ABSTRACT
Dengue virus (DENV) evolutionary dynamics are characterized by frequent DENV genotype/lineage replacements, potentially associated with changes in disease severity and human immunity. New Caledonia (NC) and Cambodia, two contrasted epidemiological settings, respectively experienced a DENV-1 genotype IV to I replacement in 2012 and a DENV-1 genotype I lineage 3–4 replacement in 2005–2007, both followed by a massive dengue outbreak. However, their underlying evolutionary drivers have not been elucidated. Here, we tested the hypothesis that these genotype/lineage switches reflected a higher transmission fitness of the replacing DENV genotype/lineage in the mosquito vector using in vivo competition experiments. For this purpose, field-derived Aedes aegypti from NC and Cambodia were orally challenged with epidemiologically relevant pairs of four DENV-1 genotype I and IV strains from NC or four DENV-1 genotype I lineage 3 and 4 strains from Cambodia, respectively. The relative transmission fitness of each DENV-1 genotype/lineage was measured by quantitative RT–PCR for infection, dissemination, and transmission rates. Results showed a clear transmission fitness advantage of the replacing DENV-1 genotype I from NC within the vector. A similar but more subtle pattern was observed for the DENV-1 lineage 4 replacement in Cambodia. Our results support the hypothesis that vector-driven selection contributed to the DENV-1 genotype/lineage replacements in these two contrasted epidemiological settings, and reinforce the idea that natural selection taking place within the mosquito vector plays an important role in DENV short-term evolutionary dynamics.
KEYWORDS: Dengue virus, genotype/lineage replacement, Aedes aegypti, transmission fitness, competition assay
Introduction
Dengue virus (DENV) is the most important arthropod-borne viral pathogen affecting humans worldwide [1]. Forms of dengue fever are broad, ranging from asymptomatic to life-threatening, sometimes resulting in hemorrhagic manifestations (even fatal) [2]. DENV is transmitted to humans through infected Aedes mosquitoes, primarily Aedes aegypti.
DENV is a positive-sense, single-stranded RNA virus belonging to the Flaviviridae family (Flavivirus genus). Four genetically distinct serotypes (DENV-1 to -4), sharing approximately 65% amino acid identity, can be distinguished [3]. Each serotype can be subdivided in several genotypes, displaying less than 6% nucleotide sequence divergence [4,5]. Phylogenetic analyses based on the DENV envelope gene sequence identified 5 genotypes for DENV-1, namely genotype I to V, representing of different geographic origins [4]. Moreover, each genotype can be subdivided into multiple lineages mainly based on the analysis of the whole genome sequence.
Phylogenetic analyses of DENV genetic diversity have revealed that DENV short-term evolutionary dynamics are characterized by rapid turnover of DENV genotypes and lineages [6–8]. These replacement events can lead to the extinction of the circulating DENV genotype/lineage, or its replacement by a new genotype or lineage [6,7,9–16]. Such mechanism has been documented for all DENV serotypes. Indeed, in the 1980s and 1990s, DENV-3 lineage replacements were observed in Sri Lanka and Thailand [6,14,15], whereas DENV-4 lineage replacements were identified in Puerto Rico [7]. Further, DENV-1 lineage or genotype replacements were observed in South-East Asia and in South/Latin America [6,9–11,17,18]. DENV-2 lineage or genotype replacements occurred in the South Pacific region in the 1970s, and in America and Asia during the last decades [12,13,16,19,20].
These genotype/lineage switches have important epidemiological implications. Indeed, both epidemiological observations and in vitro studies suggest that distinct genotypes have a different potential to cause severe dengue epidemics [21]. For example, the appearance of hemorrhagic forms in the Americas in 1981 was associated with the introduction of a DENV-2 Southeast Asian genotype that supplanted the native American genotype [12,22]. Furthermore, recent study indicated that protection from homologous DENV re-infection may sometimes be incomplete, although virological complementary studies are needed to be performed in order to confirm this observation [23]. In the same manner, another study showed that the immune response may be variable within the same serotype [24]. The mechanisms underlying the DENV genotype/lineage turnover, however, remain poorly understood. Some studies have suggested that these dynamics result from stochastic events [9,15]. Conversely, other studies proposed that this turnover arises from differences in viral fitness, such as higher viremia in humans [13] or higher transmissibility in mosquitoes [10,25–27]. Indeed, a significantly higher replicative index of the replacing DENV strain was observed in some DENV lineage replacements as early as initial infection establishment in Ae. aegypti. The increase of viral titre in the midgut was associated with a greater dissemination efficiency, suggesting a higher viral transmission rate and/or a shorter extrinsic incubation period [25,26]. Despite the complex epidemiological pattern of dengue, the turnover of DENV genotypes and lineages is observed in all affected regions irrespective of their epidemic, endemic or even hyper-endemic status. Here, we investigated these replacements in two epidemiologically contrasted countries: New Caledonia (NC) (epidemic) and Cambodia (hyper-endemic).
In NC, DENV circulation has been characterized by an epidemic transmission of a single dominant DENV serotype/genotype for three to five years, with subsequent replacement by another serotype/genotype. This epidemiological profile has evolved over the last fifteen years, with more frequent epidemics, co-circulation of several arboviruses and an unusual persistence of DENV-1 [28] (Supplementary Figure 1). Phylogenetic analyses using DENV-1 strains isolated between 2001 and 2017 showed that NC experienced a replacement of the genotype IV with the genotype I in 2012 [28,29]; this introduction being associated with a major outbreak in 2013 (more than 10,000 cases diagnosed), with uncommon complications. Since 2018, DENV-1 genotype I has been replaced by DENV-2 in NC [30].
In contrast, Cambodia is a dengue endemic country (10,000–60,000 annual cases). All four DENV serotypes co-circulate each year although the predominant serotype has alternated mainly between DENV-1, DENV-2, and DENV-3 over the last decades [31] (Supplementary Figure 2). DENV-1 co-circulated with other serotypes as a minor serotype between 2000 and 2009 [11,32], and strains isolated over this period belong to genotype I grouped into four lineages (L1 to L4) with specific dynamics [11]. Indeed, three lineage replacements were observed: in 2000–2003 L2 displaced L1; in 2002–2003 L3 displaced L2; in 2005–2007, L4 replaced L3 [11]. In 2011, DENV-1 was detected throughout the country and resulted in a high magnitude outbreak in 2012 [33]. Unlike DENV-3, which was sporadically detected after having caused a major outbreak in 2007, DENV-1 continued to circulate as a dominant serotype until 2015.
Interestingly, these genotype/lineage replacements were associated with periods of low DENV-1 circulation in both Cambodia and NC [11,29,33]. Periods of low virus transmission could favour the emergence of previously rare variants randomly replacing dominant ones, resulting in a ladder-like tree topology [7,34–36]. The genotype/lineage replacements observed in both countries are inconsistent with this scenario because a newly introduced genotype/lineage replaced the resident one after a period of co-circulation [11,29]. They are more consistent with an adaptive replacement reflecting the higher fitness of the invasive genotype/lineage relative to the resident one. However, the selective forces involved have not been identified yet. Here, we investigated whether the DENV genotype/lineage replacement events observed in these two epidemiologically contrasted settings could reflect viral fitness differences in the mosquito vector.
Materials and methods
Viruses
Thirty-three epidemic DENV-1 strains were selected in NC from 2009 to 2017 and 21 DENV-1 strains were selected in Cambodia from 2005 to 2016. Viruses were isolated from human serum by three successive passages in Aedes albopictus (C6/36) cells, maintained in Leibovitz's L-15 medium (Sigma-Aldrich, Saint Louis, MO, USA) supplemented with 5% heat-inactivated fetal bovine serum (FBS, Gibco, ThermoFisher Scientific, Paisley, UK) and 10% tryptose phosphate broth (Gibco, ThermoFisher Scientific, Paisley, UK). After five days of incubation at 28°C, supernatants were collected. For each DENV-1 strain selected for competition assays, viral titre was determined by immuno-fluorescent focus assay [37] using the anti-DENV complex antibody, clone D3-2H2-9-21 (Millipore, Temecula, CA, USA), and was expressed as focus-forming unit per milliliter (FFU/mL). If necessary, two concentration methods were employed to reach the appropriate viral titre. Viruses were either centrifuged using Vivaspin 6 centrifugal concentrator (Sartorius, Stonehouse, UK) or added with Polyethylene Glycol (PEG, Sigma-Aldrich, Germany) and centrifuged at 1800 rpm for 1 hour.
Whole-genome sequencing and phylogenetic analysis
Whole-genomes of the 54 DENV-1 strains selected in this study were obtained by high-throughput sequencing of cell-culture supernatants [38]. The resulting whole-genomes were deposited in GenBank (Supplementary Table 1). Additionally, five whole-genome sequences from Cambodia already available in GenBank (FJ639684, FJ639687, HM181944, FJ639685, and GU131893) were included in the analysis. Phylogenetic analysis of DENV-1 was conducted on whole-genome sequences from NC and Cambodia with reference sequences from GenBank representing the five genotypes of DENV-1. Alignment was done using the MAFFT software [39]. The best substitution model for the alignment sequence was determined in W-IQ-Tree tool using the Model selection option, resulting in the GTR + G4 + I as the best model. The whole-genome phylogenetic tree was constructed by the Maximum Likelihood method based on the best substitution model with 1.000 bootstrap replicates in W-IQ-Tree tool [40]. DENV-3 (AY858048) was used as the out-group.
Quantitative RT–PCR
Viral RNA from all samples (body, head/legs/wings, and saliva) was extracted with the QIAamp Viral RNA mini kit (Qiagen, Hilden, Germany). The detection and quantification of DENV-1 genotypes I and IV (DENV-1 GI and GIV) and DENV-1, genotype I, lineages 3 and 4 (DENV-1 L3 and L4) was performed by specific TaqMan quantitative RT–PCR assays with the Superscript III Platinum One-Step RT-qPCR kit (ThermoFisher Scientific, Carlsbad, CA, USA). Genotype or lineage-specific primers and probes, respectively targeting the NS2B or E gene for New Caledonian or Cambodian strains, were used (Supplementary Table 2). Amplifications were performed on either a LightCycler 480 instrument II (Roche, Basel, Switzerland) or a Bio-Rad CFX96 Real-Time Detection System (Bio-Rad, Hercules, CA, USA) using the following programme: one cycle (50°C, 15 min; 95°C, 2 min) followed by 45 cycles (95°C, 15 s; 57°C, 1 min).
Mosquitoes
Ae. aegypti larvae and pupae were collected at the end of the hot season (May 2018 in NC and April 2019 in Cambodia) and reared to the adult stage in laboratory. Adults were then maintained with access to 10% sucrose solution ad libitum at 28°C ± 1°C, 80% ± 10% relative humidity and 12:12 h light:dark cycle. F1 and F2 generations were successively produced by sib-mating and collective oviposition following blood feeding.
In vivo competition assay
Two DENV-1 strains representatives of each genotype (GI and GIV) or lineage (L3 and L4) were selected from the phylogenetic tree generated (Figure 1) to measure their relative fitness by competition assays in mosquitoes. Five- to seven-days-old nulliparous Ae. aegypti F2 females were challenged with infectious blood meals at a final concentration of 5.106 FFU/mL with two initial FFU ratios (DENV-1 GI/GIV relative percentage: 50/50 and 10/90, or DENV-1 L3/L4: 50/50) (Supplementary Table 3 and Supplementary Figure 3). For each competition Ae. aegypti from NC and Cambodia were orally challenged with the selected DENV-1 strains from NC and Cambodia respectively as previously described [37]. At 7 and 14 days post challenge (pc), 30 Ae. aegypti mosquitoes randomly collected were subjected to a forced salivation. Briefly, after removing wings and legs, their proboscis was inserted into a 20-µL filter tip filled with 5 µL FBS for 30 minutes. The mixture was subsequently supplemented with 45 µL of Leibovitz's L-15 medium and stored at −80°C. After decapitation, the body and head of each mosquito were mechanically homogenized for 2 minutes at 2000 rpm using a mini-BeadBeater 96 (BioSpecProducts, Bartlesville, OK, USA) with metal beads in 200 µL of Leibovitz medium supplemented with 2% FBS, 10% tryptose phosphate broth and antibiotics/antifungals (100 units/mL of penicillin, 0.1 mg/mL of streptomycin and 0.25 µg/mL amphotericin B, Gibco, ThermoFisher Scientific, Carlsbad, CA, USA) in NC. In Cambodia, the body and head/legs/wings were stored in 400 µL phosphate buffered saline (PBS) supplemented with 10% FBS, 1% Penicillin/Streptomycin, 1% Amphotericin B (GIBCO, Waltham MA, USA) until homogenization. The homogenization was performed using a MagNA Lyser (Roche, Basel, Switzerland) with ceramic beads at 6,500 rpm for 50 s. The lysates were then centrifuged at 10,000 g for 5 minutes and stored at −80°C. Detection of viral RNA from body, head and saliva for each competition assay was performed by two specific quantitative RT–PCR experiments as described above. The viral infection profile for the three vector competence indices (infection, dissemination and transmission rates) was determined as follows: (i) the infection rate is the number of mosquitoes displaying a DENV positive body among all tested mosquitoes, (ii) the dissemination rate is the number of mosquitoes with an infected head divided by the number of mosquitoes having a DENV positive body, and (iii) the transmission rate is the number of mosquitoes with a DENV positive saliva divided by the number of mosquitoes with an infected head (Supplementary Figure 3).
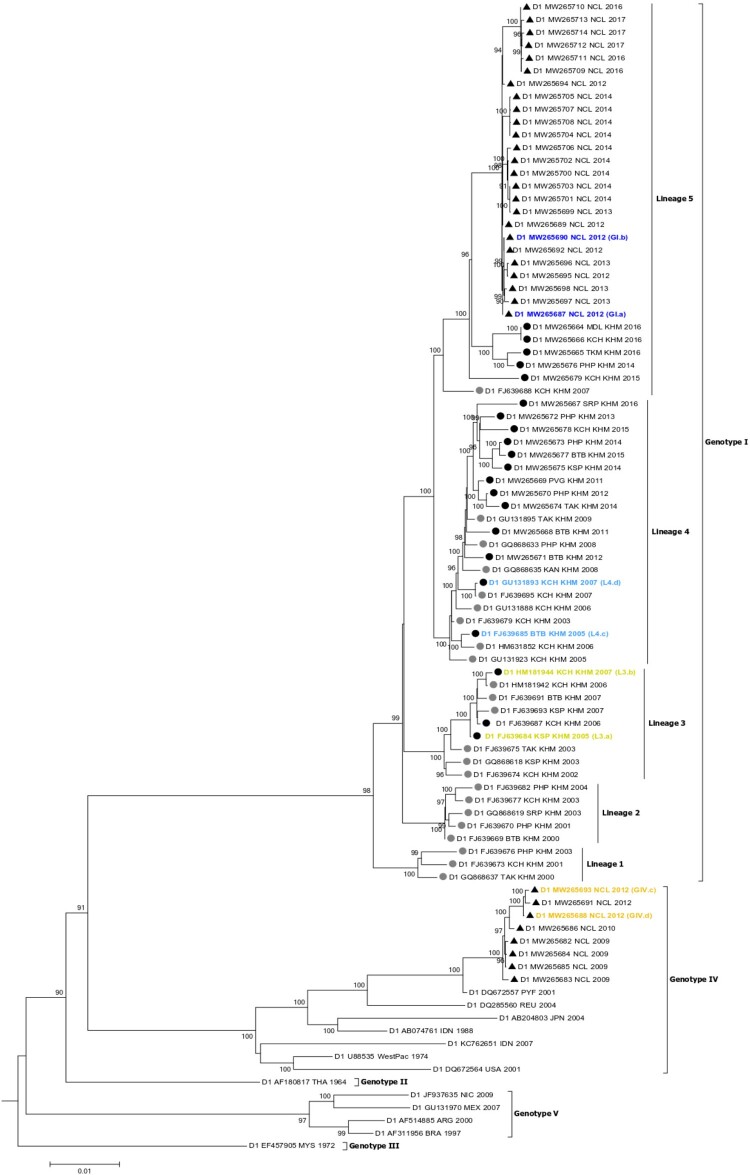
Figure 1.
Phylogenetic tree of whole genomes of DENV-1 strains. Cambodia: strains included in our study (black circle); in previous studies (grey circle). Strains from NC: black triangle. Two DENV-1 strains representatives of each genotype (GIV in orange; GI in blue) from NC or lineage (L3 in yellow; L4 in sky-blue belonging to genotype I) from Cambodia were selected for in vivo competition assays.
Statistical analysis
Competition assays were analyzed statistically by detecting each genotype/lineage in individual mosquitoes and comparing their relative frequency for infection, dissemination, and transmission indices. The 2 × 2 contingency tables of presence/absence for both competing viruses were compared by McNemar's chi-squared test for paired nominal data. All statistical analyses and data plotting of competition assays were performed with R v3.6.1 software [41], considering p-values <0.05 as statistically significant.
Ethics statement
In NC, the use of DENV strains isolated from the serum of anonymized patients was granted by the Consultative Ethics Committee of New Caledonia 26.11.2019 and by the Comité de Protection des Personnes Sud-Est II (N° Eudract 2019-A03114-53). In Cambodia, the use of DENV strains obtained from patients enrolled in the National Program for Dengue Surveillance by Ministry of Health of Cambodia was approved by National Ethics Committee for Health Research (No 264NECHR).
Results
Phylogenetic analysis
Fifty-four whole genomes of DENV-1 strains from NC and Cambodia clustered within two of the five DENV-1 genotypes with 85% (45/54) within genotype I (Figure 1). Five lineages were observed inside the genotype I. The 21 Cambodian strains from this study belonged to genotype I. They fell into three (L3, L4 and L5) of the five distinct lineages among other Cambodian strains reported previously. DENV-1 strains in lineage L3 circulated between 2002 and 2007, L4 between 2003 and 2016 and L5 between 2007 and 2016. Twenty-five New Caledonian strains collected from 2012 to 2017 clustered in lineage L5 along with five Cambodian strains collected in 2014, 2015 and 2016 (Figure 1). The last eight strains from NC collected between 2009 and 2012, fell into genotype IV, and are closely related to a virus sampled in French Polynesia in 2001.
Phylogenetic analyses indicated that the last lineage replacement occurred in Cambodia between 2005 and 2007 with an extinction of DENV-1 lineage L3 and an expansion of L4. Lineage L4 then circulated until 2016 and co-circulated with lineage L5 since 2015. However, no extinction of lineage L4 was observed during the period (Figure 1). Among the 21 Cambodian strains, five belong to this co-circulation period with 3 assigned to lineage L3 and 2 to lineage L4. Genotype replacement also occurred in NC in 2012 with a co-circulation of both genotypes during this period, as observed previously [29]. Among the 33 NC DENV strains collected for this study, nine belong to this co-circulation period with three strains falling into genotype IV and six into genotype I. For in vivo competition assays, two strains of each genotype from NC (GI and GIV) or lineage from Cambodia (L3 and L4) isolated during these periods of co-circulation (2012 for NC and 2005–2007 for Cambodia) that favoured the expansion of the newly introduced genotype (GI for NC) or lineage (L4 for Cambodia) were selected (Figure 1).
DENV-1 genotypes (GI and GIV) competition assays
Co-infection of Ae. aegypti mosquitoes with DENV-1 GI and GIV from NC were performed using two strains of each DENV-1 genotype (GI.a, GI.b, GIV.c, GIV.d). DENV-1 was detected in mosquitoes either as mono-infection of GI or GIV or as co-infection with GI and GIV from 7 days pc, regardless of the initial ratio or viral combination (Figures 2 and 3; Supplementary Figure 3).
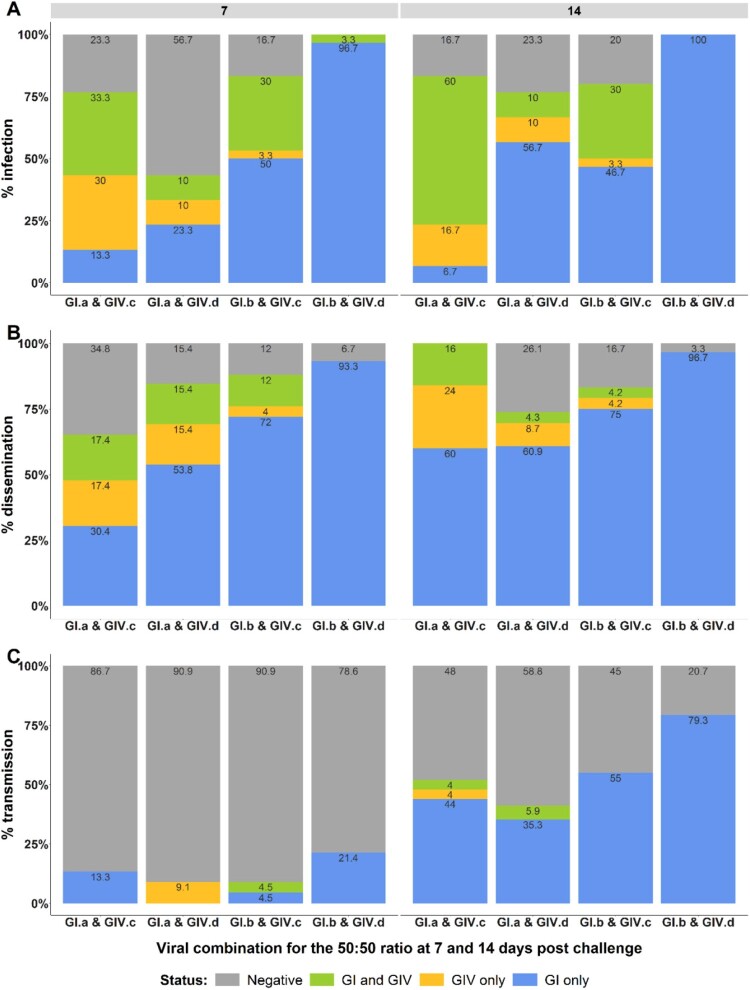
Figure 2.
Infection, dissemination and transmission rates of New Caledonian Ae. aegypti mosquitoes observed with the 50:50 infectious ratio. Infection rates (A), dissemination rates (B) and transmission rates (C) are represented. Status GI/GIV, GI or GIV corresponds to the detection by RT-qPCR of both targets, only GI or only GIV respectively in each mosquito compartment (body, head and saliva). No amplification detected for negative status.
Figure 3.
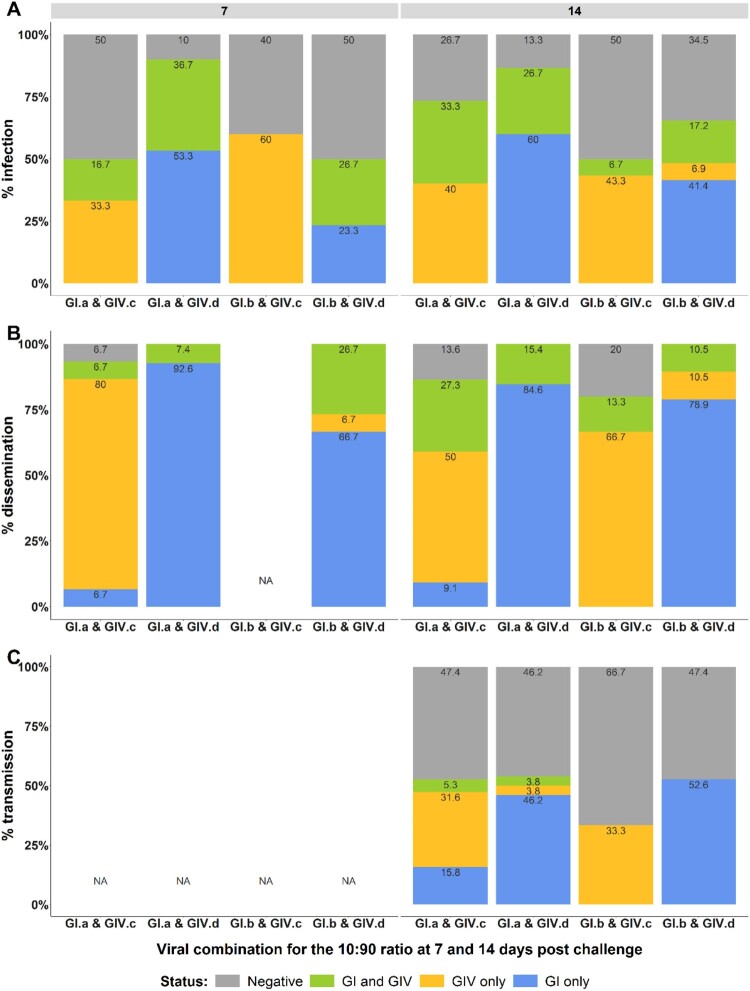
Infection, dissemination and transmission rates of New Caledonian Ae. aegypti mosquitoes measured with the 10:90 infectious ratio. Infection rates (A) dissemination rates (B) and transmission rates (C) are represented. Status GI and GIV, GI or GIV corresponds to the detection by RT-qPCR of both targets, only GI or only GIV respectively in each mosquito compartment (body, head and saliva). No amplification detected for negative status. NA: not analyzed.
With the 50:50 infectious ratio, infection rates ranged from 43.3% to 100% and from 76.7% to 100% at 7 and 14 days pc, respectively. A significant predominance of DENV-1 GI relative to DENV-1 GIV was observed at 7 days pc for the two viral combinations with the GI.b isolate (p-values = 0.001 and <0.001, for GI.b & GIV.c and GI.b & GIV.d combinations, respectively). At 14 days pc, a third viral combination also showed a significantly higher infection with DENV-1 genotype I (p-values = 0.004, =0.002 and =0.002, for GI.a & GIV.d, GI.b & GIV.c and GI.b & GIV.d, respectively) (Figure 2A). The same profile was observed for dissemination rates, with a significantly higher dissemination of DENV-1 GI for two and three out of the four viral combinations at 7 and 14 days pc, respectively (p-values <0.001 at 7 days pc for GI.b & GIV.c and GI.b & GIV.d, and =0.01, <0.001, <0.001 at 14 days pc for GI.a & GIV.d, GI.b & GIV.c and GI.b & GIV.d respectively) (Figure 2B). Viral transmission was detected at 7 and 14 days pc, ranging from 9% to 21.4% and 41.2% to 79.3%, respectively (Figure 2C). At 7 days pc, only the viral combination GI.b & GIV.d showed a transmission advantage for DENV-1 GI compared to DENV-1 GIV (p-values = 0.04). The DENV-1 GI was significantly better transmitted compared to DENV-1 GIV at 14 days pc for the four combinations (p-values = 0.01, =0.04, =0.003 and =0.003 for GI.a & GIV.c, GI.a & GIV.d, GI.b & GIV.c and GI.b & GIV.d, respectively).
With the 10:90 infectious ratio in favour of DENV-1 GIV, results were more heterogeneous. Infection rates ranged from 50% to 90% irrespective of the day pc. A significant predominance of DENV-1 GIV infection compared to DENV-1 GI was observed for the two viral combinations containing the GIV.c isolate regardless of the day pc (p-values = 0.004, <0.001 and =0.001, =0.001 for GI.a & GIV.c and GI.b & GIV.c at 7 and 14 days pc, respectively). Furthermore, no DENV-1 GI only infection was detected in these two viral combinations at 7 and 14 days pc. Conversely, the two viral combinations containing the GIV.d isolate showed a significant higher infection with DENV-1 GI irrespective of the GI isolate and the day post challenge (p-values = <0.001, =0.02 and <0.001, =0.001 for GI.a & GIV.d and GI.b & GIV.d at 7 and 14 days post challenge respectively) (Figure 3A). Although DENV-1 GIV was in excess in these two viral combinations, no DENV-1 GIV only infected mosquitoes were detected, except at 14 days pc for GI.b & GIV.d. Dissemination rates ranged from 93.3% to 100% and 80% to 100% at 7 and 14 days pc, respectively. The same profile as for the infection rates was observed for all viral combinations. Heads from the combination GI.b & GIV.c at 7 days pc were not analyzed, since only DENV-1 GIV was detected in infected mosquitoes (bodies). Interestingly, for the GI.a & GIV.c viral combination, some infected mosquito heads were only positive for DENV-1 GI despite the significant predominance of DENV-1 GIV (Figure 3B). Viral transmission was assessed at 14 days pc for the four viral combinations. Transmission rates were close to 50%, except for GI.b & GIV.c, which was 33%. DENV-1 GI was significantly better transmitted compared to DENV-1 GIV for only one out the four viral combinations (p-value = 0.01 for GI.a & GIV.d). Transmission of DENV-1 GI, however, was also detected in two other viral combinations although no significant difference was detected. No DENV-1 GI transmission was observed for GI.b & GIV.c (Figure 3C).
DENV-1 genotype I lineages (L3 and L4) competition assays
Cambodian Ae. aegypti mosquitoes were infected with a mixture of DENV-1 L3 (L3.a, L3.b) and L4 (L4.c, L4.d) at 50:50 ratio. All the four viral combinations showed an infection in mosquitoes from 7 days pc whatever the initial ratio or viral combinations (Figure 4; Supplementary Figure 3). DENV-1 was detected in mosquitoes either as mono-infection of L3 or L4 or as co-infection with L3 and L4.
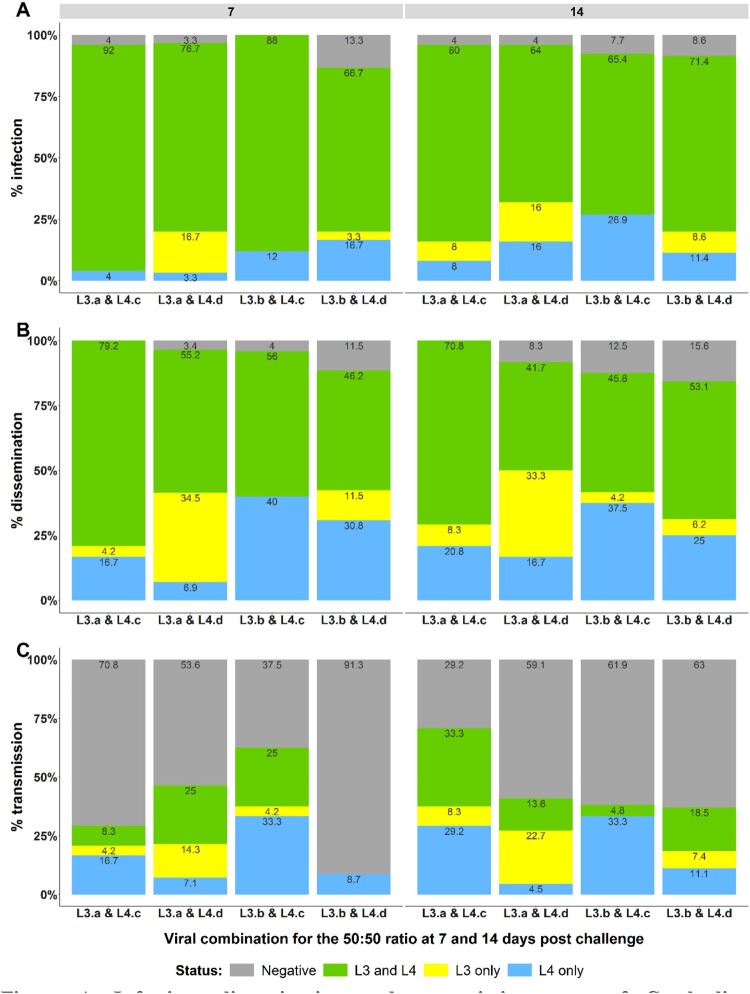
Figure 4.
Infection, dissemination and transmission rates of Cambodian Ae. aegypti mosquitoes detected with the 50:50 infectious ratio. Infection rates (A), dissemination rates (B) and transmission rates (C) are represented. Status L3 and L4, L3 or L4 corresponds to the detection by RT-qPCR of both targets, only L3 or only L4 respectively in each mosquito compartment (body, head and saliva). No amplification detected for negative status.
The infection rates were high (ranging from 86.7% to 100%) irrespective of the pc. Although DENV-1 L4 strains seemed to be mostly detected at higher percentage compared to L3 in mosquitoes at 7 days pc and at 14 days pc, the difference was significant only for combination L3.b & L4.c at 14 days pc (p-value = 0.023) (Figure 4A). The dissemination profile was similar to that of infection rate with a tendency to a greater dissemination of DENV-1 L4 in three out of the four viral combinations at 7 and 14 days pc. However, the percentage of mosquitoes infected with L4 was significantly higher than that of L3 only for the combination L3.b and L4.c (p-values = 0.004 and =0.026 at 7 and 14 days pc, respectively) (Figure 4B). Likewise, the transmission rate of DENV-1 L4 was significantly higher than that of L3 for the combination L3.b and L4.c at 7 and 14 days pc (p-values = 0.045 and =0.023, respectively) (Figure 4C).
Discussion
A typical feature of DENV epidemiology is the fluctuation in dengue incidence and serotype prevalence. In addition, rapid turnover of DENV genotypes and lineages is frequently observed in DENV evolutionary dynamics. NC and Cambodia experienced such replacements [11,29]. Detailed phylogenetic analyses performed on DENV-1 strains isolated in both countries between 2005 and 2017 confirmed that NC experienced a genotype replacement in 2012: DENV-1 GI rapidly displaced DENV-1 GIV [29]. All DENV-1 GI from NC belong to lineage 5 along with Cambodian strains isolated between 2014 and 2016. Our results also confirmed that Cambodia experienced lineages replacements in 2002–2003 (L2 displaced L1) and 2005–2007 (L4 displaced L3) [11]. Whereas Cambodia has experienced frequent DENV lineage or genotype replacements [11], the DENV-1 genotype replacement observed in 2012 is the first described in NC [28].
To date, few studies have examined the evolutionary mechanisms driving DENV genotype/lineage replacement events, and specifically focusing on the potential role of natural selection. A previous study did not detect a significant difference in infection, dissemination and transmission rates between DENV-1 GI or DENV-1 GIV in Ae. aegypti from NC orally challenged by the two genotypes separately [37]. In competition assays, however, a fitness advantage of DENV-4 on DENV-1 was shown in Ae. aegypti while no difference was observed between the corresponding mono-infections, highlighting the value of assessing viral fitness in mixed infection experiments [42]. More generally, competition assays are the gold standard in viral fitness studies because they eliminate host-to-host variation that can reduce the power of experiments with individual viral strain infections, and because the viral ratio can be assayed with more accuracy than individual virus titres [43–45]. In this study, we experimentally tested the hypothesis that genotype/lineage replacement events observed in NC and Cambodia were associated with enhanced transmissibility of the replacing genotype/lineage relative to the resident ones. Our assessment relied on competition experiments in mosquitoes in vivo, in which we monitored the relative proportions of the competing DENV genotype/lineage to determine their relative transmission fitness.
The results obtained in this study support the hypothesis that DENV-1 genotype or lineage replacement observed in NC and Cambodia have been driven, at least in part, by viral fitness differences in the vector. Indeed, a markedly higher transmission fitness was observed for DENV-1 GI relative to GIV in NC. Provided at equal titre, three out of the four viral combinations showed a competitive advantage for DENV-1 GI for the infection and dissemination rates at 14 days pc. Moreover, all four viral combinations showed a higher fitness of DENV-1 GI at 14 days pc for the ultimate stage of vector competence: the transmission rate. Interestingly, in mixed infection with 10-fold less amount of DENV-1 GI, a higher transmission fitness of DENV-1 GI relative to GIV was observed. This higher transmission fitness could result from a better replication fitness from the invasive DENV genotype at the early stage of mosquito infection. Although the difference is more subtle between DENV-1 lineage L3 and L4, a slightly higher transmission fitness for DENV-1 L4 over L3 was observed in the same manner in Cambodia. Our results are in accordance with a previous study showing that a DENV-1 clade replacement in Thailand was associated with enhanced mosquito transmission [10]. Further, a previous study showed that the lineage replacement of the DENV-2 Asian-American genotype in Nicaragua was associated with a higher replicative index of the replacing NI-2B lineage over NI-1 lineage in Ae. aegypti mosquitoes orally infected with both lineages as soon as 3 days post-infectious blood-meal [25]. A difference in the production of subgenomic flaviviral RNA fragments (sfRNA), that inhibit the interferon expression or disrupt the mosquito immunity, may explain the higher transmission fitness. Indeed, DENV lineage replacements in Puerto-Rico and Nicaragua were recently associated with increased production of sfRNAs by the invasive lineage in the human host [46]. Likewise, another study focusing on DENV lineage replacement in Puerto-Rico showed a link between higher epidemiological fitness and increased production of sfRNAs in the vector [47]. Thus, this viral factor seems to play an important role in the epidemiological fitness of DENV [48,49].
Further studies are needed to complete our results and identify the mechanisms underlying the observed fitness differences. Indeed, we demonstrated a better transmission fitness of the invasive DENV-1 genotype I in the natural course of infection in the vector. All the results, however, were expressed as RNA copies per mosquito, which do not directly reflect the concentration of infectious viruses. DENV are known to produce a large amount of viral defective genomes that could confound the results if their relative amount differed between DENV genotypes/lineages. DENV epidemiologic fitness, defined as the capacity of a DENV strain to become dominant in the field, relative to other DENV strains, depends on the combination of replicative fitness and transmission fitness [50]. Our experimental design does not allow us to investigate the replicative fitness strictly speaking as we did not measure the capacity of the DENV used in this study to produce infectious progeny in the vector. Moreover, in Cambodia, we cannot exclude the non-mutually exclusive alternative hypothesis that the lineage replacement could also result from a stochastic event due to genetic drift [9,51]. In a same way, a purifying selection or vector genetic factors may also have contributed to genotype/lineage replacement in both countries [35,51,52]. Furthermore, impact of population herd-immunity were not assessed in this study although it may have partly conditioned this genotype replacement observed in NC. Indeed recent analyses suggest that antigenic heterogeneity may exist within each DENV serotype [24]. Another study demonstrated an association between higher lineage replicative fitness and reduced antigenicity, weak B and T cell stimulation and weak host immune system interactions [53]. Finally, selective forces occurring in the human host may also be important drivers shaping the DENV epidemiologic fitness. Indeed, higher transmission fitness of the invasive genotype observed within the vector may not be preserved in the human host [54]. In NC, we can suppose, however, that the selection of DENV-1 GI in the mosquito, in relation with the enhanced mosquito transmission potential, had an impact on transmission with a higher probability of human-to-mosquito transmission as described previously [13]. Indeed, a major outbreak occurred in 2013 following this genotype replacement, and the circulation of DENV-1 GI was maintained until 2017 [28].
Although other selective and stochastic forces may also drive DENV evolution, our study provides evidence that vector-driven selection may have contributed to the DENV-1 genotype/lineage replacement that occurred in NC and to a smaller extent, in Cambodia. To our knowledge, few studies have assessed the transmission fitness of epidemiologically relevant DENV strains by their native vector. We demonstrated the higher transmissibility of the invading DENV strain using competition experiments that included analyzes of mosquito saliva, representing the ultimate step of DENV transmission from the mosquito to the human host. At the beginning of the vaccine era, our work highlights the needs for a better understanding of the evolutionary mechanisms driving DENV genotype/lineage replacements. These data are crucial as they may impact vaccine strategies by the complexity of their antigenic properties.
Author contributions
Conceptualization: OOC, VD, LL, MDR; Formal analysis: OOC, TOP; Funding Acquisition MDR; Investigation OOC, TPO, FA, SD, SR, DG, SI, MM, SL; Resources SB, PD, NP, VBS; Writing – Original Draft Preparation: OOC; Writing – Review & Editing: OOC, TPO, VD, LL, MDR. All authors carefully revised the manuscript.
Supplementary Material
Acknowledgments
We gratefully thank the Clinical Research Department of the Center for Translational Research in Paris for its support in ethics procedures. We thank Laurent Wantiez and Katie Anders for their helpful discussion on statistical approaches. We also thank Louis Cognet for his contribution to samples handling.
Funding Statement
This work was funded by the incentive grant, Inter-Pasteurian Concerted Actions (ACIP-06-2016). LL and FA were supported by the European Union’s Horizon 2020 research and innovation programme under ZikaPLAN grant agreement no. 734584 and the French Government’s Investissement d’Avenir program Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant ANR-10-LABX-62-IBEID).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Bhatt S, Gething PW, Brady OJ, et al. . The global distribution and burden of dengue. Nature. 2013 Apr 25;496(7446):504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control: New Edition. Geneva: © World Health Organization.; 2009. eng. [PubMed]
- 3.Guzman MG, Halstead SB, Artsob H, et al. . Dengue: a continuing global threat. Nat Rev Microbiol. 2010 Dec;8(12 Suppl):S7–S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weaver SC, Vasilakis N.. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol. 2009 Jul;9(4):523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes EC, Twiddy SS.. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol. 2003 May;3(1):19–28. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Mammen MP Jr, Chinnawirotpisan P, et al. . Clade replacements in dengue virus serotypes 1 and 3 are associated with changing serotype prevalence. J Virol. 2005 Dec;79(24):15123–15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett SN, Holmes EC, Chirivella M, et al. . Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003 Oct;20(10):1650–1658. [DOI] [PubMed] [Google Scholar]
- 8.Ramos-Castañeda J, Barreto Dos Santos F, Martínez-Vega R, et al. . Dengue in Latin America: systematic review of molecular epidemiological trends. PLoS Negl Trop Dis. 2017 Jan;11(1):e0005224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myat Thu H, Lowry K, Jiang L, et al. . Lineage extinction and replacement in dengue type 1 virus populations are due to stochastic events rather than to natural selection. Virology. 2005 Jun 5;336(2):163–172. [DOI] [PubMed] [Google Scholar]
- 10.Lambrechts L, Fansiri T, Pongsiri A, et al. . Dengue-1 virus clade replacement in Thailand associated with enhanced mosquito transmission. J Virol. 2012 Feb;86(3):1853–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duong V, Simmons C, Gavotte L, et al. . Genetic diversity and lineage dynamic of dengue virus serotype 1 (DENV-1) in Cambodia. Infect Genet Evol. 2013 Apr;15:59–68. [DOI] [PubMed] [Google Scholar]
- 12.Rico-Hesse R, Harrison LM, Salas RA, et al. . Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997 Apr 14;230(2):244–251. [DOI] [PubMed] [Google Scholar]
- 13.Vu TT, Holmes EC, Duong V, et al. . Emergence of the Asian 1 genotype of dengue virus serotype 2 in viet nam: in vivo fitness advantage and lineage replacement in South-East Asia. PLoS Negl Trop Dis. 2010 Jul 20;4(7):e757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messer WB, Gubler DJ, Harris E, et al. . Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003 Jul;9(7):800–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wittke V, Robb TE, Thu HM, et al. . Extinction and rapid emergence of strains of dengue 3 virus during an interepidemic period. Virology. 2002 Sep 15;301(1):148–156. [DOI] [PubMed] [Google Scholar]
- 16.Steel A, Gubler DJ, Bennett SN.. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a retrospective phylogenetic study of events in the South Pacific three decades ago. Virology. 2010 Sep 30;405(2):505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salvo MA, Aliota MT, Moncla LH, et al. . Tracking dengue virus type 1 genetic diversity during lineage replacement in an hyperendemic area in Colombia. PLoS One. 2019;14(3):e0212947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrillo-Valenzo E, Danis-Lozano R, Velasco-Hernández JX, et al. . Evolution of dengue virus in Mexico is characterized by frequent lineage replacement. Arch Virol. 2010 Sep;155(9):1401–1412. [DOI] [PubMed] [Google Scholar]
- 19.de Jesus JG, Dutra KR, Sales F, et al. . Genomic detection of a virus lineage replacement event of dengue virus serotype 2 in Brazil, 2019. Mem Inst Oswaldo Cruz. 2020;115:e190423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, Phadungsombat J, Nakayama EE, et al. . Genotype replacement of dengue virus type 3 and clade replacement of dengue virus type 2 genotype Cosmopolitan in Dhaka, Bangladesh in 2017. Infect Genet Evol. 2019 Nov;75:103977. [DOI] [PubMed] [Google Scholar]
- 21.Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubler DJ, Clark GG.. Dengue/dengue hemorrhagic fever: the emergence of a global health problem. Emerg Infect Dis. 1995 Apr-Jun;1(2):55–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forshey BM, Reiner RC, Olkowski S, et al. . Incomplete protection against dengue virus type 2 re-infection in Peru. PLoS Negl Trop Dis. 2016 Feb;10(2):e0004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katzelnick LC, Fonville JM, Gromowski GD, et al. . Dengue viruses cluster antigenically but not as discrete serotypes. Science. 2015 Sep 18;349(6254):1338–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quiner CA, Parameswaran P, Ciota AT, et al. . Increased replicative fitness of a dengue virus 2 clade in native mosquitoes: potential contribution to a clade replacement event in Nicaragua. J Virol. 2014 Nov;88(22):13125–13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanley KA, Nelson JT, Schirtzinger EE, et al. . Superior infectivity for mosquito vectors contributes to competitive displacement among strains of dengue virus. BMC Ecol. 2008 Feb 13;8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson JR, Rico-Hesse R.. Aedes aegypti vectorial capacity is determined by the infecting genotype of dengue virus. Am J Trop Med Hyg. 2006 Nov;75(5):886–892. [PMC free article] [PubMed] [Google Scholar]
- 28.Inizan C, Tarantola A, O’Connor O, et al. . Dengue in New Caledonia: knowledge and gaps. Trop Med Infect Dis. 2019 Jun 20;4(2):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupont-Rouzeyrol M, Aubry M, O’Connor O, et al. . Epidemiological and molecular features of dengue virus type-1 in New Caledonia, South Pacific, 2001–2013. Virol J. 2014 Mar 31;11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inizan C, O’Connor O, Worwor G, et al. . Molecular characterization of dengue type 2 outbreak in Pacific Islands countries and territories, 2017–2020. Viruses. 2020 Sep 25;12(10):1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huy R, Buchy P, Conan A, et al. . National dengue surveillance in Cambodia 1980–2008: epidemiological and virological trends and the impact of vector control. Bull World Health Organ. 2010 Sep 1;88(9):650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ASEAN e-Health Bulletin . The ASEAN dengue day: sustaining the united fight against dengue 2017; Special edition for ASEAN Dengue Day 2017: Available from: https://reliefweb.int/sites/reliefweb.int/files/resources/06-e-Health-Bulletin-11th-Issue_ASEAN-Website.pdf.
- 33.Cousien A, Ledien J, Souv K, et al. . Predicting dengue outbreaks in Cambodia. Emerg Infect Dis. 2019 Dec;25(12):2281–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Descloux E, Cao-Lormeau VM, Roche C, et al. . Dengue 1 diversity and microevolution, French Polynesia 2001–2006: connection with epidemiology and clinics. PLoS Negl Trop Dis. 2009 Aug 4;3(8):e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klungthong C, Zhang C, Mammen MP Jr, et al. . The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology. 2004 Nov 10;329(1):168–179. [DOI] [PubMed] [Google Scholar]
- 36.Lourenço J, Recker M.. Viral and epidemiological determinants of the invasion dynamics of novel dengue genotypes. PLoS Negl Trop Dis. 2010 Nov 23;4(11):e894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Connor O, Calvez E, Inizan C, et al. . Vector competence of Aedes aegypti from New Caledonia for the four recent circulating dengue virus serotypes. PLoS Negl Trop Dis. 2020 May;14(5):e0008303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fontaine A, Lequime S, Moltini-Conclois I, et al. . Epidemiological significance of dengue virus genetic variation in mosquito infection dynamics. PLoS Pathog. 2018 Jul;14(7):e1007187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Rozewicki J, Yamada KD.. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019 Jul 19;20(4):1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen LT, Schmidt HA, von Haeseler A, et al. . IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015 Jan;32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Foundation for Statistical Computing V, Austria. R Core Team. R: A language and environment for statistical computing. 2016.
- 42.Vazeille M, Gaborit P, Mousson L, et al. . Competitive advantage of a dengue 4 virus when co-infecting the mosquito Aedes aegypti with a dengue 1 virus. BMC Infect Dis. 2016 Jul 8;16:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Liu Y, Shan C, et al. . Role of mutational reversions and fitness restoration in Zika virus spread to the Americas. Nat Commun. 2021 Jan 26;12(1):595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grubaugh ND, Weger-Lucarelli J, Murrieta RA, et al. . Genetic drift during systemic arbovirus infection of mosquito vectors leads to decreased relative fitness during host switching. Cell Host Microbe. 2016 Apr 13;19(4):481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergren NA, Haller S, Rossi SL, et al. . “Submergence” of western equine encephalitis virus: evidence of positive selection argues against genetic drift and fitness reductions. PLoS Pathog. 2020 Feb;16(2):e1008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manokaran G, Finol E, Wang C, et al. . Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science. 2015 Oct 9;350(6257):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pompon J, Manuel M, Ng GK, et al. . Dengue subgenomic flaviviral RNA disrupts immunity in mosquito salivary glands to increase virus transmission. PLoS Pathog. 2017 Jul;13(7):e1006535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finol E, Ooi EE.. Evolution of subgenomic RNA shapes dengue virus adaptation and epidemiological fitness. iScience. 2019 Jun 28;16:94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syenina A, Vijaykrishna D, Gan ES, et al. . Positive epistasis between viral polymerase and the 3′ untranslated region of its genome reveals the epidemiologic fitness of dengue virus. Proc Natl Acad Sci U S A. 2020 May 19;117(20):11038–11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wargo AR, Kurath G.. Viral fitness: definitions, measurement, and current insights. Curr Opin Virol. 2012 Oct;2(5):538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lequime S, Fontaine A, Ar Gouilh M, et al. . Genetic drift, purifying selection and vector genotype shape dengue virus intra-host genetic diversity in mosquitoes. PLoS Genet. 2016 Jun;12(6):e1006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goncalvez AP, Escalante AA, Pujol FH, et al. . Diversity and evolution of the envelope gene of dengue virus type 1. Virology. 2002 Nov 10;303(1):110–119. [DOI] [PubMed] [Google Scholar]
- 53.Pinheiro TM, Mota MTO, Watanabe ASA, et al. . Viral immunogenicity determines epidemiological fitness in a cohort of DENV-1 infection in Brazil. PLoS Negl Trop Dis. 2018 May;12(5):e0006525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vasilakis N, Deardorff ER, Kenney JL, et al. . Mosquitoes put the brake on arbovirus evolution: experimental evolution reveals slower mutation accumulation in mosquito than vertebrate cells. PLoS Pathog. 2009 Jun;5(6):e1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.