Abstract
The complete mitochondrial genome of Polypedilum unifascium (Diptera: Chironomidae) was determined by Illumina sequencing technology. The whole mitogenome is 16,452 bp in length with an A + T bias of 79.3%, and contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), and two ribosomal RNAs (rRNAs). All PCGs start with ATN codon and use TAA as the stop codon. Gene arrangement of the 13 PCGs is identical to that of other known Chironomidae mitochondrial genomes. The resultant Bayesian inference and maximum-likelihood trees based on the sequence data of 13 PCGs support its close relationship with P. vanderplanki.
Keywords: Chironomidae, mitochondrial genome, phylogeny, Polypedilum unifascium
The aquatic chironomids, commonly known as non-biting midges are often the dominant group among freshwater benthic invertebrates. The family Chironomidae consists of diverse species which are used as biomonitors of water quality (Saether 1979). The species identification and phylogenetic analysis of chironomids mostly rely on morphologic characteristics, as well as molecular makers especially mitochondrial genomes. Nevertheless, only three valid mitochondrial genomes have been deposited in public database GenBank of NCBI. In a biodiversity survey at Lishui, China, a species of Chironomidae, Polypedilum unifascium, was found. P. unifascium was first described in Japan, and continuously recorded in Korea, China, and Russian Far East (Zhang et al. 2016; Song et al. 2017). So far, mitochondrial genome of this species has not been reported.
This article reports the complete mitochondrial genome of P. unifascium. The larvae were collected from fresh water sediment at Lishui, Zhejiang, China (27°45′16″N, 119°11′15″E) on August 2020. The specimens are deposited at the College of Life Sciences, Taizhou University (www.tzc.edu.cn, Xin Qi, qixin0612@tzc.edu.cn) under the voucher number BSZ13. Genomic DNA was extracted from the abdomen of a larva using DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) and subjected to conduct next-generation sequencing at Illumina NovaSeq 6000 platform. The raw reads were filtered by Trimmomatic version 0.39 (Bolger et al. 2014) and the clean reads were assembled by SPAdes version 3.14.1 (Bankevich et al. 2012). The assembly mitochondrial genome sequence was annotated with MITOS web server (Bernt et al. 2013) and tRNAscan-SE (Lowe and Chan 2016). Some annotations were corrected manually.
The complete mitochondrial genome of P. unifascium is 16,452 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), and two ribosomal RNAs (rRNAs). The overall base composition is 40.4% A, 38.9% T, 7.9% G, and 12.8% C. All the PCGs start with ATN codon, and use TAA as the stop codon. Gene arrangement of the 13 PCGs is identical to that of other known Chironomidae mitochondrial genomes.
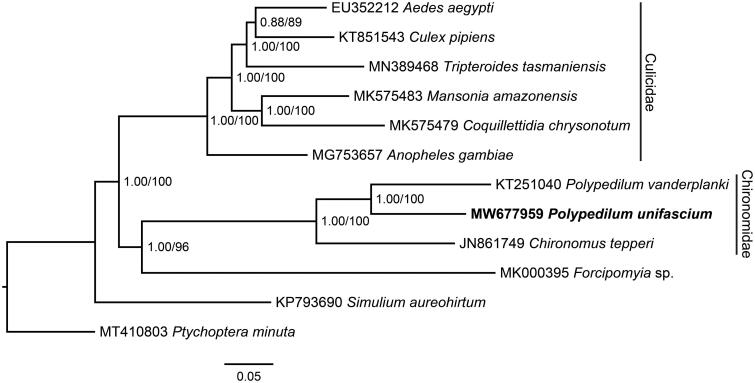
To reveal the phylogenetic position of P. unifascium, nucleotide sequences of the 13 PCGs from P. unifascium and 10 closely related Culicomorpha species, as well as an outgroup Ptychoptera minuta (Ptychopteromorpha; MT410803), were used to construct phylogenetic trees. The sequences were multiple aligned using MUSCLE (Edgar 2004) in software MEGA version X (Kumar et al. 2018), and conserved blocks were identified by Gblocks version 0.91b (Talavera and Castresana 2007). The phylogenetic relationships were reconstructed using the Bayesian inference and maximum-likelihood methods through MrBayes version 3.2.6 (Huelsenbeck and Ronquist 2001) and RAxML version VI (Stamatakis 2014). The resultant trees shared the same topologies and revealed that P. unifascium belonged to Chironomidae and was in close association with P. vanderplanki (Figure 1). This work provides molecular characterizations of P. unifascium and contributes to the phylogenetic analysis of Chironomidae.
Figure 1.
Bayesian inference and maximum-likelihood phylogenetic trees inferred from the nucleotide sequence data of mitogenomic 13 PCGs. The numbers at the nodes indicate the posterior probability values of Bayesian inference tree and the bootstrap values of maximum likelihood tree.
Funding Statement
This work was financially supported by National Natural Science Foundation of China [32070481], Zhejiang Provincial Natural Science Foundation of China [LY17C040001], and Science & Technology Project of Taizhou [1902gy23].
Disclosure statement
The authors report no conflicts of interest and are alone responsible for the content and writing of this paper.
Data availability statement
The data support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The complete mitochondrial genome of Polypedilum unifascium for this study has been deposited in GenBank with accession number MW677959. The associated BioProject, BioSample, and SRA numbers are PRJNA705522, SAMN18091136, and SRS8339075.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Chan PP.. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether OA. 1979. Chironomid communities as water quality indicators. Hol Ecol. 2(2):65–74. [Google Scholar]
- Song C, Liu W, Zhang R, Wang X.. 2017. Two gynandromorphic species of Polypedilum Kieffer, 1912 (Diptera: Chironomidae), with DNA barcodes from Oriental China. Pan-Pac Entomol. 93(2):95–104. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Zhang RL, Song C, Qi X, Wang XH.. 2016. Taxonomic review on the subgenus Tripodura Townes (Diptera: Chironomidae: Polypedilum) from China with eleven new species and a supplementary world checklist. Zootaxa. 4136(1):1–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov. The complete mitochondrial genome of Polypedilum unifascium for this study has been deposited in GenBank with accession number MW677959. The associated BioProject, BioSample, and SRA numbers are PRJNA705522, SAMN18091136, and SRS8339075.