+ T cells shows that, compared to control spleens, Mycdel spleens display a similar proportion of effector cells (CD62L−CD44+) and a decrease of memory cells (CD62L+CD44+). However, when the analysis is focused on CD8+ T cells expressing eYFP, we clearly observe an absence of CD62L+CD44+ and CD62L−CD44+ cell subsets (Figure 4C). In the same line, FACS analysis reveals a strong decrease of effector/memory CD4+eYFP+ cells, yet in most of the analyzed Mycdel mice, we distinguish a small CD62L−CD44+ effector/memory cell population (Figure 4C). However, conversely to the MYC-proficient counterpart, this cell population does not comprise a cell subset expressing CXCR3, a marker of T helper 1 (Th1) cells (Figure 4D), indicating that terminal effector differentiation does not occur in MYC-deficient CD62L−CD44+ cells.
Figure 4.
Absence of Myc-deficient effector/memory T cell subsets
(A) Barplot showing the percentage of cells from each genotype in naive and effector/memory T cells clusters (with the total of cells per genotype scaled to 100). Number of cells for each cluster are indicated on the top.
(B) Heatmap showing eYFP average expression for cells in CD8 naive (cluster 1), CD8 memory (cluster 11) and CD8 effector (cluster 19) T cells according to genotype. (C & D) Typical flow cytometry plots of spleens from control and Mycdel mice (representative of 3 mice of each genotype).
(C) CD4/CD8 expression (left plots). CD62L and CD44 expression (right plots) were assessed for cells indicated at the top of the plots. Percentages of cells in depicted gates: naive, CD62L+CD44-; central memory, CD62L+CD44+; and effector/memory, CD62L−CD44+, are indicated.
(D) CXCR3 expression in eYFP+ effector/memory CD4 T cells.
(E) Histogram reporting the percentage of splenic CD8+TCRγδ+ T cells in CD3+eYFP+ gated cells. Mean percentages are 0.5; 0.43; 2.4 & 4.6 for Control, Ptendel, Mycdel and MycdelPtendel mice respectively. Each dot represents a distinct mouse (the number of mice is indicated). Error bars represent means with standard deviation (SD). Differences with control mice were assessed using Mann-Whitney test (∗∗p < 0.01; ∗∗∗p < 0.001).
Altogether, scRNAseq data and FACS analysis show that MYC deficiency induces an important decrease of effector/memory T cells at steady state.
Expansion of TCRγδ+ T-cells in Myc-deficient mice
As mentioned above, scRNAseq data analysis indicates that effector/memory CD8 T cells (clusters 11 and 19) originating from Mycdel and MycdelPtendel backgrounds do not express eYFP (Figure 4B). We also observed that a high proportion of those cells express TCRγ/TCRδ genes (Figure S4A). In accordance with this observation, the analysis of TCRγδ expression by flow cytometry reveals a general increase of TCRγδ T cells in Myc-deleted backgrounds (Figures 4E and S4B). In particular, compared to control and Ptendel spleens, percentages of CD8+TCRγδ+ cells in Mycdel and MycdelPtendel spleens are increased by 5-fold and 10-fold factors, respectively (Figure 4E).
Discussion
Herein, we first showed that MYC is absolutely required for thymocyte leukemogenesis mediated by PTEN loss. This conclusion is consistent with a previous report in which IFN-inducible Mx1-Cre system was used to induce acute Myc and/or Pten ablation in all hematopoietic cells (Zhang et al., 2011). However, this latter study describes that Pten/Myc double-knockout mice developed lymphadenopathy featured by a very strong enlargement of lymph nodes (comprising polyclonal B and T cells). In sharp contrast, we do not observe such lymphadenopathy in MycdelPtendel mice (data not shown), in which gene deletion specifically occurs in DP thymocytes. In our study, MycdelPtendel, and Mycdel T cells phenotypes are similar. Collectively, our data indicate that the impact of MYC inactivation during pathological or physiological development of T cells, overrides the one of Pten deletion (Figures 1 and 2D), this led us to revisit the role of MYC in normal T cell differentiation.
During thymic differentiation, Myc is expressed at DN stages, then its expression level drops at DP stage and restarts weakly at the SP stage (Mingueneau et al., 2013). Accordingly, MYC plays a key role in the early stages of thymic differentiation (Dose et al., 2006). Indeed, inactivation of MYC at the DN stage inhibits thymocyte proliferation at the β-selection stage, without preventing differentiation. Overall, this leads to a severe decrease of mature thymocytes production (Dose et al., 2006; Herranz et al., 2014). In line with Myc expression profile, our study and previous published data (Dose et al., 2009) show that inactivation of MYC at the DP stage only minimally affects the final stages of conventional thymocyte differentiation.
Within the peripheral lymphoid organs, the initial response of T lymphocytes to antigenic stimulation is the conversion of small resting naive T cell into large blasts, followed by cell division. Then to sustain growth and proliferation, activated T cell increases glucose and glutamine metabolism through a metabolic reprogramming, which is controlled by MYC (Wang et al., 2011). Accordingly, it was previously reported that MYC-deficient T cells can be activated, yet they failed to proliferate (Trumpp et al., 2001; Wang et al., 2011). In line with these reports, we show that TCR stimulation induces activation of MYC-deficient T cells as revealed by surface expression of CD25 and CD69, but those cells do not actively proliferate (Figure 3A). CD25 and CD69 are early markers of activation that are expected to disappear few days after stimulation at the time when CD44 is expressed (Garcia et al., 1999). We monitored CD25 and CD69 expression at later time and showed that those markers remain expressed at the cell surface of MYC-deficient T cells 3 days and up to 5 days post-TCR stimulation (Figure 3). These data indicate that absence of MYC induces a T cell blockade in an “activated” state.
According to the classical view of T cell priming, activated naive T cell proliferates and then differentiates into effector/memory cell. Therefore, we may hypothesize that the blockade in an activation stage and the impairment of proliferation induced by MYC deficiency would further impede effector T cell differentiation. In contrast to this hypothesis, previous analyses suggest that MYC deficiency impacts marginally peripheral T lymphocytes population (Dose et al., 2009; Wong et al., 2015; Zhang et al., 2011). Herein, we report a significant decrease of CD4+ and CD8+ T lymphocytes in Mycdel and MycdelPtendel spleens. Moreover, we clearly demonstrate that those T lymphocyte populations are largely “contaminated” by MYC-proficient T cells, which likely blur all previous MYC loss-of-function analyses performed without a Cre-reporter system. The eYFP expression allowed us to track true MYC-deficient T lymphocyte populations and to uncover that these populations mostly comprise naive T cells. This result is even more striking for CD8+ T lymphocyte population for which there is an absence of effector and memory cell subsets. The differentiation of activated CD8 T cell requires various stimuli and notably the help of CD4 effector T cell (Bevan, 2004). Yet, in Mycdel mice the number of these CD4 helper cells is largely decreased (Figures 4C and 4D), as a result this may affect negatively CD8 effector differentiation. Moreover, it was previously reported that the amount of MYC protein is greater in CD8+ than in CD4+ T lymphocytes (Marchingo et al., 2020), suggesting that CD8+ T cells would be more sensitive to Myc deletion. Conversely to our result, a recent study (Heckler et al., 2021) shows that MYC inhibition (using Palbociclib, a CDK4/6 inhibitor) promotes CD8 T cell memory formation. This contrasting observation can be explained by the fact that in Heckler et al. paper only the MYC-MAX function is affected, thus some MYC functions independent of MAX, such as the transcription of RNA polymerase III targets, are not impacted (van Riggelen et al., 2010). Nevertheless, Palbociclib might represent an interesting therapeutic approach in T-ALL by having potentially two distinct actions: targeting MYC-dependent leukemic blasts and enhancing long-term protective immunity.
Herein, we also observed an increase of CD8+TCRγδ+ T-lymphocytes. Most TCRγδ+ T cells are double-negative for CD4 and CD8. Nevertheless, in the murine spleen, it exists a minority of TCRγδ+ CD8 T cells (Sato et al., 1993). We observe an over-representation of these cells in Mycdel and MycdelPtendel mice compared to Control and Ptendel mice (Figure 4E). CD8+ TCRγδ+ T cells are eYFP negative; therefore, this increase is not the result of an intrinsic MYC defect in these cells. A previous study indicates that the number of CD8+TCRγδ+ T cells increases in the context of a lymphopenic spleen (French et al., 2009). Thus, the T lymphopenia observed in Mycdel and MycdelPtendel spleens (Figure 1) is probably at the origin of the deregulation of CD8+TCRγδ+ T cells homeostasis. We concluded that the expansion in the periphery of CD8+TCRγδ+ cells is a collateral effect of MYC inactivation during thymocyte ontogeny.
In conclusion, despite numerous investigations on the role of MYC in the initial steps of T cell activation and proliferation, its impact on effector differentiation was not clearly described. Our work provides compelling evidence that the process of effector/memory T lymphocyte differentiation is fully dependent on MYC expression.
Limitations of the study
Droplet-based scRNA-seq does not detect all expressed genes in every cell. Typically, Myc mRNA is not very well captured in our scRNAseq analysis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3 APC Cy7 | BD Pharmigen | Cat#: 560590; RRID: AB_1727461 |
| CD4 V450 | BD Pharmigen | Cat#: 560468; RRID: AB_1645271 |
| CD4 APC | BD Pharmigen | Cat#: 553051; RRID: AB_398528 |
| CD4 PercP Cy 5.5 | BD Pharmigen | Cat#: 561115; RRID: AB_10563934 |
| CD8a PercP Cy5.5 | BD Pharmigen | Cat#: 561109; RRID: AB_10563417 |
| CD8a PE | BD Pharmigen | Cat#: 553033; RRID: AB_394571 |
| CD8a PE Cy7 | BD Pharmigen | Cat#: 552877; RRID: AB_394506 |
| CD8a BUV395 | BD Horizon | Cat#: 563786; RRID: AB_2732919 |
| CD25 PE | BD Pharmigen | Cat#: 561065; RRID: AB_10563211 |
| CD44 PE | BD Pharmigen | Cat#: 553134; RRID: AB_394649 |
| CD44 APC | BD Pharmigen | Cat#: 559250; RRID: AB_398661 |
| CD44 APC Cy7 | BD Pharmigen | Cat#: 560568; RRID: AB_1727481 |
| CD62L PE Cy7 | BD Pharmigen | Cat#: 560516; RRID: AB_1645257 |
| CD62L APC | BD Pharmigen | Cat#: 561919; RRID: AB_10895379 |
| CD69 PercP Cy5.5 | BD Pharmigen | Cat#: 561931; RRID: AB_10892815 |
| CD122 PE Cy7 | BioLegend | Cat#: 123215; RRID: AB_2562894 |
| CD183 BV650 | BD OptiBuild | Cat#: 740630; RRID: AB_2740325 |
| CD194 APC | BioLegend | Cat#: 131211; RRID: AB_1279135 |
| CD196 BV480 | BD OptiBuild | Cat#: 747830; RRID: AB_2872294 |
| CD197 BV421 | BD Horizon | Cat#: 562675; RRID: AB_2737716 |
| TCR Vβ5 PE | BD Pharmigen | Cat#: 553190; RRID: AB_394698 |
| TCR Vα2 APC | eBioscience | Cat#: 17-5812-80; RRID: AB_469460 |
| TCR γδ PE | BD Pharmigen | Cat#: 561997; RRID: AB_10897008 |
| TotalSeqTMA0301 anti-mouse Hastag 1 antibody | BioLegend | Cat#: 155801; RRID: AB_2750032 |
| TotalSeqTMA0302 anti-mouse Hastag 2 antibody | BioLegend | Cat#: 155803; RRID: AB_2750033 |
| TotalSeqTMA0303 anti-mouse Hastag 3 antibody | BioLegend | Cat#: 155805; RRID: AB_2750034 |
| TotalSeqTMA0304 anti-mouse Hastag 4 antibody | BioLegend | Cat#: 155807; RRID: AB_2750035 |
| TotalSeqTMA0305 anti-mouse Hastag 5 antibody | BioLegend | Cat#: 155809; RRID: AB_2750036 |
| TotalSeqTMA0306 anti-mouse Hastag 6 antibody | BioLegend | Cat#: 155811; RRID: AB_2750037 |
| TotalSeqTMA0307 anti-mouse Hastag 7 antibody | BioLegend | Cat#: 155813; RRID: AB_2750039 |
| TotalSeqTMA0308anti-mouse Hastag 8 antibody | BioLegend | Cat#: 155815; RRID: AB_2750040 |
| Cleaved Notch1 (Val1744) antibody | Cell signaling technology | Cat#: 2421; RRID: AB_2314204 |
| Antibody actin (I-19) | Santa Cruz | Cat#: sc-1616; RRID: AB_630836 |
| Donkey anti-goat IgG (H + L), highly Cross-adsorbed | Biotium | Cat#: 20277; RRID: AB_10855004 |
| Goat anti-mouse IgG (H + L), highly Cross-adsorbed | Biotium | Cat#: 20065; RRID: AB_10852820 |
| Chemicals, peptides, and recombinant proteins | ||
| CellTrace violet | Invitrogen | Cat#: C34557 |
| Dynabeads® mouse T-Activator CD3/CD28 | Life Technologies | Cat#: 11456D |
| Kit 10X 3′ version 2 | 10X Genomics | Cat#: 120267 |
| Annexin V-APC | BD Pharmingen | Cat#: 550474; RRID: AB_2868885 |
| Complete Freund's adjuvant | Sigma | Cat#: 344289 |
| EasySep™ Mouse T cell isolation kit | Life Technologies | Cat#: 19851 |
| Deposited data | ||
| Raw and pre-processed data | This paper | SRA: SRP311697 GEO: GSE169374, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169374 |
| Docker containers and R objects | This paper | Zenodo: http://doi.org/10.5281/zenodo.4636520 |
| Mouse reference MM10 (Ensembl 93) 3.0.0 | 10X Genomics | ftp://ftp.ensembl.org/pub/release-93/fasta/mus_musculus/dna/Mus_musculus.GRCm38.dna.primary_assembly.fa.gz |
| Experimental models: organisms/strains | ||
| Ptenflox/flox mice | European Mouse Mutant Archive | EM:00406 |
| Mycflox/flox mice | Andreas Trumpp (DKFZ) | Bishop JM. Nature. 414, 768-73 (2001). |
| CD4-cre mice | European Mouse Mutant Archive | EM: 01139 |
| OT-II mice | Barnden MJ. Immunol Cell Biol.76, 34–40 (1998) | MGI: 3046083 |
| ROSA26-LSL-eYFP reporter mice | Jackson Laboratory | MGI: 2449038 |
| Oligonucleotides | ||
| RQ-PCR Myc fwd: gcccctag tgctgcatgag/rev: ccacagacaccacatcaatttctt |
n/a | |
| RQ-PCR Abl fwd : Tgtggccagtggagataacactc /rev : ttcacaccattccccattgtg | n/a | |
| PCR Myc : 5’Flox: caccgcctacatcctgtccattc /3’Flox : tacagtcccaaagccccagccaag | n/a | |
| PCR Cre : fwd : agcattgctgtcacttggtcg / rev : cgtacaccaaaatttgcct | n/a | |
| PCR Pten : fwd :gccttacctagtaaagcaag /rev : ggcaaagaatcttggtgttac | n/a | |
| PCR TCRα OTII : fwd :aaagggagaaaaagctctcc /rev : acacagcaggttctgggttc | n/a | |
| PCR TCRβ OTII: fwd :gctgctgcacagacctact /rev : cagctcacctaacacgagga | n/a | |
| PCR Rosa: HL15 : aagaccgcgaagagtttgtcc / HL54 : taagcctgcccagaagactcc / HL152 : aagggagctgcagtggagata |
n/a | |
| PCR AT6/3’Flox. AT6: tttctgactcgctgtagtaattcc/3’Flox : tacagtcccaaagccccagccaag | n/a | |
| Software and algorithms | ||
| GraphPad Prism version 8.1.0 | GraphPad | https://www.graphpad.com/ |
| FlowJo version 10 | FlowJo | https://www.flowjo.com/ |
| Diva version 8.0.1 | BD Biosciences | https://www.bdbiosciences.com/ |
| Citeseq count version 1.4.3 | NYGCtech | GitHub - Hoohm/CITE-seq-Count: A tool that allows to get UMI counts from a single cell protein assay |
| R version 3.5.3 | R PROJECT | R: The R Project for Statistical Computing (r-project.org) |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dominique Payet Bornet (payet@ciml.univ-mrs.fr).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Fastq raw data and processed data are available in SRA (https://www.ncbi.nlm.nih.gov/sra?term=SRP311697) and from the NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169374) respectively.
For reproducibility, docker images and detailed scripts used for preprocessing and further analysis are available respectively at zenodo (https://zenodo.org/record/4636520) and github (https://github.com/mathisnozais/MycPten).
Experimental models and subject details
Mice
Mice were bred and housed in specific pathogen-free conditions in CIML animal facilities and are handled in accordance with French and European guidelines. Procedures of the project have been validated by French ethical committee (project APAFIS#4484-2016031113534101). Conditional Ptenflox/flox mice (Marino et al., 2002) and Mycflox/flox (Trumpp et al., 2001) were obtained from European Mouse Mutant Archives (EMMA) and from Andreas Trumpp’s laboratory, respectively. CD4-Cre mice (Lee et al., 2001) and OT-II mice (Barnden et al., 1998) were bred and maintained in CIML animal facilities. ROSA26-LSL-eYFP reporter mice (Srinivas et al., 2001) were purchased from The Jackson Laboratory. This mouse contains an eYFP gene inserted into the Gt(ROSA)26Sor locus. eYFP expression is inhibited by an upstream LoxP-flanked stop sequence, yet in presence of Cre recombinase the stop sequence is deleted allowing eYFP expression.
Method details
Flow cytometry analysis
Single-cell suspensions were stained with conjugated antibodies for 30 min at 4 °C and washed twice with FACS buffer (PBS, 2 % FCS, 1 mM EDTA). Multicolor flow cytometry analysis was performed with FACS Canto II (Becton–Dickinson Pharmingen) and data analyzed with FlowJo software (FlowJo, Becton–Dickinson).
Real-time Quantitative PCR (RQ-PCR)
RNA was extracted from cells using the column-based system RNAeasy mini kit (Qiagen) according to the manufacturer’s instructions. Reverse-transcription was performed with High-capacity cDNA reverse transcription kit (Applied Biosystems), and cDNA was analyzed by real-time PCR (RQ-PCR) on an ABI-PRISM 7500 Fast Real-Time PCR system (Applied Biosystems). PCR reactions were performed in 25 μL of diluted cDNA (10X dilution), 0.3 μmol of each primer and 12.5 μL of SYBR Green Master Mix (Roche). All RQ-PCR were performed in duplicate. To allow comparison between samples, transcript quantification was performed after normalization with ABL using the ΔCt method and calculated according to the following formula 2Δ (CtABL-Ctgene).
Immunoblotting analysis
50 × 106 cells were lysed for 30 min at 4°C in lysis buffer (100 mM Tris-HCl pH 8, 10 mM EDTA, 10 mM NaCl) supplemented with protease and phosphatase inhibitor cocktail tablets (Roche) and samples were centrifuged at 13,000 rpm for 30 min at 4°C. Protein concentrations of supernatants were determined using Bio-Rad protein assay (BIO-RAD). Equivalent protein extract (∼80μg) for each sample was separated by SDS-PAGE and transferred to nitrocellulose membrane using Iblot Gel Transfer stacks and Iblot system (Invitrogen). Membranes were blocked in TTBS (137 mM NaCl, 2 mM KCl, 25 mM Tris and 0.1% tween 20) supplemented with 5% non-fat milk and incubated with primary antibodies against Cleaved Notch1 (Val1744) Antibody (Cell signaling Technology) or Actin (clone I-19, Santa Cruz Biotechnology Inc.) overnight at 4°C with agitation. The fluorescent secondary antibodies, Anti-rabbit or -goat conjugated to CF770 (Biotium) were added for 1 hr at room temperature. Immunoblots were revealed using an Odyssey infrared imaging system (Li-Cor Biosciences) and stripped using Restore Western blot stripping buffer (Pierce).
Ex vivo and in vivo TCR stimulation assays
Splenic T cells from Mycdel and control mice were purified with EasySep Mouse T cell Isolation (Stemcell Technologies) and labeled with CellTrace Violet (CTV) Cell Proliferation Kit (Life Technologies). For ex vivo TCR stimulation assays, cells were mixed (ratio 1:1) with Dynabeads Mouse T-Activator CD3/CD28 (Life Technologies) in 96-well flat bottom plates and incubated for 24 or 72H (37°C, 5% CO2). For unstimulated controls, thymocytes were incubated in the same conditions but without anti-CD3/CD28 coated beads. Proliferation was measured using CTV and apoptosis was followed using AnnexinV labeling (BD Pharmingen) according to the manufacturer's instructions. For in vivo TCR stimulation assays, one million of CTV-labeled cells were i.v. injected in C57BL/6 mice. After two days, mice were immunized s.c. (foot pad and ear immunization) with 100 μg of Ovalbumin in the presence of complete Freund's adjuvant. Mice were sacrificed at day 5 post immunization, and cells collected from cervical and popliteal lymph nodes, were analyzed by FACS.
Single-cell RNA sequencing experiment
Thymus and spleen were dilacerated on a 70 μM nylon mesh (Corning), then treated with 1 mL RBC Lysis Buffer (ThermoFischer Scientific) and finally cells were resuspended in PBS supplemented with 2% fetal bovine serum (FCS). Splenic T cells were further purified using EasySep Mouse T cell Isolation Kit (StemCell technologies). T-cells from thymus and spleen of the different genotypes were each labeled with a distinct Hashtag oligo (HTO), TotalSeq anti-mouse Hashtag reagent (BioLegend), then pooled and finally single-cell isolation was achieved using the Chromium technology from 10X Genomics. Single-cell cDNA synthesis and sequencing libraries were prepared with Single Cell 3' Library & Gel Bead kit (10X Genomics) according to manufacturer's instructions. Libraries were sequenced using a Next-seq500 and the following parameters, Read1: 26 cycles, i7: 8 cycles, Read2: 57 cycles. We performed this assay in duplicate.
Single-cell RNA sequencing data analysis
For data preprocessing, mRNA library reads were aligned to the mm10 version of the mouse genome and quantified using CellRanger count (version 3.0.1). Antibody counts for cell hashing were quantified using CITE-seq-count (version 1.4.3, NYGCtech, https://github.com/Hoohm/CITE-seq-Count) using default parameters. The produced mRNA and HTO data matrices were imported into R (v3.5.3) and downstream analysis were performed with the Seurat package (v3.0.1) (Butler et al., 2018; Stuart et al., 2019).
In order to perform sample demultiplexing, hashtag oligos (HTOs) for each cell were normalized using a centered log ratio (CLR) transformation across cells. For both experiment cells were demultiplexed using the Seurat HTODemux function, and cell doublets and background empty droplets subsequently removed. Before mRNA expression analysis, we filtered out low quality cells from the mRNA matrix (i.e. cells expressing less than 200 genes or more than 10% of mitochondrial associated genes). Genes expressed in less than 3 cells were removed. Then, expression raw counts were normalized and scaled using respectively the Seurat NormalizeData and scaleData functions. Samples from the two experiments were integrated with the seurat FindIntegrationAnchors (using the intersection of the top 2000 variable genes) and IntegrateData functions. A dimensionality reduction was performed on the integrated object using the principal component analysis method (RunPCA function) and a UMAP was computed with the 12 first components (RunUMAP function). Cell clusters were identified using Seurat FindClusters function with Louvain Algorithm option (using 12 dimensions and a resolution of 1). Based on the expression of Cd74, Cd19 (B cells clusters), Ncr1 (NK cells) and lack of Cd3d we exclude non-T cells clusters of our analysis. UMAP dimensionality reduction was performed for a second time using the top 2000 variable genes of the selected cells using 18 dimensions and 1.8 clustering resolution.
Quantification and statistical analysis
Kaplan-Meier survival curves and statistical analyses were performed using GraphPad Prism software (GraphPad Software). Significance was evaluated by 2-tailed Mann-Whitney U test. A p value inferior to 0.05 was considered significant.
Acknowledgments
High-throughput sequencing was performed by the IPMC UCAGenomiX genomics platform, partner of the National Infrastructure France Génomique, supported by the Commissariat aux Grands Investissements (ANR-10-INBS-09-03, ANR-10-INBS-09-02). This study was partly supported by research funding from the Canceropôle PACA, Institut National Du Cancer and Région Sud. SP received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant Agreement No. 713750, and from the Regional Council of Provence-Alpes-Côte d’Azur, A∗MIDEX (No. ANR-11-IDEX0001-02). NG was supported by ‘La Fondation ARC’ (M2R20190508774). DPB received financial support from ITMO Cancer of AVIESAN (Alliance Nationale pour les Sciences de la Vie et de la Santé, National Alliance for Life Sciences & Health) within the framework of the Cancer Plan (project n° C19046S) and from CNRS ‘Osez l'interdisciplinarité!’ program-‘DMATh’ project. The authors thank the CIML animal facility and especially Elodie Pinsard for mouse care. We thank Dr. Andreas Trumpp (DKFZ) for providing Mycflox/flox mice. We acknowledge Pierre Milpied and Laurine Gil for their advices on scRNAseq assays, the cytometry and bioinformatic platforms of CIML for technical support.
Author contributions
M.N., M.L., C.G., N.G., J.Q., C.M., D.P. and D.P.B. performed the experiments and analyzed the data. M.N., S.P. and D.P. performed bioinformatics analysis. B.N., M.L., D.P. and D.P.B. wrote the paper. All authors read and approved the final manuscript.
Declaration of interestsd
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102761.
Contributor Information
Delphine Potier, Email: potier@ciml.univ-mrs.fr.
Dominique Payet-Bornet, Email: payet@ciml.univ-mrs.fr.
Supplemental information
References
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Belver L., Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2016;16:494–507. doi: 10.1038/nrc.2016.63. [DOI] [PubMed] [Google Scholar]
- Bevan M.J. Helping the CD8+ T-cell response. Nat. Rev. Immunol. 2004;4:595–602. doi: 10.1038/nri1413. [DOI] [PubMed] [Google Scholar]
- Bonnet M., Loosveld M., Montpellier B., Navarro J.-M., Quilichini B., Picard C., Di Cristofaro J., Bagnis C., Fossat C., Hernandez L. Posttranscriptional deregulation of MYC via PTEN constitutes a major alternative pathway of MYC activation in T-cell acute lymphoblastic leukemia. Blood. 2011;117:6650–6659. doi: 10.1182/blood-2011-02-336842. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopp L.B., Gopalan V., Ciucci T., Ruchinskas A., Rae Z., Lagarde M., Gao Y., Li C., Bosticardo M., Pala F. An integrated epigenomic and transcriptomic map of mouse and human αβ T cell development. Immunity. 2020;53:1182–1201.e8. doi: 10.1016/j.immuni.2020.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang C.V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013;3:a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M., Khan I., Guo Z., Kovalovsky D., Krueger A., von Boehmer H., Khazaie K., Gounari F. c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood. 2006;108:2669–2677. doi: 10.1182/blood-2006-02-005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M., Sleckman B.P., Han J., Bredemeyer A.L., Bendelac A., Gounari F. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. U S A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French J.D., Roark C.L., Born W.K., O’Brien R.L. Gammadelta T lymphocyte homeostasis is negatively regulated by beta2-microglobulin. J. Immunol. 2009;182:1892–1900. doi: 10.4049/jimmunol.0803165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S., DiSanto J., Stockinger B. Following the development of a CD4 T cell response in vivo: from activation to memory formation. Immunity. 1999;11:163–171. doi: 10.1016/s1074-7613(00)80091-6. [DOI] [PubMed] [Google Scholar]
- Girardi T., Vicente C., Cools J., De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129:1113–1123. doi: 10.1182/blood-2016-10-706465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gon S., Loosveld M., Crouzet T., Potier D., Bonnet M., Morin S.O., Michel G., Vey N., Nunès J.A., Malissen B. Fit αβ T-cell receptor suppresses leukemogenesis of Pten-deficient thymocytes. Haematologica. 2018;103:999–1007. doi: 10.3324/haematol.2018.188359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Lasky J.L., Chang C.-J., Mosessian S., Lewis X., Xiao Y., Yeh J.E., Chen J.Y., Iruela-Arispe M.L., Varella-Garcia M. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008;453:529–533. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckler M., Ali L.R., Clancy-Thompson E., Qiang L., Ventre K.S., Lenehan P., Roehle K., Luoma A., Boelaars K., Peters V. Inhibition of CDK4/6 promotes CD8 T-cell memory formation. Cancer Discov. 2021 doi: 10.1158/2159-8290.CD-20-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D., Ambesi-Impiombato A., Palomero T., Schnell S.A., Belver L., Wendorff A.A., Xu L., Castillo-Martin M., Llobet-Navás D., Cordon-Cardo C. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat. Med. 2014;20:1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S.M., Wherry E.J., Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2002;2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P.M., Hogquist K.A. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat. Rev. Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Liu X., Karnell J.L., Yin B., Zhang R., Zhang J., Li P., Choi Y., Maltzman J.S., Pear W.S., Bassing C.H. Distinct roles for PTEN in prevention of T cell lymphoma and autoimmunity in mice. J. Clin. Invest. 2010;120:2497–2507. doi: 10.1172/JCI42382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchingo J.M., Sinclair L.V., Howden A.J., Cantrell D.A. Quantitative analysis of how Myc controls T cell proteomes and metabolic pathways during T cell activation. Elife. 2020;9:e53725. doi: 10.7554/eLife.53725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S., Krimpenfort P., Leung C., van der Korput H.A.G.M., Trapman J., Camenisch I., Berns A., Brandner S. PTEN is essential for cell migration but not for fate determination and tumourigenesis in the cerebellum. Development. 2002;129:3513–3522. doi: 10.1242/dev.129.14.3513. [DOI] [PubMed] [Google Scholar]
- Milani G., Matthijssens F., Loocke W.V., Durinck K., Roels J., Peirs S., Thénoz M., Pieters T., Reunes L., Lintermans B. Genetic characterization and therapeutic targeting of MYC-rearranged T cell acute lymphoblastic leukaemia. Br. J. Haematol. 2019;185:169–174. doi: 10.1111/bjh.15425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milella M., Falcone I., Conciatori F., Cesta Incani U., Del Curatolo A., Inzerilli N., Nuzzo C.M.A., Vaccaro V., Vari S., Cognetti F. PTEN: multiple functions in human malignant tumors. Front Oncol. 2015;5:24. doi: 10.3389/fonc.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingueneau M., Kreslavsky T., Gray D., Heng T., Cruse R., Ericson J., Bendall S., Spitzer M.H., Nolan G.P., Kobayashi K. The transcriptional landscape of αβ T cell differentiation. Nat. Immunol. 2013;14:619–632. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Botting R.A., Conde C.D., Popescu D.-M., Lavaert M., Kunz D.J., Goh I., Stephenson E., Ragazzini R., Tuck E. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:eaay3224. doi: 10.1126/science.aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J., Yetil A., Felsher D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Sato K., Ohtsuka K., Watanabe H., Asakura H., Abo T. Detailed characterization of gamma delta T cells within the organs in mice: classification into three groups. Immunology. 1993;80:380–387. [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Starza R., Borga C., Barba G., Pierini V., Schwab C., Matteucci C., Lema Fernandez A.G., Leszl A., Cazzaniga G., Chiaretti S. Genetic profile of T-cell acute lymphoblastic leukemias with MYC translocations. Blood. 2014;124:3577–3582. doi: 10.1182/blood-2014-06-578856. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinquand A., Tanguy-Schmidt A., Ben Abdelali R., Lambert J., Beldjord K., Lengliné E., De Gunzburg N., Payet-Bornet D., Lhermitte L., Mossafa H. Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. J. Clin. Oncol. 2013;31:4333–4342. doi: 10.1200/JCO.2012.48.5292. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G.R., Bishop J.M. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Chen C., Wu Q., Liu Y., Zheng P. A critical role for the regulated Wnt–myc pathway in naive T cell survival. J.I. 2015;194:158–167. doi: 10.4049/jimmunol.1401238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xiao Y., Guo Y., Breslin P., Zhang S., Wei W., Zhang Z., Zhang J. Differential requirements for c-Myc in chronic hematopoietic hyperplasia and acute hematopoietic malignancies in Pten-null mice. Leukemia. 2011;25:1857–1868. doi: 10.1038/leu.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq raw data and processed data are available in SRA (https://www.ncbi.nlm.nih.gov/sra?term=SRP311697) and from the NCBI Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169374) respectively.
For reproducibility, docker images and detailed scripts used for preprocessing and further analysis are available respectively at zenodo (https://zenodo.org/record/4636520) and github (https://github.com/mathisnozais/MycPten).