Significance Statement
The current Banff classification of kidney transplant rejection is on the basis of complex and discretionary combinations of histologic scores. As a purely empiric classification, it was not primarily developed to reflect clinically meaningful outcomes such as graft failure, and allows ambiguous phenotypes to overlap. This paper describes the use of data-driven clustering methods to produce a phenotypic reclassification of kidney transplant rejection that is both histologically and clinically relevant. Six novel cluster phenotypes are validated on external data. Each of these new phenotypes is significantly associated with graft failure and overcomes the current limitations of intermediate and mixed phenotypes. The data-driven phenotypic reclassification of kidney transplant rejection is a proof of concept, opening future research directions.
Keywords: acute allograft rejection, kidney biopsy, transplant pathology, transplant outcomes, kidney transplantation
Visual Abstract
Abstract
Background
Over the past decades, an international group of experts iteratively developed a consensus classification of kidney transplant rejection phenotypes, known as the Banff classification. Data-driven clustering of kidney transplant histologic data could simplify the complex and discretionary rules of the Banff classification, while improving the association with graft failure.
Methods
The data consisted of a training set of 3510 kidney-transplant biopsies from an observational cohort of 936 recipients. Independent validation of the results was performed on an external set of 3835 biopsies from 1989 patients. On the basis of acute histologic lesion scores and the presence of donor-specific HLA antibodies, stable clustering was achieved on the basis of a consensus of 400 different clustering partitions. Additional information on kidney-transplant failure was introduced with a weighted Euclidean distance.
Results
Based on the proportion of ambiguous clustering, six clinically meaningful cluster phenotypes were identified. There was significant overlap with the existing Banff classification (adjusted rand index, 0.48). However, the data-driven approach eliminated intermediate and mixed phenotypes and created acute rejection clusters that are each significantly associated with graft failure. Finally, a novel visualization tool presents disease phenotypes and severity in a continuous manner, as a complement to the discrete clusters.
Conclusions
A semisupervised clustering approach for the identification of clinically meaningful novel phenotypes of kidney transplant rejection has been developed and validated. The approach has the potential to offer a more quantitative evaluation of rejection subtypes and severity, especially in situations in which the current histologic categorization is ambiguous.
Kidney transplant biopsies are crucial in the follow-up of patients after transplantation. Both at the time of graft dysfunction (indication biopsies) and the time of stable graft function (protocol biopsies), the histologic evaluation of these biopsies enables one to distinguish rejection mechanisms from other injury processes and orient the appropriate therapeutic interventions. Over the past decades, an international group of experts has developed a consensus classification of kidney transplant rejection phenotypes, known as the Banff classification.1–3
The Banff classification relies on the histologic evaluation of a set of well-defined lesions, further translated into semiquantitative, ordinal lesion scores.4 The diagnostic classification process consists of a set of if-then rules that map conditional clauses on the basis of lesion scores to a diagnosis category. Currently, the Banff classification encompasses five main categories3: (1) normal biopsy or nonspecific changes; (2) antibody-mediated rejection (ABMR); (3) borderline changes; (4) T cell–mediated rejection (TCMR); and (5) polyomavirus nephropathy. Several of these categories are further divided into subtypes. This classification was developed iteratively, on the basis of studies that examine the associations between lesions and risk factors such as donor-specific HLA antibodies, between lesions and graft failure, and among lesions themselves.5–7 Banff diagnostic categories are not mutually exclusive and Banff lesions are not specific for disease processes, which leads to overlapping diagnoses and mixed rejection phenotypes. Although this reflects a histologic reality, the clinical interpretation of this complex categorization process is difficult, leading to unstable clinical decisions.
Instead of this iterative consensus process for disease classification, data-driven mathematic modeling of the multidimensional histologic data could be appropriate. Such an approach could refine the thresholds for the diagnostic phenotypes, simplify the complex and discretionary if-then rules, avoid the issue of mixed phenotypes, and yield new phenotypes and disease reclassification. Categorizing data into groups without pre-existing labels is commonly referred as unsupervised clustering.8 Although the resulting clusters (reclassified disease phenotypes) might be valid from a mathematic perspective, there is no guarantee they will show relevant association with external outcome variables. To overcome this, introducing information on outcome in the clustering process could be of interest.9–11 Whether such a mathematic modeling approach would also be applicable to the classification of kidney transplant rejection has not been evaluated yet.
On the basis of these considerations, we built and externally validated a model for mathematic reclassification of acute kidney transplant rejection, on the basis of the integration of the set of inflammatory lesions in kidney transplant biopsies, informed by graft failure, in a retrospective observational cohort study.
Methods
Data
Patients and Biopsies
For the training cohort, all consecutive adult recipients of a kidney transplant at the Leuven University Hospitals between March 2004, the start of the protocol biopsy program, and February 2013 were eligible for this study (n=1137). A minimum of 5 years follow-up at time of data extraction (March 2018) was required. Recipients of combined transplantation (n=113) or kidney transplantation after another solid organ transplantation (n=24) were excluded. All transplants were performed with negative complement-dependent cytotoxicity crossmatches. The clinical data were collected during routine clinical follow-up in electronic medical records, which were used for clinical patient management and directly linked to the SAS database from which the research database was extracted. The standard immunosuppressive maintenance regimen consisted of tacrolimus, mycophenolate, and corticosteroids.12 The histologic data consisted of all 3622 kidney transplant biopsies performed at the Leuven University Hospitals between April 2004 and February 2015 in 949 patients. Biopsies were performed on medical indication (indication biopsies at time of graft dysfunction) or as part of an established follow-up protocol (protocol biopsies).13 Biopsies with missing lesion scores were excluded (n=112), due to missing HLA–donor specific anti-HLA antibodies (DSA) (n=73) and/or missing the score of C4d deposition in peritubular capillaries (n=40). A total of 3510 biopsies from 936 recipients remained available for analysis. This study was approved by the Ethical Committee of the University Hospitals Leuven (S64006).
For the validation cohort, the electronic database of Lyon University Hospitals (registration AC-2016–2706) and the Paris Transplant Group were screened with the same selection criteria as detailed above. Between January 2007 and December 2015 for the Lyon dataset, and between March 2009 and October 2019 for the Paris dataset, 1356 (from 726 transplants) and 2479 biopsies (from 1304 transplants), respectively, were included as an independent validation set, performed either for indication or as part of the routine follow-up at 3 and 12 months after transplantation. Only complete data were included. Clinical, histologic, and immunologic data were extracted from these databases, anonymized, and transmitted to Leuven to be used as an external independent validation cohort.
Histologic Scoring
In the training cohort, all post-transplant kidney allograft biopsies performed in this cohort, until the time of data extraction in December 2018, were included. One pathologist (EL) reviewed all biopsies, independent of clinical information to avoid bias. The severity of the histologic lesions was semiquantitatively scored, according to the Banff categories with a small deviation for C4d thresholds.12 The set of individual Banff lesions (n=14) represents either acute or chronic injury processes. We focused on the following seven acute Banff lesions, with semiquantitative scores reflecting disease activity, in concordance with the Banff guidelines4: tubulitis (t; score 0–3), interstitial inflammation (i; score 0–3), glomerulitis (g; score 0–3), intimal arteritis (v; score 0–3), C4d deposition in peritubular capillaries (C4d; score 0–3), peritubular capillaritis (ptc; score 0–3), and thrombotic microangiopathy (TMA; present versus absent). We considered transplant glomerulopathy (cg; score 0–3), interstitial fibrosis (ci; score 0–3), tubular atrophy (ct; score 0–3), vascular intimal thickening (cv; score 0–3), mesangial matrix increase (mm; score 0–3), arteriolar hyalinosis (ah; score 0–3), and glomerulosclerosis (gs; score 0–3) as chronic lesions and did not take them into account in the classification of acute rejection phenotypes. As the presence of HLA-DSA is a defining feature in the Banff diagnosis of ABMR, HLA-DSA was also considered in the clustering process (present versus absent), as defined previously for this cohort.14
The biopsies were classified into acute rejection categories, on the basis of the criteria as defined by the most recent Banff 2019 consensus.3 Overall, each biopsy was assigned to one of the six following categories on the basis of the Banff acute rejection phenotype: (1) no rejection, (2) borderline changes, (3) TCMR, (4) ABMR, (5) mixed borderline rejection, and (6) mixed rejection. Borderline changes were diagnosed as foci of tubulitis (t >0) with minor interstitial inflammation (i1) or moderate to severe interstitial inflammation (i2 or i3) with mild (t1) tubulitis. ABMR was diagnosed by the presence of the three Banff criteria for either acute or chronic active ABMR, according to the Banff 2019 classification, but not taking into account potential non-HLA antibodies or gene expression changes. Due to lack of information on i-IFTA and total-i scores, chronic TCMR was not considered separately. We labeled biopsies presenting an overlap of ABMR and TCMR as mixed rejection, and the biopsies with an overlap of ABMR and borderline changes as mixed borderline rejection.
Data Analysis
Semisupervised Clustering Strategy
We scaled each histologic lesion score (feature) into the unit interval. We adapted semisupervised learning from10 where additional information was used to facilitate the creation of clinically meaningful clusters. Specifically, Bair and Tibshirani10 used the Cox scores from univariate models to perform a feature selection before clustering, whereas we used the Cox score to weigh the features. We chose k-means as the core algorithm for the clustering process because of its straightforward implementation, its efficiency, its ability to accommodate the weighting of features, and the possibility to classify new biopsies into nonoverlapping clusters. The information from the death-censored kidney transplant survival outcome was introduced with a weighted Euclidean distance to provide additional guidance during the clustering process. Each feature was weighted with the normalized coefficient’s z score of univariate Cox models, adjusted for clustered data, namely, repeated biopsies from the same patients, using a sandwich variance estimate. Features with a higher weight contribute more heavily to the notion of dissimilarity between clusters than low-weight features, which will be less relevant to the definition of a cluster. Although guided by external survival information, the clustering task remains mostly unsupervised as the lesion scores patterns are the most influential driving force in the final clusters.
Consensus Clustering
We used consensus clustering15 on the basis of 400 clustering partitions of the data, with different random initializations of the k-means algorithm seed and a different subsampling (80%) of the original data, similar to the approach used by Monti.16 For the clustering process, all biopsies were considered independent. We used the nearest centroid method to assign a cluster label to the remaining 20% of out-of-bag biopsies for each partition. The final consensus clustering was achieved through majority voting along the 400 partitions. To avoid introducing biases in the clustering process by the overrepresentation of protocol biopsies, we adopted a scheme where indication biopsies and protocol biopsies were weighted on the basis of the inverse of their total proportion in the dataset. Cluster profiles were reported using the normalized mean value of lesions, or for binary features the percentage of biopsies with the feature present. We also report the proportion of each original lesion score. Where appropriate, individual lesion scores were compared between a pair of clusters with a chi-squared test. The degree of similarity between two different partitions of the data were evaluated with the adjusted rand index (ARI). This index accounts for overlapping partitions due to chance. It varies from −1 to 1, an ARI of 0 meaning random partitioning. A decision tree was trained on the cluster-labeled data to mimic the internal clustering process. The tree was generated using the Gini criterion, with a minimum of ten biopsies per leaf.
Tuning of Parameters
To define the optimal number of clusters, we used the proportion of ambiguous clustering (PAC)17 to assess the stability of our results at different values of k, namely, the number of clusters, with thresholds set at 10% and 90% of consensual clustering. Intuitively, PAC measures the proportion of all possible pairs of biopsies from the whole dataset that demonstrate inconsistent cluster attributions over the 400 partitions. The lower the PAC, the more stable the clustering across different conditions. We discarded very low values of k, because they only create a restricted number of clusters (typically no rejection versus any rejection with k=2), which is not helpful to describe different phenotypes.
Biopsy Stability
To identify biopsies that are part of pairs with an unstable cluster assignment over the set of clustering partitions, we developed an empirical individual stability score on the basis of the consensus matrix. The consensus matrix C is a n × n matrix, where n is the total number of biopsies and entries C i, represent the proportion of times biopsies i and j are clustered in the same cluster over the whole set of different partitions. The stability score s for biopsy i was defined as , where ci,j represented the value from the consensus matrix at the ith row and jth column, and N the total number of biopsies. Intuitively, a theoretic score of 1 reflects that the biopsy was consistently part of biopsy pairs that are either clustered together in the same cluster, or clustered in different clusters. A theoretic score of 0 would reflect a biopsy that forms ambiguous pairs with any other biopsy.
Survival Analysis
Graft survival times are reported as the number of days until graft failure, calculated from each biopsy date. Patients were administratively censored at the of last follow-up date or at time of death. Survival curves are plotted with Kaplan-Meier estimators along with the 95% confidence interval (95% CI). To avoid artificially increasing the incidence of transplant failure events due to repeated biopsies in a given individual, survival times from repeated biopsies in a given cluster were averaged for each patient. Pairwise comparison of survival curves was performed using Cox modeling and hazard ratio (HR) with 95% CI. Because potentially, proportional hazard assumption violations might bias the HR, we also report the restricted mean survival times (RMST)18 at 5 and 10 years, and its 95% CI. This measure can be interpreted as the mean survival time without event within a predefined time range, representing the area under the survival curve up to a predefined time point. We also report the differences in RMST (DRMST) with a baseline category, which estimates the difference in average event-free survival, in years, between a given category and the baseline group.
Visualization
Principal component analysis (PCA) was performed on the Cox score weighted acute lesions scores, and the first two components were used for two-dimensional visualization purposes. To better visualize the heterogeneity in the acute lesion scores, we developed a two-dimensional plot using polar coordinates, with the radius calculated as the sum of reweighted acute lesions scores, scaled to the unit interval (from 0 to 1), and the theta angle is a scaled version (for visual purposes) of the second component of the PCA, which is directly related to the main rejection phenotype. Because the sum of lesions is directly related to graft failure due to the individual weighting of lesions scores, this approach combines the severity and the phenotype trend in one single plot.
All analyses have been performed with Python 3.6.19 A web application where others can upload their own patient data and derive the clusters from the individual Banff lesion scores is available at https://rejectclass.pythonanywhere.com.
Results
Patient and Biopsy Characteristics
Descriptive patient (n=936) and biopsy (n=3510) data of the training cohort are shown in Table 1. On average, 3.75 biopsies (range 1–11) were performed per patient. Of the 773 indication biopsies, 644 (83.3%) were performed within the first year of transplantation (median at 22 days post-transplantation), and 129 (16.7%) after 1 year. HLA-DSA were present at the time of 468 (13.3%) biopsies.
Table 1.
Demographic, clinical, and histologic characteristics of the patients and biopsies included
| Cohort Characteristics | Total (n=936) |
|---|---|
| Donor demographics | |
| Donor type, N (%) | |
| Donation after brain death | 726 (77.6) |
| Donation after cardiac death | 153 (16.3) |
| Living donation, N (%) | 57 (6.1) |
| Age (yr), mean±SD | 47.7±14.7 |
| Male, N (%) | 497 (53.1) |
| Diabetes, N (%) | 24 (2.6) |
| Recipient demographics | |
| Age (yr), mean±SD | 53.5±13.3 |
| Male, N (%) | 572 (61.1) |
| Ethnicity, N (%) | |
| Caucasian | 920 (92.3) |
| African | 12 (1.3) |
| Asian | 3 (0.3) |
| Hispanic | 1 (0.1) |
| BMI (kg/m2), mean (range) | 25.4 (4.5) |
| Pre-transplant donor-specific HLA antibodies, N (%) | 408 (11.6%) |
| Repeat transplantation, N (%) | 141 (15) |
| Cold ischemia time (h), mean±SD | 14.2±5.7 |
| Total number of HLA A/B/DR mismatches, mean±SD | 2.8 (1.3) |
| Biopsy characteristics | Total (n=3510) |
| Banff 2019 diagnosis, N (%) | |
| No rejection | 2671 (76.1) |
| Borderline changes | 333 (9.5) |
| TCMR | 314 (8.9) |
| ABMR | 110 (3.1) |
| Mixed rejection (ABMR + TCMR) | 61 (1.7) |
| Mixed borderline rejection (ABMR + borderline changes) | 21 (0.6) |
| Indication biopsies, N (%) | n=773 (22.0) |
| Days since transplantation, median (interquartile range) | 22 (8–96) |
| eGFR at d of biopsy, median (interquartile range) | 19.8 (10.9–29.0) |
| Protocol biopsies, N (%) | n=2737 (78.0) |
| 3 mo | 823 (30.1) |
| 12 mo | 759 (27.7) |
| 24 mo | 639 (23.3) |
| 36 mo | 205 (7.5) |
| 48 mo | 22 (0.8) |
| 60 mo | 289 (7.6) |
| Days since transplant, median (interquartile range) | 377 (100–752) |
| eGFR at d of biopsy, median (interquartile range) | 46.4 (36.5–57.8) |
Semisupervised Clustering of Rejection Phenotypes
Fully unsupervised clustering of our biopsy cohort (n=3510) yielded an optimum of four different clusters, on the basis of the PAC (Supplemental Figure 1). Compared with cluster 1 (essentially normal biopsies), the three other clusters associated significantly with impaired graft survival. However, their histologic and clinical relevance were less clear, as none of these three clusters were defined on the basis of microcirculation inflammation and antibody activity (glomerulitis, peritubular capillaritis, and C4d), suggesting the number of clusters was insufficient to reflect the clinical reality and previous knowledge on the relevance of these lesions and ABMR. Increasing the number of clusters created clusters that were no longer associated with impaired graft survival compared with cluster 1 (Supplemental Table 1).
To optimize the clinical significance of the clusters, we applied a semisupervised clustering approach, weighing the histologic features with survival information. The optimal number of clusters (k) was six, on the basis of the PAC (Supplemental Table 1). We labeled the six identified clusters from 1 to 6, according to the overall association with graft failure (Figure 1, Supplemental Table 2). Biopsies in cluster 1 were dominated by 0 scores for the lesions, in cluster 2 by high g scores, and in cluster 3 by t and i. Clusters 1–3 were HLA-DSA negative, whereas all biopsies included in clusters 4–6 were in patients with HLA-DSA.
Figure 1.
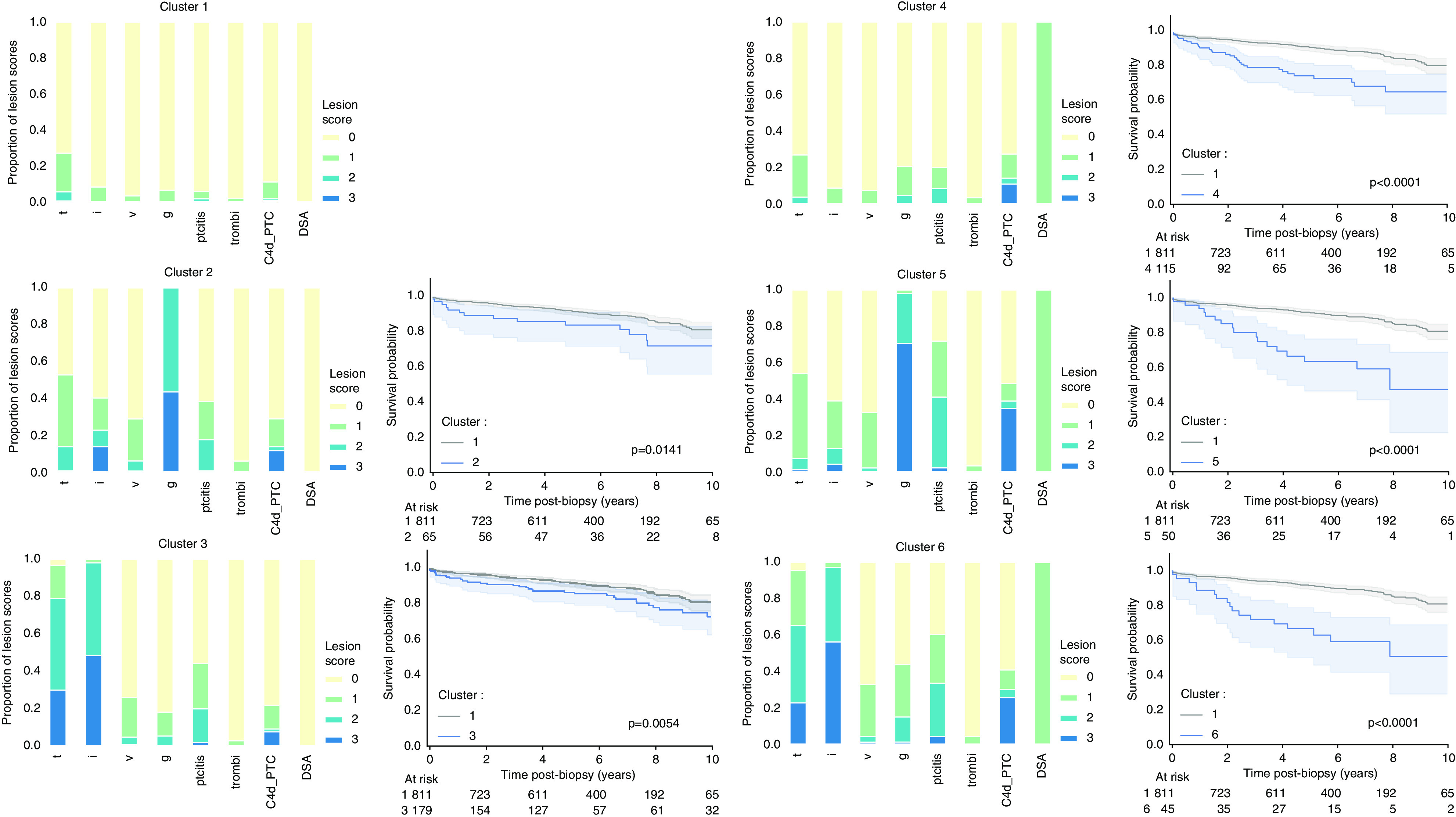
Distribution of the individual acute lesion scores in the different clusters, and postbiopsy Kaplan-Meier graft survival curves relative to cluster 1 of the derivation cohort (n=3510 biopsies). Biopsies included in cluster 1 were dominated by 0 scores (more than 90% have 0 score in all lesions, except for t [t0 in 72.9%] and C4d [C4d0 in 89.0%]). High g scores drove cluster 2 (56.4% g2 and 43.6% g3; no biopsies with g0 or g1). Compared with cluster 1, biopsies in cluster 2 had a higher proportion of score 1 or 2 acute lesions other than g. High t and i scores dominated cluster 3 biopsies (48.9% t2, 29.9% t3; 49.4% i2; and 48.5% i3). Biopsies in cluster 3 also had a higher proportion of score 1 and 2 acute lesion scores compared with cluster 1, but no g score 2 or higher. Cluster 4 was similar to cluster 1 and was dominated by low acute lesion scores. The main differences besides the presence of DSA was the higher proportion of g (g1 in 16.3% in cluster 4 versus 6.6% in cluster 1; g2 4.9% in cluster 4 versus 0.0% in cluster 1; P≤0.0001), a higher proportion of ptc (ptc1 in 11.4% in cluster 4 versus 4.2% in cluster 1; ptc2 in 8.5% in cluster 4 versus 1.2% in cluster 1; ptc3 in 0.3% in cluster 4 versus 0.1% in cluster 1; P≤0.0001), and a higher proportion of C4d (C4d1 in 13.4% in cluster 4 versus 9.5% in cluster 1; C4d2 in 3.3% in cluster 4 versus 0.5% in cluster 1; C4d3 in 11.1% in cluster 4 versus 1.0% in cluster 1, P≤0.0001). Cluster 5, similarly to cluster 2, was dominated by high g scores (27.4% g2 and 70.5% g3) and did not contain biopsies without g. As in cluster 2, we noted a higher proportion of score 1 and 2 acute lesions (ptc, t, i, v) compared with the cluster 1. Finally, in the presence of DSA, high t and i scores determined cluster 6 (42.4% t2, 22.7% t3; 40.9% i2; and 56.1% i3), and frequent presence of g and ptc. P values refer to HR from the Cox models. C4d_ptc, C4d deposition in peritubular capillaries; thrombi, thrombotic microangiopathy.
Biopsies in cluster 1 had no or very limited inflammation, a good outcome, and could be considered as no rejection. In cluster 2, all patients had moderate to severe glomerulitis in the absence of HLA-DSA, sometimes accompanied by tubulo-interstitial inflammation and peritubular capillaritis. These cases of glomerulitis in the absence of HLA-DSA are not fully understood and not reflected in the current Banff classification, yet associate with impaired graft outcome. Cluster 3 is characterized by moderate to severe degrees of tubulo-interstitial inflammation, reminiscent of acute TCMR in the Banff classification. In cluster 4, no or only very limited inflammation is noted, sometimes C4d deposition in peritubular capillaries. All patients in cluster 4 were HLA-DSA positive, which appeared to be a risk factor for graft failure in comparison to cluster 1, even in the absence of extensive inflammation. Biopsies in cluster 5 were HLA-DSA positive with high g scores and could be considered to reflect active ABMR. Biopsies in cluster 6 were HLA-DSA positive with high t and i scores, often combined with g and ptc, and could suggest mixed rejection. Biopsies in cluster 1 and 4 were most often protocol biopsies, whereas clusters 2, 3, and 5 were similarly distributed between protocol and indication biopsies (Supplemental Table 3). Cluster 6 was observed most often in indication biopsies and had worst eGFR and highest proteinuria.
Although we focused on acute histologic lesions, they often co-occurred with chronic lesions (Supplemental Figure 2). With k < or > six, we observed larger PAC (Supplemental Table 1). Increasing k did not drastically reshape previously found clusters, but rather added new clusters and conserved the similar centroids of the clusters derived at lower k. With k=7, we observed the separation of cluster 1 in two clusters on the basis of the t lesion (and, to a lesser extent, i). However, the survival curves from those two subclusters were largely overlapping (log-rank test P value=0.97), illustrating that also from clinical perspective, the optimal number of clusters was k=6. The ARI was similar between the various k values and the minimal distance between two centroids also decreased with a greater k.
On the basis of the consensus matrix, the average stability scores per cluster were 0.98, 0.98, 0.99, 0.98, and 0.99 for clusters 1, 3, 4, 5, and 6, respectively. In comparison with the other clusters, cluster 2, characterized by glomerulitis in the absence of HLA-DSA, was less stable, with an average stability score of 0.75. Overall, 44 biopsies had a low stability score (<0.5): 30 biopsies from cluster 2 (29.7%) and 14 biopsies from cluster 1 (0.5%). Because k-mean is a distance-based algorithm, it is possible to compute relative distances to the closest cluster’s boundary. If a biopsy is nearer to a cluster centroid than it is from the second closest centroid, the relative distance will be small. In contrast, a biopsy that is near the cluster’s boundary will have a relative distance approaching 1, translating to an almost equidistant position (Supplemental Figure 3). Biopsies with low stability scores were mostly found on the cluster edges.
Comparison of Disease Clusters with Banff 2019 Rules
There was important overlap between the clusters and the Banff categories with an ARI of 0.48 (Supplemental Results, Table 2). Due to its distance-based approach, the clustering algorithm led to better separation of the biopsies than the Banff 2019 classification, as also illustrated in the plots of PCA applied on the weighted acute lesion scores (Figure 2A). Although all lesions were taken into account simultaneously to assign each biopsy to a cluster, a decision tree could be derived, on the basis of the four main driving forces, g, HLA-DSA, i, and t (Supplemental Figure 4). This decision tree assigned the correct cluster with 97% of balanced accuracy, which confirmed the dominance of these four lesions in the phenotype reclassification. The 3% misclassified patients related to 24 biopsies.
Table 2.
Contingency tables comparing the Banff 2019 diagnosis and the six clusters derived from semi-supervised learning. Proportions represent the distribution in the clusters per Banff category (n=3510 biopsies)
| Banff 2019 Diagnosis | N | Cluster 1 (%) | Cluster 2 (%) | Cluster 3 (%) | Cluster 4 (%) | Cluster 5 (%) | Cluster 6 (%) |
|---|---|---|---|---|---|---|---|
| No rejection | 2659 | 2387 (89.8) | 53 (2.0) | 4 (0.2) | 215 (8.1) | 0 (0.0) | 0 (0.0) |
| Borderline changes | 327 | 261 (79.8) | 9 (2.8) | 26 (8.0) | 23 (7.0) | 0 (0.0) | 8 (2.4) |
| TCMR | 285 | 48 (16.8) | 25 (8.8) | 184 (64.6) | 5 (1.8) | 0 (0.0) | 23 (8.1) |
| ABMR | 122 | 8 (6.6) | 4 (3.3) | 0 (0.0) | 56 (45.9) | 53 (43.4) | 1 (0.8) |
| Mixed borderline rejection | 27 | 1 (3.7) | 3(11.1) | 1 (3.7) | 3 (11.1) | 15 (55.6) | 4 (14.8) |
| Mixed rejection | 90 | 5 (5.6) | 7 (7.8) | 16 (17.8) | 5 (5.6) | 27 (30.0) | 30 (33.3) |
| Total | 3510 | 2710 (77.2) | 101 (2.9) | 231 (6.6) | 307 (8.7) | 95 (2.7) | 66 (1.9) |
Figure 2.
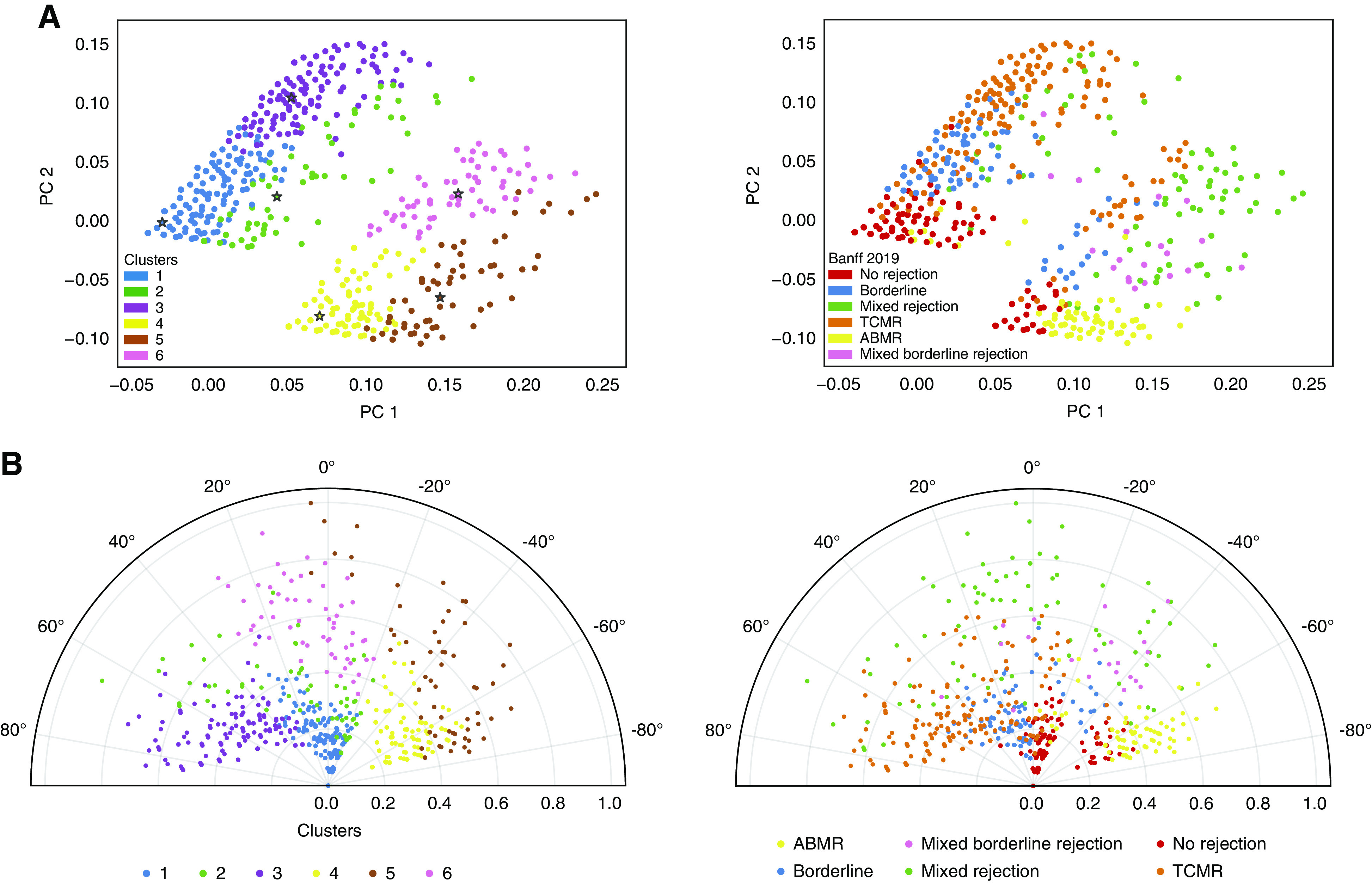
Visualization of the Banff classification and the six clusters on the whole set of kidney transplant biopsies. (A) PCA of the 3510 derivation cohort biopsies calculated from the acute lesion scores and DSA status, overlaid with the six clusters obtained from the semisupervised reclassification pipeline (left panel) and according to the Banff 2019 classification (right panel). Due to the distance-based approach of k-mean, the clusters obtained have a visually better separation than the Banff classification on two-dimensional plots. (B) Polar plot of the 3510 biopsies, with the radius representing the sum of re-weighted acute lesions scores, scaled to the unit interval (from 0 to 1), and the theta angle being directly related to the phenotype using the second semisupervised principal component, namely, the second component of PCA after weighting the lesions scores, overlaid with the six clusters obtained from the semisupervised reclassification pipeline (left panel) and according to the Banff 2019 classification (right panel).
Quantitative Visual Presentation of Disease Clusters
As expected, the superposition of the six disease clusters on the two-dimensional polar plot aligned better visually with the mathematic disease reclassification than with the different Banff 2019 phenotypes (Figure 2B). Biopsies projected with a negative angle were mostly associated with Banff TCMR, whereas those with a positive angle represented Banff ABMR. Biopsies with mixed rejection phenotypes were projected around 0°. When plotting individual lesions, and combinations of those (g+ptc = microcirculation inflammation) and (i+t = tubulo-interstitial inflammation) (Supplemental Figure 5), the PCA and the theta values associated with these two major components (microcirculation inflammation versus tubulo-interstitial inflammation), driving the disease reclassification. The radius on the plot was higher in indication biopsies compared with protocol biopsies (mean±SD: 0.22±0.23 versus 0.08±0.13 respectively, t test P<0.0001), illustrating more inflamed biopsies at time of graft dysfunction than at time of stable graft function (Supplemental Figure 6).
Association Between Disease Clusters and Graft Failure
During follow-up, 125 grafts failed, at a median of 3.67 years (1 day to 12 years) after transplantation. Of grafts, 9.1%, 22.4%, 25.0%, 30.0%, 37.7% and 50.0% failed within the first 5 years after the biopsy in cluster 1–6, respectively. The disease clusters 2–6 all associated with an increased risk of graft failure in comparison with cluster 1 (Figure 1, Table 3). Although Banff rejection categories had significant association with graft failure, except for borderline changes, the clusters’ weighted average in DRMST at 5 and 10 years were higher than the weighted average DRMST from the Banff classification (respectively 0.46 and 1.25 years for the clusters versus 0.29 and 0.72 years for the Banff categories), illustrating an overall better discrimination in terms of graft failure (Table 3). Furthermore, we observed an asymmetry between the first three and last three clusters, on the basis of HLA-DSA status. HRs on the HLA-DSA–negative/HLA-DSA–positive pair of clusters reported the following values: cluster 1 versus cluster 4: HR, 2.84; 95% CI, 1.80 to 4.30; P<0.0001; cluster 2 versus cluster 5: HR, 2.02; 95% CI, 1.00–4.12; P=0.051; and cluster 3 versus cluster 6: HR, 2.41; 95% CI, 1.35 to 4.30; P=0.003 (Supplemental Figure 7). The survival outcome of each cluster did not depend on the adjustment method for repeated biopsies per patient (Supplemental Figure 8). The clustering of biopsies led to improved prediction of graft failure, compared with the Banff classification (Supplemental Results).
Table 3.
Graft survival, RMST, and DMRST at 5- and 10-yr post-biopsy, according to each cluster and each Banff diagnostic category (n=3510)
| Banff Diagnosis | % Graft Survival At 5 Yr (%) | % Graft Survival At 10 Yr (%) | RMST At 5 Yr Postbiopsy, (95% CI) | RMST at 10 Yr Postbiopsy, (95% CI) | HR Versus No, (95% CI) | HR P Value Versus Yr | DRMST At 5 Yr Versus No Rejection, (95% CI) | DRMST At 10 Yr Versus No Rejection, (95% CI) |
|---|---|---|---|---|---|---|---|---|
| No rejection | 89.5 | 51.0 | 4.74 (4.63 to 4.85) | 9.01 (8.56 to 9.46) | — | — | — | — |
| Borderline changes | 83.1 | 42.3 | 4.66 (4.47 to 4.85) | 8.87 (8.04 to 9.7) | 1.27 (0.88 to 1.84) | 0.201 | 0.08 (-0.06 to 0.22) | 0.14 (-0.24 to 0.53) |
| TCMR | 75.5 | 41.2 | 4.50 (4.27 to 4.74) | 8.46 (7.7 to 9.22) | 1.66 (1.17 to 2.36) | 0.004 | 0.24 (0.05 to 0.42) | 0.55 (0.09 to 1.01) |
| ABMR | 70.2 | 22.0 | 4.26 (3.89 to 4.63) | 7.63 (6.25 to 9.02) | 2.63 (1.65 to 4.21) | <0.0001 | 0.48 (0.15 to 0.81) | 1.38 (0.51 to 2.25) |
| Mixed borderline rejection | 63.6 | 8.3 | 3.55 (4.06 to 4.57) | 6.74 (4.06 to 7.72) | 4.26 (2.29 to 7.94) | <0.0001 | 0.67 (0.06 to 1.28) | 2.13 (0.65 to 3.61) |
| Mixed rejection | 59.2 | 20.5 | 3.98 (3.48 to 4.48) | 7.03 (5.74 to 8.32) | 3.24 (2.08 to 5.05) | <0.0001 | 0.76 (0.34 to 1.18) | 1.98 (1.00 to 2.96) |
| Average | — | — | — | — | — | 0.45 (0.11 to 0.78) | 1.24 (0.40 to 2.07) | |
| Weighted average | — | — | — | — | — | 0.29 (0.06 to 0.53) | 0.72 (0.14 to 0.93) | |
| Cluster | % Graft Survival at 5 Yr (%) | % Graft Survival at 10 Yr (%) | RMST at 5 Yr Postbiopsy (95% CI) | RMST at 10 Yr Postbiopsy (95% CI) | HR Versus Cluster 1 | HR P Value Versus Cluster 1 | DRMST At 5 Yr Versus Cluster 1 (95% CI) | DRMST At 10 Yr Versus Cluster 1 (95% CI) |
| Cluster 1 | 90.9 | 54.6 | 4.76 (4.65 to 4.87) | 9.09 (8.63 to 9.56) | — | — | — | — |
| Cluster 2 | 77.6 | 33.3 | 4.42 (4.01 to 4.84) | 8.28 (6.98 to 9.59) | 1.98 (1.15 to 3.43) | 0.014 | 0.34 (0.02 to 0.70) | 0.81 (−0.03 to 1.65) |
| Cluster 3 | 75.0 | 39.8 | 4.54 (4.30 to 4.79) | 8.56 (7.75 to 9.36) | 1.72 (1.17 to 2.52) | 0.005 | 0.22 (0.02 to 0.41) | 0.53 (0.05 to 1.02) |
| Cluster 4 | 70.0 | 28.6 | 4.23 (3.87 to 4.58) | 7.67 (6.36 to 8.98) | 2.84 (1.88 to 4.30) | <0.0001 | 0.53 (0.24 to 0.82) | 1.42 (0.69 to 2.15) |
| Cluster 5 | 62.3 | 6.1 | 4.06 (3.48 to 4.64) | 6.84 (5.04 to 8.64) | 4.17 (2.48 to 7.03) | <0.0001 | 0.70 (0.26 to 1.14) | 2.25 (1.07 to 3.42) |
| Cluster 6 | 50.0 | 6.2 | 3.96 (3.43 to 4.48) | 6.78 (4.96 to 8.59) | 4.37 (2.59 to 7.35) | <0.0001 | 0.80 (0.31 to 1.29) | 2.31 (1.11 to 3.52) |
| Average | — | — | — | — | — | 0.52 (0.17 to 0.87) | 1.46 (0.58 to 2.35) | |
| Weighted average | — | — | — | — | — | 0.46 (0.09 to 0.69) | 1.25 (0.48 to 2.01) |
The radius on the polar plot of each biopsy associated independently with graft failure, with an Area Under the Curve of the Receiver Operating Characteristic for 2- and 5-year postbiopsy graft survival of respectively 0.70 (95% CI, 0.66 to 0.73) and 0.69 (95% CI, 0.67 to 0.72), respectively. Biopsies projected on the outer ranges of the radius had higher inflammatory lesion scores, and significantly worse survival compared with biopsies near the center of the polar plot (Figure 3A). Similar association with graft failure was obtained when we predicted graft failure for each biopsy separately, from the information available on the nearest neighborhood, calculated using the weighted Euclidean distance. For example, Figure 3B displays the survival probability at 5 years postbiopsy, estimated from local Kaplan-Meier estimates on the basis of 40 nearest neighbors. With this local approach, solely on the basis of the lesion scores and HLA-DSA status and not taking into account graft functional data or post-transplant time, the Area Under the Curve of the Receiver Operating Characteristic of the probability for graft failure were 0.72 (95% CI, 0.68 to 0.74) and 0.70 (95% CI, 0.67 to 0.73), respectively at 2 and 5 years postbiopsy.
Figure 3.
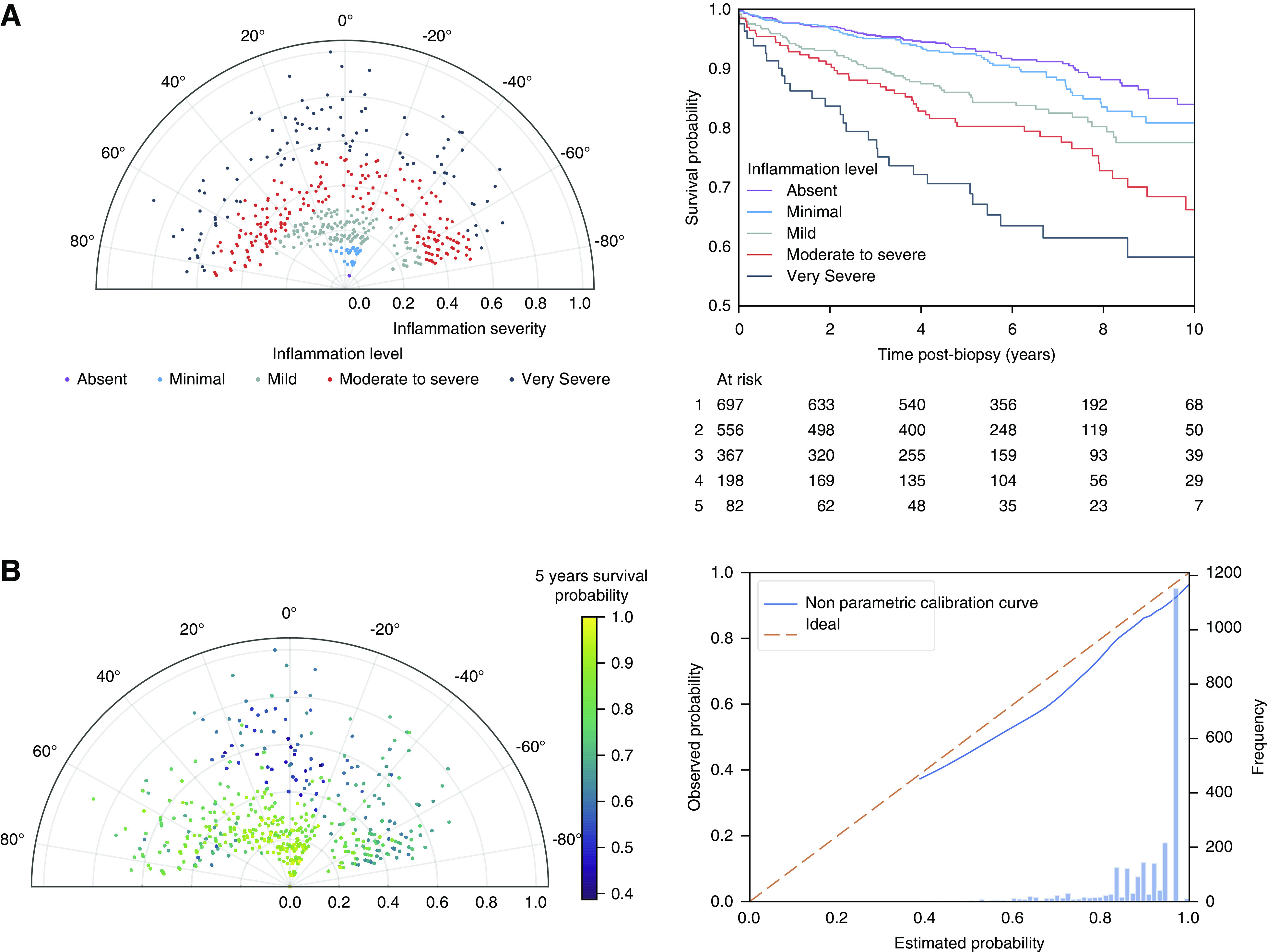
Association with graft survival in the polar plot visualization tool. (A) Association of the polar plot radius with graft survival in the derivation cohort. We stratified the 3510 biopsies along the radius axis in five strata and plotted the corresponding Kaplan-Meier survival curves. This demonstrates that the radius of the polar plot, which represents the extent of inflammation (the sum of the reweighted acute lesions scores, scaled to the unit interval from 0 to 1) is positively associated with the risk of graft failure. The different levels of inflammation correspond to the following radius: “No inflammation”: radius 0.00–0.04; “Minimal inflammation”: radius 0.04–0.10; “Mild inflammation”: radius 0.10–0.24; “Moderate to severe inflammation”: radius 0.24–42; and “Very severe inflammation”: radius 0.42–1.00. (B) Estimated graft survival probability at 5 years postbiopsy, calculated from the nearest neighborhood with k=40 (left panel) with corresponding calibration curve (right panel).
External Validation
Using the features weights and the cluster centroids obtained from the consensus clustering process, we are able to classify any new biopsy into one of the six previously described clusters. We applied this algorithm, starting from the lesion scores and HLA-DSA status only, without information on graft survival, to an external dataset of 3835 biopsies from Lyon University Hospital (n=1356) and the Paris Transplant Group (n=2479) (Supplemental Table 4). Note that this dataset did not include thrombotic microangiopathy in its variables. We therefore imputed this feature from the mean value of our training data. A comparison of the final clusters proportions between the two centers is presented in Supplemental Figure 9. Similar to the training set, biopsies from the external validation set were largely dominated by noninflamed cluster 1. The main difference in cluster distribution was a higher proportion of cluster 4 biopsies in the external dataset compared with the Leuven dataset (26.0% versus 8.7%, P<0.0001), explained by a larger prevalence of HLA-DSA–positive biopsies in the external data. Logically, the proportion of lesions within each clusters of the external validation set were very similar to the clusters obtained from the original data. There was also a similar association of the clusters with graft failure (Supplemental Figure 10).
A polar plot illustrates the full overlap in the histologic presentations between the training and validation cohorts (Supplemental Figure 11). Although the proportion of biopsies performed on indication was notably higher in the validation cohort (22.0% versus 37.7%, chi-squared test P<0.0001), the overall distribution of inflammation, estimated using the radius on the polar plot and the association with graft survival, was comparable between the training and validation datasets (Supplemental Figures 12 and 13). Comparing the clusters obtained on the validation dataset with the Banff categories, we obtained an ARI of 0.35. While maintaining a large overlap between the clustering method and the Banff classification (Supplemental Table 5), it demonstrates a higher reclassification rate in the validation dataset.
Discussion
Using a semisupervised and data-driven approach on 7345 post-transplant kidney biopsies with reweighting of acute histologic lesions, we derived and validated six distinct, clinically meaningful, phenotypic clusters. This mathematic clustering approach was fundamentally different from the iterative Banff classification process, which relies on a set of clinically derived if-then rules. Nevertheless, both in the training and the validation cohort, the novel phenotypes for kidney transplant rejection had a good degree of similarity with the Banff rejection categories, while redistributing intermediate and mixed phenotypes and maintaining the association with graft failure. The novel rejection phenotypes led to improved prediction of graft failure compared with the Banff classification. For integration of the novel phenotypic clustering with disease severity, and to move away from the black-and-white disease categorization, we proposed and validated a method for easily interpretable two-dimensional visual and quantitative presentation of the multidimensional histologic data.
Despite the similarity between the novel clusters and the Banff categories, we showed statistically improved prediction of graft failure with the clustering approach than using the Banff categories, especially in ambiguous situations, such as borderline changes or mixed rejection phenotypes. The association between (non)-inflamed clusters and graft survival remained present, even when the biopsies were stratified according to the rejection or nonrejection categories defined by Banff. An example of the clinical effect of this is, for example, the lack of cluster reflecting Banff borderline changes. Borderline changes are not reflected in a separate cluster, but most often (79.8%) classified to noninflamed cluster 1, with best post-transplant graft survival. Using this clustering approach therefore may solve the clinically difficult issue of how to deal with minimal tubulo-interstitial inflammation, below the current thresholds for TCMR. Also, the clustering algorithm proposed a novel phenotype, which is driven by glomerulitis in the absence of HLA-DSA. Although the causes of this phenotype are unknown, this resembles the phenotype described in recent publications on HLA-DSA–negative microcirculation inflammation.12,20–22 This phenotype is not recognized in the Banff classification6 and should be worked out in greater detail with respect to pathophysiology, risk factors, and clinical presentation. Finally, cluster 6 represents patients with mixed rejection phenotypes (ABMR with TCMR or borderline changes), which is not recognized as separate category in the Banff classification, but representing a clinical dilemma.23
Our clustering method directly relies on distance computation and provides a clinically relevant similarity metric to compare biopsies without concomitant clinical data besides HLA-DSA. For instance, we demonstrated and validated that local survival prediction on the basis of the nearest biopsies exhibited prognostic value solely on the basis of the histologic lesions and HLA-DSA status, thus not taking into account graft functional parameters or demographic factors relevant for outcome.24 Relying on this ad-hoc distance metric and to move beyond the black-and-white clustering approach, we developed an intuitive two-dimensional visualization tool, enabling the plotting of newly performed post-transplant biopsies and rapid assessment of the disease severity along with the dominant phenotype of neighboring biopsies. Because the k-mean algorithm is a hard-clustering algorithm, biopsies near the clusters’ boundaries are strictly allocated to one of the two neighbor clusters, preventing an overlap of diagnoses. This explains, for instance, that the mixed rejection biopsies are now split into one of the major clusters on the basis of their dominant lesions. However, contrasting with the Banff categorization, our clustering system can provide some degree of certainty regarding the classification, as expressed in terms of the relative distance to the closest cluster prototype (centroid). As a time-independent approach, our method is intended solely for reclassification of rejection (clustering algorithm and theta angle on the polar plot) and assessment of disease severity (radius on the polar plot). Our analysis on the accuracy of the local survival prediction needs to be seen as support for the clinical validity of the location of each sample on the polar plot, and does not suggest clinical utility of the local survival prediction as a prognostic tool on its own. For prognostication, more granular tools are becoming available, such as the iBox prediction score,24 which also integrate time post-transplantation and graft functional parameters into the models. Finally, diagnosis of other relevant disease phenotypes, such as GN or polyomavirus nephropathy, are on the basis of other parameters that are not included in the algorithm. These diseases should not be evaluated with our system solely intended for reclassification of rejection phenotypes.
As chronic histologic lesions in kidney transplant biopsies are nonspecific,4 we focused solely on acute inflammatory lesions to derive the novel rejection phenotypes. The evolution from active/early-stage disease, to chronic active, and finally chronic inactive forms of the same disease, was therefore not assessed and can be considered for future developments reclassification system. In addition, our approach fully depends on the quality of the histologic assessment, which is pathologist dependent and therefore not fully reproducible.25,26 More objective data, such as computerized imaging data or molecular expression data, or information on, for example, non-HLA antibodies and other immune risk factors, could further improve the reproducibility and accuracy of our system. Next, although the clusters described are sound biologically/clinically, whether treatment decisions on the basis of clusters instead of on the basis of Banff diagnosis will yield better outcome cannot be tested in this retrospective study. Similarly, data-driven algorithms do not assess pathophysiological mechanisms, hence no causal relations can be deducted from any cluster. Besides these clinical aspects, some technical limitations also warrant discussion. In concordance to the method described earlier,10 we used the whole dataset to compute the lesion score weights. The more data available to compute the weights, the more precise their estimation will be. In this semisupervised setting, weight overfitting is less detrimental than in a purely supervised approach. Despite its good performance, the k-mean algorithm remains simplistic. More elaborated core clustering algorithms, such as model-based or fuzzy clustering methods, could benefit the current approach and warrant additional studies.
Although we described a meaningful data-driven alternative to classify kidney transplant biopsies, and although our system has benefits over the current Banff categories, we do not suggest replacing the existing Banff classification with this algorithm, but use it in addition to Banff categorization, especially in patients that are difficult to categorize according to Banff. The clinical or scientific utility of our approach needs to be shown in further studies that validate the improved clinical decision making with regards to rejection treatment. Clinical implementation will depend on further external validation and detailed discussion at future Banff meetings and international consensus. The underlying risk factors and clinical presentations of each of the clusters still needs to be evaluated in greater depth, including information on HLA-DSA subtypes and profiles, non-HLA antibodies, etc. Inference on treatment decisions could not be made on our cohort, given the fact that patients with Banff rejection were treated with high-dose corticosteroids, and that patients with ABMR were treated with antibody-targeted therapies only very rarely.12 Nevertheless, this study highlights the potential of using the full scale of lesion grades for classification of kidney transplant biopsies, rather than using discretionary cutoff values. In the era of increasing availability of morphometric27 or molecular2 data, advanced statistical analysis and machine learning, with many resources to handle high-dimensional continuous variables,28 the existing expert-based consensus of if-then rules could be further improved using our approaches. We have developed and validated a semisupervised clustering approach for the identification of clinically meaningful novel phenotypes for kidney transplant rejection, on the basis of individual lesion scores. This approach has the potential to offer a more quantitative evaluation of rejection subtypes and severity, especially in situations where the current histologic categorization is ambiguous.
Disclosures
B. Sprangers reports being a scientific advisor to or member as an expert ad hoc for the European Medicines Agency. D. Kuypers reports consultancy agreements with Astellas Company, CSL Behring, and UCB; reports receiving research funding from Astellas; reports receiving honoraria from Astellas, CSL Behring, and UCB; reports being a scientific advisor to or member of associate editor Transplantation, Editorial board member Transplantation Reviews, Therapeutic Drug Monitoring, Current Clinical Pharmacology; and Speakers Bureau from Astellas. E. Van Loon reports shared inventorship of European patent “mRNA-based biomarker for antibody mediated transplant rejection,” filed January 17, 2019. M.-P. Emonds reports being a scientific advisor to or member of the Eurotransplant Tissue typing via the Advisory Committee. M. Naesens reports being a scientific advisor to or member via the Editorial Board for several journals and Advisor for the European Medicines Agency. O. Thaunat reports consultancy agreements from Novartis; reports receiving research funding from Biomerieux, Bristol Myers Squibb, Immucor, and Novartis; and reports being a scientific advisor to or membership of European Society for Organ Transplantation. All remaining authors have nothing to disclose.
Funding
This work is supported by The Research Foundation Flanders (FWO) and the Flanders Innovation and Entrepreneurship agency (VLAIO), with a TBM project (IWT.150199) and by the KU Leuven C3 internal grant (C32/17/049). M. Naesens and B. Sprangers are senior clinical investigators of the FWO (supported by grants 1844019N and 1842919N, respectively). T. Vaulet, E. Van Loon, and J. Callemeyn are supported by the FWO through holding fellowship grants (1S93918N, 1143919N, and 1196119N, respectively). O. Thaunat is supported by the Agence Nationale pour la Recherche (ANR-16-CE17-0007-01), the Fondation pour la Recherche médicale (PME20180639518), and the Etablissement Français du Sang. B. De Moor is supported by KU Leuven through the KU Leuven Research Fund (projects C16/15/059, C32/16/013, C24/18/022), Industrial Research Fund(Fellowship 13-0260) and several Leuven Research and Development bilateral industrial projects, Flemish Government Agencies, FWO (EOS Project 30468160 [SeLMA] and SBO project I013218N). B. De Moor also received funding from the Flemish Government (AI Research Program), VLAIO (City of Things COT.2018.018) Industrial Projects (HBC.2018.0405), and the European Commission. This project has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant 885682). A. Loupy is supported by Institut National de la Santé et de la Recherche Médicale grant ATIP Avenir. O. Aubert is supported by a Fondation Bettencourt Schueller fellowship grant. G. Divard is supported by a Fondation pour la Recherche Médicale fellowship grant.
Supplementary Material
Acknowledgments
The authors thank the centers of the Leuven Collaborative Group for Renal Transplantation, the clinicians and surgeons, nursing staff, and the patients. Prof. Olivier Thaunat is grateful to the Dr. Dijou and Dr. Picard from the Department of Pathology of the Hospices Civils de Lyon for their contribution to the histologic follow-up of kidney transplant patients. Prof. Olivier Thaunat is indebted to the members of the Laboratoire de Recherche Translationnelle en Immunologie des Greffes from Edouard Herriot Hospital for their help during data collection.
Dr. Thibaut Vaulet and Prof. Maarten Naesens designed the study and the analysis plan. Prof. Evelyne Lerut, Dr. Aleksandar Senev, Dr. Elisabet Van Loon, Dr. Jasper Callemeyn, Prof. Marie-Paule Emonds, Dr. Valérie Dubois, Dr. Maud Rabeyrin, Prof. Dirk Kuypers, Prof. Olivier Thaunat, Prof. Maarten Naesens were involved in clinical data collection and data quality control. Dr. Thibaut Vaulet did the statistical analyses and created the figures and tables, with input from Prof. Bart De Moor and Prof. Maarten Naesens. Dr. Thibaut Vaulet and Prof. Maarten Naesens interpreted the results and wrote the article, and all coauthors revised and approved it.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020101418/-/DCSupplemental.
Supplemental Table 1. Performance indices for a range of k values used to define the optimal number of clusters in the semisupervised clustering algorithm.
Supplemental Table 2. Details of cluster composition according to the individual Banff lesion scores and donor-specific HLA antibodies (N=3510 biopsies of the derivation cohort).
Supplemental Table 3. Distribution of biopsies among clusters and stratification into protocol versus indication biopsies (N=3510 biopsies of the derivation cohort).
Supplemental Table 4. Demographic, clinical, and histologic characteristics of the patients and biopsies included in the validation dataset.
Supplemental Table 5. Contingency tables comparing the Banff 2019 diagnosis and the six clusters obtained on the external validation dataset.
Supplemental Figure 1. Distribution of the individual acute lesion scores in the clusters using an unweighted approach, and postbiopsy Kaplan-Meier graft survival curves relative to cluster 1 of the derivation cohort.
Supplemental Figure 2. Distribution of chronic lesions in the six acute lesion clusters.
Supplemental Figure 3. Relative distances to the closest cluster’s boundary.
Supplemental Figure 4. Decision tree of the clustering process.
Supplemental Figure 5. Various combinations of lesions scores displayed on the polar plots.
Supplemental Figure 6. Comparison of indication versus protocol biopsies, as superposed on the polar plot.
Supplemental Figure 7. Postbiopsy graft survival in the three DSA-/DSA+ pair of clusters.
Supplemental Figure 8. Postbiopsy graft survival in the six clusters, according to the adjustment method for repeated biopsies per patient.
Supplemental Figure 9. Comparison of cluster proportion per center.
Supplemental Figure 10. Distribution of the individual acute lesion scores in the different clusters, and postbiopsy Kaplan-Meier graft survival curves relative to cluster 1 of the external validation cohort.
Supplemental Figure 11. Overlay of the data from Leuven and the external dataset in the polar plot, according to the six clusters identified in the derivation cohort.
Supplemental Figure 12. Distribution of the radius from the polar plot.
Supplemental Figure 13. Association with graft survival in the polar plot visualization tool on the basis of the validation data.
References
- 1. Solez K, Axelsen RA, Benediktsson H, Burdick JF, Cohen AH, Colvin RB, et al.: International standardization of criteria for the histologic diagnosis of renal allograft rejection: The Banff working classification of kidney transplant pathology. Kidney Int 44: 411–422, 1993. [DOI] [PubMed] [Google Scholar]
- 2. Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al.: The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant 18: 293–307, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Loupy A, Haas M, Roufosse C, Naesens M, Adam B, Afrouzian M, et al.: The Banff 2019 Kidney Meeting Report (I): Updates on and clarification of criteria for T cell- and antibody-mediated rejection. Am J Transplant 20: 2318–2331, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roufosse C, Simmonds N, Clahsen-van Groningen M, Haas M, Henriksen KJ, Horsfield C, et al.: A 2018 reference guide to the banff classification of renal allograft pathology. Transplantation 102: 1795–1814, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Racusen LC, Colvin RB, Solez K, Mihatsch MJ, Halloran PF, Campbell PM, et al.: Antibody-mediated rejection criteria - an addition to the Banff 97 classification of renal allograft rejection. Am J Transplant 3: 708–714, 2003. [DOI] [PubMed] [Google Scholar]
- 6. Haas M, Sis B, Racusen LC, Solez K, Glotz D, Colvin RB, et al.; Banff meeting report writing committee: Banff 2013 meeting report: Inclusion of c4d-negative antibody-mediated rejection and antibody-associated arterial lesions [published correction appears in Am J Transplant 15: 2784, 2015 10.1111/ajt.13517]. Am J Transplant 14: 272–283, 2014. [DOI] [PubMed] [Google Scholar]
- 7. Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, et al.: The Banff 2015 kidney meeting report: Current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant 17: 28–41, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hastie T, Tibshirani R, Friedman J: The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd Ed., New York: Springer Science & Business Media, 2009. [Google Scholar]
- 9. Bullinger L, Döhner K, Bair E, Fröhling S, Schlenk RF, Tibshirani R, et al.: Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 350: 1605–1616, 2004. [DOI] [PubMed] [Google Scholar]
- 10. Bair E, Tibshirani R: Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol 2: E108, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kickingereder P, Burth S, Wick A, Götz M, Eidel O, Schlemmer HP, et al.: Radiomic profiling of glioblastoma: Identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 280: 880–889, 2016. [DOI] [PubMed] [Google Scholar]
- 12. Senev A, Coemans M, Lerut E, Van Sandt V, Daniëls L, Kuypers D, et al.: Histological picture of antibody-mediated rejection without donor-specific anti-HLA antibodies: Clinical presentation and implications for outcome. Am J Transplant 19: 763–780, 2019. [DOI] [PubMed] [Google Scholar]
- 13. Coemans M, Van Loon E, Lerut E, Gillard P, Sprangers B, Senev A, et al.: Occurrence of diabetic nephropathy after renal transplantation despite intensive glycemic control: An observational cohort study. Diabetes Care 42: 625–634, 2019. 10.2337/dc18-1936 [DOI] [PubMed] [Google Scholar]
- 14. Senev A, Lerut E, Van Sandt V, Coemans M, Callemeyn J, Sprangers B, et al.: Specificity, strength, and evolution of pretransplant donor-specific HLA antibodies determine outcome after kidney transplantation. Am J Transplant 19: 3100–3113, 2019. [DOI] [PubMed] [Google Scholar]
- 15. Strehl A, Ghosh J: Cluster ensembles—a knowledge reuse framework for combining multiple partitions. J Mach Learn Res 3: 583–617, 2003. [Google Scholar]
- 16. Monti S, Tamayo P, Mesirov J, Golub T: Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn 52: 91–118, 2003. [Google Scholar]
- 17. Şenbabaoğlu Y, Michailidis G, Li JZ: Critical limitations of consensus clustering in class discovery. Sci Rep 4: 6207, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Royston P, Parmar MK: Restricted mean survival time: An alternative to the hazard ratio for the design and analysis of randomized trials with a time-to-event outcome. BMC Med Res Methodol 13: 152, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van, Rossum G: Python tutorial, Amsterdam, Centrum voor Wiskunde en Informatica (CWI), 1995. [Google Scholar]
- 20. Koenig A, Chen CC, Marçais A, Barba T, Mathias V, Sicard A, et al.: Missing self triggers NK cell-mediated chronic vascular rejection of solid organ transplants. Nat Commun 10: 5350, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bestard O, Grinyó J: Refinement of humoral rejection effector mechanisms to identify specific pathogenic histological lesions with different graft outcomes. Am J Transplant 19: 952–953, 2019. [DOI] [PubMed] [Google Scholar]
- 22. Callemeyn J, Lerut E, de Loor H, Arijs I, Thaunat O, Koenig A, et al.: Transcriptional changes in kidney allografts with histology of antibody-mediated rejection without anti-HLA donor-specific antibodies. J Am Soc Nephrol 31: 2168–2183, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madill-Thomsen K, Perkowska-Ptasińska A, Böhmig GA, Eskandary F, Einecke G, Gupta G, et al.; MMDx-Kideny Study Group: Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant 20: 1341–1350, 2019. [DOI] [PubMed] [Google Scholar]
- 24. Loupy A, Aubert O, Orandi BJ, Naesens M, Bouatou Y, Raynaud M, et al.: Prediction system for risk of allograft loss in patients receiving kidney transplants: International derivation and validation study. BMJ 366: l4923, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furness PN, Taub N, Assmann KJ, Banfi G, Cosyns JP, Dorman AM, et al.: International variation in histologic grading is large, and persistent feedback does not improve reproducibility. Am J Surg Pathol 27: 805–810, 2003. [DOI] [PubMed] [Google Scholar]
- 26. Smith B, Cornell LD, Smith M, Cortese C, Geiger X, Alexander MP, et al.: A method to reduce variability in scoring antibody-mediated rejection in renal allografts: Implications for clinical trials - a retrospective study. Transpl Int 32: 173–183, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sicard A, Meas-Yedid V, Rabeyrin M, Koenig A, Ducreux S, Dijoud F, et al.: Computer-assisted topological analysis of renal allograft inflammation adds to risk evaluation at diagnosis of humoral rejection. Kidney Int 92: 214–226, 2017. [DOI] [PubMed] [Google Scholar]
- 28. Fröhlich H, Balling R, Beerenwinkel N, Kohlbacher O, Kumar S, Lengauer T, et al.: From hype to reality: Data science enabling personalized medicine. BMC Med 16: 150, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.