Abstract
The skin epidermis is a constantly renewing stratified epithelium that provides essential protective barrier functions throughout life. Epidermal stratification is governed by a step-wise differentiation program that requires precise spatiotemporal control of gene expression. How epidermal self-renewal and differentiation are regulated remains a fundamental question. Cell-intrinsic and -extrinsic mechanisms that modify chromatin structure and interactions have been identified as key regulators of epidermal differentiation and stratification. Here, we will review the recent advances in our understanding of how chromatin modifiers, tissue-specific transcription factors, and force-induced nuclear remodeling processes function to shape chromatin and to control epidermal tissue development and homeostasis.
Introduction
The mammalian skin epidermis is a stratified epithelium that constantly self-renews to replace terminally differentiated, dying or damaged cells. During development and homeostatic turnover in adults, stem/progenitor cells located in the basal layer withdraw from the cell cycle, commit to differentiate and move upward to the suprabasal layers while undergoing defined, stepwise changes in their transcriptome, proteome, morphology, and function [1,2]. The epidermis relies on the precise activation of differentiation in response to tissue needs, while maintaining a stable pool of stem/progenitor cells. Regulation of such precisely timed cell fate transitions involve changes in chromatin organization, where chromatin remodelers, histone- and DNA-modifying enzymes, and histone chaperones collaborate with transcription factors to generate patterns of cell-type specific gene expression [3]
The epidermis is a load-bearing and mechanically challenged tissue, where the dynamic alterations in tension, as in other epithelia, have fundamental implications for morphogenesis, maintenance, and disease [4,5]. The nucleus is mechanically tethered to the extracellular environment and the contractile cytoskeleton through adaptor protein-mediated interactions at the nuclear envelope. Additional protein interactions link chromatin, in particular histone H3 lysine 9 tri-methylated (H3K9me3) constitutive heterochromatin, directly to the nuclear envelope [6]. Thus, cell-intrinsic and -extrinsic mechanical forces not only induce cellular and nuclear shape changes, but also directly deform and remodel chromatin. Consequently, mechanical signals to the nucleus can be transduced indirectly through mechanosensitive signaling pathways or directly through stress wave-like propagation of mechanical force to influence chromatin states and gene expression [7]. This review will discuss recent advances on the roles of chromatin modifications, nuclear architecture, and mechanical forces in regulation of the epidermis. We will specifically highlight the diverse biochemical and biophysical mechanisms by which chromatin modifiers regulate chromatin structure and interactions to control epidermal tissue function.
Chromatin modifiers in epidermal differentiation and development
Chromatin modifiers provide key epigenetic mechanisms by which cell identity is regulated. In addition to the H3K9me3-marked constitutive heterochromatin, histone H3 tri-methylated (H3K27me3) facultative heterochromatin plays an important role in establishing barriers to cell fate changes [8]. Central regulators of facultative heterochromatin are the Polycomb repressive complex (PRC) 1 and PRC2, which catalyze histone H2A monoubiquitination (H2AK119ub) and H3K27me3, respectively [8]. Epidermal deletion of any one of the core PRC2 subunits, EZH1/2, SUZ12, or EED, results in precocious epidermal stratification as evident by the accelerated formation of differentiated granular and conrnified suprabasal layers and epidermal barrier function during embryogenesis [9]. At the molecular level, PRC2-catalyzed deposition of H3K27me3 within the epidermal differentiation complex (EDC) locus prevents premature recruitment of the AP1 transcriptional activator and thus represses expression of key differentiation genes [10]. In addition, PRC2 prevents premature cell cycle exit of epidermal progenitors by repressing the Ink4A locus [10]. Although PRC1 and PRC2 largely co-localize at genomic targets, epidermal deletion of the core PRC1 subunits, RING1A/B does not affect the timing of epidermal differentiation during morphogenesis [11,12]. Instead, PRC1 is required for epidermal integrity and its loss leads to a skin blistering disorder similar to human skin fragility syndromes [12]. Collectively, the emerging model is that PRC1 and PRC2 co-operate to suppress a set of non-lineage genes in the epidermis, although PRC1 seems to play a more dominant role [12]. In contrast, PRC2 but not PRC1 plays a critical role in preventing premature activation of differentiation genes. Interestingly, PRC1 has a PRC2-independent role in binding to promoter regions of active epidermal cell-adhesion and lineage genes to promote their expression [11,12]. The promoters of the affected adhesion genes contain RING1B but not H3K27me3, suggesting that they are targets of non-canonical PRC1 complexes, which typically contain RYBP/YAF2 subunit instead of the canonical CBX subunit that recognizes PRC2-mediated H3K27me3 [13]. A similar activating role for non-canonical PRC1 complexes has been discovered in other systems [14,15], indicating that gene activation is likely to be a general feature of PRC1 activity.
Dual function of chromatin modifiers in epidermal transcriptional control has also been recently described for PRMT1, an arginine methyltransferase which modifies both histones and proteins [16]. PRMT1 sustains the expression of proliferation genes and maintains epidermal progenitor self-renewal. In addition, PRMT1 prevents premature differentiation by repressing epidermal differentiation genes such as GRHL3 [17]. Collectively these studies highlight multiple and diverse mechanisms by which chromatin modifiers spatially regulate epidermal tissue development and function (Fig. 1a, b).
Figure 1. Epidermal transcriptional regulation by chromatin modifiers and transcription factors.
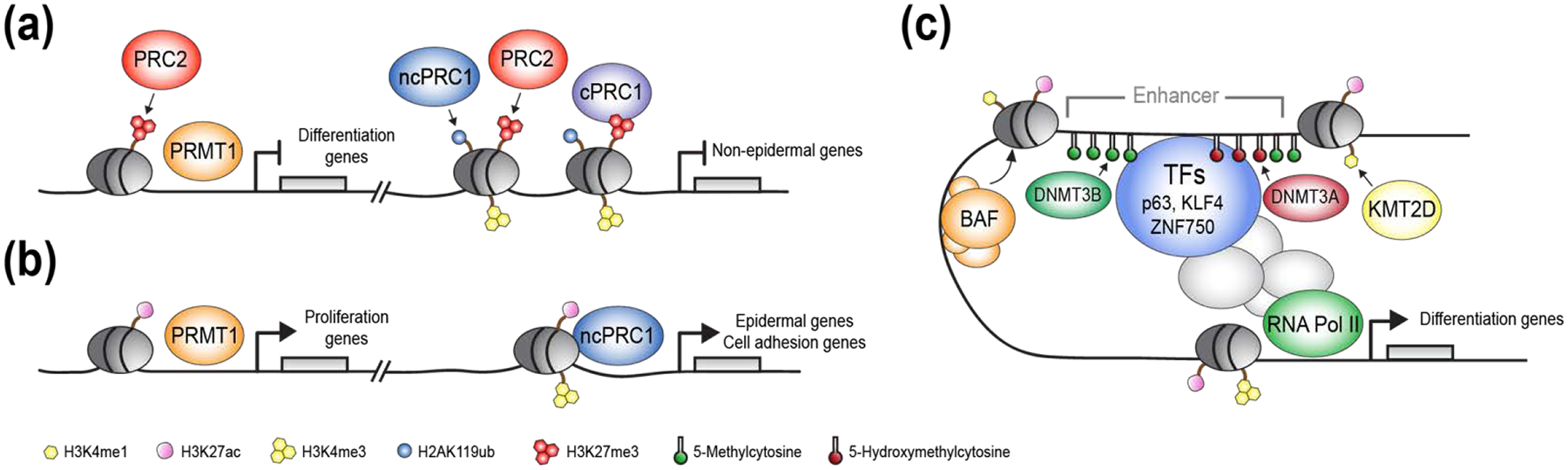
(a, right) Promoters of non-epidermal and epidermal differentiation genes are repressed by Polycomb repressive complex 2 (PRC2)-dependent histone H3 lysine 27 trimethylation (H3K27me3) and PRC1-depenendent histone H2A lysine 119 monoubiquitination (H2AK119ub) histone marks. (a, left) PRC2 and PRMT1 repress expression of epidermal differentiation genes and prevents premature epidermal differentiation. (b, right) Non-canonical PRC1 (ncPRC1) binds to promoters of epidermal lineage and cell adhesion genes and promotes gene expression, independently of PRC2 or canonical-PRC1 (cPRC1). (b, left) PRMT1 promotes expression of proliferation genes and sustains epidermal progenitor self-renewal. (c) Schematic illustration of the diverse mechanisms in which lineage-specific epidermal transcription factors work in-concert with chromatin modifiers to establish an open chromatin state and facilitate enhancer-promoter interactions, leading to activation of gene expression.
Chromatin regulation of enhancers in epidermal differentiation and homeostasis
Chromatin is organized into topologically associated domains (TADs), genomic regions characterized by DNA sequences that interact with each other more than with other genomic regions outside of the TAD [18]. The EDC locus is an excellent model system to study long-range chromosomal interactions, as it spans ~2Mb on human 1q21 (mouse chromosome 3q) and contain four clusters of gene families encoding for crucial structural and regulatory proteins, many of which are activated during epidermal differentiation [19]. In addition, several EDC proteins have been implicated in common skin diseases such as atopic dermatitis and psoriasis [20]. Chromatin Conformation Capture Carbon Copy (5C) studies on mouse epidermal keratinocytes recently identified multiple TADs within the EDC. In contrast to other cellular systems, chromatin interactions in the EDC occur not only within individual TADs but also between TADs [21]. Majority of the observed interactions are in the vicinity of gene promoters and enhancers, and are cell type-specific, indicating potential importance of these interactions for epidermal cell identify.
The interaction between enhancers and promoters is a critical feature of gene regulation. Prior to gene activation, enhancers are present at a closed chromatin state and nucleosomes at these enhancers are marked by histone H3 lysine 4 mono-methylation (H3K4me1). Upon activation, enhancers transition to an open chromatin state and gain histone-specific acetylation, including the acetylation of H3 lysine 27 (H3K27ac) [22,23]. Recently, significant progress has been made on understanding how enhancers are regulated during epidermal differentiation and homeostasis. A genome-wide promoter capture Hi-C method coupled with chromatin immunoprecipitation (ChIP) analysis of H3K27ac uncovered two classes of enhancer-promoter contacts associated with epidermal differentiation genes [24]. The first class gained H3K27ac and increased in strength during differentiation, whereas the second class was already established and marked by H3K27ac in progenitor cells. The “stable” class of enhancer-promoter contacts was associated with cohesin that stabilizes enhancer-promoter interactions via chromatin looping [24]. Reduction of critical cohesin subunit expression in epidermal progenitors led to reduced expression of self-renewal genes and spontaneous epidermal differentiation, indicating a critical role of cohesin in controlling the progenitor cell state [25]. In contrast, enhancer-promoter contacts that were gained during differentiation lacked cohesin binding [24]. These contacts were regulated by the transcription factors KLF4 and ZNF750, which both facilitated the establishment of enhancer-promoter contacts and activated enhancers. The effect of KLF4 and ZNF750 was restricted to a subset of enhancers associated with epidermal differentiation, suggesting that additional transcription factors are required to engage the full panel of differentiation gene enhancers.
Another key transcription factor that regulates epidermal proliferation and differentiation is p63 [26,27]. ChIP analyses of undifferentiated keratinocytes revealed that only a subset of p63-bound enhancers were marked by H3K27ac and active, suggesting that p63 “bookmarks” epidermal identity and differentiation gene enhancers, but additional co-regulators are required to activate gene expression [28]. Two key co-regulators of p63 have recently been identified, the BAF chromatin remodeling complex and the H3K4 mono-methyltransferase KMT2D. BAF and p63 mutually recruit each other to a subset of open chromatin regions during differentiation [29]. At these sites, BAF maintains an open chromatin state by displacing nucleosomes from p63 binding sites and by promoting recruitment of the transcriptional machinery. KMT2D and p63 co-target enhancers of genes associated with epidermal development, adhesion, and differentiation, and at these sites KMT2D promotes enhancer activity and the maintenance of H3K4me1 and H3K27ac histone marks [30].
p63 also functions to specify the surface ectoderm fate during early embryonic development [31]. To dissect mechanisms of p63 control of epidermal lineage specification, recent in vitro studies utilized retinoic acid and BMP4 to differentiate human embryonic stem cells into surface ectoderm [32]. These studies demonstrated that p63 can operate on chromatin to drive transcriptional changes only after chromatin accessibility and epigenetic landscape were established in response to morphogenic signals. One of the mechanisms by which morphogens and p63 can then regulate epidermal specification is by modifying long-range chromatin interactions and altering chromosome conformation [32], although the exact mechanisms by which the exposure to specific morphogens lead to initial changes in chromatin accessibility are not fully understood.
Collectively these studies reveal a complex regulatory network of transcription factors, chromatin remodelers and modifiers in control of stage-specific enhancer activity to establish precise epidermal programs (Fig. 1c).
Coupling nuclear morphology, mechanics and epigenetic gene regulation during differentiation
A key question is how the specific, molecular regulation of differentiation gene expression discussed above is coupled to the changing positions of the differentiating cells within the tissue. Cell shape and size changes are intimately linked to epidermal differentiation. In the highly proliferative embryonic epidermis and in vitro, crowding-induced cell compression combined with elongation triggers epidermal cell differentiation [33]. In the less dynamic adult epidermis, cell enlargement to occupy space liberated by delaminated differentiated cells triggers stem cell division to maintain homeostatic cell numbers [34]. The precise molecular mechanisms of space sensing by the progenitor cells are currently unclear. However, based on physical tethering of chromatin to the nuclear envelope, it has been demonstrated that cell shape changes directly lead to dynamic deformation of the nucleus and chromatin [35,36]. Hence, nuclear shape changes are able to remodel chromatin, which could thus directly alter gene expression in the epidermis (Fig. 2) [4,37].
Figure 2. A model describing changes in the nuclear and chromatin architecture during epidermal stratification.
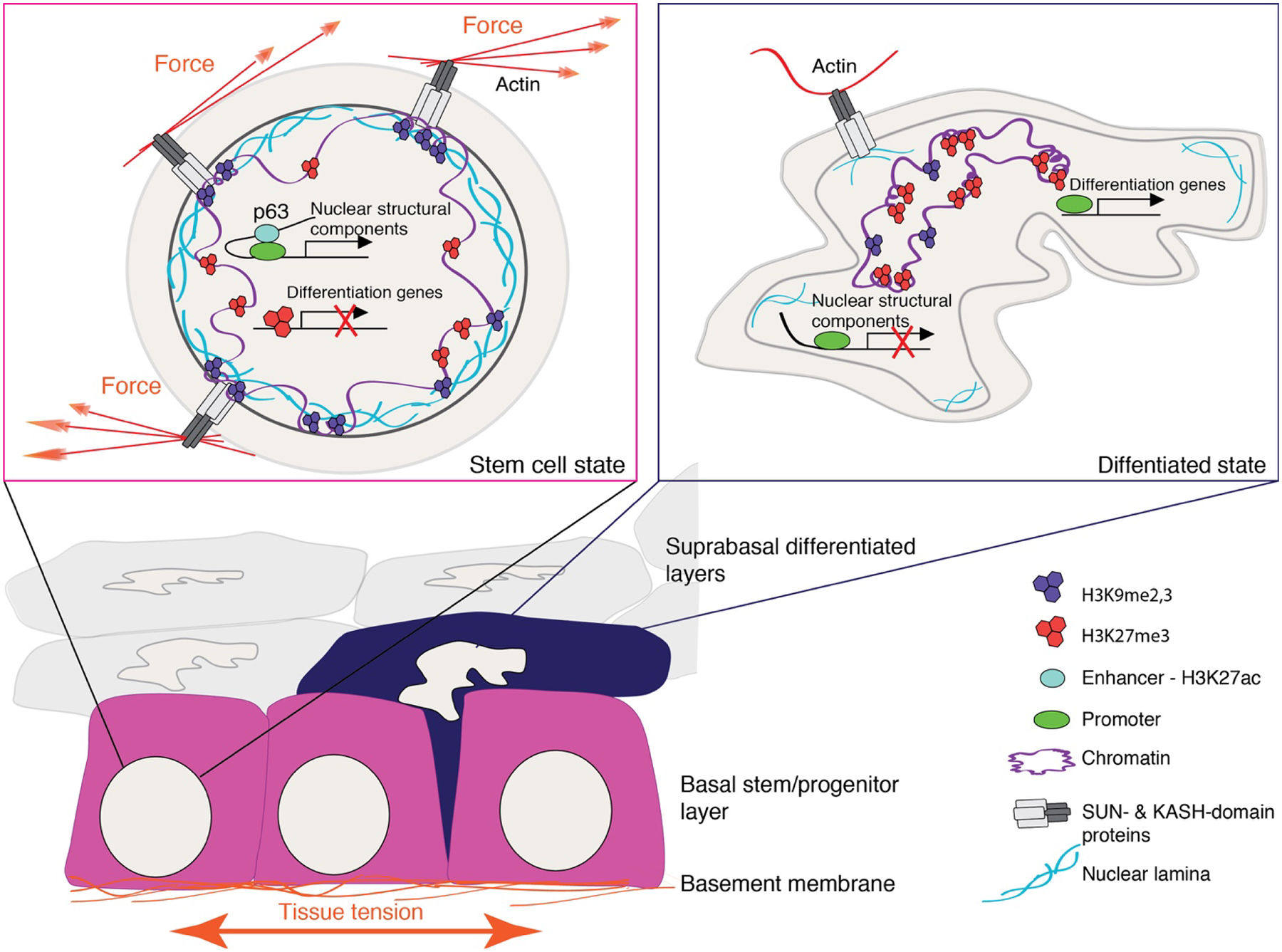
Extrinsic mechanical forces from the epidermal basement are transduced to the nucleus of basal stem/progenitor cells via members of the linker of nucleoskeleton and cytoskeleton, SUN- and KASH- domain proteins. Stem cells thus exhibit a mechanically stiff nucleus with p63-driven enhancer-promoter interactions driving expression of nuclear structural components such as A-type lamins and nesprins and anchoring of H3K9me2,3-marked heterochromatin to the nuclear lamina. Precautious differentiation is prevented by H3K27me3-mediated suppression of differentiation genes that can be reinforced by mechanical force.
As cells commit to differentiation, they withdraw from the cell cycle, delaminate from the basement membrane and move upward to the suprabasal layers. During delamination, epidermal cells decrease their cell-intrinsic tension and nuclear volume, lose p63 expression, and their H3K9me2,3-and H3K27me3-marked heterochromatin transitions to the nuclear interior. These changes are likely to elevate nuclear deformability and thus facilitate the upward movement of cells within the tightly packed cell layers.
The nuclear envelope (NE) plays a crucial role in regulating the shape and mechanical stability of the nucleus, nucleocytoplasmic transport, chromatin organization, and gene expression. The mechanosensitivity of the nucleus and the NE lies in the direct connections of the NE to the contractile cytoskeleton network. In addition to the central role of the nuclear lamina, the mobility, deformability and architecture of chromatin contribute to the mechanical properties of the nucleus [38,39].
During terminal differentiation, epidermal cells undergo marked changes in nuclear architecture and chromatin organization: their nuclear volume decreases, transcriptional activity declines, and the amount of heterochromatin increases [40,41]. Deletion of p63 results in abnormal nuclear morphology and reduced expression of structural components of the NE such as SUN1 and nesprins [42]. Nesprins are large nuclear transmembrane proteins that directly bind cytoplasmic actin and through their direct interaction with the inner nuclear membrane SUN proteins which bind lamins provide a direct mechanical link with the nuclear lamina and the cytoplasmic actin cytoskeleton (REF). p63 deletion further attenuates Ezh2 and HP1a expression and relocates H3K27me3 and H3K9me3-marked heterochromatin from the nuclear periphery towards the interior [42]. This suggests that epidermal stem cells regulate the mechanical stability of their nuclei via p63-mediated coupling of stemness with transcription of NE structural components as well as by positioning heterochromatin that is able to stiffen the nucleus [43] close to the lamina. As removal of p63 coincides with the exit of differentiating cells from the basal layer and increased nuclear deformability [44], one could envision that the enhanced deformability could be advantageous or even a prerequisite for the cell to be able to squeeze itself into the tightly packed suprabasal cell layer (Fig. 2).
How exactly do mechanical and morphological changes of the nucleus interface with gene expression changes required for terminal differentiation? Definitive answers are still missing, but studies in mesenchymal stem cells have shown that persistent mechanical stimulus acting on the nucleus induces heterochromatin formation, leading to a suppression of gene expression within heterochromatin condensed regions [45]. Consistent with this, long-term (minutes to hours) mechanical strain in epidermal progenitors leads to global transcriptional repression [4]. Although this transcriptional repression is global, it specifically facilitates accumulation of the silencing H3K27me3 mark on promoters of lineage-specific differentiation genes that are under the control of PRC2 [8,10] thereby attenuating epidermal progenitor cell differentiation (Fig. 2) [4]. Dynamic, real-time analyses of changes in nuclear shape and chromatin state during differentiation, in combination with mechanistic studies to interfere with nuclear mechano-sensing, are now required to establish the role of the nuclear lamina-chromatin linkage in geometry sensing during epidermal morphogenesis and homeostasis.
Concluding remarks
In summary, the dynamic patterns of gene expression required to form and maintain the epidermis involve controlled action of chromatin modifiers, transcription factors, and force-induced nuclear remodeling. Now that several key molecular players and pathways have been identified, a critical step will be to understand how these distinct transcription factors and chromatin regulators collaborate in space and time to control chromatin interactions and gene activity. This will require integration of 3D chromatin organization datasets with other genomic and epigenomic analyses, as well as live-cell imaging tools, to provide the necessary spatiotemporal resolution and positional context within the dynamic epidermal tissue.
Acknowledgements
We thank members of the Ezhkova and Wickström labs for discussions and Carmit Bar for Figure1 illustrations. SAW is funded by the Helsinki Institute of Life Science (University of Helsinki), Wihuri Research Institute, European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement 770877 - STEMpop), Academy of Finland, Max Planck Society, Erkko Foundation, and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; Project # 73111208 - SFB 829). EE is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under award number R01AR069078, and the Tisch Cancer Institute P30 Cancer Support Grant. YAM is a recipient of the EMBO Long Term Fellowship ALTF 728-2017. IC is supported by a Training Program in Stem Cell Biology fellowship from the New York State Department of Health (NYSTEM-C32561GG).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References and recommended reading:
Papers of particular interest, published within the period of review have been highlighted as
* of special interest
** of outstanding interest
- 1.Gonzales KAU and Fuchs E (2017) Skin and Its Regenerative Powers: An Alliance between Stem Cells and Their Niche. Dev. Cell 43, 387–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chacón‐Martínez CA et al. (2017) Hair follicle stem cell cultures reveal self‐organizing plasticity of stem cells and their progeny. EMBO J 36, 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bickmore WA and Van Steensel B (2013) Genome architecture: Domain organization of interphase chromosomes. Cell 152, 1270–1284. [DOI] [PubMed] [Google Scholar]
- 4.Le HQ et al. (2016) Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol 18, 864–75 [DOI] [PubMed] [Google Scholar]; ** This paper demonstrates how extrinsic mechanical strain impacts chromatin structure and epigenetic gene regulation. The authors show that while strain induces general chromatin decompaction in epidermal progenitor cells, transcriptional repression leads to accumulation of H3K27me3 at terminal differentiation genes, preventing their expression.
- 5.Samuel MS et al. (2011) Actomyosin-mediated cellular tension drives increased tissue stiffness and β-catenin activation to induce epidermal hyperplasia and tumor growth. Cancer Cell 19, 776–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchwalter A et al. (2019) Coaching from the sidelines: the nuclear periphery in genome regulation., Nat Rev Genet 20, 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miroshnikova YA et al. (2017) Emerging roles of mechanical forces in chromatin regulation. J. Cell Sci 130, 2243–2250 [DOI] [PubMed] [Google Scholar]
- 8.Simon JA and Kingston RE Occupying Chromatin: Polycomb Mechanisms for Getting to Genomic Targets, Stopping Transcriptional Traffic, and Staying Put. Mol. Cell 49, 808–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauber KL et al. (2016) Dissecting the Roles of Polycomb Repressive Complex 2 Subunits in the Control of Skin Development. J. Invest. Dermatol 136, 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezhkova E et al. (2009) Ezh2 Orchestrates Gene Expression for the Stepwise Differentiation of Tissue-Specific Stem Cells. Cell 20, 1122–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen I et al. (2018) PRC1 Fine-tunes Gene Repression and Activation to Safeguard Skin Development and Stem Cell Specification. Cell Stem Cell 22, 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study provides a unique and comprehensive dissection of the various PRC1 complexes in a mammalian somatic stem cell system, demonstrating that PRC1 complexes control skin epithelium development and stem cell formation by regulating both silent and active genes through diverse mechanisms.
- 12.Cohen I et al. (2019) PRC1 preserves epidermal tissue integrity independently of PRC2. Genes Dev 33, 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study provides a side-by-side analysis of PRC1 and PRC2 in the developing skin epidermis, highlighting that epidermal ablation of PRC1 or PRC2 can lead to different biological outcome. Remarkably, loss of PRC1 function, but not PRC2, leads to impaired epidermal tissue integrity similar to blistering skin observed in human skin fragility syndromes.
- 13.Gao Z et al. (2012) PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Z et al. (2014) An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 516, 349–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan HL et al. (2018) Polycomb complexes associate with enhancers and promote oncogenic transcriptional programs in cancer through multiple mechanisms. Nat. Commun 9, 3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedford MT and Clarke SG (2009) Protein Arginine Methylation in Mammals: Who, What, and Why. Molecular Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao X et al. (2017) CSNK1a1 Regulates PRMT1 to Maintain the Progenitor State in Self-Renewing Somatic Tissue. Dev. Cell 43, 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon JR et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh IY and de Guzman Strong C (2017) The Molecular Revolution in Cutaneous Biology: EDC and Locus Control., J Invest Dermatol 137, e101–e104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffjan S and Stemmler S (2007) On the role of the epidermal differentiation complex in ichthyosis vulgaris, atopic dermatitis and psoriasis. Br J Dermatol 157, 441–449 [DOI] [PubMed] [Google Scholar]
- 21.Poterlowicz K et al. (2017) 5C analysis of the Epidermal Differentiation Complex locus reveals distinct chromatin interaction networks between gene-rich and gene-poor TADs in skin epithelial cells. PLoS Genet 13, e1006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rickels R and Shilatifard A (2018)Enhancer Logic and Mechanics in Development and Disease. Trends Cell Biol 28, 608–630 [DOI] [PubMed] [Google Scholar]
- 23.Pradeepa MM et al. (2016) Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet 48, 681–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubin AJ et al. (2017) Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet 49, 1522–28 [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study used promoter capture Hi-C and H3K27ac ChIP-seq methods and identified two types of enhancer-promoter interactions associated with epidermal differentiation genes. The first type of enhancer-promoter interactions, already pre-established in epidermal progenitors, is marked by H3K27ac and is regulated by cohesin. The second type of enhancer-promoter interactions increases its contact strength and gains H3K27ac upon differentiation, and is regulated by lineage transcription factors.
- 25.Noutsou M et al. (2017) The Cohesin Complex Is Necessary for Epidermal Progenitor Cell Function through Maintenance of Self-Renewal Genes. Cell Rep 20, 3005–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koster MI et al. (2004) p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev 18, 126–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong AB et al. (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20. 3185–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouwenhoven EN et al. (2015) Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep 16, 863–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao X et al. (2015) A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol 16, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin-Shiao E et al. (2018) KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev 32, 181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study uncovered that in epidermal progenitors, KMT2D, a methyltransferase that establishes an enhancer specific H3K4me1, interacts with p63 to promote enhancer activity and expression of genes associated with epidermal development and adhesion.
- 31.Mills AA et al. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–13 [DOI] [PubMed] [Google Scholar]
- 32.Pattison JM et al. (2018) Retinoic acid and BMP4 cooperate with p63 to alter chromatin dynamics during surface epithelial commitment. Nat. Genet 50, 1658–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miroshnikova YA et al. (2018) Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat. Cell Biol 20, 69–80 [DOI] [PubMed] [Google Scholar]
- 34.Mesa KR et al. (2018) Homeostatic Epidermal Stem Cell Self-Renewal Is Driven by Local Differentiation. Cell Stem Cell 23, 677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Versaevel M et al. (2012) Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun 3, 671, 1668 [DOI] [PubMed] [Google Scholar]
- 36.Almeida FV et al. (2015) The cytolinker plectin regulates nuclear mechanotransduction in keratinocytes. J. Cell Sci 128, 4475–86 [DOI] [PubMed] [Google Scholar]
- 37.Heo S-J et al. (2016) Differentiation alters stem cell nuclear architecture, mechanics, and mechano-sensitivity. Elife 5, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spagnol ST and Dahl KN (2016) Spatially resolved quantification of chromatin condensation through differential local rheology in cell nuclei fluorescence lifetime imaging. PLoS One 11, 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chalut KJ et al. (2012) Chromatin decondensation and nuclear softening accompany Nanog downregulation in embryonic stem cells. Biophys. J 103, 2060–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Botchkarev VA et al. (2012) The molecular revolution in cutanous biology: Chromosomal territories, higher-order chromatin remodeling, and the control of gene expression in keratinocytes J. Invest. Dermatol 137, e93–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckhart L et al. (2013) Cell death by cornification. Biochim Biophys Acta 1833, 3471–80 [DOI] [PubMed] [Google Scholar]
- 42.Rapisarda V et al. (2017) p63 Transcription Factor Regulates Nuclear Shape and Expression of Nuclear Envelope-Associated Genes in Epidermal Keratinocytes. J. Invest. Dermatol 137, 2157–67 [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes how p63 regulates nuclear shape and expression of structural proteins of the nuclear envelope, linking stemness with nuclear shape and mechanics.
- 43.Stephens AD et al. (2018) Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell 29, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gdula MR et al. (2013) Remodeling of three-dimensional organization of the nucleus during terminal keratinocyte differentiation in the epidermis. J. Invest. Dermatol 133, 2191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heo SJ et al. (2016) Mechanically Induced Chromatin Condensation Requires Cellular Contractility in Mesenchymal Stem Cells. Biophys. J 111, 864–874 [DOI] [PMC free article] [PubMed] [Google Scholar]


