Abstract
Pancreatic β-cells secrete insulin commensurate to circulating nutrient levels to maintain normoglycemia. The ability of β-cells to couple insulin secretion to nutrient stimuli is acquired during a postnatal maturation process. In mature β-cells the insulin secretory response adapts to changes in nutrient state. Both β-cell maturation and functional adaptation rely on the interplay between extracellular cues and cell type-specific transcriptional programs. Here we review emerging evidence that developmental and homeostatic regulation of β-cell function involves collaboration between lineage-determining and signal-dependent transcription factors (LDTFs and SDTFs, respectively). A deeper understanding of β-cell SDTFs and their cognate signals would delineate mechanisms of β-cell maturation and functional adaptation, which has direct implications for diabetes therapies and for generating mature β-cells from stem cells.
Keywords: β-cell, transcription factors, insulin secretion, metabolism, maturation, adaptation
β-cell function and identity are regulated by distinct processes
Insulin is the major glucose-lowering hormone whose relative or absolute deficiency underlies all forms of diabetes. Insulin is exclusively produced by pancreatic β-cells, which secrete insulin in a precisely controlled manner corresponding to the nutrient state of the organism. In type 2 diabetes (T2D), β-cells fail to respond to nutrients and cannot secrete sufficient amounts of insulin to maintain normoglycemia, while in type 1 diabetes (T1D) β-cells are destroyed by an autoimmune mechanism. Understanding how β-cell function is acquired and regulated is crucial for designing therapies to restore glucose control in diabetes. In particular, the mechanisms underlying acquisition of β-cell function have been the focus of recent interest given efforts to generate functional human β-cells from pluripotent stem cell sources. While the process of β-cell differentiation is fairly well understood [1,2], less is known about the process that equips the β-cell with the ability to regulate insulin secretion. It is clear from a large body of work that the acquisition of β-cell identity (see Glossary) is necessary but not sufficient for nutrient-stimulated insulin secretion, underscoring the major knowledge gap in understanding how insulin secretion first becomes coupled to nutrients in a process termed functional maturation. There is emerging evidence that environmental signals play an important role in β-cell functional maturation. Moreover, mature β-cells constantly monitor the nutrient environment and adjust the insulin secretory response according to changes in organismal nutrient state (functional adaptation). Therefore, the insulin secretory response is not a static property of mature β-cells but rather is actively fine-tuned in response to extracellular signals. Altogether, environmental signals modulate the insulin secretory response throughout lifespan (β-cell functional plasticity) indicating that the functional output of the β-cell at a given moment results from the interplay between extracellular signals and intrinsic properties of the β-cell.
In this review, we discuss evidence for the concept that β-cell functional plasticity is governed by environmental signals that alter gene transcription through signal-dependent transcription factors (SDTFs). SDTFs are TFs with dynamic activity or expression dependent upon environmental signals, exemplified by Bmal1, Creb, NFATc1, and Pparγ (Fig. 1A and Table 1). We propose the effects of extracellular signals upon the insulin secretory response are borne out through the interplay between environment-sensing SDTFs and cell type-selective lineage-determining transcription factors (LDTFs). LDTFs are defined as TFs with restricted expression patterns that confer cell identity by promoting the expression of cell type-specific genes during differentiation [3] (Fig. 1A). These TFs function in tandem with developmental regulation of chromatin state to establish the gene regulatory landscape in a cell, thereby creating a permissive context for later transcriptional regulation [3-6]. Once established during differentiation, gene regulatory elements can be acted upon by SDTFs, which are capable of responding to changes to the extracellular environment but do not directly impact cell identity [3] (Fig. 1A, B). In essence, LDTFs provide genomic “addresses” for recruitment of SDTFs, which fine-tune transcription of cell type-specific genes in response to environmental signals [3]. While SDTFs are activated by environmental signals in diverse cell types, their recruitment to the genome by LDTFs allows environmental signals to regulate the specialized functions of a cell [3, 7] (Fig. 1A, C). Upon differentiation, LDTFs endow the β-cell with its defining features including expression of insulin-processing enzymes and the insulin gene in addition to characteristic metabolic enzymes, ion channels, and the exocytotic machinery, as exemplified by the metabolic enzyme Pcx [1] (Table 1 and Box 1). However, despite possessing the requisite machinery for insulin secretion, newly differentiated β-cells are not yet capable of coupling the insulin secretory response to plasma glucose levels [8-10]. The time period spanning birth to early adulthood is associated with extensive changes in diet, nutrient metabolism, and the hormonal milieu, and a growing body of work has indicated that β-cells mature by sensing and responding to these changes in their environment. During functional maturation, environmental signals act upon cell type-specific gene regulatory programs acquired prenatally to regulate genes involved in insulin secretion, thereby promoting acquisition of glucose stimulated insulin secretion (GSIS). Similarly, in mature β-cells, nutrient signals modulate the insulin secretory response through transcriptional regulation of β-cell-characteristic genes, as we have reviewed in detail elsewhere [11]. In this review, we synthesize evidence that the transcriptional program established by LDTFs is modified by environment-dependent SDTFs to promote β-cell maturation and adapt the insulin secretory response to changing nutrient environments (Fig. 1). Collaborative transcriptional control by LDTFs and SDTFs was originally proposed for immune cells [3] (Fig. 1). Here, we utilize the proposed model of LDTF-SDTF collaboration as a conceptual guide for the interpretation of a growing body of work regarding β-cell functional plasticity. This model can be summarized by two core properties that are largely consistent with the β-cell literature: (1) Gene regulation should occur in a stepwise fashion first involving the establishment of cell type-specific regulatory elements through the activities of LDTFs followed by the adjustment of transcription via environment-dependent SDTFs (Fig. 1B), and (2) SDTFs should be capable of directly regulating cell type-specific genes due to their recruitment being guided by LDTFs, and in this way LDTF-SDTF collaboration confers cell type-specific responses to environmental signals (Fig. 1C). Recurrent findings from studies of β-cell functional plasticity broadly support these properties of SDTF-LDTF collaboration in regulation of the insulin secretory response.
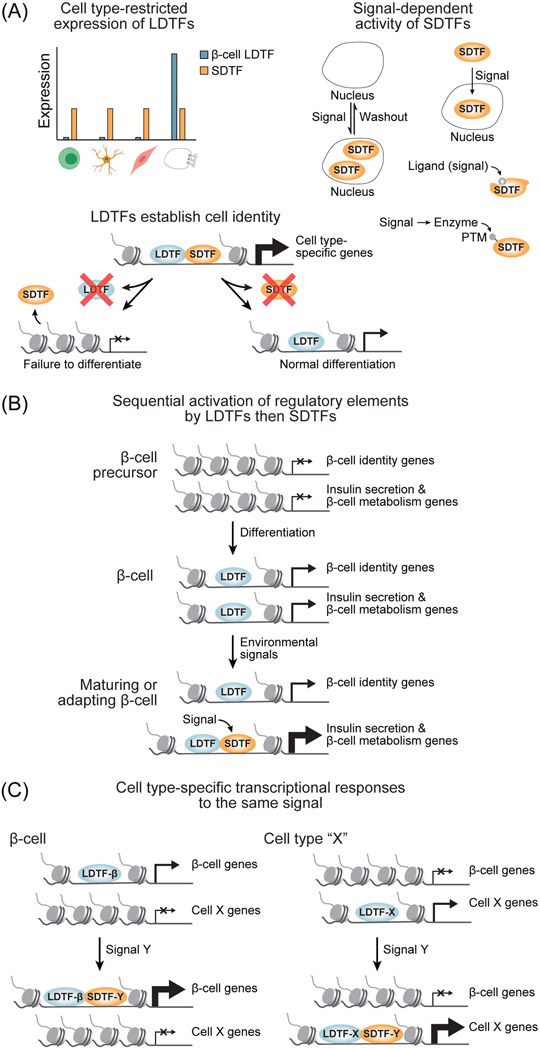
Figure 1. Mechanisms of transcriptional regulation of β-cell identity and functional plasticity.
(A) Defining properties of lineage-determining transcription factors (LDTFs) and signal-dependent transcription factors (SDTFs). Although it is possible for TFs to exhibit characteristics of both classes, these rules classify the vast majority of TFs studied in the context of β-cells (see Table 1). Top left, LDTFs are expressed in restricted numbers of cell types. Bottom left, LDTFs are required for differentiation of specific cell types. Right, activity or expression of SDTFs are dynamically regulated by extracellular signals. PTM, post-translational modification. (B) β-cell-characteristic genes are activated in a stepwise manner during development and maturation first involving establishment of gene regulatory programs by LDTFs followed by fine-tuning of transcription in response to environmental signals by SDTFs. (C) LDTFs provide cell type specificity to the response to environmental signals by directing SDTFs to cell type-specific gene regulatory elements. This figure was created using BioRender (https://biorender.com/).
Table 1.
Lineage-determining and signal-dependent transcription factors in the β-cell.
| TF | Class | Known Regulatorsa |
Process regulated in β-cell maturation |
Function regulated in adult β-cells |
References |
|---|---|---|---|---|---|
| Foxa2 | LDTF | Cell identity | Regulation of GSIS | [1, 2, 78] | |
| Glis3 | LDTF | Cell identity | Cell survival | [1, 2] | |
| Hnf1α | LDTF | GSIS | [1, 2] | ||
| Hnf4α | LDTF | Regulation of GSIS | GSIS | [1, 2, 79] | |
| Insm1 | LDTF | Cell identity | GSIS | [1, 2, 35] | |
| Isl1 | LDTF | Cell identity, proliferation | Ins1/2 transcription, GSIS | [1, 2] | |
| MafB | LDTF | Cell identity | GSIS (human only) | [1, 2, 26] | |
| NeuroD | LDTF | Cell identity | GSIS | [1, 2, 80] | |
| Nkx2.2 | LDTF | Cell identity | Cell identity | [1, 2, 81] | |
| Nkx6.1 | LDTF | Cell identity, proliferation | Insulin processing | [1, 2, 71] | |
| Pax6 | LDTF | Cell identity | Cell identity | [1, 2] | |
| Pdx1 | LDTF | Cell identity, proliferation | Cell identity | [1, 2, 82] | |
| Rfx3 | LDTF | Cell identity | [1] | ||
| Rfx6 | LDTF | Cell identity | GSIS | [1, 2, 76] | |
| Atf3 | SDTF | cAMP | Ins1/2 transcription, GSIS | [83] | |
| Atf4/5 | SDTF | ER stress | Proliferation, survival | [84] | |
| Bmal1 | SDTF | Circadian clock | GSIS | GSIS | [38, 42] |
| Creb/Crtc2 | SDTF | cAMP | GSIS, survival | [27, 56] | |
| Egr1 | SDTF | Fatty acids | Proliferation | GSIS | [85] |
| Errγ | SDTF | Wnt4 | GSIS | GSIS | [30, 43] |
| Fos/AP-1 | SDTF | cAMP, growth factors | Proliferation | GSIS | [86] |
| FoxO1 | SDTF | ROS | Cell identity, GSIS | [11, 46, 48] | |
| FoxO3/4 | SDTF | GSIS | [87] | ||
| MafA | SDTF | T3, cAMP, glucose | GSIS | GSIS | [24, 25] |
| c-Myc | SDTF | Ca2+, mTOR, PKCζ | Proliferation, basal insulin secretion | Proliferation | [74, 88] |
| NFATc1/2 | SDTF | Ca2+-Cn | Insulin processing, proliferation | Proliferation, GSIS | [21, 89-91] |
| Nr4a1 | SDTF | ER stress, fatty acids | Proliferation | GSIS | [52, 53] |
| p53 | SDTF | DNA damage | Regulation of GSIS | [92] | |
| Pparα | SDTF | Fatty acidsb | GSIS | [11] | |
| Pparγ | SDTF | Fatty acidsb | GSIS | [47] | |
| Pparδ | SDTF | Fatty acidsb | GSIS | [11, 54] | |
| Rev-erbα | SDTF | Circadian clock, hemeb | GSIS, proliferation | [93] | |
| SIX2/3 | SDTF | Glucocorticoids | GSIS (human only) | [36, 37] | |
| Smad2/3 | SDTF | Gdf11 | Insulin production, GSIS | Insulin production | [59, 94] |
| Srebp1 | SDTF | Insulin, fatty acids | GSIS | [11] | |
| Srf | SDTF | cAMP, growth factors | Proliferation | GSIS | [60] |
| ThrA/B | SDTF | T3b | Proliferation, GSIS | [25] |
GSIS, glucose-stimulated insulin secretion; ROS, reactive oxygen species; cAMP, cyclic AMP; ER, endoplasmic reticulum; T3, thyroid hormone.
Regulatory relationships between LDTFs during β-cell differentiation have been reviewed in detail elsewhere [2] and are not shown here.
Nuclear receptor ligand
Box 1. Example of a gene regulated by SDTF-LDTF collaboration.
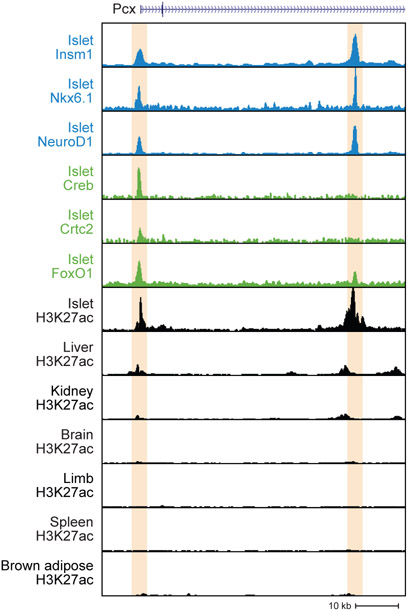
Collaboration between LDTFs and SDTFs enables cells to adjust the expression level of cell type-specific genes, with LDTFs establishing a permissive chromatin state during differentiation and SDTFs fine-tuning transcriptional output. This concept is exemplified in the β-cell by the Pyruvate carboxylase (Pcx) gene, which encodes a metabolic enzyme responsible for generating mitochondrial metabolites that promote insulin secretion. In rodents, Pcx expression increases over the course of functional maturation and during compensation for insulin resistance [8, 49]. β-cell LDTFs bind to cell type-specific enhancers of the Pcx gene (shown as a genome browser snapshot of mouse Pcx in Figure I, data from [27, 35, 46, 71, 72]) and are collectively required for Pcx expression [35, 71]. Pcx transcription is further enhanced during functional maturation by the SDTF MafA and during adaptation by several SDTFs including Pparγ and the Creb/Crtc2 complex [47, 56, 73]. MafA and Pparγ are not shown in the genome browser snapshot due to the absence of high quality ChIP-seq datasets for these SDTFs in mouse islets.
The mechanism of glucose-stimulated insulin secretion
β-cell nutrient sensing primarily occurs through regulated glucose metabolism, which enables tight coupling between circulating glucose levels and insulin release [12] (Fig. 2A, B). To achieve tight regulation of glucose metabolism, mature β-cells express metabolic enzymes and transporters whose activities are dynamic within the range of physiological substrate concentrations. Glycolysis produces ATP via substrate-level phosphorylation by pyruvate kinase [13] and also generates pyruvate, which enters the TCA cycle to stimulate oxidative phosphorylation (OxPhos). The increase of the ATP:ADP ratio resulting from OxPhos and pyruvate kinase activity closes KATP channels on the plasma membrane, thereby inhibiting K+ currents and depolarizing the cell. Voltage-gated Ca2+ channels open in response to plasma membrane depolarization, resulting in Ca2+ influx and activation of the insulin exocytotic machinery, thereby initiating insulin secretion (Fig. 2B). The effectiveness of Ca2+ in promoting insulin vesicle exocytosis is further modulated by signal transduction pathways and mitochondrial metabolites termed metabolic coupling factors (MCFs) [12, 14] (Fig. 2B).
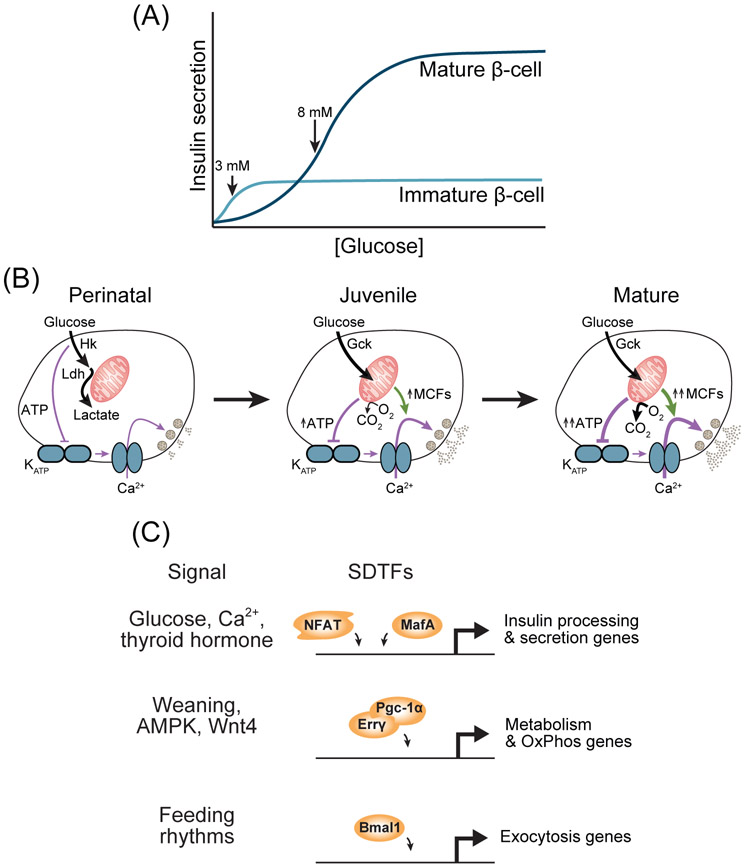
Figure 2. Transcriptional regulation of β-cell maturation.
(A) The relationship between insulin secretion and glucose concentration for mature and immature β-cells, approximated from observations in [16, 30]. (B) Schematic of the metabolic and functional changes of β-cells during maturation. MCFs, metabolic coupling factors. (C) Signals and their cognate signal-dependent transcription factors (SDTFs) involved in β-cell maturation. OxPhos, oxidative phosphorylation.
In newborns, glucose metabolism in the β-cell is constitutive rather than glucose-regulated, which causes a constitutive yet partial activation of Ca2+ influx at glucose levels spanning the physiological range [15] (Fig. 2B). Incomplete restriction of glucose metabolism to the mitochondria results in low rates of OxPhos as well as insufficient production of mitochondrial metabolites that promote insulin secretion [15] (Fig. 2B). Below we summarize current knowledge of how β-cells rewire their metabolism to acquire the glucose-sensing mechanism characteristic of mature β-cells. We posit that glucose sensing is acquired by the exposure of β-cells to environmental signals which evoke gene expression changes via activation of SDTFs. As embryonic β-cells already express a full compendium of LDTFs, such as Foxa2, NeuroD1, Nkx2.2, Nkx6.1, and Pdx1 [1], LDTFs alone are not sufficient for the acquisition of GSIS. The processes involved in conferring β-cell identity through LDTFs have been extensively reviewed elsewhere [1]. Here, we focus on SDTFs and their role in equipping the β-cell with its characteristic functional properties.
Postnatal acquisition of the nutrient-sensing machinery
β-cells first respond to glucose stimulation in the physiological range at 1-2 weeks of age in rodents [16, 17] and as early as one year of age in humans [10] (Fig. 2A). This capability requires metabolic remodeling involving acquisition of tightly regulated glucose oxidation [18] leading to reduction of basal insulin secretion and enhanced insulin secretion in stimulatory glucose (Fig. 2A, B). These functional changes coincide with upregulation of metabolic genes characteristic to the mature β-cell [8, 17, 19]. Maturation of the β-cell metabolic program requires signals from the extracellular environment that activate SDTFs (Fig. 2C and Table 1). Postnatal increases in circulating glucose are required for acquisition of GSIS in part through the Ca2+-activated SDTF NFATc1 [16, 20, 21] (Fig. 2C). NFATc1 binds to the promoters and regulates expression of β-cell-characteristic genes involved in glucose metabolism, such as Gck and Slc2a2, suggesting it is recruited by β-cell LDTFs that initiate expression of these genes. Additional fine-tuning of β-cell metabolism involves selective repression of a set of disallowed genes whose expression would enable metabolic reactions that are constitutively active at physiological nutrient levels or would shunt nutrients away from signal-generating pathways [22] (Fig. 2B and Box 2). This class of genes is exemplified by genes encoding the low Km glucose-phosphorylating enzymes Hk1-3. Gene disallowance is achieved in part by the SDTF MafA, which represses these genes [23, 24]. A spike in thyroid hormone production in the first weeks of life (in rodents) increases MafA expression in the β-cell, leading to repression of disallowed genes as well as increased expression of β-cell-characteristic metabolic genes [24, 25] (Fig. 2C). MafA directly interacts with Pdx1 [26] and colocalizes in the genome with Pdx1 along with Foxa2 and NeuroD [27, 28], suggesting these LDTFs guide recruitment of the SDTF MafA to gene regulatory elements in the β-cell. Altogether, metabolic remodeling associated with the earliest stages of β-cell functional maturation requires the concerted activities of LDTFs intrinsic to the β-cell and SDTFs responsive to changes in the postnatal environment.
Box 2. Disallowed genes.
The repression of disallowed genes is necessary to establish the β-cell-characteristic metabolic program during functional maturation. As gene disallowance involves transcriptional repression rather than activation, it is unclear whether this process adheres to the properties of SDTF-LDTF collaboration. The SDTF c-Myc is known to promote expression of disallowed genes such as Hk3 in immature β-cells, thereby contributing to the high basal insulin secretion characteristic of these cells (Fig. 2A). c-Myc protein decreases in abundance as β-cells mature, leading to a reduction of basal insulin secretion in part due to downregulation of disallowed genes [74]. Postnatal reductions in circulating amino acids are likely signals leading to degradation of c-Myc protein during β-cell functional maturation [74, 75]. However, because the absence of c-Myc protein is not specific to β-cells, it does not fully explain specific repression of β-cell disallowed genes. While several LDTFs including Insm1 [35], NeuroD [35], and Rfx6 [76] directly repress disallowed genes, these TFs are expressed much earlier than the onset of disallowed gene repression, suggesting additional mechanisms confer repressive activities to these TFs later in β-cell maturation (see Outstanding Questions). Other mechanisms of disallowed gene repression that act in tandem with repressive LDTFs include microRNAs that specifically target disallowed genes [17] and epigenetic mechanisms that render the promoters of disallowed genes refractory to further regulation in mature β-cells [77]. Due to their epigenetic repression in mature β-cells, regulation of disallowed genes is restricted to the maturation process and is not thought to contribute to β-cell functional adaptation.
Figure I.
LDTFs and SDTFs co-bind islet-specific regulatory elements of the Pcx gene.
The expression levels of OxPhos genes continuously increase throughout the juvenile period, suggesting these changes contribute to increasingly robust activation of Ca2+ influx in response to glucose [29, 30] (Fig. 2B). Prepubescent rodent or human islets secrete less insulin than adult islets, which has been attributed to changes in expression of mitochondrial metabolic genes [29, 31, 32]. The transition from a milk fat-based diet to a carbohydrate-based diet during weaning provides nutrient signals to SDTFs that promote maturation of the β-cell metabolic program. Premature weaning of mice to a chow diet accelerates β-cell functional maturation [17]. Conversely, weaning mice instead to a high fat diet - mimicking the fat content of milk - delays the acquisition of GSIS, indicating that the change in diet composition during weaning plays a key role in β-cell maturation. Weaning to a chow diet is associated with the activation of AMPK signaling, which leads to upregulation of Pgc-1α. Pgc-1μ is a coactivator of SDTFs that promotes mitochondrial function and biogenesis [33] (Fig. 2C). In β-cells, the Pgc-1α-activated SDTF Errγ has been identified as an important regulator of mitochondrial metabolic genes whose expression increases at this stage of β-cell maturation [30, 34]. In addition to its canonical target genes involved in OxPhos, Errγ regulates β-cell-specific genes involved in insulin vesicle trafficking and exocytosis [30]. How Errγ is recruited to its target sites in β-cells is still unknown. The LDTF Insm1 is a candidate, as Insm1 has been shown to bind to the promoters of exocytotic genes Rab3a and Vamp2 that are also regulated by Erry [35]. SIX2 is an SDTF shown to promote the expression of OxPhos genes in human β-cells between adolescence and adulthood [36, 37], consistent with the onset of SIX2 expression in β-cells during adolescence and the upregulation of SIX2 in adulthood [31]. SIX2 also regulates β-cell-characteristic genes involved in insulin processing and exocytosis [36, 37]. Analysis of genomic SIX2 binding sites in β-cells revealed enrichment of the binding motif for the LDTF MAFB, suggesting that MAFB could guide SIX2 recruitment.
The transition to solid food is also characterized by the onset of rhythmic feeding as opposed to constant nutrient intake during suckling. These changes in the pattern of nutrient intake provide entrainment signals to TFs in the core circadian clock that promote β-cell functional maturation [38] (Fig. 2C). Cell-intrinsic circadian rhythms are driven by the core circadian clock, which is comprised of TFs engaged in a self-sustaining feedback loop. Analysis of binding for core clock TFs (i.e., Bmal1 and Rev-erbα) in different tissues has shown highly tissue-specific patterns of recruitment [39, 40]. Motif analysis further revealed enrichment of motifs for tissue-specific LDTFs at Bmal1 and Rev-erbα binding sites [39-41]. Thus, cell type specificity of circadian gene regulation involves LDTF-mediated recruitment of core clock SDTFs resulting in tissue-specific binding distribution. The core circadian clock is not functional in β-cells of newborn mice but rather develops with the acquisition of glucose responsiveness [38]. Supporting a direct role for core circadian clock TFs in β-cell maturation, β-cell deletion of Bmal1 prevents the acquisition of GSIS [38]. In β-cells Bmal1 has been shown to bind to the genome coincident with the LDTF Pdx1 to promote rhythmic expression of genes involved in metabolism and insulin exocytosis [42] (Fig. 2C), suggesting Pdx1 guides recruitment of Bmal1 in the β-cell.
Despite recent advances, much remains unknown about how SDTFs sense and respond to changes in the environment during β-cell functional maturation. For many SDTFs, the compendium of signals that activate them in β-cells is missing or incomplete (Table 1). For example, while culture models have identified some regulators of SDTFs involved in β-cell maturation, including regulation of Erry by Wnt4 [43] and of SIX2 by glucocorticoids [44], it remains an open question whether this holds true at physiological concentrations of these signals in vivo. Identification of the physiological signals regulating SDTFs and the mechanisms whereby these signals are propagated to the nucleus will be necessary to build a complete understanding of how β-cells acquire and adapt the insulin secretory response.
Transcriptional mechanisms of β-cell functional adaptation
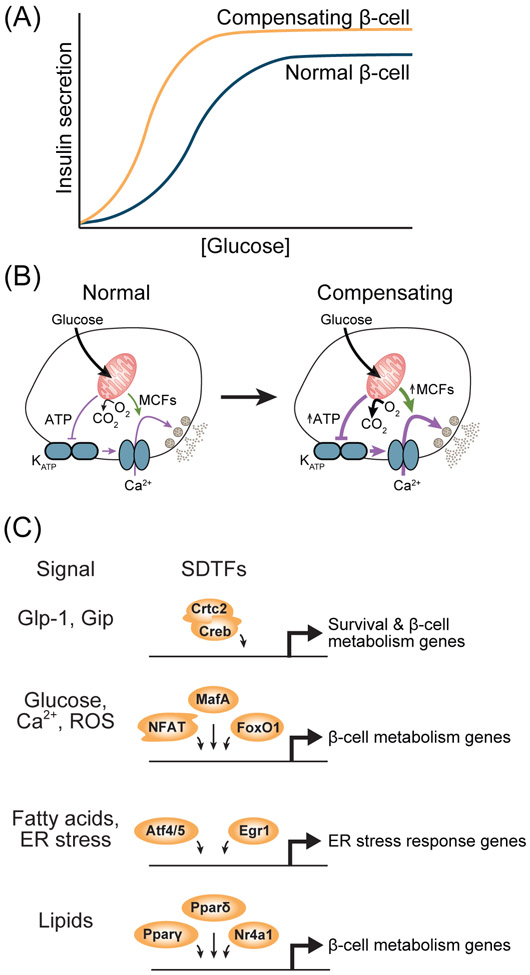
In mature β-cells, changes in nutrient state are an important environmental cue for adapting the insulin secretory response to changes in organismal insulin demand, which we have reviewed in detail elsewhere [11] (Fig. 3A). This adaptive response is necessary to avoid hypoglycemia in the fasted state and to prevent glucose intolerance during insulin resistance. Recurrent observations suggest that fluctuations of nutrient state in adulthood lead to quantitative changes in transcription through a pre-existing network of LDTFs that direct binding of SDTFs, thereby providing the β-cell with the ability to adjust its function in response to changing insulin demands (Table 1). Indeed, assessment of active chromatin in islets of high fat diet-fed mice [45] or db/db mice [46] revealed that changes in nutrient state predominantly affect preexisting regulatory elements rather than activate regulatory elements de novo. This model is consistent with epigenomic profiling of human islets indicating that enhancers unique to the β-cell are co-bound by several β-cell LDTFs and are highly enriched for binding motifs of SDTFs such as AP-1 [28] (Table 1). Mechanistic studies of the reactive oxygen species (ROS)-activated SDTF FoxO1 further support a role for SDTFs in fine-tuning transcription of LDTF-bound regulatory elements during functional adaptation. In obesity, ROS production leads to nuclear translocation of FoxO1, leading to regulation of β-cell-characteristic metabolic genes [46-48]. FoxO1 colocalizes in the genome with β-cell LDTFs such as Pdx1 and NeuroD [46], suggesting these LDTFs guide recruitment of FoxO1 to allow this SDTF to regulate genes involved in insulin secretion. Thus, analogous to β-cell maturation, β-cell functional adaptation involves regulation of genes involved in insulin secretion by environmental signals and cognate SDTFs [11]. The gene regulatory programs of β-cell maturation and functional adaptation have similarities but are not identical. Unique to β-cell maturation is the repression of disallowed genes (Box 2), whereas both processes involve changes in β-cell intracellular glucose metabolism [11, 49-51] (Fig. 3B). During obesity, moderate lipid accumulation in the β-cell activates several SDTFs that promote adaptive insulin secretion through metabolic remodeling including Nr4a1 [52, 53], Pparγ [47], and Pparδ [11, 54] (Fig. 3C).
Figure 3. Transcriptional regulation in the β-cell during functional adaptation.
(A) The relationship between insulin secretion and glucose concentration during compensation for insulin resistance relative to normal β-cells, based on observations in [50, 51]. (B) Schematic of the metabolic and functional changes in β-cells during compensation for insulin resistance. MCFs, metabolic coupling factors. (C) Signals and their cognate signal-dependent transcription factors (SDTFs) involved in compensation for insulin resistance. ROS, reactive oxygen species; ER, endoplasmic reticulum.
How an SDTF expressed in several tissues orchestrates cell type-specific transcriptional responses as a result of its recruitment by LDTFs is best illustrated by the SDTF Creb, which promotes adaptive insulin secretion in β-cells. The second messenger cAMP, which activates Creb, evokes distinct physiological responses in different cell types in part through cell type specificity of Creb target genes (Fig. 3C). For example, while cAMP enhances the insulin secretory response of β-cells, in hepatocytes cAMP promotes gluconeogenesis [27, 55, 56]. These differences have been attributed in part to cell type specificity of genomic Creb binding sites [27]. In β-cells, the LDTF NeuroD recruits Creb to gene regulatory elements, thereby enabling Creb to regulate β-cell-specific genes involved in insulin secretion [27]. Knockdown of NeuroD reduces Creb binding at β-cell-specific sites and interferes with transcriptional activation of Creb targets unique to the β-cell without disrupting overall Creb function. Ectopic NeuroD expression in pancreatic exocrine cells leads to recruitment of Creb to β-cell-specific regulatory elements [27]. The example of NeuroD and Creb illustrates that LDTFs provide genomic “addresses” to SDTFs to enable tissue-specific transcriptional responses to second messengers (Fig. 1C). Deeper investigation of SDTF-LDTF complexes in β-cells holds promise for identifying regulatory programs capable of modulating specific aspects of β-cell function.
Relevance of collaborative transcriptional regulation to therapeutic strategies in diabetes
Intensive study of pancreatic development for the purpose of developing β-cell replacement therapies has led to the design of pluripotent stem cell differentiation protocols that generate insulin-producing cells expressing nearly a full complement of β-cell LDTFs [57]. Despite these advances, β-cells produced by current differentiation protocols exhibit several functional defects, including high basal insulin secretion, lower first phase insulin secretion compared to primary β-cells, and reduced or absent second phase insulin secretion [57-59]. As reviewed here, environmental signals and their cognate SDTFs play fundamental roles in acquisition and adjustment of the insulin secretory response. However, our understanding of SDTFs in the β-cell represents a major knowledge gap in β-cell biology that lags behind that of β-cell LDTFs. The catalog of SDTFs necessary for β-cell functional plasticity is almost certainly incomplete. Unbiased analysis of genes correlating with maturation state of single β-cells suggests a role for a number of SDTFs including Atf3, Srf, and the AP-1 family TFs in β-cell functional maturation [60]. Our group recently compared motif enrichment at active chromatin sites in islets from fed and fasted mice, which revealed AP-1 and ETS families as candidate SDTFs for mediating β-cell functional adaptation [61]. The recent advent of technologies for mapping chromatin state at the single cell level [62] should aid the unbiased identification of SDTFs involved in the regulation of β-cell functional plasticity. Application of these emerging technologies to conditions associated with β-cell functional plasticity should shed light onto this process and help identify strategies for promoting maturation of β-cells derived from pluripotent stem cells or enhancing functional adaptation of endogenous β-cells.
The environment of incipient diabetes has been shown to activate SDTFs that promote β-cell dysfunction, which could be targets for diabetes prevention. Activation of gene regulatory elements in response to cytokine treatment mimicking the inflammatory environment of T1D revealed that LDTFs can direct recruitment of SDTFs mediating β-cell dysfunction such as IRF family TFs [63]. Similarly, the systemic environment of T2D can activate maladaptive transcriptional programs through SDTFs [64, 65] that impair insulin secretion. Animal models of severe T1D or T2D have additionally revealed impaired expression of LDTFs and cell type-specific genes in the β-cell together with ectopic expression of non-pancreatic hormones [66-68]. The extent to which these phenomena also occur in human diabetes is under intense investigation. Nevertheless, these findings suggest that the environments of T1D and T2D disrupt β-cell function and identity in a process distinct from a simple reversal of developmental β-cell differentiation and maturation. Further exploration of how β-cell transcriptional regulation is remodeled by the stressful environments associated with T1D and T2D has potential to reveal novel pathogenic mechanisms and therapeutic targets.
Concluding remarks
Glucose-regulated insulin secretion is acquired in a process distinct from β-cell differentiation in part through environmental signals, and insulin secretion is continually adjusted throughout lifespan in response to changes in the nutrient environment. We here reviewed evidence supporting a mechanism whereby β-cell LDTFs endow the β-cell with characteristic ion channels and the machinery to process and exocytose insulin, whereas SDTFs act upon the transcriptional program established by LDTFs to mature and adapt the insulin secretory response in response to environmental signals (Figs. 1B, 2C, and 3C).
While the conceptual model of LDTF-SDTF collaboration is intended to distill an abundance of observations from the literature, it is almost certainly a simplification of the complex transcriptional regulation that occurs in vivo, and there will likely be exceptions to the generalized rules discussed in this review (Fig. 1). The operative definitions of LDTFs and SDTFs (Fig. 1A) leave open the possibility for individual TFs to exhibit characteristics of both classes and therefore defy strict categorization. In some cases, environmental signals have been shown to activate gene regulatory elements de novo without preexisting chromatin priming or TF binding [63], indicating that not all regulatory elements undergo sequential activation (Fig. 1B). Finally, little is known about the specific mechanisms of SDTF cooperation with LDTFs in the β-cell. With the exception of Creb recruitment by NeuroD [27], there are few mechanistic studies of this process. Cooperative, rather than sequential, binding of SDTFs and LDTFs remains possible, and transient binding and dissociation of TFs to DNA could confound interpretations of sequential binding of different TFs as inferred through static assays such as chromatin immunoprecipitation sequencing (ChIP-seq) [69]. While gain- or loss-of-function experiments followed by ChIP and gene expression assays are the gold standard for delineating the logic of TF binding and direct gene regulation [3], such datasets have not been generated for many TFs discussed here. With increased sensitivity of assays to map TF binding [70], it will be possible to characterize mechanisms of SDTF recruitment by LDTFs in β-cells. In writing this review, we hope to stimulate further studies of collaborative transcriptional control in the β-cell (see Outstanding Questions) with the expectation that a deeper understanding of these transcriptional networks will lead to improved human stem cell-derived β-cell models and novel strategies for enhancing adaptive insulin secretion or preventing β-cell decompensation.
Outstanding Questions.
Are SDTFs appropriately activated during in vitro differentiation of β-cells from pluripotent stem cells? If not, what environmental signals or SDTFs are missing from current β-cell differentiation protocols?
How do TFs that normally activate gene expression repress disallowed genes during β-cell maturation?
What is the role of the epigenome in the response to environmental signals promoting β-cell functional plasticity?
What are the upstream signals that couple environmental cues to SDTF activity in β-cells?
How do SDTFs contribute to β-cell failure in T1D and T2D?
Highlights.
Lineage determining transcription factors (LDTFs) are required for β-cell differentiation, yet acquisition of β-cell identity is not sufficient for glucose-stimulated insulin secretion.
Environmental signals that regulate signal-dependent transcription factors (SDTFs) govern acquisition and adaptation of glucose-stimulated insulin secretion.
β-cell function is acquired in a stepwise manner whereby LDTFs initiate expression of cell type-characteristic genes, then SDTFs fine-tune gene expression to confer metabolic and functional properties to β-cells.
LDTFs guide the recruitment of SDTFs to provide cell type specificity to the transcriptional effects of environmental signals in neonatal and adult β-cells.
Glossary
- β-cell identity
The capability to express insulin and genes that participate in insulin processing, granule formation, and exocytosis
- Disallowed genes
Genes selectively repressed in islets or β-cells compared to other tissues
- Functional adaptation
An increase or decrease of the insulin secretory response per β-cell
- Functional maturation
Acquisition of glucose-responsive insulin secretion and reduction of basal insulin secretion by the β-cell
- Functional plasticity
A blanket term referring to both β-cell functional adaptation and functional maturation
- Lineage-determining transcription factor (LDTF)
A constitutively active, sequence-specific transcription factor (TF) exhibiting a restricted expression pattern that is typically required for differentiation and maintenance of cell identity
- Signal-dependent transcription factor (SDTF)
A TF broadly expressed or induced in diverse cell types that is activated by extracellular stimuli
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dassaye R, et al. (2016) Transcription factor regulation of pancreatic organogenesis, differentiation and maturation. Islets 8, 13–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arda HE, et al. (2013) Gene regulatory networks governing pancreas development. Developmental Cell 25, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinz S, et al. (2015) The selection and function of cell type-specific enhancers. Nature Reviews Molecular Cell Biology 16, 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang A, et al. (2015) Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 16, 386–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee K, et al. (2019) FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Reports 28, 382–393 e387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez-Dominguez JR, et al. (2020) Circadian entrainment triggers maturation of human in vitro islets. Cell Stem Cell 26, 108–122 e110 [DOI] [PubMed] [Google Scholar]
- 7.Mullen AC, et al. (2011) Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147, 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jermendy A, et al. (2011) Rat neonatal beta cells lack the specialised metabolic phenotype of mature beta cells. Diabetologia 54, 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rorsman P, et al. (1989) Failure of glucose to elicit a normal secretory response in fetal pancreatic beta cells results from glucose insensitivity of the ATP-regulated K+ channels. Proceedings of the National Academy of Sciences of the United States of America 86, 4505–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henquin JC and Nenquin M (2018) Immaturity of insulin secretion by pancreatic islets isolated from one human neonate. Journal of Diabetes Investigation 9, 270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wortham M and Sander M (2016) Mechanisms of beta-cell functional adaptation to changes in workload. Diabetes, Obesity & Metabolism 18 Suppl 1,78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell JE and Newgard CB (2021) Mechanisms controlling pancreatic islet cell function in insulin secretion. Nature Reviews Molecular Cell Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewandowski SL, et al. (2020) Pyruvate kinase controls signal strength in the insulin secretory pathway. Cell Metabolism 32, 736–750 e735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalwat MA and Cobb MH (2017) Mechanisms of the amplifying pathway of insulin secretion in the beta cell. Pharmacology & Therapeutics 179, 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boschero AC, et al. (1990) D-glucose and L-leucine metabolism in neonatal and adult cultured rat pancreatic islets. Molecular and Cellular Endocrinology 73, 63–71 [DOI] [PubMed] [Google Scholar]
- 16.Blum B, et al. (2012) Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nature Biotechnology 30, 261–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacovetti C, et al. (2015) Postnatal beta-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nature Communications 6, 8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stolovich-Rain M, et al. (2015) Weaning triggers a maturation step of pancreatic beta cells. Developmental Cell 32, 535–545 [DOI] [PubMed] [Google Scholar]
- 19.Dhawan S, et al. (2015) DNA methylation directs functional maturation of pancreatic beta cells. The Journal of Clinical Investigation 125, 2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aguayo-Mazzucato C, et al. (2006) Restructuring of pancreatic islets and insulin secretion in a postnatal critical window. PloS One 1, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, et al. (2019) In vivo imaging of beta-cell function reveals glucose-mediated heterogeneity of beta-cell functional development. eLife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pullen TJ and Rutter GA (2013) When less is more: the forbidden fruits of gene repression in the adult beta-cell. Diabetes, Obesity & Metabolism 15, 503–512 [DOI] [PubMed] [Google Scholar]
- 23.Hang Y, et al. (2014) The MafA transcription factor becomes essential to islet beta-cells soon after birth. Diabetes 63, 1994–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimura W, et al. (2015) MafA is critical for maintenance of the mature beta cell phenotype in mice. Diabetologia 58, 566–574 [DOI] [PubMed] [Google Scholar]
- 25.Aguayo-Mazzucato C, et al. (2013) Thyroid hormone promotes postnatal rat pancreatic beta-cell development and glucose-responsive insulin secretion through MAFA. Diabetes 62, 1569–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scoville DW, et al. (2015) MLL3 and MLL4 methyltransferases bind to the MAFA and MAFB transcription factors to regulate islet beta-cell function. Diabetes 64, 3772–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Velde S, et al. (2019) CREB promotes beta cell gene expression by targeting its coactivators to tissue-specific enhancers. Molecular and Cellular Biology 39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasquali L, et al. (2014) Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nature Genetics 46, 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avrahami D, et al. (2015) Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved beta cell function. Cell Metabolism 22, 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshihara E, et al. (2016) ERRgamma Is required for the metabolic maturation of therapeutically functional glucose-responsive beta cells. Cell Metabolism 23, 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arda HE, et al. (2016) Age-dependent pancreatic gene regulation reveals mechanisms governing human beta cell function. Cell Metabolism 23, 909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wortham M, et al. (2018) Integrated in vivo quantitative proteomics and nutrient tracing reveals age-related metabolic rewiring of pancreatic beta cell function. Cell Reports 25, 2904–2918 e2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaafar R, et al. (2019) mTORC1 to AMPK switching underlies beta-cell metabolic plasticity during maturation and diabetes. The Journal of Clinical Investigation 130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devarakonda S, et al. (2011) Disorder-to-order transition underlies the structural basis for the assembly of a transcriptionally active PGC-1alpha/ERRgamma complex. Proceedings of the National Academy of Sciences of the United States of America 108, 18678–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia S, et al. (2015) Insm1 cooperates with Neurod1 and Foxa2 to maintain mature pancreatic beta-cell function. The EMBO Journal 34, 1417–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velazco-Cruz L, et al. (2020) SIX2 regulates human beta cell differentiation from stem cells and functional maturation in vitro. Cell Reports 31, 107687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bevacqua RJ, et al. (2021) SIX2 and SIX3 coordinately regulate functional maturity and fate of human pancreatic beta cells. Genes & Development [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakshit K, et al. (2018) Postnatal ontogenesis of the islet circadian clock plays a contributory role in beta-cell maturation process. Diabetes 67, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. (2016) HNF6 and Rev-erbalpha integrate hepatic lipid metabolism by overlapping and distinct transcriptional mechanisms. Genes & Development 30, 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. (2015) GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 348, 1488–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beytebiere JR, et al. (2019) Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions. Genes & Development 33, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perelis M, et al. (2015) Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350, aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshihara E, et al. (2020) Immune-evasive human islet-like organoids ameliorate diabetes. Nature 586, 606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aylward A, et al. (2020) Glucocorticoid signaling in pancreatic islets modulates gene regulatory programs and genetic risk of type 2 diabetes. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nammo T, et al. (2018) Genome-wide profiling of histone H3K27 acetylation featured fatty acid signalling in pancreatic beta cells in diet-induced obesity in mice. Diabetologia 61, 2608–2620 [DOI] [PubMed] [Google Scholar]
- 46.Kuo T, et al. (2019) Identification of C2CD4A as a human diabetes susceptibility gene with a role in beta cell insulin secretion. Proceedings of the National Academy of Sciences of the United States of America 116, 20033–20042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta D, et al. (2013) Peroxisome proliferator-activated receptor gamma (PPARgamma) and its target genes are downstream effectors of FoxO1 protein in islet beta-cells: mechanism of beta-cell compensation and failure. The Journal of Biological Chemistry 288, 25440–25449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitamura YI, et al. (2005) FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metabolism 2, 153–163 [DOI] [PubMed] [Google Scholar]
- 49.Gupta D, et al. (2017) Temporal characterization of beta cell-adaptive and - maladaptive mechanisms during chronic high-fat feeding in C57BL/6NTac mice. The Journal of Biological Chemistry 292, 12449–12459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou YP, et al. (1999) Basal insulin hypersecretion in insulin-resistant Zucker diabetic and Zucker fatty rats: role of enhanced fuel metabolism. Metabolism: Clinical and Experimental 48, 857–864 [DOI] [PubMed] [Google Scholar]
- 51.Liu YQ, et al. (2002) Beta-cell adaptation to insulin resistance. Increased pyruvate carboxylase and malate-pyruvate shuttle activity in islets of nondiabetic Zucker fatty rats. The Journal of Biological Chemistry 277, 39163–39168 [DOI] [PubMed] [Google Scholar]
- 52.Briand O, et al. (2012) The nuclear orphan receptor Nur77 is a lipotoxicity sensor regulating glucose-induced insulin secretion in pancreatic beta-cells. Molecular Endocrinology 26, 399–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds MS, et al. (2016) Beta-cell deletion of Nr4a1 and Nr4a3 nuclear receptors impedes mitochondrial respiration and insulin secretion. American Journal of Physiology. Endocrinology and Metabolism 311, E186–201 [DOI] [PubMed] [Google Scholar]
- 54.Tang T, et al. (2013) Desnutrin/ATGL activates PPARdelta to promote mitochondrial function for insulin secretion in islet beta cells. Cell Metabolism 18, 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park S, et al. (2006) Exendin-4 uses Irs2 signaling to mediate pancreatic beta cell growth and function. The Journal of Biological Chemistry 281, 1159–1168 [DOI] [PubMed] [Google Scholar]
- 56.Blanchet E, et al. (2015) Feedback inhibition of CREB signaling promotes beta cell dysfunction in insulin resistance. Cell Reports 10, 1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veres A, et al. (2019) Charting cellular identity during human in vitro beta-cell differentiation. Nature 569, 368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nair GG, et al. (2019) Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived beta cells. Nature Cell Biology 21, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Velazco-Cruz L, et al. (2019) Acquisition of dynamic function in human stem cell-derived beta cells. Stem Cell Reports 12, 351–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng C, et al. (2017) Pseudotemporal ordering of single cells reveals metabolic control of postnatal beta cell proliferation. Cell Metabolism 25, 1160–1175 e1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wortham M, et al. (2019) Nutrient regulation of the islet epigenome controls adaptive insulin secretion. BioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buenrostro JD, et al. (2015) Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramos-Rodriguez M, et al. (2019) The impact of proinflammatory cytokines on the beta-cell regulatory landscape provides insights into the genetics of type 1 diabetes. Nature Genetics 51, 1588–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang E, et al. (2019) Preserving insulin secretion in diabetes by inhibiting VDAC1 overexpression and surface translocation in beta cells. Cell Metabolism 29, 64–77 e66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montemurro C, et al. (2019) IAPP toxicity activates HIF1alpha/PFKFB3 signaling delaying beta-cell loss at the expense of beta-cell function. Nature Communications 10, 2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo S, et al. (2013) Inactivation of specific beta cell transcription factors in type 2 diabetes. The Journal of Clinical Investigation 123, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sachs S, et al. (2020) Targeted pharmacological therapy restores beta-cell function for diabetes remission. Nature Metabolism 2, 192–209 [DOI] [PubMed] [Google Scholar]
- 68.Chung KM, et al. (2020) Endocrine-exocrine signaling drives obesity-associated pancreatic ductal adenocarcinoma. Cell 181, 832–847 e818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swinstead EE, et al. (2016) Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skene PJ and Henikoff S (2017) An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor BL, et al. (2013) Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Reports 4, 1262–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen Y, et al. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H, et al. (2007) MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia 50, 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Puri S, et al. (2018) Replication confers beta cell immaturity. Nature Communications 9, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Helman A, et al. (2020) A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metabolism 31, 1004–1016 e1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Piccand J, et al. (2014) Rfx6 maintains the functional identity of adult pancreatic beta cells. Cell Reports 9, 2219–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemaire K, et al. (2016) Disallowed and allowed gene expression: Two Faces of Mature Islet Beta Cells. Annu Rev Nutr 36, 45–71 [DOI] [PubMed] [Google Scholar]
- 78.Gao N, et al. (2007) Foxa2 controls vesicle docking and insulin secretion in mature Beta cells. Cell Metabolism 6, 267–279 [DOI] [PubMed] [Google Scholar]
- 79.Gupta RK, et al. (2005) The MODY1 gene HNF-4alpha regulates selected genes involved in insulin secretion. The Journal of Clinical Investigation 115, 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gu C, et al. (2010) Pancreatic beta cells require NeuroD to achieve and maintain functional maturity. Cell Metabolism 11, 298–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gutierrez GD, et al. (2017) Pancreatic beta cell identity requires continual repression of non-beta cell programs. The Journal of Clinical Investigation 127, 244–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gao T, et al. (2014) Pdx1 maintains beta cell identity and function by repressing an alpha cell program. Cell Metabolism 19, 259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zmuda EJ, et al. (2010) The roles of ATF3, an adaptive-response gene, in high-fat-diet-induced diabetes and pancreatic beta-cell dysfunction. Molecular Endocrinology 24, 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Juliana CA, et al. (2018) A PDX1-ATF transcriptional complex governs beta cell survival during stress. Molecular Metabolism 17, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheong MW, et al. (2015) Loss of Egr-1 sensitizes pancreatic beta-cells to palmitate-induced ER stress and apoptosis. Journal of molecular medicine 93, 807–818 [DOI] [PubMed] [Google Scholar]
- 86.Ray JD, et al. (2016) Nkx6.1-mediated insulin secretion and beta-cell proliferation is dependent on upregulation of c-Fos. FEBS letters 590, 1791–1803 [DOI] [PubMed] [Google Scholar]
- 87.Kim-Muller JY, et al. (2014) Metabolic inflexibility impairs insulin secretion and results in MODY-like diabetes in triple FoxO-deficient mice. Cell Metabolism 20, 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosselot C, et al. (2019) Myc Is required for adaptive beta-cell replication in young mice but is not sufficient in one-year-old mice fed with a high-fat diet. Diabetes [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goodyer WR, et al. (2012) Neonatal beta cell development in mice and humans is regulated by calcineurin/NFAT. Developmental Cell 23, 21–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heit JJ, et al. (2006) Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature 443, 345–349 [DOI] [PubMed] [Google Scholar]
- 91.Keller MP, et al. (2016) The transcription factor Nfatc2 regulates beta-cell proliferation and genes associated with type 2 diabetes in mouse and human islets. PLoS Genetics 12, e1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li X, et al. (2016) The MDM2-p53-pyruvate carboxylase signalling axis couples mitochondrial metabolism to glucose-stimulated insulin secretion in pancreatic beta-cells. Nature Communications 7, 11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vieira E, et al. (2012) The clock gene Rev-erbalpha regulates pancreatic beta-cell function: modulation by leptin and high-fat diet. Endocrinology 153, 592–601 [DOI] [PubMed] [Google Scholar]
- 94.Smart NG, et al. (2006) Conditional expression of Smad7 in pancreatic beta cells disrupts TGF-beta signaling and induces reversible diabetes mellitus. PLoS Biol 4, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]