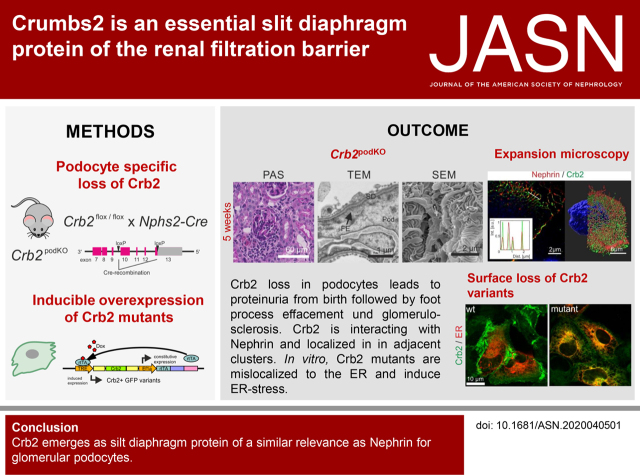
Significance Statement
Mutations in the Crumbs2 (CRB2) gene were recently identified as a cause of steroid-resistant nephrotic syndrome (SRNS), but it is unknown how Crumbs2 dysfunction damages the glomerular filtration barrier (GFB). Crumbs2 is a slit diaphragm protein, organized in clusters and able to interact with Nephrin. In mice, loss of Crumbs2 results in heavy proteinuria, accompanied by progressive podocyte foot process effacement, injury of renal cells, and inflammation, indicating that Crb2 is of similar relevance for the GFB as Nephrin. In vitro, CRB2 mutant proteins accumulate in the ER and trigger ER stress. Thus, in addition to loss of Crumbs2 at the slit diaphragm, ER stress induction could play a pivotal role in onset and disease progression in patients with SRNS.
Keywords: glomerular disease, podocyte, proteinuria, Crumbs2, slit diaphragm, ER stress, Crb2, CRB2, glomerular filtration barrier
Visual Abstract
Abstract
Background
Crumbs2 is expressed at embryonic stages as well as in the retina, brain, and glomerular podocytes. Recent studies identified CRB2 mutations as a novel cause of steroid-resistant nephrotic syndrome (SRNS).
Methods
To study the function of Crb2 at the renal filtration barrier, mice lacking Crb2 exclusively in podocytes were generated. Gene expression and histologic studies as well as transmission and scanning electron microscopy were used to analyze these Crb2 podKO knockout mice and their littermate controls. Furthermore, high-resolution expansion microscopy was used to investigate Crb2 distribution in murine glomeruli. For pull-down experiments, live cell imaging, and transcriptome analyses, cell lines were applied that inducibly express fluorescent protein–tagged CRB2 wild type and mutants.
Results
Crb2 podKO mice developed proteinuria directly after birth that preceded a prominent development of disordered and effaced foot processes, upregulation of renal injury and inflammatory markers, and glomerulosclerosis. Pull-down assays revealed an interaction of CRB2 with Nephrin, mediated by their extracellular domains. Expansion microscopy showed that in mice glomeruli, Crb2 and Nephrin are organized in adjacent clusters. SRNS-associated CRB2 protein variants and a mutant that lacks a putative conserved O-glycosylation site were not transported to the cell surface. Instead, mutants accumulated in the ER, showed altered glycosylation pattern, and triggered an ER stress response.
Conclusions
Crb2 is an essential component of the podocyte’s slit diaphragm, interacting with Nephrin. Loss of slit diaphragm targeting and increasing ER stress are pivotal factors for onset and progression of CRB2-related SRNS.
Podocytes are postmitotic cells that serve as part of a molecular sieve that establishes the permselective properties of the glomerular filtration barrier (GFB). They consist of a voluminous cell body, which bulges into the urinary space and highly branched foot processes. The foot processes of neighboring podocytes regularly interdigitate, leaving meandering filtration slits between them that are bridged by the slit diaphragm. Together with the glomerular basement membrane (GBM) and the fenestrated endothelium, the slit diaphragm builds the three-layered GFB for proper blood filtration.1–3
Studies of the past 20 years identified many components of the slit diaphragm and elucidated the central role of this unique structure. The extracellular parts of these approximately 40-nm-wide cell junction serve as a macromolecular filter that prevents loss of essential serum proteins, like albumin.1–3 The intracellular adaptors of the slit diaphragm complex mediate binding to the actin cytoskeleton.4,5 In addition to these structural tasks, the slit diaphragm functions as a hub, integrating various signals from inside and outside to control dynamic changes of the podocytes and their branched foot processes.6–8 The slit diaphragm contains elements of adherence9 and tight junctions10 and additional components that are specific to podocytes.3,6,8
Genetic studies of hereditary proteinuric diseases uncovered mutations in slit diaphragm genes.11 The best-studied slit diaphragm component is the type I transmembrane protein Nephrin (gene: NPHS1).8,12 So far, about 250 NPHS1 mutations have been identified causing congenital nephrotic syndrome of the Finnish type (CNF),13 a type of steroid-resistant nephrotic syndrome (SNRS) that leads to severe proteinuria, frequently starting in utero, and that is often associated with death within the neonatal period.14,15 Most of the CNF mutations are found within the extracellular domain (ECD) that mediates homotypic—Nephrin-Nephrin—or heterotypic Nephrin interactions with Neph1, a Nephrin-like slit diaphragm protein.12,13,16 The intracellular C-terminal domain (ICD) of Nephrin mediates binding to various adaptor proteins. The most important one is Podocin (gene: NPHS2) that binds and directs Nephrin into the lipid rafts of the slit diaphragm.17–19
In addition, the Nephrin ICD contains several phosphorylation sites for Fyn tyrosine or phosphatidylinositol 3-kinase, which modulate binding to various adaptors (e.g., Crk and Nck family) to control actin cytoskeleton dynamics of foot processes.8 Thus, Nephrin also acts as a signaling hub in various signal transduction pathways and the crosstalk between slit diaphragm signaling and podocyte attachment to the extracellular matrix.8,20
Crumbs2 (mouse: Crb2, human: CRB2) is a member of the evolutionary conserved apically localized Crumbs protein family that controls cell polarization and establishment of cell-cell contacts.21–23 Mammals express three Crumbs isoforms (Crb1, Crb2, and two splice variants of Crb3 [Crb3a/Crb3b]) in cell type– and tissue-specific patterns. Crb3 isoforms (Crb3a/b) are small proteins lacking all domains of the ECD.24–26 They are the predominant isoforms in kidney and expressed in all renal epithelia. In contrast, Crb2, which promotes cell ingression during the epithelial-to-mesenchymal transition at gastrulation, is exclusively expressed in Crb3-positive renal precursor cells that later mature into postmitotic highly branched podocytes.27,28
Interestingly, mutations in the human CRB2 gene cause a triad of phenotypes, including elevated α-fetoprotein levels, cerebral ventriculomegaly, and renal phenotypes strongly resembling CNF caused by Nephrin mutations.29–31 Crb2 is expressed during embryogenesis,28,32 in brain,33,34 in retina35,36 as well as in developing27 and mature podocytes of the kidney.29–31,37,38
Like Nephrin, Crb2 is a multidomain protein with a short ICD (37 amino acids), a transmembrane region, and a long ECD (1225 amino acids) that carries most of the so far identified variations in patients with CRB2 SRNS, indicating a special role of the CRB2 ECD for the slit diaphragm.31 Indeed, earlier studies demonstrated that knockdown of the Crb2 homolog in zebrafish (crb2b) causes disorganization of foot process architecture and loss of the slit diaphragm.37
However, so far, a mammalian model lacking CRB2 in podocytes is missing, and on a cellular level, it is still incompletely understood how CRB2 mutants cause SRNS-associated phenotypes. Here, we addressed these aspects by analyzing the phenotype of mice lacking Crb2 exclusively in podocytes (Crb2 podKO mice) as well as in podocyte cell lines expressing green fluorescent protein (GFP)–tagged Crb2 wild type and mutants. Our results show that Crb2 podKO mice developed heavy proteinuria, foot process effacement, and upregulation of inflammatory and renal injury markers. Additionally, we identified Crb2 as a novel Nephrin binding partner and elucidated by high-resolution expansion microscopy that both proteins are organized in adjacent clusters. Cell culture studies and live cell imaging revealed that CRB2 protein mutants accumulate in the endoplasmic reticulum (ER), accompanied by altered glycosylation patterns and induction of ER stress pathways.
Methods
Mice and Genotyping
All experimental protocols and methods in this work involving animals were approved by and conducted in accordance with all guidelines and regulations set forth by the German regional authorities (Az.: 84–02.04.2014 A405; LANUV). Animals were housed under standard specific pathogen–free conditions with free access to tap water and standard animal chow. The conditional Crb2 knockout mice (Crb2 flox/flox) were described earlier.36, 39 To excise the floxed Crb2 genes in the kidney, we used the Nphs2 and Six2-Cre lines.40–42 For DNA isolation and genotyping, tissue was incubated in lysis buffer (50 mM KCl, 1.5 mM MgCl2, 10 mM Tris/HCl, pH 8.3, 0.45% NP-40, and 0.45% Tween 20) containing 0.05 µg/µl proteinase K (Qiagen) at 56°C overnight (ON). The proteinase K was then inactivated by incubation at 95°C for 10 minutes, the samples were centrifuged, and the DNA-containing supernatant was used for genotyping. Mice were genotyped as described earlier using primer Crb2Fwd and Crb2Rev for floxed Crb2 and CreFwd and CreRev to identify Cre-positive mice (Supplemental Table 1). Spontaneous urine of mice was loaded on SDS gels to determine proteinuria phenotype. Protein-creatinine ratio (grams/gram) was measured in urine samples at IDEXX BioAnalytics GmbH laboratories (n=5 animals per group).
Tissue Preparation and Histologic Analyses
Kidneys were perfused with PBS or Sterofundin and 4% PFA in PBS and immediately removed. Native tissue was frozen directly for protein extraction and RNA preparation in RNAlater (Qiagen) or immersed with 10% sucrose for cryosections. Longitudinally halved kidneys were incubated with 4% PFA in PBS for 1–2 hours at room temperature (RT) for 100-µm-thick vibratome slices for expansion microscopy or processed by standard procedures for paraffin sections (2 µm) and stained with periodic acid–Schiff.
Glomeruli Isolation
For isolation of glomeruli, kidneys were cut in small pieces with a scalpel in 3 ml HBSS and treated with collagenase II (1 mg/ml) in HBSS at 37°C for 30 minutes. The tissue was transferred on a 100-µm nylon tissue sieve and rinsed with 30 ml HBSS. Afterward, the suspension was flown through a 70-µm sieve and rinsed with additional 15 ml PBS. The last sieving step was performed with a 40-µm sieve. Glomeruli retained on the sieve could be transferred with a pipette into a new 15-ml reaction tube. The flow through (tubuli fraction) and the glomeruli fraction were centrifuged for 10 minutes at 2000 rpm and were lysed for RNA isolation or protein immunoblotting.
Immunohistochemistry Analysis
For cryosections, native PBS-perfused mice kidneys were dehydrated with 10% sucrose ON before freezing them with cold 2-methyl-butane in liquid nitrogen into Tissue-Tek O.C.T. Compound (Sakura Finetek BV, Leiden, The Netherlands) blocks. Cryosections (cut with cryostat Leica CM3050S, 5 µm) were fixed for 10 minutes with 4% PFA in PBS and washed before incubation with 0.5% Triton X in PBS for 10 and 20 minutes with 100 mM NH4Cl/100 mM glycine in PBS. After washing, the kidney sections were blocked with 3% BSA/5% NGS in PBS for 45 minutes followed by incubation with primary antibodies in 1% BSA (Supplemental Table 2) ON at RT. Subsequently, sections were washed 3×10 minutes in PBS before incubation with AlexaFluor-conjugated antibodies and DAPI (Supplemental Table 2) for 45 minutes at RT. After additional washing (3×10 minutes in PBS), sections were mounted in Fluoromount (Sigma Aldrich) and imaged with an Observer Z1 microscope with Apotome 2.0, Axiocam MRm (Zeiss), and EC Plan Apochromat 63×/1.40 Oil M27 or EC Plan-Neofluar 40×/1.30 Oil DIC M27 objectives. Images were processed with ImageJ or Zen software (Zeiss GmbH, Jena, Germany).
Histopathologic Analysis
The severity of kidney injury was assessed in representative formalin-fixed, paraffin-embedded 2-µm-thick kidney sections by a renal pathologist blinded to the genotype of the mice (n≥3 animals). To evaluate the glomerular injury, the total amounts of glomeruli without or with segmental (<50%) or global (>50% of the glomerular area) increase in matrix were counted and categorized as “normal,” “segmental glomerular sclerosis,” or “global glomerular sclerosis,” respectively (≥70 glomeruli per animal). Acute tubular injury was estimated in the corticomedullary areas by determining the percentage of tubules with cast formation, dilation, and degeneration (epithelial cell necrosis or loss of apical brush border). For each of these parameters, a five-point scale according to Markó et al. 43 was used (zero: none; one: <25%; two: 25%–50%; three: 50%–75%; and four: >75% injured tissue). Slit diaphragm injury was determined by counting the number of slit diaphragms per micrometer of GBM in TEM images in at least >250 µm GBM per animal and of different glomeruli.44
Expansion Microscopy
The method of mouse tissue expansion microscopy was described earlier.45 In short, 100-µm vibratome kidney slices were first blocked with blocking buffer (3% BSA and 0.1% Trition X-100 in PBS) for 12 hours at 4°C. Nephrin, Crb2, or Podocin antibodies (Supplemental Table 2) were diluted 1:100 in blocking buffer and incubated on the slices for 24 hours at 4°C on a shaker. After three washing steps (15 minutes) in blocking buffer at RT, tissue slices were incubated with secondary antibodies and DAPI (Supplemental Table 2) in blocking buffer ON at 4°C. Slices were washed 3×15 minutes in PBS and treated with freshly diluted 1 mM MA-NHS in PBS for 2 hours at RT followed by PBS washing steps. After staining and pre-expansion treatment, tissue was embedded in an acrylamide gel. For that, slices were incubated in monomer solution (1× PBS, 11.7% NaCl, 2.5% acrylamide, 0.15% N,N′-methylenebisacrylamide, and 8.625% sodium acrylate) for 1 hour at 4°C. A gelation chamber was built using a 1.5 coverslip as described earlier.46 Each tissue slice was gently moved in between surrounding coverslips, and excess liquid was removed from the sample. The gelation solution (monomer solution containing 0.01% TEMPO, 0.2% APS, and 0.2% TEMED) was applied to the tissue on the glass surface. Another coverslip was placed on top of the tissue and the other coverslip pieces to build up the chamber. The tissue chamber was placed in a humidified environment at 37°C for 2 hours. Afterward, excess gel and glass around the tissue sample were removed before digestion. Gelled tissue slides were digested with 1–2 ml freshly prepared proteinase K digestion buffer (1× TAE buffer, 0.5% Triton X-100, 0.8 M guanidine HCl, and 8 units ml−1 proteinase K) ON at 37°C. After digestion, the samples were incubated in ddH2O for expansion with three to four water exchanges every 30 minutes. Nuclear restaining using DAPI was performed in the first washing step. Expanded gels were placed in a poly-d-lysine–covered µ-Slide 2 Ibidi well and covered with water before imaging with a Leica SP8 confocal microscope using 40× water objective with a numerical aperture of 1.10.
Constructs and Cloning
We used pENTR plasmids with codon-optimized or wild-type cDNA encoding human CRB2 wild type (amino acids 1–1285) with a GFP or SNAP tag within the 13th EGF-like repeat of the ECD (inserted after an Aspartate at position 1097). These plasmids were used for site-directed mutagenesis to obtain S267A, R610W, C629S, R633W, E792D, N800K, and R1249Q mutants or to generate Crb2 deletion mutants ΔECD (amino acids 1–107 and 1095–1285) and ΔECD + ΔICD (amino acids 1–107+1095–1259). In addition, we cloned pENTR constructs with cDNAs encoding full-length human Nephrin, Nephrin with a C-terminal GFP tag, or an N-terminal GFP-tagged Nephrin ICD. All pENTR plasmids were shuttled into pINDUCER21_Puro plasmids using LR Clonase according to the manufacturer’s instructions as described earlier.47,48 The mCherrySec61β plasmid was a gift from Gia Voeltz (#49155; Addgene).49 All details concerning constructs and primers are summarized in Supplemental Table 1.
Cell Culture and Generation of Stable Cell Lines
HEK293T and human immortalized podocytes (AB8/13)50 were cultivated as described earlier.47,48 For transient transfection, AB8 cells were transfected using Lipofectamine 2000 according to the manufacturers’ instructions or HEK293T by the calcium phosphate method as described earlier. Stable cell lines were generated as previously described.47,48 Briefly, lentivirus production was performed in HEK293T cells transiently transfected with psPAX2 and pMD2.G helper plasmids and a modified pINDUCER21_Puro47,48 encoding EGFP and human CRB2 wild type and mutants, respectively. Supernatants containing virus particles pseudotyped with VSV-G were collected and filtered through a 0.45-µm sterile filter (EMD Millipore) and added to AB8 podocyte cells. For that, we used one volume of fresh DMEM/RPMI (Gibco, Grand Island, NY) medium and one volume of the virus-containing filtrate supplemented with polybrene (final concentration 8 µg/ml). After 24 hours, the virus particle–containing medium was replaced by fresh medium, and cells were regenerated for another 24 hours. Transduced cells were selected using puromycin (4 µg/ml for HEK293T and 2 µg/ml for AB8 cells). All established stable cell lines were tested for inducible overexpression of CRB2 proteins by western blot and live cell imaging.
Preparation of Cell Lysates
Cells on cell culture plates were washed one time with 1× PBS before lysis in an appropriate volume of RIPA buffer on ice (150 mM NaCl, 50 mM Tris, pH 8.0, 1% NP-40, 1% Triton X-100, 0.1% SDS, and 0.5% Na-deoxycholate; supplemented with complete protease tablets [Roche] and phosphatase inhibitor cocktails [Sigma Aldrich]). Cells were scratched off the plate into a reaction tube, which was vortexed every 5 minutes within 30 minutes. After incubation for 10 minutes in an ultrasonic bath, lysates were centrifuged 15 minutes at 14,000 rpm at 4°C. The supernatant was transferred to a new reaction tube and mixed with 2× Laemmli buffer before immunoblotting. For immunoprecipitation, cells overexpressing GFP fusion proteins were cultivated on 10-cm cell culture plates and washed one time with 1× PBS before scratching with 500 µl of IP buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 20 mM NaCl, 50 mM NaF, 15 mM Na4P2°7, and 1% [vol/vol] Triton X-100) supplemented with protease (complete tablets; Roche) and phosphatase inhibitor cocktails (Sigma Aldrich) on ice into a reaction tube. Cells were mechanical shredded using 20-gauge cannula and vortexed every 5 minutes within 30 minutes. Lysates were centrifuged 15 minutes at 14,000 rpm at 4°C. Fifty microliters of the supernatant was transferred to a new reaction tube and mixed with 2× Laemmli buffer before immunoblotting. The rest was used for GFP bead incubation (see GFP pull-down experiments).
Glycosylation Analysis
For analysis of the glycosylation pattern, RIPA cell lysates were treated for 2–6 hours at 37°C with PNGaseF (P0704L; NEB) and Endo H (P0702L; NEB) enzymes following the manufacturer’s instructions (n≥3).
Western Blot Analysis
Western blot analysis was performed as previously described.41,47 Briefly, cell lysates were boiled for 5 minutes at 95°C, and equal volumes were separated via SDS-PAGE using 6%–10% gels (BioRad, Hercules, CA). Afterward, proteins were transferred to a PVDF membrane (EMD Millipore) and incubated in blocking buffer (5% skim milk powder dissolved in TBS containing 0.05% Tween 20 [TBS-T]) for 1 hour at RT.
Primary antibodies were diluted (Supplemental Table 2) in TBS-T with 5% BSA and incubated at 4°C ON. Next, the membrane was washed three times with TBS-T and incubated with horseradish peroxidase–coupled secondary antibodies (Jackson Immunoresearch) (Supplemental Table 2) diluted 1:3000 in blocking buffer for 45 minutes at RT. After three additional wash steps with TBS-T, the signal was detected using a chemiluminescence detection reagent (Clarity; BioRad) in an Azure biosystems imager (c600; BioRad).
GFP Pull-Down Experiments (GFP-Trap)
For immunoprecipitation of GFP fusion proteins, GFP-Trap Agarose beads (gta-20; Chromotek) were used. Each washing step was performed by centrifuging the beads for 2 minutes at 9000 rpm at 4°C and discarding the supernatant. Twenty microliters of Agarose beads were washed two times with 500 µl 1× PBS and one time with 500 µl dilution buffer (150 mM NaCl, 10 mM Tris, and 0.5 mM EDTA, pH 7.5). Washed beads were incubated with 200 µl dilution buffer and 400 µl IP lysate for 2–4 hours at 4°C on a rotator. After binding, beads were washed again four times with 1 ml dilution buffer by centrifugation steps. In between the first two washing steps, beads were rotated again for 10 minutes at 4°C. After washing, beads were suspended in 50 µl 2× Laemmeli buffer.
Live Cell Imaging of Cells
AB8 CRB2-GFP cells were seeded in Ibidi eight-well chambers (Ibidi; 80,826) and transfected with mCherry-sec61β using Lipofectamine 2000 according to the manufacturers’ instruction (Thermo Fisher Scientific) before induction (200 nM doxycycline [Dox], 48 hours). For live cell staining, cells were incubated with Hoechst 33,342 (1 µg/ml; Invitrogen) and ER-Tracker Red (1 µM; Invitrogen) 30 minutes before imaging. For surface staining, cells overexpressing CRB2-SNAP tag were seeded into Ibidi eight-well chambers, induced for 48 hours with 200 nM Dox, and incubated with 5 µM SNAP Surface 488 (S9124S; NEB) diluted in RPMI media for 30 minutes at 33°C. Cells were washed twice with medium and imaged in HBSS containing 30 mM HEPES by using an Observer Z1 microscope equipped with an Apotome 2.0, an Axiocam MRm camera (Zeiss), and Plan Apochromat 63×/1.40 oil or EC Plan-Neofluar 40×/1.30 oil objective. Images were processed with Fiji (http://fiji.sc/) or Zen software (Zeiss GmbH). For quantification, we counted at least 200 cells per cell line (≥15 independent images) whether CRB2-GFP (wild type or variants) can reach and localize to the PM (cell-cell junctions) or if they are stuck at the ER (visualized by ER-Tracker Red). Partial and complete localization of the signal at cell borders was counted for “PM” localization.
RNA Isolation and Quantitative Real-Time PCR
To analyze the transcriptome of AB8 cells (GFP-tagged S267A mutant) cells were treated with 200 nM Dox for 48 hours to induce expression. Noninduced cells served as control (woDox). Total RNA isolation and real-time quantitative RT-PCR (qRT-PCR) were done as described before.51 For that, GenElute Mammalian total RNA Miniprep Kit (Sigma Aldrich) and Superscript III Reverse transcription kit (Invitrogen) were used according to the manufacturers’ instructions. qRT-PCR was on the basis of SYBR Green dye (Applied Biosystems, Foster City, CA) and was done by the Core Facility Genomics (Medical Faculty, University Hospital Muenster, Muenster, Germany) using CFX384 Touch (BioRad). Relative gene expressions were normalized to GAPDH and compared using the comparative ΔCT method.51 Used qRT-PCR primers are listed in Supplemental Table 3.
RNA Sequencing
RNA sequencing (RNAseq) was performed as described earlier.51 In brief, library preparation of the total RNA was performed with the NEBNext Ultra II RNA directional kit, and single-read sequencing was performed using a NextSeq 500 System with a read length of 75 bp. Using a molecular barcode, the samples were demultiplexed (bcl2fastq2) to fastq data and quality controlled (FastQC). Trimmomatic was used for adapter trimming and read filtering. The resulting reads were aligned to the Ensembl GRCh38 reference genome using Hisat. The aligned reads were sorted using the same tools. The sorted and aligned reads were counted into genes using htsec counts. The test for differential expression was performed using the r-package deseq2. Subsequently, differentially expressed genes were evaluated by GO-Term enrichment analyses that were performed using the DAVID Bioinformatics Resources 6.8 tool.52 RNAseq data will be available through Gene Expression Omnibus accession number GSE148661.
Three-Dimensional Image Processing
Image processing was performed according to the procedures outlined earlier.53 In the first step, confocal images were spatially deconvolved using Huygens software (professional version 19.04; Scientific Volume Imaging, Hilversum, The Netherlands) using a theoretical point-spread function and the classic likelihood estimation algorithm, with an SNR: 40 and 100 iterations. Improving the spatial resolution of the images (Supplemental Figure 1) allowed us a better visualization of the foot processes, thus telling apart details that elucidated patterns of distribution for Nephrin, Podocin, and Crb2 through the sample.
In the second step, the deconvolved three-dimensional (3D) stacks of 16-bit images were processed using custom-written MATLAB scripts, which allowed parallel data processing. For that, the intensity histograms were adjusted to homogenize brightness and contrast throughout the complete dataset. Every 3D stack was scanned to find its minimum and maximum intensity values. With the respective values, a linear intensity adjustment was performed to cover the full dynamic range. For a final intensity adjustment and cropping of the images, Fiji (http://fiji.sc/) was used.
Intensity profiles were determined using custom-written MATLAB scripts to visually compare the intraglomerular localization of Nephrin, Podocin, and Crb2. First, we manually selected a region of interest, with a line width of two. Second, the data were fitted with a series of Gaussians with a 95% confidence bound.
The 3D representation of the data were achieved using the Surpass view in Imaris (version 9.5.1; Bitplane Inc., Zurich, Switzerland). The 3D segmentation of different regions of interest was performed with the Surface tool in Imaris using a Surface Grain Size of the two-fold pixel size and Background Subtraction (local contrast) adjusting the values accordingly. All data processing was performed on a workstation equipped with two Intel Xeon Platinum 8160 CPU (2.1 GHz, 24 cores), 512 GB memory, and an Nvidia Quadro P5000 GPU (16 GB GDDR5×) running under Windows 10 Pro.
Statistical Analyses
Tests for statistical significance of normally distributed data were performed using GraphPad PRISM software with an unpaired two-tailed t test for comparison of two groups of data or one-way ANOVA with a Bonferroni correction for comparing three or more groups of data. If not otherwise indicated, all data are given as mean of at least three independent measurements (*P=0.05, **P=0.01, and ***P<0.001).
Results
Podocyte-Specific Loss of Crb2 Leads to Heavy Proteinuria in Mice
Crb2 is the main Crumbs protein isoform in murine glomeruli, Crb3a and Crb3b are less expressed (Supplemental Figure 2). The Nephrin staining in Figure 1A showed a typical glomerular slit diaphragm protein distribution.54,55 In addition, immunofluorescence analyses showed overlap of the Nephrin and Crb2 staining patterns, indicating that Crb2 is located in glomerular podocytes (Figure 1, A and B).
Figure 1.
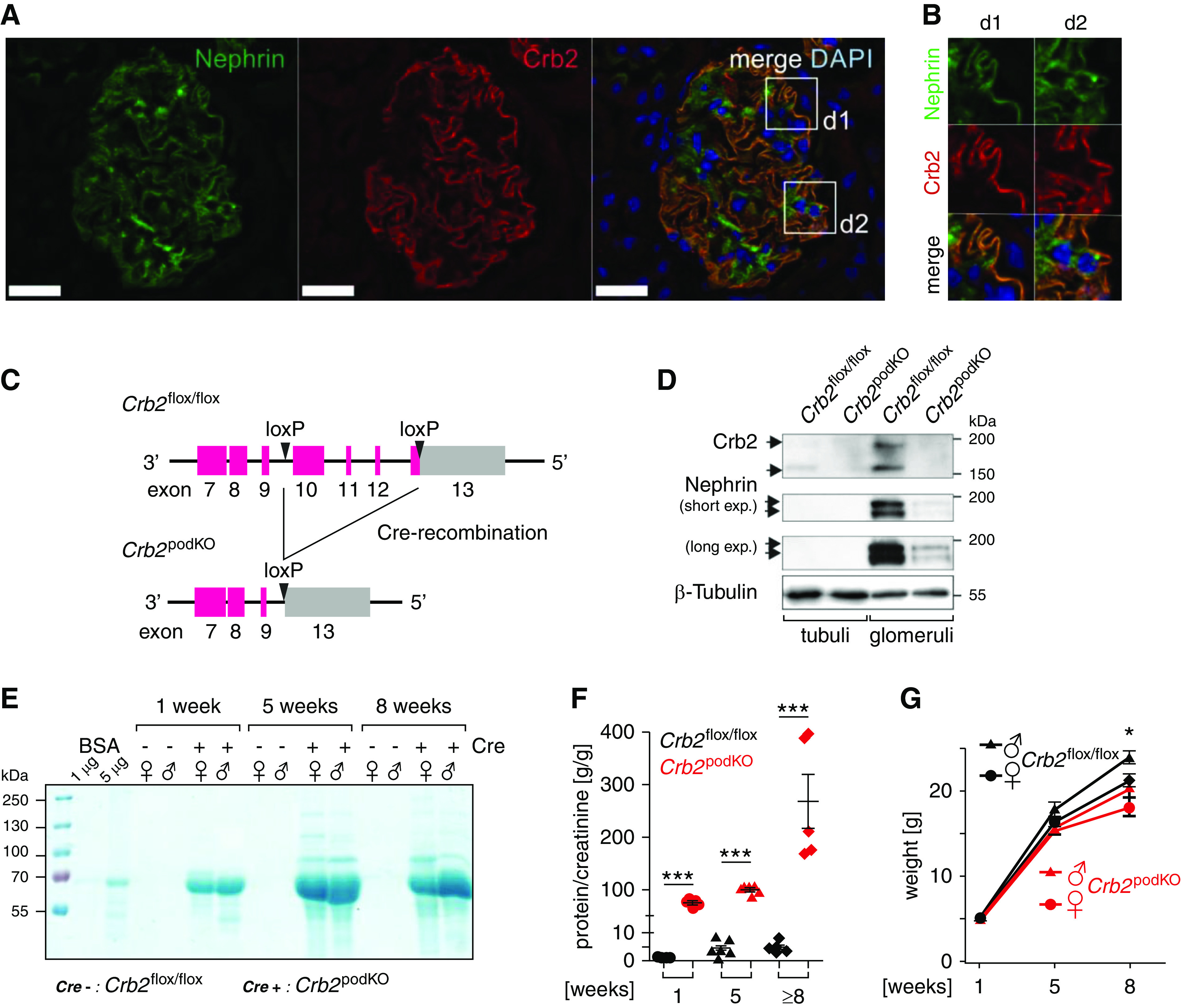
Crb2 is an essential component of the renal filtration barrier. (A) Immunofluorescence analysis of a murine glomerulus showed overlap of Nephrin (green) and Crb2 (red). (B) Details of (A) in higher magnification. Scale bar and edge length of details: 10 µm. (C) For Crb2flox/flox mice, the loxP recombination sites were introduced in intron 9 and in the 3′ noncoding region of exon 13. Upon Cre-mediated recombination, the exons 10–12 and a part of exon 13 are removed (exons: pink boxes). (D) Crb2 is only expressed in glomeruli fraction of murine kidney lysates. Crb2 podKO mice showed no Crb2 and a reduced Nephrin expression compared with control Crb2 flox/flox mice glomeruli (n=4). (E) Representative SDS-PAGE analyses of 1 µl spontaneous urine from Crb2 flox/flox and Crb2 podKO mice demonstrate the early onset of heavy proteinuria in Crb2-depleted mice. (F) Quantification of proteinuria by determination of the protein-creatinine ratio (n=5 per group) (Supplemental Figure 2C). (G) Body weight of Crb2 podKO mice is significant reduced in comparison with Crb2 flox/flox littermate controls only after 8 weeks (n≥3 per sex) (Supplemental Table 4). (F and G) Unpaired two-tailed t test. *P=0.05; ***P<0.001.
To address the role of Crb2 in podocytes in vivo, we bred conditional Crb2 knockout mice (Crb2 flox/flox)36 with transgenic mice that express Cre recombinase under the control of the Nphs2 (Podocin) promotor.42 The breeding results in mice lacking Crb2 exclusively in podocytes in expected Mendelian ratios (thereafter designated as Crb2 podKO) (Figure 1C). Because of high expression of Crb2 in podocytes, the Crb2 signal was only found in glomerular lysates of the wild type and was absent in tubular and glomerular lysates of homozygote Crb2 podKO mice (Figure 1D). All Crb2 podKO mice developed an early onset of severe proteinuria, starting in the first week after birth (Figure 1, E and F, Supplemental Figure 2C) but with minor reduction of total weight after 8 weeks (Figure 1G).
Nephron epithelia, including podocytes, are descendants of Six2-positive renal vesicle precursor cells.40,41 Therefore, we used Six2-Cre driver in combination with the Crb2 flox/flox to target very early stages of podocyte development. Surprisingly, we obtained only heterozygous but no homozygous Six2-Cre–positive Crb2 flox/flox mice, indicating early embryonic lethality (Supplemental Figure 3). This may be explained by Six2-Cre activity during early steps of embryogenesis leading to Crb2 depletion in nonrenal progenitor cells.56
Analyses of Crb2-Depleted Podocytes
Kidneys of Crb2 podKO and their littermates from 1-week-old (Supplemental Figure 4), 5-week-old (Figure 2), and ≥8-week-old (Supplemental Figure 5) mice were analyzed by histologic and ultrastructural analyses. Despite the early onset of proteinuria in Crb2 podKO mice at 1 week after birth, glomeruli showed a normal morphology at this age. Also in ultrastructural analyses, podocytes were normal, and only some podocytes showed atypical electron-dense structures (Supplemental Figure 4, C–G), which are also visible in murine podocytes lacking Nephrin or Fat1.16,57,58
Figure 2.
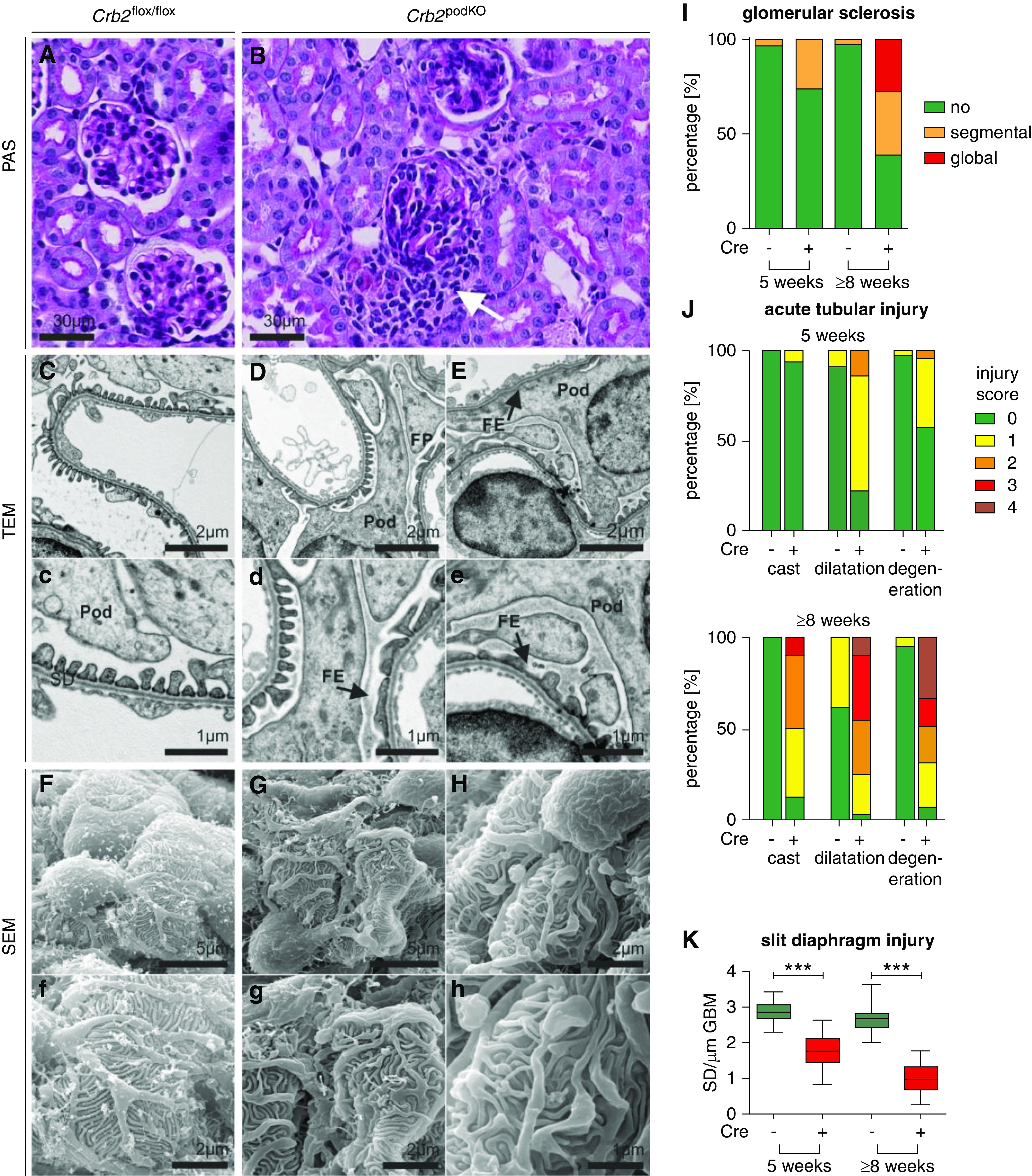
Crb2 loss results in disordered and effaced foot processes (FPs). (A and B) Periodic acid–Schiff (PAS) staining. Kidney tissue of 5-week-old Crb2 podKO infiltration of cells (white arrow). (C and D) TEM analyses. Ultrastructural analyses revealed segmental foot process effacement (FE; black arrows) in (D, d, E, and E) Crb2 podKO in comparison with (C and c) Crb2 flox/flox littermate control mice. (F–H) Scanning electron microscopy analyses. (F and f) In contrast to Crb2 flox/flox, (G, g, H, and h) podocyte FPs of Crb2 podKO mice were disordered and variable in size. Pod, podocyte cell body. (I–K) Quantification of the histopathologic changes in 5- and ≥8-week-old animals revealed increased kidney injury in Crb2 podKO mice (n=3–5) (Supplemental Tables 5 and 6). (I) Percentage of glomeruli (n≥3 animals, ≥70 glomeruli) with segmental, global, or no glomerular sclerosis. (J) Percentage of the corticomedullary area (n≥3 animals) with a given acute tubular injury score with respect to tubular cast formation, tubular dilation, and tubular degeneration. (K) Quantification of slit diaphragm number in TEM images of 5- and 8-week-old mice showed strong reduction of slit diaphragm number in Crb2 podKO as compared with Crb2flox/flox mice (n=3–4). Unpaired two-tailed t test. SD, slit diaphragm. ***P<0.001.
Contrarily, in 5-week-old Crb2 podKO mice, periodic acid–Schiff-stained kidney sections showed some cellular infiltrates surrounding the glomeruli (Figure 2, A and B), acute tubular injury, and focal expansion of the glomeruli due to incipient glomerular sclerosis (Figure 2, I and J, Supplemental Tables 5 and 6). Ultrastructural analyses by TEM and SEM elucidated that podocyte foot processes of Crb2 podKO mice were effaced (Figure 2, C–E) and disordered (Figure 2, F–H) accompanied by a strong reduction of the slit diaphragm number per GBM (Figure 2K). In addition, SEM images showed roundish ends of secondary processes in some podocytes of Crb2 podKO mice. These histopathologic findings were clearly more pronounced in the ≥8-week-old mice. Moreover, an increased formation of proteinaceous casts in the renal tubules was noted (Figure 2, I–K, Supplemental Figure 5).
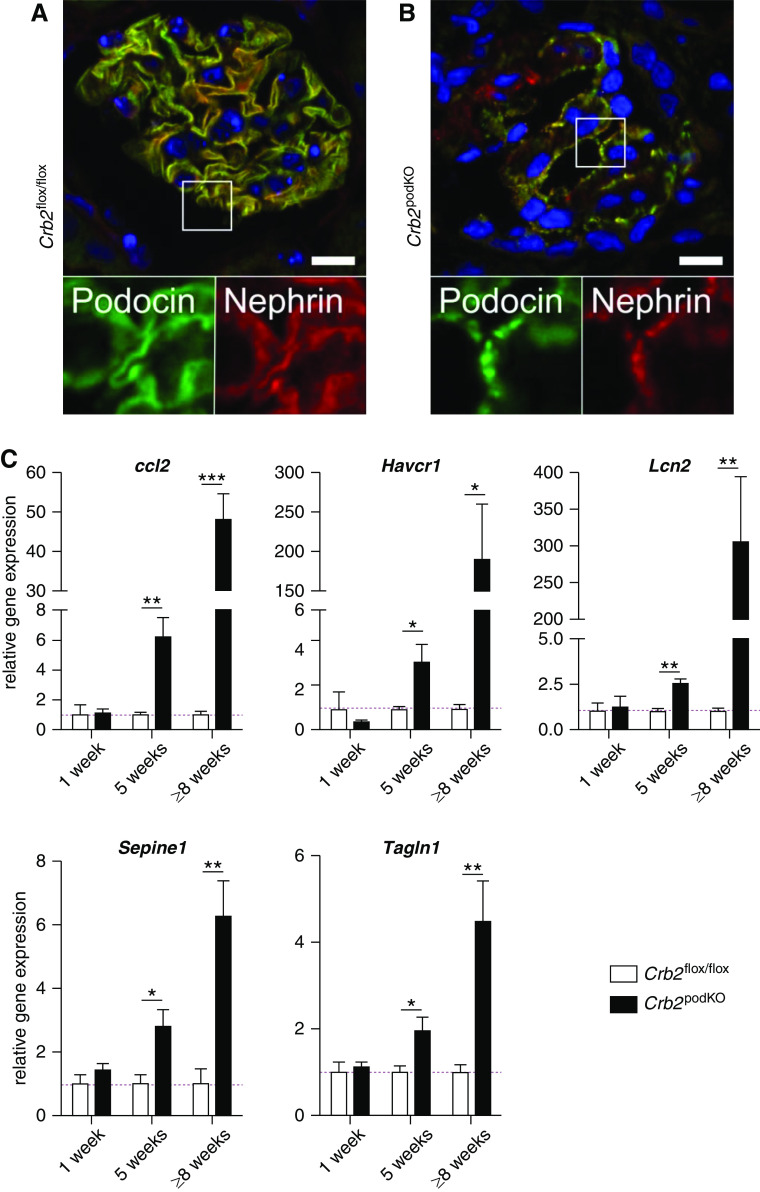
Additional immunofluorescence analyses using antibodies against Podocin and Nephrin confirmed a reduction of slit diaphragm proteins signals in Crb2 podKO mice (Figure 3, A and B) that we found already by quantification of Nephrin expression in western blots of glomerular lysates (Crb2 flox/flox: 100%±19.2% versus Crb2 podKO: 18.0%±6.2%; n=4) (Figure 1D). Next, we analyzed the expression of Ccl2, a fibrosis and inflammation marker as well as Havcr1 (Kim-1), Lcn2 (Ngal), and Serpine1 (Pai-1) as tubular and Tagln1 (Transgelin 1) as glomerular injury markers. The tested markers for inflammation (Ccl2), tubular-intestinal injury (Havcr1, Lcn2, and Pai-1), and glomerular injury (Tagln1) remained unchanged within the first weeks. After 5 weeks, we observed upregulation of all markers that dramatically increased within the next 3 weeks, in particular Ccl2, Havcr1, and Lcn2 (Figure 3C). Together, our data (Figures 1–3) show that heavy proteinuria of Crb2 podKO mice precedes obvious slit diaphragm injury, glomerular sclerosis, and acute tubular injury in histopathologic analyses as well as stepwise upregulation of renal injury markers. Hence, structural injury on the molecular level of the slit diaphragm protein composition by deleting Crb2, which might not be resolvable by this standard characterization data, could be the mechanism leading to early proteinuria followed by functional GFB damage.
Figure 3.
Crb2 depletion in podocytes results in altered expression of slit diaphragm proteins and activation of injury pathways. (A and B) Representative immunofluorescence analysis of glomeruli from Crb2 flox/flox and Crb2 podKO 8-week-old mice. (A) In contrast to Crb2 flox/flox littermate controls that showed strong expression and colocalization of slit diaphragm proteins Nephrin (red) and Podocin (green), (B) slit diaphragm protein expression was decreased in Crb2 podKO. Boxes show details in higher magnifications below the merged images. Scale bars: 10 µm. (C) Relative gene expression (normalized to Crb2 flox/flox littermates) of renal injury and inflammation markers of 1-, 5-, and ≥8-week-old animals (n=3–5 animals per group). The tested markers for inflammation (Ccl2) and tubulointerstitial (Havcr1, Lcn2, and Serpine1) and glomerular injury (Tagln1) remained unchanged within the first week and changed significantly after 5 weeks. During the next 3 weeks (8 weeks), all injury markers (particularly, Ccl2, Havcr1, and Lcn2) were strongly upregulated. Unpaired two-tailed t test. *P=0.05; **P=0.01; ***P<0.001.
CRB2 and Nephrin Are Binding Partners
Previous studies elucidated homo- or heterotypic interactions between Nephrin and other slit diaphragm membrane proteins.54,59–61 We, therefore, tested if Nephrin also binds to CRB2 and performed GFP-trap pull-down assays by cotransfection of HEK293T cells with expression constructs of human (untagged) full-length Nephrin and GFP-tagged CRB2 wild type or CRB2 truncation mutants lacking the extracellular part (∆ECD) or both the extra- and intracellular part of CRB2 (∆ECD + ∆ICD) (Figure 4A). Indeed, GFP-tagged CRB2 containing the ECD binds to full-length Nephrin as well as to PALS1, which is a well-known intracellular binding partner of CRB2.62,63 CRB2 lacking the ECD only can bind to PALS1 but is not able to interact with Nephrin. CRB2 lacking the ICD and ECD failed to bind PALS1 or Nephrin (Figure 4B). Hence, CRB2 binds to Nephrin via its ECD. We controlled this observation by a vice versa experiment using GFP-tagged full-length Nephrin and Nephrin lacking the ECD (Figure 4C). In these pull-down experiments, only full-length Nephrin binds to CRB2, whereas Nephrin without the ECD failed to interact with CRB2. In conclusion, the pull-down assays shown in Figure 4B and the vice versa experiment shown in Figure 4C demonstrate an interaction between these proteins via their ECDs (n≥3).
Figure 4.
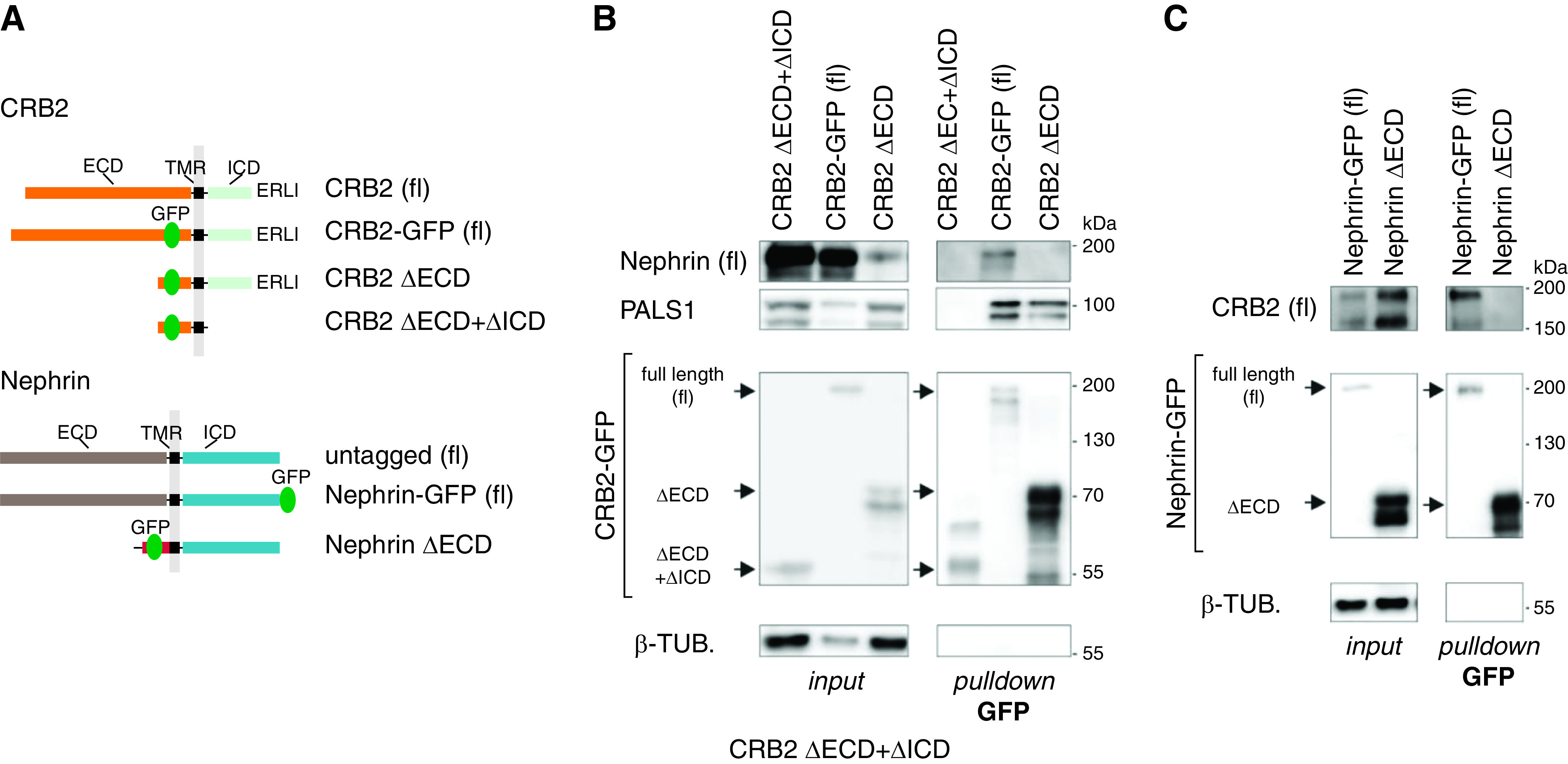
CRB2 interacts with Nephrin. (A) Scheme: CRB2 and Nephrin constructs used for pull-down assays. (B) GFP-trap pull downs: full-length (fl) CRB2 wild type binds to untagged fl Nephrin, whereas CRB2 lacking the ECD (CRB2 ΔECD) or both ECD and ICD (ΔECD + ΔICD) failed to bind. PALS1 that interacts with the ICD of CRB2 was precipitated by fl and CRB2 ΔECD (n≥3). (C) Vice versa, only fl Nephrin-GFP but not Nephrin lacking the ECD was able to bind to untagged CRB2 (fl; n=4), suggesting an interaction of CRB2 and Nephrin via the extracellular protein domains. β-TUB, β-tubulin; Δ, deletion; TMR, transmembrane region.
Crb2 and Nephrin Are Organized in Adjacent Clusters at the Slit Diaphragm
To obtain insights into the 3D intraglomerular distribution of Crb2 in vivo, we used the recently established high-resolution expansion microscopy approach45,46,64 for comparing the localization of Nephrin and Podocin with that of Crb2 and Nephrin. It should be noted that by this approach, a lateral resolution of well below 100 nm can be achieved. Expanded wild-type mouse glomeruli stained for Nephrin and Podocin showed the typical loop-like pattern of slit diaphragm proteins as previously observed.45 Optical sections taken perpendicular to the slit diaphragm membrane suggested that Podocin was located more closely to the DAPI-labeled nuclei than Nephrin according to expectation (Figure 5, A and B; magnifications are in Figure 5, b1 and b2, and a 3D rendering is in Figure 5C). Processing of image stacks obtained from expanded glomeruli resulted in 3D images (Figure 5, C and D), and movies (Supplemental Movies 1 and 2) supported this impression. The observed signal distribution supported earlier studies that Podocin serves as intracellular binding partner of Nephrin.17–19
Figure 5.
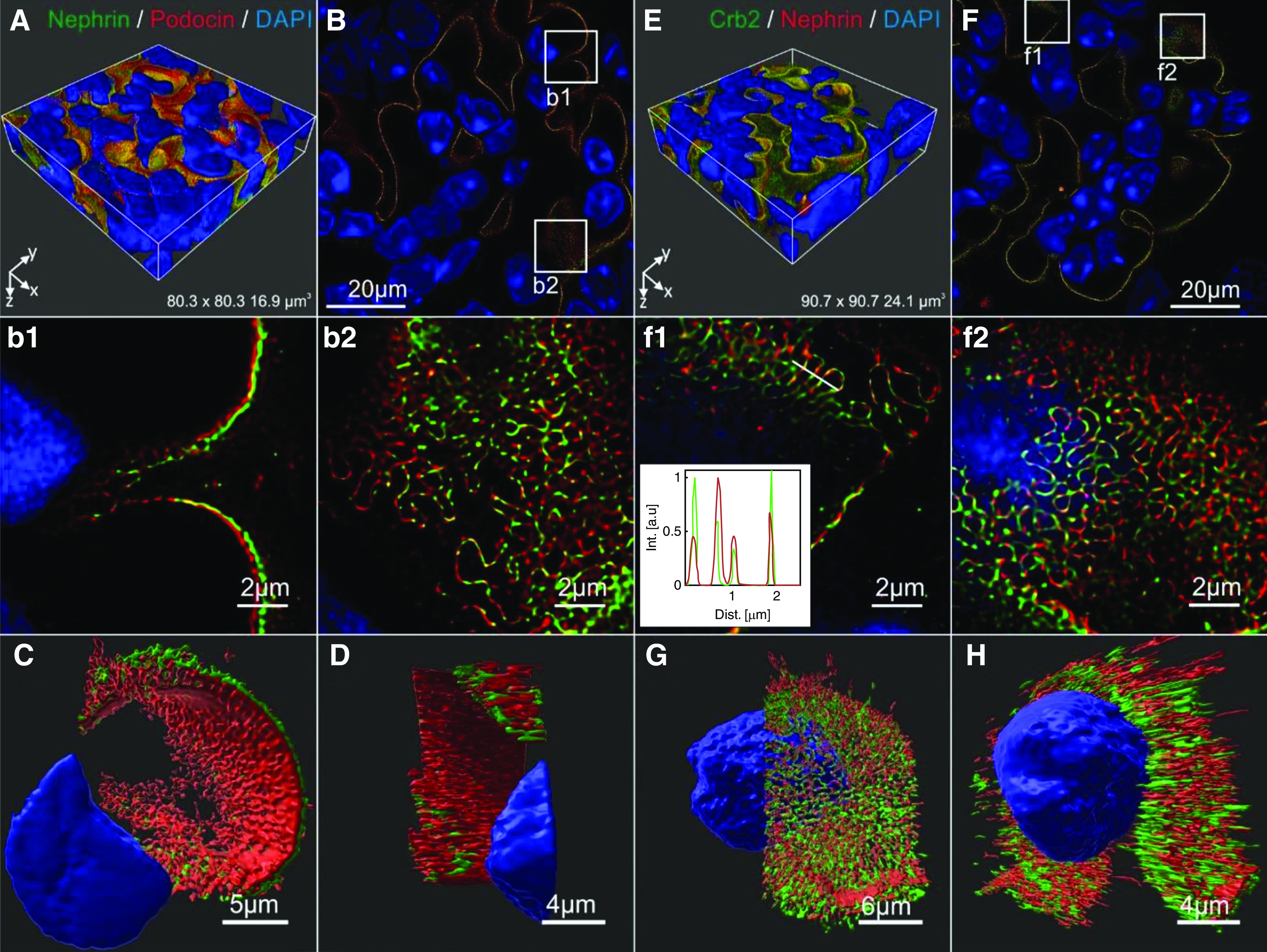
Crb2 and Nephrin are organized in adjacent clusters at the slit diaphragm. High-resolution expansion microscopy (ExM) approach to compare the intraglomerular localization of (A–D) Nephrin and Podocin with (E–H) Crb2 and Nephrin. Glomeruli sections were expanded in acrylamide gels to achieve a higher resolution in confocal imaging: (A) 3D block and (B) single confocal section (plane of the z stack at a depth of 2.70 µm, deconvolved using Huygens software) of exemplary expanded glomeruli stained for Nephrin and Podocin. (b1 and b2) Boxes in (B) indicate the localization of shown details that (b1) elucidated similar but slightly separated patterns of the Nephrin and Podocin signals in cross-section to nucleus. (b2) Both slit diaphragm proteins showed a typical loop-like distribution that has been found in earlier ExM as well. (C and D) Two 3D views of ExM image details visualized and segmented using IMARIS. These images indicated that the intracellular adapter Podocin localizes more closely to the nucleus (blue) compared with the transmembrane protein Nephrin (Supplemental Movies 1 and 2), as one would expect. (E) The 3D block and (F) single confocal section (plane of the z stack at a depth of 2.64 µm, deconvolved using Huygens software) of the expanded glomeruli stained for slit diaphragm proteins Crb2 (green) and Nephrin (red). (f1 and f2) Boxes in (F) indicate the localization of shown details. Crb2 and Nephrin show highly overlapping loop-like slit diaphragm pattern but with different intensities. (Inset in f1) Intensity line profile in arbitrary units (a.u.), including a fit of the profiles with several Gaussian functions of the white line shown in (f1). (G and H) ExM images processed by IMARIS suggested the presence of adjacent but not colocalized Crb2 and Nephrin clusters on the foot process surface. In contrast to Nephrin and Podocin in (b1 and C), there was no indication of a spatial separation of Crb2 and Nephrin in relation to the cell nucleus (Supplemental Movies 3 and 4).
For Crb2 and Nephrin, we also observed typical loop-like slit diaphragm structures in respective confocal image stacks of expanded WT mouse glomeruli (Figure 5, E and F; details are in Figure 5, f1 and f2). However, compared with Nephrin and Podocin, Nephrin and Crb2 appeared to be organized in a different way (3D representations in Figure 5, C and D versus Figure 5, G and H). The fluorescence signals of Crb2 (green) and Nephrin (red) appeared to have a similar distance to the nucleus. There was no clear spatial separation of the signals in relation to the nuclei as it appeared for the intracellular Podocin and the transmembrane protein Nephrin (Figure 5C versus Figure 5H). Interestingly, Nephrin-rich regions often exhibited weak staining of Crb2 and vice versa (Figure 5f1, inset). Deconvolution of the image data corroborated this view (Figure 5, G and H, Supplemental Movies 3 and 4) and suggested the existence of neighbored Crb2 and Nephrin clusters, presumably on the extracellular surface of podocytes. Thus, our data of expansion microscopy and pull-down experiments suggested the presence of homotypic Crb2-Crb2 and Nephrin-Nephrin interactions within and probably, heterotypic Crb2-Nephrin interactions between adjacent clusters.
CRB2 Mutants Showed Altered Intracellular Distribution and Lacked Cell Surface Localization
Recessive inheritance of CRB2 mutants in patients with SRNS and the observation that only homozygous but not heterozygous Crb2 podKO mice developed proteinuria indicate a loss of function phenotype in mice and humans. To obtain more insight into Crumbs2’s role on a cell biologic level, we selected immortalized AB8 podocytes without endogenous CRB2 expression and generated stable cell lines with a Dox-dependent expression of GFP-tagged CRB2 wild type and some representative SRNS-associated mutants (Figure 6A, Supplemental Figure 6, A and B). We selected three mutants in the ECD: two mutants (C629S and R633W) in the tenth EGF repeat and one (N800K), which lacks a putative N-glycosylation site. In addition, we used one mutant between the transmembrane region and ICD (R1249Q).29,30 As control for these cell-based assays, we also included two additional CRB2 variants that have been not linked to diseases, R610W (inside the tenth EGF-like repeat) and E792D (inside the second Laminin G domain). Live cell imaging revealed that in contrast to the wild type, all CRB2 disease-associated variants colocalized in ≥86% of the cells completely with ER markers (mCherry-tagged Sec61β or ER-Tracker) (Figure 6, B–E and G, Supplemental Table 7). Like the CRB2 wild type, R610W and E792D variants showed a localization (≥94%) at the plasma membrane, between neighboring cells (Figure 6K, Supplemental Figure 6, C–E, Supplemental Table 7).
Figure 6.
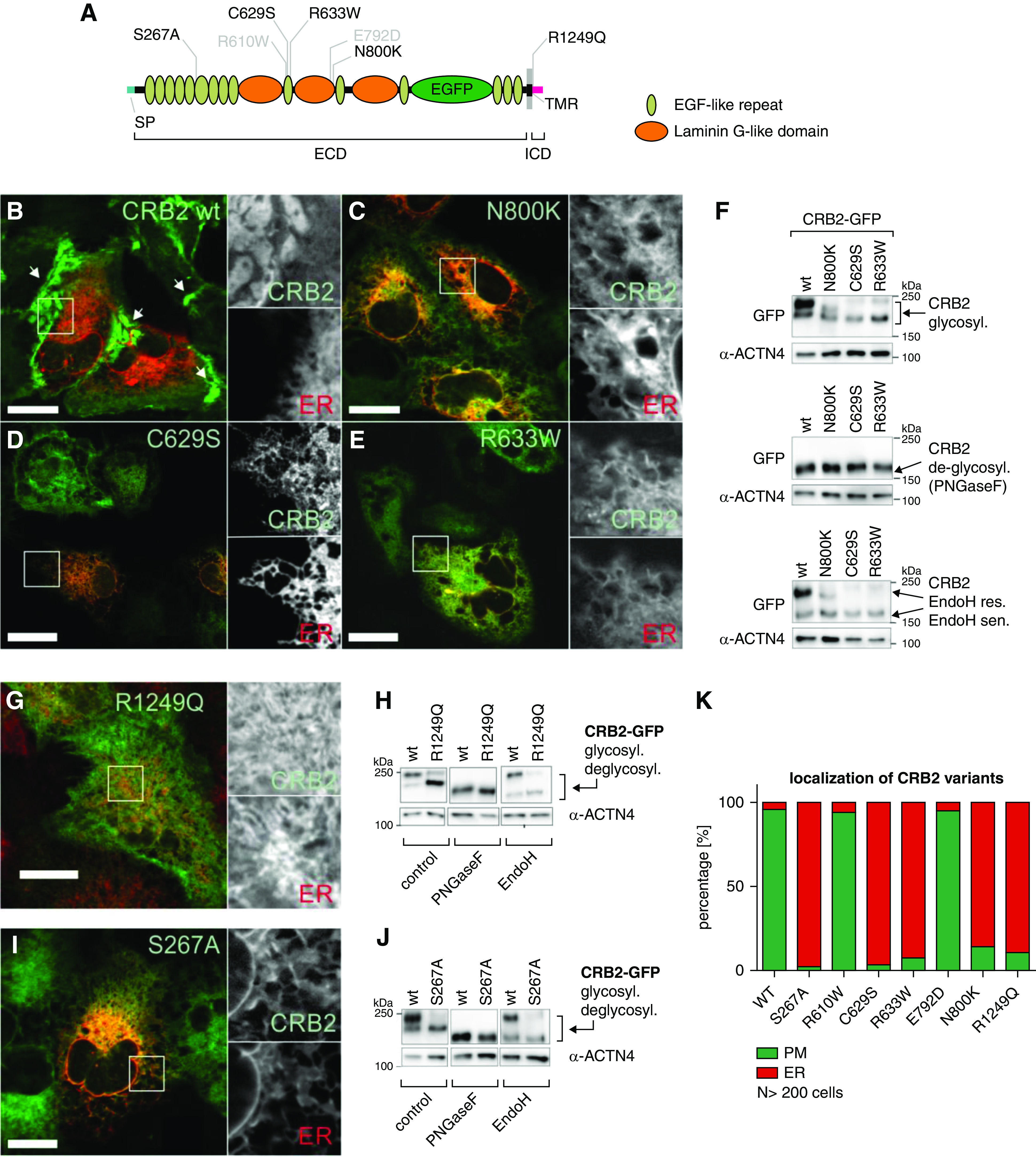
CRB2 mutations result in reduced surface localization. (A) Scheme: CRB2 GFP wild type (wt), variants (gray), and mutants (black) used in the study. (B–E, G, and I) Live cell imaging: ER was visualized by using mCherry-tagged ER marker Sec61β or ER tracker (red). CRB2 GFP wt (green) expressed in immortalized podocytes showed localization at the surface in overlapping regions between neighboring cells (white arrows in B). (C–E and G) In contrast, SRNS mutants N800K, C629S, R633W, and R1249Q and the S267A mutant accumulate at the ER. Scale bars: 20 µm. (F, H, and J) Glycosylation analyses: untreated control cells showed double bands representing two distinct glycoforms. Here, SRNS-associated and S267A mutants showed lower mol wt in SDS-PAGE analyses. PNGaseF treatment demonstrated that different migration behaviors of CRB2 wt and mutants are due to differences in the N-glycosylation pattern as deglycosylated forms of all variants have the same weight. A shift of the lower mol wt band in wt and mutant CRB2 glycoforms to the deglycosylated form after Endo H treatment indicates that the former harbor immature mannose-rich N-glycans. Mature wt CRB2 with complex N-glycans (glycosyl.; upper bands) was not affected by Endo H treatment (n=3). (K) Quantification of ≥200 cells of indicated GFP-tagged CRB2 variants showed a predominant ER localization of S267A and SRNS-associated (C629S, R633W, N800K, and R1249Q) mutants (≥86% of the cells). The used wt and nondisease-associated variants (R610W and E792D) are efficiently transported to the plasma membrane (in ≥94% of the cells) and accumulate in overlapping regions (Supplemental Figures 6 and 7). α-ACTN4, α-Actinin 4; deglycosyl., deglycosylated; PM, plasma membrane; res., resistant; sen., sensitive; SP, signal peptide; TMR, transmembrane region.
This observation indicated defects in the correct targeting of the mutants to the plasma membrane. Such transport defects of slit diaphragm proteins can be induced by impaired glycosylation.65 We, therefore, compared the glycosylation pattern of CRB2 wild type and SRNS mutants and indeed, found differences in the N-glycosylation pattern (Figure 6, F and H, Supplemental Figure 6G). In line with the live cell imaging studies, CRB2 disease variants were more sensitive to Endo H glycosidase that specifically targets mannose-rich glycosylation patterns of unprocessed proteins at the ER (Figure 6, F and H).
During embryogenesis of the mouse kidney, loss of O-glucosyltransferase 1 (Poglut1) results in an almost identical phenotype as loss of Crb2 (knockout animals), suggesting that both N- and O-glycosylation are crucial for Crb2 processing.28,66 To test the effect of O-glycosylation of CRB2 in podocytes, we mutagenized Serine 267—a conserved putative Poglut1 O-glycosylation site in mouse and human—to Alanine, rendering it resistant to O-glycosylation (Figure 6A).28 Like the SRNS-associated variants (Figure 6, C–E and G), the CRB2 S267A protein mutant failed to reach the cell surface (Figure 6, I and K, Supplemental Table 7). Moreover, the CRB2 S267A protein showed an altered N-glycosylation pattern with an elevated Endo H–sensitive fraction, like the analyzed SRNS-CRB2 mutants (Figure 6J), indicating a central role of O-glycosylation for human CRB2.
To specifically detect surface localization of CRB2, we replaced the GFP tag within the 13th EGF-like repeat of CRB2 wild type and the CRB2 mutants S267A and N800K by an SNAP tag. By using a membrane-impermeable dye (SNAP-Surface488) that only binds to SNAP-tagged proteins on the cell surface, only CRB2-SNAP wild type–expressing cells were positively stained. Cells expressing CRB2 variants S267A and N800K remained negative, indicating failure to reach the plasma membrane surface (Supplemental Figure 6, H–J).
CRB2 Mutants Trigger Expression of ER Stress Genes
Altered glycosylation combined with ER retention can cause ER stress.67 Therefore, we used the S267A variant as an example for ER-retained CRB2 mutants and analyzed the transcriptomes of podocyte cell lines overexpressing CRB2 S267A under the control of Dox. In comparison with noninduced control cells, cells expressing CRB2 S267A revealed 1067 significantly regulated genes (Figure 7A). The 557 upregulated genes were further analyzed for functional annotation enrichment by the DAVID bioinformatics tool.52 Strikingly, we found significant enrichment of genes that play a role in ER-linked processes and/or ER stress–associated pathways (Figure 7B, Supplemental Figure 7). To confirm the RNAseq data by qRT-PCR, we selected the ER stress marker genes HSPA5 (protein BiP/Grp78), CANX (Calnexin), HSP90B1 (Grp94), and HYOU1 (Orp150/Hsp12A) that have been linked to the unfolded protein response and ER stress before.68 Next, we analyzed mRNA of noninduced cells, cells expressing CRB2 wild type and the CRB2 mutants S267A and N800K. Again, only cells expressing ER-localized S267A and N800K variants showed significant upregulation of ER stress markers (Figure 7C).
Figure 7.
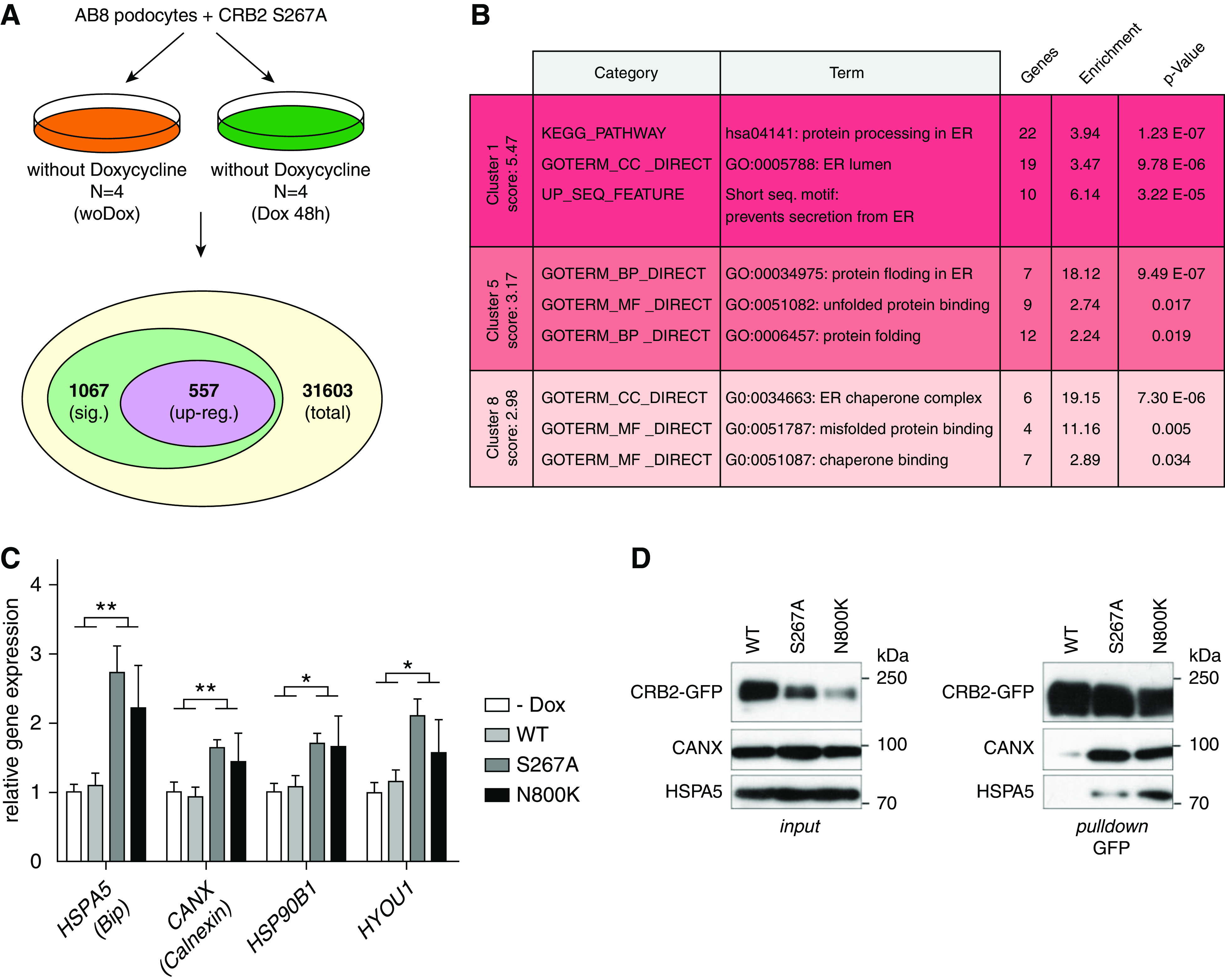
ER-localized CRB2 mutants cause ER stress. (A) RNAseq analyses of AB8 podocyte cells expressing the GFP-tagged CRB2 S267A mutant (with Dox) compared with noninduced control (woDox) resulted in 1067 differentially expressed genes (sig.). (B) Functional gene enrichment analyses (DAVID bioinformatics) of upregulated genes (upreg.; 557) revealed activation of ER stress and ER stress–associated pathways. (C) Quantitative real-time PCR analyses of ER stress genes HSPA5 (BiP, Grp78), CANX (Calnexin), HSP90B1 (Grp94) and HYOU1 (Grp170, Orp150): ER stress was only induced in cells expressing the S267A or N800K mutant. Noninduced cells (−Dox) or cells expressing wild-type CRB2 (WT) did not show upregulation of these genes. Unpaired two-tailed t test. n≥5. *P=0.05; **P=0.01. (D) GFP pull-down assays: CRB2 S267A and N800K mutants bind stronger to ER-localized protein folding chaperones HSPA5 (BiP) and CANX (Calnexin) than WT. (n=3).
HSPA5 (BiP) and CANX (Calnexin) are chaperones that support folding and processing of membrane proteins in the ER lumen.68 Indeed, pull-down assays, using GFP-tagged CRB2 wild type and mutants S267A and N800K, showed stronger binding of CRB2 mutants S267A and N800K to these chaperones than wild-type protein (Figure 7D). This indicates high amounts of unfolded and/or unprocessed CRB2 mutants in the ER and is in line with the observed induction of ER stress in these cell lines.
Discussion
To investigate the role of Crb2 at the renal filtration barrier, we generated mice lacking Crb2 exclusively in podocytes, as conventional Crb2 knockout mice die before embryonic day E12.5 due to primitive streak formation defects during epithelial-to-mesenchymal transition process at gastrulation.28,32 Crb2 depletion in podocytes results in an early onset of proteinuria, accompanied by disordered and effaced foot process formation, upregulation of renal injury markers, and glomerular sclerosis as well as strong acute tubular injury likely secondary to high protein loss. Remarkably, heavy proteinuria can be observed as first phenotype in Crb2 podKO animals directly after birth. This indicates that molecular changes in the GFB or the slit diaphragm architecture precede the glomerular and tubular changes detected upon histologic examination. The Crb2 podKO phenotype strongly resembled that of human SRNS-associated mutations within the CRB2 gene.29,30,37 Moreover, the striking clinical similarities between SRNS forms caused by a mutation in CRB2,29–31 NPHS1,8,12 FAT1,58 and KIRREL1 (encoding Neph1)69 genes and vertebrate models lacking these genes in podocytes55,58,70,71 indicate analogous or even synergistic functions of these slit diaphragm components in building up the slit diaphragm.
Previous studies identified Nephrin and Neph1 as major components of the slit diaphragm. The homo- and heterotypic interactions of these proteins provided first structural insights into how this unique cell-cell junction between two neighboring foot processes bridges an approximately 40-nm width.3,6,8 A more recent study demonstrated the existence of quasiperiodical clusters of Nephrin and Neph1.16 Here, both slit diaphragm proteins form rather homotypic—meaning Nephrin-Nephrin or Neph1-Neph1—than heterotypic Nephrin-Neph1 interactions.16 Neph1 appeared as the lower part of the slit diaphragm junction close to the GBM, whereas Nephrin molecules act more apically to form adjacent junctions with a larger width.16 In this context, it is interesting that in addition to Neph1, EphrinB1 also binds to the Nephrin ECD.54 However, it is assumed that this heterophilic interaction rather functions as scaffold for Nephrin in cis and not in trans from the opposite foot process.
In this study, we identified CRB2 as a further binding partner for the Nephrin ECD, indicating that the molecular architecture of the slit diaphragm is even more complex than previously postulated (Figure 8). Moreover, our expansion microscopy approach revealed adjacent protein clusters of Nephrin and Crb2, suggesting the presence of homotypic Crb2-Crb2 assemblies in addition to homotypic Nephrin-Nephrin and Neph1-Neph1 clusters. Homotypic interactions of Crumbs proteins, which are most likely the precondition for Crb2 clusters, have been described before in Drosophila and zebrafish.21,22,72,73 Thus, heterotypic Crb2-Nephrin interactions might have the function to connect borderlines of Crb2- and Nephrin-positive clusters.
Figure 8.
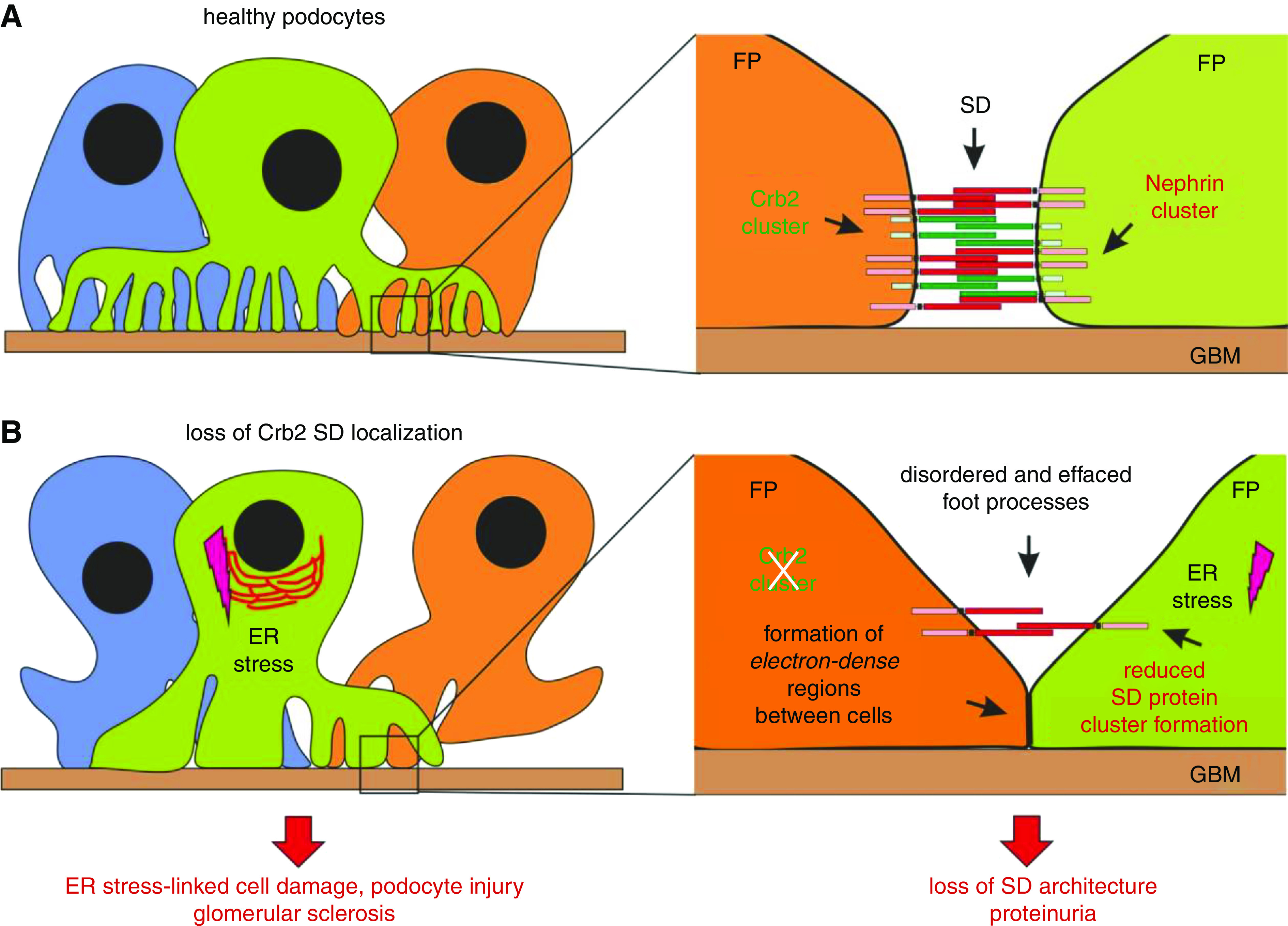
Proposed model: Loss of Crb2 expression at the SD is critical for podocyte injury in Crb2podKO and human CRB2 mutations. (A) In healthy glomeruli, podocytes foot processes (FPs) are interdigitating with each other and form adjacent Crumbs2 (green) and Nephrin (red) clusters at the slit diaphragm. (B) Crb2 loss at the slit diaphragm due to genetic inactivation (in Crb2 podKO mice) or due to dysfunctional Crb2 export to the cell surface (in patients with CRB2-related SRNS) prevents an interaction between Crb2 and Nephrin and destabilizes Crb2 cluster formation at the slit diaphragm. In podocytes, this results in increased formation of effaced and disorganized FPs, accompanied by enhanced appearance of tight junction–like (electron-dense) structures. In addition, in case of CRB2 mutants, misfolded Crb2 proteins can induce an ER stress response in podocytes (pink lightning). In summary, ER stress–linked cell damaging and slit diaphragm composition injury cause proteinuria that later results in glomerular sclerosis and renal failure. SD, slit diaphragm.
The SRNS-associated mutants that have been analyzed on a cell biologic level so far showed that Nephrin and Podocin mutants failed to be transported to the cell surface, which is—in the case of Nephrin—accompanied by an abnormal glycosylation pattern and accumulation at the ER.18,67,74,75 This indicates folding defects of missense protein variants that prevent proper processing and subsequent transport of these transmembrane proteins to the slit diaphragm. Vice versa, impaired glycosylation can result in dysfunctional surface export and cause FSGS-like phenotypes, emphasizing the effect of ER-linked glycosylation and folding of slit diaphragm proteins.65,76 Other studies27,66 and our data are in line with these results because selected SNRS-CRB2 mutants C629S, R633W, R1249Q, and N800K (carrying a putative N-glycosylation site) as well as the O-glycosylation defective mutant (S267A) accumulated at the ER and were hardly transported to the plasma membrane. There are some differences between the mutants. Whereas >97% of the S267A is at the ER, SRNS-associated R1249Q and N800K, for example, show a “rest” PM localization of about 10%. These differences in the in vitro system could be a first hint that clinical features of CRB2-associated diseases may also depend on the kind and the position of amino acids exchanges. In this context, it is worth it to mention that the pathogenic relevance of the R1249Q mutant is still unclear. This mutant was linked to pathogenic29,30 and nonpathogenic phenotypes (gnomAD/ClinVar entries) in homozygous patients.
Remarkably, Crb2 depletion in Crb2 podKO mice, the knockdown of crb2b in zebrafish, or the expression of Podocin missense mutants led to impaired transport of Nephrin, suggesting cotransport mechanisms of slit diaphragm proteins.18,37 We observed complete or partial mislocalization of Crb2 and propose that this altered distribution pattern prevents the establishment of functional Crb2 clusters and Crb2-Nephrin interactions between the boundaries of neighboring Crb2 and Nephrin clusters. Hence, our data support a loss of slit diaphragm localization of these proteins as an important pathomechanism in this kind of inherited monogenetic diseases (Figure 8).
Retention and accumulation of membrane proteins in the ER trigger the unfolded protein response and ER stress. Interestingly, ER stress itself is discussed as an aggravating or disease-promoting factor for podocyte diseases (reviewed by Cybulsky77). Our data support this assumption (Figure 8) because we detected an upregulation of ER stress–associated signaling pathways accompanied by increased binding of ER-localized CRB2 S267A and N800K variants to the ER chaperones Calnexin and Grp78/BiP. This is in agreement with previous studies showing similar results for SRNS mutants of Nephrin and of particular interest as modulators of ER stress could be attractive targets for drug discovery.67
Despite the striking and obvious similarities between Crb2 and Nephrin, there are also remarkable differences between these two slit diaphragm proteins. Crb2 showed a different expression profile compared with Nephrin,15 not only during embryo- and organogenesis28,32 but also, in developing podocytes during glomeruli maturation.27 Moreover, during S-shaped and capillary loop stages, Crb2 is also expressed in the parietal epithelium of glomeruli, whereas Nephrin expression was restricted to podocytes.8, 27
Crb2 is a member of the evolutionary conserved apically localized Crumbs protein family that controls cell polarization and establishment of cell-cell contacts in various species.21–23 These functions are mediated by an ICD that binds to a set of intracellular proteins, which differs from other slit diaphragm proteins, including PALS1, the FERM family like EPB4.1L5, or the AP-2 complex that mediates endocytosis.62,63,78 Therefore, it is likely that Crb2 recruits these proteins into the slit diaphragm complex. In addition, other studies linked Crb2 to Notch, Hippo, and mTORC1 signaling.27,39,79,80
Thus, Crumbs2 adds additional pathways to the slit diaphragm signaling hub that controls podocyte polarity and mechanosensing and regulates membrane trafficking and rearrangements of the actin cytoskeleton. Together, Crumbs2 emerges as a slit diaphragm protein of a similar relevance as Nephrin for the physiologic roles of glomerular podocytes.
Disclosures
H. Pavenstädt reports research funding from Sanofi and scientific advisor or membership with Deutsche Forschungsgemeinschaft. All remaining authors have nothing to disclose.
Funding
The work was supported by Deutsche Forschungsgemeinschaft (German Research Foundation) grants KU 2474/12-2 (to U. Kubitscheck), SFB1009-B10 (to H. Pavenstädt), WE 2550/2-2 (to T. Weide), and WE 2550/4-1 (to T. Weide). P. Boor was supported by Deutsche Forschungsgemeinschaft (German Research Foundation) grants SFB/TRR57, SFB/TRR219, BO3755/3-1, and BO3755/6-1, Bundesministerium für Bildung und Forschung (German Ministry of Education and Research) grant STOP-FSGS-01GM1901A, and Interdisziplinäres Zentrum für Klinische Forschung (IZKF grant O3-2) RWTH Aachen University. J. Wijnholds was supported by ZonMw (The Netherlands Organisation for Health Research and Development) grant 43200004.
Supplementary Material
Acknowledgments
The authors thank Kristin Doctor, Beate Naber, Stephan Rütten and Truc Van Le for excellent technical support. In addition, they thank Prof. Dr. Gia Voeltz and Prof. Dr. Michael Krahn for providing plasmids and Dr. Andreas Huge from core genomic facility in Muenster for his support for RNA-seq analyses. Thanks to Dr. Britta George and Dr. Daniela Anne Braun for critical reading the manuscript and all members of the contributing laboratories for fruitful discussions.
This work is part of the PhD thesis of A. Möller-Kerutt.
Prof. Dr. U. Kubitscheck, A. Möller-Kerutt, Prof. Dr. H. Pavenstädt, Dr. J.-P. Siebrasse, Prof. Dr. T. Weide, and Prof. Dr. J. Wijnholds designed the study; N. Boon, A. Möller-Kerutt, J. E. Rodriguez-Gatica, and K. Wacker performed the experiments; Prof. Dr. P. Boor, A. Möller-Kerutt, Prof. Dr. H. Pavenstädt, Dr. V. Van Marck, and Prof. Dr. T. Weide evaluated the histologic and electron microscopy data; Prof. Dr. U. Kubitscheck, A. Möller-Kerutt, J. E. Rodriguez-Gatica, Dr. J.-P. Siebrasse, and Prof. Dr. T. Weide performed the analyses of imaging and experimental data; Prof. Dr. U. Kubitscheck, A.Möller-Kerutt, Prof. Dr. H. Pavenstädt, Dr. J.-P. Siebrasse Prof. Dr. T. Weide, and Prof. Dr. J. Wijnholds prepared the manuscript; and all authors approved the final manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020040501/-/DCSupplemental.
Supplemental Table 1. Primers for genotyping and cloning.
Supplemental Table 2. Antibodies used in this study.
Supplemental Table 3. Primer pairs for RT-PCR.
Supplemental Table 4. Weight of observed wild-type (Cre−) and Crb2podKO (Cre+) mice.
Supplemental Table 5. Evaluation of glomerular injury in Crb2flox/flox (control) and Crb2podKO (knockout) mice.
Supplemental Table 6. Evaluation of tubular injury in wild-type Crb2flox/flox (Cre−) and Crb2podKO (Cre+) mice.
Supplemental Table 7. Localization of CRB2 variants in cells lines.
Supplemental Figure 1. Improved spatial resolution by deconvolution.
Supplemental Figure 2. Crb2 is highly expressed in mice glomeruli.
Supplemental Figure 3. Non-Mendelian inheritance distribution of Six2-Cre-Crb2wt/flox × Crb2flox/flox breeding.
Supplemental Figure 4. Analyses of 1-week-old Crb2podKO and littermate controls.
Supplemental Figure 5. Crb2 loss results in glomerular sclerosis, accompanied by disordered and effaced foot processes (≥8-week-old mice).
Supplemental Figure 6. Doxycycline-dependent expression of GFP- or SNAP-tagged CRB2.
Supplemental Figure 7. RNAseq analyses of CRB2 S267A mutant.
Supplemental Movie 1. Three-dimensional stack of an expanded glomerulus stained against Nephrin, Podocin, and DAPI.
Supplemental Movie 2. Three-dimensional segmentation of a region of an expanded glomerulus shown in Supplemental Movie 1 visualizing that the intracellular adapter Podocin localizes more closely to the nuclei than the transmembrane protein Nephrin.
Supplemental Movie 3. Three-dimensional stack of an expanded glomerulus stained against Nephrin, Podocin, and DAPI.
Supplemental Movie 4. Three-dimensional segmentation of a region of an expanded glomerulus shown in Supplemental Movie 2 revealing the presence of adjacent Crb2 and Nephrin clusters, in contrast to the Nephrin/Podocin localization.
References
- 1. Pavenstädt H, Kriz W, Kretzler M: Cell biology of the glomerular podocyte. Physiol Rev 83: 253–307, 2003. [DOI] [PubMed] [Google Scholar]
- 2. Scott RP, Quaggin SE: Review series: The cell biology of renal filtration. J Cell Biol 209: 199–210, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grahammer F, Schell C, Huber TB: The podocyte slit diaphragm--from a thin grey line to a complex signalling hub. Nat Rev Nephrol 9: 587–598, 2013. [DOI] [PubMed] [Google Scholar]
- 4. Schell C, Huber TB: The evolving complexity of the podocyte cytoskeleton. J Am Soc Nephrol 28: 3166–3174, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garg P: A review of podocyte biology. Am J Nephrol 47[Suppl 1]: 3–13, 2018. [DOI] [PubMed] [Google Scholar]
- 6. Kawachi H, Fukusumi Y: New insight into podocyte slit diaphragm, a therapeutic target of proteinuria. Clin Exp Nephrol 24: 193–204, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reiser J, Sever S, Faul C: Signal transduction in podocytes--spotlight on receptor tyrosine kinases. Nat Rev Nephrol 10: 104–115, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin CE, Jones N: Nephrin signaling in the podocyte: An updated view of signal regulation at the slit diaphragm and beyond. Front Endocrinol (Lausanne) 9: 302, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reiser J, Kriz W, Kretzler M, Mundel P: The glomerular slit diaphragm is a modified adherens junction. J Am Soc Nephrol 11: 1–8, 2000. [DOI] [PubMed] [Google Scholar]
- 10. Schnabel E, Anderson JM, Farquhar MG: The tight junction protein ZO-1 is concentrated along slit diaphragms of the glomerular epithelium. J Cell Biol 111: 1255–1263, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vivante A, Hildebrandt F: Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol 12: 133–146, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kestilä M, Lenkkeri U, Männikkö M, Lamerdin J, McCready P, Putaala H, et al.: Positionally cloned gene for a novel glomerular protein--nephrin--is mutated in congenital nephrotic syndrome. Mol Cell 1: 575–582, 1998. [DOI] [PubMed] [Google Scholar]
- 13. Guaragna MS, Cleto TL, Souza ML, Lutaif ACGB, de Castro LCG, Penido MG, et al.: NPHS1 gene mutations confirm congenital nephrotic syndrome in four Brazilian cases: A novel mutation is described. Nephrology (Carlton) 21: 753–757, 2016. [DOI] [PubMed] [Google Scholar]
- 14. Patrakka J, Kestilä M, Wartiovaara J, Ruotsalainen V, Tissari P, Lenkkeri U, et al.: Congenital nephrotic syndrome (NPHS1): Features resulting from different mutations in Finnish patients. Kidney Int 58: 972–980, 2000. [DOI] [PubMed] [Google Scholar]
- 15. Patrakka J, Tryggvason K: Nephrin--a unique structural and signaling protein of the kidney filter. Trends Mol Med 13: 396–403, 2007. [DOI] [PubMed] [Google Scholar]
- 16. Grahammer F, Wigge C, Schell C, Kretz O, Patrakka J, Schneider S, et al.: A flexible, multilayered protein scaffold maintains the slit in between glomerular podocytes. JCI Insight 1: 86177, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nishibori Y, Liu L, Hosoyamada M, Endou H, Kudo A, Takenaka H, et al.: Disease-causing missense mutations in NPHS2 gene alter normal nephrin trafficking to the plasma membrane. Kidney Int 66: 1755–1765, 2004. [DOI] [PubMed] [Google Scholar]
- 18. Huber TB, Simons M, Hartleben B, Sernetz L, Schmidts M, Gundlach E, et al.: Molecular basis of the functional podocin-nephrin complex: Mutations in the NPHS2 gene disrupt nephrin targeting to lipid raft microdomains. Hum Mol Genet 12: 3397–3405, 2003. [DOI] [PubMed] [Google Scholar]
- 19. Schwarz K, Simons M, Reiser J, Saleem MA, Faul C, Kriz W, et al.: Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2AP and nephrin. J Clin Invest 108: 1621–1629, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dlugos CP, Picciotto C, Lepa C, Krakow M, Stöber A, Eddy M-L, et al.: Nephrin signaling results in integrin β 1 activation. J Am Soc Nephrol 30: 1006–1019, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pocha SM, Knust E: Complexities of Crumbs function and regulation in tissue morphogenesis. Curr Biol 23: R289–R293, 2013. [DOI] [PubMed] [Google Scholar]
- 22. Thompson BJ, Pichaud F, Röper K: Sticking together the Crumbs - an unexpected function for an old friend. Nat Rev Mol Cell Biol 14: 307–314, 2013. [DOI] [PubMed] [Google Scholar]
- 23. Rodriguez-Boulan E, Macara IG: Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol 15: 225–242, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B: Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene 302: 21–29, 2003. [DOI] [PubMed] [Google Scholar]
- 25. Djuric I, Siebrasse JPJPJP, Schulze U, Granado D, Schlüter MA, Kubitscheck U, et al. : The C-terminal domain controls the mobility of Crumbs 3 isoforms. Biochim Biophys Acta 1863[6 Pt A]: 1208–1217, 2016. [DOI] [PubMed] [Google Scholar]
- 26. Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B: A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin β interactions. J Cell Biol 178: 387–398, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamano S, Nishibori Y, Hada I, Mikami N, Ito-Nitta N, Fukuhara D, et al.: Association of crumbs homolog-2 with mTORC1 in developing podocyte. PLoS One 13: e0202400, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramkumar N, Omelchenko T, Silva-Gagliardi NF, McGlade CJ, Wijnholds J, Anderson KV: Crumbs2 promotes cell ingression during the epithelial-to-mesenchymal transition at gastrulation. Nat Cell Biol 18: 1281–1291, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ebarasi L, Ashraf S, Bierzynska A, Gee HY, McCarthy HJ, Lovric S, et al.: Defects of CRB2 cause steroid-resistant nephrotic syndrome. Am J Hum Genet 96: 153–161, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slavotinek A, Kaylor J, Pierce H, Cahr M, DeWard SJ, Schneidman-Duhovny D, et al.: CRB2 mutations produce a phenotype resembling congenital nephrosis, Finnish type, with cerebral ventriculomegaly and raised alpha-fetoprotein. Am J Hum Genet 96: 162–169, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Slavotinek AM: The family of crumbs genes and human disease. Mol Syndromol 7: 274–281, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao Z, Patrakka J, Nukui M, Chi L, Niu D, Betsholtz C, et al.: Deficiency in Crumbs homolog 2 (Crb2) affects gastrulation and results in embryonic lethality in mice. Dev Dyn 240: 2646–2656, 2011. [DOI] [PubMed] [Google Scholar]
- 33. Dolón JF, Paniagua AE, Valle V, Segurado A, Arévalo R, Velasco A, et al.: Expression and localization of the polarity protein CRB2 in adult mouse brain: A comparison with the CRB1rd8 mutant mouse model. Sci Rep 8: 11652, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dudok JJ, Murtaza M, Henrique Alves C, Rashbass P, Wijnholds J: Crumbs 2 prevents cortical abnormalities in mouse dorsal telencephalon. Neurosci Res 108: 12–23, 2016. [DOI] [PubMed] [Google Scholar]
- 35. van den Hurk JAJM, Rashbass P, Roepman R, Davis J, Voesenek KEJ, Arends ML, et al. : Characterization of the Crumbs homolog 2 (CRB2) gene and analysis of its role in retinitis pigmentosa and Leber congenital amaurosis. Mol Vis 11: 263–273, 2005. [PubMed] [Google Scholar]
- 36. Alves CH, Sanz AS, Park B, Pellissier LP, Tanimoto N, Beck SC, et al.: Loss of CRB2 in the mouse retina mimics human retinitis pigmentosa due to mutations in the CRB1 gene. Hum Mol Genet 22: 35–50, 2013. [DOI] [PubMed] [Google Scholar]
- 37. Ebarasi L, He L, Hultenby K, Takemoto M, Betsholtz C, Tryggvason K, et al.: A reverse genetic screen in the zebrafish identifies crb2b as a regulator of the glomerular filtration barrier. Dev Biol 334: 1–9, 2009. [DOI] [PubMed] [Google Scholar]
- 38. Hochapfel F, Denk L, Mendl G, Schulze U, Maaßen C, Zaytseva Y, et al.: Distinct functions of Crumbs regulating slit diaphragms and endocytosis in Drosophila nephrocytes. Cell Mol Life Sci 74: 4573–4586, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alves CH, Bossers K, Vos RM, Essing AHW, Swagemakers S, van der Spek PJ, et al.: Microarray and morphological analysis of early postnatal CRB2 mutant retinas on a pure C57BL/6J genetic background. PLoS One 8: e82532, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, et al.: Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3: 169–181, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weide T, Vollenbröker B, Schulze U, Djuric I, Edeling M, Bonse J, et al.: Pals1 haploinsufficiency results in proteinuria and cyst formation. J Am Soc Nephrol 28: 2093–2107, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moeller MJ, Sanden SK, Soofi A, Wiggins RC, Holzman LB: Podocyte-specific expression of cre recombinase in transgenic mice. Genesis 35: 39–42, 2003. [DOI] [PubMed] [Google Scholar]
- 43. Markó L, Vigolo E, Hinze C, Park JK, Roël G, Balogh A, et al.: Tubular epithelial NF-κB activity regulates ischemic AKI. J Am Soc Nephrol 27: 2658–2669, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. George B, Fan Q, Dlugos CP, Soofi AA, Zhang J, Verma R, et al.: Crk1/2 and CrkL form a hetero-oligomer and functionally complement each other during podocyte morphogenesis. Kidney Int 85: 1382–1394, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chozinski TJ, Mao C, Halpern AR, Pippin JW, Shankland SJ, Alpers CE, et al.: Volumetric, nanoscale optical imaging of mouse and human kidney via expansion microscopy. Sci Rep 8: 10396, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bucur O, Zhao Y: Nanoscale imaging of kidney glomeruli using expansion pathology. Front Med (Lausanne) 5: 322, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Granado D, Müller D, Krausel V, Kruzel-Davila E, Schuberth C, Eschborn M, et al.: Intracellular APOL1 risk variants cause cytotoxicity accompanied by energy depletion. J Am Soc Nephrol 28: 3227–3238, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schulze U, Vollenbröker B, Braun DA, Van Le T, Granado D, Kremerskothen J, et al.: The Vac14-interaction network is linked to regulators of the endolysosomal and autophagic pathway. Mol Cell Proteomics 13: 1397–1411, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zurek N, Sparks L, Voeltz G: Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12: 28–41, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saleem MA, O’Hare MJ, Reiser J, Coward RJ, Inward CD, Farren T, et al.: A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 13: 630–638, 2002. [DOI] [PubMed] [Google Scholar]
- 51. Bonse J, Wennmann DO, Kremerskothen J, Weide T, Michgehl U, Pavenstädt H, et al.: Nuclear YAP localization as a key regulator of podocyte function. Cell Death Dis 9: 850, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. : The DAVID gene functional classification tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8: R183, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bürgers J, Pavlova I, Rodriguez-Gatica JE, Henneberger C, Oeller M, Ruland JA, et al.: Light-sheet fluorescence expansion microscopy: Fast mapping of neural circuits at super resolution. Neurophotonics 6: 015005, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fukusumi Y, Zhang Y, Yamagishi R, Oda K, Watanabe T, Matsui K, et al.: Nephrin-binding Ephrin-B1 at the slit diaphragm controls podocyte function through the JNK pathway. J Am Soc Nephrol 29: 1462–1474, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Verma R, Venkatareddy M, Kalinowski A, Li T, Kukla J, Mollin A, et al.: Nephrin is necessary for podocyte recovery following injury in an adult mature glomerulus. PLoS One 13: e0198013, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou Z, Wang J, Guo C, Chang W, Zhuang J, Zhu P, et al.: Temporally distinct six2-positive second heart field progenitors regulate mammalian heart development and disease. Cell Rep 18: 1019–1032, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Succar L, Boadle RA, Harris DC, Rangan GK: Formation of tight junctions between neighboring podocytes is an early ultrastructural feature in experimental crescentic glomerulonephritis. Int J Nephrol Renovasc Dis 9: 297–312, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gee HY, Sadowski CE, Aggarwal PK, Porath JD, Yakulov TA, Schueler M, et al.: FAT1 mutations cause a glomerulotubular nephropathy. Nat Commun 7: 10822, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barletta GM, Kovari IA, Verma RK, Kerjaschki D, Holzman LB: Nephrin and Neph1 co-localize at the podocyte foot process intercellular junction and form cis hetero-oligomers. J Biol Chem 278: 19266–19271, 2003. [DOI] [PubMed] [Google Scholar]
- 60. Gerke P, Huber TB, Sellin L, Benzing T, Walz G: Homodimerization and heterodimerization of the glomerular podocyte proteins nephrin and NEPH1. J Am Soc Nephrol 14: 918–926, 2003. [DOI] [PubMed] [Google Scholar]
- 61. Wartiovaara J, Öfverstedt LG, Khoshnoodi J, Zhang J, Mäkelä E, Sandin S, et al.: Nephrin strands contribute to a porous slit diaphragm scaffold as revealed by electron tomography. J Clin Invest 114: 1475–1483, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gosens I, Sessa A, den Hollander AI, Letteboer SJFF, Belloni V, Arends ML, et al.: FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Exp Cell Res 313: 3959–3970, 2007. [DOI] [PubMed] [Google Scholar]
- 63. Li Y, Wei Z, Yan Y, Wan Q, Du Q, Zhang M: Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proc Natl Acad Sci U S A 111: 17444–17449, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Unnersjö-Jess D, Scott L, Blom H, Brismar H: Super-resolution stimulated emission depletion imaging of slit diaphragm proteins in optically cleared kidney tissue. Kidney Int 89: 243–247, 2016. [DOI] [PubMed] [Google Scholar]
- 65. Yan K, Khoshnoodi J, Ruotsalainen V, Tryggvason K: N-linked glycosylation is critical for the plasma membrane localization of nephrin. J Am Soc Nephrol 13: 1385–1389, 2002. [DOI] [PubMed] [Google Scholar]
- 66. Ramkumar N, Harvey BM, Lee JD, Alcorn HL, Silva-Gagliardi NF, McGlade CJ, et al.: Protein O-Glucosyltransferase 1 (POGLUT1) promotes mouse gastrulation through modification of the apical polarity protein CRUMBS2. PLoS Genet 11: e1005551, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Drozdova T, Papillon J, Cybulsky AV: Nephrin missense mutations: Induction of endoplasmic reticulum stress and cell surface rescue by reduction in chaperone interactions. Physiol Rep 1: e00086, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Morito D, Nagata K: ER stress proteins in autoimmune and inflammatory diseases. Front Immunol 3: 48, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Solanki AK, Widmeier E, Arif E, Sharma S, Daga A, Srivastava P, et al.: Mutations in KIRREL1, a slit diaphragm component, cause steroid-resistant nephrotic syndrome. Kidney Int 96: 883–889, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grahammer F: New structural insights into podocyte biology. Cell Tissue Res 369: 5–10, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Putaala H, Soininen R, Kilpeläinen P, Wartiovaara J, Tryggvason K: The murine nephrin gene is specifically expressed in kidney, brain and pancreas: Inactivation of the gene leads to massive proteinuria and neonatal death. Hum Mol Genet 10: 1–8, 2001. [DOI] [PubMed] [Google Scholar]
- 72. Fletcher GC, Lucas EP, Brain R, Tournier A, Thompson BJ: Positive feedback and mutual antagonism combine to polarize Crumbs in the Drosophila follicle cell epithelium. Curr Biol 22: 1116–1122, 2012. [DOI] [PubMed] [Google Scholar]
- 73. Zou J, Wang X, Wei X: Crb apical polarity proteins maintain zebrafish retinal cone mosaics via intercellular binding of their extracellular domains. Dev Cell 22: 1261–1274, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu XL, Doné SC, Yan K, Kilpeläinen P, Pikkarainen T, Tryggvason K: Defective trafficking of nephrin missense mutants rescued by a chemical chaperone. J Am Soc Nephrol 15: 1731–1738, 2004. [DOI] [PubMed] [Google Scholar]
- 75. Liu L, Doné SC, Khoshnoodi J, Bertorello A, Wartiovaara J, Berggren PO, et al.: Defective nephrin trafficking caused by missense mutations in the NPHS1 gene: Insight into the mechanisms of congenital nephrotic syndrome. Hum Mol Genet 10: 2637–2644, 2001. [DOI] [PubMed] [Google Scholar]
- 76. Niculovic KM, Blume L, Wedekind H, Kats E, Albers I, Groos S, et al.: Podocyte-specific sialylation-deficient mice serve as a model for human FSGS. J Am Soc Nephrol 30: 1021–1035, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cybulsky AV: Endoplasmic reticulum stress, the unfolded protein response and autophagy in kidney diseases. Nat Rev Nephrol 13: 681–696, 2017. [DOI] [PubMed] [Google Scholar]
- 78. Lin YH, Currinn H, Pocha SM, Rothnie A, Wassmer T, Knust E: AP-2-complex-mediated endocytosis of Drosophila Crumbs regulates polarity by antagonizing Stardust. J Cell Sci 128: 4538–4549, 2015. [DOI] [PubMed] [Google Scholar]
- 79. Mitsuishi Y, Hasegawa H, Matsuo A, Araki W, Suzuki T, Tagami S, et al.: Human CRB2 inhibits γ-secretase cleavage of amyloid precursor protein by binding to the presenilin complex. J Biol Chem 285: 14920–14931, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pellissier LP, Alves CH, Quinn PM, Vos RM, Tanimoto N, Lundvig DMS, et al.: Targeted ablation of CRB1 and CRB2 in retinal progenitor cells mimics Leber congenital amaurosis. PLoS Genet 9: e1003976, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.